Summary
OsmY is a widely conserved but poorly understood 20 kDa periplasmic protein. Using a folding biosensor, we previously obtained evidence that OsmY has molecular chaperone activity. To discover natural OsmY substrates, we screened for proteins that are destabilized and thus present at lower steady-state levels in an osmY-null strain. The abundance of an outer membrane protein called antigen 43 was substantially decreased and its β-barrel domain was undetectable in the outer membrane of an osmY-null strain. Antigen 43 is a member of the diffuse adherence family of autotransporters. Like strains that are defective in antigen 43 production, osmY-null mutants failed to undergo cellular autoaggregation. In vitro, OsmY assisted in the refolding of the antigen 43 β-barrel domain and protected it from added protease. Finally, an osmY-null strain that expressed two members of the diffuse adherence family of autotransporters that are distantly related to antigen 43, EhaA and TibA, contained reduced levels of the proteins and failed to undergo cellular autoaggregation. Taken together, our results indicate that OsmY is involved in the biogenesis of a major subset of autotransporters, a group of proteins that play key roles in bacterial pathogenesis.
Keywords: Molecular Chaperones, Protein Folding, Proteostasis, Bacterial Outer Membrane Proteins, Bacterial Secretion Systems
Graphical abstract
Introduction
The cell envelope of Gram-negative bacteria is composed of an outer membrane and an inner membrane that enclose a compartment called the periplasm (Ruiz et al., 2006). The periplasm is an aqueous but crowded compartment that occupies ~20% of total cell volume; it contains a thin layer of peptidoglycan and ~300 different proteins (Van Wielink and Duine, 1990). Periplasmic proteins are capable of performing diverse functions, including envelope biogenesis, signal transduction, the absorption and transportation of nutrients, the efflux of toxic substances, and the determination of cell shape and virulence (Miller and Salama, 2018).
In vivo, the folding of proteins is commonly assisted by chaperones (Kim et al., 2013). This is true in the periplasm as well as in the cytoplasm (Stull et al., 2018). Periplasmic chaperones can assist protein folding independent of ATP, a remarkable feature that differentiates them from most cytoplasmic chaperones (Goemans et al., 2014). Periplasmic chaperones have been shown to be involved in two important processes: (1) the biogenesis of outer membrane β-barrel proteins (OMPs), and (2) the protection of the periplasmic proteome from unfolding and/or aggregation under stress conditions. Nascent polypeptides for OMPs are secreted into the periplasm through an inner membrane channel complex called SecYEG (Wickner et al., 1991); periplasmic chaperones then bind to them, maintain them in a partially-unfolded form, and escort them to the Bam complex, which inserts β-barrel proteins into the outer membrane (Konovalova et al., 2017, Noinaj et al., 2017). The outer membrane serves as a semi-permeable barrier between the bacteria and the external environment. The proteins embedded in this layer help control which small molecules and proteins are allowed into the periplasmic space and which are excluded. These OMPs are a diverse group of proteins that commonly fold into β-rich structures often consisting of a barrel like structure, with 8–36 β-strands integrated into the outer membrane. Some possess periplasmic or extracellular domains. In some proteins, the core is open, forming a pore, while in others the core is filled. A number of periplasmic chaperones are thought to maintain the solubility and assist in the folding of OMPs as they transit the periplasm. These include SurA, Skp, and DegP. These proteins appear to have at least somewhat overlapping and redundant roles (Missiakas et al., 1996, Lazar and Kolter, 1996, Spiess et al., 1999). The absence of SurA, Skp, or DegP results in decreased levels of certain OMPs and a minor outer membrane biogenesis defect (Vertommen et al., 2009, Denoncin et al., 2012), whereas deletions of both SurA and Skp or SurA and DegP leads to inviability (Rizzitello et al., 2001, Denoncin et al., 2012). Since the outer membrane is permeable, the periplasm is vulnerable to changes in the external environment; periplasmic proteins must therefore be able to cope with harsh conditions. Several periplasmic chaperones have been shown to function specifically under stressful conditions; these include HdeA and HdeB, which act in response to exposure to acidic conditions (Hong et al., 2005, Kern et al., 2007), and Spy, which is induced by protein unfolding agents such as butanol and tannins (Quan et al., 2011).
We recently found the 20 kDa periplasmic protein OsmY to be a molecular chaperone through a genetic selection that forces cells to optimize unstable protein folding in vivo (Lennon et al., 2015). OsmY was first discovered due to its strong induction by osmotic stress conditions (Yim and Villarejo, 1992). The OsmY sequence is composed of two repeated conserved regions, each of which contain a bacterial OsmY nodulation (BON) domain. The BON domain is a conserved domain that is typically ~60 residues long and arranged in an αββαβ fold (Yeats and Bateman, 2003). In vitro, we found OsmY could inhibit the aggregation of a number of proteins commonly used for assaying chaperone activity, including lactate dehydrogenase, luciferase, and α-lactalbumin (Lennon et al., 2015). Consistent with our findings, several studies previously reported that OsmY, when fused to the N or C terminus of difficult-to-express or poorly-folded recombinant proteins, allows large quantities of properly folded proteins to be exported into the extracellular media (Qian et al., 2008, Bokinsky et al., 2011, Kotzsch et al., 2011, Zheng et al., 2012, Gupta et al., 2013, Cheng et al., 2014). These findings suggest that OsmY can function as a chaperone in cis (i.e., when present as part of the same molecule), in keeping with our finding that OsmY can function as a chaperone in trans. The genetic selection through which OsmY was discovered to be a chaperone involves the stabilization of a tripartite fusion protein consisting of an unstable mutant of maltose binding protein that is inserted into β-lactamase; other in vivo clients for OsmY’s chaperone activity remain unknown.
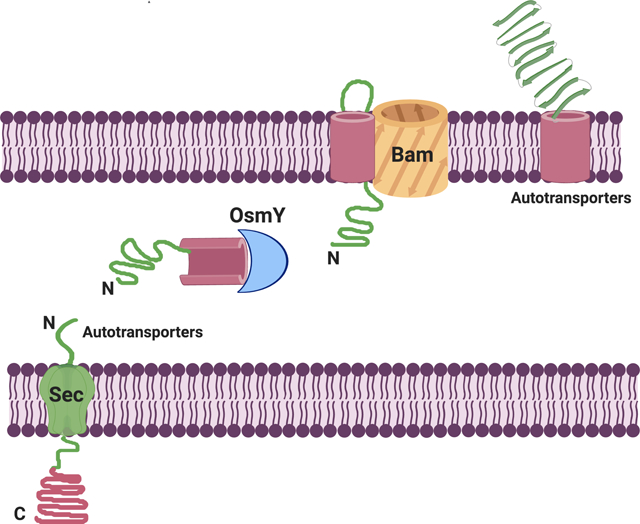
One particularly interesting class of OMPs are the so called autotransporters, a large family of virulence-linked OMPs that are present in numerous Gram-negative bacteria (Henderson et al., 2004, Dautin and Bernstein, 2007, Leyton et al., 2012). In addition to a C-terminal β-barrel domain, autotransporters contain an N-terminal extracellular (“α”) domain. The two domains are connected by a linker that traverses the pore of the β-barrel domain. Although the α domains diverge greatly in sequence, size, and function, they usually fold into a repetitive β-helical structure that promotes virulence (Henderson and Nataro, 2001, Celik et al., 2012). The C-terminal domains also show minimal sequence conservation, but form nearly superimposable 12-stranded β barrels (Oomen et al 2004; Barnard et al., 2007; van den Berg 2010, Zhai Y et al. 2011). Autotransporters in E. coli mainly fall into three groups based on homology: (1) serine protease autotransporters of the Enterobacteriaceae (SPATEs), (2) adhesins involved in diffuse adherence (AIDA-I) type autotransporters, and (3) trimeric autotransporter adhesins (TAAs) (Wells et al., 2010, Vo et al., 2017).
Autotransporters were originally thought to be self-contained secretion systems, i.e., these proteins were thought to encode all the information and machinery necessary to transport their α domain across the outer membrane (Pohlner et al., 1987). Although this is no longer thought to be the case, it is still unclear what ensemble of host factors are involved in autotransporter biogenesis. Based primarily on studies on EspP, a member of the SPATE family, there is now evidence that after being translocated by the Sec complex into the periplasm, both domains interact with molecular chaperones, including SurA, Skp, DegP, and FkpA (Ruiz-Perez et al., 2009, Ieva and Bernstein, 2009, Ieva et al., 2011). The exact function of the chaperones is unclear because individual chaperones can be deleted without affecting viability, possibly due to redundancy (Sklar et al., 2007, Rizzitello et al., 2001). Nevertheless, they are thought to maintain autotransporters in an assembly-competent conformation by preventing their misfolding and aggregation in the periplasm (Ieva et al., 2011, Bernstein, 2015, Bernstein, 2019). DegP may also be required for the inner membrane translocation of the autotransporter pertactin (Braselmann et al., 2016).
Available evidence indicates that the β-barrel domain of EspP begins to fold in the periplasm (Ieva et al., 2008; Hussain and Bernstein, 2018). The protein is subsequently targeted to the Bam complex, a five protein heterooligomer (BamABCDE) that promotes both the insertion of the β-barrel domain into the outer membrane and the translocation of the α domain across the membrane (Ieva and Bernstein, 2009). The mechanism of α domain secretion is unclear, but has been proposed to occur through a hybrid channel consisting of the β-barrels of both the autotransporter and BamA in an open conformation (Pavlova et al., 2013, Fan et al., 2016).
Other factors, however, may play important roles in the assembly of autotransporters that do not belong to the SPATE family. For example, the translocation and assembly module TamAB has been reported to contribute to the maturation of the autotransporter Ag43 (Selkrig et al., 2012). Here, we report findings indicating that OsmY plays an important role in the biogenesis of a number of AIDA-I group autotransporters, including Ag43, EhaA, and TibA.
Results
OsmY homologs are widespread within Proteobacteria
A PSI-BLAST search of the nonredundant database using E. coli MG1655 OsmY as the query sequence retrieved thousands of sequences with greater than 30% identity to OsmY. Most of these are present in Proteobacteria. E. coli OsmY contains two bacterial BON domains that share 43% identity to each other. Many organisms, however, are predicted to produce related proteins containing either just one or more than two BON domains, but the BON domain is also found in proteins with more complex protein architectures, as described in the Pfam data base entry for this protein family (http://pfam.xfam.org/family/BON)
Ag43 is poorly expressed in an osmY-null strain
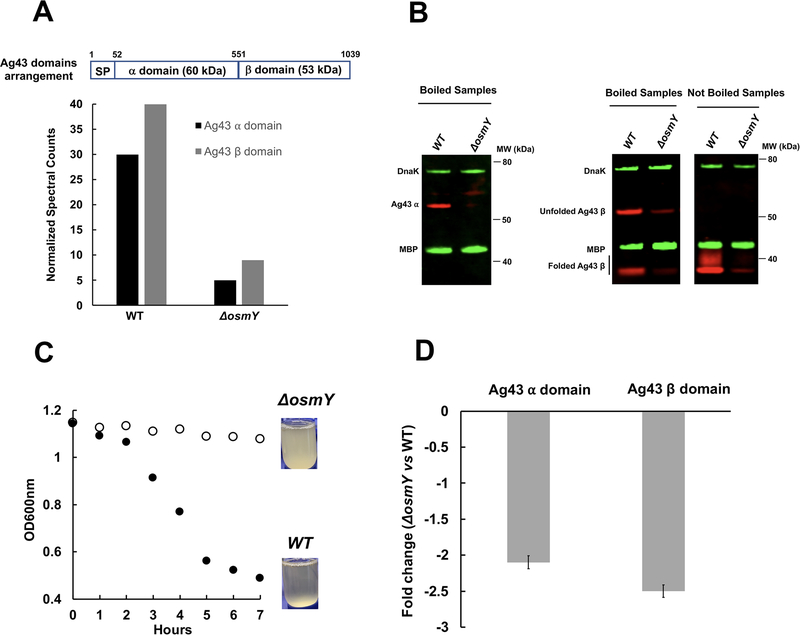
We previously isolated OsmY as a protein with chaperone activity (Lennon et al., 2015). To better define the in vivo function of OsmY and to screen for its in vivo substrates, we compared the steady-state levels of proteins expressed in wild-type (WT) and osmY-null mutants through quantitative proteomics. We reasoned that proteins that require OsmY for their proper folding may be destabilized and thus decreased in abundance in ΔosmY strains. The only cell envelope protein that we found to be significantly reduced in the ΔosmY strain was the autotransporter Ag43, encoded by a gene called “flu” originally designated for the “fluffing” phenotype it produces (Diderichsen, 1980). Presumably following the completion of assembly, Ag43 is cleaved by an unknown mechanism into a ~60 kDa N-terminal fragment that contains most of the surface exposed α domain (hereafter referred to as the “α domain”) and a ~53 kD C-terminal fragment that likely contains not only the outer membrane integrated β-barrel domain but also a portion of the extracellular domain (hereafter referred to as the “β-barrel domain”) (Owen et al., 1996). Both domains were found to decrease 5- to 6-fold in MS spectral counts in a ΔosmY strain relative to the levels found in a WT strain (Fig. 1A). Western blotting using antisera raised against Ag43’s α and β domains verified these proteomics results. Because antibody against the α domain is not sensitive enough to detect trace amounts of α domain in the ΔosmY strain, we quantified the β-domain bands. We performed three independent experiments and found that β-domain levels were decreased by 8.5 ± 1.7-fold in the ΔosmY strain compared to the WT strain. Like many other OMPs, the β-barrel domain remained folded in the absence of heat and migrated much more rapidly than its predicted molecular weight on SDS-PAGE (at 37 kD), as reported previously (Owen et al., 1996) (Fig. 1B). In addition, the ΔosmY strain showed a phenotype associated with a loss of Ag43 function in that it failed to mediate cellular autoaggregation (Fig. 1C). This phenotype is at least partially complemented by expression of osmY from a plasmid (Fig. S1). Co-expression of osmY and its down-stream gene ytjA did not increase Ag43-mediated aggregation, ruling out the possibility that the poor expression of Ag43 is due to a polar effect in the ΔosmY strain (Fig. S1). We next used quantitative RT-PCR to determine the steady-state levels of Ag43 mRNA. Although these experiments did show a 2- to 3-fold decrease in steady-state levels of Ag43 mRNA in the osmY-null mutant, this decrease was insufficient to entirely explain the decrease in protein levels and was much smaller than the decrease observed in cells that have a reduced concentration of SecA (Yap and Bernstein, 2013) (Fig. 1D). In addition, the level of two regulators of flu gene transcription for Ag43, OxyR and Dam (van der Woude and Henderson, 2008), did not significantly change in the osmY-null mutant as determined by quantitative proteomics (Table S3). We therefore conclude that Ag43 is destabilized in the osmY deletion mutant, which is consistent with the idea that OsmY is a chaperone that assists in either the folding or stabilization of Ag43.
Fig. 1.
Comparison of Ag43 steady-state levels in WT and osmY-null strains.
A. WT and ΔosmY cells were assayed for quantitative proteomics using LC-MS/MS; shown are normalized Ag43 α- and β-domain spectral counts.
B. WT and ΔosmY cell pellets were resuspended in SDS-reducing sample buffer. After boiling at 95°C for 5 min or not, equal volumes were analyzed by SDS-PAGE and western blot using antiserum raised against the indicated proteins.
C. Autoaggregation of WT and ΔosmY cells was assayed by taking samples 1 cm below the liquid surface and measuring optical density at 600 nm.
D. Graph shows the quantitative RT-PCR analysis of Ag43 α- and β-domain mRNA levels in ΔosmY relative to WT.
Overproduction of Ag43 in a ΔosmY strain results in its partial proteolysis
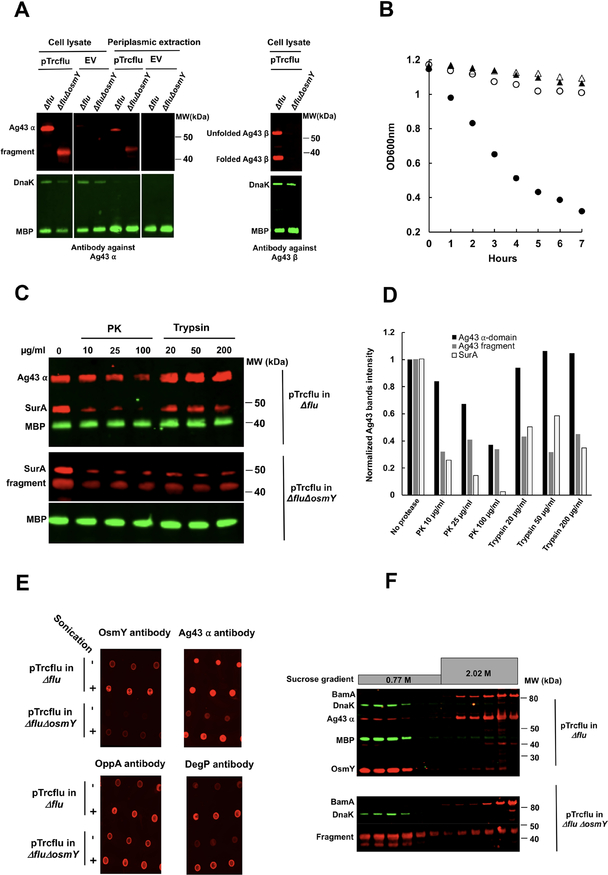
To gain insight into the role OsmY plays in Ag43 maturation and export, we cloned the Ag43 gene (flu) into a pTrc vector to generate pTrcflu and, using this vector, overexpressed Ag43 in Δflu and ΔfluΔosmY strains. The Δflu strain can be complemented by Ag43 expression of pTrcflu which restores the ability of Ag43 to mediate cellular autoaggregation (Fig. 2B), and a band that reacted to Ag43 antibody and migrated at the position of the Ag43 α domain was prominently observed using western blotting (Fig. 2A). Overexpression of pTrcflu in ΔfluΔosmY did not cause cellular aggregation (Fig. 2B), suggesting that these strains are phenotypically Ag43 minus, but a band migrating at ~42 kDa was observed in western blots using antiserum against Ag43 α-domain N-terminal peptide (Fig. 2A). This band is thus likely to be a proteolytic fragment that contains only an N-terminal portion of the Ag43 α domain. Both the α domain and its fragment were found in the periplasmic/outer membrane fraction (Quan et al., 2013) (Fig. 2A).
Fig 2.
Overproduction of Ag43 in the osmY-null mutant results in the production of a ~42 kDa α-domain proteolytic fragment. Strains deleted for Ag43 (Δflu) and those deleted for both Ag43 and OsmY (ΔfluΔosmY) harboring either pTrcflu to overexpress Ag43 or an the empty vector pTrc as a control, were cultured as described in Experimental Procedures.
A. Whole cell lysates and periplasmic preparations extracted with 1 mg/ml polymyxin were boiled at 95°C for 5 min and analyzed by SDS-PAGE and western blotting using antiserum raised against the indicated proteins.
B. Autoaggregation assays by taking samples 1 cm below the liquid surface and measuring optical density at 600 nm. ●, Ag43 overexpressing pTrcflu in Δflu. ○, Ag43 overexpressing pTrcflu in ΔfluΔosmY. ▲, empty vector (pTrc) in Δflu. △, empty vector (pTrc) in ΔfluΔosmY.
C. PK and trypsin-treated experiments were performed as described in Experimental Procedures. Samples were analyzed by SDS-PAGE and western blot using antiserum raised against the indicated proteins.
D. Ag43 α domain and fragment bands were quantified using Image J, and intensities were normalized to MBP band intensities, which were used as a protease resistant loading control. The amount of Ag43 α domain, fragment and SurA bands at no added protease were set to one.
E. Dot blot assays were performed as described in Experimental Procedures. Membranes were blocked and probed with the indicated antibodies. A high signal that is unaffected by sonication indicates surface exposure, a low signal in unsonicated cells that is enhanced by sonication indicates the protein is contained within the cell, low but visible signals in the absence of sonication can be due to some cell lysis. OppA and DegP were used as periplasmically localized control proteins.
F. Cell fractionation were performed as described in Experimental Procedures. Each fraction was then analyzed by SDS-PAGE and western blot using antiserum raised against the indicated proteins.
We then used a protease treatment approach to characterize the Ag43 α domain and its fragment. The α domain was relatively resistance to proteolysis in cell lysates, remaining undigested by trypsin at concentrations from 20–200 μg/ml and proteinase K at 10 μg/ml and was only partially digested at higher proteinase K concentrations(figure 2C and 2D), indicating it is well folded, consistent with previous reports (Babu et al., 2018). However, the α-domain fragment that is present in the ΔosmY strain was much more protease sensitive at all concentrations of trypsin and proteinase K used, indicating it is not properly folded (Fig. 2C and 2D). The small amount of α-domain fragment remaining after proteolysis is similar to the amount of the protease sensitive control protein SurA that that remains at all protease concentrations. The apparently protease resistant subpopulations of these two proteins are very likely due to a residual population of unlysed cells.
Whether the Ag43 α domain and its fragment were surface-exposed or not were determined by dot blot assays, which determine whether or not antibodies to a protein can react with unlysed cells (Cho et al., 2014, Konovalova et al., 2014). A strong signal for the α domain of Ag43 was detected in the cells of Δflu containing pTrcflu, whereas virtually no signal for the α domain fragment that was present in the ΔfluΔosmY cells containing pTrcflu was detected unless the cells were sonicated (Fig. 2E).
Cellular fractionation using ultracentrifugation revealed that most of the Ag43 α domain is found in the membrane fraction, whereas most of the fragment is found in the soluble fraction (Fig. 2F). Cells overexpressing Ag43 autoaggregated, presumably via the surface-exposure of the Ag43 α-domain, but cells overexpressing Ag43 from pTrcflu in ΔfluΔosmY strains did not autoaggregate, even though they expressed a large amount of the Ag43 fragment (Fig 2A and 2B). This indicates that the Ag43 fragment present in ΔosmY strains is not functional. No Ag43 β-domain cross-reacting material was observable when we attempted Ag43 overexpression from pTrcflu in ΔfluΔosmY strains (Fig. 2A), suggesting that the β domain undergoes complete proteolysis. This proteolysis could occur either before or after insertion into the outer membrane, thereby leaving an α-domain fragment to accumulate in the periplasm in an unfolded, misfolded, or partially-folded state. One possible scenario is that the β barrel portion of Ag43 misfolds and cannot integrate into the outer membrane. Ag43 is therefore retained in the periplasm and the N terminus adopts a non-native but relatively stable conformation. The less well-folded C terminus is then clipped off and degraded, leaving the better-folded N terminus behind in the periplasm. It seems unlikely that the N terminus is completely unfolded—if it were, it would probably be degraded by periplasmic proteases. Independent of the exact model, our results suggest that OsmY is important for the folding and/or insertion-competence of the Ag43 β domain.
OsmY specifically stabilizes the Ag43 β domain in vitro
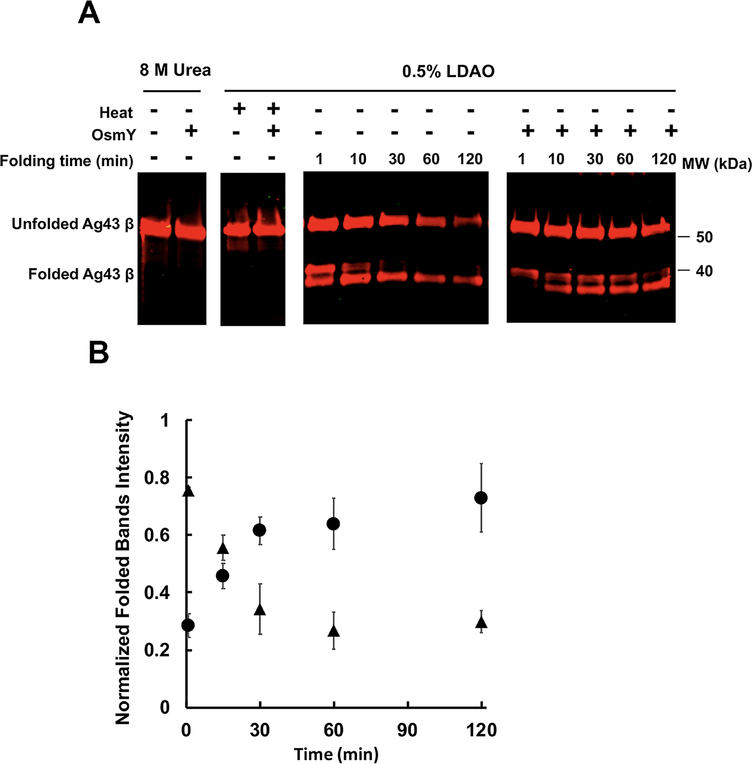
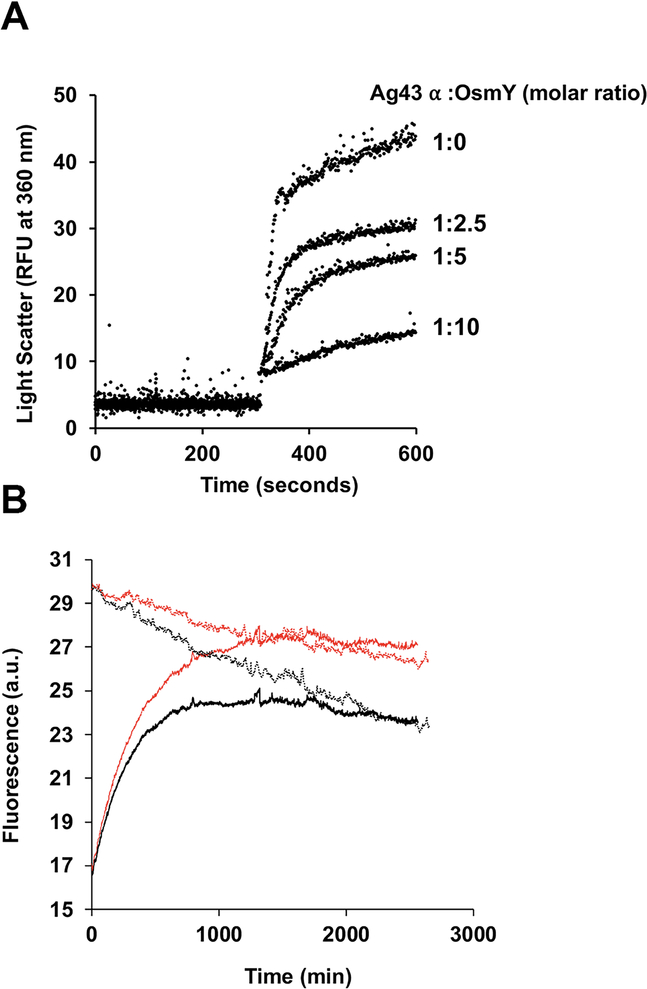
To study the refolding of the Ag43 β-barrel domain in vitro, we overexpressed it in the E. coli cytosol and purified it from inclusion bodies. We then dissolved the domain in 8 M urea and attempted to refold it in the presence or absence of OsmY. To do this, we diluted the urea-dissolved Ag43 β domain into a buffer containing 0.5% of the detergent N,N-dimethyldodecylamine N-oxide (LDAO), which has been used to refold β domains of other autotransporters (Zhai et al., 2011, Yuan et al., 2018), and monitored refolding over time using gel mobility on SDS-PAGE. This method exploits the observation that heating alters the mobility of OMPs in a conformationally dependent manner. Two forms of Ag43 migrated more rapidly than the urea-denatured form; these were detected as a prominent species by western blotting after only one minute of refolding. The upper band of these two fast migrating species decreased over time, possibly due to chasing into the lower band or proteolysis. Either possibility implies that the upper band of the doublet is less well folded than the more rapidly migrating species. However, since boiling the refolded sample caused both of these fast migrating forms to disappear, we deduce that both bands are at least partially folded (Fig. 3A). Quantification of these two refolded bands suggests that the addition of OsmY initially slows the folding of the β-barrel domain but results in a higher folded yield (Fig. 3B). The abundance of both the unfolded and folded forms decreased with time in the absence of OsmY, suggesting that proteolytic degradation may be occurring due to protease contamination of our partially pure Ag43 preparation. We reasoned that since there are probably similar amounts of protease in the samples incubated in the presence or absence of OsmY, and since degradation was much more prominent in its absence, OsmY may be protecting the Ag43 β-barrel domain from degradation. To test this hypothesis, we added a fixed amount of proteinase K into refolding mixtures that were supplemented either with OsmY or not. The results revealed that OsmY efficiently protects both the unfolded and folded Ag43 β-barrel domain from proteolysis. We then tested several well-known periplasmic chaperones to see if they could also stabilize Ag43, using lysozyme as an additional protein control. Among the chaperones shown to play roles in OMP biogenesis, only SurA and Skp were able to protect the Ag43 β-barrel domain from proteolysis as well as OsmY does (Fig. 4). Proteolysis by trypsin gave similar results (Fig. S2). Since the Ag43 β domain belongs to the β-barrel OMP family, we next examined the effect of added OsmY on the proteinase K digestion of several other unfolded OMPs in detergent micelles. OsmY was able to prevent degradation for all those tested (Fig. S3). This effect may be specific to β barrels, as OsmY was unable to mitigate the degradation of the unfolded Ag43 α domain (Fig. S4). We also found that OsmY barely affects α-domain refolding from a urea-denatured form, although it is able to inhibit the time-dependent aggregation of the well-folded α-domain (Fig. 5), as might be expected for a protein possessing broad anti-aggregation activity against a number of commonly used, but admittedly heterologous substrates (Lennon et al., 2015). In summary, our in vitro results suggest that OsmY plays a critical role in Ag43 β-barrel domain refolding and stabilization in detergent micelles, which would impact α-domain maturation in vivo.
Fig. 3.
Refolding of the Ag43 β domain into LDAO micelles.
A. Purified Ag43 β domain held in 8 M urea was diluted 10 times into 0.5% LDAO buffer to a final concentration of 5 μM at 37°C; buffer either contained 25 μM OsmY or 25 μM BSA. Equal volumes of sample were removed from the reaction mixture at the indicated time points, followed by heating at 95°C for 5 min or not, then analyzed by SDS-PAGE and western blot using antiserum raised against the β domain.
Images are representative of three independent experiments.
B. The folded Ag43 β-domain bands of three independent experiments were quantified using Image J and plotted against time. In each experiment, the intensities of the folded β domain were normalized to urea-denatured unfolded β-domain band intensities, which were first normalized to Ag43 β-domain concentrations used in each experiment. ▲, refolding of β domain without OsmY. ●, refolding of β domain with OsmY.
Fig. 4.
PK digestion of the Ag43 β domain in LDAO micelles. 5 μM samples of purified Ag43 β domain were refolded in 0.5% LDAO preincubated with 25 μM BSA, lysozyme, Spy, SurA, Skp, DegP S210A, or OsmY, respectively, for 20 min at 37°C. PK was added to the reaction mixture, and equal volumes of sample were removed into 10 mM PMSF at the indicated time points. Samples were analyzed by SDS-PAGE and western blot using antiserum raised against the β domain. In all experiments, the molar ratio of Ag43 β domain to PK was 2000:1.
Fig. 5.
OsmY prevents Ag43 α-domain aggregation following acid induced unfolding, but has no effect on its refolding in vitro.
A. 50 μM samples of purified Ag43 α domain in 50 mM Tris pH 8.0, 150 mM NaCl buffer were diluted 20 times into the same buffer at pH 4.0 that had been preincubated with different concentrations of OsmY. Acid induced protein aggregation over time was monitored by measuring light scattering at 360 nm using a fluorescence spectrophotometer at 25°C.
B. 50 μM samples of purified Ag43 α domain were denatured with 8 M urea, and then diluted 50 times into 50 mM Tris pH 8.0, 150 mM NaCl in the presence or absence of 10 μM OsmY. Refolding was monitored by measuring tryptophan fluorescence excitation at 295 nm and emission at 350 nm using a Cary Eclipse fluorimeter at 25°C. Refolding of 1 μM samples of purified Ag43 α domain (native form) was also monitored at the same time using the same refolding assay in the presence or absence of 10 μM OsmY. Solid black, Ag43 α-domain refolding. Solid red, Ag43 α-domain refolding with OsmY. Dotted black, native Ag43 α domain. Dotted red, native Ag43 α domain with OsmY.
Display to cell surface of AIDA-I type autotransporters is impaired in the osmY-null mutant
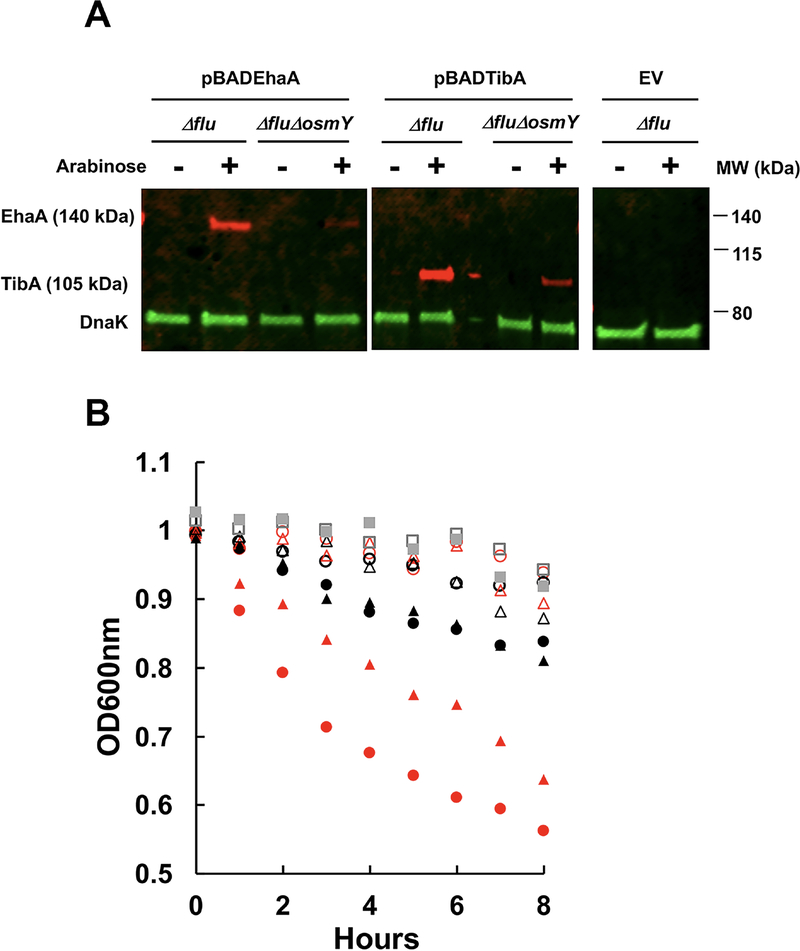
To determine whether any AIDA-I family autotransporters besides Ag43 are also dependent on OsmY for their activity, we expressed EhaA from enterohemorrhagic E. coli and TibA from enterotoxigenic E. coli in Δflu and ΔfluΔosmY strains. We found similar defects for these autotransporters in an osmY deletion strain as we did for Ag43, both in terms of expression, as detected by western blot, and the cell aggregation phenotype, as detected by assaying static culture optical density (Fig. 6). Because three AIDA-I autotransporters showed defects in an osmY-null mutant, but EspP, which does not belong to the AIDA-I group, did not show biogenesis defects in the ΔosmY strain (Fig. S5), we tentatively conclude that OsmY affects the biogenesis of a range of AIDA-I type autotransporters.
Fig. 6. Deletion of the osmY gene inhibits EhaA- and TibA-mediated cellular autoaggregation.
Δflu and ΔfluΔosmY strains harboring pBADEhaA, pBADTibA with a C-terminal MycHis tag, and empty vector (EV), respectively, were cultured in LB media with 100 μg/ml ampicillin until the OD600 reached 0.6. L-arabinose (0.002%) was added or not and cells were induced for 3 h.
A. Whole cell lysates were analyzed by SDS-PAGE followed by western blotting using antiserum raised against DnaK and the C-Myc tag.
B. Autoaggregation of different cells was assayed by taking samples 1 cm below the liquid surface for optical density readings at 600 nm. ○, pBADEhaA in Δflu. ●, pBADEhaA in Δflu with arabinose. ○, pBADEhA in ΔfluΔosmY. ●, pBADEhA in ΔfluΔosmY with arabinose. △, pBADTibA in Δflu. ▲, pBADTibA in Δflu with arabinose. △, pBADTibA in ΔfluΔosmY. ▲, pBADTibA in ΔfluΔosmY with arabinose. □, EV in Δflu. ■, EV in ΔfluΔosmY.
Discussion
In this report, we show that OsmY plays a key role in the assembly of Ag43 and other members of the AIDA-I family of bacterial autotransporters. Whereas Ag43 is normally cleaved into two stable fragments after the β-barrel domain is inserted into the outer membrane and the α domain is secreted, disruption of the osmY gene leads to the accumulation of an N-terminal α-domain fragment in the periplasm and the almost complete disappearance of the β-barrel domain. This phenotype is striking given that: (1) proteolytic fragments of autotransporters have not been reported to accumulate following the depletion of BamA (Jain and Goldberg, 2007), and (2) the loss of a single periplasmic chaperone does not always strongly affect autotransporter assembly (Ruiz-Perez et al., 2009, Ieva and Bernstein, 2009). The results suggest a scenario in which OsmY is specifically required to maintain the Ag43 β-barrel domain in an insertion-competent state. In the absence of OsmY, Ag43 remains in the periplasm where the C terminus of the protein is eventually digested by proteases. Presumably because the protein resides in the periplasm for an abnormally long time, an N-terminal segment that may correspond to one arm of the “L” structure of the α domain (Heras et al., 2014) has an opportunity to fold into a protease-resistant conformation. Consistent with this interpretation, we found that OsmY promotes the refolding of the purified Ag43 β-barrel domain in vitro. Furthermore, OsmY protects the β-barrel domain, but not the α domain, from digestion by exogenous proteases. OsmY was also required for the stable expression of two other members of the AIDA-I family of autotransporters, EhaA and TibA. Our results are noteworthy because they may shed some light on the cellular factors that could promote autotransporter assembly.
Given the multiplicity of periplasmic chaperones and their apparent functional redundancy, it should be interesting to determine why Ag43 specifically requires OsmY for assembly. Unlike the assembly of Ag43, the assembly of the SPATE protein EspP was not impaired in a ΔosmY strain (Fig. S5). The two proteins are also distinct in that the efficient assembly of only Ag43 appears to require TamA/TamB (Selkrig et al., 2012, Kang’ethe and Bernstein, 2013). In addition to having an unusual L-shaped α domain, Ag43 has a β-barrel domain that is very distantly related to the EspP β-barrel domain (< 20% identity). It is conceivable that the Ag43 β-barrel domain has structural elements that distinguish it from the C-terminal domain of other autotransporters. Perhaps unique features of the α domain and/or the β-barrel domain require the recruitment of additional assembly factors. As previously proposed, TamB might modulate α-domain folding in the periplasm (Bamert et al., 2017, Babu et al., 2018) and might maintain the Ag43 α domain in a secretion-competent conformation. Indeed, it is possible that the stable α-domain fragment that we observed in the absence of OsmY results from its interaction with TamB. Consistent with current models (Albenne and Ieva, 2017), BamA and TamA might also function consecutively or cooperatively to catalyze the efficient insertion of the Ag43 β-barrel domain. Interestingly, an interaction between OsmY and the Tam complex has been reported (Babu et al., 2018). This observation suggests that OsmY may target proteins to TamA/TamB. In any case, it seems reasonable to speculate that OsmY and TamA/TamB function in a pathway that is parallel to the canonical pathway (i.e., the chaperones SurA, Skp, and DegP plus the Bam complex) and that is required for the biogenesis of a subset of autotransporters.
Our finding that OsmY protects a variety of E. coli OMPs from degradation in vitro suggests that it has a broad affinity for β barrels. Although the structure of OsmY is unknown, it is conceivable that the hydrophobic regions of the BON domains interact with exposed hydrophobic surfaces of partially-folded β-barrel proteins and prevent them from aggregating. The interaction of OsmY and the hydrophobic amino acid phenylalanine has been reported previously (Piazza et al., 2018). OsmY may be analogous to Skp in forming a cage that provides a protective environment for OMPs (Walton et al., 2009, Burmann et al., 2013, Schiffrin et al., 2016). However, the finding that the level of most OMPs is similar in WT and ΔosmY strains strongly suggests that other chaperones can effectively substitute for OsmY under physiological conditions, perhaps analogous to how Skp and SurA can substitute for each other depending on the protein and growth conditions (Stull et al., 2018).
The high conservation of OsmY in Proteobacteria strongly suggests that it may facilitate the biogenesis of other AIDA-I family autotransporters, which are also widespread in Proteobacteria. Whether OsmY is responsible for biogenesis of autotransporters besides those belonging to the AIDA-I family requires further study. Autotransporters are intimately involved in virulence (Henderson et al., 2004). Our finding that OsmY is an indispensable factor in Ag43 maturation is interesting in light of several recent reports that OsmY is also involved in bacterial virulence. One study showed that an osmY-null mutant of Yersinia ruckeri failed to be infectious in fish (Mendez et al., 2018), and a second study suggested that osmY is linked to virulence factors that promote biofilm formation and flagellar motility in Cronobacter sakazakii (Ye et al., 2015). OsmY has also been proposed to be indirectly associated with virulence in Salmonella typhimurium and E. coli (Bader et al., 2003, Dong and Schellhorn, 2009). Taken together with our results, these studies suggest that OsmY plays a common role in the assembly of a subset of specialized OMPs that differ considerably in structure from the generic porins that dominate the outer membrane of laboratory strains of E. coli.
Experimental procedures
Bacterial strains and plasmids
All the strains and plasmids used in this study are listed in Table S1. Deletion of the osmY gene was performed as previously described (Datsenko and Wanner, 2000). The inserted antibiotic cassette generated using this procedure was excised from the chromosome using pCP20. The chromosomal insertion/deletion of the flu gene encoding Ag43 was transferred from the strain BW25133 (Keio collection) to MC4100 and ΔosmY strains by P1 transduction (Baba et al., 2006). Vectors expressing Ag43 and OsmY were made by amplifying their respective genes with PCR from MC4100 and directly cloning into pTrc- or pBAD-based vectors. The genes for EhaA and TibA were amplified from E. coli O157:H7 and H10407 genomic DNA, respectively. All the vectors were constructed using In-Fusion HD Cloning kits (TaKaRa); all the oligo primers used are listed in Table S2.
Cell growth
Liquid cultures were grown in Luria-Bertani (LB) media at 37°C shaking at 200 r.p.m. Overnight cultures were diluted 1:100 into fresh LB media. If necessary, 100 μg/ml ampicillin was added to maintain the pTrc- and pBAD-based vectors. Expression from the lac promoter on pTrc-based vectors was done by adding IPTG to 0.5 mM final concentration followed by 3 h of induction prior to harvesting. Expression from the arabinose promoter in pBAD-based vectors was similarly done but by using 0.002% arabinose final concentration.
Quantification of protein levels
WT and ΔosmY strains were grown until the OD600 reached 1.0. 1.0 ml of the cells was centrifuged at 3000 × g for 10 min, washed by resuspending in PBS buffer (Na2HPO4 10 mM, KH2PO4 1.8 mM, KCl 2.7 mM, NaCl 137 mM, pH 7.4) of a volume equal to the culture volume, and then recentrifuged. The cell pellets were provided to MS Bioworks (Ann Arbor) who performed Mass Spec analysis as follows. The cell pellets were lysed by resuspension in 500 μl of 2% SDS, 150 mM NaCl, 50 mM Tris pH 8.0 containing one tablet of Roche Complete Protease Inhibitor Cocktail, followed by sonication for 3 cycles of 20 sec on ice (Fisherbrand Model 505). The amount of protein present in the lysate was quantified by Qubit fluorometry (Invitrogen). Lysates containing 10 μg of protein were processed by SDS-PAGE using a 10% Bis-Tris NuPAGE Novex mini gel (Thermo) and the supplied MES buffer system. The region of the gel containing stained proteins was excised and then processed by in-gel digestion with trypsin using a ProGest robot and the following protocol: (1) The gel slice was washed twice with 50 μl of 25 mM ammonium bicarbonate followed by a wash with 50 μl of acetonitrile; (2) proteins in the gel slice were reduced using 40 μl of 10 mM dithiothreitol at 60°C followed by alkylation using 40 μl of 50 mM iodoacetamide at room temperature; (3) proteins were digested by addition of 200 ng of sequencing grade trypsin (Promega) at 37°C for 4 h; (4) digestion was stopped by the addition of 30 μl of trifluoroacetic acid. Each gel digest was then analyzed by nano LC-MS/MS with a Waters NanoAcquity HPLC system interfaced to a ThermoFisher Q Exactive. Peptides were loaded on a trapping column and eluted over a 75 μm analytical column. Both columns were packed with Luna C18 resin (Phenomenex). Peptides were eluted at 350 nl/min with a 2 h binary reverse phase gradient. Buffer A was 0.1% formic acid; buffer B was 0.1% formic acid in acetonitrile. The gradient was at 0 min 98% A, at 1 min 95% A, at 95 min 75% A, at 110 min 65% A, at 112 min 10% A, at 113 min 98% A, at 120 min maintained at 98% A. The mass spectrometer was operated in data-dependent mode, with the Orbitrap operating at 70,000 full width at half maximum (FWHM) and 17,500 FWHM for MS and MS/MS respectively. The fifteen most abundant ions were selected for MS/MS.
SDS-PAGE and western blots
The indicated volumes of cells were centrifuged at 3000 × g for 10 min and washed with amounts of PBS buffer equal to that of the culture volume. Cells were resuspended in SDS-reducing sample buffer and, if indicated, boiled at 95°C for 5 min. SDS-PAGE gels were done using NuPAGE 4–12% Bis-Tris gel (Invitrogen) or 4–20% Mini-PROTEAN TGX stain free gels (Bio-Rad) as specified in the figure legends. After electrophoresis, gels were transferred to a turbo polyvinylidene difluoride (PVDF) membrane (Bio-Rad) using a Trans-Blot Turbo apparatus (Bio-Rad). The blotted PVDF membranes were then blocked using 5% nonfat dried milk in TBST (20 mM Tris, 150 mM NaCl, 0.1% Tween 20) for 1 h at room temperature and probed with the following primary antibodies at these dilutions in 5% nonfat dried milk for 1 h at room temperature: rabbit-derived OsmY polyclonal antibody (Pacific Immunology), 1:5000; rabbit-derived Ag43 α-domain polyclonal antibody (a gift from Begona Heras, La Trobe University), 1:3000; rabbit-derived Ag43 β-domain polyclonal antibody (Pacific Immunology), 1:5000; mouse-derived maltose binding protein (MBP) monoclonal antibody (Biolabs), 1:15000; and mouse-derived DnaK monoclonal antibody (Enzo), 1:15000. The membranes were then washed 3 times (10 min each) with shaking by TBST and probed with fluorescence dye conjugated goat anti-rabbit and goat anti-mouse secondary antibodies (1:15000) (LI-COR Biosciences). Imaging was performed using LI-COR Odyssey CLx.
Dot blot assay
Bacterial strains were grown until the OD600 reached ~1.0. 1 ml of these cells was centrifuged at 3000 × g for 10 min and resuspended in one volume of PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4). Half of these cells were then lysed by 3 cycles of 20 sec sonication on ice (Fisherbrand Model 505), with the other half serving as the intact cell sample. 2 μl of intact cells or lysed cells was spotted on a nitrocellulose membrane and air-dried. Membranes were blocked with 2% nonfat dried milk in PBS for 30 min at room temperature and probed with the following primary antibodies in 5% nonfat dried milk for 1 h at room temperature: rabbit-derived OsmY polyclonal antibody, 1:5000; rabbit-derived Ag43 α-domain polyclonal antibody, 1:3000; rabbit-derived OppA polyclonal antibody, 1:5000 (Pacific Immunology); rabbit-derived DegP polyclonal antibody, 1:5000 (a gift from Michael Erhmann). The procedures for probing with secondary antibodies and imaging were done using the same protocol as described for the western blot.
Membrane fractionation
100 ml of Δflu and ΔfluΔosmY cells harboring pTrcflu were centrifuged at 3000 × g for 10 min, washed with 10 mM HEPES buffer pH 7.5 and 2 mM MgCl2, and resuspended in 20 ml of the same buffer supplemented with 1 mg of DNase and 1 mg of RNase. Cells were lysed by passing through a French press at 12,000 psi. Cell debris was removed by centrifugation at 4,200 × g at 4°C for 5 min. 16 ml of the supernatant was then loaded on top of a two-step sucrose gradient (2.3 ml 2.02 M sucrose and 6.6 ml 0.77 M sucrose). The samples were centrifuged at 130,000 × g at 4°C for 3 h in a Type 70.1 Ti rotor (Beckman Coulter). After centrifugation, 500 μl samples were removed from the top to the bottom of the gradient and analyzed by SDS-PAGE and western blotting.
Quantitative RT-PCR
WT and ΔosmY strains were grown to the late log phase (OD600 of 1.0) then harvested by centrifugation at 3000 × g for 10 min. Total RNA was then isolated using a NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany). DNA contamination was eliminated by the use of DNase treatment and removal reagents in a DNA removal kit (Ambion by Life Technologies, AM1906). cDNAs were then synthesized with a primeScript 1st strand cDNA synthesis kit (Takara) using the supplied mixture of random primers. Quantitative PCRs were performed using the Eppendorf Realplex® PCR detection system in a triplicate reaction. The reaction mixture contained Radiant™ Green qPCR Mix Lo-ROX, 400 nM primers, and 100 ng cDNA. PCR was preformed using the following program: 95°C for 2 min followed by 40 cycles of 95°C for 5 sec and 60°C for 1 min. The threshold cycle (CT) was determined using the manufacturer’s software. Primers 11 and 12, 13 and 14, and 15 and 16 were used to amplify Ag43, Ag43 α-domain, Ag43 β domain, and gapA, respectively. mRNA levels of Ag43 were normalized to the reference gene gapA and calculated using the comparative CT method.
Refolding of outer membrane β-barrel proteins into detergent micelle and protease digestions
To initiate refolding, the purified Ag43 β domain held in 8 M urea was diluted 10 times into 0.5% LDAO buffer to a final concentration of 5 μM at 37°C. To test the effect of OsmY on refolding, the buffer contained 25 μM OsmY, 25 μM BSA, or neither. Folding progress was followed by removing aliquots of this folding reaction at various time intervals and adding them into SDS-reducing sample buffer. Folding status was determined by comparing the migration on SDS-PAGE with and without heating at 95°C for 5 min. Many folded OMPs, including Ag43, are known to migrate more rapidly on SDS-PAGE than unfolded versions (Owen et al., 1996) when analyzed by SDS-PAGE and western blotting using antiserum raised against Ag43’s β domain. Folded status was further determined by proteolysis. In these experiments, purified Ag43 β domain, EspP β domain, OmpA, OmpC, and OmpT were refolded in 0.5% LDAO for 20 min, respectively after preincubation with 25 μM of known chaperones or control proteins, including BSA, lysozyme, OsmY, Spy, SurA, Skp, or the chaperone-active protease-inactive DegP variant DegP-S210A. Following various intervals of folding, proteinase K or trypsin was added to the reaction mixture. For the Ag43 β-barrel folding reaction, an Ag43: proteinase K ratio of 2000:1 was used, and for the other OMPs, a 1000:1 protein: proteinase K ratio was used. Equal volumes of sample were removed from the reaction mixture into 10 mM phenylmethylsulfonyl fluoride (PMSF) to stop protease activity, then analyzed by SDS-PAGE and western blotting using antiserum raised against the corresponding OMPs.
Protein expression and purification
Expression and purification of OsmY was performed as reported previously (Lennon et al., 2015). His-tagged SurA and various OMPs (EspPΔ5, OmpA, OmpC, and OmpT) lacking signal sequences were expressed and purified as previously described (Roman-Hernandez et al., 2014, Hussain and Bernstein, 2018). Skp was purchased directly from https://MyBiosource.com./ Ag43 β domain was cloned into pET21a with a C-terminal His6 tag. The recombinant proteins were overexpressed in E. coli BL21(DE3) strain and isolated from inclusion bodies using buffer A (8M urea, 25 mM Tris, 150 mM NaCl, pH 8.0). The proteins were purified on a 5 ml HisTrap HP column (Amersham Biosciences) equilibrated with buffer A. After washing with buffer A containing 50 mM imidazole, the proteins were eluted with buffer A containing 500 mM imidazole. The degP gene was cloned into pET28b with a C-terminal His6 tag. The QuikChange site directed mutagenesis kit (Stratagene) was then used to introduce the S210A mutation into degP. Recombinant DegP S210A was overexpressed in E.coli BL21(DE3) and isolated from cell lysates using buffer B (PBS containing 20 mM imidazole). The protein was purified on a Ni-NTA (Qiagen) column equilibrated with buffer B. After washing with buffer B containing 400 mM NaCl and 850 mM NaCl, the protein was eluted with buffer B containing 250–500 mM imidazole. Elution fractions containing DegP S210A were pooled and buffer-exchanged into PBS using PD-10 Sephadex desalting columns (GE Healthcare). The purified protein was then concentrated using Amicon 30 kD centrifugal filter units (Millipore) and the concentration was determined using the DC Protein Assay (Bio-Rad).
Supplementary Material
Acknowledgements
We thank Patricia Clark for advice and Ke Wan in the Bardwell lab for protein purification. JCAB is a Howard Hughes Medical Institute Investigator. This work was also supported by Wacker Chemie AG, and the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases. The authors have no conflicts of interest.
Footnotes
Data Sharing
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Albenne C, Ieva R (2017) Job contenders: roles of the beta-barrel assembly machinery and the translocation and assembly module in autotransporter secretion. Mol Microbiol 106, 505–517. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M, Bundalovic-Torma C, Calmettes C, Phanse S, Zhang Q, Jiang Y, et al. (2018) Global landscape of cell envelope protein complexes in Escherichia coli. Nat Biotechnol 36, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, et al. (2003) Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol 50, 219–230. [DOI] [PubMed] [Google Scholar]
- Bamert RS, Lundquist K, Hwang H, Webb CT, Shiota T, Stubenrauch CJ, et al. (2017) Structural basis for substrate selection by the translocation and assembly module of the beta-barrel assembly machinery. Mol Microbiol 106, 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein HD (2015) Looks can be deceiving: recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol Microbiol 97, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein HD (2019) Type V Secretion in Gram-Negative Bacteria. EcoSal Plus 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, et al. (2011) Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci U S A 108, 19949–19954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braselmann E, Chaney JL, Champion MM, Clark PL (2016) DegP Chaperone Suppresses Toxic Inner Membrane Translocation Intermediates. PLoS One 11, e0162922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Wang C, Hiller S (2013) Conformation and dynamics of the periplasmic membrane-protein-chaperone complexes OmpX-Skp and tOmpA-Skp. Nat Struct Mol Biol 20, 1265–1272. [DOI] [PubMed] [Google Scholar]
- Celik N, Webb CT, Leyton DL, Holt KE, Heinz E, Gorrell R, et al. (2012) A bioinformatic strategy for the detection, classification and analysis of bacterial autotransporters. PLoS One 7, e43245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CM, Tzou SC, Zhuang YH, Huang CC, Kao CH, Liao KW, et al. (2014) Functional production of a soluble and secreted single-chain antibody by a bacterial secretion system. PLoS One 9, e97367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Szewczyk J, Pesavento C, Zietek M, Banzhaf M, Roszczenko P, et al. (2014) Detecting envelope stress by monitoring beta-barrel assembly. Cell 159, 1652–1664. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautin N, Bernstein HD (2007) Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol 61, 89–112. [DOI] [PubMed] [Google Scholar]
- Denoncin K, Schwalm J, Vertommen D, Silhavy TJ, Collet JF (2012) Dissecting the Escherichia coli periplasmic chaperone network using differential proteomics. Proteomics 12, 1391–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderichsen B (1980) flu, a metastable gene controlling surface properties of Escherichia coli. J Bacteriol 141, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Schellhorn HE (2009) Global effect of RpoS on gene expression in pathogenic Escherichia coli O157:H7 strain EDL933. BMC Genomics 10, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan E, Chauhan N, Udatha DB, Leo JC, Linke D (2016) Type V Secretion Systems in Bacteria. Microbiol Spectr 4. [DOI] [PubMed] [Google Scholar]
- Goemans C, Denoncin K, Collet JF (2014) Folding mechanisms of periplasmic proteins. Biochim Biophys Acta 1843, 1517–1528. [DOI] [PubMed] [Google Scholar]
- Gupta S, Adlakha N, Yazdani SS (2013) Efficient extracellular secretion of an endoglucanase and a beta-glucosidase in E. coli. Protein Expr Purif 88, 20–25. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Nataro JP (2001) Virulence functions of autotransporter proteins. Infect Immun 69, 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D (2004) Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 68, 692–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, et al. (2014) The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc Natl Acad Sci U S A 111, 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Jiao W, Hu J, Zhang J, Liu C, Fu X, et al. (2005) Periplasmic protein HdeA exhibits chaperone-like activity exclusively within stomach pH range by transforming into disordered conformation. J Biol Chem 280, 27029–27034. [DOI] [PubMed] [Google Scholar]
- Hussain S, Bernstein HD (2018) The Bam complex catalyzes efficient insertion of bacterial outer membrane proteins into membrane vesicles of variable lipid composition. J Biol Chem 293, 2959–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R, Bernstein HD (2009) Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci U S A 106, 19120–19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R, Tian P, Peterson JH, Bernstein HD (2011) Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proc Natl Acad Sci U S A 108, E383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Goldberg MB (2007) Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J Bacteriol 189, 5393–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang’ethe W, Bernstein HD (2013) Charge-dependent secretion of an intrinsically disordered protein via the autotransporter pathway. Proc Natl Acad Sci U S A 110, E4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern R, Malki A, Abdallah J, Tagourti J, Richarme G (2007) Escherichia coli HdeB is an acid stress chaperone. J Bacteriol 189, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82, 323–355. [DOI] [PubMed] [Google Scholar]
- Konovalova A, Kahne DE, Silhavy TJ (2017) Outer Membrane Biogenesis. Annu Rev Microbiol 71, 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A, Perlman DH, Cowles CE, Silhavy TJ (2014) Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of beta-barrel proteins. Proc Natl Acad Sci U S A 111, E4350–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzsch A, Vernet E, Hammarstrom M, Berthelsen J, Weigelt J, Graslund S, Sundstrom M (2011) A secretory system for bacterial production of high-profile protein targets. Protein Sci 20, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar SW, Kolter R (1996) SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol 178, 1770–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon CW, Thamsen M, Friman ET, Cacciaglia A, Sachsenhauser V, Sorgenfrei FA, et al. (2015) Folding Optimization In Vivo Uncovers New Chaperones. J Mol Biol 427, 2983–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyton DL, Rossiter AE, Henderson IR (2012) From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol 10, 213–225. [DOI] [PubMed] [Google Scholar]
- Mendez J, Cascales D, Garcia-Torrico AI, Guijarro JA (2018) Temperature-Dependent Gene Expression in Yersinia ruckeri: Tracking Specific Genes by Bioluminescence During in Vivo Colonization. Front Microbiol 9, 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Salama NR (2018) The gram-negative bacterial periplasm: Size matters. PLoS Biol 16, e2004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiakas D, Betton JM, Raina S (1996) New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol 21, 871–884. [DOI] [PubMed] [Google Scholar]
- Noinaj N, Gumbart JC, Buchanan SK (2017) The beta-barrel assembly machinery in motion. Nat Rev Microbiol 15, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen P, Meehan M, de Loughry-Doherty H, Henderson I (1996) Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol Med Microbiol 16, 63–76. [DOI] [PubMed] [Google Scholar]
- Pavlova O, Peterson JH, Ieva R, Bernstein HD (2013) Mechanistic link between beta barrel assembly and the initiation of autotransporter secretion. Proc Natl Acad Sci U S A 110, E938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza I, Kochanowski K, Cappelletti V, Fuhrer T, Noor E, Sauer U, Picotti P (2018) A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication. Cell 172, 358–372 e323. [DOI] [PubMed] [Google Scholar]
- Pohlner J, Halter R, Beyreuther K, Meyer TF (1987) Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325, 458–462. [DOI] [PubMed] [Google Scholar]
- Qian ZG, Xia XX, Choi JH, Lee SY (2008) Proteome-based identification of fusion partner for high-level extracellular production of recombinant proteins in Escherichia coli. Biotechnol Bioeng 101, 587–601. [DOI] [PubMed] [Google Scholar]
- Quan S, Hiniker A, Collet JF, Bardwell JC (2013) Isolation of bacteria envelope proteins. Methods Mol Biol 966, 359–366. [DOI] [PubMed] [Google Scholar]
- Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, et al. (2011) Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat Struct Mol Biol 18, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzitello AE, Harper JR, Silhavy TJ (2001) Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol 183, 6794–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Hernandez G, Peterson JH, Bernstein HD (2014) Reconstitution of bacterial autotransporter assembly using purified components. Elife 3, e04234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ (2006) Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol 4, 57–66. [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP (2009) Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J Bacteriol 191, 6571–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffrin B, Calabrese AN, Devine PWA, Harris SA, Ashcroft AE, Brockwell DJ, Radford SE (2016) Skp is a multivalent chaperone of outer-membrane proteins. Nat Struct Mol Biol 23, 786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkrig J, Mosbahi K, Webb CT, Belousoff MJ, Perry AJ, Wells TJ, et al. (2012) Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol 19, 506–510, S501. [DOI] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Kahne D, Silhavy TJ (2007) Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21, 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Beil A, Ehrmann M (1999) A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97, 339–347. [DOI] [PubMed] [Google Scholar]
- Stull F, Betton JM, Bardwell JCA (2018) Periplasmic Chaperones and Prolyl Isomerases. EcoSal Plus 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Henderson IR (2008) Regulation and function of Ag43 (flu). Annu Rev Microbiol 62, 153–169. [DOI] [PubMed] [Google Scholar]
- Van Wielink JE, Duine JA (1990) How big is the periplasmic space? Trends Biochem Sci 15, 136–137. [DOI] [PubMed] [Google Scholar]
- Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF (2009) Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9, 2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo JL, Martinez Ortiz GC, Subedi P, Keerthikumar S, Mathivanan S, Paxman JJ, Heras B (2017) Autotransporter Adhesins in Escherichia coli Pathogenesis. Proteomics 17. [DOI] [PubMed] [Google Scholar]
- Walton TA, Sandoval CM, Fowler CA, Pardi A, Sousa MC (2009) The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc Natl Acad Sci U S A 106, 1772–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells TJ, Totsika M, Schembri MA (2010) Autotransporters of Escherichia coli: a sequence-based characterization. Microbiology 156, 2459–2469. [DOI] [PubMed] [Google Scholar]
- Wickner W, Driessen AJ, Hartl FU (1991) The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem 60, 101–124. [DOI] [PubMed] [Google Scholar]
- Ye Y, Gao J, Jiao R, Li H, Wu Q, Zhang J, Zhong X (2015) The Membrane Proteins Involved in Virulence of Cronobacter sakazakii Virulent G362 and Attenuated L3101 Isolates. Front Microbiol 6, 1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats C, Bateman A (2003) The BON domain: a putative membrane-binding domain. Trends Biochem Sci 28, 352–355. [DOI] [PubMed] [Google Scholar]
- Yim HH, Villarejo M (1992) osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J Bacteriol 174, 3637–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Johnson MD, Zhang J, Lo AW, Schembri MA, Wijeyewickrema LC, et al. (2018) Molecular basis for the folding of beta-helical autotransporter passenger domains. Nat Commun 9, 1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Zhang K, Huo Y, Zhu Y, Zhou Q, Lu J, et al. (2011) Autotransporter passenger domain secretion requires a hydrophobic cavity at the extracellular entrance of the beta-domain pore. Biochem J 435, 577–587. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Chen T, Zhao M, Wang Z, Zhao X (2012) Engineering Escherichia coli for succinate production from hemicellulose via consolidated bioprocessing. Microb Cell Fact 11, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.