Abstract
The benefits of opioid-based treatments to mitigate chronic pain can be hindered by the side effects of opioid-induced hyperalgesia (OIH) that can lead to higher consumption and risk of addiction. The present study advances the understanding of the molecular mechanisms associated with OIH by comparing mice presenting OIH symptoms in response to chronic morphine exposure (OIH treatment) relative to control mice (CON treatment). Using RNA-Seq profiles, gene networks were inferred in the trigeminal ganglia (TG), a central nervous system region associated with pain signaling, and in the nucleus accumbens (NAc), a region associated with reward dependency. The biological process of nucleic acid processing was over-represented among the 122 genes that exhibited a region-dependent treatment effect. Within the 187 genes that exhibited a region-independent treatment effect, circadian rhythm processes were enriched among the genes over-expressed in OIH relative to CON mice. This enrichment was supported by the differential expression of the period circadian clock 2 and 3 genes (Per2 and Per3). Transcriptional regulators in the PAR bZip family that are influenced by the circadian clock and that modulate neurotransmission associated with pain and drug addiction, were also over-expressed in OIH relative to CON mice. Also notable was the under-expression in OIH relative to CON mice of the Toll-like receptor, nuclear factor-kappa beta, and interferon gamma genes and enrichment of the adaptive immune processes. The results from the present study offer insights to advance the effective use of opioids for pain management while minimizing hyperalgesia.
Keywords: opioid-induced hyperalgesia, morphine, RNA-Seq, circadian rhythm, adaptive immune response
Introduction
Chronic pain affects 30 to 40% of the population in the United States and Europe [1-3]. Supervised short-term opioid-based therapies can help patients ameliorate pain while minimizing the risk to develop analgesic tolerance and addiction. The chronic use of opioids can elicit opioid-induced hyperalgesia (OIH), a paradoxical hypersensitivity to stimuli triggered by the same stimuli originally prescribed to alleviate pain. Thus, opioid use can engender a more fierce sense of pain instead of alleviating the pain and in turn may lead to higher opioid consumption [4,5]. A human (f)MRI study demonstrated that subjects who developed hyperalgesia to noxious thermal stimuli post-opioid infusion also presented activation of the midbrain reticular formation that transmits pain signals during the hour-long infusion [6]. The molecular mechanisms supporting this activation could be the foundation of pro-nociceptive systems.
The intersection between OIH and development of opioid tolerance together with the complexity of the OIH pathophysiology have challenged our understanding of molecular processes associated with OIH and identification of therapeutic targets [7]. Candidate gene studies have associated the desensitization of mu opioid receptor (MOR) triggered by repeated opioid exposure with both OIH and loss of opioid treatment effectiveness due to the development of tolerance to analgesia [8,5]. MORs are expressed in sensory neurons in the trigeminal ganglia (TG) and the nucleus accumbens (NAc), and the levels of expression are modulated by morphine treatment [9]. These two central nervous system (CNS) regions are involved in nociception, pain processing and reward dependence, respectively [10,11].
The TG neurons relay cephalic sensations of pain, touch, or pressure to the central nervous system [12], and many types of opioid receptors are abundantly expressed in these sensory neurons [13]. In addition to relaying sensory information to the brain, the TG neurons can trigger vasodilation and neurogenic inflammation, and elicit hyperalgesia, sensitization, and allodynia in response to certain chronic stimuli [10,14]. Opioid use increases the release of dopamine in the NAc, and the disruption in NAc processes plays a role in hyperalgesia comorbidities including depression, irritability, fatigue, sleepiness, and loss of appetite [15]. The chronic administration of opioids to rodents is a proven construct to advance the characterization of the neurological basis of OIH [16-18]. Despite the known involvement of TG and NAc in processes associated with chronic pain and the potential to be affected by chronic opioid exposure, the simultaneous molecular response of these two CNS regions to chronic opioid use in a model of OIH has not been reported.
The overall objective of this study is to advance the understanding of the molecular interactions underlining OIH. High-throughput RNA-Seq technology was used to profile the NAc and TG transcriptome of mouse that presented OIH triggered by chronic opioid exposure. Three supporting objectives were explored: a) to detect and characterize region-dependent and - independent profiles and networks associated with OIH; b) to identify gene networks and biological processes that are predominantly dysregulated in the TG or NAc of individuals experiencing OIH; and c) to uncover regulatory factors and networks associated with OIH. The findings from these complementary analyses offered insights that can be used in the development of effective strategies to prevent, cure, or manage OIH while benefiting from the analgesic effects of controlled opioid-based therapies.
Materials and Methods
Animal experiments
Male C57BL6/J mice (Jackson Laboratories, Bar Harbor, ME), between 9-12 weeks old were studied. Mice were group-housed in a 12-12 light-dark cycle, and the food was available ad libitum. All experimental procedures were approved by the University of Illinois at Chicago Office of Animal Care and Institutional Biosafety Committee in accordance with AALAC guidelines, the Animal Care Policies of the University of Illinois at Chicago and the European Union directive on the subject of animal rights. Animals were weighed daily during treatment, and no adverse effects of treatment were observed on body weight or visibly healthy performance.
Twenty samples distributed on a two-by-two experimental design including the factors of treatment and region were studied. Mice in the OIH treatment group were injected with morphine dissolved in 0.9% saline solution whereas mice in the CON treatment group were injected with saline vehicle administered as a 10mL/kg volume. Mice were injected morphine or vehicle twice daily (s.c.) for 4 days [19].On days 1-3 the dose of morphine was 20 mg/kg, and on day 4 the dose was 40 mg/kg. Baselines were determined before the morning injection of morphine or vehicle.
Hyperalgesia was confirmed using hind paw and cephalic mechanical sensitivity tests. The mice were counterbalanced into groups following the first basal response for mechanical sensitivity taken on day 1. The mechanical sensitivity was assessed in the plantar surface of the left hind paw (n=5 mice/treatment) on days 1-4, and these mice were used in the RNA-Seq experiment. Complementary mechanical sensitivity was assessed in the cephalic/periorbital region caudal to the eyes and near the midline on a separate group of mice (n=6 mice/treatment) on days 1, 3 and 5. Following protocols previously described [20], mice were habituated to the testing boxes for 2 days before testing, and the experimenter was blinded to the drug condition being tested. Mechanical response testing occurred in low-light conditions in the morning prior to the first injection of the day and the threshold for response to punctate mechanical stimuli (withdraw) was tested according to the up-and-down method [20,21] The testing regions were stimulated with a series of 8 von Frey filaments with a bending force ranging from 0.008 to 2 g [22-24].
A two-way ANOVA with interaction model was used to describe the mechanical threshold measurements and to test for the effects of treatment, day, and interaction. A significant interaction effect (P-value < 0.05) was followed with a Holm-Sidak post-hoc correction of the pair-wise contrasts between each treatment-day group and the CON-day 1 group of mice.
Region and RNA extraction and sequencing
The collection of samples for RNA-Seq analysis started on day 5 of the experiment after mechanical response testing and at approximately 15h following the last morphine or vehicle injection. Mice were anesthetized with pentobarbital (Somnosol), euthanized and intracardially perfused with ice-cold saline following our proven protocols [22-24]. Brains were extracted, and the TG and NAc were rapidly dissected, snap-frozen, and stored at −80 °C following our protocols [20,25]. The CNS regions from individual mice were homogenized with TRIzol (Invitrogen, Carlsbad, CA) and ceramic beads (MO BIO, Carlsbad, CA), and RNA was isolated using the RNA-kit (Omega Biotek, Norcross, GA).
The RNA integrity numbers of the 20 samples corresponding to five animals each for two treatments (OIH and CON) and two CNS regions (TG and NAc) were above 7.5. Libraries were constructed from these samples and were individually analyzed using a HiSeq 4000 (Illumina, San Diego, CA) platform that generated 100nt paired-end reads. The FASTQ data files from this RNA-Seq analysis are available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (GSE126662). The average Phred quality score of the reads assessed using FastQC [26] was > 30 across all read positions, and therefore no reads were trimmed.
RNA quantification and differential expression analysis
The paired-end reads from the individual samples were aligned to the C57B1/6J mouse genome (version GRCm38, downloaded in October 2016 from NCBI [27] using Kallisto [28] with default settings. The normalized (trimmed mean of M-values) gene expression values were described using a generalized linear model encompassing the main effects of treatment group (OIH or CON) and CNS region (TG or NAc) and the corresponding interaction and analyzed using edgeR v. 3.14.0. in the R v. 3.3.1 environment [29]. Genes supported by > 5 reads per treatment-region combination were analyzed to ensure adequate representation across mouse groups.
Orthogonal pairwise contrasts between treatment-region groups were evaluated in addition to testing for the effects of treatment-region interaction and main effects of treatment and region. The groups compared in the contrasts are identified by the treatment followed by the region and encompassed: OIH_TG, OIH_NA, CON_TG and CON_NA. The test-statistic P-values were adjusted for multiple testing using the Benjamini-Hochberg False Discovery Rate (FDR) approach [30].
Functional enrichment and network inference
Two complementary approaches were used to identify over-represented functional categories among the genes exhibiting differential expression across treatment and region groups [31-34]. Functional categories investigated included Gene Ontology (GO) biological processes (BPs) and molecular functions (MFs) and KEGG pathways. The Gene Set Enrichment Analysis (GSEA) approach implemented in the software package GSEA-P 2.0 [35] was used to identify category over-representation with gene over- and under-expressed while considering all genes analyzed, sorted by the signed and standardized logarithm-transformed fold change between the groups compared. The enrichment score of each functional category in the Mus musculus Molecular Signature Database (MSigDB) was calculated using the maximum deviation of the cumulative sum based on the signed and standardized fold-change. The statistical significance of the category enrichment was assessed using the FDR-adjusted P-value computed from 1000 permutations.
The over-representation of functional categories was also studied among the genes that exhibited a significant interaction or main effect using the Database for Annotation, Visualization and Integrated Discovery (DAVID 6.8). The enrichment of Direct GO categories in the DAVID database was evaluated [36]. The Mus musculus genome was used as the background for enrichment testing, and enrichment is reported using the Expression Analysis Systematic Explorer (EASE) score that was computed using a one-tailed jackknifed Fisher hypergeometric exact test. Functional categories were clustered to ease the interpretation of enriched categories, and the statistical significance of each cluster is summarized as the geometric mean of the −log10 EASE scores from the categories in the cluster [37-39].
Differences among genes differentially expressed between treatment-region groups were studied by reconstructing gene networks using the Bisogenet package [40] in the Cytoscape platform [41]. Information from gene and protein interactions annotated in databases including BIOGRID, HPRD, DIP, BIND, INTACT, and MINT was used to visualize relationships between genes [42-47]. The network framework includes genes that exhibited a significant interaction effect (FDR-adjusted P-value < 0.1) and that were differentially expressed in at least one of the 3 orthogonal contrasts (OIH_TG-CON_TG, OIH_NA-CON_NA or OIH_TG-OIH_NA) at FDR-adjusted P-value < 0.1. The study of the previously described orthogonal contrasts enabled the direct comparison of treatment effect across networks supported by gene-to-gene connectivity and intersectionality between CNS regions. The genes differentially expressed between treatments within each region were used as network framework genes and are identified by full nodes with size commensurate with the fold change between treatments. In addition to the framework genes, directly connected genes in the Bisogenet database that did not reach the differential expression threshold were included in the network. The comparison of these networks enabled the detection of shared and distinct co-regulation pattern between OIH and CON across CNS regions
Additional understanding of the differences in gene regulation between OIH and CON across and within CNS region was gained by testing for the over-representation of potential transcription factors of the genes that exhibited differential expression between treatments and regions. The iRegulon plugin within the Cytoscape environment was used to identify transcriptional regulators of differentially expressed target genes [48]. Transcription factor binding motifs over-represented among the differentially expressed genes were identified using the position weight matrix (PWM) method [48] and the over-representation is expressed in terms of a normalized enrichment score (NES). Motif match was assigned at FDR-adjusted P-value < 0.001 and with a minimum Receiving Operating Curve threshold for estimation of the Area under the Curve > 0.03.
Results
Mechanical pain threshold and chronic morphine exposure
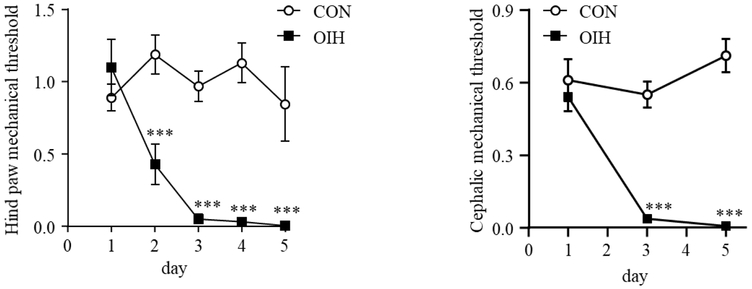
Figures 1A and 1B present the profile of hind paw and cephalic mechanical threshold across treatment (OIH and CON) and days, respectively The interaction between treatment and day, the main effect of treatment and the main effect of day were significant (P-value < 0.001) in both regions. Post-hoc contrasts of the mechanical threshold between the OIH mice at days 2-5 and the CON-day 1 group were significant at P-value < 0.001 and are denoted with “***”.
Fig 1.
Chronic treatment with morphine elicited mechanical hyperalgesia. Mice were treated with vehicle (CON) or morphine (OIH) twice daily (20 mg/kg/injection on days 1-3, and 40 mg/kg/injection on day 4). Mechanical responses were assessed in the morning before the first injection of the day. (A) Two-way ANOVA of hind paw mechanical threshold (n=5/group), P-value < 0.001 day, treatment, and interaction effect; *** P-value < 0.001 contrast to day 1 CON. (B) Two-way ANOVA of cephalic/periorbital mechanical threshold on an independent experiment (n=6/group), P-value < 0.001 day, treatment and interaction effects; *** P-value < 0.001 contrast to day 1 CON
Sequencing metrics
The sequencing of the 20 RNA samples produced approximately 3.1 billion sequenced reads and approximately 76.6 million paired-end reads per sample. The number of reads was consistent across treatment-region groups (coefficient of variation < 3.4%) and the average percentage of mapped reads was approximately 84%. The effects of the interaction and the main effects of treatment and CNS region were tested on 23,349 genes that surpassed the minimum number of reads per treatment-region combination.
Transcriptome changes associated with opioid-induced hyperalgesia that are region-dependent
Overall, 89 genes exhibited a significant (FDR-adjusted P-value < 0.05) opioid-induced hyperalgesia treatment-by-CNS region interaction effect. The 35 genes that showed the most significant (FDR-adjusted P-value < 0.001) interaction effect are presented in Table 1. An extended list of the 122 genes that exhibited an interaction effect at FDR-adjusted P-value < 0.1) can be found in File S1 Table A. Among these, an approximately even number of genes were differentially expressed between the treatment groups in the NAc and TG, respectively. Eight genes showed opioid treatment effect both in NAc and TG, and 6 of these genes displayed opposite expression patterns in the 2 CNS regions studied (Table 1) including sodium channel, voltage-gated, type VII, alpha (Scn7a), adipocyte enhancer binding protein 1 (Aebp1), integrin subunit beta 4 (Itgb4), and plasmolipin (Pllp). The rest of the genes exhibiting an interaction effect are substantially more differentially expressed in response to the opioid treatment in one CNS region than the other such as phosphoinositide-interacting regulator of transient receptor potential channels (Pirt) and LMBR1 domain containing 2 (Lmbrd2).
Table 1.
Genes exhibiting significant (FDR-adjusted P-value < 0.005) opioid-induced hyperalgesia treatment-by-central nervous system region interaction effect.
| Gene Symbol |
P-value |
aFDR P- value |
bOIH NA- CON NA |
OIH TG- CON TG |
CON TG- CON NA |
OIH TG- OIH NA |
OIH TG- CON NA |
CON TG- OIH NA |
|---|---|---|---|---|---|---|---|---|
| Tusc5 | 1.7E−14 | 3.9E−10 | −1.33 | 0.04 | 8.02 | 9.41 | 8.09 | 9.36 |
| Lnpep | 4.2E−14 | 4.9E−10 | −0.13 | 1.35 | 1.44 | 2.95 | 2.82 | 1.58 |
| Pirt | 7.0E−12 | 5.4E−08 | −0.96 | −0.01 | 8.10 | 9.07 | 8.12 | 9.06 |
| Itgb4 | 1.5E−10 | 8.7E−07 | −0.36 | 0.15 | 2.84 | 3.37 | 3.01 | 3.21 |
| Serpinb1a | 4.1E−10 | 1.9E−06 | −0.59 | −0.03 | 2.91 | 3.49 | 2.90 | 3.51 |
| Gm33320 | 6.4E−10 | 2.5E−06 | 0.31 | −0.33 | 1.78 | 1.15 | 1.47 | 1.47 |
| Acpp | 7.9E−10 | 2.6E−06 | −1.14 | 0.05 | 8.10 | 9.31 | 8.17 | 9.25 |
| Scn7a | 2.5E−09 | 7.4E−06 | −0.94 | 0.17 | 7.25 | 8.38 | 7.44 | 8.20 |
| Cysltr2 | 6.4E−09 | 1.6E−05 | −1.31 | −0.08 | 2.69 | 3.94 | 2.64 | 4.01 |
| Gm33449 | 6.9E−09 | 1.6E−05 | 0.63 | −1.51 | 0.60 | −1.52 | −0.88 | −0.03 |
| Aebp1 | 1.2E−08 | 2.5E−05 | −0.50 | 0.54 | 1.81 | 2.86 | 2.37 | 2.31 |
| Prelp | 2.2E−08 | 4.3E−05 | −0.64 | 0.02 | 0.53 | 1.21 | 0.57 | 1.18 |
| Cldn19 | 2.9E−08 | 5.3E−05 | −1.28 | 0.13 | 7.70 | 9.16 | 7.85 | 9.01 |
| Pllp | 3.2E−08 | 5.3E−05 | −0.32 | 0.18 | 1.57 | 2.08 | 1.76 | 1.89 |
| Arfgef3 | 6.3E−08 | 9.8E−05 | −0.01 | 0.61 | 0.23 | 0.88 | 0.87 | 0.25 |
| Mpz | 1.0E−07 | 1.5E−04 | −1.26 | 0.01 | 8.36 | 9.65 | 8.39 | 9.63 |
| Myo1d | 1.1E−07 | 1.5E−04 | −0.24 | 0.20 | 1.73 | 2.19 | 1.96 | 1.98 |
| Tlx2 | 1.8E−07 | 2.3E−04 | −2.46 | −0.03 | 8.32 | 10.77 | 8.31 | 10.78 |
| Prx | 2.4E−07 | 3.0E−04 | −0.80 | 0.10 | 7.79 | 8.70 | 7.90 | 8.59 |
| Zbed6 | 4.3E−07 | 5.0E−04 | 0.17 | 1.58 | −0.35 | 1.08 | 1.25 | −0.51 |
| Avil | 5.2E−07 | 5.7E−04 | −0.92 | −0.03 | 8.03 | 8.94 | 8.02 | 8.96 |
| Sncg | 5.6E−07 | 5.9E−04 | −1.27 | −0.07 | 7.63 | 8.84 | 7.58 | 8.90 |
| Tmem233 | 7.9E−07 | 8.0E−04 | −1.62 | −0.01 | 8.48 | 10.09 | 8.49 | 10.09 |
| Ppp1r1c | 8.4E−07 | 8.2E−04 | −0.88 | 0.00 | 7.97 | 8.87 | 7.99 | 8.86 |
| BC005561 | 9.7E−07 | 9.1E−04 | 0.12 | 0.93 | 0.12 | 0.96 | 1.08 | 0.01 |
| Gm42411 | 1.4E−06 | 1.3E−03 | −0.08 | 1.25 | −6.92 | −5.53 | −5.62 | −6.85 |
| Pmp22 | 2.9E−06 | 2.5E−03 | −0.55 | 0.03 | 6.76 | 7.36 | 6.81 | 7.32 |
| Lmbrd2 | 3.6E−06 | 3.0E−03 | 0.03 | 0.57 | 0.26 | 0.82 | 0.85 | 0.24 |
| A2m | 4.4E−06 | 3.5E−03 | −0.79 | 0.00 | 4.57 | 5.38 | 4.59 | 5.37 |
| Dok6 | 4.9E−06 | 3.7E−03 | −0.17 | 1.05 | −1.50 | −0.26 | −0.43 | −1.32 |
| Slc22a8 | 4.9E−06 | 3.7E−03 | −0.14 | 0.91 | −1.39 | −0.32 | −0.45 | −1.24 |
| Dbp | 5.3E−06 | 3.9E−03 | 0.45 | 1.15 | −2.48 | −1.76 | −1.31 | −2.93 |
| Apln | 6.3E−06 | 4.4E−03 | −0.52 | 0.26 | −3.45 | −2.65 | −3.17 | −2.93 |
| Prrxl1 | 7.0E−06 | 4.8E−03 | −0.70 | 0.06 | 7.60 | 8.39 | 7.68 | 8.32 |
| Ahnak2 | 7.2E−06 | 4.8E−03 | −0.95 | 0.08 | 8.09 | 9.14 | 8.19 | 9.04 |
False Discovery rate adjusted P-value.
Log2(fold change) between two treatment-brain region groups: OIH= opioid-induced hyperalgesia mice; CON=Control mice; TG=trigeminal ganglia; and NA=nucleus accumbens.
Functional and network analysis of genes that exhibit region-dependent associations with opioid-induced hyperalgesia
Genes expressing significant OIH treatment-by-region interaction effect can exhibit one of many patterns. Therefore, a GSEA study of the genes over- or under-expressed between OIH and CON groups within CNS region was explored to gain further insights into the functional categories predominantly impacted by treatment in a region-dependent manner. RNA and DNA processing including GO BP mRNA metabolic process (GO:0016071), RNA processing (GO:0006396), and regulation of DNA metabolic process (GO:0051052) were enriched (FDR-adjusted P-value < 0.05; NES > 1.9) among the genes presenting a significant region-dependent treatment effect and over-expressed in the NAc of OIH relative to CON mice. Additional categories borderline enriched included: neuron projection development (P-value < 0.001) among genes under-expression in the NAc of OIH relative to CON mice; GTPase binding (P-value < 0.006) among the genes over-expressed in the TG of OIH relative to CON mice; and angiogenesis (P-value < 0.007) among the genes under-expressed in the NAc of OIH relative to CON mice.
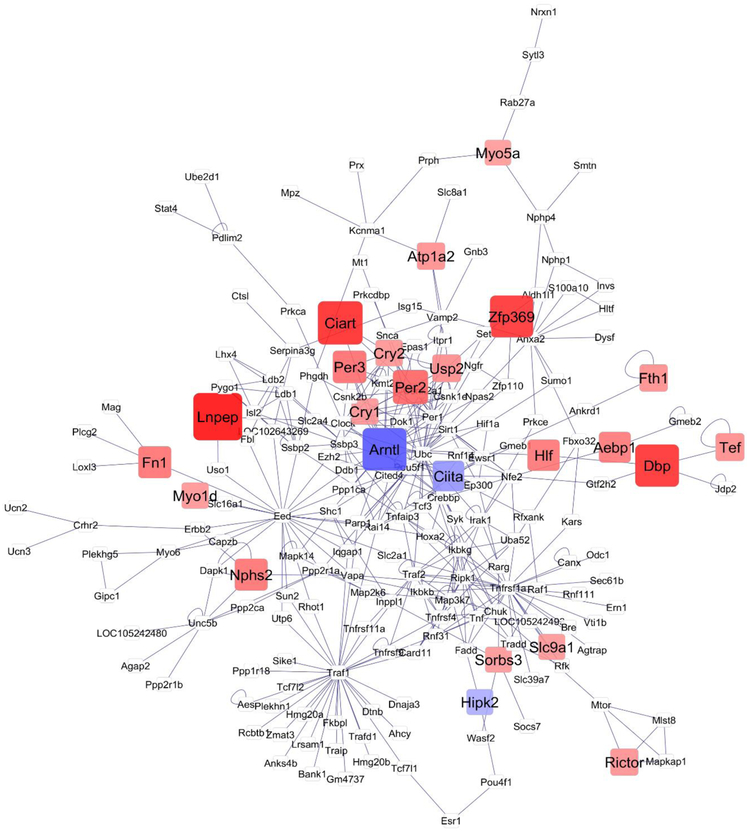
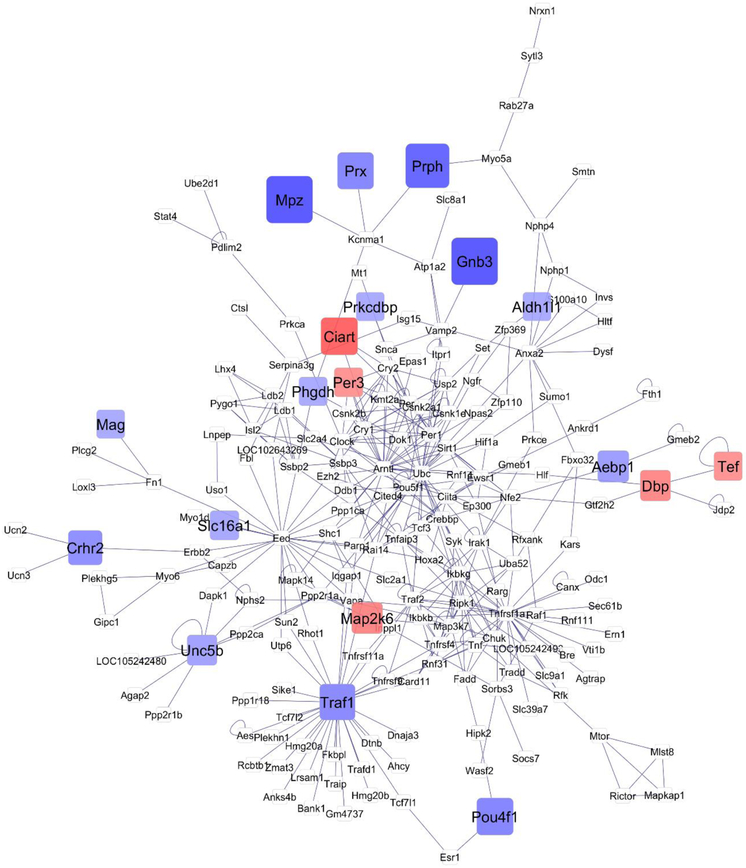
Network visualization further the understanding of the relationships among the 122 genes that exhibited significant (FDR-adjusted P-value < 0.1) treatment-by-region interaction effect. This interaction effect encompassed one or more significant ((FDR-adjusted P-value < 0.1) orthogonal contrast (OIH_NA-CON_NA, or OIH_TG-CON_TG or OIH_NA-OIH_TG). The networks that depict the relationship between genes for the OIH_NA-CON_NA and OIH_TG-CON_TG contrasts are depicted in Figures 2 and 3, respectively. The network of the remaining orthogonal contrast (OIH_NA-OIH_TG) is included for completeness in File S1 Figure 1. The simultaneous study of these genes that form the network framework, the consideration of the differential expression patterns and connectivity helped us uncover region-dependent transcript changes associated with OIH. Red and blue rectangular nodes represent framework genes and edges represent the known associations between genes based on curated databases of molecular interactions. White nodes represent genes directly connecting framework genes.
Fig 2.
Network of differential gene expression associated with opioid-induced hyperalgesia in the nucleus accumbens (contrast OIH_NA-CON_NA). Node color: red -framework genes over-expressed in OIH; blue –framework gene under-expressed in OIH; white -genes connecting framework genes but not differentially expressed in this study; node size indicates fold change
Fig 3.
Network of differential gene expression associated with opioid-induced hyperalgesia in the nucleus accumbens (contrast OIH TG-CON TG). Node color: red -framework genes over-expressed in OIH; blue –framework gene under-expressed in OIH; white -genes connecting framework genes but not differentially expressed in this study; node size indicates fold change
Transcriptome changes associated with opioid-induced hyperalgesia shared across regions
Overall, 97 genes exhibited significant (FDR-adjusted P-value < 0.05) differential expression between OIH and CON mice that was consistent across central nervous system regions. Of these, Table 2 presents the 32 genes exhibiting the highest OIH effect (FDR-adjusted P-value < 0.005). The 187 genes differentially expressed between OIH and CON mice at FDR-adjusted P-value < 0.1) are listed in S1 File Table B). Notable, among the genes over-expressed in OIH relative to CON mice (Table 2 and S1 File Table B) are three transcription factors in the circadian rhythm pathway, D site-binding protein (Dbp), thyrotroph embryonic factor (Tef) and hepatic leukemia factor (Hlf). Other genes over-expressed in OIH relative to CON mice included: circadian associated repressor of transcription (Ciart), period circadian clock 3 (Per3), dimethylaniline monooxygenase (Fmo2), nuclear receptor subfamily 1 group D member 2 (Nr1d2), and collagen alpha-1(III) chain preproprotein (Col3a1).
Table 2.
Genes differentially expressed (FDR-adjusted P-value < 0.005) between opioid-induced hyperalgesia (OIH) and control (CON) mice.
| Gene symbol |
Gene Name |
aOIH- CON |
P-value |
bFDR P- value |
|---|---|---|---|---|
| Ciart | circadian associated repressor of transcription | 0.99 | 1.0E−37 | 2.4E−33 |
| Per3 | period circadian clock 3 | 0.49 | 3.0E−18 | 3.5E−14 |
| Fmo2 | dimethylaniline monooxygenase | 0.69 | 1.8E−13 | 1.2E−09 |
| Nr1d2 | nuclear receptor subfamily 1 group D member 2 | 0.27 | 2.0E−13 | 1.2E−09 |
| Col3a1 | collagen alpha-1(III) chain preproprotein | −0.54 | 5.0E−13 | 2.1E−09 |
| Tef | thyrotroph embryonic factor | 0.32 | 7.6E−13 | 2.5E−09 |
| Ankrd12 | ankyrin repeat 12 | 0.26 | 7.7E−12 | 2.2E−08 |
| Fabp7 | fatty acid-binding protein, brain | −0.56 | 1.4E−10 | 3.6E−07 |
| Cd93 | complement component C1q receptor precursor | −0.32 | 9.6E−09 | 2.2E−05 |
| Rhoj | rho-related GTP-binding protein RhoJ precursor | −0.41 | 4.3E−08 | 9.0E−05 |
| Gm3417 | t-complex-associated testis expressed 3-like | 1.64 | 8.9E−08 | 1.7E−04 |
| Gm10863 | predicted gene 10863 | −0.52 | 1.0E−07 | 1.7E−04 |
| Nr1d1 | nuclear receptor subfamily 1 group D member 1 | 0.34 | 1.0E−07 | 1.7E−04 |
| Per2 | period circadian clock 2 | 0.52 | 1.3E−07 | 2.0E−04 |
| Arntl | aryl hydrocarbon receptor nuclear translocator-like protein 1 | −0.73 | 1.5E−07 | 2.1E−04 |
| Bhlhe41 | class E basic helix-loop-helix protein 41 | 0.25 | 1.5E−07 | 2.1E−04 |
| Cldn5 | claudin-5 | −0.35 | 2.8E−07 | 3.7E−04 |
| Setx | senataxin | 0.19 | 4.5E−07 | 5.4E−04 |
| Plin4 | perilipin 4 | 0.81 | 4.6E−07 | 5.4E−04 |
| Fam76a | family with sequence similarity 76, member A | 0.16 | 6.1E−07 | 6.7E−04 |
| Hspa1a | heat shock 70 kDa protein 1A | −0.70 | 1.1E−06 | 1.2E−03 |
| Hmgcs2 | hydroxymethylglutaryl-CoA synthase, mitochondrial precursor | −0.29 | 1.4E−06 | 1.5E−03 |
| Fam107a | family with sequence similarity 107, member A | 0.33 | 1.6E−06 | 1.6E−03 |
| Ly86 | lymphocyte antigen 86 precursor | −0.44 | 1.7E−06 | 1.6E−03 |
| Fkbp5 | peptidyl-prolyl cis-trans isomerase FKBP5 | 0.27 | 2.0E−06 | 1.8E−03 |
| Tmem45a | transmembrane protein 45A | −0.59 | 2.0E−06 | 1.8E−03 |
| Gm40853 | predicted gene 40853 | 1.31 | 2.3E−06 | 1.9E−03 |
| Hspa1b | heat shock 70 kDa protein 1B | −0.82 | 2.8E−06 | 2.2E−03 |
| Vegfa | vascular endothelial growth factor A | −0.15 | 3.4E−06 | 2.7E−03 |
| Mpzl1 | myelin protein zero-like protein 1 | −0.20 | 4.8E−06 | 3.6E−03 |
| Plat | tissue-type plasminogen activator preproprotein | −0.20 | 5.0E−06 | 3.7E−03 |
| Gm35908 | predicted gene 35908 | −0.70 | 6.1E−06 | 4.3E−03 |
Log2(fold change) between OIH and CON mice.
False Discovery rate.
Functional analysis of gene exhibiting region-independent associations with opioid-induced hyperalgesia
The DAVID analysis of genes differentially expressed between OIH and CON groups across regions uncovered enrichment (ES=3.568) and irrespective to pattern uncovered the enrichment of GO BP myelination (GO:0042552) and ensheathment of neurons (GO:0007272). Within the enriched category ensheathment of neurons, the genes that exhibited an interaction effect include Prx, Fa2h, Pllp, Cldn11, and Pmp22 and the genes that exhibited a treatment effect include Serinc5, Hexa, cldn5, Mir138-1, Lgi4, Lpar1, and Gjc3. Other categories detected by the DAVID analysis that did not reach the statistical threshold included protein folding (GO:0006457, P-value < 0.001); negative regulation of cellular metabolic process (GO:0031324, P-value < 0.00068); cell adhesion molecule binding (GO:0050839, P-value < 0.002) and regulation of circadian rhythm (GO:0042752, P-value 0.00018).
Enrichment results from the GSEA study within gene under- and over-expressed patterns complemented the findings from DAVID. The most enriched informative categories (FDR-adjusted P-value < 0.05 and normalized enrichment score > ∣1.8∣ are presented in Table 3 and the extended list of categories (FDR-adjusted P-value < 0.1) is presented in S1 File Table C.
Table 3.
Enriched informative categories among the genes under- or over-expressed in the opioid-induced hyperalgesia relative to control mice using GSEA.
|
aExpression/ bCategory |
GO Identifier | GO name | cNES | P-value |
dFDR P- value |
|---|---|---|---|---|---|
| Over | |||||
| BP | GO:0043153 | entrainment of circadian clock by photoperiod | 2.18 | 0.0E+00 | 3.1E−02 |
| BP | GO:0009648 | photoperiodism | 2.08 | 0.0E+00 | 3.8E−02 |
| BP | GO:0042752 | regulation of circadian rhythm | 2.1 | 0.0E+00 | 4.0E−02 |
| Under | |||||
| KEGG | cell adhesion molecules cams | −2.18 | 0.0E+00 | 0.0E+00 | |
| BP | GO:0006458 | de novo protein folding | −2.11 | 0.0E+00 | 9.7E−04 |
| BP | GO:0002224 | toll-like receptor signaling pathway | −2.06 | 0.0E+00 | 4.0E−03 |
| BP | GO:0007266 | rho protein signal transduction | −2.03 | 0.0E+00 | 6.3E−03 |
| MF | GO:0005201 | extracellular matrix structural constituent | −2 | 0.0E+00 | 6.9E−03 |
| BP | GO:0032675 | regulation of interleukin 6 production | −1.98 | 0.0E+00 | 8.1E−03 |
| BP | GO:0060333 | interferon gamma mediated signaling pathway | −1.92 | 0.0E+00 | 1.7E−02 |
| BP | GO:0032653 | regulation of interleukin 10 production | −1.9 | 1.6E−03 | 2.2E−02 |
| BP | GO:0032757 | positive regulation of interleukin 8 production | −1.88 | 1.6E−03 | 2.8E−02 |
| BP | GO:0001819 | positive regulation of cytokine production | −1.84 | 0.0E+00 | 3.7E−02 |
| BP | GO:0032735 | positive regulation of interleukin 12 production | −1.82 | 3.3E−03 | 4.3E−02 |
| BP | GO:0002250 | adaptive immune response | −1.81 | 0.0E+00 | 4.8E−02 |
Expression: Over- and under-expressed in opioid-induced hyperalgesia relative to control mice.
BP: biological process; MF: molecular function.
normalized enrichment score.
False Discovery Rate adjusted P-value.
Notable is the enrichment of circadian rhythm categories among the genes over-expressed in OIH relative to CON mice. Also notable is the enrichment among the genes under-expressed in OIH relative to CON mice of genes that participate on adaptive immune regulation including toll-like receptor pathway and several cytokines including interleukin 6, 8, 10 and 12.
Transcriptome differences between trigeminal ganglia and nucleus accumbens independent of treatment
Overall, 738genes were differentially expressed between TG and NAc (log2(fold change) > 2 and FDR-adjusted P-value <1.0E-5) and are listed in the S1 File Table D. Among the genes that present region effect but failed to present treatment-by-region interaction effect, the 18 genes that presented the most extreme fold change are listed in Table 4. Genes over-expressed in TG relative to NAc included: transient receptor potential cation channel subfamily V member 1 (Trpv1), transient receptor potential cation channel, subfamily A, member 1 (Trpa1), Piezo-type mechanosensitive ion channel component 2 (Piezo2), calcitonin-related polypeptide alpha (Calca), neuromedin B (Nmb), and MAS-related GPR, member a3 (Mrgpra3). Genes over-expressed in NAc relative to TG included: dopamine receptor D1 (Drd1), opioid receptor Kappa 1 (Oprk1), neuropeptide Y (Npy), glutamate decarboxylase 1 and 2 (Gad1 and Gad2).
Table 4.
Genes presenting the most extreme fold change of expression between trigeminal ganglia (TG) and nucleus accumbens (NAc) at FDR-adjusted P-value <1.0E−8.
| Gene Symbol |
Gene Name | aTG-NAc |
|---|---|---|
| Mrgpra3 | mas-related G-protein coupled receptor member A3 | 9.24 |
| Slc32a1 | vesicular inhibitory amino acid transporter | −9.06 |
| Foxd3 | forkhead box protein D3 | 8.98 |
| Ankrd63 | ankyrin repeat domain-containing protein 63 | −8.96 |
| Isl2 | insulin gene enhancer protein ISL-2 | 8.91 |
| Tlx3 | T-cell leukemia homeobox protein 3 | 8.86 |
| Rprml | reprimo-like protein | −8.82 |
| Six1 | homeobox protein SIX1 | 8.81 |
| Mrgprx1 | mas-related G-protein coupled receptor member X1 | 8.76 |
| Mrgprd | mas-related G-protein coupled receptor member D | 8.75 |
| Gad2 | glutamate decarboxylase 2 | −8.67 |
| Hoxd8 | homeobox protein Hox-D8 | 8.60 |
| Scn10a | sodium channel, voltage-gated, type X, alpha | 8.52 |
| Arx | homeobox protein ARX | −8.49 |
| Avil | advillin | 8.46 |
| Mrgprb4 | mas-related G-protein coupled receptor member B4 | 8.46 |
| Slc36a2 | solute carrier family 36 (proton/amino acid symporter), member 2 | 8.43 |
| Hpcal4 | hippocalcin-like protein 4 | −8.42 |
| Gad1 | glutamate decarboxylase 1 | −8.42 |
Log2(fold change) between the TG and NAc regions.
The clusters of informative categories with highest DAVID enrichment scores (enrichment score >1.7) among the genes differentially expressed between CNS regions (log2(fold change) > 2 and FDR-adjusted P-value <1.0E-5) are listed in Table 5. Notable enriched categories among the genes differentially expressed between CNS regions are related to addiction (e.g., nicotine, amphetamine), glutamate-related activities receptor activities (KEGG mmu04724, GO:0004970).
Table 5.
Clusters of enriched functional categories (enrichment score ES > 1.7) among the genes differentially expressed between the trigeminal ganglia and nucleus accumbens identified using DAVID.
| aCategory | Category Identifier and Name | P-value |
bFDR P- value |
|---|---|---|---|
| Cluster 1 | ES=3.89 | ||
| KEGG | mmu04512:ECM-receptor interaction | 1.2E−05 | 1.5E−02 |
| KEGG | mmu04510:Focal adhesion | 3.7E−05 | 4.8E−02 |
| Cluster 2 | ES=2.89 | ||
| BP | GO:0006813~potassium ion transport | 1.7E−05 | 3.0E−02 |
| MF | GO:0005244~voltage-gated ion channel activity | 1.2E−04 | 1.8E−01 |
| BP | GO:0034765~regulation of ion transmembrane transport | 1.3E−04 | 2.3E−01 |
| Cluster 3 | ES=2.89 | ||
| MF | GO:0004970~ionotropic glutamate receptor activity | 6.2E−04 | 9.6E−01 |
| MF | GO:0005234~extracellular-glutamate-gated ion channel activity | 6.2E−04 | 9.6E−01 |
| Cluster 4 | ES=2.35 | ||
| KEGG | mmu04015:Rap1 signaling pathway | 4.8E−04 | 6.2E−01 |
| Cluster 5 | ES=2.03 | ||
| KEGG | mmu05033:Nicotine addiction | 1.8E−04 | 2.3E−01 |
| Cluster 6 | ES=1.79 | ||
| KEGG | mmu04713:Circadian entrainment | 2.0E−06 | 2.6E−03 |
| KEGG | mmu04724:Glutamatergic synapse | 3.9E−06 | 5.1E−03 |
| KEGG | mmu04720:Long-term potentiation | 6.7E−05 | 8.6E−02 |
| KEGG | mmu05031:Amphetamine addiction | 3.7E−04 | 4.7E−01 |
| KEGG | mmu04911: Insulin secretion | 7.3E−04 | 9.4E−01 |
BP: biological process; MF: molecular function; KEGG: KEGG pathway.
False Discovery Rate adjusted P-value.
Regulation of genes associated with opioid-induced hyperalgesia treatment
The regulatory analysis enabled us to detect transcription factors that target many of the genes dysregulated by chronic morphine treatment. Transcription factor motifs were searched in the regulatory region of genes that exhibited differential expression between OIH and CON mice within TG, within NAc, and across both regions. The targets include 75 genes that were over-expressed and 112 genes that were under-expressed in OIH relative to CON within or across CNS regions.
Table 6 highlights the most enriched transcription factors (normalizing enrichment score > 3, associated with > 20% of the genes differentially expressed between the treatments considered. In this table, the transcription factors are sorted by the expression pattern indicated by the log2(fold change) in OIH relative to CON mice. Among the seven highly enriched transcription factors three factors were also differentially expressed (FDR-adjusted P-value < 0.1) in OIH relative to CON mice: D site-binding protein (Dbp), hepatic leukemia factor (Hlf), and Class E basic helix-loop-helix protein 41 (Bhlhe41). The individual transcription factors were identified by more than 11 motifs. The transcription factor MLX interacting protein like (Mlxipl) in Table 6 shares all the target genes with Dbp and Hlf and therefore this factor also reaches the enrichment level cutoff.
Table 6.
Most enriched transcription factors (TFs) and corresponding target genes that exhibited differential over- or under-expression between opioid-induced hyperalgesia (OIH) and control (CON) mice within and across central nervous system regions (normalized enrichment score >3.0; > 20% of target genes).
| OIH vs CON | TF | aNES |
bGene Count |
cMotif | 4Log2(OIH/CON) | P-value | eFDR |
|---|---|---|---|---|---|---|---|
| Over | Dbp | 8.26 | 37 | 61 | 0.79 | 5.4E−13 | 2.1E−09 |
| Hlf | 5.40 | 30 | 12 | 0.41 | 3.5E−05 | 1.6E−02 | |
| Bhlhe41 | 4.83 | 46 | 49 | 0.25 | 1.5E−07 | 2.1E−04 | |
| Mlxipl | 3.26 | 23 | 1 | 0.02 | 8.3E−01 | 9.8E−01 | |
| Under | Hsf1 | 6.37 | 54 | 20 | −0.06 | 1.8E−01 | 8.0E−01 |
| Bcl3 | 4.31 | 38 | 2 | −0.36 | 1.3E−02 | 3.5E−01 | |
| Nfatc1 | 3.86 | 37 | 2 | −0.18 | 5.4E−02 | 5.9E−01 | |
| Srf | 3.71 | 26 | 4 | −0.03 | 6.3E−01 | 9.7E−01 | |
| E2f6 | 4.29 | 23 | 4 | −0.06 | 4.6E−02 | 5.6E−01 |
NES: normalized enrichment score or maximal enrichment score for a transcription factor
Number of target genes
Number of motifs per transcription factor.
Average log2(fold change) of the target genes between OIH and CON mice.
False Discovery Rate adjusted P-value of the transcription factor differential expression between OIH and CON
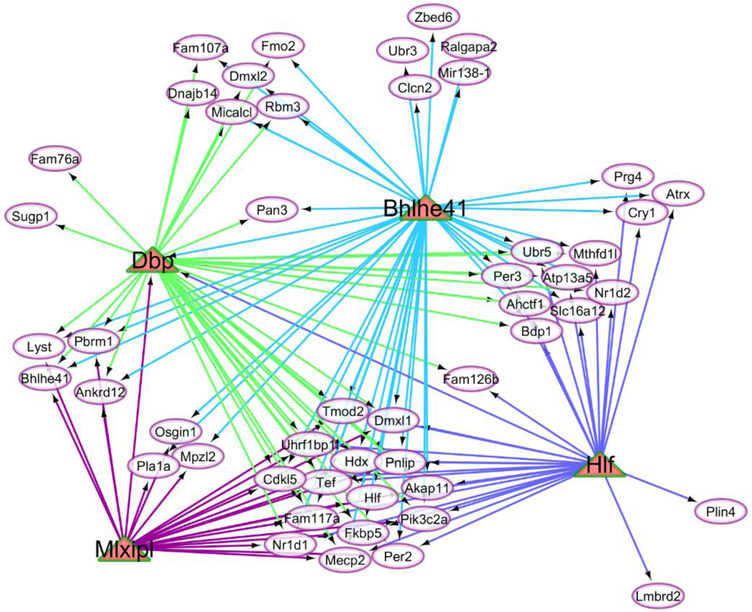
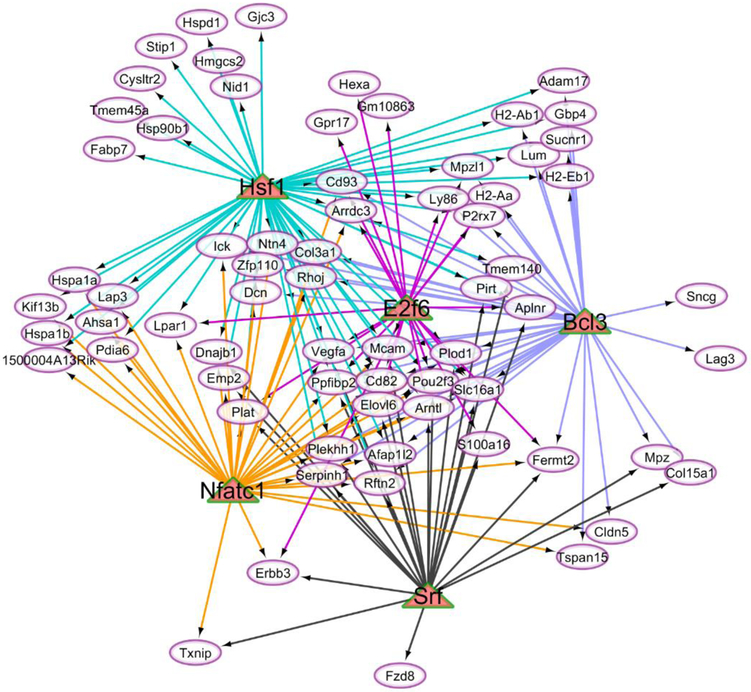
The visualization of the regulatory networks including the enriched transcription factors and target genes demonstrated the different connectivity of these molecules. The relationship between the target genes (ovals) over-expressed in OIH relative to CON mice at FDR-adjusted P-value < 0.1 and the matched regulatory transcription factors (triangle) is depicted in Figure 4 whereas Figure 5 depicts the same relationship for the transcription factors that were under-expressed in OIH relative CON mice.
Fig 4.
Network of enriched transcription factors (triangles) over-expressed in the opioid-induced hyperalgesia relative to control mice and associated with target genes (ovals) differentially expressed between treatments at FDR-adjusted P-value < 0.1
Fig 5.
Network of enriched transcription factors (triangles) under-expressed in the opioid-induced hyperalgesia relative to control mice and associated with target genes (ovals) differentially expressed between treatments at FDR-adjusted P-value < 0.1
Discussion
Chronic pain afflicts a large percentage of the population and managed opioid-based therapies can effectively reduce pain sensation while controlling for the development of opioid tolerance and addiction. However, chronic use of opioids like morphine can elicit OIH in some patients, a paradoxical hypersensitivity to stimuli, exacerbating rather than alleviating pain. Understanding the molecular mechanisms associated with OIH is essential to develop therapies that can maximize the benefit of managed opioid treatment. In this study, we advanced the knowledge on the transcriptome profiles associated with OIH using proven validated mouse model of this disorder [16].
The significantly lower mechanical threshold for withdrawal of OIH relative to CON mice both in the peripheral and cephalic regions as early as the second day of morphine treatment confirms the robustness of the model used to study hyperalgesia elicited by chronic opioid exposure (Figure 1). Our results are in agreement with previous reports of nociceptive sensitivity in response to chronic morphine administration [16,49,17]
The TG and NAc transcriptomes were profiled because of the neurological processes associated with pain and addiction encompassed by these CNS regions. The TG encompasses sensory neurons with abundant opioid receptors and plays a role in pain signaling [50], while NAc participates in motivation and reward-dependent processes including development and maintenance of addiction to drugs of abuse [51]. We uncovered both region-dependent and region-independent differences between OIH and CON mice in gene expression, and gene and regulatory networks and enrichment of functional categories among the impacted genes. Our work offers novel insights into molecular mechanisms underlying OIH and the identification of altered networks of genes provides potential targets to block the development of OIH.
Transcriptome changes associated with opioid-induced hyperalgesia that are region-dependent
Insights into the region-dependent association of OIH with transcript profiles were gained from the study of the 122 genes presenting significant (FDR-adjusted P-value < 0.1) region-by-treatment interaction effect (Table 1, and Table A in File S1). Many of these genes have been associated with hyperalgesia and pain sensation. The majority of the genes exhibited a higher fold change between OIH and CON in the NAc than in the TG, and this finding supports the more amply study of the participation of NAc in nociception [52]. A review of the genes presenting the most common patterns and supporting literature is presented by CNS region.
Genes over-expressed in the TG of OIH relative to CON mice included: docking protein 6 (Dok6), leucyl-cystinyl aminopeptidase (Lnpep), LMBR1 domain-containing protein 2 (Lmbrd2), and the transcriptional repressor Aebp1, and their patterns are consistent with previous studies (Table 1). Related to our findings, polymorphisms in Dok6 were significantly associated with morphine dose requirements for pediatric day surgery and postoperative pain scores [53] and Dok6 promotes neurite growth in sensory ganglia [54]. M1 aminopeptidases such as Lnpep play a role in pain [55] and consistent with our findings, aminopeptidase gene-deficient mice exhibit impaired pain response [56]. The pattern of Lnpep can be related to hyperalgesia through a complementary mechanism because the coded enzyme cleaves vasopressin, oxytocin and other neuropeptide hormones that participate in blood pressure regulation [57] and antigen presentation [58]. The profile observed in the present study advances the understanding of the role of TG neurons triggering vasodilation and neurogenic inflammation and eliciting hyperalgesia, sensitization, and allodynia in response to certain chronic stimuli [10].
The over-expression of Lmbrd2 in the TG of OIH relative to CON mice is consistent with the potential role as a regulator of adrenoceptor signaling [59] in consideration that opioid use can evoke complex regional pain syndrome through the action of adrenoreceptors [60]. The genes D site albumin promoter binding protein (Dbp) and zinc finger BED domain-containing protein 6 (Zbed6) were significantly over-expressed in TG and to a lesser extent in the NAc of OIH relative to CON mice. Dbp is involved in rhythmic processes and circadian rhythm disorders have been associated with headache [61]. Only aryl hydrocarbon receptor nuclear translocator-like protein 1 (Arntl) exhibits under-express “consistent pattern”. Arntl is a transcriptional activator that forms a core component of the circadian clock and also plays a role in mood-related behavior [62]. The under-expression of giant axonal neuropathy (Gan) in OIH relative to CON in the TG (S1 File Table A) is consistent with reports that this gene was under-expressed in mice that present tactile allodynia and that mutation in this gene is linked to sensorimotor neuropathy [63].
The genes that presented significant under-expression in the NAc of OIH relative to CON mice included: Galanin (Gal), acid phosphatase prostate (Acpp); myelin protein P0 (Mpz); synuclein gamma (Sncg); phosphoinositide-interacting regulator of transient receptor potential channels (Pirt); integrin subunit beta 4 (Itgb4); serine (or cysteine) peptidase inhibitor, clade B, member 1a (Serpinb1a); trafficking regulator of GLUT4 (Trarg1 or Tusc5); guanine nucleotide-binding protein subunit beta-3 (Gnb3); Periaxin (Prx); Claudin-19 (Cldn19); sodium channel, voltage-gated, type VII, alpha (Scn7a); and Pllp. The profiles observed for these genes are in agreement with previous studies of comparable models and experiments (Table 1). For example, Gal was 20 fold under-expressed in the NAc OIH relative to CON mice (S1 File Table A), and this finding agrees with the antinociception function of this gene in the NAc of rats presenting inflammatory pain [64]. Gal has been proposed as a molecule participating in the neuroinflammatory mechanism associated with opioid tolerance and paradoxical hyperalgesia [65]. The under-expression of Acpp in the NAc of OIH relative to CON mice agrees with reports that a product of this gene, transmembrane prostatic acid phosphatase (TMPAP), suppresses pain sensation in healthy animals and conversely, mice deficient in TMPAP present an increased synthesis of striatal dopamine [66]. Our results may suggest that the lower expression of Gal and Acpp observed in this study lowers the capacity of these gene products to block the perception of pain by sensory neurons resulting in hyperalgesia
The pattern of expression of several genes suggests an alteration in the molecular mechanisms that may ameliorate pain sensation and hyperalgesia. The under-expression of Pirt and Itgb4 in the NAc of OIH relative to CON mice (Table 1) is consistent with previous reports. Pirt knockout mice present lower mechanical allodynia and thermal hyperalgesia [67], and the inhibition of Itgb4 alleviates neuropathic pain [68]. We propose that the under-regulation of Pirt and Itgb4 does not completely block OIH, but the pattern may act as a defense to counter OIH.
The under-expression of Serpinb1a, Sncg, and Tusc5 in the NAc of OIH relative to CON mice (Table 1) is supported by previous studies. Serpinb1a was under-expressed in a rat model of peripheral neuropathy experiencing chronic pain relative to controls [69]. Both Sncg and Tusc5 are considered sensory neuron markers [70,71]. The over-expression of Sncg in sensory neurons in culture disrupts neurofilament structure, a key component to neuronal plasticity associated with addiction [72]. Serpinb1a was under-expressed in a rat model of peripheral neuropathy experiencing chronic pain relative to controls [69]. The protein coded by Mpz is necessary for normal myelination in the peripheral nervous system [73], and our finding is in line with a study of mutations in Mpz leading to rapidly progressive and painful axonal neuropathy [74].
The observed under-expression of Prx in the NAc of OIH relative to CON mice (Table 1) is in agreement with reports that Prx deficiency in mice can lead to mechanical allodynia, thermal hyperalgesia, and demyelination whereas formation and maintenance of myelin is essential for normal transmission of nerve impulses and perception of sensory stimuli [75]. The under-expression of Pllp in binge-drinking rats is consistent with the observed under-expression of this gene in the NAc of OIH relative to CON mice [76].
Sodium channel protein type 7 subunit alpha (Scn7a) was under-expressed in the NAc of OIH relative to CON mice (Table 1). This pattern is consistent with the positive association between the enhanced expression of Scn7a, a gene that encodes an atypical sodium channel termed (Nax) and increased excitability of dorsal root ganglion neurons and bone cancer pain [77]. The Nax channel is considered an important determinant of sodium sensing in the brain [78] and the Scn7a knockdown prevented the onset of hyperalgesia in dorsal root ganglion [79]. Likewise, the under-expression of Itgb4 in the NAc and over-expression in the TG of OIH relative to CON mice and reverse pattern in the TG is consistent with reports of the role of integrins in pathological pain [80].
Cldn19 was under-expressed in the NAc of OIH relative to CON mice, and this trend agrees with electrophysiological analyses showing that deficiency in this protein affected nerve conduction of peripheral myelinated fibers. Also, a transcriptome study of inherited sensory neuropathies that have high prevalence of neuropathic pain identified significant differential expression of Cldn19 in sciatic nerve [81]. Guanine nucleotide-binding protein subunit beta-3 (Gnb3) was under-expressed in the NAc of OIH relative to CON mice (S1 File Table A) and this trend is consistent with reports that polymorphisms that compromise the expression of this gene are implicated in enhanced signal transduction and pain sensation in the head [82].
Functional analysis followed the identification of region-dependent gene profiles between OIH and CON mice. The enrichment of nucleotide (DNA and RNA) processing and metabolic categories among the genes presenting a significant region-dependent treatment effect and over-expressed in the NAc of OIH relative to CON mice is consistent with previous reports. Acute morphine administration is associated with lower incorporation and higher catabolism of uridine into the RNA precursor in the brains of mice without altering the concentration of brain RNA. Narcotics such as opium and its derivatives inhibit RNA synthesis in glial cell cultures and morphine administration altered the metabolism of pyrimidine nucleosides and nucleotides and the incorporation of precursors into the brain RNA [83]. Also, thermal hyperalgesia has been associated with histone 3 hypermethylation in the dorsal horn of the spinal cord of rats, and this methylation was associated with interference of RNA processing [84].
The enrichment of the biological process neuron projection development among the genes under-expressed in the NAc of OIH relative to CON mice is consistent with increased release of dopamine in the neuron projections of the ventral tegmental area into the NAc that participate in morphine-induced antinociception signals [85]. The prevalence of genes annotated to the biological process angiogenesis that were under-expressed in the TG of OIH relative to CON mice supports reports that chronic morphine stimulates angiogenesis in mice [86]. The enrichment of the biological process GTPase binding among the genes over-expressed in the TG of OIH relative to CON mice may be related to GPTases contribution to the modulation of hypoalgesia in the spine [87]. The investigation of the association between OIH-associated transcriptome changes in the NAc (e.g., genes annotated to nucleic acid processing) and in the TG (e.g., genes annotated to GTPase binding) regions offered novel insights into the interactions between the paradoxical sensitivity to stimuli and pain that can be triggered by opioid exposure.
Region-dependent gene networks
Further understanding of the distinct interplay between genes annotated to the circadian rhythm, neuroimmune response, transcriptional regulation was gained from the comparison of the gene networks that consider treatment effect within the CNS region studied. A notable finding is that the majority of the genes that were differentially expressed in both networks (Ciart, Per3, Tef, Dbp) were over-expressed in OIH relative to CON mice, albeit at different levels. Ciart, Per3, Tef, Dbp are annotated to circadian rhythm processes and Dbp and Tef are transcription factors. The exception was the transcriptional repressor Aebp1 that was under-expressed in the NAc (Figure 2) and over-expressed in the TG (Figure 3) of OIH relative to OIH mice.
A reversal of Aebp1 differential expression between OIH and CON across regions may be associated with the multiple contributions to different processes. The over-expression of Aebp1 in the TG of OIH relative to CON may be associated with the capacity of the coded protein to positively regulate the activity of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and promote inflammatory responsiveness [88]. Conversely, inhibitors of NF-κB can alleviate morphine withdrawal-induced allodynia and hyperalgesia [89]. The under-expression of Aebp1 in the NAc of OIH relative to CON mice corresponds with the under-expression of this gene in the prelimbic cortex of mice chronically treated with the psychostimulant cocaine when comparing knockouts of serotonin transporter and wild type groups [90].
The majority of the framework genes in the networks were differentially expressed in one of the regions studied. The distribution of profiles of genes uniquely differentially expressed in one region confirm the predominance of gene over-expression in the TG and under-expression in the NAc of OIH relative to CON mice. Several genes differentially expressed in one of the two regions studied are annotated to circadian processes and regulation of transcription including, for NAc: Gnb3, Prkcdbp, Pou4f1, Traf1, Map2k6, and Per3; and for TG: Per2, Cry1, Cry2, Arntl, Usp2, Cry1, Cry2, Hlf, Hipk2, Slc9a1, Sorbs3, and Zfp369.
The peripheral distribution of high differentially expressed genes between OIH and CON mice in the NAc (Figure 2) and more centric distribution in the TG network (Figure 3) offered insights into the interaction between nervous system region and OIH treatment. The networks suggest that OIH is associated with predominant change in expression of more central genes in TG that generates a sense of pain whereas change in less central genes is favored in NAc. Core components of a network are usually conserved and central in terms of functions, while periphery components are usually more variable and specialized [91]. In the context of this study, the former components may be central to the fundamental process of pain signal transmission in the TG whereas the latter components may be associated to a wide range of rewards and motivations that the NAc can respond.
Region-independent associations between opioid-induced hyperalgesia and transcriptome changes
The identification of 187 genes that exhibited differential expression (FDR-adjusted P-value < 0.1) between OIH and CON mice across TG and NAc advances the understanding of molecules that can present a consistent response to therapies for the amelioration of OIH across NAc and TG regions (Table 2 and S1 File Table B). Multiple genes annotated to circadian rhythm processes were over-expressed in OIH relative to CON mice including period circadian clock 2 (Per2), Per3, circadian-associated transcriptional repressor (Ciart), nuclear receptor subfamily 1 group D member 1 and 2 (Nr1d1 and Nr1d2, respectively). Pain sensation is modulated by circadian processes and consistent with our findings, withdrawal from opioid consumption was associated with lower levels of Per2 and Per3 gene expression in the NAc of rats [92] and the opioid fentanyl affected Per gene expression in the hamster suprachiasmatic nuclei [93]. Mice exhibiting hind paw and cephalic hyperalgesia after repeated treatments with nitroglycerin presented over-expression of Per3 in the NAc and TG [25]. Per3 has been associated with sensitivity to pain and regulates nervous and immune cell activity and function [94]. Variants in Per3 have been associated with opioid addiction or the pharmacological effects of treatment of addiction [95]. The expression of Nr1d1 has been associated with neuropathic pain [96]. The under-expression of the photoperiod gene aryl hydrocarbon receptor nuclear translocator-like protein 1 (Arntl), in OIH relative to CON mice (Table 2) is in agreement with the observed over-expression of its repressors: Per2, Per3, Nr1d1 and Nr1d2 [97].
A motivating finding is the set of transcription factors associated with circadian rhythm and neurotransmission processes that were over-expressed in OIH relative to CON mice including Dbp, Hlf and Tef, and Bhlhe41 (Table 2 and S1 File Table B). Bhlhe41 participates in the entrainment of the circadian system to altered external cues in mammals [98] and Dbp, Hlf and Tef are members of the proline and acidic amino acid-rich basic leucine zipper (PAR bZip) family. Members of the PAR bZip family of transcription factors are controlled by the circadian clock and can influence the homeostasis of neurotransmitter in the brain, such as those associated with pain and drug addiction [99]. PAR bZip-deficient mice had lower levels of pyridoxal phosphate, serotonin, and dopamine in the brain [99]. These trends support our findings of higher levels of PAR bZip members correlated with OIH.
Most of the differential expression patterns between OIH and CON groups support hypersensitivity to stimuli whereas the altered expression of other genes suggest the activation of molecular mechanisms that can reduce stimuli sensation, processing and hyperalgesia. Examples of these latter genes include flavin containing monooxygenase (Fmo2), several heat shock proteins (Hspas), and claudin-5 (Cldn5). The enzyme coded by Fmo2 metabolizes drugs [100] such that the over-expression of Fmo2 in OIH may enhance the metabolization of morphine, therefore lowering the impact of the opioid treatment.
The differential expression of heat shock genes observed in the present study may also be involved in ameliorating the effect of opioids to induce hyperalgesia. Heat shock 70 kDa protein 1A (Hspa1a), 70 kDa protein 1B (Hspa1b) and 70kDa protein 5 (Hspa5) were all under-expressed in OIH relative to CON mice across regions (Table 2, S1 File Table B). This pattern is consistent with the suppression of neuropathic pain achieved by inhibitors of heat shock proteins [101]. Hspa1a and Hspa1b present circadian rhythmicity in agreement with the enrichment of the circadian rhythm process among the genes differentially expressed between OIH and CON mice uncovered in the present study [102].
Cldn5, a gene that participates in cell-cell adhesion processes, was under-expressed in OIH relative to CON mice (Table 2) in a pattern parallel to Cldn19 within the NAc. Consistent with the present finding, Cldn5 was over-expressed in the midbrain and medulla of cholecystokinin knockout mice that do not display allodynia induced by chronic constriction injury [103]. Another gene that presents cell-cell adhesion function, myelin protein zero-like protein 1 (Mpzl1), was also under-expressed in OIH relative to CON mice.). The previous patterns could be indicative of the activation of anti-nociceptive systems.
Functional analysis of genes presenting region-independent associations with opioid-induced hyperalgesia
Functional analysis of gene patterns between OIH and CON across TG and NAc using complementary DAVID and GSEA advanced the understanding of molecular mechanisms associated with OIH. Among the genes differentially expressed, DAVID analysis identified the enrichment of myelination, axon ensheathment and ensheathment of neurons categories. Most of the genes supporting the enrichment results were under-expressed in OIH relative to CON mice, and this pattern is consistent with reports that demyelination and unstable ensheathment resulted in neuropathic pain [75,104]. Results from the GSEA analysis identified the enrichment of circadian rhythm and immune response-related categories among the genes over-expressed in OIH relative to CON (Table 3 and S1 File Table C) similar to the previously discussed region-dependent enriched categories (Tables 1 and 2). These results further confirm the connection between circadian rhythm, neuroinflammatory response, neuropathic pain and hyperalgesia [105,106,61,107]. The same processes that cause opioid use paradoxically to elicit hyperalgesia (rather than the expected amelioration of sensitivity to pain stimuli) also may paradoxically depress the expression of pro-inflammatory (rather than the expected heighten).
The enrichment of the biological process interferon gamma (INFG) -related immune response (GO:0060333; Table 3) and of the KEGG peroxisome proliferators-activated receptor (PPAR) signaling pathway (S1 File Table C) among the genes over-expressed in OIH mice is in line with reports that that PPAR-gamma agonist can attenuate inflammatory pain in the presence of lipopolysaccharide/interferon-γ [108]. This finding also agrees with reports that the PPAR signal pathway participates in the regulation of inflammatory responses in the CNS associated with neuropathic pain [109-111].
The enrichment of the biological process toll-like receptors (TLRs) signaling among genes under-expressed between OIH and CON mice (Table 3) is in agreement with accumulating evidence that the inflammatory consequences TLR activation on sensory neurons can have an impact on nociceptive processing and progress to exaggerated and unresolved pain [107]. Moreover, morphine induces TLR4 oligomerization that, in turn, results in NF-κB -mediated release of IL-1β, TNF, and nitric oxide. The paradoxical opioid-induced hyperalgesia may be mediated by TLR signaling [107].
Within the TLR functional category, the myeloid differentiation primary response gene 88 (MyD88) -dependent TLR signaling pathway was also over-represented (S1 File Table C). MyD88 is an adaptor protein associated with nearly all TLR-mediated signaling including TLR2/1/6, TLR4, TLR5, TLR7/8, and TLR9. Signal transduction triggered by MyD88 adaptor protein leads to nuclear translocation of activated transcription factors and the production of inflammatory cytokines, immune mediators, and interferons. In nociceptive pathways such as dorsal root ganglia, chronic constriction injury elicits higher expression of MyD88 along with elevated levels of phosphorylated NF-κB, phosphorylated extracellular signal-regulated kinase (ERK), and interleukin 1 beta downstream [107].
Our study confirms the previous association through the enrichment (FDR-adjusted P-value < 0.077) of the BP positive regulation of kinase within NF-κB signaling among the genes under-expressed in OIH relative to CON mice. The multiple profiles of genes annotated to INFG, TLR, and NF-κB and associated with OIH offer novel support to the proposition that opioids evoke neuroinflammatory responses underlying hyperalgesia. Our study of the association between OIH and transcriptome changes advances the understanding of the complex interplay between the paradoxical sensitivity to stimuli and pain that can be associated with opioids, circadian processes, and multi-layered neuroinflammatory response.
Main effect of nervous system region on gene expression and functional enrichment
Genes that are markers or highly expressed in the CNS regions studied region were confirmed in the present study. Genes, that were highly over-expressed in the TG included: Trpv1 [112], Trpa1, Piezo2 [113], Calca [114], Nmb [112], Mrgpra3 [115] whereas genes highly over-expressed in the NAc included: Drd1 [116], Kappa-type opioid receptor (Oprk1), pro-neuropeptide Y preproprotein (Npy) [116,117], Gad1, and Gad2 [118]. The functional categories enriched among the differentially expressed genes agreed with the known role of TG in the context of transmission of sensory signals, pain processing and of NAc in the context of motivation, reward dependence, and opioid addiction.
The categories enriched among the genes differentially expressed between TG and NAc included ECM-receptor interaction, ion channel activity, glutamate receptor activity, and glutamatergic synapse, nicotine and amphetamine addiction and circadian entrainment (Table 5). These results are in agreement with a report that chronic opiate exposure enhances the release of glutamate by primary afferent neurons and hyperexcitability of second-order neurons that modulate nociceptive sensitization and hyperalgesia [119].
Regulatory networks
Motivated by our finding of multiple transcriptional regulators differentially expressed in OIH relative to CON mice (Tables 1, 2 and 3, Figure 2 and 3, F1 File Tables A, B, and C), an analysis of regulatory elements was undertaken. The identification of transcription factors including motifs that corresponded to differentially expressed genes offered insights into potential regulatory mechanisms associated with OIH. The associations between the enriched transcription factors Dbp, Hlf, and Bhlhe41 transcription factors and circadian rhythm processes were previously reviewed. Consistent with the detected enrichment of carbohydrate-responsive element-binding protein (Mlxipl), this long non-coding RNA was implicated in the pathogenesis of neuropathic pain and was differentially expressed in the spinal cords of rats modeling spared nerve injury [120], Mlxipl was also differentially expressed in human sural nerves of patients presenting diabetic neuropathy [121].
Target genes that were under-expressed in OIH relative to CON mice offered evidence of the enrichment of four transcription factors: heat shock factor protein 1 (Hsf1), B-cell lymphoma 3 protein homolog (Bcl3), serum response factor (Srf), and nuclear factor of activated T-cells, cytoplasmic 1 (Nfatc1). These transcription factors were previously associated with immune response processes, and this association is consistent with the corresponding category enrichment identified in the present study (Table 3 and S1 File Table C).
Hsf1 is a circadian clock gene and consistent with the under-expression in OIH relative to CON mice, this protein participates in the negative regulation of tumor necrosis factor production and response to lipopolysaccharide challenge [122]. Likewise, Bcl3 participates in NF-κB processes and in lowering the inflammatory response to lipopolysaccharide injection [123]. Consistent with the pattern observed in the present study, RNA interference of Bcl3 attenuated thermal hyperalgesia in neuropathic rats and reversed the suppression of neuroinflammatory cytokines triggered by an intrathecal IL-10 injection and also [124]. Also, Bcl3 was over-expressed in the spinal cord of morphine-tolerant rats [125].
The under-expression of serum response factor (Srf) in OIH relative to CON mice is consistent with reports that SRF knockdown rats presented lower mechanical hyperalgesia priming [126]. The regulation of pro-nociceptive genes via the activation of NFAT-dependent transcription has been reported [127]. The under-expression of nuclear factor of activated T-cells, cytoplasmic 1 (Nfatc1) observed in this study may be a mechanism to ameliorate OIH.
The comparison of the regulatory networks between over- and under-expressed offered evidence of different regulatory mechanisms. The over-expressed transcription factors tended to share target genes with one other over-expressed transcription factor (Figure 4). On the other hand, the transcription factors under-expressed in OIH relative to CON mice (Figure 5) tended to share multiple target genes with multiple other transcription factors and this topological difference may be associated with the high number of motifs among the over-expressed enriched transcription factors and the overlap across factors. The difference in the level of differential expression between over- and under-expressed transcription factors may also affect the regulatory network connections depicted.
Methodological Considerations
Nociceptive sensitization and morphine effects are influenced by several factors in rodents. For example, a significant decrease in the pain threshold of rats in response to mechanical stimuli was observed across age and females were more sensitive than males [128]. The effect of morphine also decreases in intensity with age in adult mice. The results from our study focused on adult male mice should be complemented with additional studies including the comparison of hyperalgesia indicators and transcriptome patterns associated with OIH in females and males, and young and aged individuals.
The present study identified and characterized changes in the TG and NAc transcriptome associated with OIH. Recent reviews hypothesize that OIH is associated with the upregulation of the non-opioid-dependent ascending sensory systems such as divisions of the thalamocortical and spinothalamic tracts [129]. Complementary studies of the previous tracts will further advance the understanding of the processes associated with OIH. Neuronal processes originated in the microglia of the spinal cord can influence the development of opioid-induced hyperalgesia [130]. A focused study of microglia in the NAc may offer complementary insights into the OIH mechanisms.
The effects of opioid use were evaluated in this study using thresholds to hind paw and cephalic mechanical hyperalgesia. Additional characterization of OIH to other stimuli including heat or cooling test of thermal sensitivity or light touch can advance the characterization of the increased sensitivity to noxious or typically nonnoxious stimulation that can be elicited by opioid use. The information offered by (f)MRI and PET studies may offer a more comprehensive or accurate characterization of differences in pain experience, and aid in the characterization of metabolic changes and modulation of glutamate and GABA concentrations in the TG, NAc and the somatosensory cortex [131].
Conclusions
The present network, regulatory and functional analyses of the TG and NAc transcriptome enabled the identification of central nervous region-independent and dependent changes associated with OIH. Noteworthy findings are the enrichment of circadian rhythm categories and transcriptional regulators including Dbp, Tef, Hlf, Bhlhe41, and Ciart among the genes over-expressed in OIH relative to CON mice. Members of the PAR bZip family (e.g., Dbp, Tef and Hlf) are controlled by the circadian clock and can influence the homeostasis of neurotransmitter in the brain, such as those associated with pain and drug addiction processes and OIH.
Also notable was the enrichment of adaptive immune regulation categories including TLR, NF-κB, IFNG and cytokine processes among the genes under-expressed in OIH relative to CON mice. The enrichment of TLRs supports accumulating evidence that the inflammatory consequences of TLR activation on sensory neurons can lead to nociceptive processing and evolve into hyperalgesia and OIH. Moreover, the simultaneous enrichment IFNG categories and of the PPAR signaling pathway is in line with reports of an association between PPAR and inflammatory and pain. The comparison of gene and regulatory networks further the understanding of the distinct interplay between genes annotated to the circadian rhythm, neuroimmune response, and transcriptional regulation between TG and NAc. The findings from this study can be used in the advancement of controlled and effective opioid use for pain management while minimizing paradoxical OIH.
Supplementary Material
Table A. Extended list of genes exhibiting opioid-induced hyperalgesia treatment-by-central nervous system region interaction effect (FDR-adjusted P-value < 0.1).
Table B. Extended list of genes differentially expressed (FDR-adjusted P-value < 0.1) between opioid-induced hyperalgesia (OIH) and control (CON) mice.
Table C. Extended list of enriched informative categories among the genes under- or over-expressed in the opioid-induced hyperalgesia relative to control mice using GSEA (FDR-adjusted P-value < 0.1)
Table D. Extended list of genes differentially expressed between TG and NAc (log2(fold change) > 2 and FDR-adjusted P-value <1.0E-5)
Figure A. Network of differential gene expression between nucleus accumbens and trigeminal ganglia in mice from the opioid-induced hyperalgesia mice (contrast OIH_NA-OIH_TG).
Acknowledgements
This research was supported by the National Institute of Health [grant numbers P30 DA018310-14 (SRZ, BS, JS), and DA031243 (AP)], the Department of Defense [grant number PR100085 (AP)], and US Department of Agriculture NIFA ILLU-538-909. We would like to thank Dr. Dennis Grayson for helpful discussion regarding experimental procedures and manuscript preparation. We are also grateful to Ying Chen for helping us with the RNA isolation and Hyeonsoo Jeong for preliminary data evaluation. Laura Moye is a member of the UIC Graduate Program in Neuroscience.
Funding: This study was funded by the National Institute of Health [grant numbers P30 DA018310-14 (SRZ, BS, JS), and DA031243 (AP)], the Department of Defense [grant number PR100085 (AP)], and US Department of Agriculture NIFA ILLU-538-909.
Footnotes
Disclosure of potential conflicts of interest
Conflict of Interest: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Dahlhamer J, Lucas J, Zelaya C, Nahin R, Mackey S, DeBar L, Kerns R, Von Korff M, Porter L, Helmick C (2018) Prevalence of Chronic Pain and High-Impact Chronic Pain Among Adults - United States, 2016. MMWR Morb Mortal Wkly Rep 67 (36):1001–1006. doi: 10.15585/mmwr.mm6736a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breivik H, Eisenberg E, O'Brien T, Openminds (2013) The individual and societal burden of chronic pain in Europe: the case for strategic prioritisation and action to improve knowledge and availability of appropriate care. BMC Public Health 13:1229. doi: 10.1186/1471-2458-13-1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breivik H, Collett B, Ventafridda V, Cohen R, Gallacher D (2006) Survey of chronic pain in Europe: prevalence, impact on daily life, and treatment. Eur J Pain 10 (4):287–333. doi: 10.1016/j.ejpain.2005.06.009 [DOI] [PubMed] [Google Scholar]
- 4.Patch Iii RK, Eldrige JS, Moeschler SM, Pingree MJ (2017) Dexmedetomidine as Part of a Multimodal Analgesic Treatment Regimen for Opioid Induced Hyperalgesia in a Patient with Significant Opioid Tolerance. Case Rep Anesthesiol 2017:9876306. doi: 10.1155/2017/9876306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayhurst CJ, Durieux ME (2016) Differential Opioid Tolerance and Opioid-induced Hyperalgesia: A Clinical Reality. Anesthesiology 124 (2):483–488. doi: 10.1097/ALN.0000000000000963 [DOI] [PubMed] [Google Scholar]
- 6.Wanigasekera V, Lee MC, Rogers R, Hu P, Tracey I (2011) Neural correlates of an injury-free model of central sensitization induced by opioid withdrawal in humans. J Neurosci 31 (8):2835–2842. doi: 10.1523/JNEUROSCI.5412-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SH, Stoicea N, Soghomonyan S, Bergese SD (2015) Remifentanil-acute opioid tolerance and opioid-induced hyperalgesia: a systematic review. Am J Ther 22 (3):e62–74. doi: 10.1097/MJT.0000000000000019 [DOI] [PubMed] [Google Scholar]
- 8.Roeckel LA, Le Coz GM, Gaveriaux-Ruff C, Simonin F (2016) Opioid-induced hyperalgesia: Cellular and molecular mechanisms. Neuroscience 338:160–182. doi: 10.1016/j.neuroscience.2016.06.029 [DOI] [PubMed] [Google Scholar]
- 9.Johnson EE, Chieng B, Napier I, Connor M (2006) Decreased mu-opioid receptor signaling and a reduction in calcium current density in sensory neurons from chronically morphine-treated mice. Br J Pharmacol 148 (7):947–955. doi: 10.1038/sjbjp.0706820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akerman S, Holland PR, Goadsby PJ (2011) Diencephalic and brainstem mechanisms in migraine. Nat Rev Neurosci 12 (10):570–584. doi: 10.1038/nrn3057 [DOI] [PubMed] [Google Scholar]
- 11.Thalakoti S, Patil VV, Damodaram S, Vause CV, Langford LE, Freeman SE, Durham PL (2007) Neuron–glia signaling in trigeminal ganglion: implications for migraine pathology. Headache: The Journal of Head and Face Pain 47 (7): 1008–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaPaglia DM, Sapio MR, Burbelo PD, Thierry-Mieg J, Thierry-Mieg D, Raithel SJ, Ramsden CE, Iadarola MJ, Mannes AJ (2017) RNA-Seq investigations of human postmortem trigeminal ganglia. Cephalalgia:333102417720216. doi: 10.1177/0333102417720216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li JL, Ding YQ, Li YQ, Li JS, Nomura S, Kaneko T, Mizuno N (1998) Immunocytochemical localization of mu-opioid receptor in primary afferent neurons containing substance P or calcitonin gene-related peptide. A light and electron microscope study in the rat. Brain Res 794 (2):347–352 [DOI] [PubMed] [Google Scholar]
- 14.Bellamy J, Bowen EJ, Russo AF, Durham PL (2006) Nitric oxide regulation of calcitonin gene-related peptide gene expression in rat trigeminal ganglia neurons. European Journal of Neuroscience 23 (8):2057–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burstein R, Jakubowski M (2005) Unitary hypothesis for multiple triggers of the pain and strain of migraine. J Comp Neurol 493 (1):9–14 [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Yang C, Wang ZJ (2010) Ca2+/calmodulin-dependent protein kinase II alpha is required for the initiation and maintenance of opioid-induced hyperalgesia. J Neurosci 30 (1):38–46. doi: 10.1523/JNEUROSCI.4346-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corder G, Tawfik VL, Wang D, Sypek El, Low SA, Dickinson JR, Sotoudeh C, Clark JD, Barres BA, Bohlen CJ, Scherrer G (2017) Loss of μ opioid receptor signaling in nociceptors, but not microglia, abrogates morphine tolerance without disrupting analgesia. Nat Med 23 (2):164–173. doi: 10.1038/nm.4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elhabazi K, Ayachi S, Ilien B, Simonin F (2014) Assessment of morphine-induced hyperalgesia and analgesic tolerance in mice using thermal and mechanical nociceptive modalities. J Vis Exp (89):e51264. doi: 10.3791/51264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang DY, Shi X, Li X, Li J, Clark JD (2007) The beta2 adrenergic receptor regulates morphine tolerance and physical dependence. Behav Brain Res 181 (1): 118–126. doi: 10.1016/j.bbr.2007.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moye LS, Novack ML, Tipton AF, Krishnan H, Pandey SC, Pradhan AA (2018) The development of a mouse model of mTBI-induced post-traumatic migraine, and identification of the delta opioid receptor as a novel therapeutic target. Cephalalgia:333102418777507. doi: 10.1177/0333102418777507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL (1994) Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods 53 (1):55–63 [DOI] [PubMed] [Google Scholar]
- 22.Tipton AF, Tarash I, McGuire B, Charles A, Pradhan AA (2016) The effects of acute and preventive migraine therapies in a mouse model of chronic migraine. Cephalalgia 36 (11): 1048–1056. doi: 10.1177/0333102415623070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moye LS, Pradhan AAA (2017) Animal Model of Chronic Migraine-Associated Pain. Curr Protoc Neurosci 80:9.60.61–69.60.69. doi: 10.1002/cpns.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pradhan AA, Smith ML, McGuire B, Tarash I, Evans CJ, Charles A (2014) Characterization of a novel model of chronic migraine. PAIN® 155 (2):269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong H, Moye LS, Southey BR, Hernandez AG, Dripps I, Romanova EV, Rubakhin SS, Sweedler JV, Pradhan AA, Rodriguez-Zas SL (2018) Gene Network Dysregulation in the Trigeminal Ganglia and Nucleus Accumbens of a Model of Chronic Migraine-Associated Hyperalgesia. Front SystNeurosci 12:63. doi: 10.3389/fnsys.2018.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. [Google Scholar]
- 27.Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic acids research 35 (suppl 1):D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34 (5):525–527 [DOI] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1): 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological):289–300 [Google Scholar]
- 31.Caetano-Anollés K, Rhodes JS, Garland T Jr, Perez SD, Hernandez AG, Southey BR, Rodriguez-Zas SL (2016) Cerebellum transcriptome of mice bred for high voluntary activity offers insights into locomotor control and reward-dependent behaviors. PloS one 11 (11):e0167095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Pena D, Nixon SE, Southey BR, Lawson MA, McCusker RH, Hernandez AG, Dantzer R, Kelley KW, Rodriguez-Zas SL (2016) Differential Transcriptome Networks between IDO 1-Knockout and Wild-Type Mice in Brain Microglia and Macrophages. PLoS One 11 (6):e0157727. doi: 10.1371/journal.pone.0157727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Pena D, Nixon SE, O’Connor JC, Southey BR, Lawson MA, McCusker RH, Borras T, Machuca D, Hernandez AG, Dantzer R (2016) Microglia transcriptome changes in a model of depressive behavior after immune challenge. PloS one 11 (3):e0150858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caetano-Anolles K, Mishra S, Rodriguez-Zas SL (2015) Synergistic and Antagonistic Interplay between Myostatin Gene Expression and Physical Activity Levels on Gene Expression Patterns in Triceps Brachii Muscles of C57/BL6 Mice. PloS one 10 (2):e0116828. doi: 10.1371/journal.pone.0116828 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP (2007) GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23 (23):3251–3253 [DOI] [PubMed] [Google Scholar]
- 36.Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4 (1):44. [DOI] [PubMed] [Google Scholar]
- 37.Serao NV, Delfino KR, Southey BR, Beever JE, Rodriguez-Zas SL (2011) Cell cycle and aging, morphogenesis, and response to stimuli genes are individualized biomarkers of glioblastoma progression and survival. BMC medical genomics 4:49-8794-8794-8749. doi: 10.1186/1755-8794-4-49 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delfino KR, Serao NV, Southey BR, Rodriguez-Zas SL (2011) Therapy-, gender- and race-specific microRNA markers, target genes and networks related to glioblastoma recurrence and survival. Cancer genomics & proteomics 8 (4):173–183. doi:8/4/173 [pii] [PMC free article] [PubMed] [Google Scholar]
- 39.Delfino KR, Rodriguez-Zas SL (2013) Transcription factor-microRNA-target gene networks associated with ovarian cancer survival and recurrence. PloS one 8 (3):e58608. doi: 10.1371/journal.pone.0058608 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin A, Ochagavia M, Rabasa L, Miranda J, Fernandez-de-Cossio J, Bringas R (2010) BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics 11 (1):1–9. doi: 10.1186/1471-2105-11-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13 (11):2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D (2004) The Database of Interacting Proteins: 2004 update. Nucleic Acids Res 32 (Database issue):D449–451. doi: 10.1093/nar/gkh086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Bobechko B, Boutilier K, Burgess E, Buzadzija K, Cavero R, D'Abreo C, Donaldson I, Dorairajoo D, Dumontier MJ, Dumontier MR, Earles V, Farrall R, Feldman H, Garderman E, Gong Y, Gonzaga R, Grytsan V, Gryz E, Gu V, Haldorsen E, Halupa A, Haw R, Hrvojic A, Hurrell L, Isserlin R, Jack F, Juma F, Khan A, Kon T, Konopinsky S, Le V, Lee E, Ling S, Magidin M, Moniakis J, Montojo J, Moore S, Muskat B, Ng I, Paraiso JP, Parker B, Pintilie G, Pirone R, Salama JJ, Sgro S, Shan T, Shu Y, Siew J, Skinner D, Snyder K, Stasiuk R, Strumpf D, Tuekam B, Tao S, Wang Z, White M, Willis R, Wolting C, Wong S, Wrong A, Xin C, Yao R, Yates B, Zhang S, Zheng K, Pawson T, Ouellette BF, Hogue CW (2005) The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res 33 (Database issue):D418–424. doi: 10.1093/nar/gki051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34 (Database issue):D535–539. doi: 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, Castagnoli L, Cesareni G (2012) MINT, the molecular interaction database: 2012 update. Nucleic Acids Res 40 (Database issue):D857–861. doi: 10.1093/nar/gkr930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B, Thorneycroft D, Zhang Y, Apweiler R, Hermjakob H (2007) IntAct--open source resource for molecular interaction data. Nucleic Acids Res 35 (Database issue):D561–565. doi: 10.1093/nar/gkl958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, Menon S, Hanumanthu G, Gupta M, Upendran S, Gupta S, Mahesh M, Jacob B, Mathew P, Chatterjee P, Arun KS, Sharma S, Chandrika KN, Deshpande N, Palvankar K, Raghavnath R, Krishnakanth R, Karathia H, Rekha B, Nayak R, Vishnupriya G, Kumar HG, Nagini M, Kumar GS, Jose R, Deepthi P, Mohan SS, Gandhi TK, Harsha HC, Deshpande KS, Sarker M, Prasad TS, Pandey A (2006) Human protein reference database--2006 update. Nucleic Acids Res 34 (Database issue):D411–414. doi: 10.1093/nar/gkj141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, Svetlichnyy D, Kalender Atak Z, Fiers M, Marine JC, Aerts S (2014) iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 10 (7):e1003731. doi: 10.1371/journal.pcbi.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang D, Shi X, Qiao Y, Angst MS, Yeomans DC, Clark JD (2008) Chronic morphine administration enhances nociceptive sensitivity and local cytokine production after incision. Mol Pain 4:7. doi: 10.1186/1744-8069-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarlani E, Balciunas BA, Grace EG (2005) Orofacial pain--Part I: Assessment and management of musculoskeletal and neuropathic causes. AACN Clin Issues 16 (3):333–346 [DOI] [PubMed] [Google Scholar]
- 51.Grueter BA, Rothwell PE, Malenka RC (2012) Integrating synaptic plasticity and striatal circuit function in addiction. Curr Opin Neurobiol 22 (3):545–551. doi: 10.1016/j.conb.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gear RW, Levine JD (2011) Nucleus accumbens facilitates nociception. Exp Neurol 229 (2):502–506. doi: 10.1016/j.expneurol.2011.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cook-Sather SD, Li J, Goebel TK, Sussman EM, Rehman MA, Hakonarson H (2014) TAOK3, a novel genome-wide association study locus associated with morphine requirement and postoperative pain in a retrospective pediatric day surgery population. Pain 155 (9):1773–1783. doi: 10.1016/j.pain.2014.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crowder RJ, Enomoto H, Yang M, Johnson EM Jr., Milbrandt J (2004) Dok-6, a Novel p62 Dok family member, promotes Ret-mediated neurite outgrowth. J Biol Chem 279 (40):42072–42081. doi: 10.1074/jbc.M403726200 [DOI] [PubMed] [Google Scholar]
- 55.Maynard KB, Smith SA, Davis AC, Trivette A, Seipelt-Thiemann RL (2014) Evolutionary analysis of the mammalian M1 aminopeptidases reveals conserved exon structure and gene death. Gene 552 (1):126–132. doi: 10.1016/j.gene.2014.09.025 [DOI] [PubMed] [Google Scholar]
- 56.Osada T, Ikegami S, Takiguchi-Hayashi K, Yamazaki Y, Katoh-Fukui Y, Higashinakagawa T, Sakaki Y, Takeuchi T (1999) Increased anxiety and impaired pain response in puromycin-sensitive aminopeptidase gene-deficient mice obtained by a mouse gene-trap method. J Neurosci 19 (14):6068–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W, Shi L, You Y, Gong Y, Yin B, Yuan J, Peng X (2010) Downstream of tyrosine kinase/docking protein 6, as a novel substrate of tropomyosin-related kinase C receptor, is involved in neurotrophin 3-mediated neurite outgrowth in mouse cortex neurons. BMC Biol 8:86. doi: 10.1186/1741-7007-8-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanioka T, Hattori A, Masuda S, Nomura Y, Nakayama H, Mizutani S, Tsujimoto M (2003) Human leukocyte-derived arginine aminopeptidase. The third member of the oxytocinase subfamily of aminopeptidases. J Biol Chem 278 (34):32275–32283. doi: 10.1074/jbc.M305076200 [DOI] [PubMed] [Google Scholar]
- 59.Paek J, Kalocsay M, Staus DP, Wingler L, Pascolutti R, Paulo JA, Gygi SP, Kruse AC (2017) Multidimensional Tracking of GPCR Signaling via Peroxidase-Catalyzed Proximity Labeling. Cell 169 (2):338–349 e311. doi: 10.1016/j.cell.2017.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vo L, Hood S, Drummond PD (2016) Involvement of Opioid Receptors and alpha2-Adrenoceptors in Inhibitory Pain Modulation Processes: A Double-Blind Placebo-Controlled Crossover Study. J Pain 17 (11):1164–1173. doi: 10.1016/j.jpain.2016.07.004 [DOI] [PubMed] [Google Scholar]
- 61.Ofte HK, Berg DH, Bekkelund SI, Alstadhaug KB (2013) Insomnia and periodicity of headache in an arctic cluster headache population. Headache 53 (10):1602–1612. doi: 10.1111/head.12241 [DOI] [PubMed] [Google Scholar]
- 62.Bering T, Carstensen MB, Wortwein G, Weikop P, Rath MF (2017) The Circadian Oscillator of the Cerebral Cortex: Molecular, Biochemical and Behavioral Effects of Deleting the Arntl Clock Gene in Cortical Neurons. Cereb Cortex. doi: 10.1093/cercor/bhw406 [DOI] [PubMed] [Google Scholar]
- 63.Dorsey SG, Leitch CC, Renn CL, Lessans S, Smith BA, Wang XM, Dionne RA (2009) Genome-wide screen identifies drug-induced regulation of the gene giant axonal neuropathy (Gan) in a mouse model of antiretroviral-induced painful peripheral neuropathy. Biol Res Nurs 11 (1):7–16. doi: 10.1177/1099800409332726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Y, Zhang Y, Li XH, Li Y, Qian R, Li J, Xu SL (2015) Involvements of galanin and its receptors in antinociception in nucleus accumbens of rats with inflammatory pain. Neurosci Res 97:20–25. doi: 10.1016/j.neures.2015.03.006 [DOI] [PubMed] [Google Scholar]
- 65.Grace PM, Maier SF, Watkins LR (2015) Opioid-induced central immune signaling: implications for opioid analgesia. Headache 55 (4):475–489. doi: 10.1111/head.12552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nousiainen HO, Quintero IB, Myohanen TT, Voikar V, Mijatovic J, Segerstrale M, Herrala AM, Kulesskaya N, Pulkka AE, Kivinummi T, Abo-Ramadan U, Taira T, Piepponen TP, Rauvala H, Vihko P (2014) Mice deficient in transmembrane prostatic acid phosphatase display increased GABAergic transmission and neurological alterations. PLoS ONE 9 (5):e97851. doi: 10.1371/journal.pone.0097851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang C, Gu L, Ruan Y, Gegen T, Yu L, Zhu C, Yang Y, Zhou Y, Yu G, Tang Z (2018) Pirt Together with TRPV1 Is Involved in the Regulation of Neuropathic Pain. Neural Plast 2018:4861491. doi: 10.1155/2018/4861491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Capasso KE, Manners MT, Quershi RA, Tian Y, Gao R, Hu H, Barrett JE, Sacan A, Ajit SK (2015) Effect of histone deacetylase inhibitor JNJ-26481585 in pain. J Mol Neurosci 55 (3):570–578. doi: 10.1007/s12031-014-0391-7 [DOI] [PubMed] [Google Scholar]
- 69.Hong QX, Xu SY, Dai SH, Zhao WX (2016) Expression profiling of spinal genes in peripheral neuropathy model rats with type 2 diabetes mellitus. Int J Clin Exp Med 9 (3):6376–6384 [Google Scholar]
- 70.Nagy V, Cole T, Van Campenhout C, Khoung TM, Leung C, Vermeiren S, Novatchkova M, Wenzel D, Cikes D, Polyansky AA, Kozieradzki I, Meixner A, Bellefroid EJ, Neely GG, Penninger JM (2015) The evolutionarily conserved transcription factor PRDM12 controls sensory neuron development and pain perception. Cell Cycle 14 (12):1799–1808. doi: 10.1080/15384101.2015.1036209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, Lam T, Kim JY, Kim TH, Zhang MQ, Dussor G, Price TJ (2018) Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 159 (7):1325–1345. doi: 10.1097/j.pain.0000000000001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buchman VL, Adu J, Pinon LG, Ninkina NN, Davies AM (1998) Persyn, a member of the synuclein family, influences neurofilament network integrity. Nat Neurosci 1 (2):101–103. doi: 10.1038/349 [DOI] [PubMed] [Google Scholar]
- 73.Miyamoto T, Morita K, Takemoto D, Takeuchi K, Kitano Y, Miyakawa T, Nakayama K, Okamura Y, Sasaki H, Miyachi Y, Furuse M, Tsukita S (2005) Tight junctions in Schwann cells of peripheral myelinated axons: a lesson from claudin-19-deficient mice. J Cell Biol 169 (3):527–538. doi: 10.1083/jcb.200501154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dacci P, Taroni F, Bella ED, Milani M, Pareyson D, Morbin M, Lauria G (2012) Myelin protein zero Arg36Gly mutation with very late onset and rapidly progressive painful neuropathy. J Peripher Nerv Syst 17 (4):422–425. doi: 10.1111/j.1529-8027.2012.00443.x [DOI] [PubMed] [Google Scholar]
- 75.Gillespie CS, Sherman DL, Fleetwood-Walker SM, Cottrell DF, Tait S, Garry EM, Wallace VC, Ure J, Griffiths IR, Smith A, Brophy PJ (2000) Peripheral demyelination and neuropathic pain behavior in periaxin-deficient mice. Neuron 26 (2):523–531 [DOI] [PubMed] [Google Scholar]
- 76.McClintick JN, McBride WJ, Bell RL, Ding ZM, Liu Y, Xuei X, Edenberg HJ (2016) Gene Expression Changes in Glutamate and GABA-A Receptors, Neuropeptides, Ion Channels, and Cholesterol Synthesis in the Periaqueductal Gray Following Binge-Like Alcohol Drinking by Adolescent Alcohol-Preferring (P) Rats. Alcohol Clin Exp Res 40 (5):955–968. doi: 10.1111/acer.13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen SP, Zhou YQ, Liu DQ, Zhang W, Manyande A, Guan XH, Tian YK, Ye DW, Omar DM (2017) PI3K/Akt Pathway: A Potential Therapeutic Target for Chronic Pain. Curr Pharm Des 23 (12):1860–1868. doi: 10.2174/1381612823666170210150147 [DOI] [PubMed] [Google Scholar]
- 78.Nehme B, Henry M, Mouginot D, Drolet G (2012) The Expression Pattern of the Na(+) Sensor, Na(X) in the Hydromineral Homeostatic Network: A Comparative Study between the Rat and Mouse. Front Neuroanat 6:26. doi: 10.3389/fnana.2012.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ke CB, He WS, Li CJ, Shi D, Gao F, Tian YK (2012) Enhanced SCN7A/Nax expression contributes to bone cancer pain by increasing excitability of neurons in dorsal root ganglion. Neuroscience 227:80–89. doi: 10.1016/j.neuroscience.2012.09.046 [DOI] [PubMed] [Google Scholar]
- 80.Maruyama K (2012) Integrins and Nitric Oxide in the Regulation of Glia Cells: Potential Roles in Pathological Pain. Journal of Anesthesia & Clinical Research S7 (01). doi: 10.4172/2155-6148.s7-008 [DOI] [Google Scholar]
- 81.Sapio MR, Goswami SC, Gross JR, Mannes AJ, Iadarola MJ (2016) Transcriptomic analyses of genes and tissues in inherited sensory neuropathies. Exp Neurol 283 (Pt A):375–395. doi: 10.1016/j.expneurol.2016.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schurks M (2010) Genetics of cluster headache. Curr Pain Headache Rep 14 (2):132–139. doi: 10.1007/s11916-010-0096-8 [DOI] [PubMed] [Google Scholar]
- 83.Harris RA, Harris LS, Dunn A (1975) Effect of Narcotic Drugs on Ribonucleic-Acid and Nucleotide-Metabolism in Mouse-Brain. J Pharmacol Exp Ther 192 (2):280–287 [PubMed] [Google Scholar]
- 84.Tsai RY, Shen CH, Feng YP, Chien CC, Lee SO, Tsai WY, Lin YS, Wong CS (2012) Ultra-low-dose naloxone enhances the antinociceptive effect of morphine in PTX-treated rats: regulation on global histone methylation. Acta Anaesthesiol Taiwan 50 (3):106–111. doi: 10.1016/j.aat.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 85.Altier N, Stewart J (1998) Dopamine receptor antagonists in the nucleus accumbens attenuate analgesia induced by ventral tegmental area substance P or morphine and by nucleus accumbens amphetamine. J Pharmacol Exp Ther 285 (1):208–215 [PubMed] [Google Scholar]
- 86.Smith HS (2008) Combination opioid analgesics. Pain Physician 11 (2):201–214 [PubMed] [Google Scholar]
- 87.Ito T, Ohtori S, Hata K, Inoue G, Moriya H, Takahashi K, Yamashita T (2007) Rho kinase inhibitor improves motor dysfunction and hypoalgesia in a rat model of lumbar spinal canal stenosis. Spine (Phila Pa 1976) 32 (19):2070–2075. doi: 10.1097/BRS.0b013e318145a502 [DOI] [PubMed] [Google Scholar]
- 88.Majdalawieh A, Zhang L, Ro HS (2007) Adipocyte enhancer-binding protein-1 promotes macrophage inflammatory responsiveness by up-regulating NF-kappaB via IkappaBalpha negative regulation. Mol Biol Cell 18 (3):930–942. doi: 10.1091/mbc.E06-03-0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bai L, Zhai C, Han K, Li Z, Qian J, Jing Y, Zhang W, Xu JT (2014) Toll-like receptor 4-mediated nuclear factor-kappaB activation in spinal cord contributes to chronic morphine-induced analgesic tolerance and hyperalgesia in rats. Neurosci Bull 30 (6):936–948. doi: 10.1007/s12264-014-1483-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simmler LD, Anacker AMJ, Levin MH, Vaswani NM, Gresch PJ, Nackenoff AG, Anastasio NC, Stutz SJ, Cunningham KA, Wang J, Zhang B, Henry LK, Stewart A, Veenstra-VanderWeele J, Blakely RD (2017) Blockade of the 5-HT transporter contributes to the behavioural, neuronal and molecular effects of cocaine. Br J Pharmacol 174 (16):2716–2738. doi: 10.1111/bph.13899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gaiteri C, Sibille E (2011) Differentially expressed genes in major depression reside on the periphery of resilient gene coexpression networks. Front Neurosci 5:95. doi: 10.3389/fnins.2011.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li SX, Liu LJ, Jiang WG, Sun LL, Zhou SJ, Le Foll B, Zhang XY, Kosten TR, Lu L (2010) Circadian alteration in neurobiology during protracted opiate withdrawal in rats. J Neurochem 115 (2):353–362. doi: 10.1111/j.1471-4159.2010.06941.x [DOI] [PubMed] [Google Scholar]
- 93.Vansteensel MJ, Magnone MC, van Oosterhout F, Baeriswyl S, Albrecht U, Albus H, Dahan A, Meijer JH (2005) The opioid fentanyl affects light input, electrical activity and Per gene expression in the hamster suprachiasmatic nuclei. Eur J Neurosci 21 (11):2958–2966. doi: 10.1111/j.1460-9568.2005.04131.x [DOI] [PubMed] [Google Scholar]
- 94.Segal JP, Tresidder KA, Bhatt C, Gilron I, Ghasemlou N (2018) Circadian control of pain and neuroinflammation. J Neurosci Res 96 (6):1002–1020. doi: 10.1002/jnr.24150 [DOI] [PubMed] [Google Scholar]
- 95.Proudnikov D, Yuferov V, Randesi M, Kreek MJ (2013) Genetics of Opioid Addiction. Biological Research on Addiction: Comprehensive Addictive Behaviors and Disorders, Vol 2:509–521. doi: 10.1016/B978-0-12-398335-0.00050-9 [DOI] [Google Scholar]
- 96.Burish MJ, Chen Z, Yoo SH (2019) Emerging relevance of circadian rhythms in headaches and neuropathic pain. Acta Physiol (Oxf) 225 (1):e13161. doi: 10.1111/apha.13161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guillaumond F, Dardente H, Giguere V, Cermakian N (2005) Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms 20 (5):391–403. doi: 10.1177/0748730405277232 [DOI] [PubMed] [Google Scholar]
- 98.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K (2002) Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419 (6909):841–844. doi: 10.1038/nature01123 [DOI] [PubMed] [Google Scholar]
- 99.Gachon F (2007) Physiological function of PARbZip circadian clock-controlled transcription factors. Ann Med 39 (8):562–571. doi: 10.1080/07853890701491034 [DOI] [PubMed] [Google Scholar]
- 100.Krueger SK, Williams DE (2005) Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther 106 (3):357–387. doi: 10.1016/j.pharmthera.2005.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Watkins LR, Hutchinson MR, Rice KC, Maier SF (2009) The "toll" of opioid-induced glial activation: improving the clinical efficacy of opioids by targeting glia. Trends Pharmacol Sci 30 (11):581–591. doi: 10.1016/j.tips.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sandstrom ME, Madden LA, Taylor L, Siegler JC, Lovell RJ, Midgley A, McNaughton L (2009) Variation in basal heat shock protein 70 is correlated to core temperature in human subjects. Amino Acids 37 (2):279–284. doi: 10.1007/s00726-008-0144-4 [DOI] [PubMed] [Google Scholar]
- 103.Koks S, Fernandes C, Kurrikoff K, Vasar E, Schalkwyk LC (2008) Gene expression profiling reveals upregulation of Tlr4 receptors in Cckb receptor deficient mice. Behav Brain Res 188 (1):62–70. doi: 10.1016/j.bbr.2007.10.020 [DOI] [PubMed] [Google Scholar]
- 104.Ko MH, Hsieh YL, Hsieh ST, Tseng TJ (2015) Nerve demyelination increases metabotropic glutamate receptor subtype 5 expression in peripheral painful mononeuropathy. Int J Mol Sci 16 (3):4642–4665. doi: 10.3390/ijms16034642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pringsheim T (2002) Cluster headache: evidence for a disorder of circadian rhythm and hypothalamic function. Can J Neurol Sci 29 (1):33–40 [DOI] [PubMed] [Google Scholar]
- 106.Plein LM, Rittner HL (2018) Opioids and the immune system - friend or foe. Br J Pharmacol 175 (14):2717–2725. doi: 10.1111/bph.13750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lacagnina MJ, Watkins LR, Grace PM (2018) Toll-like receptors and their role in persistent pain. Pharmacol Ther 184:145–158. doi: 10.1016/j.pharmthera.2017.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hasegawa-Moriyama M, Kurimoto T, Nakama M, Godai K, Kojima M, Kuwaki T, Kanmura Y (2013) Peroxisome proliferator-activated receptor-gamma agonist rosiglitazone attenuates inflammatory pain through the induction of heme oxygenase-1 in macrophages. Pain 154 (8):1402–1412. doi: 10.1016/j.pain.2013.04.039 [DOI] [PubMed] [Google Scholar]
- 109.Petrosino S, Palazzo E, de Novellis V, Bisogno T, Rossi F, Maione S, Di Marzo V (2007) Changes in spinal and supraspinal endocannabinoid levels in neuropathic rats. Neuropharmacology 52 (2):415–422. doi: 10.1016/j.neuropharm.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 110.Kostadinova R, Wahli W, Michalik L (2005) PPARs in diseases: control mechanisms of inflammation. Curr Med Chem 12 (25):2995–3009 [DOI] [PubMed] [Google Scholar]
- 111.D'Agostino G, La Rana G, Russo R, Sasso O, Iacono A, Esposito E, Mattace Raso G, Cuzzocrea S, Loverme J, Piomelli D, Meli R, Calignano A (2009) Central administration of palmitoylethanolamide reduces hyperalgesia in mice via inhibition of NF-kappaB nuclear signalling in dorsal root ganglia. Eur J Pharmacol 613 (1-3):54–59. doi: 10.1016/j.ejphar.2009.04.022 [DOI] [PubMed] [Google Scholar]
- 112.Goswami SC, Thierry-Mieg D, Thierry-Mieg J, Mishra S, Hoon MA, Mannes AJ, Iadarola MJ (2014) Itch-associated peptides: RNA-Seq and bioinformatic analysis of natriuretic precursor peptide B and gastrin releasing peptide in dorsal root and trigeminal ganglia, and the spinal cord. Mol Pain 10:44. doi: 10.1186/1744-8069-10-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Flegel C, Schobel N, Altmuller J, Becker C, Tannapfel A, Hatt H, Gisselmann G (2015) RNA-Seq Analysis of Human Trigeminal and Dorsal Root Ganglia with a Focus on Chemoreceptors. PLoS ONE 10 (6):e0128951. doi: 10.1371/journal.pone.0128951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lopes DM, Denk F, McMahon SB (2017) The Molecular Fingerprint of Dorsal Root and Trigeminal Ganglion Neurons. Front Mol Neurosci 10:304. doi: 10.3389/fnmol.2017.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nguyen MQ, Wu Y, Bonilla LS, von Buchholtz LJ, Ryba NJP (2017) Diversity amongst trigeminal neurons revealed by high throughput single cell sequencing. PLoS ONE 12 (9):e0185543. doi: 10.1371/journal.pone.0185543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Delbes AS, Castel J, Denis RGP, Morel C, Quinones M, Everard A, Cani PD, Massiera F, Luquet SH (2018) Prebiotics Supplementation Impact on the Reinforcing and Motivational Aspect of Feeding. Front Endocrinol (Lausanne) 9:273. doi: 10.3389/fendo.2018.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van den Heuvel JK, Furman K, Gumbs MC, Eggels L, Opland DM, Land BB, Kolk SM, Narayanan NS, Fliers E, Kalsbeek A, DiLeone RJ, la Fleur SE (2015) Neuropeptide Y activity in the nucleus accumbens modulates feeding behavior and neuronal activity. Biol Psychiatry 77 (7):633–641. doi: 10.1016/j.biopsych.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen H, Liu Z, Gong S, Wu X, Taylor WL, Williams RW, Matta SG, Sharp BM (2011) Genome-Wide Gene Expression Profiling of Nucleus Accumbens Neurons Projecting to Ventral Pallidum Using both Microarray and Transcriptome Sequencing. Front Neurosci 5:98. doi: 10.3389/fnins.2011.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee M, Silverman SM, Hansen H, Patel VB, Manchikanti L (2011) A comprehensive review of opioid-induced hyperalgesia. Pain Physician 14 (2):145–161 [PubMed] [Google Scholar]
- 120.Zhou J, Fan Y, Chen H (2017) Analyses of long non-coding RNA and mRNA profiles in the spinal cord of rats using RNA sequencing during the progression of neuropathic pain in an SNI model. RNA Biol 14 (12):1810–1826. doi: 10.1080/15476286.2017.1371400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hur J, Sullivan KA, Pande M, Hong Y, Sima AA, Jagadish HV, Kretzler M, Feldman EL (2011) The identification of gene expression profiles associated with progression of human diabetic neuropathy. Brain 134 (Pt 11):3222–3235. doi: 10.1093/brain/awr228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ (1999) HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J 18 (21):5943–5952. doi: 10.1093/emboj/18.21.5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wessells J, Baer M, Young HA, Claudio E, Brown K, Siebenlist U, Johnson PF (2004) BCL-3 and NF-kappaB p50 attenuate lipopolysaccharide-induced inflammatory responses in macrophages. J Biol Chem 279 (48):49995–50003. doi: 10.1074/jbc.M404246200 [DOI] [PubMed] [Google Scholar]
- 124.Wu HY, Mao XF, Tang XQ, Ali U, Apryani E, Liu H, Li XY, Wang YX (2018) Spinal interleukin-10 produces antinociception in neuropathy through microglial beta-endorphin expression, separated from antineuroinflammation. Brain Behav Immun 73:504–519. doi: 10.1016/j.bbi.2018.06.015 [DOI] [PubMed] [Google Scholar]
- 125.Shao J, Wang J, Huang J, Liu C, Pan Y, Guo Q, Zou W (2018) Identification of lncRNA expression profiles and ceRNA analysis in the spinal cord of morphine-tolerant rats. Mol Brain 11 (1):21. doi: 10.1186/s13041-018-0365-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gomez R, Kohler DM, Brackley AD, Henry MA, Jeske NA (2018) Serum response factor mediates nociceptor inflammatory pain plasticity. Pain Rep 3 (3):e658. doi: 10.1097/PR9.0000000000000658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Groth RD, Mermelstein PG (2003) Brain-derived neurotrophic factor activation of NFAT (nuclear factor of activated T-cells)-dependent transcription: a role for the transcription factor NFATc4 in neurotrophin-mediated gene expression. J Neurosci 23 (22):8125–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jourdan D, Boghossian S, Alloui A, Veyrat-Durebex C, Coudore MA, Eschalier A, Alliot J (2000) Age-related changes in nociception and effect of morphine in the Lou rat. Eur J Pain 4 (3):291–300. doi: 10.1053/eujp.2000.0188 [DOI] [PubMed] [Google Scholar]
- 129.Goldberg JS (2013) Chronic opioid therapy and opioid tolerance: a new hypothesis. Pain Res Treat 2013:407504. doi: 10.1155/2013/407504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ferrini F, Trang T, Mattioli TA, Laffray S, Del'Guidice T, Lorenzo LE, Castonguay A, Doyon N, Zhang W, Godin AG, Mohr D, Beggs S, Vandal K, Beaulieu JM, Cahill CM, Salter MW, De Koninck Y (2013) Morphine hyperalgesia gated through microglia-mediated disruption of neuronal Cl(−) homeostasis. Nat Neurosci 16 (2):183–192. doi: 10.1038/nn.3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bruehl S, Apkarian AV, Ballantyne JC, Berger A, Borsook D, Chen WG, Farrar JT, Haythornthwaite JA, Horn SD, Iadarola MJ, Inturrisi CE, Lao L, Mackey S, Mao J, Sawczuk A, Uhl GR, Witter J, Woolf CJ, Zubieta JK, Lin Y (2013) Personalized medicine and opioid analgesic prescribing for chronic pain: opportunities and challenges. J Pain 14 (2):103–113. doi: 10.1016/j.jpain.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. Extended list of genes exhibiting opioid-induced hyperalgesia treatment-by-central nervous system region interaction effect (FDR-adjusted P-value < 0.1).
Table B. Extended list of genes differentially expressed (FDR-adjusted P-value < 0.1) between opioid-induced hyperalgesia (OIH) and control (CON) mice.
Table C. Extended list of enriched informative categories among the genes under- or over-expressed in the opioid-induced hyperalgesia relative to control mice using GSEA (FDR-adjusted P-value < 0.1)
Table D. Extended list of genes differentially expressed between TG and NAc (log2(fold change) > 2 and FDR-adjusted P-value <1.0E-5)
Figure A. Network of differential gene expression between nucleus accumbens and trigeminal ganglia in mice from the opioid-induced hyperalgesia mice (contrast OIH_NA-OIH_TG).