Abstract
Introduction
Non-small cell lung cancer (NSCLC) is a common cause of deaths all over the world. Emerging evidence has indicated that microRNA (miR) play key roles in NSCLC progression. We aimed to determine the functions of miR-129 in NSCLC. miR-129 was dramatically downregulated in NSCLC tissue samples and cells. The decreased miR-129 was found to be associated with poorer prognosis and malefic phenotype of NSCLC patients. We demonstrated that miR-129 upregulation could inhibit NSCLC cell growth. Furthermore, we also sought the molecular mechanism by which miR-129 repressed NSCLC development.
Methods
QRT-PCR was applied to detect the expressions of miR-129 in 51 pairs of NSCLC tissue samples. We further performed the Kaplan–Meier analysis to determine the association between miR-129 expressions and the survival rate of NSCLC patients. We then measured the expression levels of miR-129 in NSCLC cell lines. After that, MTT assays were performed to determine the influence of miR-129 on A549 cell proliferation. Transwell assay was then conducted to explore the biological functions of miR-129 in invasion and migration of NSCLC cells.
Results
Results showed that ZEB2 was directly targeted by miR-129 in NSCLC cell lines. Moreover, miR-129 restoration could inhibit EMT and Wnt/β-catenin in NSCLC cell lines.
Conclusion
In short, all these results indicated that miR-129/ZEB2 axis maybe a useful diagnostic and prognostic biomarker for NSCLC treatment.
Keywords: miR-129, ZEB2, NSCLC, Wnt/β-catenin, EMT
Introduction
Lung cancer has the highest morbidities and mortalities worldwide.1 Particularly, 85% of the cases with lung cancer are non-small cell lung cancer (NSCLC), which is the leading cause for lung cancer deaths.2 Currently, surgery is the main treatment for NSCLC, and adjuvant chemotherapy has gradually become common in patients with proper indications post-operation.3 Moreover, after surgical resections and other interventions, the 5-year survival rate for NSCLC remains below 50%.4 NSCLC is still one of the most challenging cancers in the clinic, despite the emergence of targeted biological agents and novel cytotoxic drugs.5 Therefore, it is necessary to search the factors of NSCLC pathogenesis for the improvement of clinical therapies.
MicroRNAs (miRNAs/miRs) could regulate gene expressions by targeting the 3ʹ UTRs in different kinds of cellular processes.6 It has been shown that miRNA plays key roles in tumorigenesis. For example, miR-411 was found to inhibit the malignant behaviors in colorectal carcinoma via regulation of PIK3R3;7 dysregulation of miR-567 could contribute to carcinogenesis of breast cancer;8 miR-544 promoted colorectal cancer progression by targeting forkhead box O1.9 Moreover, functional research of miRNAs shows that miRNAs are almost involved in all biological processes, such as cell metastasis, growth, differentiation and apoptosis.10–12 Therefore, in cancer progression, over-expressed or down-regulated miRNAs may be potential candidates for therapeutic interventions. In addition, miRNA, which regulates the responses of tumor cells to chemotherapy, could be inhibited or over-expressed as an adjuvant for tumor therapy. Importantly, recent studies have indicated the underlying significance of miR-129 in diagnosis and prognosis predictions of NSCLC.13,14 However, the mechanism of miR-129 remains largely unknown. In the current study, we explored the bio-functions of miR-129 in NSCLC to identify new biomarkers for effective diagnosis and prediction of prognosis in tumor treatments, which may exhibit significant implications in the clinic.
Zinc finger E-box binding homeobox 2 (ZEB2) is a member of the ZEB family of transcription factors.15 Studies have reported that ZEB2 was a regulator of epithelial-to-mesenchymal transition (EMT).16 In EMT, cells in epithelial phenotypes are converted into mesenchymal phenotypes with increased invasion and migration capacities. In this process, mesenchymal marker is upregulated while E-cadherin marker is downregulated.17 The overexpression of ZEB2 has been reported in different cancer types and has been suggested as a candidate biomarker for poor prognosis.18,19 Therefore, suppressing ZEB2 activation is a promising approach for suppressing cancer by inhibiting EMT. The signaling pathways known to be activated in NSCLC included Wnt/β-catenin signaling pathway, which regulated multiple processes involved in tumor growth, survival, migration, differentiation, and apoptosis.20–22 Consequently, this study investigated the roles of miR-129 in NSCLC Wnt/β-catenin and EMT, to provide new ideas for the efficacious developments of clinical therapy of NSCLC.
Materials And Methods
Tissue Samples
From October 2011 to June 2012, 51 pairs of NSCLC tissue samples and matched para-carcinoma tissue samples were collected from the Jinan City People’s Hospital after receiving written informed consent. All enrolled patients underwent no prior radiation therapy or chemotherapy. The collected tissue samples were frozen in liquid nitrogen and reserved at −80°C. Our study gains approval from the Ethics Committee of Jinan City People’s Hospital. All patients provided written informed consent. This study was conducted in accordance with the Declaration of Helsinki.
Cell Lines
Human NSCLC cells (NCI-H460, NCI-H1299, and A549) and normal bronchial epithelium cell line BEAS-2B were obtained from American Type Culture Collection (ATCC). The spca1 cell line was obtained from Shanghai Sure Shengwu Technology Co., LTD (Shanghai, China). All the cells were maintained in RPMI-1640 (Gibco; Thermo Fisher Scientifc, Inc., Waltham, MA, USA) including 10% FBS (Invitrogen, Carlsbad, CA, USA) at 37°C.
Cell Transfections
MiR-129 mimics, inhibitor or negative controls (NC) used in current study were obtained from GenePharma (Shanghai, China). miRNAs were transfected into NSCLC cells by Lipofectamine® 2000 (Invitrogen) following the manufacturer’s protocols.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated from cells and tissues with TRIzol (Invitrogen, Carlsbad, CA, USA), and the PrimeScript reverse transcription reagent kit (Thermo Fisher Scientific) was utilized for cDNAs synthesis. qPCR was conducted with SYBR® Premix Ex Taq™ II (Takara, Dalian, China) using a 7500 Fast Real-Time PCR detection system (Thermo Fisher Scientifc). 2−ΔΔCt method was utilized to calculate relative expressions. U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were internal controls. The primer sequences are list in Table 1.
Table 1.
Primer Sequences For qRT-PCR
| Primer | Sequence |
|---|---|
| miR-129 forward | 5ʹ-GTTGGGGAGATTTAGTTTGTT-3ʹ |
| miR-129 reverse | 5ʹ-CCTACTCCAATTCCCCCTATAATAC-3ʹ |
| U6 forward | 5ʹ-CTCGCTTCGGCAGCACA-3ʹ |
| U6 reverse | 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ |
| ZEB2 forward | 5ʹ-CAAGAGGCGCAAACAAGCC −3ʹ |
| ZEB2 reverse | 5ʹ-GGTTGGCAATACCGTCATCC-3ʹ |
| GAPDH forward | 5ʹ-ACCTGACCTGCCGTCTAGAA-3ʹ |
| GAPDH reverse | 5ʹ-TCCACCACCCTGTTGCTGTA-3ʹ |
Notes: U6: small nuclear RNA, snRNA.
Abbreviations: ZEB2, zinc finger E-box binding homeobox 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
MTT Assay
The cell proliferation ability was detected by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assays. In brief, NSCLC cells with different transfections (miR-129 mimics/inhibitor) were plated into 96-well plates and incubated for 0, 24, 48, 72hrs. Then, MTT (5 mg/mL) solution was appended into each well and incubated for another 4hrs. After that, 150 μL dimethyl sulfoxide (DMSO) was added. The OD490 values were determined by a microplate (BioTek, Winooski, VT, USA).
Transwell Assays
Transwell assay was carried out to explore the roles of miR-129 in invasive and migratory capacities of NSCLC cells with 8-µm pore sized transwell chamber (BD Biosciences, San Jose, CA, USA). For invasion assay, the upper chambers were precoated with a matrigel (BD Biosciences). The transfected cells at 1 × 105 cells/well resuspended in FBS-free medium were plated into the top chamber. Medium containing 10% FBS was added to the bottom chambers. After incubated for 48 hrs, the non-invading or -migrating cells were scraped whereas cells adhering to the bottom surface of the upper chamber were fixed and stained. Finally, the results were quantified by counting five independent visual fields under the microscope (Olympus).
Western Blot
The cell lysates were prepared with radioimmunoprecipitation assay (RIPA) buffer (Thermo Scientific) and a BCA Protein Assay Kit (Pierce, Rockford, IL, USA) was used to examine the protein concentrations. Then, the protein was loaded on 10% SDS-PAGE for separation and then transferred onto polyvinylidene fluoride (PVDF) membranes (Invitrogen) which were blocked with 5% skim milk in TBST at room temperature for 2hrs. Subsequently, the membrane was incubated overnight with primary first antibodies at 4°C. After that, the membranes were then incubated with HRP-labeled secondary antibody (1:3000, Abcam) at room temperature for 2hrs. The protein band was analyzed with ECL reagent (Beyotime). The following primary antibodies were used: antibodies against cyclin D1 (1:1000, Abcam Cambridge, MA, USA), c-Myc (1:1000, Abcam), β-catenin (1:1000, Abcam), p-GSK3β (1:1000, Abcam), total GSK3β (1:1000, Abcam), E-cadherin (1:2000, Abcam), N-cadherin (1:2000, Abcam), Vimentin (1:1000, Abcam), ZEB2 (1:1000, Abcam) and GAPDH (1:1000, Abcam). GAPDH was an internal reference.
Luciferase Reporter Assay
The mutant (MUT) or wild-type (WT) sequences containing the predicted target site of miR-129 in the 3ʹUTRs of ZEB2 mRNA were inserted into the pmir-GLO vectors (Promega Corporation, Madison, WI, USA). Then, 50 nM miR-129 mimics and the ZEB2 −3ʹUTR-MUT or ZEB2 −3ʹUTR-WT were cotransfected into NSCLC cells using Lipofectamine 2000; 5 ng of pRL‐SV40 was added per 80 ng of plasmid; 48 hrs after transfections, luciferase activity was detected by the Dual-Luciferase Reporter Assay system (Promega) in line with the manufacturers’ proposals.
Statistical Analysis
All data were from at least 3 independent experiments. Statistical analysis was performed using SPSS software version 17.0 (SPSS Inc., Chicago, IL). Differences between two groups were analyzed by using the Student’s t-test. Comparison between groups was done using one-way ANOVA test followed by Post Hoc Test (Least Significant Difference). The survival rate was analyzed by Kaplan–Meier analysis and log-rank test. P<0.05 was statistically significant difference.
Results
Downregulated miR-129 In NSCLC Tissue Samples Indicated A Poor Survival Rate
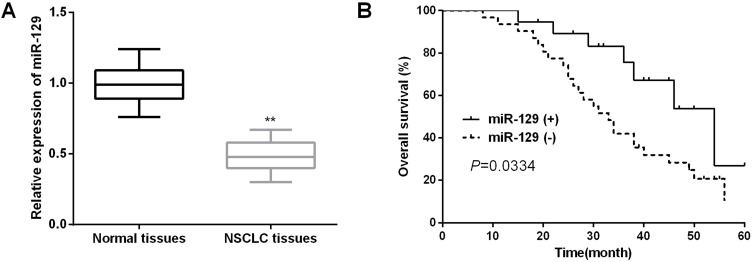
QRT-PCR was applied to detect the expressions of miR-129 in NSCLC tissue samples. As shown in Figure 1A, miR-129 expression in NSCLC tissues was dramatically decreased compared to the adjacent non-tumor tissue samples. We then explored the correlation between miR-129 level and clinical characteristics of NSCLC patients. As itemized in Table 2, the downregulated miR-129 was associated with the adverse clinicopathologic parameters of NSCLC patients. We further performed the Kaplan–Meier analysis to determine the association between miR-129 expressions and the survival rate of NSCLC patients. Results indicated that patients with low miR-129 group had a significantly poor overall survival (OS) than those in high miR-129 group (Figure 1B).
Figure 1.
Decreased miR-129 in NSCLC tissues indicated poor prognosis of NSCLC patients. (A) miR-129 expression in NSCLC tissues was detected by qRT-PCR. (B) Kaplan–Meier analysis was used to detect the OS of NSCLC patients with different miR-129 expressions. **P<0.01.
Table 2.
Correlation Of miR-129 Expression With The Clinicopathological Characteristics Of The NSCLC Patients
| Clinicopathological Features | Cases (n=51) | miR-129a Expression | P-Value | |
|---|---|---|---|---|
| High (n=20) | Low (n=31) | |||
| Age (years) | 0.4356 | |||
| >60 | 29 | 12 | 17 | |
| ≤60 | 22 | 8 | 14 | |
| Gender | 0.4632 | |||
| Male | 27 | 9 | 18 | |
| Female | 24 | 11 | 13 | |
| Tumor size (cm) | 0.2723 | |||
| ≥5.0 | 25 | 6 | 19 | |
| <5.0 | 26 | 14 | 12 | |
| Lymph node metastasis | 0.0014* | |||
| Yes | 21 | 16 | 5 | |
| No | 30 | 4 | 26 | |
| Histology | 0.2932 | |||
| Squamous cell carcinoma | 26 | 11 | 15 | |
| Adenocarcinoma | 25 | 9 | 16 | |
| TNM stage | 0.0021* | |||
| I+II | 23 | 17 | 6 | |
| III+IV | 28 | 3 | 25 | |
| Smoker | 0.5636 | |||
| Yes | 27 | 12 | 15 | |
| No | 24 | 8 | 16 | |
Notes: *Statistically significant. aThe mean expression level of miR-129 was used as the cutoff.
Abbreviations: NSCLC, non-small cell lung cancer; TNM, tumor-node-metastasis.
MiR-129 Overexpression Repressed NSCLC Cell Proliferation
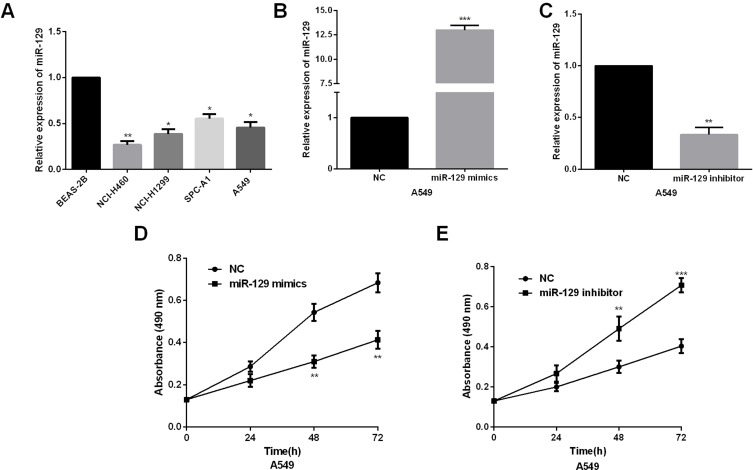
We then measured the expression levels of miR-129 in NSCLC cell lines. As expected, miR-129 was prominently downregulated in NSCLC cells in comparison with that in bronchial epithelium cell (Figure 2A). The effects of miR-129 on the biological phenotypes were investigated; 48hrs after transfection of miR-129 mimics or inhibitors, the transfection efficiencies in A549 cells were detected by qRT-PCR analysis. miR-129 expression was evidently increased by miR-129 mimics whereas decreased by miR-129 inhibitor in A549 cells (Figure 2B and C). After that, MTT assays were performed to determine the influence of miR-129 on A549 cell proliferation. miR-129 overexpression significantly inhibited A549 cell proliferation (Figure 2D). On the other hand, results also shown that A549 cell proliferation ability was dramatically promoted by miR-129 inhibition (Figure 2E).
Figure 2.
miR-129 overexpression inhibited NSCLC cell proliferation ability. (A) miR-129 expressions were detected in human NSCLC cells (NCI-H460, NCI-H1299, and A549) and normal bronchial epithelium cell line BEAS-2B by qRT‐PCR. (B and C) qRT‐PCR showed the expression of miR-129 in A549 cell transfected with miR-129 mimic or inhibitors. (D and E) MTT assay showed proliferation of A549 transfected with miR-129 mimic or inhibitors. *P<0.05, **P<0.01, ***P<0.001.
MiR-129 Upregulation Suppressed NSCLC Cell Invasion And Migration
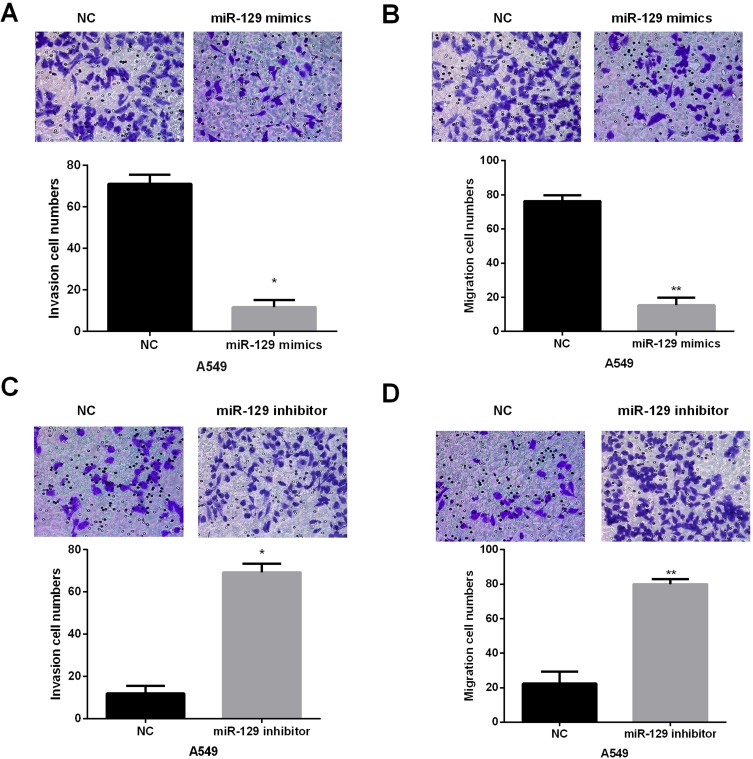
Transwell assay was then conducted to explore the biological functions of miR-129 in invasion and migration of NSCLC cells. Results indicated that miR-129-overexpressed A549 cells showed remarkably decreased invasion and migration abilities (Figure 3A and B). In contrast, the invasion and migration abilities were significantly promoted by miR-129 inhibition in A549 cells (Figure 3C and D). The above findings suggested that miR-129 served as an anti-tumor miRNA in NSCLC.
Figure 3.
miR-129 overexpression suppressed A549 cell migration and invasion. (A and B) The influence of miR-129 upregulation on A549 cell migration and invasion was detected by Transwell assays. (C and D) miR-129 inhibition facilitated A549 cell migration and invasion. *P<0.05, **P<0.01.
ZEB2 Was A Target Of miR-129 In NSCLC Cell Lines
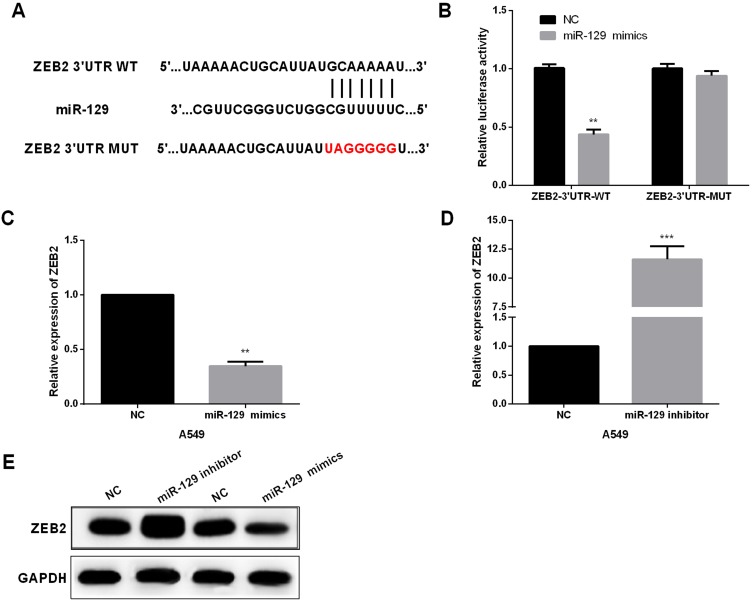
To investigate the potential mechanisms underlying miR-129-induced regulation of NSCLC biology, TargetScan was utilized to explore the targets of miR-129. ZEB2 was one candidate target of miR-129 (Figure 4A). Next, we performed luciferase reporter assays to confirm the above hypothesis. A549 cell lines were cotransfected with ZEB2-3ʹUTR-WT or -MUT and miR-129 mimics. As shown in Figure 4B, the luciferase activity was significantly decreased by miR-129 mimics in the ZEB2-3ʹUTR-WT group, while luciferase activity of ZEB2-3ʹUTR-MUT exhibited no obvious changes. Next, the regulatory functions of miR-129 in ZEB2 expressions were investigated. Results demonstrated that miR-129 upregulation led to a prominent repression of ZEB2 expressions, and on the other hand, transfection of miR-129 inhibitor remarkably promoted the ZEB2 expression (Figure 4C–E). Therefore, ZEB2 was a potential target of miR-129 in NSCLC cell lines.
Figure 4.
ZEB2 was directly targeted by miR-129 in NSCLC cell lines. (A) The binding sites of miR-129 on the 3ʹUTRs of ZEB2. (B) Luciferase reporter assays were carried out to confirm the correlation between miR-129 and ZEB2. (C–E) The expression of ZEB2 was detected by qRT‐PCR and Western blot in A549 after transfection with miR-129 mimic or inhibitors. **P<0.01, ***P<0.001.
MiR-129 Inhibited NSCLC Cell EMT And Wnt/β-Catenin Signaling Pathway
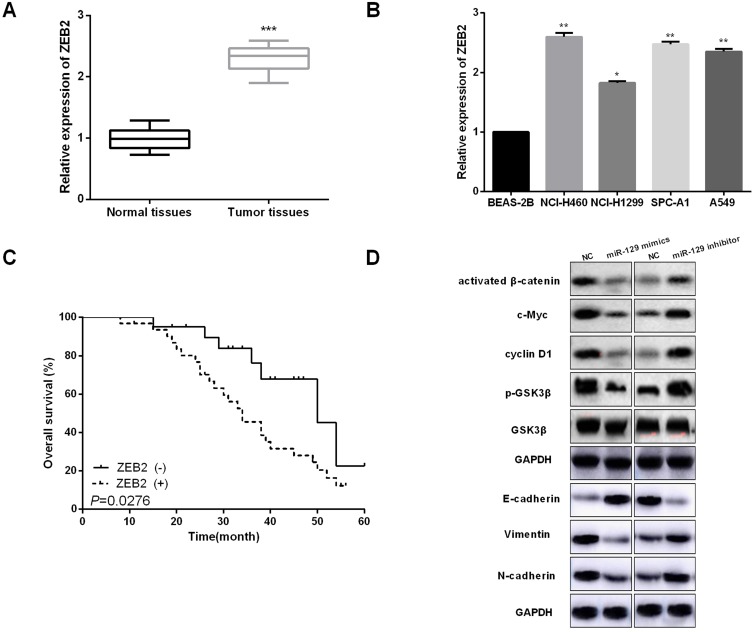
We then performed qRT-PCR to determine the expressions of ZEB2 in NSCLC tissues. Results indicated that ZEB2 in NSCLC tissues was mainly located in the nucleus (Figure 5A). Positive rates of ZEB2 protein expressions in adjacent normal tissue samples were evidently lower than that in NSCLC tissue samples (Figure 5B). Subsequently, we analyzed the influence of ZEB2 on the OS of NSCLC patients. It was found that patients with higher ZEB2 expression level presented lower OS than patients with lower ZEB2 expression level (Figure 5C). Next, Western blot analysis was used to detect the functions of miR-129 in NSCLC cell EMT and Wnt/β-catenin. As shown in Figure 5D, miR-129 upregulation significantly promoted E-cadherin expression while inhibited Vimentin and N-cadherin expressions in A549 cells, and on the other hand, miR-129 inhibition exhibited the opposite functions. Moreover, Western blotting results showed that the expressions of activated β-catenin, c-Myc, cyclin D1, and p-GSK3β were significantly decreased by miR-129 overexpression and were notably increased in cells with low miR-129 expressions (Figure 5D). Findings indicated that miR-129 participated in regulating NSCLC cell EMT and Wnt/β-catenin signaling pathway.
Figure 5.
miR-129 upregulation inactivated Wnt/β-catenin and EMT in NSCLC cell lines. (A and B) ZEB2 expressions in NSCLC tissues and cell lines were detected by qRT-PCR. (C) High ZEB2 expressions presented shorter OS of NSCLC patients by Kaplan–Meier analysis. (D) miR-129 overexpression suppressed Wnt/β-catenin and EMT in A549 cell. *P<0.05, **P<0.01, ***P<0.001.
MiR-129 Inhibited NSCLC Cell Progression Via Regulating ZEB2
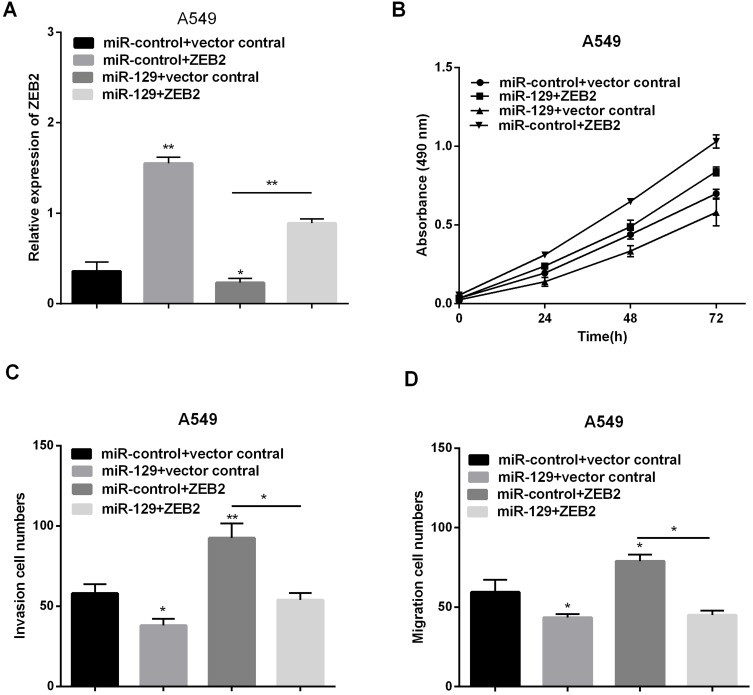
Since above results have indicated ZEB2 was direct target of miR-129 in NSCLC, ZEB2 might take part in miR-129-mediated inhibition of NSCLC cells. Therefore, to detect whether ZEB2 affects miR-129-induced function, we used miR-129 mimic to over express miR-129 in A549 cells and then transfect with ZEB2 plasmid in these cells to reverse ZEB2 expression. The mRNA level of ZEB2 was reduced by miR-129 mimic, and then increased after transfection with ZEB2 plasmid in A549 cells (Figure 6A). As estimated, miR-129 up-regulation inhibited NSCLC cells proliferation, migration and invasion, and ZEB2 plasmid evidently reversed miR-129 inhibited cell proliferation, migration and invasion as shown in Figure 6B–D. Overall, we deducted that ZEB2 was involved in miR-129-mediating inhibition of NSCLC progression.
Figure 6.
MiR-129 inhibited NSCLC proliferation, migration and invasion by targeting ZEB2. (A) RT-qPCR was used to detect ZEB2 expression after transfection with ZEB2 plasmid or miR-129 mimics. (B–D) Proliferation, migration and invasion were detected via MTT and transwell assays. *P<0.05, **P<0.01.
Discussion
NSCLC remains one of the main causes for cancer deaths all around the world due to a lack of understanding about the mechanism of NSCLC tumorigenesis.23 Novel biomarkers which may effectively assess the risks of tumor metastasis and recurrence, are worthy of exploration for NSCLC patients. Emerging studies have confirmed the dysregulation of miRNAs in NSCLC, indicating that aberrantly expressed miRNAs (or the target genes) play key roles in NSCLC progression.24 Therefore, to seek more effective treatments; contemporary research is centralized on mechanism of epigenetic or genetic NSCLC processes. For instance, Liao XH et al found that miR-500a-3p suppressed NSCLC cell invasion and proliferation by regulating LY6K.25 Chen Y et al reported that miR-148a served as a prognostic factor and suppressed migration and invasion through Wnt1 in NSCLC.26 Peng X et al revealed that miR-19 promoted NSCLC cell proliferation via inhibiting CBX7 expression.27
Our study provided functional evidence that miR-129 was important in NSCLC tumorigenesis. In particular, miR-129 expression was obviously decreased in NSCLC. Moreover, low miR-129 expression was related to malignant phenotypes and poor prognosis of patients with NSCLC. Findings also indicated that miR-129 overexpression prominently repressed NSCLC cell proliferation, invasion and migration capacities. Moreover, upregulation of miR-129 repressed EMT and Wnt/β-catenin in NSCLC cells. Additionally, miR-129 overexpression decreased the in vivo NSCLC growth. Altogether, these results demonstrated miR-129 exerted tumor-suppressive role in NSCLC by regulating EMT and Wnt/β-catenin.
The dysregulation of miR-129 is frequently identified in multiple types of human cancers, playing important roles in tumor occurrence and development.miR-129 was involved in the occurrence of uterine fibroid through inhibiting TET1.28 In colorectal cancer, miR-129 enhanced chemosensitivity and promoted apoptosis to 5-fluorouracil.29 miR-129 potentiated chemosensitivity and inhibited growth in neuroblastoma via regulation of MYO10.30 miR-129 inhibited prostate cancer development via PI3K/AKT/mTOR and targeting ETS1.31 These findings suggested that miR-129 can be developed into an effective therapeutic target for these types of cancer. In addition, multiple targets of miR-129 have been validated, including CDK1 and iASPP in Burkitt lymphoma,32 PAK5 in hepatocellular carcinoma,33 and MAL2 in papillary thyroid carcinoma.34 These studies suggested that miR-129 exhibited tissue-specific functions. ZEB2 confirmed as a functional target of miR-129 in NSCLC. Furthermore, ZEB2 was involved in miR-129-mediating inhibition of NSCLC progression. Given the important roles of ZEB2 in NSCLC, regulation of the miR-129/ZEB2/Wnt/β-catenin axis is a potential novel therapeutic strategy for NSCLC patients.
Conclusion
In conclusion, miR-129 was significantly down-regulated in NSCLC. Elevated miR-129 inhibited tumorigenesis and malignant progression of NSCLC. Moreover, we confirmed that the anti-NSCLC roles of miR-129 were regulated by ZEB2 and Wnt/β-catenin. Therefore, our findings suggest that miR-129 is a potential molecular target in NSCLC treatment.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou YW, Li R, Duan CJ, Gao Y, Cheng YD, Zhang CF. Overexpressed C14orf166 associates with disease progression and poor prognosis in non-small-cell lung cancer. Biosci Rep. 2018;38(5). doi: 10.1042/BSR20180479 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Giaccone G. Clinical impact of novel treatment strategies. Oncogene. 2002;21(45):6970–6981. doi: 10.1038/sj.onc.1205565 [DOI] [PubMed] [Google Scholar]
- 5.Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer Am Cancer Soc. 2015;121(5):664–672. doi: 10.1002/cncr.29098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Xu J, Zhang R. MicroRNA-411 inhibits malignant biological behaviours of colorectal cancer cells by directly targeting PIK3R3. Oncol Rep. 2018;39(2):633–642. doi: 10.3892/or.2017.6135 [DOI] [PubMed] [Google Scholar]
- 8.Bertoli G, Cava C, Diceglie C, et al. MicroRNA-567 dysregulation contributes to carcinogenesis of breast cancer, targeting tumor cell proliferation, and migration. Breast Cancer Res Treat. 2017;161(3):605–616. doi: 10.1007/s10549-016-4079-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao GD, Zhang YF, Chen P, Ren XB. MicroRNA-544 promotes colorectal cancer progression by targeting forkhead box O1. Oncol Lett. 2018;15(1):991–997. doi: 10.3892/ol.2017.7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G, Zhou H. MiRNA-708/CUL4B axis contributes into cell proliferation and apoptosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(17):5452–5459. doi: 10.26355/eurrev_201809_15805 [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Li F, Wang W, Wang X, Li S, Liu J. The effect of antisense inhibitor of miRNA 106b approximately 25 on the proliferation, invasion, migration, and apoptosis of gastric cancer cell. Tumour Biol. 2016;37(8):10507–10515. doi: 10.1007/s13277-016-4937-x [DOI] [PubMed] [Google Scholar]
- 12.Yu Y, Luo W, Yang ZJ, et al. miR-190 suppresses breast cancer metastasis by regulation of TGF-beta-induced epithelial-mesenchymal transition. Mol Cancer. 2018;17(1):70. doi: 10.1186/s12943-018-0818-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Wang H, Ke H, Ni S. MiR-129 regulates MMP9 to control metastasis of non-small cell lung cancer. Tumour Biol. 2015;36(8):5785–5790. doi: 10.1007/s13277-015-3247-z [DOI] [PubMed] [Google Scholar]
- 14.Liu MX, Zhou KC, Cao Y. MCRS1 overexpression, which is specifically inhibited by miR-129*promotes the epithelial-mesenchymal transition and metastasis in non-small cell lung cancer. Mol Cancer. 2014;13:245. doi: 10.1186/1476-4598-13-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santiago JJ, Dangerfield AL, Rattan SG, et al. Cardiac fibroblast to myofibroblast differentiation in vivo and in vitro: expression of focal adhesion components in neonatal and adult rat ventricular myofibroblasts. Dev Dyn. 2010;239(6):1573–1584. doi: 10.1002/dvdy.22280 [DOI] [PubMed] [Google Scholar]
- 16.Jacob S, Nayak S, Fernandes G, et al. Androgen receptor as a regulator of ZEB2 expression and its implications in epithelial-to-mesenchymal transition in prostate cancer. Endocr Relat Cancer. 2014;21(3):473–486. doi: 10.1530/ERC-13-0514 [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Yang C, Gao W, et al. FOXA2 attenuates the epithelial to mesenchymal transition by regulating the transcription of E-cadherin and ZEB2 in human breast cancer. Cancer Lett. 2015;361(2):240–250. doi: 10.1016/j.canlet.2015.03.008 [DOI] [PubMed] [Google Scholar]
- 18.Li MZ, Wang JJ, Yang SB, et al. ZEB2 promotes tumor metastasis and correlates with poor prognosis of human colorectal cancer. Am J Transl Res. 2017;9(6):2838–2851. [PMC free article] [PubMed] [Google Scholar]
- 19.Dai YH, Tang YP, Zhu HY, et al. ZEB2 promotes the metastasis of gastric cancer and modulates epithelial mesenchymal transition of gastric cancer cells. Dig Dis Sci. 2012;57(5):1253–1260. doi: 10.1007/s10620-012-2042-6 [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Duan N, Zhang Q, et al. DNA methylation mediated down-regulation of miR-370 regulates cell growth through activation of the Wnt/beta-catenin signaling pathway in human osteosarcoma cells. Int J Biol Sci. 2017;13(5):561–573. doi: 10.7150/ijbs.19032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Lu Q, Xie W, Wang Y, Wang G. Anti-tumor effects of triptolide on angiogenesis and cell apoptosis in osteosarcoma cells by inducing autophagy via repressing Wnt/beta-Catenin signaling. Biochem Biophys Res Commun. 2018;496(2):443–449. doi: 10.1016/j.bbrc.2018.01.052 [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Ji L, Jiang G, et al. FL118, a novel camptothecin analogue, suppressed migration and invasion of human breast cancer cells by inhibiting epithelial-mesenchymal transition via the Wnt/beta-catenin signaling pathway. Biosci Trends. 2018;12(1):40–46. doi: 10.5582/bst.2017.01288 [DOI] [PubMed] [Google Scholar]
- 23.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 24.Wang K, Chen M, Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World J Surg Oncol. 2017;15(1):175. doi: 10.1186/s12957-017-1244-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao XH, Xie Z, Guan CN. MiRNA-500a-3p inhibits cell proliferation and invasion by targeting lymphocyte antigen 6 complex locus K (LY6K) in human non-small cell lung cancer. Neoplasma. 2018;65(5):673–682. doi: 10.4149/neo_2018_170516N355 [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Min L, Ren C, et al. miRNA-148a serves as a prognostic factor and suppresses migration and invasion through Wnt1 in non-small cell lung cancer. PLoS One. 2017;12(2):e171751. doi: 10.1371/journal.pone.0171751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng X, Guan L, Gao B. miRNA-19 promotes non-small-cell lung cancer cell proliferation via inhibiting CBX7 expression. Onco Targets Ther. 2018;11:8865–8874. doi: 10.2147/OTT.S181433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu JL, Zhao L, Han SC, et al. MiR-129 is involved in the occurrence of uterine fibroid through inhibiting TET1. Eur Rev Med Pharmacol Sci. 2018;22(14):4419–4426. doi: 10.26355/eurrev_201807_15492 [DOI] [PubMed] [Google Scholar]
- 29.Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013;4:e659. doi: 10.1038/cddis.2013.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Li J, Xu X, Zheng J, Li Q. miR-129 inhibits tumor growth and potentiates chemosensitivity of neuroblastoma by targeting MYO10. Biomed Pharmacother. 2018;103:1312–1318. doi: 10.1016/j.biopha.2018.04.153 [DOI] [PubMed] [Google Scholar]
- 31.Xu S, Ge J, Zhang Z, Zhou W. MiR-129 inhibits cell proliferation and metastasis by targeting ETS1 via PI3K/AKT/mTOR pathway in prostate cancer. Biomed Pharmacother. 2017;96:634–641. doi: 10.1016/j.biopha.2017.10.037 [DOI] [PubMed] [Google Scholar]
- 32.Zou H, Zou R, Chen K, et al. miR-129 targets CDK1 and iASPP to modulate Burkitt lymphoma cell proliferation in a TAp63-dependent manner. J Cell Biochem. 2018;119(11):9217–9228. doi: 10.1002/jcb.27189 [DOI] [PubMed] [Google Scholar]
- 33.Zhai J, Qu S, Li X, et al. miR-129 suppresses tumor cell growth and invasion by targeting PAK5 in hepatocellular carcinoma. Biochem Biophys Res Commun. 2015;464(1):161–167. doi: 10.1016/j.bbrc.2015.06.108 [DOI] [PubMed] [Google Scholar]
- 34.Gao X, Chen Z, Li A, Zhang X, Cai X. MiR-129 regulates growth and invasion by targeting MAL2 in papillary thyroid carcinoma. Biomed Pharmacother. 2018;105:1072–1078. doi: 10.1016/j.biopha.2018.06.050 [DOI] [PubMed] [Google Scholar]