Summary:
Nicotinic acetylcholine receptors are pentameric ion channels that mediate fast chemical neurotransmission. The α3β4 nicotinic receptor subtype forms the principal relay between the central and peripheral nervous systems in the autonomic ganglia. This receptor is also expressed focally in brain areas that affect reward circuits and addiction. Here we present structures of the α3β4 nicotinic receptor in lipidic and detergent environments, using functional reconstitution to define lipids appropriate for structural analysis. Structures of the receptor in complex with nicotine, as well as the α3β4-selective ligand AT-1001, complemented by molecular dynamics, suggest principles of agonist selectivity. The structures further reveal much of the architecture of the intracellular domain, where mutagenesis experiments and simulations define residues governing ion conductance.
Keywords: nicotinic receptor, cryo-EM, ligand-gated ion channel, lipids, ganglionic, addiction, nicotine
eTOC blurb
Gharpure et al. combine single-particle cryo-electron microscopy, electrophysiology, and molecular dynamics simulations to interrogate the structure and function of the α3β4 nicotinic acetylcholine receptor in a lipidic environment.
Introduction
The autonomic nervous system comprises sympathetic and parasympathetic pathways and facilitates all involuntary control of visceral organs. The sympathetic branch classically mediates “fight or flight” activity in response to acute stress, whereas the parasympathetic branch maintains peripheral homeostasis. Despite their antagonistic effects, the two branches share a generally conserved disynaptic architecture. Neurons in the autonomic ganglia that innervate downstream targets receive cholinergic input from the central nervous system (Wehrwein et al., 2016). The predominant neurotransmitter receptor in these neurons is the α3β4 nicotinic acetylcholine receptor, commonly referred to as the ganglionic nicotinic receptor (Conroy and Berg, 1995; Couturier et al., 1990; Skok, 2002).
While this receptor forms a critical relay in the autonomic nervous system, its expression is not limited to the periphery. The α3β4 receptor is also found in abundance in the habenulo-interpeduncular tract (Grady et al., 2009; Mulle et al., 1991; Quick et al., 1999), which modulates the mesolimbic dopamine system, the main reward pathway in the brain (McCallum et al., 2012; Nishikawa et al., 1986; Sutherland, 1982). Functional antagonists of α3β4, such as 18-methoxycoronaridine and AT-1001, have been shown to decrease self-administration of a wide range of drugs, including morphine, cocaine, alcohol, and nicotine in rodent models (Glick et al., 1996; Glick et al., 2000; Rezvani et al., 1997; Toll et al., 2012). Consequently, this subtype has received considerable interest as a potential target for anti-addiction therapeutics (Glick et al., 2002).
The nicotinic receptor family consists of many different subtypes, each with distinct physiological functions and biophysical properties (Albuquerque et al., 2009). Thus far, high-resolution information has been restricted to structures of the α4β2 subtype (Morales-Perez et al., 2016b; Walsh et al., 2018), limiting our understanding of the diversity in this family. Moreover, the α4β2 structures were determined in the presence of detergent and were missing much of the intracellular domain (ICD). Here we go beyond previous structural studies of nicotinic receptors by determining agonist-bound complexes of the human α3β4 nicotinic receptor reconstituted in a functionally supportive lipidic environment. This work reveals molecular mechanisms underlying ligand selectivity, as well as ion permeation through the ICD.
Results and Discussion
Biochemistry, structure determination, and receptor architecture
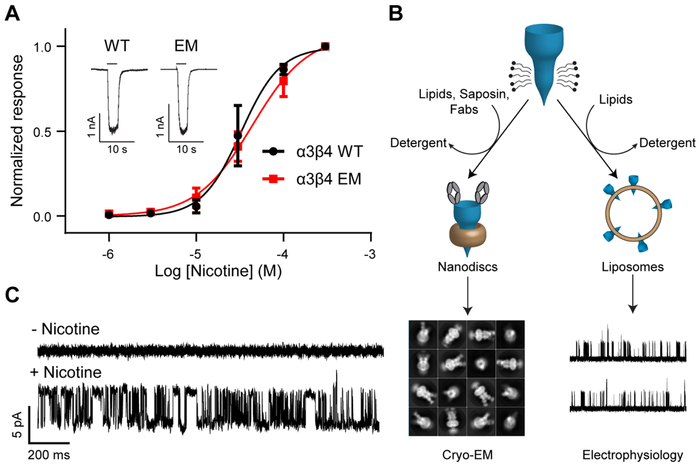
We optimized biochemical stability of the human α3 and β4 genes by replacing a portion of the intracellular M3-M4 loop that was predicted to be disordered with BRIL, a thermostable fusion partner (EM construct; see Methods). The EM construct retains function qualitatively equivalent to that of wild-type α3β4 (Fig. 1A). To better simulate the native environment of nicotinic receptors, we reconstituted receptors into lipid nanodiscs using saposin (Frauenfeld et al., 2016), soy lipids, and cholesteryl hemisuccinate (CHS), a soluble cholesterol derivative and functional proxy (Addona et al., 1998). This lipid mixture was selected for its ability to support channel activity, as demonstrated by proteoliposome patch-clamp experiments using receptor purified as for structural analysis (Fig. 1B-C). We raised monoclonal antibodies to the α3β4 receptor and purified receptor-Fab complexes to break the pseudo-symmetry of the heteromeric protein and facilitate particle alignment in cryo-EM studies (Walsh et al., 2018). The Fab had a small positive effect on ligand binding (Fig. S1C-F), but little to no effect on the functional response to nicotine (Fig. S1B).
Fig. 1. Construct modification and functional reconstitution.
(A) Whole-cell electrophysiology dose-response experiments comparing WT and EM constructs. WT EC50 = 32.7 μM (95% CI: 25.6-44.8 μM; n = 3). EM EC50 = 43.9 μM (95% CI: 35.7-57.7 μM; n = 5). Inset shows representative responses of WT and EM constructs to 30 μM nicotine. (B) Flowchart describing reconstitution for electrophysiology and cryo-EM sample preparation. (C) Proteoliposome patch-clamp recording shows single channel currents from a representative patch (n = 3) before and after application of nicotine to the bath solution.
See also Figure S1.
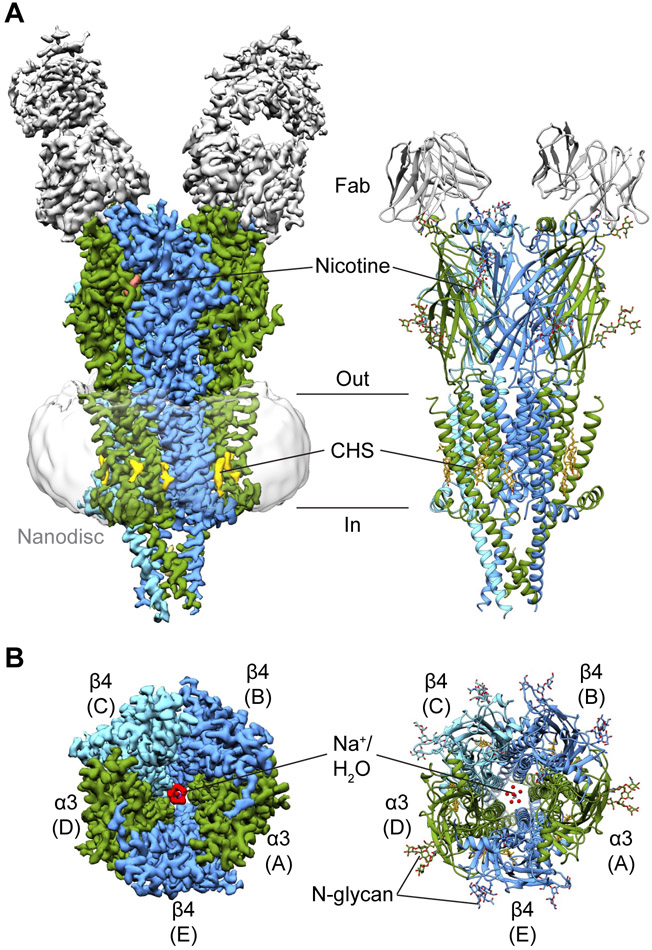
Single particle analysis of the receptor-Fab-nanodisc complex in the presence of nicotine revealed an assembly with two Fabs bound to the top of the receptor (Fig. 2A). The final reconstruction was resolved to approximately 3.3 Å (Table S1), allowing us to build a model for the receptor and the variable domains of the Fab fragments. We determined that the Fabs bind to α3 subunits, indicating a 2α:3β stoichiometry. Interestingly, the structural epitopes recognized by these Fabs are homologous to the main immunogenic region (MIR) of α1 subunits in muscle-type receptors, the target of the majority of autoantibodies that cause myasthenia gravis (Luo et al., 2009) (Fig. S1G-H). The α3β4 receptor itself resembles a cone with α3 (chains A and D) and β4 (chains B, C, and E) subunits arranged pseudo-symmetrically about the channel axis in an alternating manner (Fig. 2B). Each subunit adopts a fold characteristic of the Cys-loop receptor superfamily, which includes the cationic nicotinic acetylcholine and 5-HT3 receptors, as well as the anionic glycine and GABAA receptors (Nemecz et al., 2016). All subunits contain an extracellular N-terminal domain composed of a β-sandwich followed by four transmembrane helices (M1-M4) with M2 helices lining the ion-conducting pore. The ICD of cationic members of the superfamily is primarily composed of the membrane-associated (MA) helices, which form the tip of the cone. In addition to density for the receptor, we observed nicotine and water at the orthosteric binding sites, CHS at the periphery of the transmembrane domain, and waters and ions in the pore (Fig. 2). Density for BRIL was notably absent in all cryo-EM maps.
Fig. 2. Architecture of the α3β4 receptor.
(A) Side views of cryo-EM map and atomic model of α3β4-nicotine complex. α3 subunits are colored in green, β4 subunits in blue, Fabs in gray, nicotine in salmon, and CHS in yellow. (B) Top views of cryo-EM map and atomic model. Coloring is as indicated before. Water is in red, sodium is in purple.
See also Figures S3-S5 and Table S1.
In the presence of AT-1001, the sample suffered from profound aggregation in nanodiscs, evident in both gel filtration and in micrographs (Fig. S2A-D). This dataset produced a density map with an overall resolution of 4.6 Å (Fig. S3D, Table S1). When purified in detergent, the sample displayed markedly improved biochemical behavior and yielded a 3.9 Å map (Table S1). Local resolution in the orthosteric pocket approached 3.0 Å with clear density for sidechains and the ligand, allowing us to confidently model this region (Fig. S3B, S4B). Importantly, we were able to compare this structure to the lower-resolution lipid-reconstituted map and did not observe conformational differences that could be attributed to detergent artifacts (Fig. S2E-F). The general architecture of the AT-1001-bound structure is consistent with the nicotine complex (r.m.s.d. = 0.75 Å Fig. S5A).
Ligand binding and selectivity
The neurotransmitter-binding sites of heteromeric neuronal nicotinic acetylcholine receptors are found extracellularly at α-β interfaces, with the α subunit forming the principal face and the β subunit forming the complementary face. Each face contributes three loops to the binding site: loops A-C from the principal side and loops D-F from the complementary side. α3β4 and α4β2 receptor subtypes share high sequence identity at these binding sites, yet affinities for many classical nicotinic receptor agonists, such as nicotine, acetylcholine, and epibatidine are ~10-100 fold lower for α3β4 than for α4β2 (Eaton et al., 2003; Xiao et al., 1998). Previous studies investigating the low affinity of these ligands to other subtypes, such as the α7 and muscle-type nicotinic receptors, identified an intrasubunit hydrogen bond between loops B and C that conferred high affinity (Grutter et al., 2003; Xiu et al., 2009). However, the residues involved in this interaction are conserved between α4 and α3, leaving the cause of the differential affinities unclear.
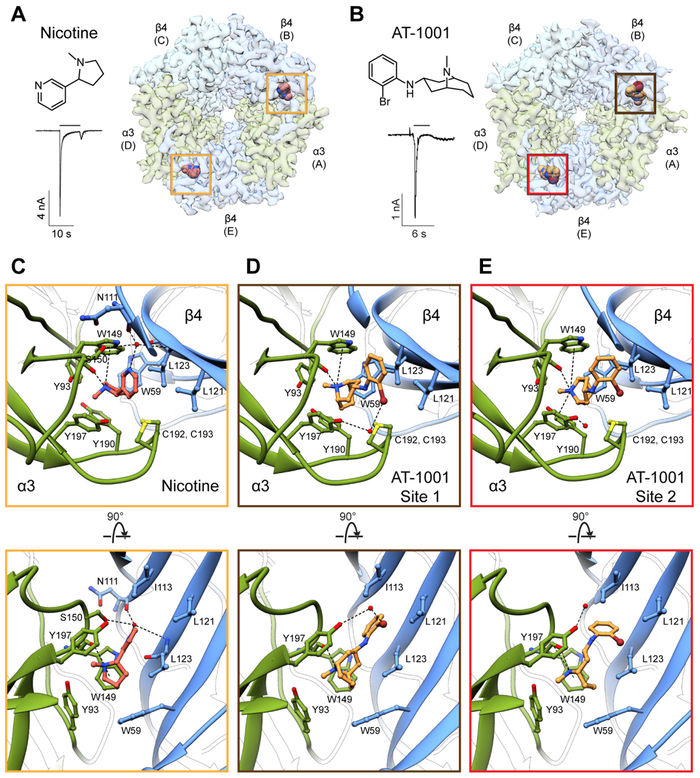
To gain insight into this question, we determined a structure of the α3β4 receptor bound to nicotine. We observed clear density corresponding to the ligand at both α-β interfaces: the A-B interface (subsequently referred to as site 1), and the D-E interface (site 2) (Fig. 3A, S6A, Video S1). The orientation of nicotine is equivalent at both sites and is analogous to that seen in structures of the α4β2 receptor and the acetylcholine binding protein (AChBP) (Celie et al., 2004; Morales-Perez et al., 2016b; Walsh et al., 2018). Nicotine is enveloped in a nest of highly conserved aromatic residues, namely Y93 from loop A, W149 from loop B, Y190 and Y197 from loop C, and W59 from loop D (Fig. 3C). The basic nitrogen in the pyrrolidine ring is oriented such that it can form a hydrogen bond with the backbone carbonyl of W149, as well as a cation-π interaction with the aromatic sidechain of this residue. As expected in the presence of an agonist, loop C is in a closed conformation, occluding the binding pocket from solvent and allowing the vicinal disulfide to form hydrophobic interactions with the ligand. Consistent with the structure of the AChBP-nicotine complex (Celie et al., 2004), we observed density for an ordered water bridging the pyridine nitrogen and the backbone carbonyl oxygen of N111, S150 sidechain, and L123 amide nitrogen (Fig. 3C, S6A). We further investigated the stability of these binding sites during triplicate 500-ns molecular dynamics simulations, during which both nicotine molecules remained within 2 Å of their initial positions (Fig. S6I).
Fig. 3. Ligand-binding sites.
(A) Structure of nicotine, whole-cell response to 1 mM nicotine (concentration used for EM sample preparation), and top view of the α3β4-nicotine complex. Yellow boxes indicate nicotine binding sites. (B) Structure of AT-1001, whole-cell response to 50 μM (concentration used for EM sample preparation), and top view of the α3β4-AT-1001 complex. The brown box indicates site 1 and the red box indicates site 2. (C) Top and side views of nicotine binding site. For clarity, loop C has been removed in the side view. Putative hydrogen bonding and cation–π interactions are represented as dashed lines. (D) Top and side view of binding site 1 for AT-1001. (E) Top and side views for binding site 2 for AT-1001.
Next, we sought to understand mechanisms underlying agonist selectivity by determining a structure of α3β4 in complex with AT-1001. AT-1001 is a partial agonist that acts functionally as an antagonist by causing rapid desensitization (Cippitelli et al., 2015) (Fig. 3B). This compound displays sub-nanomolar affinity to the human α3β4 receptor and is ~200-fold selective for this subtype over α4β2 (Tuan et al., 2015). Our structure revealed that AT-1001 binds at the orthosteric sites (Fig. 3B). Although density for the bromine atom was underrepresented, we were able to confidently model the ligand due to clear signal from the large bicyclic ring and the smaller halo-phenyl group (Fig. S6B-C). To our surprise, we found that AT-1001 unambiguously adopts different poses at the two binding sites (Videos S2-S3). At site 1, the halo-phenyl moiety is oriented roughly parallel to the long axis of the bicyclic ring (Fig. 3D). The bromine points outwards, away from the channel axis, coordinating a water in conjunction with the hydroxyl group of Y197, and the phenyl ring is slotted in between I113 and L121 on loop E. The positively-charged bridging nitrogen of the bicyclic ring (pKa 9.35) mimics the basic nitrogen of nicotine, forming a hydrogen bond and a cation-π interaction with W149. At site 2, AT-1001 assumes a distinctly kinked pose, with the halo-phenyl ring bent and rotated toward the complementary face (Fig. 3E). Here, the bromine positions in between L121 and L123 on loop E, disrupting the hydrogen bond with the water, which moves closer to Y197. This orientation of the top ring forces an outward rotation of the bicyclic ring, positioning the tertiary nitrogen to form a cation-π interaction with Y197 in addition to W149. At both sites, the aromatic nest remains intact, with constituent residues oriented similarly as in the nicotine structure, with loop C in the closed conformation (Fig. 3C-E). Similar to nicotine, AT-1001 remained bound within 2 Å of its initial position in simulations (Fig. S6J).
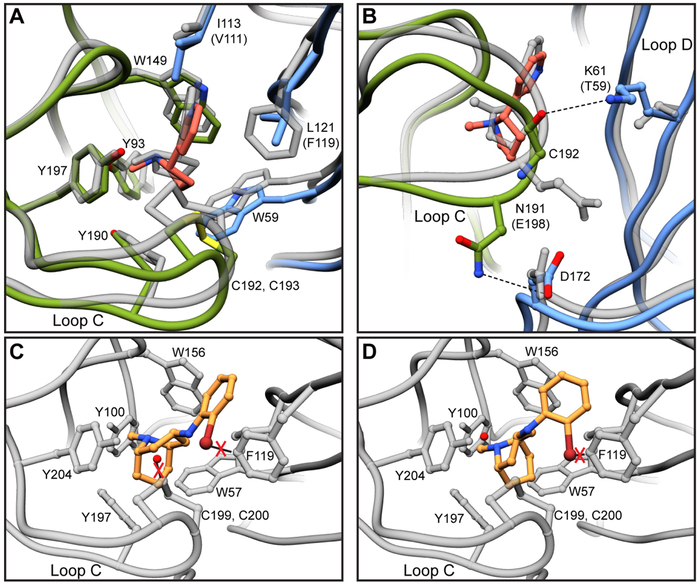
These studies allow us to address questions regarding ligand selectivity between nicotinic receptor subtypes. By comparing the nicotine-bound structure to previous structures of the α4β2 receptor (Morales-Perez et al., 2016b; Walsh et al., 2018), we sought to understand the relatively low affinity of non-selective agonists for the α3β4 receptor. Superposition of the α4β2 and α3β4 binding sites revealed that not only are the majority of residues contacting nicotine conserved, but they are also similarly oriented (Fig. 4A). The only two substitutions directly in the binding pocket are I113 (V111 in β2) and L121 (F119) on loop E. Of these two, the L121F substitution is of greater interest, as the aromatic sidechain of phenylalanine could engage in a π-π interaction with the pyridine of nicotine. We also noticed an outward displacement of ~2.1 Å in loop C when compared to α4β2 (Fig. 4A, S6G). These shifts result in a less compact binding pocket in α3β4 (Fig. S6E-F), potentially leading to a loss in affinity for nicotine by weakening Van der Waals contacts with surrounding residues. We found key differences between α3β4 and α4β2 that may account for the divergence in loop C positioning. The residue immediately prior to the vicinal cysteines on loop C is an asparagine in α3 and a glutamate in α4. Both residues appear poised to interact with a conserved aspartate on loop F. While N191 in α3 can hydrogen bond with the aspartate, pulling loop C downward, the negative charge of E198 in α4 is repelled away, potentially pushing loop C toward the ligand binding site (Fig. 4B). K61 on loop D is positioned to form a hydrogen bond with the backbone carbonyl of C192 in α3β4, propping open loop C. This hydrogen bond cannot form in α4β2, as the lysine is replaced by a threonine, whose sidechain is not long enough to interact with loop C (Fig. 4B). Next, we looked to explain the selectivity of AT-1001 for α3β4. We speculate that the more spacious binding pocket of α3β4 may better accommodate the larger ligand. Superpositions revealed potential clashes of the bromine on AT-1001 with F119 on β2 at both sites, and a clash of the water with loop C at site 1 (Fig. 4C-D). These structural comparisons provide initial insights into the different ligand sensitivities of these subtypes; however, a thorough mutational analysis will be required to draw definitive conclusions.
Fig. 4. Comparisons of α3β4 and α4β2 binding pockets.
(A) Overlay of α3β4 and α4β2 binding sites. α4β2 structure is shown in gray. Residue numbering is for α3β4 and substitutions between subtypes are indicated in parentheses. (B) Hydrogen bonds between α3 and β4 that may account for differential loop C structures and corresponding residues in α4β2. (C) Overlay of AT-1001 site 1 orientation in α4β2 binding site. Potential clashes are indicated with red X’s. (D) Overlay of AT-1001 site 2 orientation in α4β2 binding site.
See also Figures S5 and S6.
Permeation pathway
Recently, there has been considerable controversy surrounding structures of Cys-loop receptors in detergent micelles and whether or not they represent physiologically relevant conformations (Cerdan et al., 2018; Laverty et al., 2019). Decades of studies on the nicotinic receptor from the Torpedo ray electric organ highlight strong lipid dependence for function of this prototypical family member (Chak and Karlin, 1992; daCosta and Baenziger, 2013). Membranes lacking essential lipidic components may stabilize an uncoupled receptor conformation that is unable to undergo agonist-induced conformational changes (daCosta and Baenziger, 2009). Previous structures of the α4β2 receptor in complex with nicotine were characterized to be in non-conducting desensitized states but were determined in the absence of phospholipids. In this study, we determined the structure of a nicotine-bound receptor reconstituted into a bilayer that we show supports channel function. We also solved a structure of the same protein bound to AT-1001 both in the presence of dodecylmaltoside (DDM) and in lipid nanodiscs, granting us the opportunity to directly compare the pore conformations of lipid-reconstituted and detergent-solubilized receptors. We found that the pore architectures for the two complexes are nearly identical and are congruent with previous structures of the α4β2 receptor (Morales-Perez et al., 2016b; Walsh et al., 2018) (Fig. 5A-B). Because all of these structures were determined in the presence of desensitizing agonists, we suggest that this shared pore architecture represents a desensitized state.
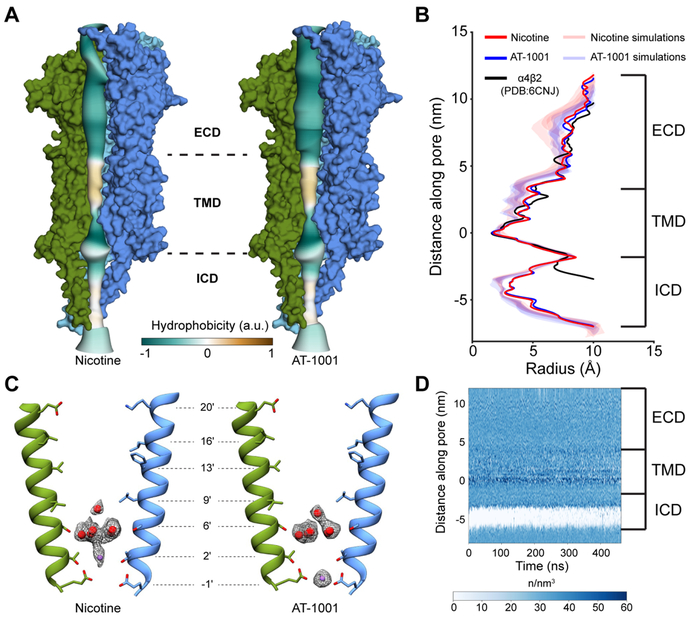
Fig. 5. Channel axis and permeation.
(A) Radius profiles of nicotine and AT-1001 structures colored by hydrophobicity. For clarity, chains A and E are not shown. (B) Pore radius profiles comparing nicotine, AT-1001, and α4β2 structures, and simulations in the presence of nicotine or AT-1001. (C) Positions of ordered waters and ions in the pore and corresponding density. Chains D and B are shown. (D) Water density profile along channel axis. In (B) and (D), zero point on y-axis corresponds to Glu-1′.
See also Figure S7.
The transmembrane portion of the α3β4 channel resembles a funnel that narrows as it approaches the cytoplasm, consistent with structural and functional evidence that the desensitization gate lies toward the intracellular end (Gielen and Corringer, 2018). The pore ultimately reaches a constriction point formed by the sidechains of Glu-1' with a radius of ~1.7 Å (Fig. 5A-C). Density for acidic sidechains is typically underrepresented due to inherent flexibility, radiation damage, and negative atomic scattering factors at low resolutions, but high-pass filtered maps have been used to resolve such sidechains (Yonekura et al., 2015). Using a 5 Å high-pass filtered map from the nicotine dataset, we observed clear density for these key residues and were able to confirm their orientation (Fig. 6A, S7A-B). The pore radius at the hydrophobic girdle, corresponding to the 9' and 13' positions, is ~3.8 Å, large enough to pass a hydrated sodium ion. Simulations of the Torpedo receptor have led to a proposed hydrophobic gating mechanism, in which the pore is not fully occluded (~3 Å radius) but a steep energetic barrier due to desolvation prevents ion flux in the resting state (Beckstein and Sansom, 2006; Ivanov et al., 2007). Throughout molecular dynamics simulations of the α3β4 channel, the pore radius profile remained stable, with a constriction point at the −1' position (Fig. 5B, S7F); however, the entire transmembrane region of the pore remained solvated (Fig. 5D). This result suggests that the observed pore conformation for α3β4 is inconsistent with consensus models of a resting state, further supporting our conclusion that these structures represent a desensitized state.
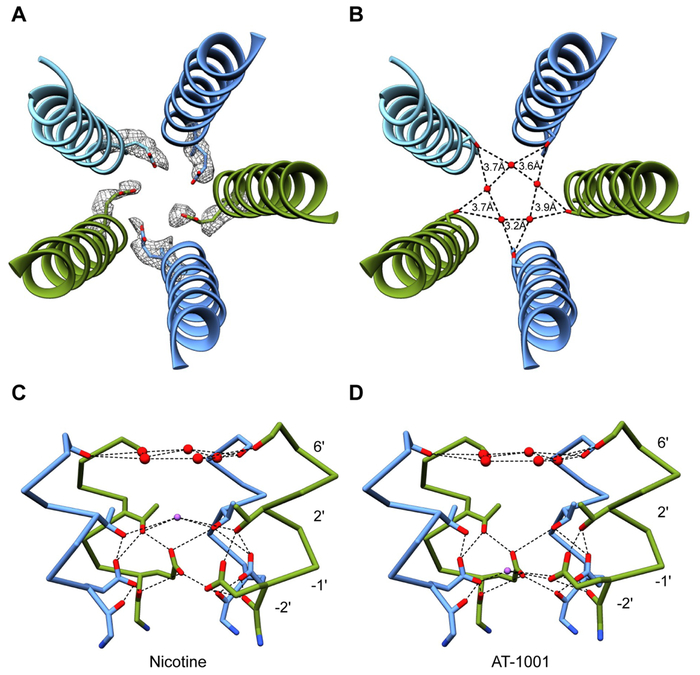
Fig. 6. Pore features.
(A) Density for Glu-1' from 5 A high-pass filtered map. (B) Coordination of pentagonal water ring from Ser6'. (C) Hydrogen bonding network showing interactions between pentagonal water ring, sodium ion, and surrounding residues in nicotine structure. Glu-1' is stabilized by Thr2' and Gly-2'. Chain C has been removed for clarity. Mean distances for dashed lines-Ser6' hydroxyl/water: 4.3 Å, Na+/Thr2' hydroxyl: 4.4 Å, Thr2' hydroxyl/Glu-1' carboxyl: 3.6 Å, Glu-1' carboxyl/Gly-2' carbonyl: 3.0 Å. (D) Same as in panel C, but for AT-1001 structure. Sodium ion may help stabilize Glu-1' in this position. Mean distances for dashed lines- Ser6' hydroxyl/water: 4.4 Å, Thr2' hydroxyl/Glu-1' carboxyl: 3.5 Å, Na+/Glu-1' carboxyl: 3.8 Å, Glu-1' carboxyl-Gly-2' carbonyl: 3.1 Å.
See also Figure S7.
Within the pore, we observed density for a pentagonal ring of waters coplanar with the hydroxyl group of Ser6' (Fig. 5C, 6B-D). Similar water pentagons have been found at this position in higher-resolution structures of GLIC (Sauguet et al., 2013), and the polar nature of 6' residues is essential for channel conductance (Imoto et al., 1991). The pore diameter in our structures is wider in this region than in GLIC, and consequently we find that the water molecules form a looser ring (mean distance of 3.6 Å vs 2.8 Å (Sauguet et al., 2013)) (Fig. 6B). We also observed densities along the pore axis above and below the pentagon. Without very high-resolution structural information or anomalous X-ray crystallographic data, the assignments for these densities remain speculative. However, molecular dynamics simulations suggested water occupies the pore near Ser6' more often than sodium ions (Fig. S7C-D); thus, we modeled the upper axial density as a water. The more cytosolic axial density differs in height between the nicotine and AT-1001 complexes, but in both cases was modeled as sodium due to similarly identified cation binding sites in GLIC (Hilf et al., 2010; Sauguet et al., 2013) and proximity to Glu-1' sidechains. The sodium ions potentially contribute to the stabilization of Glu-1' sidechains in the observed “up” (mm) rotamer (Fig. 6C-D). During 500 ns simulations, these sidechains predominantly remained in this conformation and sodium ions preferentially occupied nearby positions in the pore (Fig. S7C-E). We thus speculate that sodium ions at these positions occupy structurally significant cation binding sites that enable the negatively charged carboxyl groups of Glu-1' to orient inward toward the channel pore and form the desensitization gate.
Structure and function of the intracellular domain
The ICD of nicotinic receptors plays crucial roles in cellular trafficking, gating kinetics, and channel conductance (Bouzat et al., 1994; Hales et al., 2006; St John, 2009). This domain consists of a short post-M3 loop, an amphipathic MX helix that lies parallel to the plane of the membrane, a poorly conserved and disordered cytoplasmic loop, and the MA helix. Previous structures of the α4β2 receptor utilized aggressive deletions in this region to promote crystallization and biochemical stability. Thus, comprehensive structural information on nicotinic ICDs has been limited to homologous domains of homomeric 5-HT3A receptors (Basak et al., 2018a; Hassaine et al., 2014; Polovinkin et al., 2018) and lower-resolution reconstructions from the Torpedo receptor (Unwin, 2005). By retaining all ordered components of this domain in our EM construct, we were able to resolve a heteromeric assembly of the ICD and explore its role in ion permeation.
Most notably, this study allowed us to examine the structure and function of the MA helices. These helices are continuous extensions of M4, protruding ~40 Å into the cytosol and forming a conical intracellular vestibule at the bottom tip of the receptor. The helices diverge as they approach the membrane, forming five distinct portals that are predominantly lined by polar and acidic residues. Toward the constriction point at the bottom of the vestibule, we observed a strong oblong density along the central axis of the receptor, lined by several rings of conserved hydrophobic residues (Fig. 7A-B). We were curious about the identity of this “hydrophobic plug,” and initially suspected it to be CHS. To test this hypothesis, we collected an EM dataset from a sample that did not include CHS during the preparation (Fig. S3C, Table S1). Density for the plug was still present in the reconstruction (Fig. S8A-B), while densities we attribute to CHS along the periphery of the transmembrane domain disappeared. Furthermore, MD simulations suggested a lipid molecule is needed at this site to promote stability of the ICD (Fig. S8J). We thus speculate that the density represents the tail of a detergent molecule or a lipid and we modeled this feature as a lipid hydrocarbon tail. This site may correspond to a promiscuous hydrophobic site also occupied physiologically. Noteworthy is that a subset of density maps from recent 5-HT3A receptor structures contain unmodeled features in this region or close by (Basak et al., 2018a; Polovinkin et al., 2018). The presence of this hydrophobic plug affirms the current consensus that ions permeate through the lateral portals of the intracellular vestibule (Basak et al., 2018a; Miyazawa et al., 1999). Even without a direct physical obstruction, the narrow radius (~2.7 Å) and highly hydrophobic nature of the central axis of the ICD would likely prevent ion flux through this region. Simulations support this concept, showing continuous hydration pathways through the portals (Fig. 7C).
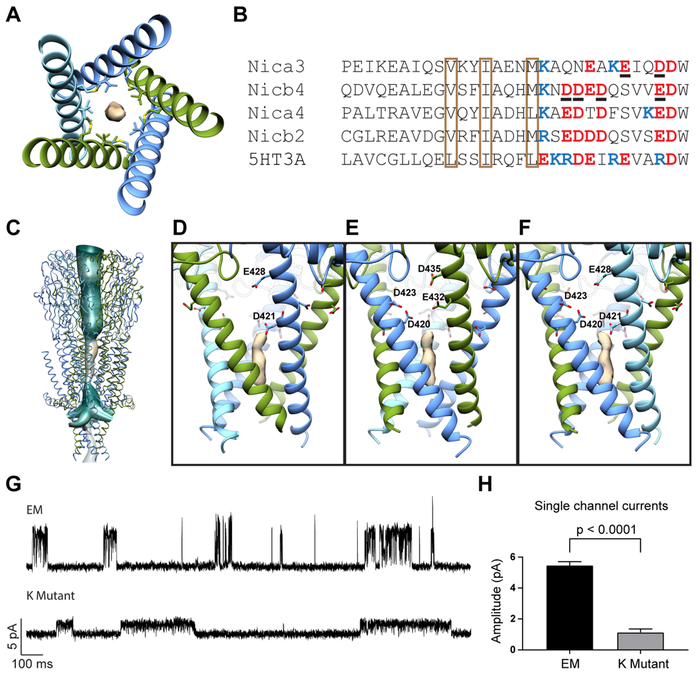
Fig. 7. Intracellular domain.
(A) Top view of ICD showing density of hydrophobic plug. Surrounding residues are shown as sticks. (B) Sequence alignment of ICD. Conserved hydrophobic residues are in brown boxes. Negatively-charged and positively-charged residues near portals are indicated in red and blue respectively. Residues mutated to lysines for the K mutant are underlined. (C) Continuous hydration pathway through the channel pore and five intracellular portals. (D) Side view of α-β portal. Mutated residues are shown as sticks. (E) Side view of β-α portal. (F) Side view of β-β portal. (G) Representative single channel recordings from EM construct and K mutant. (H) Comparison of single channel current amplitudes at 100 mV from EM construct and K mutant (n = 3 for each).
See also Figure S8.
To verify the importance of the portals in ion permeation, we explored the contribution of portal-lining residues to channel conductance. Guided by the structure, we identified negatively charged residues whose sidechains project into the lateral windows. We found two residues in α3, E432 and D435, and four in β4, D420, D421, D423, and E428 (Fig. 7D-F). We flipped the charges of these amino acids by mutating them to lysines and made single-channel recordings from cells transfected with the mutated subunits. The “K mutant” exhibited significantly attenuated single-channel responses in comparison with the EM construct (Fig. 7G-H). This work expands on seminal studies of 5-HT3A homomers that identified three regularly spaced arginines that underlie this channel’s remarkably low conductance (Kelley et al., 2003). These studies first highlighted the functional importance of charged residues in MA helices. Here, we used direct structural information to decrease the electronegativity in the intracellular portals and showed that this causes a substantial reduction in channel conductance.
Finally, we investigated the positioning of the post-M3 loop and MA helix. Structures of the 5-HT3A receptor suggest that channel gating involves conformational changes in these intracellular substructures (Basak et al., 2018a; Basak et al., 2018b). A putative open 5-HT3A structure reveals an upward translation of MX, pulling the post-M3 loop away from the lateral portals (Fig. S8C). This movement, along with a kinking of the MA-M4 helix, widens the proposed exit pathway for ions. In the α3β4 receptor, this region more closely resembles 5-HT3A in the closed state. The MX helix lies along the membrane-cytoplasm interface and the post-M3 loop extends even further down into the lateral window (Fig. S8C). The position of the post-M3 loop may be stabilized by a hydrogen bond between the hydroxyl group of T303 in α3 or S304 in β4 with the sidechain of an acidic residue on the neighboring MA helix (Fig. S8D-F). However, this sunken orientation of the loop does not appear to fully obstruct ion permeation. Fenestrations of ~6 Å are present in portals at α-β and β-β interfaces (Fig. S8G, S8I). K431 of α3 partially occludes this opening at α-β interfaces (Fig. S8H), although flexibility of this and surrounding sidechains may still permit ion passage. Further structural studies elucidating alternate conformational states of nicotinic receptors will provide additional insight into the role of this region in channel gating.
Conclusion
Here we present the first structures of the α3β4 ganglionic nicotinic acetylcholine receptor. A foundation of this work is the lipid reconstitution and electrophysiology experiments, which allowed us to obtain high resolution structural information in an environment we demonstrate supports channel function; a first in the Cys-loop receptor family for recombinantly-produced protein. We analyzed structures of the receptor in complex with the non-selective agonist nicotine and the selective partial agonist AT-1001 to suggest that selectivity and overall lower agonist affinity at the ganglionic receptor stem from a less compact neurotransmitter site. These structures further reveal the architecture of the ICD and support conclusions from homologous protein structures that ions permeate through side portals and not through the hydrophobic axial pathway. Computational analyses complement all aspects of the structure-function study to examine pore hydration, ion permeation, and ligand site stability, building from the rigid structural snapshots toward a dynamic understanding of receptor mechanisms.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources including α3 and β4 subunit expression constructs should be directed to and will be fulfilled by the Lead Contact, Ryan E. Hibbs (ryan.hibbs@utsouthwestern.edu).
Experimental Model and Subject Details
Cultured Cells
HEK293S GnTI− cells were used for protein production and electrophysiology experiments. Cells were obtained from ATCC and were not further authenticated. Adherent cells were grown at 37°C with 5% CO2 in DMEM medium (Corning) supplemented with 10% fetal bovine serum (EMD Millipore). Suspension cells were grown at 37°C with 8% CO 2 in FreeStyle 293 expression medium (Gibco) supplemented with 2% fetal bovine serum (EMD Millipore).
Sf9 cells were used to produce baculovirus. Cells were obtained from ATCC and were not further authenticated. Cells were grown at 27°3 in SF900-III SFM expression medium (Gibco).
Method Details
Construct design
Human α3 (UniProtKB P32297) and β4 (P30926) genes were codon optimized, synthesized, and cloned into the pEZT-BM expression vector (Morales-Perez et al., 2016a). Enhanced green fluorescent protein (EGFP) was inserted into the M3-M4 loop of both subunits and a Strep-tag was placed at the C-terminus of the β4 subunit. Constructs were initially screened via co-transfection of HEK293S GnTI− cells (ATCC CRL-3022) with combinations of EGFP-tagged and untagged subunits via Lipofectamine 2000 (Invitrogen). Cells were pelleted, solubilized with 20 mM Tris, pH 7.4, 150 mM NaCl (TBS buffer), 40 mM n-dodecyl-β-D-maltopyranoside (DDM; Anatrace), and 1 mM phenylmethanesulfonyl fluoride (PMSF; Sigma-Aldrich), and analyzed by fluorescence-detection size-exclusion chromatography (FSEC) (Kawate and Gouaux, 2006). Viable candidates without EGFP were then co-transfected in GnTI− cells in small-scale purification experiments (1-2 mL culture scale). Cells were pelleted, solubilized as before, and allowed to bind to high-capacity Strep-Tactin (IBA) affinity resin. Resin was washed with TBS containing 1 mM DDM, and protein was eluted with the same buffer supplemented with 5 mM desthiobiotin (Sigma-Aldrich) before being evaluated by FSEC monitoring tryptophan fluorescence. Screens revealed that robust expression and stable pentamer formation required deletions in the M3-M4 loop and were aided by the inclusion of soluble fusion partners in this region. Thus, in the final EM constructs, residues Asn348-Ser402 in α3 and Pro341-Ser395 in β4 were replaced with apocytochrome b(562)RIL (BRIL), a thermostabilized four-helical bundle that has been used to promote crystallization in G protein-coupled receptors (Chun et al., 2012).
Receptor expression and purification
Bacmam viruses for each subunit were produced as described for the α4β2 receptor (Morales-Perez et al., 2016a). Briefly, constructs in the pEZT-BM vector were transformed in DH10-Bac cells to produce recombinant bacmid. 2 mL of Sf9 cells (ATCC CRL-1711) were transfected with purified bacmid DNA to generate “P1” virus. 500 μL of this virus was then added to 1 L of Sf9 cells at a cell density of 1 × 106 cells/ml to produce a second generation of amplified virus (“P2”). Suspension cultures of GnTI− cells were grown at 37°C, 8% CO 2 and were transduced with α3 and β4 P2 viruses at a cell density of 4 × 106 cells/ml. At the time of transduction, 1 mM sodium butyrate (Sigma-Aldrich) was added to the culture and temperature was dropped to 30°C to boost protein expression. Af ter 72 hours, cells were harvested by centrifugation, resuspended in TBS and 1 mM PMSF, and disrupted using an Avestin Emulsiflex. Lysed cells were centrifuged for 20 minutes at 10,000 g, and the resulting supernatants were centrifuged for 2 hours at 186,000 g to isolate membranes. Membrane pellets were mechanically homogenized and solubilized for 1 hour at 4°C with 40 mM DDM in TBS. Solubilized membranes were centrifuged for 40 minutes at 186,000 g then passed over high capacity Strep-Tactin resin. The resin was washed with TBS, 1 mM DDM, 0.2 mM cholesteryl hemisuccinate (CHS, Anatrace), and 1 mM TCEP (Thermo Fisher Scientific), and protein was eluted in the same buffer containing 5 mM desthiobiotin. For the nicotine-bound structure, 1 mM nicotine (Sigma) was included during affinity purification and for the AT-1001-bound structure, 10 μM AT-1001 (generous gift from Astrea Therapeutics) was included.
α3β4 receptors have been proposed to assemble in multiple subunit stoichiometries: (α3)2(β4)3 and (α3)3(β4)2 (Covernton et al., 1994; Grishin et al., 2010; Krashia et al., 2010). To bias expression towards one stoichiometry, a fluorescence-based assay was used, as described previously for the α4β2 nicotinic receptor (Morales-Perez et al., 2016a). Briefly, bacmam viruses were made and titered for α3-mCherry and β4-EGFP constructs. Viruses were used to transduce one liter of GnTI- cells. Protein was purified as described above, and molar concentrations of each subunit were calculated by measuring absorbance at the maxima for the two fluorophores (GFP, 488 nm; mCherry, 587 nm) and dividing by their respective extinction coefficients (GFP, 56,000 M−1 cm−1; mCherry, 72,000 M−1 cm−1). Protein was analyzed by FSEC measuring GFP and mCherry fluorescence, allowing scale factors to be calculated relating known molar concentrations to fluorescence signal. This medium-scale purification was followed by small-scale experiments where different viral ratios were used to transduce 1 mL of GnTI-cells. Cells were solubilized and analyzed by FSEC, and previously calculated scale factors were used to determine molar ratios of α3 and β4 subunits. A 1:1 ratio of α3 and β4 viruses was found to produce a homogenous population of pentamers containing two α3 and three β4 subunits, and thus this ratio of viruses was used to produce protein for structural studies, reconstitutions for electrophysiology, and for binding assays.
Saposin nanodisc reconstitution
The Saposin A expression plasmid was provided by Salipro Biotech AB. Reconstitution of α3β4 into saposin nanoparticles was modified from Nguyen et al. (Nguyen et al., 2018) The reaction contained a 1:20:100 molar ratio of α3β4: saposin: soy polar lipid extract (Avanti). Lipids and saposin were mixed in TBS and 14 mM DDM and allowed to rotate at 4°C for 1 hour. Affinity-purified α3β4 was concentrated to ~20 μM, added to the saposin/lipid mixture, and rotated for 30 min at 4°C. 200 mg/mL Bio-Beads SM-2 (BioRad) were added to the mixture and rotation was continued overnight. The following morning, Bio-Beads were removed and replaced with 150 mg/mL fresh Bio-Beads for 2 hours.
Generation of monoclonal antibodies and Fab fragments
The 4G9 monoclonal antibody (mAb) (IgG2b, κ) was raised using standard methods following immunization of mice with α3β4 in detergent (Monoclonal Core, Vaccine and Gene Therapy Institute, Oregon Health & Science University). High affinity and specificity of the antibody for properly folded receptor was assayed by FSEC with EGFP-tagged receptor (shift in elution volume) and western blot (no binding). Fab fragments were generated by papain cleavage of whole antibody at a final concentration of 0.5 mg/ml for 2 hours at 37°C in 50 mM NaPO4, pH 7.0, 1 mM EDTA, 10 mM cysteine and 1:10 (w/w) papain. Digestion was quenched using 30 mM iodoacetamide at 25°C for 30 min. Fab w as purified by anion exchange using a HiTrap Q HP (GE Healthcare) column in 10 mM Tris, pH 8.0 and a NaCl gradient elution. Cloning and sequencing of Fab antibody regions were performed from mouse hybridoma cells.
Cryo-EM sample preparation and data collection
Affinity-purified α3β4 receptors reconstituted in nanodiscs were mixed with 4G9 Fab in a 1:1 (w/w) ratio and injected over a Superose 6 Increase 10/300 GL column (GE Healthcare) equilibrated in TBS, 1 mM TCEP, and ligand (1 mM nicotine or 50 μM AT-1001). Receptors purified in detergent followed the same protocol, but the buffer included 1 mM DDM and 0.2 mM CHS. Peak fractions were evaluated by analytical SEC, monitoring tryptophan fluorescence, and concentrated to an A280 of ~6. Samples in nanodiscs were supplemented with 1 mM Fos-Choline-8, fluorinated (Anatrace) immediately prior to freezing to induce random orientations in the grid holes. Protein sample (3 μL) was applied to glow-discharged gold R1.2/1.3 300 mesh holey carbon grids (Quantifoil) and immediately blotted for 4 s at 100% humidity and 4°C before being plunge-frozen into liquid ethane cooled by liquid nitrogen using a Vitrobot Mark IV (FEI).
Cryo-EM data were collected on a 300 kV Titan Krios microscope (FEI) equipped with a K2 Summit direct electron detector (Gatan) and a GIF quantum energy filter (20 eV) (Gatan) using EPU (FEI) and a 200 kV Talos Arctica (FEI) equipped with a K3 direct electron detector (Gatan) using Serial EM (Mastronarde, 2005). Sample-specific details are included in Table S1.
Cryo-EM data processing
All datasets were processed using the same general workflow in RELION 3.0 (Zivanov et al., 2018). Dose-fractionated images were gain normalized, 2 x Fourier binned, aligned, dose-weighted, and summed with MotionCor2 (Zheng et al., 2017). Contrast transfer function correction and defocus value estimation were done with GCTF (Zhang, 2016). Several hundred particles were manually picked and subjected to 2D classification to generate templates for auto-picking. Auto-picked particles were subjected to 2D classification to remove false positives. Ab initio models were generated in RELION and used for 3D classification. 3D classes with strong ICD density were selected for 3D refinement. An initial round of 3D refinement using the best 3D class as an initial model (low-pass filtered to 60 A) was followed by a second round with finer angular sampling using the map from the first refinement low-pass filtered to 10 Å as the initial model. Next, per-particle CTF refinement and beam tilt estimation were performed before another round of 3D classification with no image alignment/angular searches. Particles from the best classes were selected, polished, and used for 3D refinement to generate the final maps. In the AT-1001 detergent dataset, particles with defocus values greater than −3.0 μm were removed from the final reconstruction to improve resolution. Local resolution was estimated with ResMap (Kucukelbir et al., 2014).
Model building, refinement, and validation
A homology model for the α3β4 receptor was generated from the cryo-EM structure of the 2α:3β assembly of the α4β2 nicotinic acetylcholine receptor (PDB ID:6CNJ) (Walsh et al., 2018) via Swiss-Model (Schwede et al., 2003). A homology model for the Fab fragment was made using PDB entry 4WFE (Brohawn et al., 2014) for the light chain and 3MXV (Maun et al., 2010) for the heavy chain. The receptor and one copy of the Fab were docked into the density map using UCSF-Chimera (Pettersen et al., 2004). Manual adjustments of the models were then done in Coot (Emsley et al., 2010). The ECD and TMD of each individual subunit, as well as the variable domain of the Fab fragment were rigid body fitted into the density map. The density in the region of the constant domain of the Fab was not sufficiently ordered to allow accurate building of an atomic model; therefore, only the variable domain was included in the final model. Once the variable domain was rebuilt into the density, it was copied into the second site and manually adjusted. MA helices, which were not included in the α4β2 structure, were built de novo, and M4 helices, which are continuous with MA, were rebuilt into the map. Well-ordered N-linked glycans were built along the surface of the ECD. In many cases, density for these glycans was clearer in a 6 Å low-pass filtered map (Fig. S4C-D), and this map was used to assist building. Likewise, the water molecule in the ligand-binding pocket was apparent in the full map but showed stronger density in a 5 Å high-pass filtered map, which was used for accurate placement of this water. A portion of the M3-M4 loop (including the bril fusion protein) in both subunits was unresolved and the following residues were not modeled: 328-409 in α3 and 329-399 in β4. Additionally, residues 1-3 were not modeled in chain C (β4) due to weak density. After manual building in Coot, global real space coordinate and B-factor refinement were performed in Phenix (Adams et al., 2010).
Sequences used in alignments were retrieved from the UnitProtKB database (UniProt Consortium, 2018). Sequence alignments were made using PROMALS3D (Pei et al., 2008). Pore radius profiles and hydrophobicity plots were made using CHAP (Klesse et al., 2019). Structural figures were made using UCSF-Chimera and PyMOL (Schrodinger, LLC). Structural biology software packages were compiled by SBGrid (Morin et al., 2013).
Electrophysiology
Whole cell voltage-clamp recordings were made from cells transiently transfected with the constructs used in structural analysis. For the patch-clamp experiments, adherent HEK293S GnTI- cells were transiently transfected with pEZT-based plasmids 2-3 days before recording. Each 35 mm dish of cells was transfected with the DNA of α3 and β4 subunits in a 1:1 ratio. Upon transfection, cells were moved to 30°C. On the day of recording, cells were washed with bath solution, which contained (in mM): 140 NaCl, 2.4 KCl, 4 MgCl2, 4 CaCl2, 10 HEPES pH 7.3, and 10 glucose. Borosilicate pipettes were pulled and polished to a resistance of 2-4 MΩ. The pipette solution contained (in mM): 150 CsCl, 10 NaCl, 10 EGTA, and 20 HEPES pH 7.3. Cells were clamped at −75 mV. The recordings were made with an Axopatch 200B amplifier, sampled at 5 kHz, and low-pass filtered at 2 kHz using a Digidata 1440A (Molecular Devices) and analyzed with pClamp 10 software (Molecular Devices). The nicotine and AT-1001 solutions were prepared in bath solution from concentrated stocks. A stock solution of 1 M nicotine was prepared in water and the stock solution of 100 mM AT-1001 was prepared in DMSO. Solution exchange was achieved using a gravity driven RSC-200 rapid solution changer (Bio-Logic).
Cell-attached single channel recordings were made from cells 1-2 days post-transient transfection, following the same procedure for transfection as above. On the day of recording, cells were washed with bath solution containing (in mM): 142 KCl, 5.4 NaCl, 1.8 CaCl2, 1.7 MgCl2, and 10 HEPES pH 7.4 (adjusted with KOH) (Mukhtasimova et al., 2016). Borosilicate pipettes were pulled and polished to initial resistances of 8-12 MΩ. The pipette solution contained (in mM): 80 KF, 20 KCl, 40 potassium aspartate, 2 MgCl2, 1 EGTA, 10 HEPES 7.4 (adjusted with KOH), and 0.05 nicotine (Mukhtasimova et al., 2016). Currents were recorded at 100 mV, sampled at 50 kHz, and filtered at 10 kHz.
For proteoliposome patch-clamp experiments, receptors were first affinity-purified in DDM/CHS, as described above. Soy polar lipids in chloroform (Avanti) were dried in a test tube under a stream of argon while rotating the tube to make a homogeneous lipid film. The lipid film was further dried under vacuum for 2 hours and resuspended to 10 mg/mL with TBS. To make uniform lipid vesicles, the lipid resuspension solution was sonicated for 15 min. Purified receptors (4 μg) were added into lipid vesicles in a protein to lipid mass ratio of 1:500 (w/w). The mixture was rotated at room temperature for 1 hour to allow the protein to incorporate into lipid vesicles. Detergent was removed by incubating with Bio-Beads SM-2 and the resultant liposomes were collected by ultracentrifugation, 4°C, 30 min at 186,000 g. The pellet was resuspended in 6 μL TBS buffer. 2 μL of the suspension was spotted on a glass coverslip, and then desiccated overnight under vacuum at 4°C. Desi ccated liposomes were rehydrated with 5 μL of buffer (320 mM sucrose, 10 mM KCl, and 2 mM MgCl2, 5 mM Hepes pH 8.0) for at least 2 hours at 4°C, and then used for patch-clamp recordi ng. Channel activity of α3β4 was examined in excised liposome patches. Data were acquired at 70 mV at a sampling rate of 50 kHz with a 10 kHz filter. The bath solution contained (in mM): 200 KCl, 40 MgCl2, 2 CaCl2, and 5 HEPES pH 7.3. Pipettes were filled with the same bath solution and initial pipette resistances ranged from 4-8 MΩ. After a stable baseline was observed, 10 mM nicotine in bath solution was added to the bath to achieve a final concentration of ~0.5 mM.
Radioligand binding
Experiments to measure binding of [3H]-epibatidine (PerkinElmer) to the α3β4 receptor were performed with protein purified in TBS with 1 mM DDM, 0.2 mM CHS, and 1 mM TCEP in the absence of agonists. The concentration of binding sites was 0.3 nM. For the binding experiments in the presence of Fab, Fab was added in large excess (1 μM). In addition to the receptor, the binding assay conditions included 20 mM Tris pH 7.4, 150 mM NaCl, 1 mM DDM, and 1 mg/mL streptavidin-YiSi scintillation proximity assay beads (SPA; GE Healthcare Life Sciences). Non-specific signal was determined in the presence of 1 mM [1H]-nicotine. All data shown are from background-subtracted measurements. For radioligand competition experiments, binding site concentration was also 0.3 nM and the concentration of [3H]-epibatidine was 1 nM.
Molecular dynamics simulations
Deposited coordinates for the cryo-electron microscopy structure of the α3β4 receptor bound to nicotine in nanodiscs were used as a starting model for molecular dynamics simulations. Nicotine and AT-1001 parameters were generated using STaGE (Lundborg and Lindahl, 2015), and virtual sites were added. Nicotine, CHS, and ions and water resolved in the channel pore were placed as in the deposited structure. To resolve instabilities observed in the partially resolved intracellular domain, 1-palmitoyl 2-oleoyl phosphatidylcholine (POPC) was docked using AutoDock Vina (Trott and Olson, 2010) in a 37.5 Å × 36 Å × 39.75 Å box surrounding the MA helical bundle. For simulations with AT-1001, the partial agonist was substituted for nicotine at both binding sites. The Amber99sb-ildn force field (Lindorff-Larsen et al., 2010) was used to describe each protein, which was embedded in a bilayer of 300 POPC molecules modeled with Slipids-extended force field parameters (Jämbeck and Lyubartsev, 2012, 2013). Each system was solvated in a cubic box using CHARMM-GUI (Jo et al., 2008; Wu et al., 2014) and the TIP3P water model (Jorgensen et al., 1983), and NaCl was added to bring the system to neutral charge and an ionic strength of 0.15 M.
All simulations were performed with GROMACS 2018 (Abraham et al., 2015). Each system was energy-minimized with a velocity rescaling thermostat (Bussi et al., 2007) set to 300 K, then equilibrated for 50 ps, both with a constant number of particles, volume, and temperature. Virtual interaction sites were used for hydrogens to enable 5-fs time steps. Each was then equilibrated with a constant number of particles, pressure, and temperature for at least 60 ns, during which the position restraints on the protein were gradually released. Agonists, CHS, and resolved ions and water in the channel pore were restrained until the final 15 ns of equilibration. For each equilibrated system containing POPC in the intracellular domain, three replicates of 500-ns unrestrained simulations were generated. An additional 200-ns unrestrained simulation was performed for the nicotine-bound model in the absence of intracellular POPC. Parrinello-Rahman pressure coupling (Parrinello and Rahman, 1980) ensured constant pressure, the particle mesh Ewald algorithm (Essmann et al., 1995) was used for long-range electrostatic interactions, and bond lengths were constrained using the LINCS algorithm (Hess, 2008). Analyses were performed using VMD (Humphrey et al., 1996), CHAP, and MDTraj (McGibbon et al., 2015).
Quantification and Statistical Analysis
Statistical analyses were performed using Prism 8 (GraphPad). To quantify differences in single channel currents between EM and mutant constructs, mean and standard deviations were calculated from three independent patches for each group. Statistical significance was determined with an unpaired t-test. To quantify differences in binding affinities and hill slopes, each set of binding reaction experiments was performed three to five times. For each independent experiment, measurements were taken in triplicate. From these triplicate measurements, mean and standard error were calculated, and Kd and nH values were determined by nonlinear regression. For competition experiments, IC50 and nH values were determined by nonlinear regression and Ki values were calculated using experimental Kd values for [3H]-epibatidine. Statistical significance between +Fab and −Fab groups was determined with unpaired t-tests.
Data and Code Availability
The structures and EM density maps generated in this study are available in the PDB and EMDB respectively. The accession numbers for the reported data are PDB: 6PV7, EMDB: EMD-20487 (α3β4EM-Nicotine complex); PDB: 6PV8, EMDB: EMD-20488 (α3β4EM-AT-1001 complex in DDM); EMDB: EMD-20489 (α3β4EM-Nicotine complex without CHS); EMDB: EMD-20490 (α3β4EM-AT-1001 complex in nanodiscs).
Supplementary Material
Video S1: Density map for nicotine and water, related to Figure 3.
Video S2: Density map for AT-1001 at Site 1, related to Figure 3.
Video S3: Density map for AT-1001 at Site 2, related to Figure 3.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-α3β4 (4G9) | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| n-Dodecyl-β-D-Maltopyranoside (DDM) | Anatrace | Cat#D310 |
| Cholesteryl hemisuccinate (CHS) | Anatrace | Cat#CH210 |
| Soy extract polar lipids | Avanti | Cat#541602 |
| Fos-choline-8, fluorinated | Anatrace | Cat#F300F |
| (−)-Nicotine | Sigma-Aldrich | Cat#N3876 |
| AT-1001 | Astrea Therapeutics | N/A |
| d-Desthiobiotin | Sigma-Aldrich | Cat#D1411 |
| Sodium butyrate | Sigma-Aldrich | Cat#303410 |
| Lipofectamine 2000 | Invitrogen | Cat#11668019 |
| Freestyle 293 expression medium | Gibco | Cat#12338-018 |
| Sf-900 III SFM | Gibco | Cat#12658-027 |
| DMEM Media | Corning | Cat#10-013-CV |
| Fetal bovine serum | EMD Millipore | Cat#TMS-013-B |
| Critical Commercial Assays | ||
| Superose 6 Increase 10/300 GL | GE Healthcare | Cat#29-0915-96 |
| Strep-Tactin Superflow resin | IBA | Cat#2-1206-025 |
| Deposited Data | ||
| Coordinates of α3β4EM-Nicotine complex | This paper | PDB: 6PV7 |
| Cryo-EM map of α3β4EM-Nicotine complex | This paper | EMDB: EMD-20487 |
| Coordinates of α3β4EM-AT-1001 complex in DDM | This paper | PDB: 6PV8 |
| Cryo-EM map of α3β4EM-AT-1001 complex in DDM | This paper | EMDB: EMD-20488 |
| Cryo-EM map of α3β4EM-Nicotine complex without CHS | This paper | EMDB: EMD-20489 |
| Cryo-EM map of α3β4EM-AT-1001 complex in nanodiscs | This paper | EMDB: EMD-20490 |
| Coordinates of the α4β2 nicotinic receptor | Walsh et al., 2018 | PDB: 6CNJ |
| Coordinates of the Torpedo nicotinic receptor | Unwin, 2005 | PDB: 2BG9 |
| Coordinates of the human TRAAK K+ channel | Brohawn et al., 2014 | PDB: 4WFE |
| Coordinates of the Fab of anti-Shh 5E1 chimera | Maun et al., 2010 | PDB: 3MXV |
| Experimental Models: Cell Lines | ||
| HEK293S GnTI- | ATCC | Cat#CRL-3022 |
| Sf9 | ATCC | Cat#CRL-1711 |
| Recombinant DNA | ||
| pEZT-BM-α3EM and mutants | This paper | N/A |
| pEZT-BM-β4EM and mutants | This paper | N/A |
| pNIC-Bsa4 N term His6 TEV SMG Saposin A | Salipro Biotech | N/A |
| Software and Algorithms | ||
| EPU | FEI | https://www.fei.com/software/epu-automated-single-particles-software-for-life-sciences/ |
| Serial EM | Mastronarde, 2005 | http://bio3d.colorado.edu/SerialEM/ |
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/cryoem-software |
| GCTF | Zhang, 2016 | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| Relion 3.0 | Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| ResMap | Kucukelbir et al., 2014 | http://resmap.sourceforge.net/ |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | Adams et al., 2010 | https://www.phenix-online.org/ |
| Pymol | PyMOL | http://www.pymol.org |
| CHAP | Klesse et al., 2019 | https://www.channotation.org/ |
| pClamp 10 | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| STaGE | Lundborg & Lindahl, 2015 | https://gerrit.gromacs.org/admin/repos/STaGE |
| AutoDock Vina | Trott & Olson, 2010 | http://vina.scripps.edu/ |
| CHARMM-GUI | Jo et al., 2008 | http://www.charmm-gui.org/ |
| GROMACS 2018 | Abraham et al., 2015 | http://www.gromacs.org/ |
| VMD | Humphrey et al., 1996 | https://www.ks.uiuc.edu/Research/vmd/ |
| MDTraj | McGibbon et al., 2015 | http://mdtraj.org/1.9.3/ |
| Other | ||
| Quantifoil R1.2/1.3 Au 300 mesh | Electron Microscopy Sciences | Cat#Q3100AR1.3 |
Highlights.
The α3β4 ganglionic nicotinic receptor is a target for anti-addiction therapeutics
Receptor structures determined in a functionally supportive lipidic environment
Comparisons with the α4β2 subtype suggest principles underlying ligand selectivity
Lateral portals in the intracellular domain facilitate ion permeation
Acknowledgements:
We thank D. Cawley for antibody production, X. Bai for EM discussion, P. Blount, R. Syeda and W. Zeng for liposome patch advice, J. Frauenfeld (Salipro Biotech) for the saposin expression construct, and all members of the Hibbs Lab for discussion. Single-particle cryo-EM data were collected at the University of Texas Southwestern Medical Center Cryo-Electron Microscopy Facility, which is supported by the CPRIT Core Facility Support Award RP170644. A.G. acknowledges support from the NIH (T32DA07290). R.W. acknowledges support from the Sara and Frank McKnight Fund for Biochemical Research and the NIH (T32GM008203). This work was supported by a McKnight Scholar Award, The Welch Foundation (I-1812) and grants from the NIH (DA037492, DA042072, and NS095899) to R.E.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests: N.T.Z. is the founder and an employee of Astraea Therapeutics and a member of its scientific advisory board.
References:
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, and Lindahl E (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25. [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addona GH, Sandermann H Jr., Kloczewiak MA, Husain SS, and Miller KW (1998). Where does cholesterol act during activation of the nicotinic acetylcholine receptor? Biochim Biophys Acta 1370, 299–309. [DOI] [PubMed] [Google Scholar]
- Albuquerque EX, Pereira EF, Alkondon M, and Rogers SW (2009). Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev 89, 73–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Gicheru Y, Rao S, Sansom MSP, and Chakrapani S (2018a). Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3A receptor. Nature 563, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Gicheru Y, Samanta A, Molugu SK, Huang W, Fuente M, Hughes T, Taylor DJ, Nieman MT, Moiseenkova-Bell V, et al. (2018b). Cryo-EM structure of 5-HT3A receptor in its resting conformation. Nat Commun 9, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstein O, and Sansom MS (2006). A hydrophobic gate in an ion channel: the closed state of the nicotinic acetylcholine receptor. Phys Biol 3, 147–159. [DOI] [PubMed] [Google Scholar]
- Bouzat C, Bren N, and Sine SM (1994). Structural basis of the different gating kinetics of fetal and adult acetylcholine receptors. Neuron 13, 1395–1402. [DOI] [PubMed] [Google Scholar]
- Brohawn SG, Campbell EB, and MacKinnon R (2014). Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature 516, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, and Parrinello M (2007). Canonical sampling through velocity rescaling. J Chem Phys 126, 014101. [DOI] [PubMed] [Google Scholar]
- Celie PH, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, and Sixma TK (2004). Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41, 907–914. [DOI] [PubMed] [Google Scholar]
- Cerdan AH, Martin NE, and Cecchini M (2018). An Ion-Permeable State of the Glycine Receptor Captured by Molecular Dynamics. Structure 26, 1555–1562 e1554. [DOI] [PubMed] [Google Scholar]
- Chak A, and Karlin A (1992). Purification and reconstitution of nicotinic acetylcholine receptor. Methods Enzymol 207, 546–555. [DOI] [PubMed] [Google Scholar]
- Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, Kunken J, Xu F, Cherezov V, Hanson MA, et al. (2012). Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 20, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cippitelli A, Wu J, Gaiolini KA, Mercatelli D, Schoch J, Gorman M, Ramirez A, Ciccocioppo R, Khroyan TV, Yasuda D, et al. (2015). AT-1001: a high-affinity alpha3beta4 nAChR ligand with novel nicotine-suppressive pharmacology. Br J Pharmacol 172, 1834–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy WG, and Berg DK (1995). Neurons can maintain multiple classes of nicotinic acetylcholine receptors distinguished by different subunit compositions. J Biol Chem 270, 4424–4431. [DOI] [PubMed] [Google Scholar]
- Couturier S, Erkman L, Valera S, Rungger D, Bertrand S, Boulter J, Ballivet M, and Bertrand D (1990). Alpha 5, alpha 3, and non-alpha 3. Three clustered avian genes encoding neuronal nicotinic acetylcholine receptor-related subunits. J Biol Chem 265, 17560–17567. [PubMed] [Google Scholar]
- Covernton PJ, Kojima H, Sivilotti LG, Gibb AJ, and Colquhoun D (1994). Comparison of neuronal nicotinic receptors in rat sympathetic neurones with subunit pairs expressed in Xenopus oocytes. J Physiol 481 ( Pt 1), 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daCosta CJ, and Baenziger JE (2009). A lipid-dependent uncoupled conformation of the acetylcholine receptor. J Biol Chem 284, 17819–17825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daCosta CJ, and Baenziger JE (2013). Gating of pentameric ligand-gated ion channels: structural insights and ambiguities. Structure 21, 1271–1283. [DOI] [PubMed] [Google Scholar]
- Eaton JB, Peng JH, Schroeder KM, George AA, Fryer JD, Krishnan C, Buhlman L, Kuo YP, Steinlein O, and Lukas RJ (2003). Characterization of human alpha 4 beta 2-nicotinic acetylcholine receptors stably and heterologously expressed in native nicotinic receptor-null SH-EP1 human epithelial cells. Mol Pharmacol 64, 1283–1294. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, and Pedersen LG (1995). A smooth particle mesh Ewald method. J Chem Phys 103, 8577. [Google Scholar]
- Frauenfeld J, Loving R, Armache JP, Sonnen AF, Guettou F, Moberg P, Zhu L, Jegerschold C, Flayhan A, Briggs JA, et al. (2016). A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods 13, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, and Corringer PJ (2018). The dual-gate model for pentameric ligand-gated ion channels activation and desensitization. J Physiol 596, 1873–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick SD, Kuehne ME, Maisonneuve IM, Bandarage UK, and Molinari HH (1996). 18-Methoxycoronaridine, a non-toxic iboga alkaloid congener: effects on morphine and cocaine self-administration and on mesolimbic dopamine release in rats. Brain Res 719, 29–35. [DOI] [PubMed] [Google Scholar]
- Glick SD, Maisonneuve IM, and Dickinson HA (2000). 18-MC reduces methamphetamine and nicotine self-administration in rats. Neuroreport 11, 2013–2015. [DOI] [PubMed] [Google Scholar]
- Glick SD, Maisonneuve IM, Kitchen BA, and Fleck MW (2002). Antagonism of alpha 3 beta 4 nicotinic receptors as a strategy to reduce opioid and stimulant self-administration. Eur J Pharmacol 438, 99–105. [DOI] [PubMed] [Google Scholar]
- Grady SR, Moretti M, Zoli M, Marks MJ, Zanardi A, Pucci L, Clementi F, and Gotti C (2009). Rodent habenulo-interpeduncular pathway expresses a large variety of uncommon nAChR subtypes, but only the alpha3beta4* and alpha3beta3beta4* subtypes mediate acetylcholine release. J Neurosci 29, 2272–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin AA, Wang CI, Muttenthaler M, Alewood PF, Lewis RJ, and Adams DJ (2010). Alpha-conotoxin AuIB isomers exhibit distinct inhibitory mechanisms and differential sensitivity to stoichiometry of alpha3beta4 nicotinic acetylcholine receptors. J Biol Chem 285, 22254–22263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutter T, Prado de Carvalho L, Le Novere N, Corringer PJ, Edelstein S, and Changeux JP (2003). An H-bond between two residues from different loops of the acetylcholine binding site contributes to the activation mechanism of nicotinic receptors. EMBO J 22, 1990–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales TG, Dunlop JI, Deeb TZ, Carland JE, Kelley SP, Lambert JJ, and Peters JA (2006). Common determinants of single channel conductance within the large cytoplasmic loop of 5-hydroxytryptamine type 3 and alpha4beta2 nicotinic acetylcholine receptors. J Biol Chem 281, 8062–8071. [DOI] [PubMed] [Google Scholar]
- Harpole TJ, and Grosman C (2014). Side-chain conformation at the selectivity filter shapes the permeation free-energy landscape of an ion channel. Proc Natl Acad Sci USA 111, E3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassaine G, Deluz C, Grasso L, Wyss R, Tol MB, Hovius R, Graff A, Stahlberg H, Tomizaki T, Desmyter A, et al. (2014). X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 512, 276–281. [DOI] [PubMed] [Google Scholar]
- Hess B (2008). P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. Journal of Chemical Theory and Computation 4, 116–122. [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Bertozzi C, Zimmermann I, Reiter A, Trauner D, and Dutzler R (2010). Structural basis of open channel block in a prokaryotic pentameric ligand-gated ion channel. Nat Struct Mol Biol 17, 1330–1336. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J Mol Graph 14, 33-38, 27. [DOI] [PubMed] [Google Scholar]
- Imoto K, Busch C, Sakmann B, Mishina M, Konno T, Nakai J, Bujo H, Mori Y, Fukuda K, and Numa S (1988). Rings of negatively charged amino acids determine the acetylcholine receptor channel conductance. Nature 335, 645–648. [DOI] [PubMed] [Google Scholar]
- Imoto K, Konno T, Nakai J, Wang F, Mishina M, and Numa S (1991). A ring of uncharged polar amino acids as a component of channel constriction in the nicotinic acetylcholine receptor. FEBS Lett 289, 193–200. [DOI] [PubMed] [Google Scholar]
- Ivanov I, Cheng X, Sine SM, and McCammon JA (2007). Barriers to ion translocation in cationic and anionic receptors from the Cys-loop family. J Am Chem Soc 129, 8217–8224. [DOI] [PubMed] [Google Scholar]
- Jämbeck JPM, and Lyubartsev AP (2012). An Extension and Further Validation of an All-Atomistic Force Field for Biological Membranes. Journal of Chemical Theory and Computation 8, 2938–2948. [DOI] [PubMed] [Google Scholar]
- Jämbeck JPM, and Lyubartsev AP (2013). Another piece of the membrane puzzle: extending slipids further. Journal of Chemical Theory and Computation 9, 774–784. [DOI] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, and Im W (2008). CHARMM-GUI: a web-based graphical user interface for CHARMM. Journal of Computational Chemistry 29, 1859–1865. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, and Klein ML (1983). Comparison of simple potential functions for simulating liquid water. J Chem Phys 79, 926. [Google Scholar]
- Kawate T, and Gouaux E (2006). Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681. [DOI] [PubMed] [Google Scholar]
- Kelley SP, Dunlop JI, Kirkness EF, Lambert JJ, and Peters JA (2003). A cytoplasmic region determines single-channel conductance in 5-HT3 receptors. Nature 424, 321–324. [DOI] [PubMed] [Google Scholar]
- Klesse G, Rao S, Sansom MS, and Tucker SJ (2019). CHAP: a versatile tool for the structural and functional annotation of ion channel pores. bioRxiv, 527275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashia P, Moroni M, Broadbent S, Hofmann G, Kracun S, Beato M, Groot-Kormelink PJ, and Sivilotti LG (2010). Human alpha3beta4 neuronal nicotinic receptors show different stoichiometry if they are expressed in Xenopus oocytes or mammalian HEK293 cells. PLoS One 5, e13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarca C, Nowak MW, Zhang H, Tang L, Deshpande P, and Lester HA (1995). Channel gating governed symmetrically by conserved leucine residues in the M2 domain of nicotinic receptors. Nature 376, 514–516. [DOI] [PubMed] [Google Scholar]
- Laverty D, Desai R, Uchanski T, Masiulis S, Stec WJ, Malinauskas T, Zivanov J, Pardon E, Steyaert J, Miller KW, et al. (2019). Cryo-EM structure of the human alpha1beta3gamma2 GABAA receptor in a lipid bilayer. Nature 565, 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, and Shaw DE (2010). Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC, Word JM, Richardson JS, and Richardson DC (2000). The penultimate rotamer library. Proteins 40, 389–408. [PubMed] [Google Scholar]
- Lundborg M, and Lindahl E (2015). Automatic GROMACS topology generation and comparisons of force fields for solvation free energy calculations. The Journal of Physical Chemistry B 119, 810–823. [DOI] [PubMed] [Google Scholar]
- Luo J, Taylor P, Losen M, de Baets MH, Shelton GD, and Lindstrom J (2009). Main immunogenic region structure promotes binding of conformation-dependent myasthenia gravis autoantibodies, nicotinic acetylcholine receptor conformation maturation, and agonist sensitivity. J Neurosci 29, 13898–13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Maun HR, Wen X, Lingel A, de Sauvage FJ, Lazarus RA, Scales SJ, and Hymowitz SG (2010). Hedgehog pathway antagonist 5E1 binds hedgehog at the pseudo-active site. J Biol Chem 285, 26570–26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum SE, Cowe MA, Lewis SW, and Glick SD (2012). alpha3beta4 nicotinic acetylcholine receptors in the medial habenula modulate the mesolimbic dopaminergic response to acute nicotine in vivo. Neuropharmacology 63, 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernández CX, Schwantes CR, Wang L-P, Lane TJ, and Pande VS (2015). Mdtraj: A modern open library for the analysis of molecular dynamics trajectories. Biophysical Journal 109, 1528–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Stowell M, and Unwin N (1999). Nicotinic acetylcholine receptor at 4.6 A resolution: transverse tunnels in the channel wall. J Mol Biol 288, 765–786. [DOI] [PubMed] [Google Scholar]
- Morales-Perez CL, Noviello CM, and Hibbs RE (2016a). Manipulation of Subunit Stoichiometry in Heteromeric Membrane Proteins. Structure 24, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Perez CL, Noviello CM, and Hibbs RE (2016b). X-ray structure of the human alpha4beta2 nicotinic receptor. Nature 538, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, and Sliz P (2013). Collaboration gets the most out of software. Elife 2, e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtasimova N, daCosta CJ, and Sine SM (2016). Improved resolution of single channel dwell times reveals mechanisms of binding, priming, and gating in muscle AChR. J Gen Physiol 148, 43–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle C, Vidal C, Benoit P, and Changeux JP (1991). Existence of different subtypes of nicotinic acetylcholine receptors in the rat habenulo-interpeduncular system. J Neurosci 11, 2588–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecz A, Prevost MS, Menny A, and Corringer PJ (2016). Emerging Molecular Mechanisms of Signal Transduction in Pentameric Ligand-Gated Ion Channels. Neuron 90, 452–470. [DOI] [PubMed] [Google Scholar]
- Nguyen NX, Armache JP, Lee C, Yang Y, Zeng W, Mootha VK, Cheng Y, Bai XC, and Jiang Y (2018). Cryo-EM structure of a fungal mitochondrial calcium uniporter. Nature 559, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Fage D, and Scatton B (1986). Evidence for, and nature of, the tonic inhibitory influence of habenulointerpeduncular pathways upon cerebral dopaminergic transmission in the rat. Brain Res 373, 324–336. [DOI] [PubMed] [Google Scholar]
- Parrinello M, and Rahman A (1980). Crystal structure and pair potentials: A molecular-dynamics study. Phys Rev Lett 45, 1196–1199. [Google Scholar]
- Pei J, Kim BH, and Grishin NV (2008). PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Polovinkin L, Hassaine G, Perot J, Neumann E, Jensen AA, Lefebvre SN, Corringer PJ, Neyton J, Chipot C, Dehez F, et al. (2018). Conformational transitions of the serotonin 5-HT3 receptor. Nature 563, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick MW, Ceballos RM, Kasten M, McIntosh JM, and Lester RA (1999). Alpha3beta4 subunit-containing nicotinic receptors dominate function in rat medial habenula neurons. Neuropharmacology 38, 769–783. [DOI] [PubMed] [Google Scholar]
- Rezvani AH, Overstreet DH, Yang Y, Maisonneuve IM, Bandarage UK, Kuehne ME, and Glick SD (1997). Attenuation of alcohol consumption by a novel nontoxic ibogaine analogue (18-methoxycoronaridine) in alcohol-preferring rats. Pharmacol Biochem Behav 58, 615–619. [DOI] [PubMed] [Google Scholar]
- Sauguet L, Poitevin F, Murail S, Van Renterghem C, Moraga-Cid G, Malherbe L, Thompson AW, Koehl P, Corringer PJ, Baaden M, et al. (2013). Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J 32, 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, and Peitsch MC (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 31, 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skok VI (2002). Nicotinic acetylcholine receptors in autonomic ganglia. Auton Neurosci 97, 1–11. [DOI] [PubMed] [Google Scholar]
- St John PA (2009). Cellular trafficking of nicotinic acetylcholine receptors. Acta Pharmacol Sin 30, 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland RJ (1982). The dorsal diencephalic conduction system: a review of the anatomy and functions of the habenular complex. Neurosci Biobehav Rev 6, 1–13. [DOI] [PubMed] [Google Scholar]
- Tapia L, Kuryatov A, and Lindstrom J (2007). Ca2+ permeability of the (alpha4)3(beta2)2 stoichiometry greatly exceeds that of (alpha4)2(beta2)3 human acetylcholine receptors. Mol Pharmacol 71, 769–776. [DOI] [PubMed] [Google Scholar]
- Toll L, Zaveri NT, Polgar WE, Jiang F, Khroyan TV, Zhou W, Xie XS, Stauber GB, Costello MR, and Leslie FM (2012). AT-1001: a high affinity and selective alpha3beta4 nicotinic acetylcholine receptor antagonist blocks nicotine self-administration in rats. Neuropsychopharmacology 37, 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, and Olson AJ (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan EW, Horti AG, Olson TT, Gao Y, Stockmeier CA, Al-Muhtasib N, Bowman Dalley C, Lewin AE, Wolfe BB, Sahibzada N, et al. (2015). AT-1001 Is a Partial Agonist with High Affinity and Selectivity at Human and Rat alpha3beta4 Nicotinic Cholinergic Receptors. Mol Pharmacol 88, 640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium, T. (2018). UniProt: the universal protein knowledgebase. Nucleic Acids Res 46, 2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N (2005). Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol 346, 967–989. [DOI] [PubMed] [Google Scholar]
- Walsh RM Jr., Roh SH, Gharpure A, Morales-Perez CL, Teng J, and Hibbs RE (2018). Structural principles of distinct assemblies of the human alpha4beta2 nicotinic receptor. Nature 557, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrwein EA, Orer HS, and Barman SM (2016). Overview of the Anatomy, Physiology, and Pharmacology of the Autonomic Nervous System. Compr Physiol 6, 1239–1278. [DOI] [PubMed] [Google Scholar]
- Wu EL, Cheng X, Jo S, Rui H, Song KC, Dávila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, et al. (2014). CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. Journal of Computational Chemistry 35, 1997–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Meyer EL, Thompson JM, Surin A, Wroblewski J, and Kellar KJ (1998). Rat alpha3/beta4 subtype of neuronal nicotinic acetylcholine receptor stably expressed in a transfected cell line: pharmacology of ligand binding and function. Mol Pharmacol 54, 322–333. [DOI] [PubMed] [Google Scholar]
- Xiu X, Puskar NL, Shanata JA, Lester HA, and Dougherty DA (2009). Nicotine binding to brain receptors requires a strong cation-pi interaction. Nature 458, 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K, Kato K, Ogasawara M, Tomita M, and Toyoshima C (2015). Electron crystallography of ultrathin 3D protein crystals: atomic model with charges. Proc Natl Acad Sci U S A 112, 3368–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Density map for nicotine and water, related to Figure 3.
Video S2: Density map for AT-1001 at Site 1, related to Figure 3.
Video S3: Density map for AT-1001 at Site 2, related to Figure 3.
Data Availability Statement
The structures and EM density maps generated in this study are available in the PDB and EMDB respectively. The accession numbers for the reported data are PDB: 6PV7, EMDB: EMD-20487 (α3β4EM-Nicotine complex); PDB: 6PV8, EMDB: EMD-20488 (α3β4EM-AT-1001 complex in DDM); EMDB: EMD-20489 (α3β4EM-Nicotine complex without CHS); EMDB: EMD-20490 (α3β4EM-AT-1001 complex in nanodiscs).