Abstract
Background
Growing evidence has demonstrated immune reactivity as a confirmed important carcinogenesis and therapy efficacy for clear cell renal cell carcinoma (ccRCC). Aquaporin 9 (AQP9) is involved in many immune-related signals; however, its role in ccRCC remains to be elucidated. This study investigated AQP9 expression in tumor tissues and defined the prognostic value in ccRCC patients.
Methods
A total of 913 ccRCC patients with available RNA-sequence data from the Cancer Genome Atlas (TCGA) database and Fudan University Shanghai Cancer Center (FUSCC) were consecutively recruited in analyses. Differential transcriptional and proteome expression profiles were obtained and validated using multiple datasets. A partial likelihood test from Cox regression analysis was developed to address the influence of independent factors on progression-free survival (PFS) and overall survival (OS). The Kaplan–Meier method and log-rank test were performed to assess survival. Receiver operating characteristic (ROC) curves were used to describe binary classifier value of AQP9 using area under the curve (AUC) score. Functional enrichment analyses and immune infiltration analysis were used to describe significantly involved hallmark pathways of hub genes.
Results
Significantly elevated transcriptional and proteomic AQP9 expressions were found in ccRCC samples. Increased AQP9 mRNA expression was significantly associated with advanced clinicopathological parameters and correlated with shorter PFS and OS in TCGA and FUSCC cohorts (p < 0.001). ROC curves suggested the significant diagnostic and prognostic ability of AQP9 (PFS, AUC = 0.823; OS, AUC = 0.828). Functional annotations indicated that AQP9 is involved in the most significant hallmarks including complement, coagulation, IL6/JAK–STAT3, inflammatory response and TNF-alpha signaling pathways.
Conclusion
Our study revealed that elevated AQP9 expression was significantly correlated with aggressive progression, poor survival and immune infiltrations in ccRCC patients, and we validated its prognostic value in a real-world cohort. These data suggest that AQP9 may act as an oncogene and a promising prognostic marker in ccRCC.
Keywords: Clear cell renal cell carcinoma, AQP9, Biomarker, Bioinformatics, Prognosis
Background
Renal cell carcinoma (RCC) is one of the most common malignant urinary tumors in the world, with an incidence rate that is increasing 2% each year, especially in developed regions [1]. The incidence and mortality rates of RCC in China are also increasing, with an estimated 66,800 new cases and 23,400 deaths in 2015 [2]. Clear cell RCC (ccRCC) is the most common and aggressive type of RCC in adults. According to the World Health Organization, ccRCC is one of the deadliest urinary tumors, with a global annual mortality rate of approximately 90,000 [3]. Although extensive research has explored the mechanism of recurrence and metastasis, the etiology and tumorigenesis of ccRCC remain unclear. A variety of indicators, such as genetic aberrations and tumor environment, have been reported to be associated with the development and progression of ccRCC [4–6]. Considering the high morbidity and mortality of RCC, it is essential to explore its causes and potential molecular mechanisms to identify potential molecular biomarkers for early diagnosis, prevention, and personalized treatment.
Aquaporins (AQPs), also called water channels, were first discovered in 1992 by Agre et al. [7], and 13 AQP family members have been identified in humans, including AQP0–12 [8]. Accumulating studies have shown that AQPs not only regulate rapid water movement in various epithelial and non-epithelial tissues [9, 10], but also participate in the pathological process of several diseases such as glaucoma, cancer, inflammation, immunity, and obesity [11]. Several AQPs are over-expressed in tumors samples and serve notable roles in cancer progression [12]. A previous study showed that AQP1 was a unique non-invasive biomarker for screening and diagnosing malignant clear cells or papillary RCC [13]. In addition, Chen et al. also found that AQP3 promoted prostate cancer cell invasion through extracellular signal-regulated kinase 1/2-mediated MMP-3 secretion [14]. Interestingly, AQP9 was significantly correlated with immune activity. For example, IL-7 induces glycerol channel AQP9 expression in CD8+ T cells and AQP9 is required for memory CD8+ T cell survival and self-renewal [15]. In addition, AQP9 was demonstrated to promote astrocytoma cell invasion and motility via the AKT pathway [16]. Therefore, understanding of the regulation and molecular function of AQP9 may identify potential targets for the diagnosis and treatment of ccRCC.
To investigate the differential AQP9 transcriptional and proteomics expression and clarify the potential prognostic value in ccRCC patients, we analyzed gene expression profiles, as well as the underlying biological interaction networks and the prognostic value. We hypothesized that the possible oncogenic activity of AQP9 may impact prognosis of ccRCC patients. Our findings may reveal potential therapeutic targets and provide insights into the molecular mechanisms of ccRCC.
Materials and methods
Ethics statement
All of the study designs and test procedures were performed in accordance with the Helsinki Declaration II. Study protocols were obtained by Fudan University Shanghai Cancer Center (FUSCC) (Shanghai, China) included in this work.
Patients and transcriptional expression profile
A total of 533 ccRCC patients with available RNA-sequence data from the Cancer Genome Atlas (TCGA) database were consecutively recruited in analyses [17]. The gene expression profile was measured experimentally using the Illumina HiSeq 2000 RNA Sequencing platform by the University of North Carolina TCGA genome characterization center. Level 3 data was downloaded from TCGA data coordination center. X-tile software was utilized to take the cut-off value of mRNA expression of AQP9, in concordance of which overall participants were divided to two groups, respectively. Student’s t tests were used to compare differential transcriptional expressions levels of AQP9 between paired AJCC stages or ISUP grades, marked in asterisk. The overall statistical expression difference of AJCC stages or ISUP grades was measured using One-way ANOVA test.
We next enrolled a total of 380 ccRCC patients from the Department of Urology, Fudan University Shanghai Cancer Center (FUSCC; Shanghai, China) from Aug 2009 to May 2018 in analyses. Tissue samples, including ccRCC and normal tissues, were collected during surgery and available from FUSCC tissue bank.
Oncomine database
In this study, transcriptional expression profiles of AQP9 in ccRCC patients were obtained from Oncomine database using Oncomine online database (http://www.oncomine.com) [18]. Difference of transcriptional expression was compared by Students’t-test. Cut-off of p value and fold change were as following: p-value = 0.01, fold change = 1.5, gene rank = 10%, Data type: mRNA.
The Human Protein Atlas
The Human Pathology Atlas project (https://www.proteinatlas.org) contains immunohistochemistry (IHC) data using a tissue microarray-based analysis on 44 different normal tissue types, and proteome analysis of 17 major cancer types [19]. Staining intensity, quantity, location and patients’ information in patients with the respective cancer types were available online. In this study, representative proteins expressions of IHC images of AQP9 were detected in ccRCC and normal tissues in Human Protein Atlas.
Real-Time Quantitative PCR (RT-qPCR) analysis
Total RNA sequence was extracted using TRIzol® reagent (Invitrogen Life Technologies, USA) from 380 paired tumor and para-carcinoma normal samples. Primers were diluted in ddH2O with SYBR Green PCR Master Mix (Applied Biosystems, Japan). Transcriptional expression was determined as the fold change of AQP9 relative to β-Actin. PCR primers sequence for AQP9 are as follows: forward are 5′-TTGCCCAAGCTATTCTCAGTCGA-3′ and reverse are 5′-CAGAGACACCGCCAGCCACAT-3′. The AQP9 mRNA expression was represented as ΔCt = Ct(AQP9) − ΔCt(β-actin). Relative expression in ccRCC was represented using the ratio of AQP9 expression in Tumor/Normal tissues (T/N). “Low AQP9 expression” and “High AQP9 expression” denote the T/N ratio of AQP9 mRNA expression with median cutoff in FUSCC cohort.
Immunohistochemical (IHC) staining and evaluation
Immunostaining of AQP9 was performed using a mouse monoclonal anti- AQP9 antibody (1:100, Cat. ab84828, Abcam, USA). Positive or negative staining of a certain protein in one FFPE slide was independently assessed by two experienced pathologists, and determined as follows. The overall IHC score grading from 0 to 12 was evaluated according to the multiply of the staining intensity and extent score, as previously described [20].
Statistical analysis
Phenotype and expression profiles of hub genes in 533 ccRCC patients from TCGA were analyzed and displayed. Survival comparison between distinct mRNA expression groups of AQP9 was analyzed in ccRCC patients. The primary end point for patients was progression-free survival (PFS), and overall survival (OS) was the secondary end point, which was evaluated from the date of first therapy to the date of death or last follow-up. The follow-up duration was estimated using the Kaplan–Meier method with 95% confidence intervals (95% CI) and log-rank test in separate curves. Univariate and multivariate analysis were performed with Cox logistic regression models to find independent variables, including age at diagnosis, age (ref. < 60 years), gender (ref. Male), pT stage (ref. T1–T2), pN stage (ref. N0), pM stage (ref. M0), AJCC stage (ref. I–II), ISUP grade (ref. 1–2) and AQP9 expression (ref. Low). X-tile software was utilized to take the cut-off value [21]. All hypothetical tests were two-sided and p-values less than 0.05 were considered significant in all tests. Integrated score was identified as sum of the weight of AQP9 and significant clinicopathological prognostic indicators.
Protein–protein interaction (PPI) network construction
Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org) (version 10.0) online database was used to predict PPI network of co-regulated hub genes and analyzing the functional interactions between proteins [22]. An interaction with a combined score > 0.4 was considered statistically significant.
Functional annotations
Subsequently, the gene ontology (GO): BP (biological process), GO: CC (cellular component), GO: MF (molecular function) and KEGG pathways analyses for hub genes in this module were performed using Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov; version 6.8) online database [23], and then visualized in bubble chart. p-value < 0.05 was considered statistically significant. Cytoscape (version 3.5), an open source bioinformatics software platform, was used to visualize molecular interaction networks [24]. ClueGO is a Cytoscape plug-in that visualizes the non-redundant biological terms for large clusters of genes in a functionally grouped network [25]. The biological process from GO and KEGG pathway analysis of hub genes was performed and visualized using ClueGO (version 2.5.3) and CluePedia (version 1.5.3), a Cytoscape plug-in that visualizes the non-redundant biological terms for large clusters of genes in a functionally grouped network [26]. Gene set enrichment analysis (GSEA) was used to predict potential hallmarks using transcriptional sequences in TCGA database. A permutation test with 1000 times was used to identify the significantly changed pathways [27]. Adj. p less than 0.01 and FDR less than 0.25 were identified as significant related genes. Statistical analysis and graphical plotting were conducted using R software (version 3.3.2).
Immune infiltration analysis
Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) was used to perform comprehensive correlation analysis between tumor-infiltrating immune cells signatures and selected hub genes. An integrated repository portal for tumor-immune system interactions (TISIDB, http://cis.hku.hk/TISIDB/index.php) [28] was utilized to examine tumor and immune system interactions in 28 types of TILs across human cancers. The relative abundance of TILs were inferred by using gene set variation analysis based on AQP9 expression profile. Spearman’s test was used to measure correlations between AQP9 and TILs. All hypothetical tests were two-sided and p-values less than 0.05 were considered significant in all tests. All of these statistical analyses were performed in R or corresponding R packages survival and survminer.
Results
This study consisted of four stages. We first screened and compared the mRNA expression of AQP family members in ccRCC and adjacent normal tissues in the TCGA database. We then examined the prognostic value of the expressions of the AQP family members in ccRCC and found that patients with high AQP9 expression had poor survival. In the second stage, we assessed differential AQP9 expression at the transcriptional and protein level according to datasets hosted on the Oncomine, TCGA and FUSCC platforms. In the third stage, survival analysis based on distinct comparison expression of AQP9 was evaluated in the TCGA and FUSCC cohorts. In the fourth stage, significantly involved hub genes of AQP9 were screened and corresponding functional annotations were performed.
Clinicopathological characteristics and AQP9 expression in ccRCC patients from TCGA and FUSCC
To first assess the association of AQP family member expressions with prognosis, we obtained follow-up and transcriptional expression data from TCGA and evaluated the impact of AQP0–11 expressions on the prognosis of ccRCC patients (Additional file 1: Figure S1). It suggested that high expression of AQP9 significantly predicted poor OS (p < 0.001) and PFS (p < 0.001).
We next examined the correlations of AQP9 expression and clinicopathological characteristics in TCGA and FUSCC cohorts. As shown in Table 1, increased AQP9 mRNA expression in ccRCC patients significantly correlated with advanced pT (p < 0.001), pN (p < 0.001), and pM stage (p < 0.001), AJCC stage (p < 0.001) and ISUP grade (p = 0.004) in the FUSCC cohort. In the TCGA cohort, increased AQP9 mRNA expression significantly correlated with advanced pT (p < 0.001), pN (p = 0.004), and pM stage (p < 0.001), AJCC stage (p < 0.001) and ISUP grade (p < 0.001) (Additional file 3: Table S1).
Table 1.
Clinicopathological characteristics baseline in relation to AQP9 expression level in FUSCC cohort
| Characteristics | FUSCC cohort (N = 380) |
AQP9 expression | χ2 | p | |
|---|---|---|---|---|---|
| High (N = 190) |
Low (N = 190) |
||||
| N (%) | |||||
| Age | 0.296 | 0.587 | |||
| < 60 years | 253 (66.6) | 124 (65.3) | 129 (67.9) | ||
| ≥ 60 years | 127 (33.4) | 66 (34.7) | 61 (32.1) | ||
| Gender | 0.435 | 0.510 | |||
| Male | 258 (67.9) | 132 (69.5) | 126 (66.3) | ||
| Female | 122 (32.1) | 58 (30.5) | 64 (33.7) | ||
| BMI | 0.541 | 0.462 | |||
| < 25 kg/m2 | 231 (60.8) | 112 (58.9) | 119 (62.6) | ||
| ≥ 25 kg/m2 | 149 (39.2) | 78 (41.1) | 71 (37.4) | ||
| pT stage | 34.336 | < 0.001 | |||
| T1–T2 | 307 (80.8) | 131 (68.9) | 176 (92.6) | ||
| T3–T4 | 73 (19.2) | 59 (31.1) | 14 (7.4) | ||
| pN stage | 14.246 | < 0.001 | |||
| N0 | 334 (87.9) | 155 (81.6) | 179 (94.2) | ||
| N1 | 46 (12.1) | 35 (18.4) | 11 (5.8) | ||
| pM stage | 40.347 | < 0.001 | |||
| M0 | 310 (81.6) | 131 (68.9) | 179 (94.2) | ||
| M1 | 70 (18.4) | 59 (31.1) | 11 (5.8) | ||
| AJCC stagea | 34.072 | < 0.001 | |||
| I–II | 292 (76.8) | 122 (64.2) | 170 (89.5) | ||
| III–IV | 88 (23.2) | 68 (35.8) | 20 (10.5) | ||
| ISUP grade | 4.218 | 0.040 | |||
| G1–G2 | 182 (47.9) | 81 (42.6) | 101 (53.2) | ||
| G3–G4 | 198 (52.1) | 109 (57.4) | 89 (46.8) | ||
FUSCC Fudan University Shanghai Cancer Center, BMI body mass index
aThe AJCC staging system is a classification system developed by the American Joint Committee on Cancer for describing the extent of disease progression in cancer patients. It utilizes in part the TNM scoring system: Tumor size, Lymph Nodes affected, Metastases
Differential expression of AQP9 in ccRCC patients in multiple cohorts
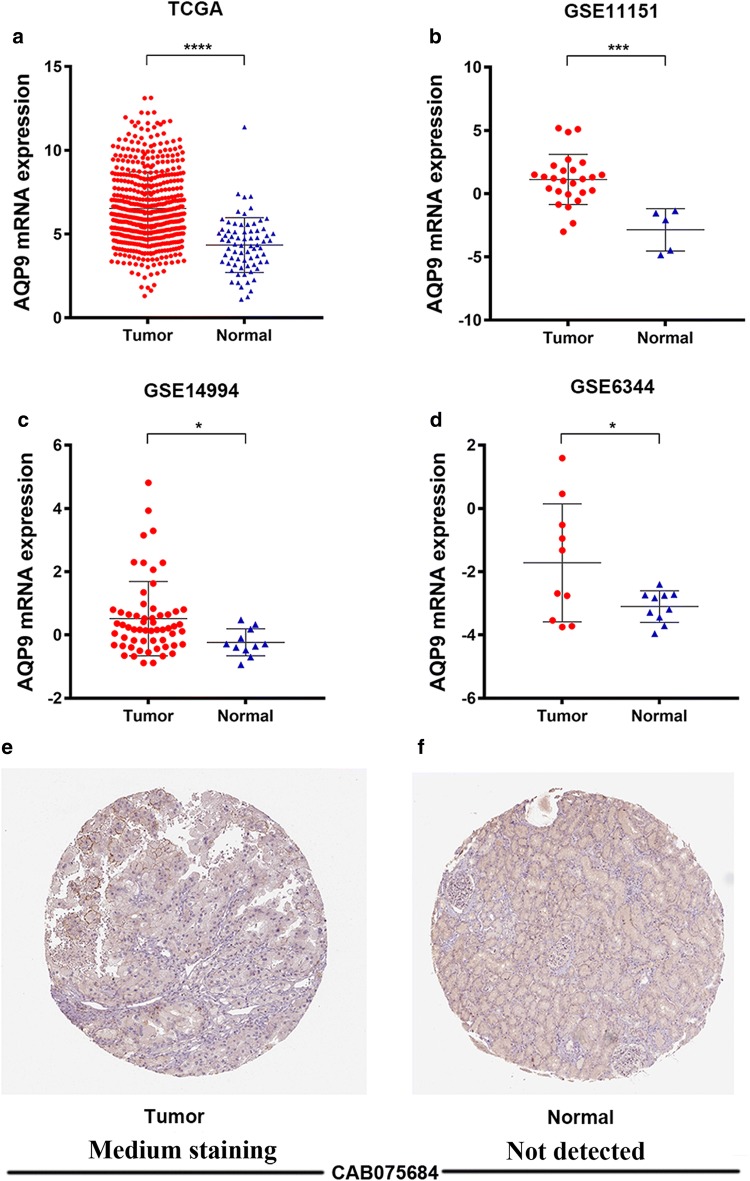
We compared the mRNA expression of AQP9 between ccRCC samples and adjacent normal tissues based on RNA-sequence data from TCGA and independent cohorts in silico. AQP9 mRNA was highly expressed in 533 ccRCC tissues compared with 72 healthy tissues (p < 0.0001), as shown in Fig. 1a. AQP9 expression was also significantly higher in ccRCC primary tumors in comparison with adjacent normal tissues in GSE11151 (Yusenko Renal dataset; ***p < 0.001) [29], GSE14994 (Beroukhim Renal dataset; *p < 0.05) [30] and GSE6344 (Gumz Renal dataset; *p < 0.05) [31] (Fig. 1b–d).
Fig. 1.
Differential AQP9 expression in ccRCC tumor tissues and adjacent normal tissues based on multiple cohorts. a Transcriptional level of AQP9 expression was found highly expressed in 533 ccRCC tissues compared with 72 normal tissues in TCGA cohort (****p < 0.0001). b–dAQP9 expression was significantly higher in ccRCC primary tumors in comparison with adjacent normal tissues in GSE11151 (Yusenko Renal dataset; ***p < 0.001) [26], GSE14994 (Beroukhim Renal dataset; *p < 0.05) [27] and GSE6344 (Gumz Renal dataset; *p < 0.05) [28] (b–d). f AQP9 expression is detected in ccRCC tissues while not detected in normal tissues using online database
IHC staining indicated that AQP9 staining was not detected in normal kidney tissues, while medium levels of expression (as defined in Methods) were observed in ccRCC tumor tissues (Fig. 1e, f). Taken together, these results suggested that AQP9 was highly expressed at transcriptional and proteomic levels in ccRCC tissues compared with normal tissues.
AQP9 mRNA expression correlated with advanced clinicopathological parameters for ccRCC patients in TCGA cohort
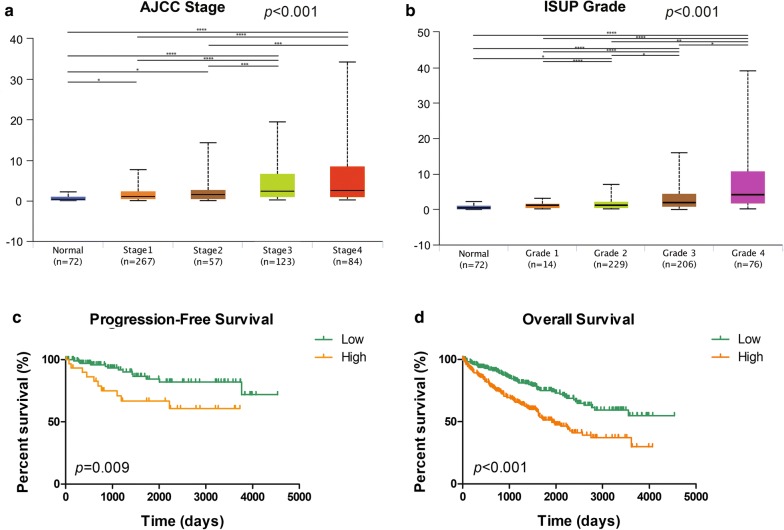
After integrating clinicopathological and survival data from TCGA, we found significantly elevated AQP9 mRNA expression in ccRCC samples compared with normal samples. As shown in Fig. 2a, AQP9 mRNA expression in ccRCC samples was significantly correlated with advanced clinical stage (p < 0.001), and the highest AQP9 mRNA expression was found in stage 4 cases. Figure 2b shows the relationship between AQP9 mRNA expression and different pathological grades, and the results suggested that AQP9 mRNA expressions were significantly correlated with pathological grade (p < 0.001). Similarly, the highest AQP9 mRNA expressions were found in grade 4 cases. Survival analysis using the Kaplan–Meier method showed that elevated AQP9 expression was significantly correlated with shorter PFS (p = 0.009) and OS (p < 0.001) in TCGA cohorts (Fig. 2c, d). Overall, elevated AQP9 mRNA expression was significantly associated with advanced clinicopathological parameters and poor prognosis in ccRCC patients from TCGA cohort.
Fig. 2.
Transcriptional expressions of AQP9 significantly correlated with advanced clinicopathological parameters and poor survival outcomes in ccRCC patients from TCGA cohort. a Transcriptional expression of AQP9 was significantly correlated with AJCC stages, patients who were in more advanced stages tended to express higher mRNA expression of AQP9. b Transcriptional expression of AQP9 was significantly correlated with ISUP grade, patients who were in more advanced grade score tended to express elevated mRNA expression of AQP9. Highest mRNA expressions of AQP9 were found in stage 4 or grade 4. *p < 0.05, **p < 0.01, ***p < 0.001. c Survival analysis in Kaplan–Meier method indicated that AQP9 was significantly correlated with shorter PFS (p = 0.009). d Survival curves suggested that patients with elevated AQP9 mRNA levels showed poorer OS in 533 included ccRCC patients (p < 0.001)
Validation of elevated AQP9 expression in ccRCC tissues from the FUSCC cohort
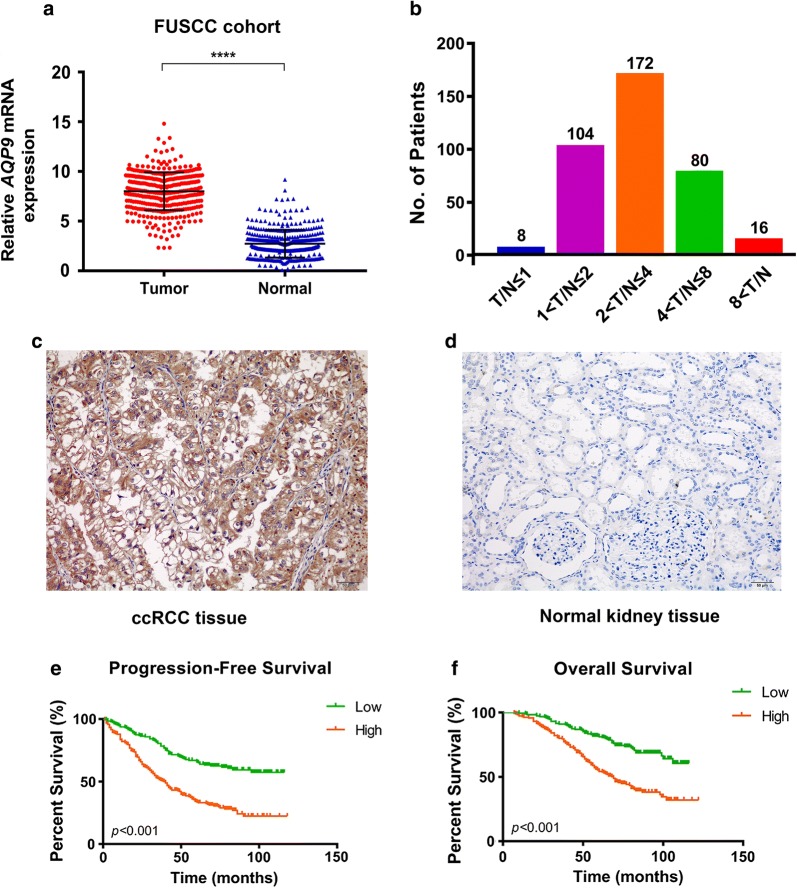
To validate AQP9 mRNA expression in ccRCC tissues, we performed RT-qPCR in 380 paired tumor and normal samples with available clinical follow-up data from FUSCC cohort. We found dramatically increased AQP9 mRNA expression in ccRCC samples: 97.9% of ccRCC patients had higher levels of AQP9 expression in tumor tissues than normal tissues (Fig. 3a, b). To assess the level of AQP9 protein expression in FUSCC tumor samples, we performed IHC staining and found significant elevated AQP9 expression in terms of density and intensity in ccRCC tissues compared with adjacent normal kidney tissues in FUSCC cohort (p < 0.001, Fig. 3c, d).
Fig. 3.
AQP9 mRNA expression and prognostic implication in FUSCC cohort. a The differential AQP9 mRNA expression in patients 380 with different T/N, which was define as the ratio of AQP9 expression in 380 paired tumor and normal tissues. b Survival analysis in Kaplan–Meier method indicated that AQP9 was significantly correlated with shorter PFS (p = 0.009). c, d IHC staining indicated significantly elevated AQP9 expression in terms of density and intensity in ccRCC tissues compared with adjacent normal kidney tissues in FUSCC cohort. e, f Survival curves suggested that patients with elevated AQP9 mRNA levels correlated with poorer PFS and OS in 380 included ccRCC patients (p < 0.001)
Cox regression analyses of TCGA and FUSCC cohorts
In univariate Cox regression analysis models, traditional prognostic factors such as pTNM stage, AJCC stage, and ISUP grade were significantly relevant to PFS (p < 0.05; Additional file 3: Table S2) and OS (p < 0.001,; Additional file 3: Table S3) in ccRCC patients in both the TCGA and FUSCC cohorts. Importantly, AQP9 amplification markedly correlated with poor PFS (TCGA: hazard ratio [HR] = 8.141, p < 0.001; FUSCC: HR = 2.593, p < 0.001) and poor OS (TCGA: HR = 2.262, p < 0.001, FUSCC: HR = 2.774, p < 0.001).
In multivariate Cox regression analysis, traditional prognostic factors, specifically pM stage, were still relevant to PFS (TCGA: HR = 2.690, p = 0.043; FUSCC: HR = 2.593, p = 0.018; Table 2) and OS (TCGA: HR = 1.763, p < 0.001; FUSCC: HR = 1.895, p = 0.014; Table 3) in ccRCC patients. In addition, pT stage, pN stage, AJCC stage and ISUP grade were significant both in PFS (pT stage: p = 0.023, pN stage: p = 0.003, AJCC stage: p = 0.006, ISUP grade: p < 0.001) and OS (pT stage: p = 0.045, pN stage: p = 0.008, AJCC stage: p < 0.001, ISUP grade: p = 0.004) in the FUSCC cohort. Importantly, elevated AQP9 expression was significantly associated with poor PFS (TCGA: HR = 3.443, p = 0.040; FUSCC: HR = 1.714, p = 0.001) and poor OS (TCGA: HR = 1.714, p = 0.026; FUSCC: HR = 1.514, p = 0.026) in both cohorts of ccRCC patients.
Table 2.
Multivariate Cox logistic regression analysis of PFS in TCGA and FUSCC cohort
| Covariates | TCGA | FUSCC | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | |
| Age | – | – | – | 1.008 | 0.996–1.020 | 0.207 |
| Gender (ref. male) | 0.563 | 0.157–2.022 | 0.379 | – | – | – |
| pT stage (ref. T1–T2) | 0.467 | 0.093–2.340 | 0.354 | 1.782 | 1.084–2.930 | 0.023 |
| pN stage (ref. N0) | – | – | – | 1.937 | 1.250–3.001 | 0.003 |
| pM stage (ref. M0) | 2.690 | 1.034–7.000 | 0.043 | 1.763 | 1.104–2.813 | 0.018 |
| AJCC stage (ref. I–II) | 4.283 | 0.703–26.095 | 0.115 | 2.425 | 1.292–4.552 | 0.006 |
| ISUP grade (ref. 1–2) | 2.269 | 0.817–6.301 | 0.116 | 1.812 | 1.330–2.471 | < 0.001 |
| AQP9 expression (ref. negative) | 3.443 | 1.058–11.205 | 0.040 | 1.714 | 1.258–2.335 | 0.001 |
p value less than 0.05 are in italics
PFS progression-free survival, TCGA The Cancer Genome Atlas, FUSCC Fudan University Shanghai Cancer Center
Table 3.
Multivariate Cox logistic regression analysis of OS in TCGA and FUSCC cohort
| Covariates | TCGA | FUSCC | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | |
| Age | 1.297 | 0.849–1.982 | 0.229 | 1.011 | 0.998–1.025 | 0.094 |
| pT stage (ref. T1–T2) | 1.692 | 0.740–3.871 | 0.213 | 1.692 | 1.011–2.830 | 0.045 |
| pN stage (ref. N0) | 1.488 | 0.740–2.993 | 0.265 | 1.837 | 1.173–2.877 | 0.008 |
| pM stage (ref. M0) | 2.629 | 1.578–4.381 | < 0.001 | 1.895 | 1.137–3.159 | 0.014 |
| AJCC stage (ref. I–II) | 1.279 | 0.510–3.207 | 0.600 | 3.553 | 1.814–6.956 | < 0.001 |
| ISUP grade (ref. 1–2) | 1.509 | 0.916–2.486 | 0.106 | 1.751 | 1.196–2.562 | 0.004 |
| AQP9 expression (ref. negative) | 1.707 | 1.067–2.731 | 0.026 | 1.514 | 1.050–2.183 | 0.026 |
p value less than 0.05 are in italics
OS overall survival, TCGA: The Cancer Genome Atlas, FUSCC Fudan University Shanghai Cancer Center
Prognostic value of AQP9 in TCGA and FUSCC cohorts
In TCGA cohorts, survival analysis showed that elevated AQP9 expression was significantly correlated with shorter PFS (p = 0.009) and OS (p < 0.001). In FUSCC cohort, survival curves suggested that elevated AQP9 mRNA levels in patients significantly correlated with poorer PFS and OS (p < 0.001; Fig. 3e, f). For high AQP9 expression patients, the median PFS was 39.5 months and the median OS was 59.5 months. For low AQP9 expression patients, the median PFS was 66 months and the median OS was 72 months. ROC curves were generated to identify the ability of the gene model to predict prognosis events.
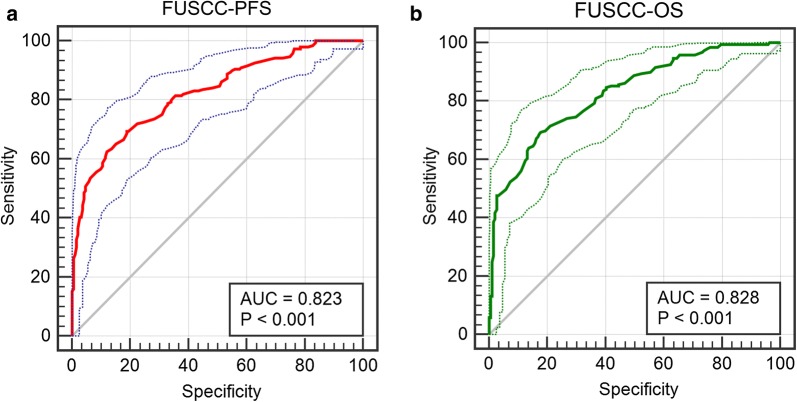
After integrating all the significant clinicopathological parameters and gene expression profiles in the Cox regression models (Table 2), we generated the formula: 1.782 × pT stage (ref. T1–T2) + 1.937 × pN stage (ref. N0) + 1.763 × pM stage (ref. M0) + 2.425 × AJCC stage (ref. I–II) + 1.812 × ISUP grade (ref. 1–2) + 1.714 × AQP9 expression (ref. Low) for PFS; and another formula: 1.692 × pT stage (ref. T1–T2) + 1.837 × pN stage (ref. N0) + 1.895 × pM stage (ref. M0) + 3.553 × AJCC stage (ref. I–II) + 1.751 × ISUP grade (ref. 1–2) + 1.514 × AQP9 expression (ref. Low) for OS. The AUC indices for the FUSCC-PFS and FUSCC-OS were 0.823 and 0.828, respectively (p < 0.001; Fig. 4a, b).
Fig. 4.
ROC curves were generated to validate the ability of the logistic model to predict prognosis. a The AUC index for the FUSCC-PFS were 0.823 (p < 0.001). b The AUC index for the FUSCC-OS were 0.828 (p < 0.001)
Functional annotations and predicted signaling pathways
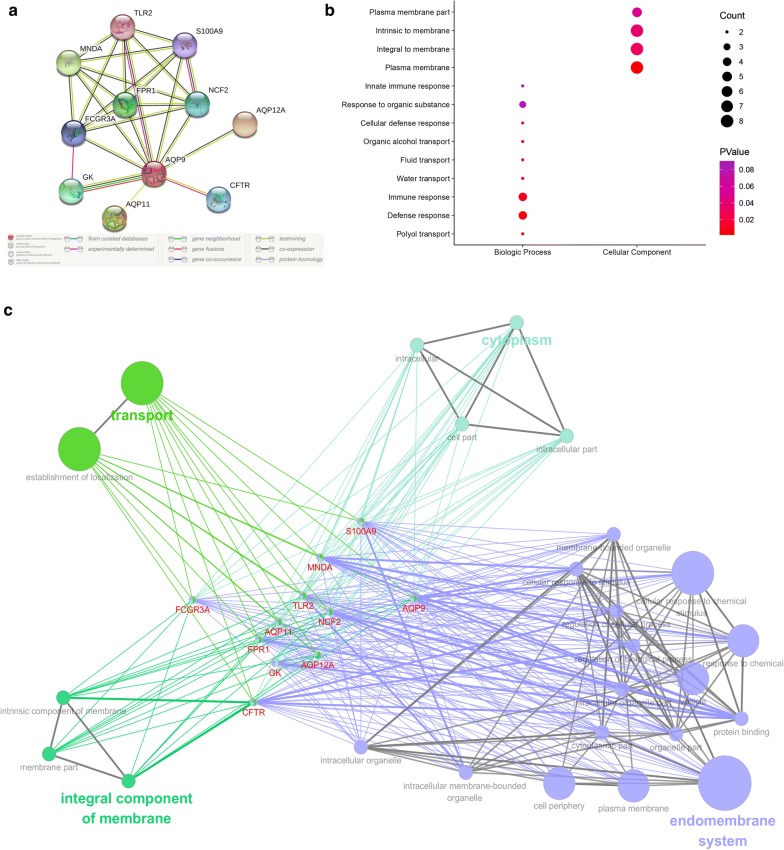
A network of AQP9 and its co-expression genes is shown in Fig. 5a. As illustrated in Fig. 5b, functional enrichment analyses of 11 involved genes were performed and the results are visualized in a bubble chart. Significant genes were significantly involved in polyol transport, defense response, and immune response, markedly participated in plasma membrane and were integral to membrane and intrinsic to membrane and the plasma membrane part. As shown in Fig. 5c, functional annotation using ClueGO indicated that changes in the biological processes of the AQP9 were significantly associated with the transport, integral component of membrane and the endomembrane system. Detailed functional annotations information and the percentage of each term were illustrated in Additional file 2: Figure S2.
Fig. 5.
Functional annotations and predicted signaling pathways. a The PPI network of AQP9 was constructed. A network of AQP9 and its co-expression genes was set up visually. b Functional enrichment analyses of a total of 11 involved genes were performed and visualized in bubble chart. Significant genes were significantly involved in polyol transport, defense response, immune response, and markedly participated in plasma membrane, integral to membrane, intrinsic to membrane and plasma membrane part. c Functional annotation using ClueGO indicated that changes in the biological processes of the AQP9 were significantly associated with the transport, integral component of membrane and endomembrane system
Significant genes and pathways obtained by GSEA
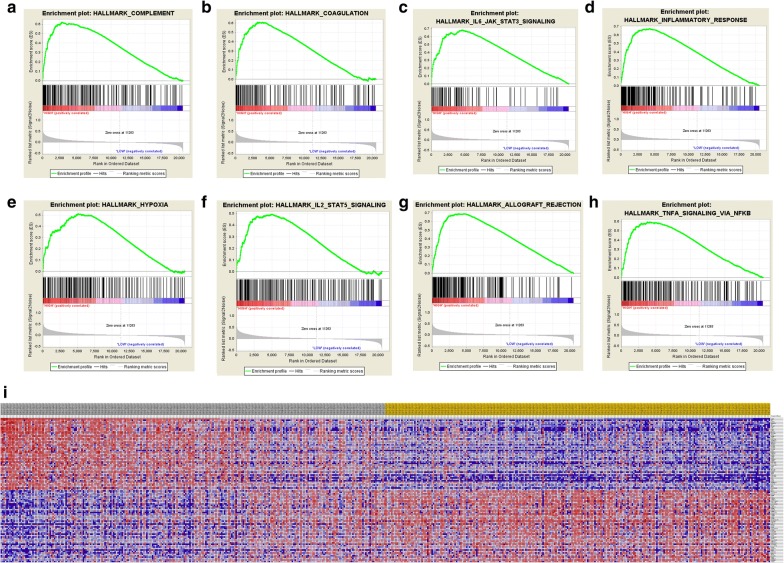
A total of 100 significant genes were obtained by GSEA with positive and negative correlations. GSEA was used to perform hallmark analysis for AQP9. The results suggested that the most of the involved significant pathways included complement, coagulation, IL6/JAK–STAT3 signaling, inflammatory response, hypoxia, IL2–STAT5 signaling, allograft rejection and TNF-A signaling via NF-κB. The details are shown in Fig. 6a–h. In addition, transcriptional expression profiles of the 100 significant genes are shown by heat map in Fig. 6i.
Fig. 6.
Significant related genes and hallmarks pathways in ccRCC obtained by GSEA. a–h The most involved significant pathways included complement, coagulation, IL6/JAK/STAT3 signaling, inflammatory response, hypoxia, IL2/STAT5 signaling, allograft rejection and TNF-alpha signaling via NFKB. i Transcriptional expression profiles of a total of 100 significant genes with positive and negative correlation were performed in a heat map
Correlation of AQP9 and immune infiltration level
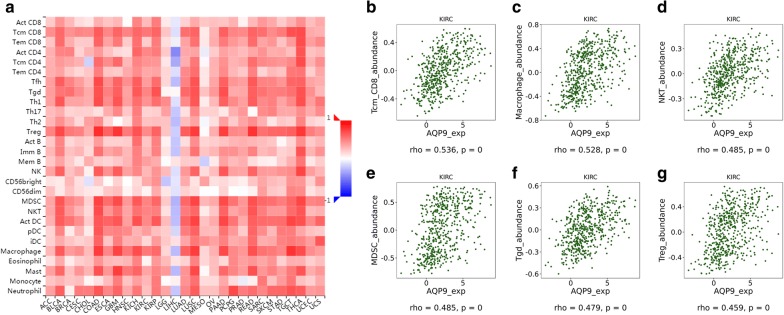
After determining the prognostic value of AQP9, we performed correlation analysis between AQP9 and immune infiltration level for ccRCC. Elevated AQP9 was significantly associated with B cell, T cell, monocyte, macrophage, tumor-associated macrophage, and neutrophil cell infiltration (p < 0.05), leading to a general increase in immune infiltration. The Spearman’s correlation showed estimated statistical significance between AQP9 expression and immune cell signature infiltration in Additional file 3: Table S4. Partial correlation and correlation adjusted by tumor purity are also provided. Important signatures of a variety of immune cells including CD8 + T cells, T cells (general), B cells, monocytes, tumor-associated macrophages, M1 macrophages, M2 macrophages, neutrophils, natural killer cells, dendritic cells, Th1, Th2, Tfh, Th17, regulatory T cells (Treg), T cell exhaustion are illustrated in Additional file 3: Table S4. Additionally, we also found significant correlations of AQP9 with 28 types of TILs across human heterogeneous cancers (Fig. 7a). AQP9 significantly correlated with abundance of central memory CD8 T cells (Tcm_CD8 T cells; rho = 0.536, p < 0.001), macrophage (rho = 0.528, p < 0.001), natural killer T cells (NK T cells; rho = 0.485, p < 0.001), myeloid derived suppressor cells (MDSC; rho = 0.485, p < 0.001), gamma delta T cells (Tgd cells; rho = 0.479, p < 0.001) and Treg (rho = 0.459, p < 0.001) in Fig. 7b–g.
Fig. 7.
Correlations between expression of AQP9 and TILs across human cancers. a Relations between expression of AQP9 and 28 types of TILs across human heterogeneous cancers. b–g AQP9 significantly correlated with abundance of central memory CD8 T cells (Tcm_CD8 T cells; rho = 0.536, p < 0.001), macrophage (rho = 0.528, p < 0.001), natural killer T cells (NK T cells; rho = 0.485, p < 0.001), myeloid derived suppressor cells (MDSC; rho = 0.485, p < 0.001), gamma delta T cells (Tgd cells; rho = 0.479, p < 0.001) and Treg (rho = 0.459, p < 0.001)
Discussion
Cancer genetics as well as abnormal epigenetic regulation have been found to participate in the progression and tumor environment for ccRCC [6]. The AQP family plays an important role in the development and progression of tumors such as breast cancer [32], nasopharyngeal carcinoma [33], and cervical cancer [34]. Although some members of the AQP family were demonstrated to be carcinogenic in many neoplasms, the prognostic value of AQP9 in ccRCC had remained to be elucidated. In this study, the expression levels and prognostic values of AQP9 in ccRCC were evaluated. We observed a significant increase in AQP9 expression in RCC that was associated with malignant behavior. Our data also indicated that high levels of AQP9 protein expression correlated with a high risk of recurrence and reduction in patient survival. These results reveal a new way for AQP9 expression to influence the pathogenesis of RCC through potential DNA damage variants. Functional enrichment and GSEA analysis illustrated that AQP9 was significantly involved in the most significant hallmarks pathways including inflammatory response, IL6/JAK–STAT3 signal pathway, IL2–STAT5 signal pathway, complement, and TNF-alpha signal pathway in RCC samples.
Inflammation is observed in basic physiological processes and is one of the hallmarks of many neoplasms [35]. Cancer-associated inflammation involves crosstalk between malignant and non-malignant cells in an autocrine and paracrine manner through mediators such as cytokines, chemokines and prostaglandins [36]. In combination with genetic alterations, the inflammatory tumor environment ultimately leads to tumor progression and metastasis [37]. For example, in the absence of the p53 tumor suppressor gene, the inflammatory response that is correlated with epithelial cell senescence significantly promotes transformation and carcinogenesis, which can be inhibited by anti-inflammatory drugs [38]. Treatment with the anti-inflammatory drug dexamethasone also markedly inhibits tumor cell transmission by inhibiting epithelial-to-mesenchymal transition (EMT), a process by which epithelial cells acquire migration and invasion properties [39].
AQP9 in RCC regulates a range of inflammation-related signaling pathways such as IL6/JAK–STAT3, IL2–STAT5 and TNF-alpha signal pathways. Previous research demonstrated that the IL-6/JAK–STAT3 pathway is aberrantly hyperactivated in many carcinomas, and hyperactivation was generally associated with unfavorable clinical prognosis [40]. In the tumor microenvironment, IL-6/JAK–STAT3 signaling promotes proliferation, invasiveness, and metastasis of tumor cells, while strongly suppressing the antitumor immune response [40]. Interleukin-6 (IL-6) is the major cytokine that induces transcriptional acute and chronic inflammation responses and a recurrence prognostic marker for localized ccRCC [41]. In addition, STAT3 is the main mediator of IL-6-induced RCC proliferation [42].
The ability of IL-2 to expand T cells with maintenance of functional activity has been translated into the first reproducible effective human cancer immunotherapies [43]. The use of cytokines from the IL-2 family (also known as the common gamma chain cytokine family) such as IL-2, IL-7, IL-15 and IL-21 to activate the immune system of cancer patients is currently the one of the most important fields of cancer immunotherapy research [43]. Infusion of IL-2 in multiple cycles at distinct doses in patients with metastatic melanoma and RCC has led to the first success in cancer immunotherapy, demonstrating that the immune system can completely eradicate tumor cells under certain conditions [44]. In this study, GSEA analysis indicated that AQP9 regulated the IL2–STAT5 signaling pathway in ccRCC patients. The IL2–STAT5 signaling pathway is involved in immune-related anti-tumor effects, promotes cancer cell proliferation, and interacts with other core cancer-related pathways. Clinical application of IL-2 to exert anti-tumor effects while inhibiting the STAT5 signaling pathway may be an effective treatment strategy for renal cancer.
Decades of research have shown that TNF is a core player in a complex network of cytokines that not only regulates pro-inflammatory responses, but also regulates cellular communication, cell differentiation and cell death including apoptosis and necroptosis [45]. The TNF family receptor Fas was recently found to promote terminal differentiation of CD4+ and CD8+ T cells, while non-apoptotic Fas signaling induces tumor cell growth and impairs the efficacy of T cell adoptive immunotherapy [46]. The AQP family is also involved in multiple TNA-alpha-induced events [47]. Blocking the non-apoptotic function of these receptors may be a new strategy to enhance anti-tumor immunity.
The relationship between AQP9 expression and carcinogenesis or prognosis of RCC has been rarely reported. However, it is worth noting that AQP9 promotes a series of immune responses and tumor environment, which are estimated to be highly expressed in many cancers [6, 48, 49]. Thus, here we used the TCGA database to evaluate the differential AQP9 expression between tumor and normal tissues, and we validated the prognostic value of AQP9 in the FUSCC cohort with long follow-up information. Furthermore, to uncover the prognostic significance of AQP9, co-regulatory proteins were included in the PPI network. Functional enrichment analysis was measured in hub gene panels. In addition, data from public databases was implemented by GSEA analysis to identify important genes and hallmark pathways, which may shed light on the association that triggers carcinogenesis.
This study has several limitations. First, only transcriptomics expression of AQP9 with clinical data was analyzed to predict PFS and OS in this study. Although differential AQP9 expression was detected between tumor and normal tissues, the prognostic implication of this finding has not been demonstrated. Second, the underlying mechanisms of signaling pathways in RCC remain unclear, while a serious of function annotations and enrichment analysis were investigated. Future research is required to explore the detailed mechanism between distinct AQP9 and carcinogenesis of ccRCC and reveal the mechanism of AQP9 in other carcinomas.
Conclusions
Our study demonstrated that elevated AQP9 expression was significantly correlated with cancer progression, poor survival and immune infiltrations in ccRCC patients from multiple cohorts. This study provides new and promising insights for subsequent research to elucidate the molecular pathogenesis of ccRCC. Randomized clinical trials and further studies are required to identify the underlying mechanism and clinical applications for ccRCC patients.
Supplementary information
Additional file 1: Figure S1. Differential expression and prognostic value of AQPs (0-11) family number for ccRCC patients from TCGA cohort.
Additional file 2: Figure S2. Functional annotations using CluePedia of Cytoscope for AQP9 and its 10 neighbor genes. A. List of the genes count number in different functions in the form of histogramns. B. The proportion of different functional categories, displayed in the form of a pie chart.
Additional file 3: Table S1. Clinicopathological characteristics baseline in relation to AQP9 expression status in TCGA cohort. Table S2. Univariate Cox logistic regression analysis of PFS in TCGA and FUSCC cohort (PFS: progression-free survival; TCGA: the Cancer Genome Atlas; FUSCC: Fudan university shanghai cancer center). Table S3. Univariate Cox logistic regression analysis of OS in TCGA and FUSCC cohort (OS: overall survival; TCGA: the Cancer Genome Atlas; FUSCC: Fudan university shanghai cancer center). Table S4. Correlation analysis between AQP9 and immune cell infiltrations in ccRCC samples using TIMER.
Acknowledgements
We thank Liwen Bianji, Edanz Editing China (http://www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Abbreviations
- RCC
renal cell carcinoma
- ccRCC
clear cell renal cell carcinoma
- FUSCC
Fudan University Shanghai Cancer Center
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
protein–protein interaction
- TCGA
the Cancer Genome Atlas
- GEO
gene expression omnibus
- PFS
progression-free survival
- OS
overall survival
- HR
hazard ratio
- CI
confidence interval
- GSEA
gene set enrichment analysis
Authors’ contributions
The work presented here was carried out in collaboration among all authors. YWD, ZHL and QYY defined the research theme, discussed analyses, interpretation, and presentation. XWH and SSN drafted the manuscript, analyzed the data, developed the algorithm and interpreted the results. XY co-worked on associated data collection, cohort validation and helped to draft the manuscript. WHK, CDL and SGH helped to perform the statistical analysis and reference collection. All authors read and approved the final manuscript.
Funding
This work is supported by Grants from the National Natural Science Foundation of China (No. 81802525) and Shanghai “Rising Stars of Medical Talent” Youth Medical Talents–Specialist Program.
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The Ethics approval and consent to participate of the current study was approved and consented by the ethics committee of Fudan University Shanghai Cancer center.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wen-Hao Xu, Shen-Nan Shi and Yue Xu contributed equally to this work
Contributor Information
Wen-Hao Xu, Email: xwhao0407@163.com.
Shen-Nan Shi, Email: 525958018@qq.com.
Yue Xu, Email: 654800813@qq.com.
Jun Wang, Email: 270801543@qq.com.
Hong-Kai Wang, Email: whhkkk123@163.com.
Da-Long Cao, Email: fudan_kenly@163.com.
Guo-Hai Shi, Email: guohaishi@126.com.
Yuan-Yuan Qu, Phone: 86-21-64175590, Email: quyy1987@163.com.
Hai-Liang Zhang, Phone: 86-21-64175590, Email: zhanghl918@163.com.
Ding-Wei Ye, Phone: 86-21-64175590, Email: 2406841044@qq.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12967-019-2113-y.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Baldewijns MM, et al. Genetics and epigenetics of renal cell cancer. Biochim Biophys Acta. 2008;1785(2):133–155. doi: 10.1016/j.bbcan.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Linehan WM, et al. The metabolic basis of kidney cancer. Cancer Discov. 2019;9(8):1006–1021. doi: 10.1158/2159-8290.CD-18-1354. [DOI] [PubMed] [Google Scholar]
- 5.Xu WH, et al. Procollagen-lysine, 2-oxoglutarate 5-dioxygenases 1, 2, and 3 are potential prognostic indicators in patients with clear cell renal cell carcinoma. Aging (Albany NY) 2019;11(16):6503–6521. doi: 10.18632/aging.102206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu WH, et al. Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment. Aging (Albany NY) 2019;11(17):6999. doi: 10.18632/aging.102233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agre P, et al. Aquaporin CHIP: the archetypal molecular water channel. Am J Physiol. 1993;265(4 Pt 2):F463–F476. doi: 10.1152/ajprenal.1993.265.4.F463. [DOI] [PubMed] [Google Scholar]
- 8.Castle NA. Aquaporins as targets for drug discovery. Drug Discov Today. 2005;10(7):485–493. doi: 10.1016/S1359-6446(05)03390-8. [DOI] [PubMed] [Google Scholar]
- 9.Verkman AS, Mitra AK. Structure and function of aquaporin water channels. Am J Physiol Renal Physiol. 2000;278(1):F13–F28. doi: 10.1152/ajprenal.2000.278.1.F13. [DOI] [PubMed] [Google Scholar]
- 10.Verkman AS. Aquaporin water channels and endothelial cell function. J Anat. 2002;200(6):617–627. doi: 10.1046/j.1469-7580.2002.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verkman AS. Aquaporins in clinical medicine. Annu Rev Med. 2012;63:303–316. doi: 10.1146/annurev-med-043010-193843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi YH, et al. Significance and expression of aquaporin 1, 3, 8 in cervical carcinoma in Xinjiang Uygur women of China. Asian Pac J Cancer Prev. 2012;13(5):1971–1975. doi: 10.7314/APJCP.2012.13.5.1971. [DOI] [PubMed] [Google Scholar]
- 13.Morrissey JJ, et al. Evaluation of urine Aquaporin-1 and Perilipin-2 concentrations as biomarkers to screen for renal cell carcinoma: a prospective cohort study. JAMA Oncol. 2015;1(2):204–212. doi: 10.1001/jamaoncol.2015.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, et al. Aquaporin 3 promotes prostate cancer cell motility and invasion via extracellular signal-regulated kinase 1/2-mediated matrix metalloproteinase-3 secretion. Mol Med Rep. 2015;11(4):2882–2888. doi: 10.3892/mmr.2014.3097. [DOI] [PubMed] [Google Scholar]
- 15.Cui G, et al. IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity. Cell. 2015;161(4):750–761. doi: 10.1016/j.cell.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv Y, et al. AQP9 promotes astrocytoma cell invasion and motility via the AKT pathway. Oncol Lett. 2018;16(5):6059–6064. doi: 10.3892/ol.2018.9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19(1A):A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asplund A, et al. Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research. Proteomics. 2012;12(13):2067–2077. doi: 10.1002/pmic.201100504. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, et al. Elevated MRE11 expression associated with progression and poor outcome in prostate cancer. J Cancer. 2019;10(18):4333–4340. doi: 10.7150/jca.31454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 22.Franceschini A, et al. STRING v91: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang DW, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smoot ME, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bindea G, et al. ClueGO: a Cytoscape plug-into decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29(5):661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ru B, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 29.Yusenko MV, et al. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beroukhim R, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69(11):4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gumz ML, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13(16):4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 32.Li X, et al. Effect of AQP-5 silencing by siRNA interference on chemosensitivity of breast cancer cells. Onco Targets Ther. 2018;11:3359–3368. doi: 10.2147/OTT.S160313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, et al. The AQP-3 water channel is a pivotal modulator of glycerol-induced chloride channel activation in nasopharyngeal carcinoma cells. Int J Biochem Cell Biol. 2016;72:89–99. doi: 10.1016/j.biocel.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Shen Q, et al. Differential expression of Aquaporins in cervical precursor lesions and invasive cervical cancer. Reprod Sci. 2016;23(11):1551–1558. doi: 10.1177/1933719116646202. [DOI] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12(10):584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 37.Singh R, Mishra MK, Aggarwal H. Inflammation, Immunity, and Cancer. Mediators Inflamm. 2017;2017:6027305. doi: 10.1155/2017/6027305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pribluda A, et al. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell. 2013;24(2):242–256. doi: 10.1016/j.ccr.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Q, et al. Prognostic value of interleukin-6 and interleukin-6 receptor in organ-confined clear-cell renal cell carcinoma: a 5-year conditional cancer-specific survival analysis. Br J Cancer. 2015;113(11):1581–1589. doi: 10.1038/bjc.2015.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horiguchi A, et al. STAT3, but not ERKs, mediates the IL-6-induced proliferation of renal cancer cells, ACHN and 769P. Kidney Int. 2002;61(3):926–938. doi: 10.1046/j.1523-1755.2002.00206.x. [DOI] [PubMed] [Google Scholar]
- 43.Rosenberg SA. IL-2: the first effective immunotherapy for human cancer. J Immunol. 2014;192(12):5451–5458. doi: 10.4049/jimmunol.1490019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sim GC, Radvanyi L. The IL-2 cytokine family in cancer immunotherapy. Cytokine Growth Factor Rev. 2014;25(4):377–390. doi: 10.1016/j.cytogfr.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 45.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3(9):745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 46.Yi F, et al. Beyond cell death: new functions for TNF family cytokines in autoimmunity and tumor immunotherapy. Trends Mol Med. 2018;24(7):642–653. doi: 10.1016/j.molmed.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagahara M, et al. TNF-alpha-induced aquaporin 9 in synoviocytes from patients with OA and RA. Rheumatology (Oxford) 2010;49(5):898–906. doi: 10.1093/rheumatology/keq028. [DOI] [PubMed] [Google Scholar]
- 48.Huang D, et al. AQP9-induced cell cycle arrest is associated with RAS activation and improves chemotherapy treatment efficacy in colorectal cancer. Cell Death Dis. 2017;8(6):e2894. doi: 10.1038/cddis.2017.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu L, et al. Significant prognostic values of aquaporin mRNA expression in breast cancer. Cancer Manag Res. 2019;11:1503–1515. doi: 10.2147/CMAR.S193396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Differential expression and prognostic value of AQPs (0-11) family number for ccRCC patients from TCGA cohort.
Additional file 2: Figure S2. Functional annotations using CluePedia of Cytoscope for AQP9 and its 10 neighbor genes. A. List of the genes count number in different functions in the form of histogramns. B. The proportion of different functional categories, displayed in the form of a pie chart.
Additional file 3: Table S1. Clinicopathological characteristics baseline in relation to AQP9 expression status in TCGA cohort. Table S2. Univariate Cox logistic regression analysis of PFS in TCGA and FUSCC cohort (PFS: progression-free survival; TCGA: the Cancer Genome Atlas; FUSCC: Fudan university shanghai cancer center). Table S3. Univariate Cox logistic regression analysis of OS in TCGA and FUSCC cohort (OS: overall survival; TCGA: the Cancer Genome Atlas; FUSCC: Fudan university shanghai cancer center). Table S4. Correlation analysis between AQP9 and immune cell infiltrations in ccRCC samples using TIMER.
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.