Abstract
Deregulated expression of circular RNAs (circRNAs) is associated with various human diseases, including many types of cancer. Despite their growing links to cancer, there has been limited characterization of circRNAs in metastatic castration-resistant prostate cancer, the major cause of prostate cancer mortality. Here, through the analysis of an exome-capture RNA-seq dataset from 47 metastatic castration-resistant prostate cancer samples and ribodepletion and RNase R RNA-sequencing of patient-derived xenografts (PDXs) and cell models, we identified 13 circRNAs generated from the key prostate cancer driver gene-androgen receptor (AR). We validated and characterized the top four most abundant, clinically relevant AR circRNAs. Expression of these AR circRNAs was upregulated during castration-resistant progression of PDXs. The upregulation was not due to global increase of circRNA formation in these tumors. Instead, the levels of AR circRNAs correlated strongly with that of the linear AR transcripts (both AR and AR-variants) in clinical samples and PDXs, indicating a transcriptional mechanism of regulation. In cultured cells, androgen suppressed the expression of these AR circRNAs and the linear AR transcripts, and the suppression was attenuated by an antiandrogen. Using nuclear/cytoplasmic fractionation and RNA in-situ hybridization assays, we demonstrated predominant cytoplasmic localization of these AR circRNAs, indicating likely cytoplasmic functions. Overall, this is the first comprehensive characterization of circRNAs arising from the AR gene. With greater resistance to exoribonuclease compared to the linear AR transcripts and detectability of AR circRNAs in patient plasma, these AR circRNAs may serve as surrogate circulating markers for AR/AR-variant expression and castration-resistant prostate cancer progression.
Keywords: circular RNA, androgen receptor, circAR, prostate cancer, castration resistance
Introduction
Arising from backsplicing of a 3’ splice donor to an upstream 5’ splice acceptor, circular RNAs (circRNAs) are covalently closed transcripts that are resistant to exoribonuclease degradation and generally more stable than linear transcripts [1]. Early occasional reports of circular RNAs were thought to represent splicing noise until the recent availability of non-polyA-selected RNA-seq data and novel backsplicing detection algorithms led to the discovery of a high prevalence and abundance of circRNAs expressed from throughout the genome [1]. The gradual acceptance that circRNAs play important functional effectors of cellular processes has accrued with a steadily increasing stream of reports showing roles of circRNAs in various cellular signaling and metabolic processes [1–11]. Although our understanding of the functions and mechanisms of action of circRNAs is still in its infancy, many circRNAs have been implicated to have non-coding functions, such as cis regulation of their gene of origin [2, 3], epigenetic alteration of gene transcription [4, 5], sponging of miRNAs [1, 6], and modulation of RNA splicing [7]. Furthermore, a small subset of circRNAs has been shown to encode proteins/peptides in a cap-independent manner [12–14].
Aberrant expression of circRNAs has been observed in many types of cancer (summarized in [15]). Significantly, a growing number of circRNAs have already been reported to be involved in cancer etiology and progression, and therapeutic implications have been suggested [15]. Further, because circRNAs have higher stability than linear RNAs and can be secreted into body fluids, significant efforts have been devoted to developing them as a promising class of non-invasive markers for cancer diagnosis and prognosis [16, 17]. In prostate cancer, the study of circRNAs has been mainly centered around the identification of circRNAs that are deregulated in primary prostate cancers compared to benign prostate tissues. Three single-circRNA centered studies reported higher expression of circMYLK [18], circ-102004 [19], and circSMARCA5 [20] in prostate cancer tissues compared to benign prostate tissues and a role for these circRNAs in promoting cell proliferation. Using an unbiased microarray approach analyzing four pairs of prostate cancer and paracancerous tissues, a recent study identified 1,021 differentially expressed circRNAs, validated elevated expression of circ_0057558 and circ_0062019 in cancer tissues, and demonstrated the potential diagnostic values of these two circRNAs [21]. While this manuscript was in preparation, two additional studies reporting global analyses of circRNAs in clinical prostate cancer samples were published [11, 17]. One study profiled circRNAs in 144 localized prostate cancer samples using ultra-deep ribosomal RNA-depleted RNA-seq (ribodepletion-seq) and identified 76,311 distinct circRNAs [11]. A functional screen using an shRNA-based loss-of-function approach identified 171 circRNAs that are essential for prostate cancer cell proliferation [11]. In the other study, exome-capture RNA-seq was performed on more than 2,000 clinical cancer samples across 40 cancer types [17]. Focused analysis of 25 pairs of matched prostate cancer and normal prostate tissues included in this study led to the identification of 652 differentially expressed circRNAs [17]. Together, these studies catalogued circRNAs that are deregulated in primary prostate cancers compared to benign prostate tissues and those that are differentially expressed in primary prostate cancers of different aggressiveness. Despite the importance of these studies, there has been limited characterization of circRNAs in metastatic castration-resistant prostate cancer (mCRPC), the major cause of prostate cancer mortality [22].
The androgen receptor (AR) drives the development and progression of prostate cancer (reviewed in [23, 24]). Even in mCRPC patients, the AR locus is indispensable for disease progression (reviewed in [22, 25, 26]). Particularly evident in mCRPC, where the AR gene is generally heavily transcribed, is extensive alternative splicing at the AR gene locus that produces more than 20 AR gene isoforms (reviewed in [27]). Accumulating evidence suggests that alternatively spliced AR variants (AR–Vs) contribute to resistance of mCRPCs to AR-directed therapies and disease progression through constitutive activation of target promoters (i.e. independent of androgen binding) (reviewed in [27]). As such, this transcript diversity, which is especially well manifested in mCRPCs, is of great importance to understanding disease progression and the pursuit of therapeutics. Here, we show even greater transcript diversity at the AR locus with our findings of 13 circular RNA isoforms. Further, with the expression of these circRNA isoforms roughly paralleling the increased expression of the linear variants in mCRPC, these new AR transcripts may similarly support certain aspects of the mCRPC phenotype and/or serve as surrogate disease markers.
Results
Detection of circRNAs derived from the AR locus in mCRPC specimens
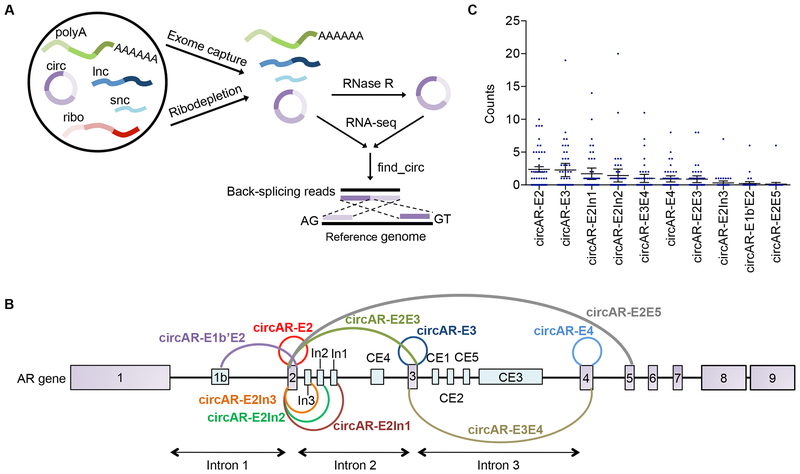
As an initial step to identify clinically relevant circRNAs that are expressed in mCRPC, we utilized publicly available RNA-seq datasets from mCRPC specimens. Although mCRPC RNA-seq datasets have become increasingly available, only a cohort of 47 mCRPC samples from the SU2C project [28] was suitable for circRNA detection since the respective libraries were generated from exome-captured RNA, an RNA enrichment method that retains circRNAs (Fig. 1A). We identified 10,520 unique backsplice junctions (supported by more than four reads in at least one sample) from this cohort with the number of backsplice junctions detected varying substantially among different samples.
Figure 1. Detection of 10 AR backsplice junctions in mCRPC specimens.
(A) Schematic diagram showing the pipeline for exome-capture RNA-seq, ribodepletion-seq, and RNase R-seq as well as subsequent backsplice junction identification. (B) Schematic diagram of 10 AR backsplice junctions that have more than four reads in at least one of the mCRPC samples in the SU2C cohort. The presumed circRNAs corresponding to these 10 backsplice junctions were named according to the exons bearing the splice donor and acceptor sites. (C) Read counts for each of the 10 AR backsplice junctions in the 47 mCRPC samples from the SU2C cohort. Error bars, mean with SEM. polyA, polyA RNA; circ, circRNA, lnc, long noncoding RNA; snc, small noncoding RNA; ribo, ribosomal RNA.
The AR primary transcript is subjected to extensive alternative splicing in mCRPC (reviewed in [27]). Adding to this complexity of transcript isoforms, we detected 10 unique AR backsplice junctions that have more than 4 unique reads in at least one sample (Fig. 1B & 1C). The top two most abundant backsplice junctions represent the circularization of a single exon, while the next two most abundant backsplice junctions result from the joining of a cryptic splice donor in intron 2 with the splice acceptor of exon 2. To confirm that the detected backsplice junctions represent true backsplicing events rather than misalignment, RNA-seq reads were re-aligned to conjoined backsplice junctions, and the coverage across each junction was visually evaluated on the Integrative Genomics Viewer (IGV). All ten AR backsplice junctions showed perfect alignment to the conjoined sequences (alignments of the top four most abundant AR backsplice junctions are presented in Supplementary Fig. S1). Strikingly, 6 of these backsplices utilized the splice acceptor of exon 2, and 2 used the splice acceptor of exon 3 (Fig. 1B). Intron 2 was found to contain 3 cryptic backsplicing donors (Fig. 1B). The high prevalence of backsplicing donors and acceptors in exon 2 to exon 3 regions indicated that these regions might contain strong backsplicing signals.
Detection of AR circRNAs (circARs) in preclinical models
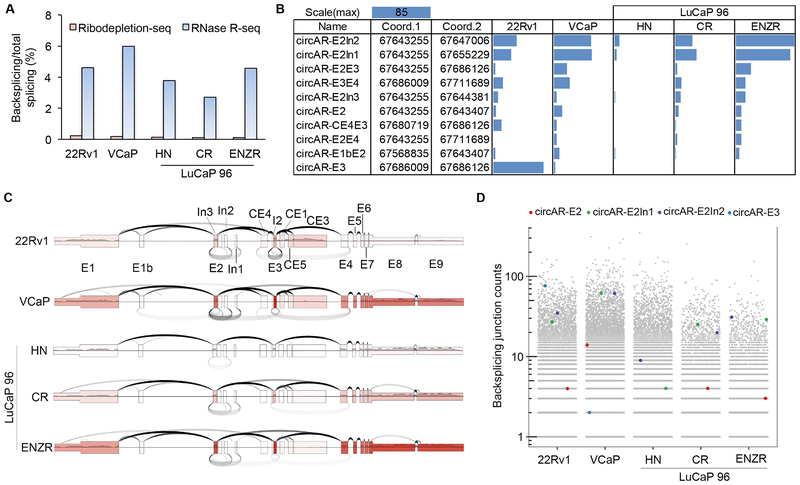
To validate the detection of the circARs in preclinical models, we performed RNase R circRNA sequencing (RNase R-seq) in two human prostate cancer cell lines that exhibit extensive AR alternative splicing, 22Rv1 and VCaP, as well as the hormone-naïve LuCaP 96 patient-derived xenograft (PDX) and its castration-resistant and enzalutamide-resistant PDX sublines (LuCaP 96CR and LuCaP 96CR–ENZR). For RNase R-seq, ribodepleted RNAs were treated with RNase R to digest linear transcripts prior to library preparation (Fig. 1A) to reduce background and to increase the accuracy and sensitivity of circRNA detection. To help assess the performance of RNase R-seq, we also carried out ribodepletion-only RNA-seq for the LuCaP 96 samples, and we downloaded ribodepletion-seq data for 22Rv1 and VCaP cells. Compared to the ribodepletion-seq data, the ratio of backsplicing to total splicing junction reads was increased by more than 20 fold in the RNase R-seq data, confirming enrichment of circRNAs after RNase R treatment (Fig. 2A). Another important finding was that less than half (31% to 45%) of the backsplice junction calls from the ribodepletion-seq data were enriched by more than 4 fold after RNase R treatment (Supplementary Fig. S2). This suggested that a sizable proportion of backsplice junction calls made using ribodepletion-seq data could be false positives, further supporting the improved specificity of RNase R-seq for circRNA detection.
Figure 2. circAR expression in prostate cancer cell lines and LuCaP 96 PDXs by RNase R-seq method.
(A) Backsplicing to total splicing read counts showing enrichment of circRNAs in RNase R-seq data compared to ribodepletion-seq data. (B) Bar graph showing the read counts for AR backsplice junctions that have more than 4 reads in at least 1 sample in RNase R-seq data. (C) Plot showing backsplicing (under arches, from RNase R-seq), forward splicing (over arches, from polyA-seq), and exon coverage (exon color intensity, from polyA-seq) at the AR locus. The number of arches corresponds to the number of junction-spanning reads. (D) Scatter plot displaying the read counts of the most abundant clinically relevant circARs in the context of all circRNA (grey dots) read counts.
Using the RNase R-seq data from the preclinical models, we also detected 10 AR backsplice junctions that were enriched in RNase R-seq data and were represented by more than 4 reads in at least one sample (Fig. 2B). These AR backsplice junctions are represented graphically in Fig. 2C, with coverage and forward-splicing read counts from polyA-selected RNA-seq (polyA-seq) data shown to provide context of linear transcript expression and forward splicing. Notably, the LuCaP 96 PDX series nicely illustrates the relationship between linear transcript expression and circAR expression in the progression to castration resistance, and then to enzalutamide resistance.
Of the 10 backsplice junctions detected in our preclinical models, seven were also detected in the exome-capture RNA-seq data from the SU2C cohort (circAR–E2In1, circAR–E2In2, circAR–E2E3, circAR–E3E4, circAR–E2In3, circAR–E2, and circAR–E3). CircAR–E2In1 and circAR–E2In2, which were the third and fourth most abundant in the SU2C cohort (Fig. 1C), were the top two most abundant circARs detected in the preclinical models (Fig. 2C). They ranked in the top 1st percentile in abundance among all detected circRNAs in LuCaP 96CR-ENZR xenograft, the top 1.6th percentile in VCaP cells, and the top 6th percentile in 22Rv1 cells (Fig. 2D). CircAR-E2, the most abundant circAR detected in the SU2C cohort, ranked in the top 37th percentile among all circRNAs in the LuCaP 96CR-ENZR xenograft and in the top 23rd percentile in VCaP cells (Fig. 2D). Together, this analysis indicated that circARs are expressed at considerable levels among all circRNAs detected, with circARE2, circAR–E2In1 and circAR–E2In2 showing especially high expression in multiple models.
Surprisingly, while circAR–E3 was the second most abundant circAR in the SU2C cohort, it was not detected in the RNase R-seq data from any of the three LuCaP 96 PDXs, and only 2 reads were detected in VCaP cells (Fig. 2C & 2D). This lack of detection is likely due to exclusion of this 117bp circRNA during the size fractionation of the Illumina Truseq library preparation (lower limit 120 bases) since circAR–E3 is readily detected in LuCaP 96 PDXs by RT–qPCR (see below, Fig. 4B). Conversely, it should be noted that at least some of the E3 splice donor to E3 splice acceptor spanning reads detected in 22Rv1 cells likely arose from forward splicing from the previously demonstrated duplication of E3 in these cells [29] (Fig. 2C & 2D).
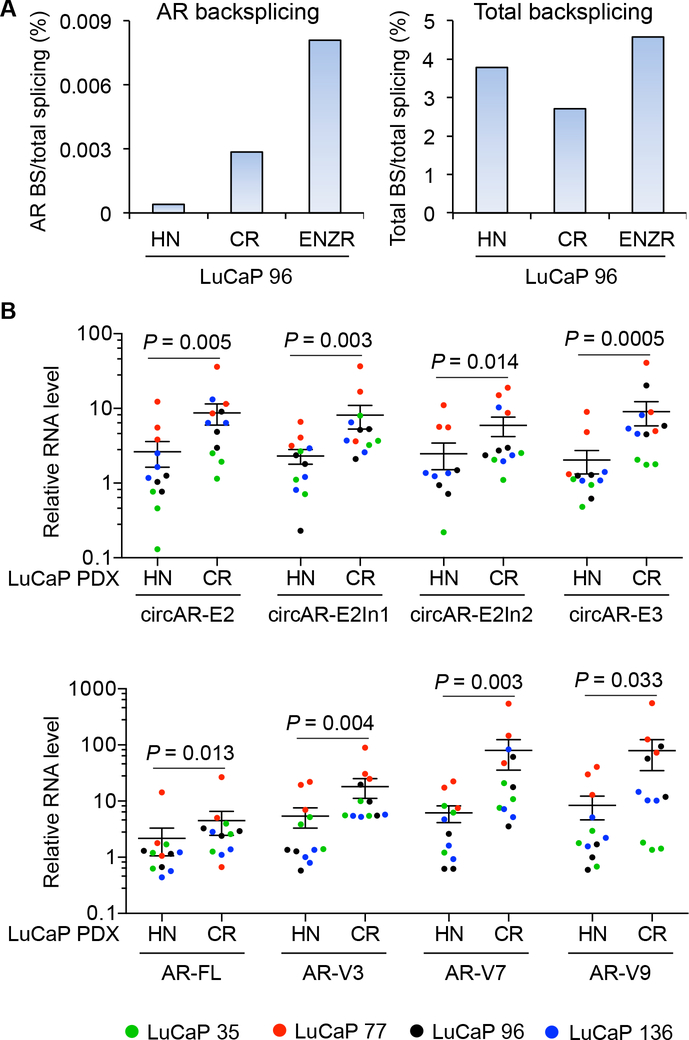
Figure 4. Increased expression of circARs during castration-resistant (CR) progression of LuCaP tumors.
(A) RNase R-seq analysis showing that the increase in the percentage of total AR backsplicing (BS) reads in total splicing reads in LuCaP 96-CR and LuCaP 96-CR-ENZR tumors compared to hormone-naïve (HN) LuCaP 96 tumors was not due to change in total backsplicing (BS) reads. (B) RT-qPCR showing that the levels of circARs were increased when LuCaP 35, 77, 96, and 136 PDXs progress to castration-resistant (upper panel) and correlated with the levels of linear AR transcripts (lower panel). The normalized levels of each AR transcript are expressed relative to the mean level of that AR transcript in HN LuCaP 96 tumors. n = 3 different xenograft passages per PDX model. Of note, since the Y axis is logarithmic, only values >0 can be plotted. Two circAR-E2In2 values of HN LuCaP 35 tumors are zero and are therefore not included on the graph. P, from Mann-Whitney test for difference between HN and CR tumors. Error bars, mean with SEM.
Resolution of AR circRNA structures
With circAR–E2, circAR-E3, circAR–E2In1, and circAR–E2In2 being the most abundantly expressed circARs in clinical samples, we focused on these circARs in follow-up studies. Using divergent primers (Supplementary Fig. S1), RT–PCR produced a single band in each case in VCaP and 22Rv1 cells (Fig. 3A). The resulting PCR amplicons were sequenced by Sanger sequencing, verifying the respective backsplice junctions in each case. To determine the structures of the In1 and In2 cryptic exons in circAR–E2In1 and circAR–E2In2, we first performed a gross analysis through visualization of the RNase R-seq read alignments using IGV (Supplementary Fig. S3A). We then identified the paired-end reads representing fragments that span both the backsplice junction as well as the forward-splice junctions that connect exon 2 to the cryptic exon. These analyses along with subsequent RT–PCR verification allowed us to delineate the entire structures of circAR–E2, circAR-E3, circAR–E2In1, and circAR–E2In2 (Supplementary Fig. S3B).
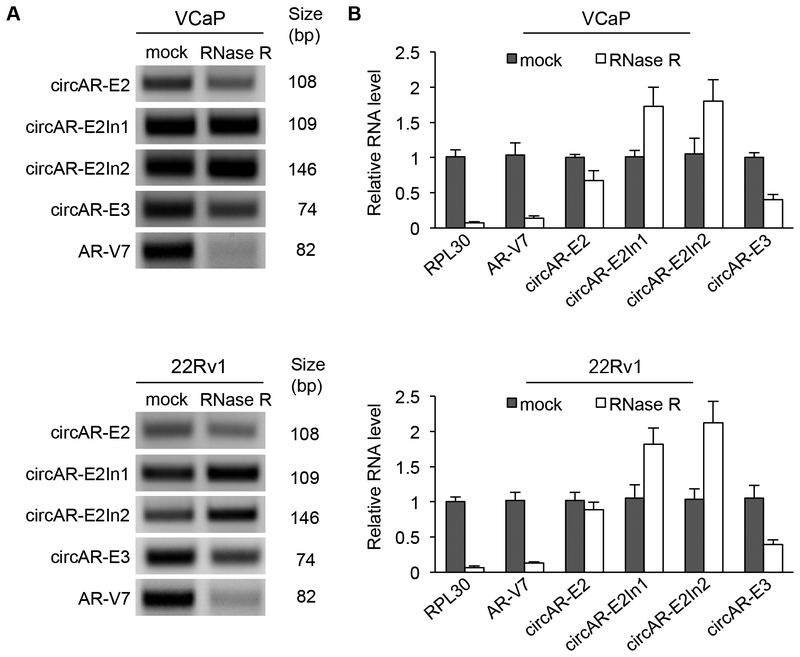
Figure 3. Resistance of circARs to RNase R.
RT-PCR (A) and RT-qPCR (B) showing that circARs are more resistant to RNase R treatment than the linear AR-V7 and RPL30 transcripts in VCaP and 22Rv1 cells.
Treatment of RNAs with RNase R prior to RT–PCR confirmed resistance of circAR–E2, circAR–E3, circAR–E2In1, and circAR–E2In2 to RNase R digestion, demonstrating their closed circular nature (Fig. 3A). We then quantitated the extents of their RNase R resistance by RT–qPCR and found that, while all 4 circARs exhibited at least 5-fold greater RNase R resistance than the RPL30 linear transcript, circAR–E2In1 and circAR–E2In2 displayed the greatest resistant, followed by circARE2 and then circAR–E3 (Fig. 3B). CircRNAs have been reported to have a greater half-life in cells than linear RNAs, presumably as a result of resistance to exoribonuclease digestion [1]. We assessed the stability of circAR–E2In1, circAR–E2In2, circAR–E3, and circAR–E2 in VCaP and 22Rv1 cells and found that although circAR-E2In1, circAR-E2In2, and circAR-E3 showed greater stability than linear AR–V7, the stability of circAR–E2 was similar to that of linear AR–V7 transcript (Supplementary Fig. S4). This analysis indicates that general exoribonuclease resistance does not always equate to higher stability of circRNAs, perhaps due to specific transcript processing mechanisms following the backsplice reactions that generate novel, potentially functional RNAs.
Upregulated expression of circARs during castration-resistant prostate cancer progression
Analysis of our RNase R-seq data revealed an ascending trend in circAR levels in the progression of LuCaP 96 PDXs to castration resistance and further to enzalutamide resistance (Fig. 4A, left panel). Notably, this was not due to an overall increase of circular RNA production but was specific to circARs (Fig. 4A, right panel). To corroborate this finding, we performed RT–qPCR analysis in hormone-naïve and castration-resistant LuCaP 35, LuCaP 77, LuCaP 96, and LuCaP 136 PDX pairs and analyzed 3 different xenograft passages per PDX. Consistent with the RNase R-seq data, all four circARs were significantly upregulated in castration-resistant PDXs compared to their hormone-naïve counterparts (Fig. 4B, top panel; Fig. S5). Notably, the upregulation of these circARsin castration-resistant PDXs mirrored the changes in AR-FL and AR-V levels (Figs. 4B and S5), indicating that the increased expression of circARs in castration-resistant tumors results most likely from increased transcription of the AR gene.
Androgen suppressing circAR expression
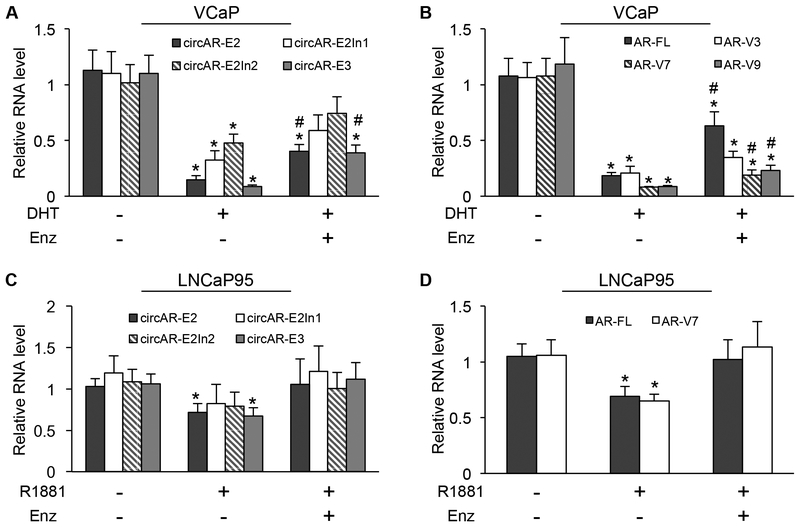
Androgen-bound AR is known to suppress the transcription of the AR gene through a negative-feedback mechanism [30, 31]. To assess whether circAR expression is also subject to androgen modulation, we treated VCaP and LNCaP95 cells with androgen in the absence or presence of enzalutamide and analyzed the levels of circARs and linear AR transcripts by RT–qPCR. Similar to AR–FL and –Vs, circARs were downregulated significantly after androgen treatment in VCaP cells, and the downregulation was attenuated by enzalutamide (Fig. 5A & 5B). A similar trend was observed in LNCaP95 cells (Fig. 5C & 5D). These data provide further support for the involvement of a transcriptional mechanism in circAR upregulation after androgen deprivation.
Figure 5. Androgen downregulating the expression of circular and linear AR transcripts.
RT-qPCR showing that circARs (A & C) and AR-FL and AR-V mRNAs (B & D) were downregulated by androgen in VCaP and LNCaP95 cells and that the downregulation was attenuated by enzalutamide (Enz, 10 μM). DHT (dihydrotestosterone), 10 nM; R1881, 1 nM; Duration, 24 h; *, P < 0.05 from the untreated control group; #, P < 0.05 from androgen-treatment group.
Positive correlation between circARs and linear AR transcripts in clinical mCRPC samples
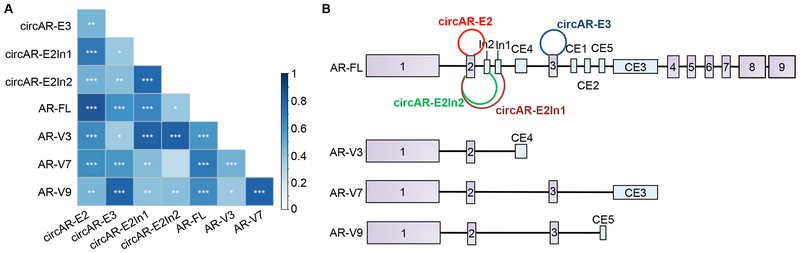
To investigate whether the correlation between circARs and linear AR transcripts also exists in clinical samples, we analyzed the exome-capture RNA-seq data from the SU2C cohort for expression of the 4 circARs, AR–FL, and three of the more abundantly expressed AR–Vs in these samples, ARV3, –V7, and –V9 [28]. Consistent with our data from preclinical models, the levels of all AR isoforms (linear and circular) were positively correlated in mCRPC tissues (Fig. 6A). Interestingly, circAR-E3 showed a higher concordance with AR-V9 than with other transcripts, and circAR-E2In1 and circARE2In2 correlated stronger to AR–V3 than to other transcripts (Fig. 6A). Noticing that the splice junctions that are specific to circAR–E3 and AR–V9 are in close proximity and that this is also the case for circAR–E2In1/circAR-E2In2 and AR–V3 (Fig. 6B), we hypothesize that the backsplicing of circARs is mechanistically linked to the forward splicing of the neighboring AR–Vs.
Figure 6. Positive correlation of circARs with AR-FL and -V transcripts.
(A) Pearson correlation analysis showing strong positive correlations between circARs with AR-FL/-Vs. The blue gradient bar indicates the the scale of correlation coefficiency scores. (B) Schematic diagrams showing that the splice junctions that are specific to the most highly concordant circAR/AR-V pairs, circAR-E3/AR-V9, circAR-E2In1/AR-V3, and circAR-E2In2/AR-V3, are in close proximity.
Subcellular localization of circARs
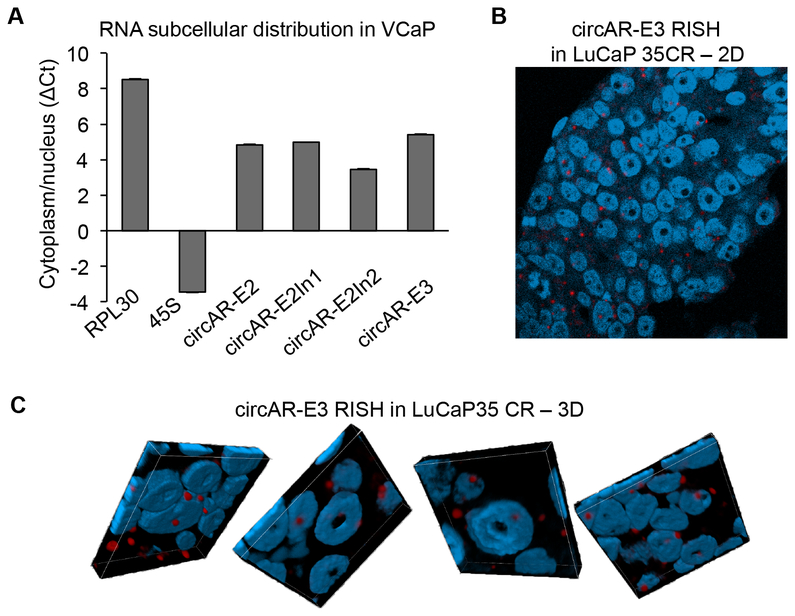
To begin to appreciate how these circARs might function in cells, we assessed their subcellular localization. We first fractionated cytoplasmic and nuclear RNAs from VCaP cells and quantified circAR levels in each fraction by RT–qPCR. circAR–E2In1, circAR–E2In2, circAR–E3, and circAR–E2 all exhibited predominant cytoplasmic localization, with circAR-E3 and circAR-E2In2 having the highest and lowest cytoplasmic to nuclear ratio, respectively (Fig. 7A). We further validated circARE3 subcellular localization using the BaseScope RNA in-situ hybridization (RISH) assay with a probe targeting the backsplice junction. The specificity of the BaseScope probe was evident from the lack of RISH signal in LNCaP cells transfected with a mutant circAR-E3 expression construct bearing mutations at the E3 splice donor and acceptor sites but strong punctate signals in LNCaP cells transfected with a wildtype circAR–E3-expression vector (Supplementary Fig. S6) and in VCaP cells (Supplementary Fig. S7A). In concordance with the cytoplasmic and nuclear fractionation result, confocal microscopy analysis of RISH-stained LuCaP 35CR PDX tissue showed predominant cytoplasmic localization of circAR-E3 (Fig. 7B & 7C).
Figure 7. Predominant cytoplasmic localization of circARs.
(A) RT-qPCR analysis of nuclear and cytoplasmic RNA fractions showing predominant cytoplasmic localization of circARs in VCaP cells. Cytoplasmic-localized RPL30 and nuclear-localized 45S were used for comparison. The cytoplasmic to nuclear RNA ratios are plotted as ΔCt between the cytoplasmic and nuclear Ct values. (B & C) Two-dimensional (B) and three-dimensional (C) confocal fluorescence microscopy of circAR-E3 RISH showing predominant cytoplasmic localization of circAR-E3 in a LuCaP 35CR tumor. DAPI was used for nuclear staining. 3D images were composited from z-stack analysis.
RISH-based confirmation of circAR upregulation during castration-resistant prostate cancer progression and circAR detection in plasma samples from mCRPC patients
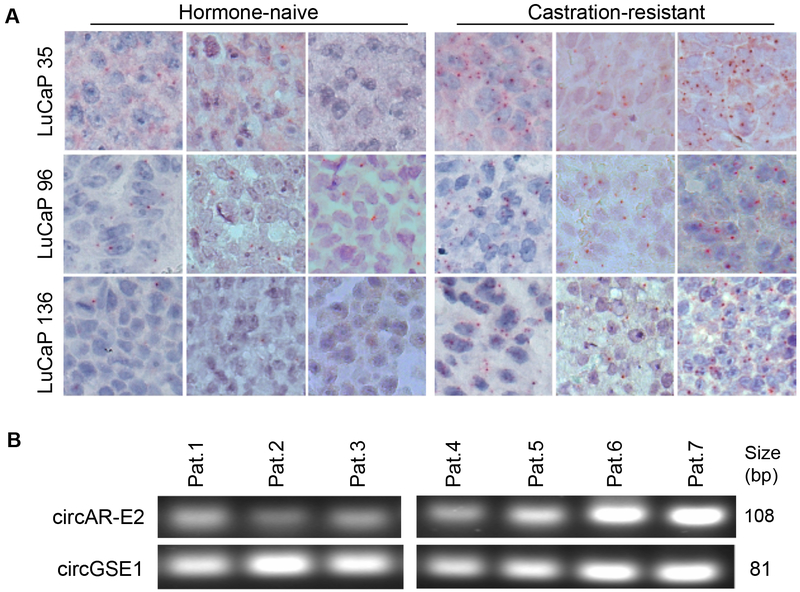
We also conducted circAR–E3 RISH analysis of hormone-naïve, castration-resistant, and enzalutamide-resistant LuCaP 96 PDX tumors that were analyzed above in the RNase R-seq analysis as well as in an additional 3 pairs of hormone-naïve and castration-resistant PDX models, LuCaP 35, LuCaP 96, and LuCaP 136. In agreement with the RNase R and RT-qPCR data (Fig. 4 and Supplementary Fig. S5), RISH signals were considerably stronger in castration-resistant PDXs than in their hormone-naïve counterparts (Fig. 8A) and were further increased in the enzalutamide-resistant PDX (Supplementary Fig. S7B). Together, these data provided solid evidence for an upregulated circAR expression during castration-resistant progression. CircRNAs can be secreted into body fluids (reviewed in [32, 33]). To explore whether circARs are potential candidate as non-invasive biomarkers for castration-resistant progression, we isolated RNA from plasma samples of 7 mCRPC patients and analyzed circAR expression by RT–PCR. Since the detection of circAR–E2 was more robust than the other circARs, we focused on the analysis of circAR–E2 in these samples. All samples showed detectable levels of circAR–E2 (Fig. 8B), and the PCR products were verified by Sanger sequencing. These data indicate that detecting circARs in plasma is attainable.
Figure 8. RISH confirmation of circAR-E3 upregulation during castration-resistant progression of LuCaP PDX models and detection of circAR-E2 in plasma samples from mCRPC patients.
(A) circAR-E3 is upregulated during castration-resistant progression of LuCaP PDX models. RISH was conducted on 3 different xenograft passages per PDX model. (B) Detection of circAR-E2 in plasma samples from 7 mCRPC patients. RT-PCR analysis was performed on RNA samples isolated from 0.25 ml of patient plasma. circGSE1 was used as a control circRNA that is present in the plasma.
Discussion
Although a number of AR backsplice junctions have been captured in several circRNA databases (Supplementary Table S2) and one has been reported in a recent publication [17], these backsplice junctions have not been validated or characterized previously. Here, through the analysis of a publicly available exome-capture RNA-seq dataset from 47 mCRPC clinical samples and our RNase R-seq data from prostate cancer cell lines and PDX models, we identified a total of 13 unique AR backsplice junctions that were supported by more than 4 reads in at least one sample. Seven of these AR backsplice junctions were detected in both clinical samples and preclinical models, and we further validated and characterized the 4 most abundantly expressed circARs in clinical samples and linked their expression to castration-resistant progression of prostate cancer.
CircRNAs have been reported to be secreted into the circulation and to be readily detectable in plasma and urine (reviewed in [32, 33]). With their generally increased stability compared to linear transcripts, circRNAs are receiving considerable attention as potential liquid biopsy biomarkers (reviewed in [32, 33]). Increased expression of AR–Vs has been ascribed as an important mechanism of resistance to second-generation AR-directed therapies, including enzalutamide and abiraterone (reviewed in [27]), and AR–V7 expression in circulating tumor cells is being developed as a prognostic test to predict patients’ response to these drugs [34]. In this study, we demonstrated strong correlation between the levels of circARs and that of AR–Vs in mCRPC tissues and PDXs. With greater resistance to exonuclease degradation than AR–Vs and detectability in plasma samples from mCPRC patients, circARs may serve as surrogate circulating markers for AR–V expression, and testing of plasma circARs may outperform AR–V7 analysis of circulating tumor cells to predict response to AR-directed therapies.
All the four circARs that we characterized here arise from the exon 2 to exon 3 regions. Notably, both exon 2 and exon 3 are flanked by long intronic sequences that harbor a number of reverse complementary SINE/LINE repeats (Supplementary Fig. S3A). These intronic repeats, which have been reported to facilitate circRNA production [35], may bring the backsplicing sites into close proximity to facilitate circAR biogenesis. Interestingly, a correlation analysis between circARs and AR-Vs in mCRPC tissues showed a high concordance between circARs and AR-Vs that share splice donors and/or acceptors unique to the respective AR–V/circAR pairings. It is possible that the backsplicing of circARs and forward splicing of the neighboring AR–Vs are mechanistically linked and/or with the biogenesis of circARs possibly mediating the splicing of the AR–Vs and vice versa.
AR gene rearrangement has been reported to contribute to the generation of certain AR-Vs (e.g. ARv567es) [36]. However, among all the cell line and PDX models that were included in this study, only the 22Rv1 cell model is known to harbor AR structural changes. This indicates that the formation of circARs does not require AR structural alterations. In addition, in the recent publication that reported the identification of one circAR, which is backspliced from exon 4 to exon 3, the authors suggested AR amplification as a likely mechanism of circAR generation [17]. This is consistent with our data supporting the involvement of a transcriptional mechanism in circAR generation, as AR gene amplification is known to lead to increased transcription of the AR gene. Nevertheless, we found that the expression of circARs was not exclusive to AR-amplified mCRPC samples (Supplementary Fig. S8), suggesting that AR gene amplification is not the sole mechanism for circAR upregulation during castration-resistant progression of prostate cancer.
Although ribodepletion-seq and RNase R-seq are considered the gold-standard methods for sequencing circRNAs, the inability to detect circAR–E3 highlighted a potential limitation of the standard protocols for these methods in some cases. As these methods adopt the Illumina Trueseq library preparation approach, circRNAs that are smaller than the library insert size (< 120 bp) may not be reliably captured. Given that 22Rv1 cells harbor duplicated AR exon 3s, we initially attributed the circAR–E3 junction reads detected in these cells in the ribodepletion-seq and RNase R-seq datasets to reads spanning the duplicated exon 3s in linear AR transcripts. However, a comparison of the ribodepletion-seq and RNase R-seq data revealed an enrichment of these reads after RNase R treatment of RNA from 22Rv1 cells, indicating that a good portion of these reads likely represent the circular transcript. Further, it is possible that this circular transcript is composed of the duplicated exon 3s, making it more easily detectable by ribodepletion-seq and RNase R-seq because it doubles the size of circAR–E3.
Compared to the other circARs analyzed here, we found that circAR-E3 is less resistant to RNase R treatment, and we hypothesize that this may also be related to its small size. Theoretically, RNase R only degrades linear RNAs, but longer incubation at high concentrations can result in reduced levels of circRNAs (data not shown). For relatively large circRNAs, after they are cut by RNase R, it is likely to have linearized fragments spanning the backsplice junctions with adequate flanking sequences that are detectable by PCR. In contrast, for a circRNA like circAR–E3 that only comprises of 100 bp, one single cut by RNase R would likely to fall inside of the PCR amplicon, making PCR detection less plausible. Thus, cautions should be exercised while designing RNase R resistance tests to minimize the concentration and duration of RNase R treatment, and the sizes of circRNAs should be considered when interpreting the data.
Many circRNAs have been shown to interact with microRNAs (miRNAs) to either sponge or stabilize miRNAs [1, 6, 11, 37]. By running find_circ on miRNA-targeting sequences detected by AGO-PAR-CLIP sequencing in prostate cancer cell lines [38], we only detected around 70 unique backsplice junctions associated with the Argonaute protein, and none of the circAR-associated backsplice junctions was present. While a number of linear AR fragments from exon 2, exon 3, and other regions were incorporated into the Argonaute complexes (Supplementary Fig. S9), it was not possible to distinguish whether these fragments were from the linear or circular AR transcripts. When we ectopically expressed circARs in LNCaP and LNCaP95 cells, we did not detect any changes in the levels of AR–FL or AR–V7 (data not shown). While we cannot yet rule out the possibility that circARs bind to miRNAs that regulate other transcripts, the co-regulated expression of circARs and linear AR transcripts is most likely not due to sponging of shared miRNAs by circARs. Some circRNAs have been reported to act in cis to regulate host gene transcription through interacting with the transcriptional and/or splicing machinery [2]. However, to function in cis, the circRNAs would need to be localized in the nucleus. The predominant cytoplasmic localization of circARs refutes their possible cis regulation of the AR parental gene.
Besides noncoding functions, some circRNAs have been shown to encode proteins/peptides [12–14]. Significantly, circAR–E3 has an open reading frame starting with a putative AUG in frame with the AR DNA-binding domain, and the translation may continue in-frame without termination due to circularization. Previous studies have provided evidence for the production of repetitive peptides from circRNAs with an infinite open reading frame [39, 40]. Based on its open reading frame and its cytoplasmic localization, we hypothesize that circAR–E3 may encode a peptide repeat that represents a portion of the AR DNA-binding domain. Data from our preliminary polysome fractionation analysis is supportive of this coding potential of circAR–E3 (not shown). However, additional evidence, such as mass spectrometry identification, is necessary to confirm the expression of such peptides.
In conclusion, this is the first comprehensive characterization of circRNAs arising from the AR gene. Upregulation of these circARs during castration-resistant progression, their strong correlation with AR–FL and -Vs, and their high stability highlight the potential value of these circARs as a new species of circulating biomarker for mCRPC patients.
Materials and Methods
Cell culture
22Rv1, VCaP, LNCaP, and 293T cells were obtained from ATCC. LNCaP95 cells were provided by Dr. Alan Meeker at Johns Hopkins University. VCaP and 293T cells were cultured in DMEM medium (ATCC, Catalog No. 30–2002) with 10% fetal bovine serum (Gibco, Catalog No.10437–028). LNCaP95 cells were cultured in phenol-red free RPMI–1640 medium (Gibco, Catalog No. 11835–030) with 10% charcoal-stripped fetal bovine serum (Atlanta, Catalog No. S11650). 22Rv1 and LNCaP cells were grown in RPMI-1640 medium (Hyclone, Catalog No. SH30027.02) with 10% fetal bovine serum. Cells used in all experiments were within 3 months of resuscitation of frozen cell stocks established within 3 passages after receipt of the cells. Cell authentication was performed at the Genetica DNA Laboratories, and cells were evaluated monthly for mycoplasma contamination.
Generation of PDX tissues
Animal procedures were performed in accordance with National Institutes of Health guidelines and approved by the University of Washington Institutional Animal Care and Use Committee. LuCaP PDX samples used for analyses were generated from PDX tumors grown subcutaneously in gonad-intact (hormone-naïve PDXs) or castrated (CR and CR-ENZR PDXs) male SCID CB17 mice (Charles River) as appropriate. The CR-ENZR (enzalutamide-resistant) PDX tumors were generated from CR (castration-resistant) PDX tumors grown in castrated male mice upon prolonged pressure of enzalutamide (50 mg/kg, oral gavage, 5 days/week, Medchem Express). When tumors reached ~500–800 mm3, animals were sacrificed and tumors processed for analyses.
Publicly available datasets
The exome-capture RNA-seq data from 47 mCRPC samples in the SU2C cohort was downloaded from dbGaP (dbGaP accession pht004946.v1.p1) [28]. The cDNA libraries for this RNA-seq dataset were prepared using Agilent SureSelect Human All Exon V4 platform, omitting the polyA selection step [28]. For VCaP and 22Rv1 cells, the ribodepletion-seq dataset was downloaded from GEO (accession: GSE92574), and the polyA-seq dataset was downloaded from the CCLE–PRAD project in the GDC legacy archive. The raw data for the AGO-PAR-CLIP dataset [38] was downloaded from the NIH SRA (bioproject accession: SRP075075), and the processed AGO-PAR-CLIP atlas files were downloaded from https://www.synapse.org/#!Synapse:syn5479902. Splicing junction analysis was performed for these datasets as described below.
RNA sequencing
Total RNA was isolated using the TRIzol reagent (Thermo Fisher, Catalog No. 15596018) following the manufacturer’s protocol, and RNA integrity was assessed using an Agilent Bioanalyzer. RNA samples were sent to Beijing Genomics Institute (BGI) for polyA-seq, ribodepletion-seq, and/or RNase R-seq. PolyA RNAs were selected using an oligo(dT) column for polyA-seq, and ribosomal RNAs (rRNAs) were depleted by hybridization capture of rRNAs for ribodepletion-seq. For RNase R-seq, RNA samples were first depleted of DNA and rRNAs and then subjected to RNase R treatment to remove linear RNAs (Fig. 1A). All sequencing was 100-base paired-end using an Illumina HiSeq 4000 system.
Splice junction and expression analysis
Backsplice junctions were identified using find_circ [1] with default parameters (Fig. 1A). The counts of unique backsplice junction reads in each sample were compiled from the .bed files in the find_circ output. For in silico validation of the identified backsplice junctions, the sequences of conjoined backsplice junctions were combined with the human hg38 genome to generate a new genome index for STAR alignment [41]. The raw RNA-seq reads were mapped to the new genome using STAR (--clip5pNbases 6 --outFilterMultimapNmax 20 --outSAMtype BAM). The reads mapped to the backsplice junctions were selected out for visualization in IGV and the number of reads was quantified.
For forward-splice junction analysis, the raw RNA-seq reads were aligned to the human hg38 genome using STAR (--clip5pNbases 6 --outFilterMultimapNmax 20 --outSAMtype BAM). The counts of the canonical junction reads were compiled from the .SJ files in the STAR output. The .sam alignment files were converted to .wiggle files using IGV for read coverage visualization in IGV.
SpliceV [42] was used to incorporate and visualize the backsplicing, forward-splicing, and exon expression information as described previously [43, 44].
RNase R resistance assay
Five μg of DNase-treated total RNA was incubated with or without 7.5 unit of RNase R (Lucigen, Catalog No. RNR07250) for 10 minutes at 37°C. RNA was purified and concentrated as described previously [43, 44] before subjecting to RT–PCR or RT–qPCR analysis.
RT-PCR and primers
cDNA was synthesized using the iScript™ Reverse Transcription Supermix (Bio-Rad, Catalog No. 1708840). For regular PCR, cDNA was amplified using the GoTaq® Green Master Mix (Promega, Catalog No. M712B-C) following the manufacturer’s protocol. After electrophoresis, the PCR products were excised from agarose gel, purified, cloned into a TA vector, and Sanger sequenced. qPCR analysis was performed as described [45], and the primer-probe sets were purchased from IDT. The sequences of PCR primer pairs and qPCR primer-probe sets are listed in Supplementary Table S1.
Plasmids and transfection
The wildtype circAR-E3 expression plasmid was constructed by inserting DNA sequences corresponding to circAR–E3 along with 50 nt of its endogenous flanking sequences on both sides into the pLVX-puro lentiviral expression vector (Clontech, Catalog No. 632164). In addition, a 40–nt (aaagtgctgagattacaggcgtgagccaccacccccggcc) and a 36-nt (ggctcggcacggtagctcacacctgtaatcccagca) inverted Alu repeats [35] were inserted 5’ to the 5’-flanking sequences and 3’ to the 3’-flanking sequences, respectively. The mutant circAR-E3 expression plasmid contains mutated splice donor and acceptor to prevent circularization. All plasmids were synthesized at Synbio Technologies and sequence-verified. The plasmids were transfected into LNCaP cells using the Lipofectamine 3000 reagent (Thermo Fisher, Catalog No. L3000015) per instruction of the manufacturer.
BaseScope RISH assay
Paraffin-embedded cell blocks (prepared as described previously [43]) and tissue blocks were sectioned at 4-micron thickness. BaseScope RISH assays were performed according to the BaseScope™ v2 Detection Reagent Kit protocol (Advanced Cell Diagnostics, Catalog No. 322910). The custom-designed circAR–E3 probe (Catalog No. 715141) targeting the backsplice junction along with the positive (Catalog No. 710171) and negative (Catalog No. 701021) control probes were purchased from Advanced Cell Diagnostics.
Supplementary Material
Acknowledgments
We are grateful to Dr. Alan Meeker at Johns Hopkins University for providing LNCaP95 cells. We appreciate the support from the Tulane Cancer Next Generation Sequence Analysis core for utilization of resources and expertise for this work.
Competing interests: This work was supported by the following grants: the National Institutes of Health grants R01CA188609, R01AI101046, R01AI106676, P01CA214091, PNW Prostate Cancer SPORE P50CA097186, P01CA163227, RCMI 2U54MD007595, and 5P20GM103424–17; Department of Defense grants W81XWH-15-1-0439, W81XWH-16-1-0317, and W81XWH-16-1-0318; The Prostate Cancer Foundation. The Richard M Lucas Foundation supported the development of the LuCaP models. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
References
- 1.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–338. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22: 256–264. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH et al. Circular intronic long noncoding RNAs. Mol Cell 2013; 51: 792–806. [DOI] [PubMed] [Google Scholar]
- 4.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010; 6: e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011; 30: 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495: 384–388. [DOI] [PubMed] [Google Scholar]
- 7.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56: 55–66. [DOI] [PubMed] [Google Scholar]
- 8.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 2017; 14: 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016; 7: 12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015; 5: 12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J et al. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell 2019; 176: 831–843 e822. [DOI] [PubMed] [Google Scholar]
- 12.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017; 66: 22–37 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L et al. Translation of CircRNAs. Mol Cell 2017; 66: 9–21 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 2017; 27: 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018; 37: 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015; 25: 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y et al. The Landscape of Circular RNA in Cancer. Cell 2019; 176: 869–881 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai Y, Li D, Chen X, Tan X, Gu J, Chen M et al. Circular RNA Myosin Light Chain Kinase (MYLK) Promotes Prostate Cancer Progression through Modulating Mir-29a Expression. Med Sci Monit 2018; 24: 3462–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Si-Tu J, Cai Y, Feng T, Yang D, Yuan S, Yang X et al. Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. International Journal of Biological Macromolecules 2019; 122: 1235–1243. [DOI] [PubMed] [Google Scholar]
- 20.Kong Z, Wan X, Zhang Y, Zhang P, Zhang Y, Zhang X et al. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun 2017; 493: 1217–1223. [DOI] [PubMed] [Google Scholar]
- 21.Xia Q, Ding T, Zhang G, Li Z, Zeng L, Zhu Y et al. Circular RNA Expression Profiling Identifies Prostate Cancer- Specific circRNAs in Prostate Cancer. Cell Physiol Biochem 2018; 50: 1903–1915. [DOI] [PubMed] [Google Scholar]
- 22.Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O. Castration-resistant prostate cancer: adaptive responses in the androgen axis. Cancer Treat Rev 2014; 40: 426–433. [DOI] [PubMed] [Google Scholar]
- 23.Huang H, Tindall DJ. The role of the androgen receptor in prostate cancer. Crit Rev Eukaryot Gene Expr 2002; 12: 193–207. [DOI] [PubMed] [Google Scholar]
- 24.Shand RL, Gelmann EP. Molecular biology of prostate-cancer pathogenesis. Curr Opin Urol 2006; 16: 123–131. [DOI] [PubMed] [Google Scholar]
- 25.Kahn B, Collazo J, Kyprianou N. Androgen receptor as a driver of therapeutic resistance in advanced prostate cancer. Int J Biol Sci 2014; 10: 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan X, Cai C, Chen S, Chen S, Yu Z, Balk SP. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene 2014; 33: 2815–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao S, Zhan Y, Dong Y. Emerging data on androgen receptor splice variants in prostate cancer. Endocr Relat Cancer 2016; 23: T199–T210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene 2012; 31: 4759–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 2011; 20: 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnstein KL. Regulation of androgen receptor levels: implications for prostate cancer progression and therapy. J Cell Biochem 2005; 95: 657–669. [DOI] [PubMed] [Google Scholar]
- 32.Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids 2019; 16: 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Han L. Circular RNAs as promising biomarkers in cancer: detection, function, and beyond. Genome Med 2019; 11: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markowski MC, Silberstein JL, Eshleman JR, Eisenberger MA, Luo J, Antonarakis ES. Clinical Utility of CLIA-Grade AR-V7 Testing in Patients With Metastatic Castration-Resistant Prostate Cancer. JCO Precis Oncol 2017; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28: 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci USA 2013; 110: 17492–17497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016; 7: 11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamilton MP, Rajapakshe KI, Bader DA, Cerne JZ, Smith EA, Coarfa C et al. The Landscape of microRNA Targeting in Prostate Cancer Defined by AGO-PAR-CLIP. Neoplasia 2016; 18: 356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abe N, Hiroshima M, Maruyama H, Nakashima Y, Nakano Y, Matsuda A et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem Int Ed Engl 2013; 52: 7004–7008. [DOI] [PubMed] [Google Scholar]
- 40.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep 2015; 5: 16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ungerleider NA, Flemington EK. SpliceV: Analysis and publication quality printing of linear and circular RNA splicing, expression and regulation. BMC Bioinformatics 2019; 20: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S et al. The Epstein Barr virus circRNAome. PLoS Pathog 2018; 14: e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ungerleider N, Jain V, Wang Y, Maness NJ, Blair RV, Alvarez X et al. Comparative analysis of gammaherpesvirus circRNA repertoires: conserved and unique viral circRNAs. J Virol 2019; 93: pii: e01952–01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong Y, Lee SO, Zhang H, Marshall J, Gao AC, Ip C. Prostate specific antigen expression is down-regulated by selenium through disruption of androgen receptor signaling. Cancer Res 2004; 64: 19–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.