Abstract
Metagenomics is the study of gene pool of an entire community in a particular niche. This provides valuable information about the functionality of host-microbe interaction in a biological ecosystem. Efficient metagenomic DNA extraction is a critical pre-requisite for a successful sequencing run in a metagenomic study. Although isolation of human stool metagenomic DNA is fairly standardized, the same protocol does not work as efficiently in fecal DNA from other organisms. In this study, we report a comparison of manual and commercial DNA extraction methods for diverse samples such as human stool, fish gut and soil. Fishes are known to have variable microbial diversity based on their food habits, so the study included two different varieties of fishes. A modified protocol for effective isolation of metagenomic DNA from human milk samples is also reported, highlighting critical precautions. Recent studies have emphasized the importance of studying functionality of human milk metagenome to understand its influence on infants’ health. While manual method works well with most samples and therefore can be a method of choice for testing new samples, broad-range commercial kit offers advantage of high purity and quality. DNA extraction of different samples would go a long way in unraveling the unexplored association between microbes and host in a biological system.
Keywords: Metagenomic DNA, Stool, Fecal, Milk, Fish gut, Soil
Introduction
It is an undeniable fact that the microorganisms have played a crucial role in the sustenance of life on earth, since its formation. The diversity and abundance of these microscopic entities vary as per their surrounding environment [1–3]. So, the information about flourishing microbes in a particular environment is the key to understand the characteristics and the functional attributes of a particular habitat, and are often used as bioindicators [4–9]. This knowledge has greatly advanced with use of the next-generation DNA sequencing (NGS) technology. As the genome characterization of an organism offers insight into its unique characteristics (genomics); similarly, metagenomic characterization reflects the community ecosystem and offers clues about its functional composition [10, 11]. The prerequisite for a successful NGS run remains isolation of high-quality metagenomic DNA from the desired samples. Extensive work on the NGS over the past few years has led to introduction of variety of protocols to extract DNA from samples; nonetheless, most of these protocols are limited to soil DNA and human fecal DNA extraction. Protocols for efficient DNA extraction from samples such as fish gut, sputum, milk still remain a major challenge [12–14]. Several commercial DNA extraction kits are available in the market (Qiagen, MP Biomedicals, MoBio DNA), but these kits are mostly limited to the commonly studied samples such as soil and stool. Additionally, the manufacturers recommend use of separate kits for different samples for efficient recovery. Thus, any study involving a unique sample type requires testing with multiple kits to standardize extraction adding significantly to the study cost. Furthermore, variables such as sampling strategy, transport of samples and storage conditions may also produce different biases in DNA extraction process, resulting in variation within the same sample [15]. Other studies in the past have reported different methods for DNA extraction depending on the type of study sample [16–20]. Therefore, it is important to optimize the extraction protocol, to make it cost-effective and rapid to yield high-quality DNA fit for downstream applications such as polymerase chain reaction (PCR), 16S rRNA sequencing and whole genome sequencing from both human and environmental samples. Although, recent efforts have led to improvement of DNA purification methods after sampling, still, there are many existing problems that ultimately leads to the misidentification of different species within the microbial community [21–23].
Our study compares three metagenomic DNA extraction methods by comparing the quality of the isolated DNA and compatibility with downstream applications such as PCR. The two most commonly used commercial column-based kits from Qiagen and the manual lysozyme/phenol–chloroform method, commonly used for bacterial genomic DNA isolation method were tested for diverse samples such as human stool, fish gut and soil. We have also standardized isolation of metagenomic DNA from milk sample by these methods by introducing few modifications to the procedure.
Materials and Methods
Sample Collection
Four different types of samples were collected for the study namely: (1) garden soil (2) fish gut from silver carp (Hypophthalmichthys molitrix) and Himalayan golden mahseer (Tor putitora) (3) stool from healthy adult volunteers (4) milk from healthy nursing mother.
(a) Soil
5 g of sample were collected in sterile falcons, air dried and 200 mg sample was taken for further processing.
(b) Fish Gut
Silver carp (herbivorous fish, 25 inches weighing about 3.8 kg) and T. putitora (omnivorous fish, 23 inches weighing about 2.4 kg) samples were collected in sterile falcons from Gobind Sagar reservoir, Bhakra barrage, Bilaspur District, Himachal Pradesh. The fish were dissected aseptically and the gut contents were squeezed off from the alimentary canal and kept at − 80 °C till further processing.
(c) Milk
10 ml breast milk sample from a healthy nursing (1 week) mother was collected in a sterile container after cleaning the skin surface, at Maulana Azad Medical College and associated Hospital, Delhi, India. Samples were immediately stored in a refrigerator and then frozen to −20 °C within 2 h of collection after aliquoting. For DNA extraction, the samples were thawed in ice and transferred to Eppendorf microcentrifuge tubes, and centrifuged at 2000g for 20 min at 4 °C to separate the fat layer.
(d) Stool
5 g specimen was collected in a sterile container and within 4 h of collection, samples were kept at − 80 °C refrigeration till further processing.
For human samples informed written consent were collected from the volunteers.
Enzymes and Reagents
The commercial kits, Qiagen DNeasy Blood & Tissue Kit (Qiagen India Pvt. Ltd, India) and QIAamp DNA stool mini kit (Qiagen India Pvt. Ltd, India) were used in the extraction of metagenomic DNA. Homogenization buffer (100 mM Tris–HCl pH 8.0; 50 mM EDTA pH 8.0 and 1 M NaCl), sodium phosphate buffer (0.1 M, pH 7.4), and enzymes lysozyme (10 mg/ml), proteinase K (20 mg/ml), RNase A (10 mg/ml) were used in the manual method. The enzymes were purchased from Thermo Fisher Scientific.
DNA Extraction
DNA preparation of stool, milk, fish gut, and soil samples by Method A
Fresh environmental samples [1 g soil, 200 mg fish-gut samples] and human samples [200 mg stool, 1 ml milk (aqueous phase)] were aliquoted into a 2 ml tube (pre-chilled) and resuspended in 1 ml homogenization buffer. The samples were vortexed at regular intervals for complete mixing of sample with the buffer. From each homogenized sample, 750 µl was transferred to a new microcentrifuge tube. The samples were incubated with lysozyme at 60 °C for 1 h in a thermomixer (Eppendorf ThermoMixer® C) with mild shaking (350 rpm). The lysate was then incubated with 10 µl of RNase (1U/µl) at 37 °C for 30 min (RNase treatment may be given after total DNA extraction, but in the manual method, this may lead to a considerable loss of sample). 10 µl proteinase K (20 mg/mL) and 100 µl SDS (10% w/v) were successively added to the tubes, and again incubated at 60 °C for 1 h (with mild shaking). For phase extraction, phenol, chloroform and isoamyl alcohol (25:24:1) emulsion was added in equal volume and an aqueous phase was obtained after centrifugation at 8000g for 15 min. After transfer to a new microcentrifuge tube, this extraction step was repeated. The resultant supernatant was separated and 1/10th volume of 7.5 M sodium acetate (pH 5.4) and an equal volume of ice-cold isopropanol was added to it, followed by precipitation at − 80 °C for 20 min. This was followed by centrifugation at 8000g for 15 min to obtain the extracted DNA as a pellet, which was subjected to two washes with cold 70% ethanol at 8000g for 5 min each and the pellet was left for overnight air-drying. The following day, the DNA was resuspended in pre-heated 50 μl nuclease-free water. For milk sample, the aqueous phase collected after centrifugation was processed similarly as described above.
-
(b)
DNA preparation of stool, milk, fish gut, and soil samples by Method B
This method employed the use of Qiagen DNeasy Blood & Tissue Kit (Qiagen India Pvt. Ltd, India) for DNA isolation, as per manufacturer’s recommendations. Slight modifications were introduced in method B for efficient recovery of milk DNA and referred as modified method B. The milk aqueous phase was extracted with pre-warmed ATL buffer provided in the kit, with continuous vortexing. The homogenized sample was incubated at 75 °C for 10 min on a thermomixer (Eppendorf ThermoMixer® C), and centrifuged at 10,000g for 2 min. The clean supernatant (400 µl) was transferred to a fresh microcentrifuge tube and treated with Proteinase K (10 µl). The sample was not treated with inhibit®Ex tablets/buffers provided with the kit, rather directly proceeded to the next step of addition of AL buffer, followed by DNA binding, washing, and elution.
-
(c)
DNA preparation of stool, milk, fish gut, and soil samples by Method C
This method employed the use of QIAamp DNA Stool mini kit (Qiagen India Pvt. Ltd, India) for DNA isolation, as per manufacturer’s instructions. Slight modifications were introduced in method C for efficient recovery of milk DNA and referred as modified method C. The milk aqueous phase was extracted with pre-warmed ASL buffer provided in the kit, with continuous vortexing. This homogenate was further processed as the steps mentioned for modified method B protocol.
Assessment of Quantity and Quality of Isolated DNA Samples
The metagenomic DNA isolated from various samples were analysed by agarose gel electrophoresis system at 80 V in 0.8% gel with 5 mg/mL ethidium bromide in 1 × TAE buffer. The gel image was acquired using gel documentation system (Amersham 600, GE). The purity and concentration of isolated DNA were estimated by using Nanodrop™ 1000 spectrophotometer (Thermo Scientific). The DNA purity was determined by A260/A280 depicting DNA/protein and A260/A230 ratios which determine protein/phenol/salt contamination.
PCR
The quality of the isolated metagenomic DNA were assessed using 16S rRNA PCR amplification. The reaction was performed in a thermal cycler (Bio-Rad, USA) in a mixture containing 1 × KOD Buffer, 100 ng of DNA template, 2.5 mM dNTPs and 20 µM of each primer and 1U KOD Hot Start DNA Polymerase. The gene-specific primers (F-5′-CGATCCCTAGCTGGTCTGAG-3′, R-5′-GTTAGCCGGTGCTTCTTCTG-3′) amplified a 230 bp region of 16S ribosomal RNA (V3). The reaction conditions were: Initial denaturation at 95 °C for 5 min, followed by 35 cycles at 95 °C for 20 s, annealing at 64 °C for 20 s, and extension at 72 °C for 10 s with final extension at 72 °C for 10 s. The amplicons thus obtained were electrophoresed on a 1.2% agarose gel and visualized using a gel documentation system (Amersham 600, GE).
Restriction Digestion
The isolated DNA samples were subjected to restriction digestion in a reaction mixture containing 1 µg of DNA, 10 × Fast digestion buffer and BamHI restriction endonuclease (Thermo Fisher Scientific) at 37 °C for 60 min.
Statistical Analysis
All the reported methods were repeated twice for each sample and R software was used for statistical analysis to check the mean and standard deviation of independent experiments.
Result and Discussion
Variation in DNA Yield and Purity Depends on the Sample Type and DNA Extraction Method
The most critical step in any DNA extraction method is efficient cell lysis, since DNA of high purity and quantity is required for downstream applications such as PCR, restriction endonuclease mapping and NGS [24–26]. In the present study, we have used both environmental and human samples (soil, fish gut, human stool and human milk) for comparative analysis of DNA extraction in diverse samples. DNA isolation was done by manual method based on the principle of lysozyme/phenol–chloroform extraction (Method A) and commercial method based on silica-column based adsorption of DNA (Method B and C). As the microbial composition varies significantly with different niches, different samples require specialized modifications in protocols to extract DNA of high quality and quantity. Previous studies on microbial diversity from samples containing low bacterial diversity such as meconium, placenta and milk were reported to contain high contamination in the isolated DNA. This leads to data bias in diversity estimation and functional analysis [27–29]. As we go on exploring diverse niches to find microbes, it would be advantageous to find a standard procedure which works well with diverse samples. The assessment of quality and quantity of DNA was done by visualizing it on gel electrophoresis to note any degradation pattern and by spectrophotometric evaluation (Fig. 1 and Table 1).
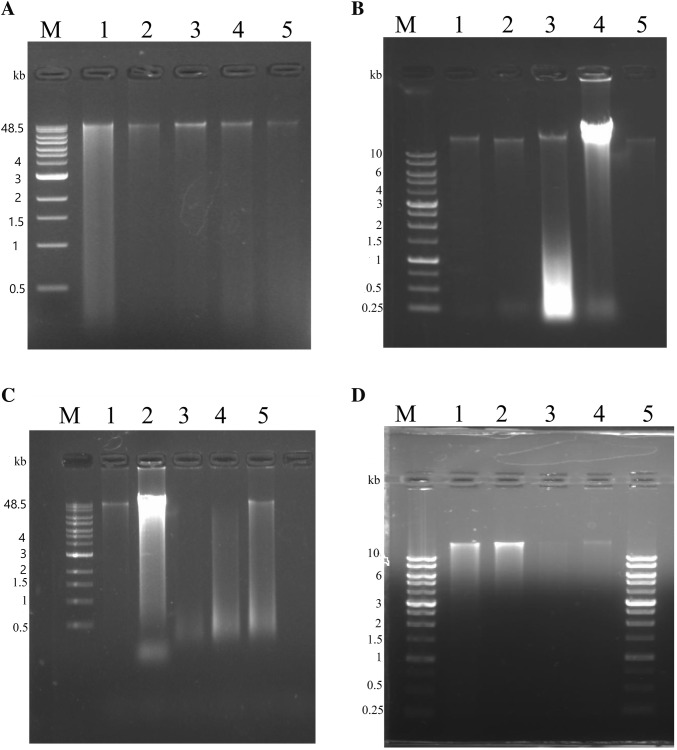
Fig. 1.
Gel electrophoresis of metagenomic DNA isolated from stool, fish gut, soil and milk samples. Metagenomic DNA was electrophoresed on a 0.8% agarose gel, stained with ethidium bromide and photographed in a gel documentation system. a Metagenomic DNA isolation by method A. Lane M: Quick-Load® 1 kb Extend DNA Ladder; lane 1: DNA isolated from stool sample; lane 2: DNA isolated from soil sample; lane 3: DNA isolated from fish gut (silver carp); lane 4: DNA isolated from fish gut (T. putitora); lane 5: DNA isolated from healthy mother’s milk. b Metagenomic DNA isolation by method B. Lane M: 1 Kb DNA Ladder RTU (GenedireX); lane 1: DNA isolated from fish gut (silver carp); lane 2: DNA isolated from fish gut (T. putitora); lane 3: DNA isolated from healthy mother’s milk; lane 4: DNA isolated from soil sample; lane 5: DNA isolated from stool sample. c Metagenomic DNA isolation by method C. Lane M: Quick-Load® 1 kb Extend DNA Ladder; lane 1: DNA isolated from soil; lane 2: DNA isolated from stool; lane 3: DNA isolated from fish gut (silver carp); lane 4: DNA isolated from fish gut (T. putitora); lane 5: DNA isolated from healthy mother’s milk. d Metagenomic DNA isolated from healthy mother’s milk. Lane M and lane 5: 1 kb DNA Ladder RTU (GenedireX); lane 1: DNA isolated from method A; lane 2: DNA isolated from modified method B; lane 3–4: DNA isolated from modified method C
Table 1.
Concentration of metagenomic DNA isolated from different human and environmental samples
| S. no. | Sample | Methods | Concentration in ng/µl (mean ± SD) | 260/280 (mean ± SD) | 260/230 (mean ± SD) |
|---|---|---|---|---|---|
| Method_A | |||||
| 1 | Stool | 141.9 ± 35.6 | 1.775 ± 0.06 | 1.285 ± 0.07 | |
| 2 | Soil | 430.85 ± 82.80 | 1.665 ± 0.04 | 1.24 ± 0.16 | |
| 3 | Fish-gut (silver carp) | 238.6 ± 64.48 | 1.7 ± 0.07 | 1.37 ± 0.07 | |
| 4 | Fish-gut (T. putitora) | 92.1 ± 4.10 | 1.475 ± 0.50 | 1.15 ± 0.37 | |
| 5 | Milk | 104.1 ± 36.48 | 1.72 ± 0.33 | 1.215 ± 0.10 | |
| Method_B | |||||
| 1 | Stool | 30.75 ± 7.99 | 1.905 ± 0.09 | 0.63 ± 0 | |
| 2 | Soil | 561.3 ± 65.19 | 1.915 ± 0.07 | 1.55 ± 0.19 | |
| 3 | Fish-gut (silver carp) | 271 ± 32.80 | 1.98 ± 0.09 | 1.11 ± 0.29 | |
| 4 | Fish-gut (T. putitora) | 72.2 ± 23.75 | 1.42 ± 0.11 | 0.875 ± 0.13 | |
| 5 | Milk | 168.095 ± 16.83 | 1.73 ± 0.06 | 1.31 ± 0.00 | |
| Method_C | |||||
| 1 | Stool | 964.95 ± 60.8 | 2.145 ± 0.02 | 1.89 ± 0.12 | |
| 2 | Soil | 76.35 ± 20.7 | 1.66 ± 0.22 | 0.97 ± 0.00 | |
| 3 | Fish-gut (silver carp) | 186.85 ± 6.15 | 2.025 ± 0.23 | 1.41 ± 0.38 | |
| 4 | Fish-gut (T. putitora) | 56.6 ± 2.12 | 1.5 ± 0 | 0.98 ± 0.10 | |
| 5 | Milk | 20.35 ± 7.51 | 1.60 ± 0.18 | 0.6 ± 0.46 |
sd standard deviation
The samples which showed most variability in DNA concentrations by tested methods were human stool, human milk and soil sample. The manual method A worked well for most of the studied samples in terms of DNA quantity and quality (Table 1 and Fig. 1a). Among the two most commonly used commercial kits, method B offered more wide applicability as it worked well for fish gut and soil but had limited use in human stool sample. With removal of use of Inhibit®Ex buffer/tablet in method B, recovery of milk DNA was also standardized (Table 1). Method C worked best for human stool samples but had limited use in human milk samples even after modifications (Table 1 and Fig. 1b, c). Recovery of soil DNA was good by both method A and B (Fig. 2). As observed in previous studies [19], for human feces, the highest DNA concentrations were obtained using Method C. For fish gut, DNA recovery from silver carp was better than T. putitora for all the three methods and in similar range (Fig. 2). These two fishes have different food preferences as silver carp primarily displays herbivory, while T. putitora has omnivorous food habit. Study of microbial diversity by sequencing is required to validate total microbial composition in these fishes. While recovery of milk DNA was standardized by introducing modifications in method A and method B, it worked better with method B consistently, although the DNA amount was relatively low, when compared to other samples. Milk sample harbors limited number of bacterial cells thus good quality of DNA is required for correct estimation of DNA community in such samples [13, 22]. While commercial methods did not recommend addition of RNase in manufacturer’s instructions, introduction of RNase treatment led to better purity in terms of A260/A230 ratio.
Fig. 2.
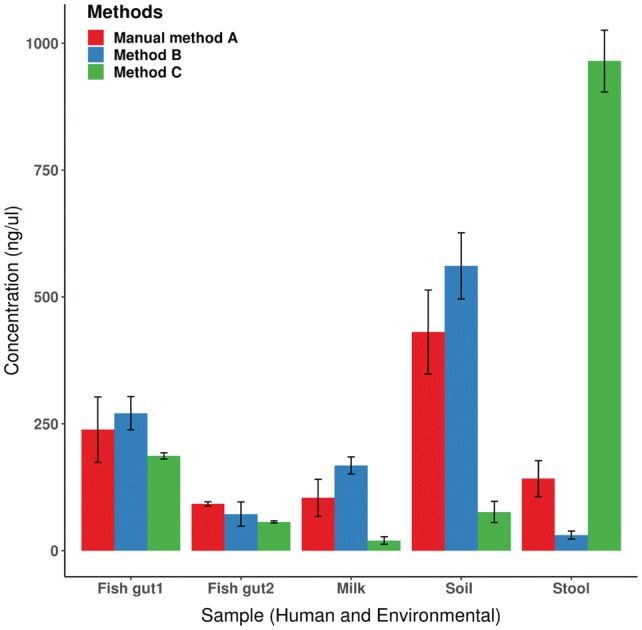
Assessment of different methods used for metagenomic DNA extraction from different samples. The plot shows mean and standard deviation
Notably, a decrease in DNA concentration over time was observed when extraction was done with samples stored for a long time. Among the samples, soil, human stool and fish gut samples were relatively stable up to a year of storage at − 80 °C. However, milk samples need to be stored after the removal of fat layer and preferably used within a month.
Suitability of Isolated DNA in Downstream Applications
To assess the purity of isolated DNA from the various methods, the samples were used for PCR amplification of 16S rRNA gene (Fig. 3). The desired amplicon of 230 bp in all lanes showed the suitability of isolated metagenomic DNA for sensitive downstream processing. In addition, restriction digestion by BamHI was also carried out which showed complete digestion indicated by a smear on gel (Fig. 4), indicating absence of impurities after purification.
Fig. 3.
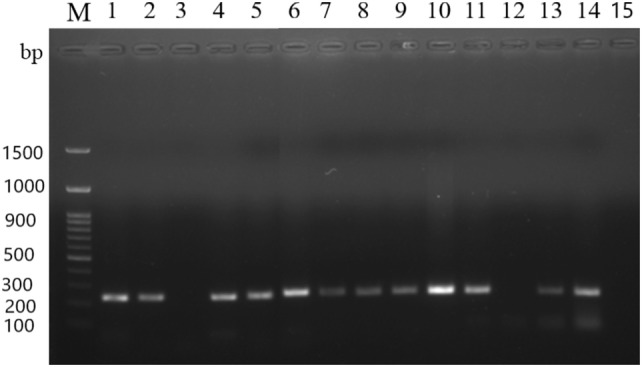
PCR amplification of 16S rRNA of metagenomic DNA isolated from stool, fish gut, soil and milk samples. The PCR products were resolved on a 1.2% agarose gel, stained with ethidium bromide and photographed in a gel documentation system. Lane M: 100 bp DNA Ladder RTU (GenedireX); lanes 1–4; 16S rRNA gene PCR amplification of DNA isolated by method A—soil, fish gut (silver carp), fish gut (T. putitora), stool, respectively; lanes 5–8; 16S rRNA gene PCR amplification of DNA isolated by method B- soil, fish gut (silver carp), fish gut (T. putitora), stool, respectively; lanes 9–12: 16S rRNA gene PCR amplification of DNA isolated by method C—soil, stool, fish gut (silver carp), fish gut (T. putitora), respectively; lanes 13–15: 16S rRNA gene PCR amplification of milk DNA, isolated by method A, modified method B, modified method C
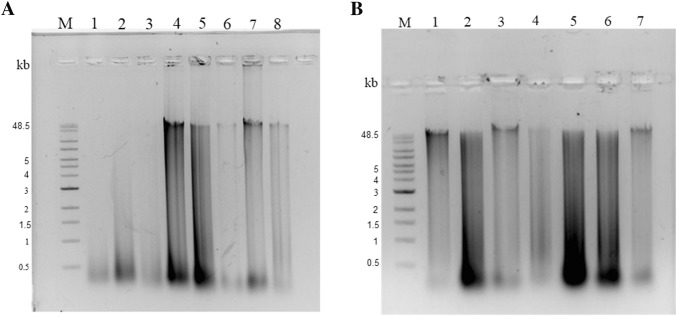
Fig. 4.
Restriction endonuclease (BamHI) digestion of DNA isolated from environmental and human samples. a Lane M, Quick-Load® 1 kb Extend DNA ladder; lanes 1–4, BamHI-digested metagenomic DNA isolated using method A- stool, fish gut (silver carp), fish gut (T. putitora), soil, respectively; lanes 5–8, BamHI-digested metagenomic DNA sample isolated by method B- stool, fish gut (silver carp), fish gut (T. putitora), soil, respectively. b Lane M, Quick-Load® 1 kb Extend DNA ladder; lanes 1–4, BamHI-digested metagenomic DNA isolated by method C- stool, fish gut (silver carp), fish gut (T. putitora), soil, respectively; lanes 5–7, BamHI-digested metagenomic DNA isolated from milk sample—method A, modified method B and modified method C
In conclusion, our results will help in assisting the selection of a suitable DNA extraction method for a particular sample. We also offer an optimized strategy for DNA isolation from milk, which is a relatively less studied microbiome niche. In our observation, the manual method has wide applicability and reproducibility in diverse samples and gives results in a time- and cost-efficient manner. The commercial column extraction methods are specialized for different samples and expensive but provide high molecular weight metagenomic DNA. Exploration of the complex microbiome associated with different hosts and environment by genomics-based studies will offer greater insights into the functional relevance of microbial community composition in different environments [30].
Acknowledgements
We thank the volunteers who provided samples and the staff at Maulana Azad Medical College and Associated Hospital. This work was funded by SERB (JC Bose research fellowship to YS). This work was supported by Brain Pool grant (NRF-2019H1D3A2A01060226) by National Research Foundation (NRF), South Korea to work at Konkuk University (VCK). This research was supported by Basic Science Research Program (2013M3A6A8073184) through the NRF funded by the Ministry of Education, Science and Technology, South Korea (JKL). The work was also supported by Science and Engineering Research Board (GIA/3186/2019-20). The funding organization had no role in the study design or manuscript preparation.
Compliance with Ethical Standards
Conflict of interest
All authors declare that they have no conflict of interests.
Ethical Standard
All procedures performed in the study involving human samples were in accordance with the ethical standards of the institute.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Richa Misra, Email: richamisra@svc.ac.in.
Yogendra Singh, Email: ysinghdu@gmail.com.
References
- 1.Cavalier-Smith T, Brasier M, Embley TM. Introduction: how and when did microbes change the world? Philos Trans R Soc Lond B Biol Sci. 2006;361:845–850. doi: 10.1098/rstb.2006.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalia VC. Microbes: the most friendly beings? In: Kalia VC, editor. Quorum sensing vs quorum quenching: a battle with no end in sight. New Delhi: Springer; 2015. pp. 1–5. [Google Scholar]
- 3.Kalia VC, Raju SC, Purohit HJ. Genomic analysis reveals versatile organisms for quorum quenching enzymes: acyl-homoserine lactone-acylase and-lactonase. Open Microbiol J. 2011;5:1–11. doi: 10.2174/1874285801105010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalia VC. In search of versatile organisms for quorum-sensing inhibitors: acyl homoserine lactones (AHL)-acylase and AHL-lactonase. FEMS Microbiol Lett. 2014;359:143. doi: 10.1111/1574-6968.12585. [DOI] [PubMed] [Google Scholar]
- 5.Diaz S, Purvis A, Cornelissen JH, Mace GM, Donoghue MJ, Ewers RM, Jordano P, Pearse WD. Functional traits, the phylogeny of function, and ecosystem service vulnerability. Ecol Evol. 2013;3:2958–2975. doi: 10.1002/ece3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalia VC. Mining metagenomes for novel bioactive molecules. In: Kalia VC, Shouche Y, Purohit HJ, Rahi P, editors. Mining of microbial wealth and metagenomics. Singapore: Springer; 2017. pp. 1–9. [Google Scholar]
- 7.Purohit HJ, Tikariha H, Kalia VC. Current scenario on application of computational tools in biological systems. In: Purohit HJ, Kalia VC, More RP, editors. Soft computing for biological systems. Singapore: Springer; 2018. pp. 1–12. [Google Scholar]
- 8.Misra R, Virmani R, Dhakan D, Maji A. Tackling the antibiotic resistance: the “gut” feeling. In: Arora G, Sajid A, Kalia VC, editors. Drug resistance in bacteria, fungi, malaria, and cancer. Cham: Springer International Publishing; 2017. pp. 325–338. [Google Scholar]
- 9.Singh A, Gaur M, Misra R. Understanding the connect of quorum sensing and CRISPR-Cas system: potential role in biotechnological applications. In: Kalia VC, editor. Quorum sensing and its biotechnological applications. Singapore: Springer; 2018. pp. 231–247. [Google Scholar]
- 10.Purohit HJ, Khurana JP, Sharma R, Lal SK, Kalia VC. Bacterial diversity, genomics and metagenomics. Indian J Microbiol. 2008;48:151. doi: 10.1007/s12088-008-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma P, Kumari H, Kumar M, Verma M, Kumari K, Malhotra S, Khurana J, Lal R. From bacterial genomics to metagenomics: concept, tools and recent advances. Indian J Microbiol. 2008;48:173–194. doi: 10.1007/s12088-008-0031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bag S, Saha B, Mehta O, Anbumani D, Kumar N, Dayal M, Pant A, Kumar P, Saxena S, Allin KH, Hansen T. An improved method for high quality metagenomics DNA extraction from human and environmental samples. Sci Rep. 2016;6:26775. doi: 10.1038/srep26775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lima SF, Bicalho MLS, Bicalho RC. Evaluation of milk sample fractions for characterization of milk microbiota from healthy and clinical mastitis cows. PLoS One. 2018;13:e0193671. doi: 10.1371/journal.pone.0193671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talwar C, Nagar S, Lal R, Negi RK. Fish gut microbiome: current approaches and future perspectives. Indian J Microbiol. 2018;58:397–414. doi: 10.1007/s12088-018-0760-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knudsen BE, Bergmark L, Munk P, Lukjancenko O, Prieme A, Aarestrup FM, Pamp SJ. Impact of sample type and DNA isolation procedure on genomic inference of microbiome composition. mSystems. 2016;1:e00095-16. doi: 10.1128/mSystems.00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boers SA, Jansen R, Hays JP. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur J Clin Microbiol Infect Dis. 2019;38:1059–1070. doi: 10.1007/s10096-019-03520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim MY, Song EJ, Kim SH, Lee J, Nam YD. Comparison of DNA extraction methods for human gut microbial community profiling. Syst Appl Microbiol. 2018;41:151–157. doi: 10.1016/j.syapm.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Verma SK, Singh H, Sharma PC. An improved method suitable for isolation of high-quality metagenomic DNA from diverse soils. 3 Biotech. 2017;7:171. doi: 10.1007/s13205-017-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maji A, Misra R, Dhakan DB, Gupta V, Mahato NK, Saxena R, Mittal P, Thukral N, Sharma E, Singh A, Virmani R, Gaur M, Singh H, Hasija Y, Arora G, Agrawal A, Chaudhry A, Khurana JP, Sharma VK, Lal R, Singh Y. Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environ Microbiol. 2018;20:402–419. doi: 10.1111/1462-2920.14015. [DOI] [PubMed] [Google Scholar]
- 20.Salonen A, Nikkila J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, Kekkonen RA, Palva A, de Vos WM. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Pollock J, Glendinning L, Wisedchanwet T, Watson M. The madness of microbiome: attempting to find consensus “best practice” for 16S microbiome studies. Appl Environ Microbiol. 2018;84:e02627-17. doi: 10.1128/aem.02627-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahlberg J, Sun L, Waller KP, Östensson K, McGuire M, Agenäs S, Dicksved J. Microbiota data from low biomass milk samples is markedly affected by laboratory and reagent contamination. PLoS One. 2019;14:e0218257. doi: 10.1371/journal.pone.0218257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks JP, Edwards DJ, Harwich MD, Rivera MC, Fettweis JM, Serrano MG, Reris RA, Sheth NU, Huang B, Girerd P, Strauss JF. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Ashram S, Al Nasr I, Suo X. Nucleic acid protocols: extraction and optimization. Biotechnol Rep. 2016;12:33–39. doi: 10.1016/j.btre.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali N, Rampazzo RCP, Costa ADT, Krieger MA. Current nucleic acid extraction methods and their implications to point-of-care diagnostics. Biomed Res Int. 2017;2017:9306564. doi: 10.1155/2017/9306564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, Leite R, Elovitz MA, Parry S, Bushman FD. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016;4:29. doi: 10.1186/s40168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhushan A, Mukherjee T, Joshi J, Shankar P, Kalia VC. Insights into the origin of Clostridium botulinum strains: evolution of distinct restriction endonuclease sites in rrs (16S rRNA gene) Indian J Microbiol. 2015;55:140–150. doi: 10.1007/s12088-015-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quigley L, O’Sullivan O, Beresford TP, Paul Ross R, Fitzgerald GF, Cotter PD. A comparison of methods used to extract bacterial DNA from raw milk and raw milk cheese. J Appl Microbiol. 2012;113:96–105. doi: 10.1111/j.1365-2672.2012.05294.x. [DOI] [PubMed] [Google Scholar]
- 29.Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 2019;27:105–117. doi: 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Vakhlu J, Sudan AK, Johri BN. Metagenomics: future of microbial gene mining. Indian J Microbiol. 2008;48:202–215. doi: 10.1007/s12088-008-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]