Abstract
The ten types of nicotinic acetylcholine receptor α-subunits show substantial sequence homology, yet some types confer high affinity for α-bungarotoxin, whereas others confer negligible affinity. Combining sequence alignments with structural data reveals three residues unique to α-toxin-refractory α-subunits that coalesce within the 3D structure of the α4β2 receptor, and are predicted to fit between loops I and II of α-bungarotoxin. Mutating any one of these residues, Lys189, Ile196 or Lys153, to the α-toxin-permissive counterpart fails to confer α-bungarotoxin binding. However, mutating both Lys189 and Ile196, affords α-bungarotoxin binding with an apparent dissociation constant of 104 nM, while combining mutation of Lys153 reduces the dissociation constant to 22 nM. Analogous residue substitutions also confer high affinity α-bungarotoxin binding upon α-toxin-refractory α2 and α3 subunits. α4β2 receptors engineered to bind α-bungarotoxin exhibit slow rates of α-toxin association and dissociation, and competition by cholinergic ligands typical of muscle nicotinic receptors. Receptors engineered to bind α-bungarotoxin co-sediment with muscle nicotinic receptors on sucrose gradients, and mirror single channel signatures of their α-toxin-refractory counterparts. Thus inability of α-bungarotoxin to bind to neuronal nicotinic receptors arises from three unique and interdependent residues that coalesce within the receptor’s 3D structure.
Keywords: neuronal nicotinic acetylcholine receptor, α-subunit, α-bungarotoxin, radio-ligand binding, sucrose gradient analysis, single channel recording, 3D structure, inter-residue interactions
1. Introduction
A defining pharmacological signature of the muscle nicotinic AChR is its high affinity for three-finger snake α-toxins (Chang and Lee, 1963). The prototypical snake α-toxin, α-bungarotoxin (α-BTX), prevents binding of ACh and subsequent opening of the receptor-coupled ion channel (Changeux, et al., 1970). α-BTX dissociates from the muscle AChR extremely slowly, enabling biochemical purification (Lindstrom, et al., 1978), studies of subunit folding and assembly (Blount and Merlie, 1988; Blount, et al., 1990; Green and Claudio, 1993), and tracking of AChRs within the membrane of intact muscle cells (Anderson, et al., 1977; Axelrod, et al., 1978). In the muscle AChR, the α1 subunit is the principal determinant of α-BTX binding (Haggerty and Freohner, 1981), yet despite the high degree of sequence identity among the ten types of α-subunits, major classes of neuronal AChRs show negligible affinity for α-BTX (Boulter, et al., 1987).
Within the α1 subunit of the muscle AChR, a structural motif known as loop C is the major determinant of α-BTX binding (Wilson, et al., 1985; McLane, et al., 1994). Loop C is a hairpin structure formed by vicinal cysteine residues at its apex, and also harbors conserved tyrosine residues that stabilize cholinergic ligands (Abramson, et al., 1989; Galzi, et al., 1991; Sine, et al., 1994). Positioned at the entrance to the ligand binding pocket, loop C is flexible and adopts a closed-in conformation that traps the ligand within the aromatic-rich binding pocket (Celie, et al., 2004; Gao, et al., 2005). Structure determination of α-BTX bound to the extracellular domain of the α1 subunit (Dellisanti, et al., 2007), or to a ligand binding domain derived from the homo-pentameric α7 AChR (Huang, et al., 2013), shows that fingers I and II of the toxin wrap around loop C and establish multiple electrostatic, polar and hydrophobic interactions. Notably, in muscle AChRs from α-BTX resistant snake and mongoose, loop C contains sites for N-linked glycosylation that prevent high affinity binding of α-BTX (Kreienkamp, et al., 1994). However, α-subunits of neuronal AChRs refractory to α-BTX lack such glycosylation sites, indicating differences in amino acid sequences among types of α-subunits determine whether α-BTX binds.
To understand why major classes of neuronal AChRs do not bind α-BTX, the present work identifies candidate determinants of α-BTX binding from sequence alignments of AChR α-subunits in light of recently determined 3D structures (Morales-Perez, et al., 2016; Walsh, et al., 2018), mutates the candidate determinants, and measures the ability of radiolabeled α-BTX to bind to the mutant AChRs. The results reveal multiple residues unique to α-BTX-null neuronal AChRs that account for their negligible affinity for α-BTX. These residues converge within a region of the receptor’s 3D structure predicted to fit between loops I and II of bound α-BTX. The generality of these determinants is tested by installing residues from equivalent positions of α-BTX-permissive AChRs into α-BTX-null α2, α3 and α4 AChR subunits, followed by co-expression with neuronal β-subunits. The resulting heteromeric AChRs are then analyzed by sucrose gradient sedimentation to test whether they form pentamers, and by single channel recording to test whether mutations that confer α-BTX binding affect receptor function.
2. Materials and methods
2.1. Molecular biology
cDNAs encoding wild type α2, α3, α4, β2 and β4 AChR subunits were kindly provided by Dr. Isabel Bermudez, Oxford-Brooks University, UK. The α4, β2 and β4 subunit cDNAs were used as provided, in the mammalian expression vector pCI (Promega), whereas the α2 and α3 cDNAs were sub-cloned into the CMV-based vector pRBG4 (Lee, et al., 1991). To facilitate introduction of mutations of candidate α-BTX binding determinants, for each α-subunit, a synthetic double-stranded oligonucleotide encoding loops B through C was generated (GeneScript, Piscataway, NJ). These synthetic oligonucleotides were designed to bridge unique restriction sites within each cDNA-expression vector combination, and also contained nucleotide substitutions of silent restriction sites to allow ligation of shorter double-stranded nucleotides harboring mutations of multiple candidate α-BTX binding determinants following digestion with the respective restriction enzymes. For each construct, the presence of intended mutations and absence of unintended mutations was determined by sequencing the region including the synthetic oligonucleotide. A cDNA encoding the endoplasmic resident protein nAChO (Gu, et al., 2016) was synthesized (GeneScript) and sub-cloned into the expression vector pRBG4.
2.2. Mammalian cell expression
A variant of the 293 HEK cell line known as peak-rapid (American Type Culture Collection) was maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 10% fetal bovine serum, and transfected at a confluence of 50-75 % by calcium phosphate precipitation. For radio-ligand binding experiments, the quantities of α, β and nAChO cDNAs were, respectively, 5, 10, and 1 μg for each 100 mm culture dish of cells. For patch clamp experiments, the quantities of α, β and nAChO cDNAs were, respectively, 1, 2, and 0.2 μg for each 35 mm culture dish of cells. Transfections were carried out for 12 to 16 hours, followed by medium exchange. Cells were incubated for an additional 72 hours at 37°C, and in some experiments, for another 24 hours at 30°C, prior to radio-ligand binding or patch clamp experiments.
2.3. 125I-α-BTX binding determinations
To determine the apparent dissociation constant for 125I-α-btx binding, transfected cells were harvested by gentle agitation in phosphate buffered saline, centrifuged at 2500 rpm for 1 min and re-suspended in the following extracellular bathing solution: 140 mM KCl, 5.4 mM NaCl, 1.8 mM CaCl2, 1.7 mM MgCl2, 25 mM HEPES, pH 7.4. Cell suspensions were incubated with specified concentrations of 125I-labeled-α-btx (PerkinElmer) for 4 hours at 21°C, with or without 2 mM (−) nicotine, and toxin-receptor complexes were separated from unbound toxin by filtration using a Brandel M-48T cell harvester. To determine non-specific binding, identical procedures were applied to cells transfected with a cDNA encoding the β2 subunit. After subtracting non-specific binding, total 125I-α-BTX binding was expressed as the fraction of that determined in the presence of 100 nM 125I-α-BTX. The difference between total binding and binding determined in the presence of (−) nicotine was fitted by an equation for a single class of binding sites using GraphPad Prism 5 software.
To determine the time course of α-BTX association, a specified concentration of 125I-α-BTX was added to a cell suspension at 21°C, and aliquots of the suspension were rapidly filtered through type A/E glass fiber filters (Gelman Sciences) at specified times. After subtracting non-specific binding, an equation describing bi-molecular association of toxin and receptor to form a toxin-receptor complex was fitted simultaneously to data obtained for multiple concentrations of 125I-α-BTX using Prism 5 software, yielding the association rate constant.
To determine the time course of α-BTX dissociation, 100 nM 125I-α-BTX was added to a cell suspension and incubated for 1 h at 21°C. The suspension was centrifuged at 2500 rpm for 1 min, the supernatant was removed, and the cell pellet was re-suspended in 30 ml of extracellular bathing solution. Aliquots of the suspension were then rapidly filtered through type A/E glass fiber filters (Gelman Sciences) at specified times. After subtracting non-specific binding, an equation describing radio-ligand dissociation from a single class of sites was fitted to the data using GraphPad Prism 5 software.
Binding of small cholinergic ligands was determined by competition against 125I-α-BTX binding, as described (Sine, et al., 1994). Briefly, transfected cells were harvested by gentle agitation, centrifuged at 2500 rpm for 1 min, and re-suspended in extracellular bathing solution. Replicate aliquots of cells were incubated with a series of increasing concentrations of competing ligand for 30 minutes, a concentration of 125I-α-BTX (10 to 30 nM) sufficient to occupy half of the available sites in 15 minutes was added, and radio-ligand bound to the cells was determined by filtration using a cell harvester. After subtracting binding in the presence of a saturating concentration of competing ligand, the fraction of sites bound was determined from the ratio of binding in the presence of competing ligand divided by that in the absence of competing ligand. The following form of the Hill equation was fitted to the data: 1-Fraction bound = 1 - [L]nH/([L]nH + KdnH), where [L] is ligand concentration, Kd is the apparent dissociation constant, and nH is the Hill coefficient.
2.4. Sucrose gradients
Cells transfected with specified AChR cDNAs were incubated 72 hours at 37°C followed by 24 hours at 30°C, and harvested in phosphate buffered saline by gentle agitation. Following centrifugation (2500 rpm for 1 min), the cell pellet was re-suspended in extracellular bathing solution followed by addition of 125I-α-BTX to a final concentration of 100 nM. Following incubation for 60 minutes at 21°C, the cells were centrifuged (2500 rpm for 1 min), the supernatant was removed, and the cell pellet was placed on ice. Cells were solubilized in an ice-cold lysis buffer containing 150 mM NaCl, 50 mM Tris, 5 mM EDTA, pH 7.5, 5 mM n-dodecyl-β-maltopyranoside (DDM, Anatrace, Maumee, Ohio), a general protease inhibitor cocktail (2 mM AEBSF, 0.3 μM Aprotinin, Bestatin, 14 μM E-64, 1 μM Leupeptin, 1 mM EDTA, Sigma) at a 20-fold final dilution. The lysate was centrifuged at 40,000 rpm for 10 min at 4°C, and the cleared lysate was layered on a continuous sucrose gradient (3-30%) containing lysis buffer plus 1 mM DDM. Gradients were centrifuged at 40,000 rpm at 4°C for 23 hours using a Beckman SW41 rotor and a Beckman Optima centrifuge. Fractions were collected from the top of the gradient, and radioactivity in each fraction was determined using a γ-counter.
2.5. Drugs
Acetylcholine (ACh), (−)nicotine, (±) epibatidine and dihydro-β-erythroidine (DHβE) were purchased from Sigma-Aldrich (St Louis, MO, USA).
2.6. Patch clamp recording
Single-channel currents were recorded in the cell-attached patch configuration at a membrane potential of −70 mV and a temperature of 21°C. The extracellular bathing solution contained (mM): 142 KCl, 5.4 NaCl, 1.8 CaCl2, 1.7 MgCl2, and 10 HEPES, adjusted to pH 7.4 with NaOH. Recording pipettes were filled with the same solution, without CaCl2, plus a specified concentration of ACh. Concentrated stock solutions of ACh were made in pipette solution and stored at −80°C until the day of each experiment. Patch pipettes were pulled from glass capillary tubes (No.7052, Garner Glass) and coated with Sylgard (Dow Corning).
2.7. Data Analysis
Single-channel currents were recorded using an Axopatch 200B patch-clamp amplifier (Molecular Devices), with a gain of 100 mV/pA and the internal Bessel filter at 10 kHz. Currents were sampled at intervals of 10 μs using a PCI-6111E acquisition card (National Instruments), and recorded to hard disk using the program Acquire (Bruxton Corporation). Channel opening and closing transitions were determined using the program TAC 4.2.0 (Bruxton Corporation), which digitally filters the data (Gaussian response, final effective bandwidth 5 kHz), interpolates the digitized points using a cubic spline function, and detects channel openings using the half-amplitude threshold criterion, as described (Colquhoun and Sigworth, 1983).
To determine single channel current amplitudes, the variable amplitude option within TAC was used, whereas to determine open and closed dwell times, the fixed amplitude option was used. Dwell time histograms were plotted using a logarithmic abscissa and square root ordinate (Sigworth and Sine, 1987), with a uniformly imposed dead time of 40 μs, and the sum of exponentials was fitted to the data by maximum likelihood using the program TACFit 4.2.0. The probability a channel, once open, will reopen was quantified by plotting the fraction of channel opening episodes with greater than N openings against the number of openings per episode, as described (Mazzaferro, et al., 2019). A channel opening episode was defined as one or more openings separated by closings shorter than τcrit, which was determined from the point of intersection between major brief and long duration closed time components, and was 1 ms for α4β2 and 2 ms for α3β4 receptors. An exponential decay function containing one or two components was fitted to each reopening distribution, and an F-test was used to determine whether a single or a bi-exponential decay best described the reopening data; a single-exponential decay was preferred unless the sum-of-squares F-test had a p-value less than 0.01. Both the exponential fitting and F-test were carried out using Prism 5 software (GraphPad). The mean number of openings per episode was calculated as the reciprocal of the decay constant, and an F-test was used to determine whether the fitted decay rates and fractional areas differed significantly between pairs of recordings from wild type and mutant AChRs. Parameters were considered significantly different if the p-value was less than 0.01.
3. Results
3.1. Identification of key residues in the a4 subunit
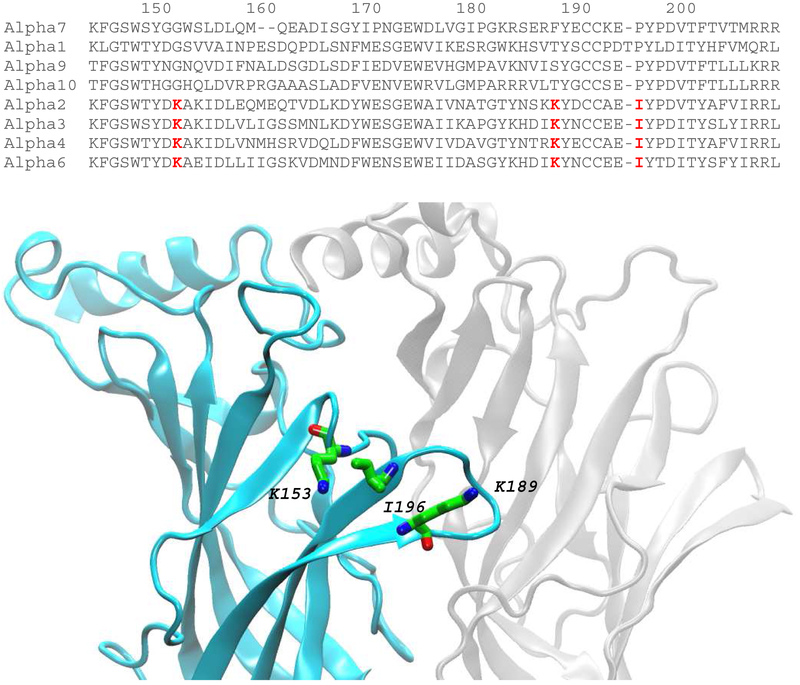
Fig. 1 shows a sequence alignment that includes loops B through C of the ligand binding domains from eight types of human AChR α-subunits, four that confer binding of α-BTX (α1, α7, α9, α10) and four that do not (α2, α3, α4, α6). Three residues, one in loop B and two in loop C (highlighted in red), are unique to α-subunits null for α-BTX binding and are chemically very different from residues at equivalent positions of α-subunits permissive for α-BTX binding. Based on recent x-ray (Morales-Perez, et al., 2016) and cryo-EM (Walsh, et al., 2018) structures of the α4β2 AChR, the three residues, Lys153, Lys189 and I196, congregate in a region equivalent to that in the α1 and α7 subunits proximal to bound α-BTX (Dellisanti, et al., 2007; Huang, et al., 2013).
Fig. 1.
Sequence alignment of α-subunits and structure of the ligand binding site of the α4β2 AChR- Upper panel shows alignment of the indicated α-subunits encompassing loops B (residues 148-155) and C (residues 184-200) from the principal face of the ligand binding site. Three residues common to α-BTX null α-subunits are highlighted in red. Lower panel shows the structure of the α4 (cyan) and β2 (grey) subunits that form one of two ligand binding sites of the α4β2 AChR (PDB code: 5KXI). The three residues highlighted in the upper panel are shown in stick representation (α-carbon atoms in green).
To determine whether the three candidate residues account for the inability of α-BTX to bind to α4β2 AChRs, each residue in the α4 subunit was mutated to the α-BTX permissive counterpart from the α7 AChR, and the mutant α4 subunit was co-expressed with the β2 subunit in 293 HEK cells. Intact cells were then incubated with a series of increasing concentrations of 125I-α-BTX, and radio-ligand bound to the cell surface was measured by filtration; bound radio-ligand is expressed relative to that determined for cells transfected with an equivalent amount of cDNA encoding the β2 subunit. For the wild type α4β2 AChR, the ratio of total to nonspecific binding is close to one for each of the 125I-α-BTX concentrations tested, confirming that the wild type AChR has negligible affinity for the toxin (Fig. 2). Similarly, for the single residue mutations, K189F (abbreviated F) and I196P (abbreviated P), and the double mutations, K153G plus I196P (abbreviated GP) and K153G plus K189F (abbreviated GF), the ratio of total to nonspecific binding is close to one, again indicating negligible affinity for the toxin. However, for the double mutation K189F plus I196P (abbreviated FP), the ratio of total to nonspecific binding increases abruptly, and after addition of the mutation K153G to form the triple mutation GFP, the ratio increases further. For the mutant receptors that bind α-BTX, the ratio of total to nonspecific binding is greater for an α-BTX concentration of 10 nM compared to that for 100 nM; this difference arises because specific binding of the toxin is saturable, whereas nonspecific binding increases with increasing toxin concentration. Thus the three residues identified by sequence alignment, combined with 3D structural data, account for the inability of α-BTX to bind to the α4β2 AChR.
Fig. 2.
Mutagenesis of key residues in the α4 subunit confers α-BTX binding- On the y-axis, α-BTX bound represents the ratio of 125I-α-BTX bound to cells expressing α4β2 AChRs containing the indicated mutations in the α4 subunit relative to that by control cells expressing the β2 subunit alone. Each column is the mean of 2-6 determinations with the error bar indicating the S.D. For each mutation, three columns are displayed corresponding to the indicated concentrations of 125I-α-BTX. Each mutation is abbreviated by the single letter amino acid code for the substituted residue: F (K189F), P (I196P), G (K153G), R (Y185R), E (T187E).
Inspection of the structure of α-BTX bound to a ligand binding domain derived from the α7 AChR reveals a pair of oppositely charged residues, Arg185 and Glu187, that borders the three unique determinants just identified (Huang, et al., 2013). However, unlike the unique determinants, these two residues vary among different types of α-subunits (Fig. 1). To test for possible contributions of these variable residues, equivalent residues in the α4 subunit, Tyr185 and Thr187, were mutated to their counterparts from the α7 subunit, Arg185 and Glu187. Combining mutations of these two residues with the mutation K189F, forming the triple mutation REF, yields a ratio of total to nonspecific α-BTX binding close to one (Fig. 2), indicating negligible affinity for the toxin. However, combining the mutation I196P, to yield the quadruple mutation REFP, affords substantial high affinity α-BTX binding that exceeds that of the α-toxin-permissive double mutation K189F plus I196P (FP). Finally, combining mutations of the three unique and two variable residues, yielding the α4 quintuple mutant GREFP, further increases high affinity α-BTX binding. Owing to their high affinity for α-BTX, heteromeric receptors containing the α4 GREFP quintuple mutant were chosen for in depth analyses via ligand binding, hydrodynamic and single channel measurements.
The expression studies in Fig. 2 were carried out with co-transfection of cDNA encoding the endoplasmic reticulum resident protein nAChO, which enhances expression of both α7 and α4β2 AChRs (Gu, et al., 2016). Thus before further analyzing the α4 quintuple mutant AChR, the amount of nAChO cDNA included in the transfection was optimized. Increasing the amount of nAChO increases cell surface α-BTX binding, revealing an optimum beyond which expression declines (Fig. 3). A further increase in cell surface binding is achieved by following incubation at 37°C with additional incubation at 30°C (Fig. 3, shaded bars). In the presence of the lowest amount of nAChO, the agonist nicotine blocks virtually all cell surface α-BTX binding, but as nAChO is increased, nicotine-resistant binding increases. Because agonists bind at subunit interfaces, agonist affinity can be exquisitely sensitive to the subunits forming the interfaces. Thus increasing the amount of nAChO may alter either the stoichiometric ratio of α4 to β2 subunits or their arrangement within the pentamer, potentially reducing affinity for nicotine. Thus the results in Fig. 3 define the amount of nAChO and cell culture incubation temperatures that achieve optimal expression of nicotine displaceable α-BTX binding to cell surface α4 quintuple mutant AChRs; in the subsequent experiments, all transfections included 1 μg of nAChO cDNA per 10 cm plate of cells.
Fig. 3.
Cell surface expression of α4β2 AChRs containing the α4 quintuple mutant subunit is enhanced by nAChO and incubation at 30°C. α-BTX bound represents 125I-α-BTX binding to cells expressing α4β2 AChRs containing the α4 quintuple mutant (abbreviated GREFP in Fig. 2) minus that determined for cells expressing the β2 subunit alone. For each amount of nAChO cDNA included in the transfection, the left bar indicates α-BTX binding in the presence of toxin alone and the right bar indicates that in the presence of 1 mM nicotine. The unfilled bars are results from cells incubated for 3 d at 37°C, whereas the shaded bars are results from cells incubated an additional 24 h at 30°C.
3.2. α-BTX binds with high affinity to AChRs comprised of α4 quintuple and β2 subunits
To estimate the affinity of α-BTX for AChRs comprised of the α4 quintuple mutant and the β2 subunit, intact cells were incubated with increasing concentrations of 125I-α-BTX, and radio-ligand bound to the cell surface was determined by filtration. In the presence of α-BTX alone, binding exhibits a robust high and a smaller low affinity component (Fig. 4A). After subtracting binding determined in the presence of nicotine, the resulting specific 125I-α-BTX binding is well described by a single class of binding sites with an apparent dissociation constant of 3.7 nM (Fig. 4B). This apparent dissociation constant is substantially smaller than those for receptors with mutations of either two (104 nM) or three (22 nM) of the unique residues in the α4 subunit (Table 1). By comparison, parallel measurements for the homomeric α7 AChR reveal an apparent dissociation constant of 26 nM (Table 1). Thus α-BTX binds specifically and with high affinity to heteromeric AChRs comprised of α4 quintuple mutant and β2 subunits.
Fig. 4.
Concentration dependence of α-BTX binding to cell surface α4β2 AChRs containing the quintuple mutant α4 subunit. Panel A shows binding in the presence of 125I-α-BTX alone (filled circles) and in the presence of 2 mM nicotine (crosses); binding to cells transfected with the β2 subunit alone is subtracted. Total binding is expressed relative to that determined in the presence of 100 nM 125I-α-BTX. Smooth curves are fits to an equation for two binding sites in which the fraction of each site is variable. Panel B shows specific binding determined as the difference between total 125I-α-BTX binding and that in the presence of nicotine. The smooth curve is the result of fitting an equation for binding to a single class of sites to the data, yielding an apparent dissociation constant of 3.7 nM (Table 1).
Table 1.
Apparent dissociation constant of mutant AChRs for 125I-α-BTX
| Subunit, mutant residue (s) | Kapp α-BTX, nM |
|---|---|
| α4, FP (K189F, I196P) | 103 ± 13 |
| α4, GFP (K153G, K189F, I196P) | 22 ± 3 |
| α4, REFP (Y185R, T187E, K153G, K189F, I196P) | 50 ± 10 |
| α4, GREFP (K153G, Y185R, T187E, K189F, I196P) | 3.7 ± 0.6 |
| α7 | 26 ± 4 |
Apparent dissociation constants were determined as in Fig. 4, ± 95 % confidence interval.
α-BTX is renowned for its slow rates of association with and dissociation from AChRs in skeletal muscle and the electric organ (Weber, et al., 1974; Weiland, et al., 1976; Sine and Taylor, 1979). To determine the rate constant for α-BTX association with quintuple mutant α4β2 AChRs, a specified concentration of 125I-α-BTX was added to intact cells, and bound α-BTX was measured at increasing times following addition. After subtracting binding to cells expressing only the β2 subunit, specific 125I-α-BTX binding is seen to increase with increasing time, approaching a maximum, and also to increase more rapidly with a higher compared to a lower α-BTX concentration (Fig. 5A). The association time courses are well described by association of α-BTX with a single class of sites with a rate constant of 5 × 104 M−1s−1, which is several orders of magnitude slower than that for a diffusion limited association reaction.
Fig. 5.
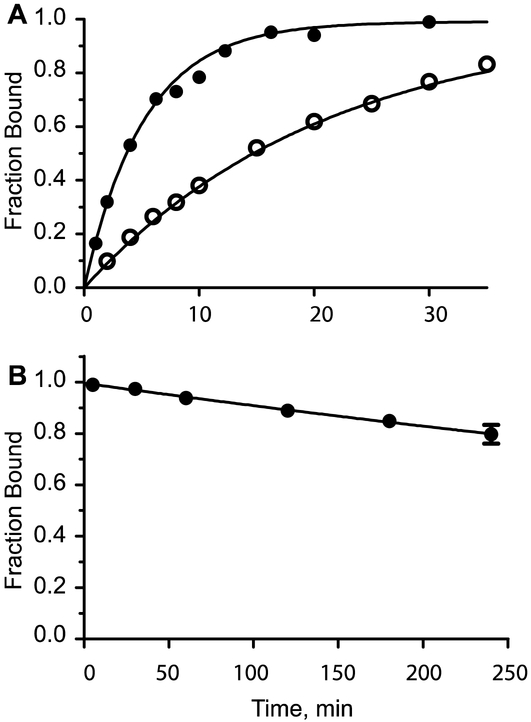
Kinetics of α-BTX binding to cell surface α4β2 AChRs containing the α4 quintuple mutant subunit. Panel A shows time courses of 125I-α-BTX association with cell surface receptors in the presence of either 15 nM (open symbols) or 60 nM (filled symbols) 125I-α-BTX. The smooth curves are the result of a simultaneous fit of an equation describing bimolecular association of receptor and toxin to the data; the fitted rate constant is 5.31 ± 0.18 × 104 M−1s−1. Panel B shows a time course of 125I-α-BTX dissociation from cell surface receptors. Symbols are the mean of 2-4 determinations with the error bars indicating ± S.E. The smooth curve is a fit of an equation describing first order dissociation of the toxin-receptor complex to the data; the fitted rate constant is 1.5 ± 0.2 × 10−5 s−1 (see Table 2).
To determine the rate constant for α-BTX dissociation from quintuple mutant α4β2 AChRs, intact cells were incubated with a concentration of 100 nM 125I-α-BTX, unbound α-BTX was removed following centrifugation, the cell pellet was rapidly diluted, and 125I-α-BTX bound to the cells was determined at specified times after dilution. The results show that 125I-α-BTX bound to cell surface receptors dissociates very slowly with a rate constant of 1.5 × 10−5 s−1, or a half-life of 12.5 h (Fig. 5B). The ratio of dissociation to association rate constants gives a dissociation constant of 0.3 nM, or some 10-fold lower than the apparent dissociation constant obtained under conditions of steady state incubation of toxin and receptor (Fig. 4B). The difference likely arises because the slow association and dissociation rate constants do not allow true steady state to be achieved during the finite incubation period. Overall, the kinetic measurements confirm that α-BTX binds with high affinity to heteromeric AChRs comprised of α4 quintuple mutant and wild type β2 subunits.
3.3. Identification of key residues in the a2 and a3 subunits
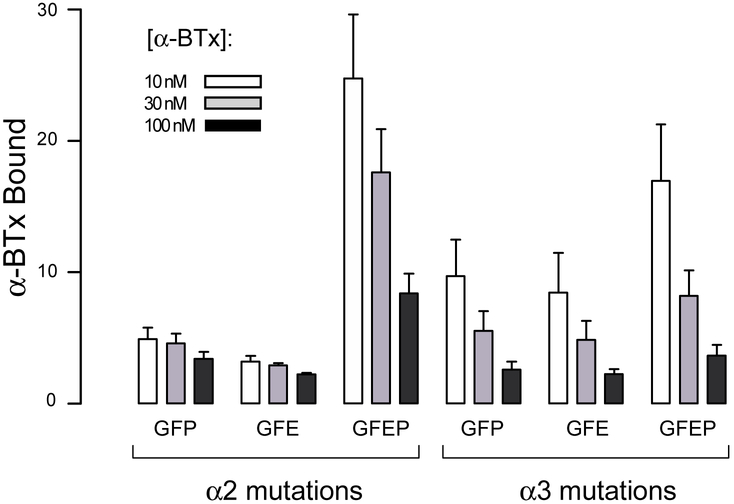
To test the generality of the three unique determinants that prevent α-BTX from binding to α4β2 AChRs, residues at equivalent positions of the α2 and α3 subunits were mutated to their α-BTX permissive counterparts from the α7 AChR; α6 was not included because, until the recent discovery of new chaperone proteins, heteromeric α6 AChRs could not be expressed on the cell surface (Gu, et al., 2019). Each triple mutant α2 or α3 subunit was then co-expressed with the β2 subunit, and the cells were incubated with a series of increasing concentrations of 125I-α-BTX, as in Fig. 2. For cells expressing either the α2 or α3 triple mutant subunits, total binding substantially exceeds non-specific binding (Fig. 6), but the ratio of total to specific binding is less than observed for the α4 quintuple mutant subunit (see Fig. 2); in addition the ratio decreases modestly from low to high α-BTX concentrations, suggesting low apparent affinity for the toxin.
Fig. 6.
Mutations in the α2 and α3 subunits confer α-BTX binding- On the y-axis, α-BTX bound represents the ratio of 125I-α-BTX bound to cells expressing either α2β2 or α3β2 AChRs containing the indicated mutations in the α2 or α3 subunits relative to that by control cells expressing the β2 subunit alone. Each column indicates the mean of 1-3 determinations with the error bars indicating ± S.D. For each mutation, three columns are displayed corresponding to the indicated concentrations of 125I-α-BTX. Each mutation is abbreviated by the single letter amino acid code for the substituted residue: F (K189F), P (I196P), G (K153G), E (D191E for α2 and N191E for α3).
A possible explanation for the low apparent affinity is that a previously identified determinant, Glu191 in loop C of the α4 and α7 subunits (Sine, et al., 2013), also contributes to α-BTX affinity. The equivalent residue in the α2 subunit is Asp, and in the α3 subunit is Asn (Fig. 1), suggesting these residues account for the reduced apparent affinity conferred by the triple mutant subunits. Thus, a fourth mutation, D187E in α2 and N187E in α3, was installed in each triple mutant α-subunit to form the quadruple mutant, abbreviated GFEP. Upon co-expressing each quadruple mutant α-subunit with the β2 subunit, measurements of α-BTX binding show a marked increase in the ratio of total to nonspecific binding (Fig. 6), approaching that conferred by the α4 quintuple mutant subunit; in addition, the ratio of specific to nonspecific binding decreases substantially from low to high α-BTX concentrations, indicating the quadruple mutant α-subunits increase affinity for α-BTX. Notably, substituting Glu at position 191 does not increase affinity if the unique determinant Ile196 is not also mutated to Pro, as can be seen by comparing the triple mutations (abbreviated GFE) with the quadruple mutations (GFEP), indicating Glu191 and Pro196 are interdependent in contributing to high affinity binding (Fig. 6). Thus in addition to the three conserved determinants, a fourth non-conserved determinant contributes to the inability of α-BTX to bind to AChRs containing either the α2 or α3 subunit.
To assess the stability of α-BTX bound to heteromeric AChRs comprised of mutant α- and wild type β-subunits, time courses of α-BTX dissociation were determined (Table 2), as in Fig. 5B. For each combination of α- and β-subunits, the toxin-receptor complexes exhibit half-lives on par with or greater than that of the muscle AChR (5.7 h), and less than that of the α7 AChR (133 h). Notably, receptors comprised of α3 quadruple mutant and β4 subunits form a highly stable complex with α-BTX, showing a dissociation half-life of 76 h. Thus heteromeric AChRs containing mutant α2, α3 or α4 subunits form stable complexes with α-BTX.
Table 2.
Dissociation of 125I-α-BTX from mutant AChRs
| Subunit, mutant residue (s) | Non-α-subunit | kdissoc, s−1 | Half-life, h |
|---|---|---|---|
| α4 GREFP | β2 | 1.5 ± 0.2 × 10−5 | 12.6 |
| α4 GREFP | β4 | 1.0 ± 0.1 × 10−5 | 19 |
| α3 GFEP | β4 | 2.5 ± 0.9 × 10−6 | 77 |
| α2 GFEP | β2 | 3.2 ± 0.3 × 10−5 | 6.0 |
| α2 GFEP | β4 | 2.0 ± 0.3 × 10−5 | 9.6 |
| α1 | β, ε, δ | 3.5 ± 0.4 × 10−5 | 5.4 |
| α7 | none | 1.5 ± 0.3 × 10−6 | 133 |
3.4. Hydrodynamic properties of AChRs comprised of mutant α- and wild type β-subunits
In the preceding experiments, α-BTX binding was measured to AChRs expressed on the surface of intact cells, implying that the oligomers formed by a mutant α- and wild type β-subunit are pentamers. To determine whether the mutated cell-surface AChRs are pentamers, cells transfected with mutant α- and wild type β-subunits were incubated with 125I-α-BTX, unbound toxin was removed following centrifugation, the cells were solubilized in the detergent β-dodecyl-maltoside, and following centrifugation to remove insoluble material, the cleared cell lysate was layered on a sucrose gradient. Following ultra-centrifugation, fractions were collected and radioactivity in each fraction was measured. To provide a frame of reference for a cell-surface pentameric AChR, cells expressing the adult muscle AChR were subjected to identical procedures. For receptors comprised of the α4 quintuple mutant and β2 subunits, a sharp peak is observed that coincides with that of the pentameric muscle AChR (Fig. 7A). In addition, for both the muscle and α4 quintuple mutant AChRs, a small peak near the top of the gradient is observed, corresponding to unbound α-BTX, and the fractions between the small and large peaks contain low but measureable amounts of radioactivity, likely due to dissociation of a portion of the α-BTX-AChR complexes during the 23 h centrifugation procedure. Similarly, when the α4 quintuple mutant subunit is co-expressed with the β4 subunit, a sharp peak is observed that coincides with that of the muscle AChR standard (Fig. 7B). Likewise, when either the α3 or α2 quadruple mutant subunit is co-expressed with the β4 subunit, a sharp pentameric peak is observed in each case (Fig. 7B). Notably for the α3 quadruple mutant plus β4 subunit, very little radioactivity is detected in the top and intermediate fractions of the gradient, indicating a highly stable α-BTX-AChR complex, as also revealed by the α-BTX dissociation time course (Table 2). Thus the various combinations of mutant α- and wild type β-subunits form stable complexes with α-BTX that co-sediment with that of the pentameric muscle AChR.
Fig. 7.
Sucrose gradient sedimentation of cell surface AChRs labeled with 125I-α-BTX. Each gradient profile is normalized to the fraction with maximum radioactivity. Fraction 1 corresponds to the top of the gradient. Cell surface AChRs were labeled with 125I-α-BTX, unbound toxin was removed by centrifugation, and solubilized AChRs were prepared as described in Materials and Methods. Mutations are abbreviated as in the legends to Figs. 2 and 6. The adult human muscle AChR (α1βεδ) is the standard for a cell-surface pentamer.
3.5. Ligand recognition by AChRs comprised of mutant α- and wild type β-subunits
To assess the ability of AChRs comprised of α4 quintuple mutant and β2 subunits to recognize small cholinergic ligands, 125I-α-BTX binding to cell surface receptors was determined in the presence of increasing concentrations of ACh, nicotine, epibatidine, or dihydro-β-erythroidine using previously described methods (Sine, et al., 1994). All four ligands compete against α-BTX binding, with apparent dissociation constants spanning a range of ~100-fold (Fig. 8A). The three agonists exhibit the rank order of potency, nicotine ≈ epibatidine > ACh, previously established for α4β2 AChRs, while the antagonist dihydro-β-erythroidine is some 10-fold more potent than reported for antagonism of ACh-elicited macroscopic currents (Chavez-Noriega, et al., 1997). Thus small cholinergic ligands compete against 125I-α-BTX binding to the α4 quintuple mutant AChR with potencies that depend on the particular ligand.
Fig. 8.
Competition of unlabeled ligands against binding of 125I-α-BTX. Panel A- Intact cells expressing α4 quintuple mutant and β2 subunits were incubated with 125I-α-BTX (10-30 nM) for 15 minutes in the presence of specified concentrations of the following competing ligands: ACh (filled circles), nicotine (open circles), epibatidine (open squares), dihydro-β-erythrodine (open triangles). Panel B- Intact cells expressing the α4 quintuple mutant (filled circles), the α3 quadruple mutant (open circles), or the α2 quadruple mutant (triangles), each with the β4 subunit, were incubated with 125I-α-BTX for 15 minutes in the presence of the indicated concentrations of nicotine. Smooth curves are fits of the Hill equation to the data, with the fitted parameters given in Table 3.
Previous work showed that different types of neuronal α-subunits confer different apparent affinities for cholinergic agonists (Chavez-Noriega, et al., 1997). Thus to test for subunit-specificity in agonist recognition, the different types of mutant α-subunits were co-expressed with the β4 subunit, and the resulting AChRs were tested for the ability of nicotine to compete against 125I-α-BTX binding. Nicotine competes against 125I-α-BTX binding, showing highest apparent affinity for AChRs containing either α2 or α4 subunits, and some 70-fold lower apparent affinity for AChRs containing the α3 subunit. This subunit dependence of nicotine binding parallels that observed from whole cell dose-response measurements (Chavez-Noriega, et al., 1997), indicating the subunits with mutations engineered to confer α-BTX binding retain subunit-specificity in ligand recognition.
3.6. Function of AChRs comprised of mutant α- and wild type β-subunits
To assess whether the mutations that confer α-BTX binding affect receptor function, patch clamp recordings were made from cells expressing AChRs comprised of either wild type or mutant α-subunits plus a wild type β-subunit; heteromeric AChRs comprised of either α4 and β2 or α3 and β4 subunits were chosen for in depth functional studies. For the α4β2 AChR, single channel currents appear predominantly as individual current pulses flanked by long periods of baseline current, with a minor contribution of two or more pulses in quick succession (Fig. 9A); this kinetic signature is observed for AChRs containing either the wild type α4 or GREFP quintuple mutant α4 subunit. Histograms of open channel dwell times, plots of single channel current amplitude against applied voltage, and probability of channel re-opening are indistinguishable between wild type and mutant AChRs. Notably, the kinetic and conductance signatures of the wild type and mutant AChRs correspond to those previously described for the low conductance (α4)2(β2)3 stoichiometry (Mazzaferro, et al., 2016); the higher conductance (α4)3(β2)2 stoichiometry was not observed, likely because the cells were transfected with an excess of the β2 over the α4 subunit.
Fig. 9.
Single channel currents recorded from wild type and mutant neuronal AChRs. Panel A- Upper, single channel currents from wild type or quintuple mutant α4β2 AChRs recorded in the cell attached patch configuration with 10 μM ACh in the pipette solution are displayed at a bandwidth of 4 kHz. Channel openings are upward deflections. Lower left, histograms of channel open dwell times are displayed with the wild type in black and mutant in orange; the smooth curves represent the overall fit of the sum of three exponentials to each data set. Lower middle, a plot of single channel current amplitude against pipette voltage is fitted by linear regression, yielding a slope conductance of 49 pS (47-51 pS, 95% confidence interval). Lower right, probability distribution of the number of channel re-openings per burst is fitted by a single exponential with the following means and 95% confidence intervals in parenthesis: wild type, 0.44 (0.30-0.57) re-openings/burst; mutant, 0.46 (0.30-0.54) re-openings/burst. Panel B- Upper, single channel currents from wild type or quadruple mutant α3β4 AChRs recorded in the cell attached patch configuration with 1 μM ACh in the pipette solution are displayed at a bandwidth of 4 kHz. Lower left, histograms of channel open dwell times are displayed with the wild type in black and mutant in orange, and the smooth curves represent the fit of the sum of three exponentials to each data set. Lower middle, a plot of single channel current amplitude against pipette voltage is fitted by linear regression, yielding a slope conductance of 56 pS (53-57 pS, 95% confidence interval). Lower right, probability distribution of the number of channel re-openings per burst fitted by the sum of two exponentials with the following means and fractional weights with 95% confidence intervals in parenthesis: wild type, component 1: 2.0 (1.9-2.2) re-openings/burst, 0.29 (0.27-0.31); component 2: 10.7 (10.5-10.9) re-openings/burst, 0.71 (0.69-0.73); mutant, component 1: 1.9 (1.8-2.1) re-openings/burst, 0.45 (0.42-0.48); component 2: 8.7 (8.4-9.1) re-openings/burst, 0.55 (0.53-0.58).
For the α3β4 AChR, ACh-induced single channel currents appear as episodes of several pulses in quick succession, with each episode flanked by long periods of baseline current (Fig. 9B); this kinetic signature contrasts with that of the α4β2 receptor, and is observed for AChRs containing either the wild type α3 or GFEP quadruple mutant α3 subunit. Histograms of open channel dwell times and plots of single channel current amplitude against applied voltage are indistinguishable between wild type and mutant AChRs. The probability of channel reopening is similar between the wild type and mutant AChRs, but a small but significant decrease in the number of openings per episode is observed for the α3 GFEP mutant. This decrease in channel reopening is unlikely to arise from slowing of rate constants for ACh association because channel openings were elicited by a low concentration of ACh (1 μM). Alternatively, the decrease in channel reopening could arise from either an increase in the rate of ACh dissociation, or slowing of transitions from intermediate closed to open states. In summary, the mutations that confer α-BTX binding do not affect unitary current amplitude or stability of the open channel, and show either no or modest effects on the kinetics of channel reopening.
4. Discussion
The present work identifies residues that render neuronal nicotinic AChRs containing α2, α3 or α4 subunits refractory to α-BTX binding. Sequence alignment reveals three residues unique to these subunits that differ from their counterparts in α-BTX permissive α-subunits. The three residues, Lys153, Lys189 and Ile196 of the α4 subunit, are not the only residues unique to α-BTX null α-subunits, but recent structures of the α4β2 AChR show that only these three residues congregate in 3D space and localize to a region predicted to contact the toxin (Figs. 1, 10). Mutation of any one of these residues does not confer α-BTX binding upon the α4β2 AChR, indicating the contribution of each residue depends on one or more of the other residues. Of the three possible pairwise mutations, only the pair Lys189Phe and Ile196Pro confers affinity for α-BTX in the nanomolar range, while addition of the third mutation, Lys153Gly, increases affinity another five-fold. Combining mutations of two neighboring residues that vary among α-subunits, Tyr185Arg and Thr187Glu, while having no effect alone, increases affinity another five-fold. Thus in the resulting quintuple mutant α4 subunit, mutation of two unique residues imparts α-BTX recognition, while mutation of one unique and two variable residues further enhances α-BTX affinity.
Fig. 10.
Structural relationship between bound α-BTX and key binding determinants. Upper, X-ray structure of the complex between α-BTX (magenta) and a ligand binding domain (four subunits cyan; one subunit off-white) derived from the α7 AChR (Huang, et al., 2013; PDB code: 4HQP). For clarity, only one of five α-BTX molecules is shown. Lower, close-up view showing key determinants of α-BTX binding; the three residues equivalent to those unique to α-BTX permissive α-subunits are shown in van der Waals representation (Ser149, Phe184, Pro190), while the three variable residues are shown in stick representation (Arg179, Glu181, Glu186).
Mutational analyses of the α2 and α3 subunits confirm that the three unique residues identified in the α4 subunit confer the α-BTX-null pharmacological signature, and also reveal a fourth residue, variable among α-subunits, that contributes to the inability of these subunits to bind α-BTX. The fourth residue is Glu at position 191 of the α4 and α7 subunits, but is Asp in α2 and Asn in α3. Substituting Glu at position 191 of either the α2 or α3 subunits that also harbor mutations of the three unique determinants markedly enhances affinity for α-BTX. The significance of this determinant was previously discovered in studies of a chimeric AChR containing α4 sequence in loop C and α7 sequence in the remainder of the ligand binding domain; in that construct substituting Asn for Glu191 markedly reduced affinity for α-BTX (Sine, et al., 2013).
The crystal structure of α-BTX bound to a ligand binding domain derived from the pentameric α7 AChR illustrates structural relationships between α-BTX and residues equivalent to the three unique and three variable determinants (Huang, et al., 2013; Fig. 10). The hairpin structure of loop C brings the two unique determinants, Phe189 and Pro196, into close spatial alignment. In addition, conformational restriction of the protein main chain by Pro196 likely contributes to this alignment, which positions the aromatic side chain of Phe189 within a hydrophobic crevice between fingers I and II of the α-toxin. The third unique determinant, Gly153 in loop B of α7, aligns in close register with Phe189 and Pro 196, and apparently contributes to the spatial relationship between loops B and C that enhances affinity for α-BTX; Gly153 is the first determinant of α-BTX binding identified within loop B. The side chain of the variable determinant, Glu191 in α4 and α7, extends along the base of finger II of the toxin. The length of the Glu191 side chain is decisive because a shorter Asp side chain, naturally present in the α2 subunit, reduces affinity for α-BTX. The two other variable determinants, Arg185 and Glu187 at the beginning of loop C, form an ion pair between the three unique determinants and finger I of the toxin, and may provide steric constraints that stabilize both structures. Altogether, the inability of α-BTX to bind to neuronal AChRs arises from two unique residues essential for α-BTX recognition, plus a third unique and three variable residues that affect affinity for the α-toxin. An emerging theme is that the determinants of α-BTX binding are synergistic in contributing to α-toxin recognition and affinity.
Pioneering studies established that loop C within the AChR α1-subunit is decisive in conferring α-BTX binding. Small peptides that include amino acid sequences from loop C bind α-BTX in a manner displaced by small cholinergic ligands (Chaturvedi, et al., 1993), whereas peptides from other regions of the α1-subunit do not (Wilson, et al., 1985; Conti-Tronconi, et al., 1990). Apparent dissociation constants of the peptides for α-BTX were some 3-4 orders of magnitude greater than that of the intact pentameric AChR, indicating loop C conformation, and possibly other regions of the subunit, contribute to nanomolar dissociation constants for the toxin.
Studies of AChRs naturally resistant to α-BTX revealed residues within loop C that contribute to high affinity binding. In loop C from the snake and mongoose, substitutions of Asn, together with consensus Ser or Thr residues, allows glycosylation that imparts α-BTX resistance to these species (Neumann, et al., 1986; Kreienkamp, et al., 1994). Mutational analyses of the neuronal α3 subunit also identified a stretch of consecutive residues in loop C that restored α-BTX recognition, although bound toxin dissociated from the mutant receptors within minutes to tens of minutes (Levandoski, et al., 1995). The overall conclusion from these studies was that multiple residues within loop C contribute to high affinity for α-BTX. However, the key residues and structural bases of high affinity binding remained elusive. A major limitation was lack of atomic resolution structures of AChRs without and with bound α-toxin.
Toward structure determination of AChR-α-toxin complexes, both NMR and x-ray crystallography were applied to α-BTX bound to small peptides that included amino acid sequences of loop C or variants thereof (Basus, et al., 1993; Scherf, et al., 1997; Harel, et al., 2001; Samson, et al., 2002). The structures revealed that loop C peptides lodged between fingers I and II of the toxin. The crystal structure of acetylcholine binding protein bound to cobra α-toxin, at 4.2 Å resolution, confirmed the spatial relationship between loop C and fingers I and II of the toxin, and also revealed relationships between fingers I and III of the toxin and the principal and complementary faces of the subunits that form the ligand binding pocket (Bourne, et al., 2005). In particular, finger II of the toxin inserts deeply within the subunit interface where it is poised to interact with conserved aromatic residues from the principal face, as well as aromatic, polar and hydrophobic residues from the complementary face. In addition, finger I abuts the base of loop C exposed to the protein surface, and finger III orients toward the complementary face. Crystal structures of α-BTX bound to ligand binding domains from either the monomeric α1 or pentameric α7 subunits confirmed the spatial relationship between the receptor and toxin, but the higher resolution of both structures (1.9 and 3.5 Å, respectively) revealed key inter-residue contacts between finger II of the toxin and aromatic residues within the binding pocket (Dellisante, 2007; Huang, et al., 2013). In particular, conserved Arg and Phe residues from the tip of finger II of the toxin form a cation-π stack that orients edge-to-face with the aromatic ring of a conserved Tyr residue from loop C, while the aromatic hydroxyl group from the same Tyr forms a hydrogen bond to a conserved Asp residue from finger II of the toxin. Mutational analyses showed that substitution of this conserved Tyr with Thr abolished high affinity binding of α-BTX (Huang, et al., 2013), and also prevented ACh-mediated channel opening (Bouzat, et al., 2008). The conserved Arg from finger II of the toxin also established hydrogen bonds to aromatic hydroxyl groups from a second conserved Tyr in loop C and a conserved Tyr in loop A. This finger-in-trap relationship between finger II of the toxin and the subunit interface that forms the ligand binding site is clearly essential for α-toxin binding. However, the results herein demonstrate that in AChRs null for α-BTX binding, disruption of contacts at the solvent exposed surface of the receptor is enough to prevent high affinity binding. From a design standpoint, it makes sense that resistance to α-BTX binding does not arise from residue substitutions within the subunit interface that envelops finger II of the toxin, as many of these residues are conserved and crucial for binding ACh and subsequent opening of the ion channel. Instead, resistance arises from residue substitutions within portions of loops B and C exposed to the protein surface; the altered residues are not obligatory for ACh binding or channel opening, as illustrated by naturally occurring residue substitutions in neuronal and α-toxin-refractory muscle AChRs.
The α2-α4 subunits engineered to bind α-BTX assemble with neuronal β-subunits to form heteropentamers. Their high affinity for α-BTX allowed them to be labeled with radio-iodinated α-BTX while still in the plasma membrane of intact cells, and following cell lysis and detergent solubilization, to be tracked by sucrose gradient sedimentation. The resulting α-BTX-AChR complexes co-sediment with adult human muscle AChRs bound to radio-iodinated α-BTX, confirming they are pentamers. The α-BTX permissive AChRs retain the ability to recognize small cholinergic ligands in a manner that depends on the particular ligand as well as the combination of α- and β-subunits that form the ligand binding site. The α-BTX permissive α4β2 and α3β4 receptors are functional, exhibiting single channel current amplitudes and open state stabilities indistinguishable from those of their α-BTX null counterparts. In each case only a single conductance class of channel openings was observed, indicating a single stoichiometry of α- and β-subunits. For the α4β2 receptor, the conductance signature corresponds to the stoichiometry (α4)2(β2)3 (Mazzaferro, et al., 2017), perhaps because the cells were transfected using an excess of the β- over the α-subunit cDNA. Further studies will be required to define experimental conditions to selectively express pentamers with different subunit stoichiometries.
In summary, we identify key residues that render neuronal AChRs refractory to α-BTX binding, describe mutations of residues that allow these AChRs to bind α-BTX, show that the mutations are interdependent in conferring α-BTX binding, and demonstrate that the α-BTX permissive α-subunits assemble as pentamers with essentially normal functional properties. Given that α-BTX is a high affinity, membrane-impermeable ligand, the α-BTX permissive α-subunits described herein will facilitate future studies of function, pharmacology, subunit assembly, and cellular distribution of these physiologically crucial neuronal AChRs.
Table 3.
Apparent dissociation constants for cholinergic ligands determined by competition against 125I-α-BTX binding.
| α-subunit | β-subunit | Ligand | Kapp | nH |
|---|---|---|---|---|
| α4 GREFP | β2 | ACh | 5.3 ± 0.6 × 10−7 | 0.81 ± 0.07 |
| α4 GREFP | β2 | Nicotine | 1.1 ± 0.1 × 10−8 | 1.4 ± 0.1 |
| α4 GREFP | β2 | Epibatidine | 4.5 ± 0.3 × 10−9 | 1.3 ± 0.1 |
| α4 GREFP | β2 | DHβE | 7.7 ± 0.4 × 10−9 | 1.4 ± 0.1 |
| α4 GREFP | β4 | Nicotine | 2.1 ± 0.2 × 10−7 | 0.80 ± 0.1 |
| α3 GFEP | β4 | Nicotine | 9.2 ± 1.5 × 10−6 | 0.75 ± 0.1 |
| α2 GFEP | β4 | Nicotine | 2.1 ± 0.1 × 10−7 | 0.96 ± 0.1 |
Highlights.
α-Bungarotoxin binds to some members of the nicotinic receptor family but not to others.
Sequence alignments and 3D structures identify candidate residues of member-selective α-bungarotoxin binding.
In α-bungarotoxin-null receptors, mutating candidate residues confers toxin binding.
Mutant receptors that bind α-bungarotoxin mimic native receptors structurally and functionally.
Acknowledgements
We thank Dr. Isabel Bermudez for generously providing cDNAs encoding α2, α3, α4, β2 and β4 subunits.
Funding
Supported by NIH grants NS031744 to S.M.S.
Footnotes
Dedication- This paper is dedicated to the memory of George M. Sine who inspired others to ask questions and to find the answers with their own hands.
Disclosures of interest
S. M. Sine, PhD reports no disclosures.
J. R. Strikwerda reports no disclosures.
S. Mazzaferro reports no disclosures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abramson SN, Li Y, Culver P, Taylor P, 1989. An analog of lophotoxin reacts covalently with Tyr190 in the α-subunit of the nicotinic acetylcholine receptor. J. Biol. Chem. 264, 12666–12672. [PubMed] [Google Scholar]
- 2.Anderson MJ, Cohen MW, Zorychta E, 1977. Effects of innervation on the distribution of acetylcholine receptors on cultured muscle cells. J. Physiol 268, 731–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axelrod D, Ravdin PM, Podleski TR, 1978. Control of acetylcholine receptor mobility and distribution in cultured muscle membranes. A fluorescence study. Biochim. Biophys. Acta. 511, 23–38. [DOI] [PubMed] [Google Scholar]
- 4.Basus VJ, Song G, Hawrot E, 1993. NMR solution structure of an α-bungarotoxin/nicotinic receptor peptide complex. Biochemistry 32, 12290–12298. [DOI] [PubMed] [Google Scholar]
- 5.Blount P, and Merlie J, 1988. Native folding of an acetylcholine receptor α-subunit expressed in the absence of other receptor subunits. J. Biol. Chem. 263, 1072–1080. [PubMed] [Google Scholar]
- 6.Blount P, Smith MM, and Merlie J, 1990. Assembly intermediates of the mouse muscle nicotinic acetylcholine receptor in stably transfected fibroblasts. J. Cell Biol. 111, 2601–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulter J, Connolly J, Deneris E, Goldman D, Heinemann S, Patrick J, 1987. Functional expression of two neuronal nicotinic acetylcholine receptors from cDNA clones identifies a gene family. Proc Natl. Acad. Sci. USA 84, 7763–7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourne Y, Talley TT, Hansen SB, Taylor P, Marchot P, 2005. The crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake α-neurotoxins and nicotinic receptors. EMBO J. 24, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouzat C, Bartos M, Corradi J, Sine SM, 2008. Binding-pore interface of homomeric Cys-loop receptors governs open channel lifetime and rate of desensitization. J. Neurosci. 28, 7808–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celie PH, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK, 2004. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41, 907–914. [DOI] [PubMed] [Google Scholar]
- 11.Chang CC, and Lee CY, 1963. Isolation of neurotoxins from the venom of bungarus multicinctus and their modes of neuromuscular blocking action. Arch. Int. Pharmacodyn. Ther_144, 241–57. [PubMed] [Google Scholar]
- 12.Changeux JP, Kasai M, Lee CY, 1970. Use of a snake venom toxin to characterize the cholinergic receptor protein. Proc. Natl. Acad. Sci. USA 67, 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaturvedi V, Donnelly-Roberts DL, Lentz TL, 1993. Effects of mutations of Torpedo acetylcholine receptor α-subunit residues 184-200 on α-bungarotoxin binding in a recombinant fusion protein. Biochemistry 32, 9570–9576. [DOI] [PubMed] [Google Scholar]
- 14.Chavez-Noriega LE, Crona JH, Wahsburn MS, Urrutia A, Elliott KJ, Johnson EC, 1997. Pharmacological characterization of recombinant human neuronal nicotinic acetylcholine receptors hα2β2, hα2β4, hα3β2, hα3β4, hα4β2, hα4β4 and hα7 expressed in Xenopus oocytes. J. Pharmacol. Exp. Ther. 280, 346–356. [PubMed] [Google Scholar]
- 15.Colquhoun D and Sigworth FJ, 1983. Fitting and statistical analysis of single channel records In Single Channel Recording. Sakmann B and Neher E, editors. Plenum Publishing Corp; New York: 191–264. [Google Scholar]
- 16.Conti-Tronconi BM, Tang F, Diethelm BM, Spencer SR, Reinhardt-Maelicke S, Maelicke A, 1990. Mapping of a cholinergic binding site by means of synthetic peptides, monoclonal antibodies, and α-bungarotoxin. Biochemistry 29, 6221–6230. [DOI] [PubMed] [Google Scholar]
- 17.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L, 2007. Crystal structure of the extracellular domain of nAChR α1 bound to α-bungarotoxin at 1.94 Å resolution. Nat. Neurosci. 10, 953–962. [DOI] [PubMed] [Google Scholar]
- 18.Galzi JL, Bertrand D, Devillers-Thiery A, Revah F, Bertrand S, Changeux JP, 1991. Functional significance of aromatic amino acids from three peptide loops of the α7 neuronal nicotinic receptor site investigated by site-directed mutagenesis. FEBS Lett. 294, 198–202. [DOI] [PubMed] [Google Scholar]
- 19.Gao F, Burghardt T, Bren N, Hansen SB, Henchman R, Taylor P, McCammon JA, Sine SM, 2005. Acetylcholine-mediated conformational changes in acetylcholine-binding protein revealed by simulation and intrinsic tryptophan fluorescence. J. Biol. Chem. 280, 8443–8451. [DOI] [PubMed] [Google Scholar]
- 20.Green WN, and Claudio T, 1993. Acetylcholine receptor assembly: subunit folding and oligomerization occur sequentially. Cell 74, 57–69. [DOI] [PubMed] [Google Scholar]
- 21.Gu S, Matta JA, Lord B, Harrington AW, Sutton SW, Davini WB, Bredt DS, 2016. Brain α7 Nicotinic Acetylcholine Receptor Assembly Requires NACHO. Neuron 89, 948–955. [DOI] [PubMed] [Google Scholar]
- 22.Gu S, Matta JA, Davini WB, Dawe GB, Lord B, Bredt DS, 2019. α6-Containing Nicotinic Acetylcholine Receptor Reconstitution Involves Mechanistically Distinct Accessory Components. Cell Rep. 26, 866–874. [DOI] [PubMed] [Google Scholar]
- 23.Haggerty JG, and Freohner SC, 1981. Restoration of 125I-α-bungarotoxin binding activity to the α-subunit of Torpedo acetylcholine receptor isolated by gel electrophoresis in sodium dodecyl sulfate. J. Biol. Chem. 256, 8294–8297. [PubMed] [Google Scholar]
- 24.Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S, 2001. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between α-bungarotoxin and a mimotope peptide. Neuron 32, 265–275. [DOI] [PubMed] [Google Scholar]
- 25.Huang S, Li S-X, Bren N, Cheng K, Gomoto R, Chen L, Sine SM, 2013. Complex between α-bungarotoxin and an α7 nicotinic receptor ligand binding domain. Biochem. J. 454, 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreienkamp H, Sine SM, Maeda R, Taylor P, 1994. Glycosylation sites selectively interfere with α-toxin binding to the nicotinic acetylcholine receptor. J. Biol. Chem. 269, 8108–8114. [PubMed] [Google Scholar]
- 27.Lee BS, Gunn RB, Kopito RR, 1991. Functional differences among nonerythroid anion exchangers expressed in a transfected human cell line. J. Biol. Chem. 266,11448–11454. [PubMed] [Google Scholar]
- 28.Levandoski M, Lin Y, Moise L, McLaughlin J Cooper E, Hawrot E, 1999. Chimeric analysis of a neuronal nicotinic receptor reveals amino acids conferring sensitivity to α-bungarotoxin. J. Biol. Chem. 274, 26113–26119. [DOI] [PubMed] [Google Scholar]
- 29.Lindstrom J, Einarson B, and Merlie J, 1978. Immunization of rats with polypeptide chains from torpedo acetylcholine receptor causes an autoimmune response to receptors in rat muscle. Proc. Natl. Acad. Sci. USA 75, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazzaferro S, Bermudez I, Sine SM, 2017. α4β2 nicotinic acetylcholine receptors: relationships between subunit stoichiometry and function at the single channel level. J. Biol. Chem. 292, 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazzaferro S, Bermudez I, Sine SM, 2019. Potentiation of a neuronal nicotinic receptor via pseudo-agonist site. Cell Mol. Life Sci. 10.1007/s00018-018-2993-7. (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLane KE, Wu X, Conti-Tronconi BM, 1994. An α-bungarotoxin-binding sequence on the Torpedo nicotinic acetylcholine receptor α-subunit: conservative amino acid substitutions reveal side-chain specific interactions. Biochemistry 33, 2576–2585. [DOI] [PubMed] [Google Scholar]
- 33.Morales-Perez CL, Noviello CM, Hibbs RE, 2016. X-ray structure of the human α4β2 nicotinic receptor. Nature 538, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann D, Barchan D, Fridkin M, Fuchs S, 1986. Analysis of ligand binding to the synthetic dodecapeptide 185-196 of the acetylcholine receptor α-subunit. Proc. Natl. Acad. Sci. USA 83, 9250–9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samson A, Scherf T, Eisenstein M, Chill J, Angilister J, 2002. The mechanism for acetylcholine receptor inhibition by α-neurotoxins and species-specific resistance to α-bungarotoxin revealed by NMR. Neuron 35, 319–332. [DOI] [PubMed] [Google Scholar]
- 36.Scherf T, Balass M, Fuchs S, Katchalski-Katzir E, Angilister J, 1997. Three-dimensional solution structure of the complex of α-bungarotoxin with a library-derived peptide. Proc. Natl. Acad. Sci USA 94, 6059–6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sine SM and Taylor P, 1979. Functional consequences of agonist-mediated state transitions in the cholinergic receptor. J. Biol. Chem. 254, 3315–3325. [PubMed] [Google Scholar]
- 38.Sine SM, Quiram P, Papanikolaou F, Kreienkamp H-J, Taylor P, 1994. Conserved tyrosines in the α-subunit of the nicotinic acetylcholine receptor stabilize quaternary ammonium groups of agonists and curariform antagonists. J. Biol. Chem 269, 8808–8816. [PubMed] [Google Scholar]
- 39.Sine SM, Huang S, Li S-X, daCosta CJB, Chen L, 2013. Inter-residue coupling contributes to high affinity, subtype selective binding of α-bungarotoxin to nicotinic receptors. Biochem. J. 454, 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh RM, Roh SH, Gharpure A, Morales-Perez CL, Teng J, Hibbs RE, 2018. Structural principles of distinct assemblies of the human α4β2 nicotinic receptor. Nature 557, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber M, and Changeux JP, 1974. Binding of Naja nigricollis (3H)α-toxin to membrane fragments from Electrophorus and Torpedo electric organs. I. Binding of the tritiated α-neurotoxin in the absence of effector. Mol. Pharmacol. 10, 1–14. [PubMed] [Google Scholar]
- 42.Weiland G, Georgia B, Wee VT, Chignell CF, Taylor P, 1976. Ligand interactions with cholinergic receptor-enriched membranes from Torpedo: influence of agonist exposure on receptor properties. Mol. Pharmacol. 12, 1091–1105. [PubMed] [Google Scholar]
- 43.Wilson PT, Lentz TL, Hawrot E, 1985. Determination of the primary amino acid sequence specifying the α-bungarotoxin binding site on the α-subunit of the acetylcholine receptor from Torpedo californica. Proc. Natl. Acad. Sci. USA 82, 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]