Abstract
To probe into the mechanism of antibiotic-associated diarrhea (AAD), the bacterial diversity and composition in the intestinal mucosa of AAD mice were investigated. Twelve specific pathogen-free Kunming mice were divided into control group and model group. The mouse model of AAD was established by gavaging with antibiotics (mixture of gentamycin sulfate and cefradine) at a total dose of 23.33 ml kg−1 day−1 for 5 days continuously, twice a day. The mice in the control group were given with an equal amount of sterile water. Then, the intestinal mucosa DNA was extracted for 16S rRNA gene sequence analysis by high-throughput sequencing. The results showed that the alpha diversity of the two groups did not differ significantly from each other, while the composition of intestinal mucosa bacteria differed dramatically between the two groups. The model group showed a higher abundance of Proteobacteria and Actinobacteria. More importantly, Lactobacillus was significantly less abundant (p = 0.000), while Enterococcus was significantly more abundant (p = 0.019) in the model group than in the control group. Furthermore, antibiotic treatment increased the abundance of Citrobacter, Stenotrophomonas, and Glutamicibacter,whereas antibiotics decreased the abundance of Mycoplasma and Helicobacter. In addition, 6 and 11 unique genera were found in the control group and model group, respectively. The combination of gentamycin sulfate and cefradine changed the intestinal mucosa bacterial composition, reduced colonization resistance and damaged the intestinal mucosal barrier by reducing the abundance of Lactobacillus.
Keywords: Antibiotics, Intestinal mucosa, Bacterial diversity, 16S rRNA, Antibiotic-associated diarrhea
Introduction
Antibiotics are used for treating infections. However, extensive evidence supports that antibiotic exposure dramatically disturbs the normal gut microbiota, and stimulates pathogenic microorganism overgrowth (Hogenauer et al. 1998; Su et al. 2013). Then, intestinal microbial dysbiosis increases susceptibility to infections, and interferes with glucose metabolism and bile acid metabolism (increases polysaccharide and decreases short chain fatty acid contents) (Rodrigues et al. 2017; Jin et al. 2016). Furthermore, antibiotics are involved in the allergic and toxic effects on the intestinal mucosa (Hogenauer et al. 1998). These changes in the gut due to antibiotic exposure usually induce diarrhea, called antibiotic-associated diarrhea (AAD). Almost all antibiotics are associated with diarrhea, but the incidence of AAD differs with the antibiotics and varies in occurrence between 5% and 25% (Anand et al. 2017). Cephalosporins, amoxicillin clavulanate and clindamycin are more likely to induce AAD than other antibiotics (Silverman et al. 2017). Although previous studies have reported that several bacterial species, such as Clostridium difficile, Clostridium perfringens, Staphylococcus aureus, Candida species and Klebsiella oxytoca, were responsible for the initiation of AAD (Hogenauer et al. 1998; Anand et al. 2017), the majority of bacteria that characterize the dysbiosis of AAD induced by different antibiotics have not been fully identified.
The intestinal microbiota refers to the microorganisms in intestinal content and mucosa. The content microbiota is variable according to the diet, drugs, diseases, and environment, while the mucosal microbiota is relatively stable in individuals (Borgo et al. 2018). An abundance of microorganisms colonize the outer mucus layer, directly modulate epithelial and mucosal function (Nishino et al. 2018), and may be deeply involved in the occurrence of AAD. Mucosal probiotics, especially Lactobacillus, play a part in establishing the intestinal mucosa barrier by inducing epithelial cell proliferation, reducing intestinal mucosa permeability, and promoting mucous secretion (Brandtzaeg 2003; De Kivit et al. 2014; Fuccio and Guido 2013). Probiotics not only prevent pathogenic microorganisms from invading but also stimulate the maturation of the intestinal immune system and modulate metabolism (Jirillo et al. 2012; Calder and Hall 2012; Tian et al. 2018). Furthermore, the mucosal microbiota is considered to be related to the inflammatory response. Given these ideas, the mucosal microbiota is vital to study the relationship between disease and the gut microbiota.
In our previous studies, the effects of antibiotics on the diversity of intestinal content bacteria and the structure of intestinal mucosa were investigated (Zhang et al. 2013, 2014; Guo et al. 2014; Long et al. 2017). However, the effects on intestinal mucosa bacteria remain elusive. The objectives of this study were to investigate the impact of antibiotics on the diversity and composition of intestinal mucosa bacteria in AAD mice. These findings could be of value in illuminating the characterization of the gut microbiota in mice with AAD induced by gentamycin sulfate and cefradine.
Materials and methods
Animal preparation
Twelve specific pathogen-free (SPF) Kunming (KM) mice (six males and six females) weighing 20 ± 2 g were purchased from Hunan Slaccas Jingda Laboratory Animal Co., Ltd. with license number SCXK (Xiang) 2016-0002. All procedures involving animals were in accordance with the Institutional Animal Care and Use Committee of Hunan University of Chinese Medicine.
Reagents
Gentamicin sulfate (batch number: 5150307) and cefradine (batch number: 151101) were purchased from Yichang Renfu Pharmaceutical Co., Ltd. and Suzhou Zhonghua Pharmaceutical Industry Co., Ltd., respectively. Then, the antibiotic mixture was prepared at a concentration of 62.5 g L−1 (Zhang et al. 2014). Protease K, lysozyme, Tris-saturated phenol–chloroform-isoamyl alcohol (25:24:1), TE buffer and acetone were purchased from Beijing Dingguo Biotechnology Co., Ltd. Other reagents were prepared in the laboratory.
Animal treatment and sample collection
After being adaptive fed (2 days) under controlled conditions (temperature 23–25 °C, humidity 50–70%), mice were randomly divided into control group (mck) and model group (mmd), six mice (three males and three females) per group. Mice in the model group were gavaged with the mixture of gentamycin sulfate and cefradine (23.33 ml kg−1 day−1) , while mice in the control group were gavaged with an equal amount of sterile water, twice a day for 5 days (Zeng et al. 2012). At the end of the 5 days of antibiotic treatment, diarrhea symptoms occurred in the mice of the model group. All the mice were killed by cervical dislocation, and intestinal segments from the jejunum to the ileum were collected immediately. Intestinal surfaces were gently rinsed with saline to remove ingesta. Mucosal samples were obtained by coverslip. The mucosal samples of one male mouse and one female mouse in the same group were mixed and stored at 4 °C for further use (Jin et al. 2012).
PCR amplification and sequencing
Total DNA of the mucosal sample was extracted according to the protocol of our previous report (Wu et al. 2012). The V3+V4 variable region of bacterial 16S rRNA was amplified using the primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′).
The PCR amplification system (20 µL) contained 10 × Buffer (2.0 µL), 2.5 mmol L−1 dNTPs (2.0 µL), forward primer (5 µmol L−1) 0.8 µL, reverse primer (5 µmol L−1) (0.8 µL), rTaq Polymerase (0.2 µL), BSA (0.2 µL), template DNA (10 ng), and ddH2O (4 µL). The cycling parameters were as follows: 95 °C for 3 min, followed by 29 cycles at 95 °C for 30 s, 55 °C for 30 s and 72 °C for 45 s, and then 72 °C for 10 min. The PCR products were evaluated by electrophoresis in 2% agarose gel and purified using AxyPred Gel Extraction Kit (Axygen, Scientific Inc., Union City, CA, USA). After purification, the PCR products were quantified by QuantiFluor™-ST (Promega Corp., Madison, WI, USA), and then sequenced by the Illumina MiSeq sequencing platform (Illumina, San Diego, CA, USA). Sequencing was completed by Wuhan Frasergen Genetic Information Company.
Bioinformatics and statistical analyses
Raw DNA sequences were analyzed by quantitative insights into microbial ecology (QIIME) (http://qiime.org/). Selected sequences were clustered into operational taxonomic units (OTUs), with a threshold similarity of 97%, using USEARCH (version 7.1, http://drive5.com/uparse/). Taxonomical information of representative sequences was assigned using ribosomal database project (RDP) classifier with a bootstrap cutoff of 70%. Alpha diversity indices and rarefaction were calculated by MOTHUR (version v.1.30.1, http://www.mothur.org/) (Schloss et al. 2011). Principal component analysis (PCA) (Wang et al. 2012), principal coordinates analysis (PCoA) (Jiang et al. 2013), nonmetric multidimensional scaling (NMDS) (Noval Rivas et al. 2013), and linear discriminant analysis effect size (LEfSe) (Segata et al. 2011) were carried out by R software to analyze the diversity and similarity of bacterial species.
SPSS 21.0 software (IBM Corp., Armonk, NY, USA) was used for statistical analysis. The results were expressed as mean ± SE. To compare the significance of differences, the independent-samples t test was used, with p values < 0.05 indicating significance.
Results
Characteristics of sequencing
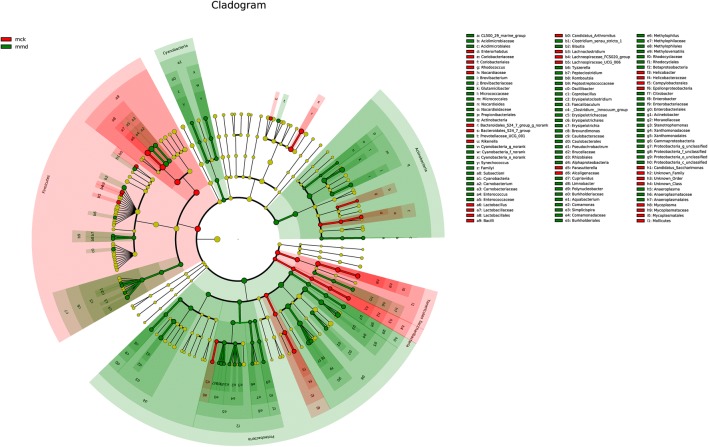
All the raw data were submitted to NCBI (Accession number: SRP223542, https://www.ncbi.nlm.nih.gov/Traces/sra_sub/). After quality control, 228,668 high-quality sequences (443.7 bp on average) were obtained from samples. The coverage of each sample reached 99.9%, which indicated that the sequences can reflect the actual situation of intestinal mucosa bacteria in the sample. Using the USEARCH software platform, OTU clusters are determined on non-repetitive sequences (excluding single sequences) according to 97% similarity. 325 and 341 OTUs were found from the control group and model group, respectively, and that 313 identical OTUs were shared by the two groups (Fig. 1). These findings showed that there was no significant difference in the DNA sequences of intestinal mucosa bacteria between the two groups.
Fig. 1.
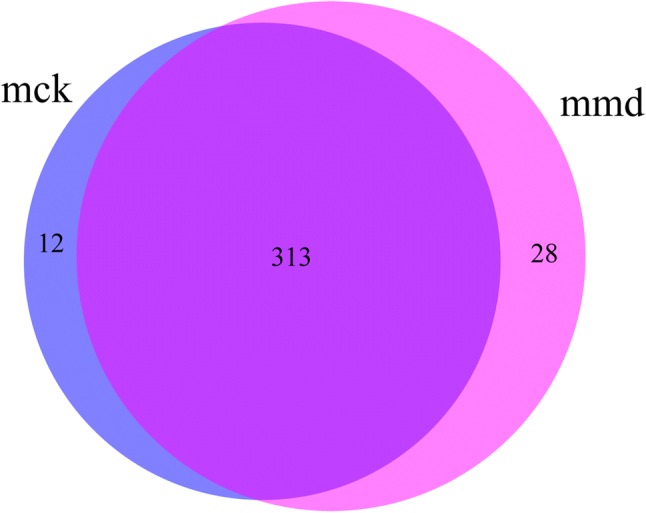
Effects of antibiotics on the intestinal mucosa bacterial OTU number. Venn diagram of OTUs based on the sequences with a threshold similarity of 97%. mck control group, mmd model group
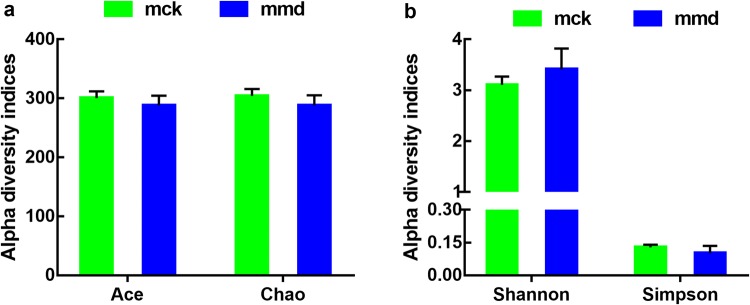
Alpha diversity analysis
Alpha diversity indices can reveal the richness and diversity of microbial communities. Generally, Chao and ACE reflect community richness, while Shannon and Simpson represent the diversity of species. In this study, Chao, Ace, and Simpson index values were higher, while the Shannon index value was lower in the control group than in the model group (p > 0.05, Fig. 2). The results indicated that antibiotics reduced the richness and increased the diversity of intestinal mucosa bacteria. However, the alpha diversity of the two groups did not differ significantly from each other.
Fig. 2.
Effects of antibiotics on the intestinal mucosa bacterial diversity. The alpha diversity indices used were Chao, ACE, Simpson and Shannon. Data are mean ± SE, n = 6. mck control group, mmd model group
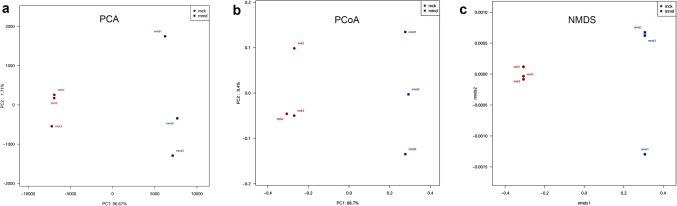
Beta diversity analysis
PCA was used to measure the similarity of sample composition, and to find the influencing factors on microbial composition in different groups. The percentage contributed to the variations of PC1 and PC2 was 96.67% and 1.71%, respectively (Fig. 3a). Furthermore, the samples in the control group were relatively concentrated, whereas the samples in the model group were discrete, and the distance between the two groups was far. The results revealed that antibiotics changed the composition of the intestinal mucosa.
Fig. 3.
Effects of antibiotics on the intestinal mucosa bacterial structure. a Principal component analysis (PCA); b principal coordinates analysis (PCoA); c nonmetric multidimensional scaling analysis (NMDS). Red points represent the control group (mck), and blue points represent the model group (mmd)
PCoA deeply investigated the homogeneity of microbial communities. The results showed that the percentage attributed to variations in PC1 and PC2 was 88.7% and 9.4%, respectively (Fig. 3b). PCoA highlighted a clear clustering of the microbial population of the model group away from that of the control group. This was further confirmed using NMDS analysis (Fig. 3c).
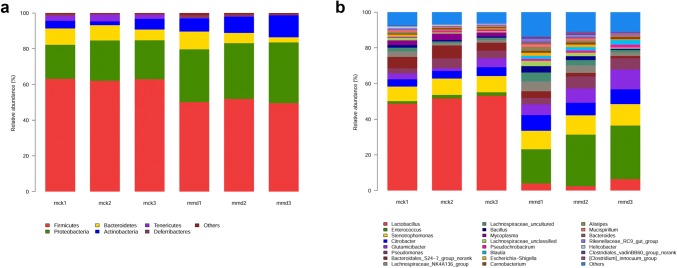
Comparison of intestinal mucosa bacteria at the phylum level
Taxon-based analysis was performed to assess specific changes in intestinal mucosa bacteria in response to antibiotic treatment. Significant differences in the composition of intestinal mucosa bacteria were found at different taxonomic levels. The microbial composition of each group at the phylum level is shown in Fig. 4a. Overall, fourteen phyla were identified. Firmicutes was the most predominant phylum in the control group and model group, accounting for 62.7% and 50.5%, respectively. Proteobacteria accounted for 21.2% and 31.6% of the control group and model group, respectively. In addition, Bacteroidetes (7.9% vs. 6.2%) and Tenericutes (2.9% vs. 0.2%) had a higher abundance in the control group than in the model group. In contrast, Actinobacteria (4.2% vs. 9.6%) and Deferribacteres (0.5% vs. 1.0%) were more abundant in the model group than in the control group. The relative abundance of Firmicutes (p = 0.000), Proteobacteria (p = 0.003), Actinobacteria (p = 0.041), and Tenericutes (p = 0.003) was significantly different between the two groups.
Fig. 4.
Microbiota structures are shown in histograms at the phylum level (a) and genus level (b). Each color represents a different taxonomy. Poorly represented taxa are grouped as others
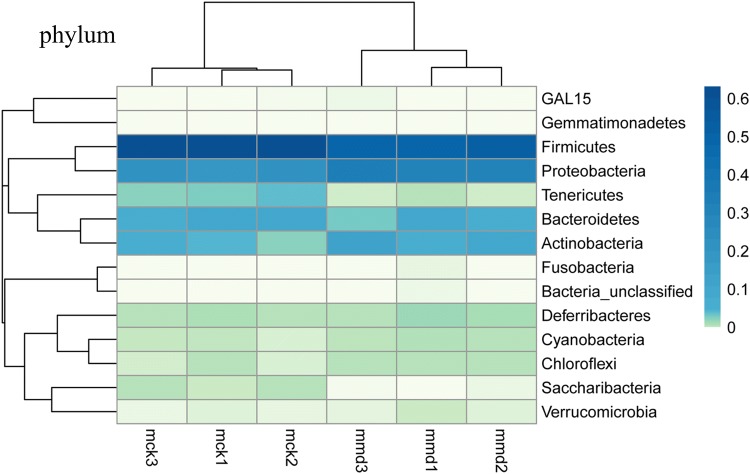
To further investigate the similarity and difference of microbial composition among samples, heatmap analysis was used. Both the diversity and abundance of bacterial species were clearly shown (Fig. 5). Species were clustered into dominant microbiota, rare microbiota and middle microbiota by the color gradient and similarity degree. Rare microbiota, including GAL15 and Gemmatimonadetes, were in the upper portion of the heatmap. Dominant microbiota, such as Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria and Tenericutes, were in the middle portion of the heatmap. Middle microbiota refers to Deferribacteres, Chloroflexi, Cyanobacteria, Saccharibacteria, Verrucomicrobia, Bacteria_unclassified, and Fusobacteria.
Fig. 5.
Effects of antibiotics on the intestinal mucosa bacterial abundance at the phylum level are shown in a heatmap. The darker the color is, the larger the values of the relative abundance
Comparison of intestinal mucosa bacteria at the genus level
By comparing with Silva (http://www.arb-silva.de) database, 151 genera were clarified. Genera in the control group were dominated by Lactobacillus (51.2%), Stenotrophomonas (8.8%), Bacteroidales_S24-7_group_norank (6.0%), Citrobacter (4.5%) and Pseudomonas (4.2%). Meanwhile, Enterococcus (26.0%), Stenotrophomonas (11.1%), Glutamicibacter (8.3%), Citrobacter (8.0%), and Pseudomonas (5.6%) were the main genera in the model group (Figs. 4b, 6). Bergeyella, Elioraea, Gemmatimonadetes_norank, Lachnospiraceae_FCS020_group, Odoribacter and WCHB1-41_norank were found only in the control group, whereas Campylobacter, Cetobacterium, Desulfatiglans, Fusobacterium, Haemophilus, Limnohabitans, Oricola, Pedobacter, Peptoclostridium, Proteiniborus and Rhodobacteraceae_unclassified were found only in the model group. Lactobacillus abundance (51.2% vs. 4.3%) was sharply reduced, while Enterococcus abundance (1.8% vs. 26.0%) conspicuously increased after antibiotics were administered.
Fig. 6.
Effects of antibiotics on the intestinal mucosa bacterial abundance at the genus level according to LEfSe. The red points represent the important bacteria in the control group (mck), and the green points represent the important bacteria in the model group (mmd)
A majority of the genera had significant differences in their relative abundance between the two groups. The relative abundance of Lactobacillus (51.2% vs. 4.3%, p = 0.000), Citrobacter (4.5% vs. 8.0%, p = 0.004), Carnobacterium (0.7% vs. 1.2%, p = 0.009), Enterococcus (1.8% vs. 26.0%, p = 0.019), Stenotrophomonas (8.8% vs. 11.1%, p = 0.019), Mycoplasma (2.8% vs.0.0%, p = 0.022), Glutamicibacter (3.2% vs. 8.3%, p = 0.045), Bacteroides (0.4% vs. 1.0%, p = 0.027), Helicobacter (0.8% vs. 0.1%, p = 0.018), Blautia (0.1% vs. 2.0%, p = 0.017) and [Clostridium]_innocuum_group (0.0% vs. 0.8%, p = 0.028) showed significant differences.
Discussion
Antibiotics and their adverse effects have caused wide public concern. Various studies have described antibiotics, AAD and AAD-inducing microorganisms. However, the relationship between the intestinal mucosa microbiota and AAD induced by specific antibiotic has not been fully explored. In this study, antibiotic mixture, which including a broad-spectrum antibiotic (gentamicin sulfate) and a narrow-spectrum antibiotic (cefradine), was selected to treat mice, and changes in the intestinal mucosa bacteria were determined. Gentamicin sulfate acts predominantly against Escherichia, Proteus and Enterobacter, but Enterococcus is resistant to it. However, cefradine targets Gram-positive cocci. Synergism has been observed when the two antibiotics are administered together (Zeng et al. 2012).
Contrary to previous reports (Puhl et al. 2012), we found that the alpha diversity increased slightly after antibiotic treatment. A dysbiotic microbiota is an imbalance in the intestinal microbial community characterized by quantitative and qualitative changes in the composition of the microbiota (Stecher et al. 2013). According to this concept, the diversity of the gut microbiota may increase or decrease during a state of dysbiosis. A higher diversity was also found in individuals with colorectal adenoma and mice with diabetes (Lu et al. 2017; Zhang et al. 2017). In this study, the slight change may be due to the ‘blooms’ of low-abundance bacteria, multidrug-resistant bacteria and pathogens (Kim et al. 2016).
Intestinal mucosa bacteria were dominated by Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria at the phylum level, while their abundances were significantly different (Borgo et al. 2018). An increased prevalence of Proteobacteria is a signature of dysbiosis and risk of disease (Shin et al. 2015). The ratio of Firmicutes to Bacteroidetes (F/B) was used to measure intestinal homeostasis. The ratio of Bifidobacterium to Enterobacterium (B/E) was an index of intestinal colonization resistance. In general, increasing F/B and decreasing B/E indicate that the intestinal microbiota was imbalanced and that the risk of intestinal pathogen invasion was greatly increased. In this study, the relative abundance of Proteobacteria increased sharply in the model group (21.2% vs. 31.6%, p = 0.003). Furthermore, the F/B value was higher (8.23 ± 1.94 vs. 10.46 ± 6.26) but the B/E value was lower (1.12 ± 0.56 vs. 0.29 ± 0.23) in the model group than in the control group. The results indicated that pathogenic bacteria multiplied and invaded the intestinal mucosa after antibiotic treatment and the intestinal mucosa microorganisms were disordered.
In this study, significant differences in the composition of intestinal mucosa bacteria were found at the genus levels. Among them, Lactobacillus sharply decreased from 51.2 to 4.3% after antibiotic treatment, which was in agreement with the results of our cultivation experiment (Guo et al. 2014; Zhang et al. 2013). Lactobacillus is one of the dominant bacteria in the healthy intestine. It can inhibit intestinal pathogenic bacteria, promote the development of auxiliary T cells, induce the production of cytokines, and enhance cellular immune function. Loss of Lactobacillus implies that colonization resistance and intestinal mucosal barrier were weakened, which then opened niches for opportunistic pathogenic bacteria (e.g., Enterococcus, Stenotrophomonas, Bacteroides) as well as inflammation-associated bacteria (e.g., Clostridium, Campylobacter, Fusobacterium), to expand. Some species of the above-mentioned genera are related to diseases, especially infections (Goh et al. 2017; Hellming et al. 2005; Larcombe et al. 2016; Fitzgerald 2015; Lee et al. 2016). The results implied that diarrhea induced by gentamicin sulfate and cefradine may be due to intestinal infections.
In addition to altering the bacterial composition, antibiotics also affect the gene expression, protein activity, and metabolism of the gut microbiota (Francino 2016). Expression of genes involved in antibiotic resistance, stress response and phage induction increased after antibiotic treatment (Francino 2016). In this study, the abundance of multidrug-resistant bacteria, such as some species of Enterococcus (Goh et al. 2017), Stenotrophomonas (Abbott and Peleg 2015) and Clostridium (Spigaglia et al. 2018), increased significantly after antibiotic treatment. We also focused on the functional genes expressed by the gut microbiota. Intestinal microbial function enzyme (lactase) gene was observed in our previous studies. The main lactase-producing strains differed in the intestinal content and mucosa. Stenotrophomonas is the main lactase-producing strain in the intestinal mucosa. Specifically, the diversity and abundance of bacterial lactase genes in the intestinal mucosa increased in the antibiotic treatment group compared with those in the control group, but the result in the intestinal content was the opposite (Long et al. 2017; Long et al. 2018). These findings showed that the comprehensive analysis of the content microbiota and mucosal microbiota from composition to function is important to explain the pathogenesis of AAD. In the next stage, we plan to apply the traditional Chinese medicine (TCM) formula for treating AAD, investigate the change in the content microbiota and mucosal microbiota, and discover how TCM works on AAD.
In summary, gentamicin sulfate and cefradine gently changed the diversity and severely destroyed the composition of intestinal mucosa bacteria. More importantly, gentamicin sulfate and cefradine significantly reduced the abundance of Lactobacillus and increased the risk of infection. The exact strains that are associated with AAD induced by gentamicin sulfate and cefradine require further research.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81573951; 81804076).
Author contributions
ZT designed the study; KT and MP performed the experiments; GX and CL analyzed the data; GX wrote the paper; DL checked the paper. The decision to submit the manuscript for publication was made by all the authors.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Human and animal rights
The study was approved by the Animal Ethics and Welfare Committee of Hunan University of Chinese Medicine.
Contributor Information
Dandan Li, Phone: (+86) 18874866037, Email: 48797696@qq.com.
Zhoujin Tan, Phone: (+86) 13974954942, Email: 003794@hnucm.edu.cn.
References
- Abbott IJ, Peleg AY. Stenotrophomonas, Achromobacter, and nonmelioid Burkholderia species: antimicrobial resistance and therapeutic strategies. Semin Respir Crit Care Med. 2015;36(1):99–110. doi: 10.1055/s-0034-1396929. [DOI] [PubMed] [Google Scholar]
- Anand A, Bharadwaj R, Pol S. Antibiotic associated diarrhea with special reference to Clostridium difficile. IJTDH. 2017;24(4):1–10. doi: 10.9734/IJTDH/2017/34541. [DOI] [Google Scholar]
- Borgo F, Garbossa S, Riva A, Severgnini M, Luigiano C, Benetti A. Body mass index and sex affect diverse microbial niches within the gut. Front Microbiol. 2018;9:213–227. doi: 10.3389/fmicb.2018.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandtzaeg P. Mucosal immunity: integration between mother and the breast-fed infant. Vaccine. 2003;21(24):3382–3388. doi: 10.1016/s0264-410x(03)00338-4. [DOI] [PubMed] [Google Scholar]
- Calder P, Hall V. Understanding gut-immune interactions in management of acute infectious diarrhea. Nurs Older People. 2012;24(9):29–37. doi: 10.7748/nop2012.11.24.9.29.c9367. [DOI] [PubMed] [Google Scholar]
- De Kivit S, Tobin MC, DeMeo MT, Fox S, Garssen J, Forsyth CB, et al. In vitro evaluation of intestinal epithelial TLR activation in preventing food allergic responses. Clin Immunol. 2014;154(2):91–99. doi: 10.1016/j.clim.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Fitzgerald C. Campylobacter. Clin Lab Med. 2015;35(2):289–298. doi: 10.1016/j.cll.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Francino MP. Antibiotics and the human gut microbiome: dysbiosis and accumulation of resistances. Front Microbiol. 2016;6:1543–1553. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccio L, Guido A. Probiotics supplementation for the prevention of gastrointestinal radiation-induced side effects: the time is now. Am J Gastroenterol. 2013;108(2):277. doi: 10.1038/ajg.2012.418. [DOI] [PubMed] [Google Scholar]
- Goh HMS, Yong MHA, Chong KKL, Kline KA. Model systems for the study of Enterococcal colonization and infection. Virulence. 2017;8(8):1525–1562. doi: 10.1080/21505594.2017.1279766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo KX, Yin KK, Wang H, Guo C, Zhao XP, Cao R. The effect of modelling dysbacterial diarrhea with antibiotics on molecular diversity of intestinal microbiota in mice. Chin J Microecol. 2014;26(3):249–252. doi: 10.13381/j.cnki.cjm.201403001. [DOI] [Google Scholar]
- Hellming S, Ott S, Musfeldt M, Kosmahl M, Rosenstiel P, Stüber E, et al. Life-threatening chronic enteritis due to colonization of the small bowel with Stenotrophomonas maltophilia. Gastroenterology. 2005;129(2):706–712. doi: 10.1016/j.gastro.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Hogenauer C, Hammer HF, Krejs GJ, Reisinger EC. Mechanisms and management of antibiotic-associated diarrhea. Clin Infect Dis. 1998;27(4):702–710. doi: 10.1086/514958. [DOI] [PubMed] [Google Scholar]
- Jiang XT, Peng X, Deng GH, Sheng HF, Wang Y, Zhou HW, et al. Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb Ecol. 2013;66(1):96–104. doi: 10.1007/s00248-013-0238-8. [DOI] [PubMed] [Google Scholar]
- Jin L, Yang XH, Ren JL, Li JL, Guo XY, Cao P, et al. Effect of dietary compound probiotics on disaccharidase in small intestine mucosa of layer breeders. Chin Poultry. 2012;34(12):14–17. [Google Scholar]
- Jin Y, Wu Y, Zeng Z, Jin C, Wu S, Wang Y, et al. From the cover: exposure to oral antibiotics induces gut microbiota dysbiosis associated with lipid metabolism dysfunction and low-grade inflammation in mice. Toxicol Sci. 2016;154(1):140–152. doi: 10.1093/toxsci/kfw150. [DOI] [PubMed] [Google Scholar]
- Jirillo E, Jirillo F, Magrone T. Healthy effects exerted by prebiotics, probiotics and symbiotics with special reference to their impact on the immune system. Int J Vitam Nutr Res. 2012;82(3):200–208. doi: 10.1024/0300-9831/a000112. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Choi YS, Baek KJ, Yoon SH, Park HK, Choi Y. Mucosal and salivary microbiota associated with recurrent aphthous stomatitis. BMC Microbiol. 2016;16(Suppl 1):57. doi: 10.1186/s12866-016-0673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcombe S, Hutton ML, Lyras D. Involvement of bacteria other than Clostridium difficile in antibiotic-associated diarrhoea. Trends Microbiol. 2016;24(6):463–476. doi: 10.1016/j.tim.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Lee Y, Eun CS, Lee AR, Park CH, Han DS. Fusobacterium isolates recovered from colonic biopsies of inflammatory bowel disease patients in Korea. Ann Lab Med. 2016;36(4):387–389. doi: 10.3343/alm.2016.36.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CX, He L, Guo YF, Liu YW, Xiao NQ, Tan ZJ. Diversity of bacterial lactase genes in intestinal contents of mice with antibiotics-induced diarrhea. World J Gastroenterol. 2017;23(42):7584–7593. doi: 10.3748/wjg.v23.i42.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CX, Liu YW, He L, Tan Q, YU Z, Xiao NQ, et al. Bacterial lactase genes diversity in intestinal mucosa of mice with dysbacterial diarrhea induced by antibiotics. 3 Biotech. 2018;8(3):176. doi: 10.1007/s13205-018-1191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YY, Zeng Y, Hu GY, Wang XP. High-throughput sequencing for analysis of structural change of intestinal microbiota in patients with colorectal adenoma. J South Med Univ. 2017;37(9):1156–1163. doi: 10.3969/j.issn.1673-4254.2017.09.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K, Nishida A, Inoue R, Kawada Y, Ohno M, Sakai S. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol. 2018;53(1):95–106. doi: 10.1007/s00535-017-1384-4. [DOI] [PubMed] [Google Scholar]
- Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Llore M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131(1):201–212. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl NJ, Uwiera RR, Yanke LJ, Selinger LB, Inglis GD. Antibiotics conspicuously affect community profiles and richness, but not the density of bacterial cells associated with mucosa in the large and small intestines of mice. Anaerobe. 2012;18(1):67–75. doi: 10.1016/j.anaerobe.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Rodrigues RR, Greer RL, Dong X, DSouza KN, Gurung M, Gurung Y, et al. Antibiotic-induced alterations in gut microbiota are associated with changes in glucose metabolism in healthy mice. Front Microbiol. 2017;8:2306. doi: 10.3389/fmicb.2017.02306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6(12):e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Silverman MA, Konnikova L, Gerber JS. Impact of antibiotics on necrotizing enterocolitis and antibiotic-associated diarrhea. Gastroenterol Clin North Am. 2017;46(1):61–76. doi: 10.1016/j.gtc.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spigaglia P, Mastrantonio P, Barbanti F. Antibiotic resistances of Clostridium difficile. In: Mastrantonio P, Rupnik M, editors. Updates on Clostridium difficile in Europe. Berlin: Springer; 2018. [Google Scholar]
- Stecher B, Maier L, Hardt WD. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11(4):277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- Su L, Zhang YB, Zhou RB. Treatment and prevention of antibiotic associated diarrhea. J Clin Emerg. 2013;14(5):240–242. doi: 10.1093/toxsci/kfw150. [DOI] [Google Scholar]
- Tian H, Zhao HL, Yang L, Wang N. Research progress of the microecology and intestinal mucosal barrier. Basic Clin Med. 2018;38(3):418–421. doi: 10.16352/j.issn.1001-6325.2018.03.026. [DOI] [Google Scholar]
- Wang Y, Sheng HF, He Y, Wu JY, Jiang YX, Tam NF, et al. Comparison of the levels of bacterial diversity in fresh water, intertidal wetland, and marine sediments by using millions of Illumina tags. Appl Environ Microbiol. 2012;78(23):8264–8267. doi: 10.1128/AEM.01821-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhou SN, Guo C, Tan ZJ, Cai GX, Zeng A, et al. A metagenome DNA extracting method of intestinal flora in mice for molecular diversity analysis based on PCR technology. Chin J Microecol. 2012;24(7):648–651. doi: 10.13381/j.cnki.cjm.2012.07.003. [DOI] [Google Scholar]
- Zeng A, Zhang HL, Tan ZJ, Cai Y, Cai GX, Zhou SN. The construction of mice diarrhea model due to dysbacteriosis and curative effect of ultra-micro Qiweibaizhusan. Microbiol Chin. 2012;39(9):1341–1348. doi: 10.13344/j.microbiol.china.2012.09.012. [DOI] [Google Scholar]
- Zhang HL, Zhou SN, Cai R, Guo KX, She Y, Tan ZJ, et al. The effect of ultra-micro Qiweibaizhusan on the mucous membrane of small intestine of diarrheal mice with dysbacteriosis. Chin J Microecol. 2013;25(1):9–14. doi: 10.13381/j.cnki.cjm.2013.01.007. [DOI] [Google Scholar]
- Zhang HL, Cai Y, Tan ZJ, Zhou SN, Guo KX, She Y. Effects of ultra-micro powder Qiweibaizhusan on metabolism diversity of intestinal microflora in diarrhea mice with dysbacteriosis. Chin J Appl Environ Biol. 2014;20(1):93–100. doi: 10.3724/SP.J.1145.2014.00093. [DOI] [Google Scholar]
- Zhang Q, Zhou ZK, Ren XC, Wang XF. Comparision of faecal microbiota in rats with type 2 diabetes and non-diabetic rats using MiSeq high-throughput sequencing. J Chin Instit Food Sci Technol. 2017;17(6):232–239. doi: 10.16429/j.1009-7848.2017.06.031. [DOI] [Google Scholar]