Abstract
The flea’s lumen gut is a poorly documented environment where the agent of flea-borne plague, Yersinia pestis, must replicate to produce a transmissible infection. Here, we report that both the acidic pH and osmolarity of the lumen’s contents display simple harmonic oscillations with different periods. Since an acidic pH and osmolarity are two of the three known stimuli of the OmpR-EnvZ two-component system in bacteria, we investigated the role and function of this Y. pestis system in fleas. By monitoring the in vivo expression pattern of three OmpR-EnvZ-regulated genes, we concluded that the flea gut environment triggers OmpR-EnvZ. This activation was not, however, correlated with changes in pH and osmolarity but matched the pattern of nutrient depletion (the third known stimulus for OmpR-EnvZ). Lastly, we found that OmpR-EnvZ and the OmpF porin are needed to produce the biofilm that ultimately obstructs the flea’s gut and thus hastens the flea-borne transmission of plague. Taken as a whole, our data suggest that the flea gut is a complex, fluctuating environment in which Y. pestis senses nutrient depletion via OmpR-EnvZ. Once activated, the latter triggers a molecular program (including at least OmpF) that produces the biofilm required for efficient plague transmission.
Keywords: Yersinia pestis, flea, Xenopsylla cheopis, plague, virulence two-component systems, OmpR/EnvZ
Graphical Abstract
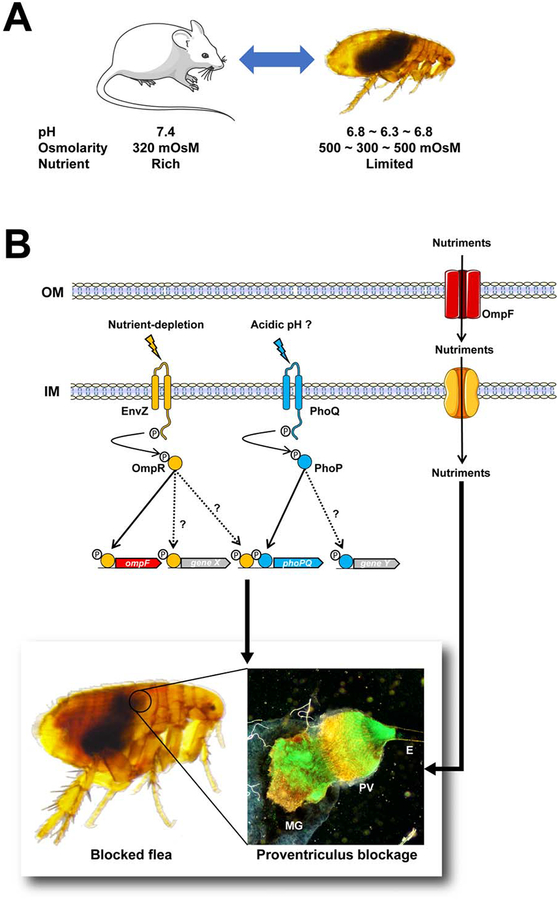
The flea gut is a complex, fluctuating environment in which Y. pestis may sense nutrient depletion via OmpR-EnvZ. Once activated, the latter triggers a molecular program (including at least OmpF) that produces the biofilm required for efficient plague transmission.
INTRODUCTION
Yersinia pestis is the bacillus responsible for plague, a disease that circulates among mammalian hosts via their associated fleas (Simond, 1898, Butler, 1983, Yersin, 1894). The hematophagous insect ingests Y. pestis during a blood meal on a bacteremic host (Ogata, 1897, Simond, 1898). During ingestion, the bacillus passes successively through the flea’s esophagus, proventriculus (a valve covered with spines and that opens rhythmically during feeding) and midgut (Bacot & Martin, 1914). Throughout the period of infection, Y. pestis remains confined to the lumen of the flea’s digestive tract, where it replicates and adapts to the insect’s physiology and defenses (Hinnebusch et al., 2002, Vadyvaloo et al., 2007, Vadyvaloo et al., 2010, Vadyvaloo et al., 2015, Tam et al., 2014, Zhou et al., 2012, Rempe et al., 2012, Rebeil et al., 2013). In some flea species, the bacterium starts to produce a biofilm after a few days or weeks of incubation; this biofilm ultimately blocks the proventriculus (Bacot & Martin, 1914, Hinnebusch et al., 1996, Jarrett et al., 2004, Bobrov et al., 2015, Sun et al., 2011, Eisen et al., 2012). This blockage of the flea’s foregut does not prevent the flea from biting the host and drawing a fresh blood sample. However, the incoming blood only reaches the blocked proventriculus, where it is contaminated by Y. pestis, and is regurgitated into the fleabite site (Bacot & Martin, 1914).
From Y. pestis’ point of view, the flea gut is probably a very dynamic, changing environment. More specifically, the passage from the host’s bloodstream to the flea’s gut is the first abrupt environmental change encountered by Y. pestis. Next, the bacterium is exposed to the sudden entry of several waves of fresh blood into the flea’s gut, followed by a digestion process initiated soon after the blood meal ingestion. Within six hours of feeding, the flea hemolyzes the blood and almost certainly uses a variety of digestive enzymes to rapidly absorb the nutrients required for survival (Vaughan & Azad, 1993, Terra & Ferreira, 1994, Hinnebusch, 2004). At the end of the digestive process, the processed meal consists of a viscous, thick, dark, nutrient-depleted mass that the flea defecates. In other words, the flea gut is an active environment for Y. pestis where the viscosity and amount of available nutrients fluctuate with the blood meal ingestion and digestion cycles. It is not known whether fundamental parameters other than nutrient concentrations oscillate with blood meals. Indeed, our knowledge of the physiology of the flea gut and the associated environment is very limited. In the literature, a pH of 6 to 7 has been cited (Wigglesworth, 1972), along with a Mg2+ concentration of ~0.1 mM, a Ca2+ concentration of ~0.1 to 0.3 mM, and an Fe concentration of ~2 to 4 mM (Rebeil et al., 2013).
Even though we have little knowledge on the composition and the physicochemical parameters of the flea’s gut after blood meals, it is clear that Y. pestis must face and adapt to rapid changes in this environment to produce a transmissible infection in the flea. In bacteria, two-component regulatory systems (2CSs) are key factors for detecting the absence or presence of environmental stimuli and appropriately controlling the transcription of genes whose products enable adaptation to environmental cues (Krell et al., 2010). These systems are canonically composed of an inner membrane receptor (the sensor) and a cytoplasmic effector (the response regulator) (Jung et al., 2012). In response to specific environmental cues, the sensor self-phosphorylates and then transfers the phosphoryl group to its cognate cytoplasmic regulator, which controls gene expression and thus adaptation.
To date, the PhoP-PhoQ system is the only 2CS with a proven role in Y. pestis’ adaptation to the flea (Rebeil et al., 2013, Vadyvaloo et al., 2015, Fukuto et al., 2018). Furthermore, this system and the regulatory system OmpR-EnvZ, are the only two of Y. pestis’ 23 2CSs required for virulence in rodent models of plague (Reboul et al., 2014, Oyston et al., 2000, Marceau, 2005). In other words, the same 2CS can be involved in both host and vector colonization - presumably because it can sense and respond to various stimuli encountered in specific environments (Clarke & Voigt, 2011). Like PhoP-PhoQ, the OmpR-EnvZ 2CS senses and responds to a variety of stimuli, including osmolarity, pH, and starvation (Yuan et al., 2017, Vescovi et al., 1997, Garcia Vescovi et al., 1996, Bader et al., 2005, Prost et al., 2007, Mizuno & Mizushima, 1990, Liu & Ferenci, 1998, Chakraborty et al., 2017, Foster & Hall, 1990). However, only the role of osmolarity has been studied in Y. pestis to date (Gao et al., 2011b, Gao et al., 2011a). In both Y. pestis and E. coli, the OmpR-EnvZ system modulates the expression of many genes, including ompF and ompC. These genes encode two major outer membrane porins involved in the diffusion of low-molecular-weight nutrients (Chopra & Eccles, 1978, Mortimer & Piddock, 1993, Nikaido, 1994, Nikaido, 2003, Yoshimura & Nikaido, 1985, Gao et al., 2011a, Gao et al., 2011b, Gao et al., 2011c, Liu et al., 2015, Cowan et al., 1992). Interestingly, when the osmolarity of the medium increases, OmpR-EnvZ upregulates ompF and ompC in Y. pestis but downregulates them in E. coli (Forst et al., 1988, Mizuno & Mizushima, 1990, Pratt et al., 1996, Gao et al., 2011a, Gao et al., 2011b) - indicating a differing role for the porins and the regulatory network in the two species.
Here, we first sought to study the pH and osmolarity of the flea gut lumen during ingestion/digestion cycles, in order to (i) provide physiologic reference data, and (ii) determine (using the OmpR-EnvZ system as a biological reporter) whether Y. pestis senses the pH and osmolarity in the flea gut. Lastly, we investigated the role of the OmpR-EnvZ system in the production of a transmissible infection in fleas, since at least one stimulus known to trigger this 2CS in E. coli (starvation) (Liu & Ferenci, 1998) is also present in the flea gut.
RESULTS
The osmolarity and pH of the flea gut lumen fluctuate with the blood meal.
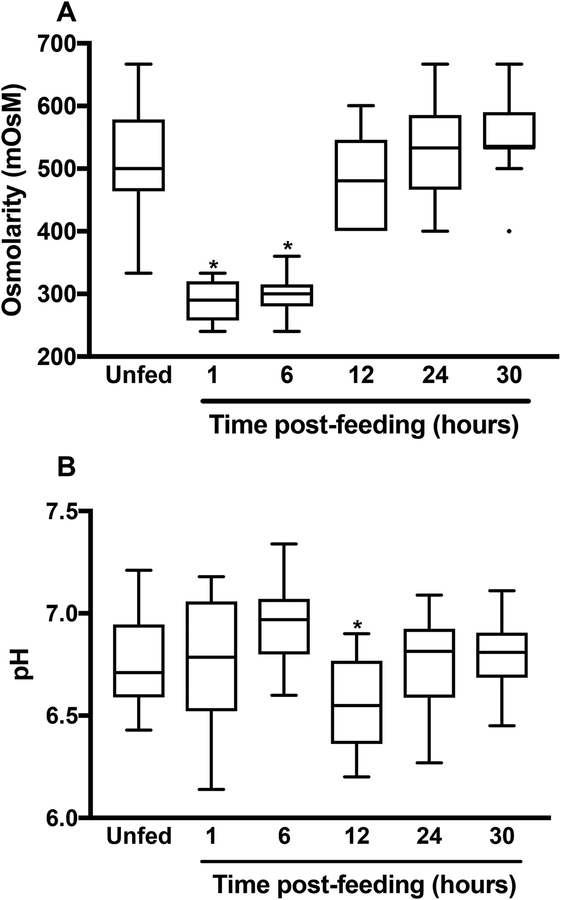
The flea gut environment has not been extensively characterized (Hinnebusch, 2004). Hence, We sought to study the osmolarity and the pH before and after a blood meal because these two basic parameters are known to (i) fluctuate with meals in the tick and mosquito guts (Bontemps-Gallo et al., 2016, Yang et al., 2000, Nepomuceno et al., 2017, Santos et al., 2011, Ramsay, 1950, Grandjean, 1983) and (ii) trigger bacterial two-component systems like PhoP-PhoQ and OmpR-EnvZ (Yuan et al., 2017, Chakraborty et al., 2017, Chakraborty & Kenney, 2018, Hall & Silhavy, 1981, Garcia Vescovi et al., 1996). We found that both osmolarity and pH displayed a biphasic profile within 24 hours of meal ingestion (Fig. 1). More specifically, the osmolarity in the flea’s gut fell from ~500 to ~300 mOsM (a value similar to that of the blood used to feed the insects) immediately after the meal had been ingested. By 12 hours post-feeding, the osmolarity had returned to its initial value, whereas the pH rose up to 6 hours, fell slightly (to 6.3) by hour 12, and then returned to its initial value within the next 12 hours. Lastly, by 24-h post-feeding, the osmolarity and pH were stable and had returned to their pre-feed values.
Figure 1. The flea gut is a fluctuating environment.
The box-and-whiskers (Tukey) plots represent the osmolarity (A) and pH (B) in the gut of X. cheopis fleas at different times after a blood meal. The osmolarity and pH of the blood used to feed the fleas are also shown. The mean ± SEM values were based on 16–21 samples, except for the osmolarity measured at 24 and 30h for which 9 and 12 samples were used, respectively. The osmolarity immediately after and 6 hours after the blood meal and the pH value 12 hours after the blood meal differed significantly from all the other measured values (*, p<0.008 in a one-way analysis of variance with Holm-Sidak’s correction for multiple comparisons).
Nutrient depletion (but neither osmolarity nor pH) may trigger the OmpR-EnvZ system in the flea gut.
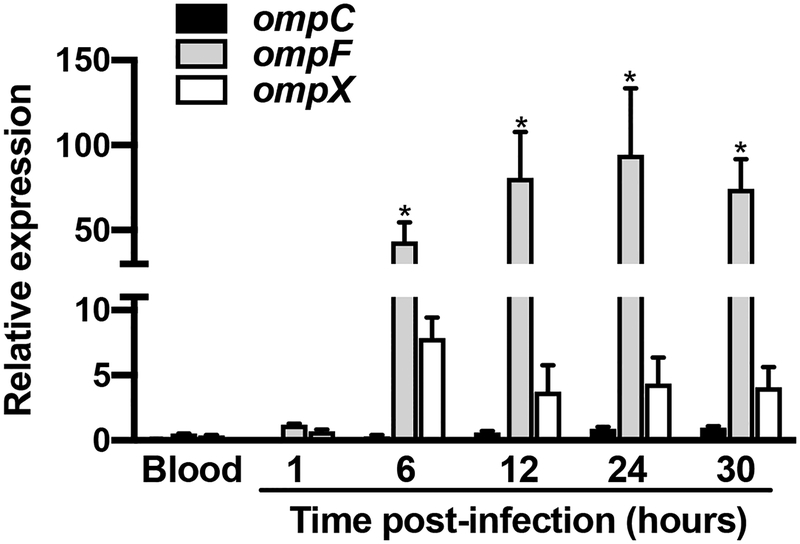
The data reported above and the fact that mouse blood is digested by X. cheopis in less than 12 hours suggest that in the flea gut, Y. pestis encounters change in osmolarity, pH, and starvation during the first 24 hours after blood ingestion. In other words, Y. pestis might sense and respond to variations in osmolarity and pH to establish an infection. Since the OmpR-EnvZ system senses osmolarity, pH, and starvation in bacteria (Chakraborty et al., 2017, Hall & Silhavy, 1981, Liu & Ferenci, 1998), we assessed the activation of this system in the flea gut at different time points post-infection and in the blood used to infect fleas. To this end, we quantified the relative expression of three OmpR-activated Y. pestis genes: ompF, ompC, and ompX (Fig. 2) (Gao et al., 2011a). We found that all three were activated in the flea but that only the activation of ompF transcription was statistically significant - presumably due to the variation between the two independent biological samples used. Interestingly, the omp genes were transcribed to a greater extent 6 hours after feeding and remained active 24 hours later. This expression profile did not match the osmolarity or pH oscillations (Fig. 1B). Hence, the results of our analysis of flea physiology and Y. pestis gene transcription suggested that the EnvZ-OmpR system senses and responds to nutrient depletion (rather than osmolarity and pH) in the flea gut from 6 hours post-feeding onwards.
Figure 2. The Y. pestis OmpR-EnvZ system is induced after a delay in the flea’s gut.
We determined the relative expression of ompC (black bars), ompF (grey bars) and ompX (white bars) in fleas at different hours post-feeding and in the blood on which they fed on. The mean ± SEM values from two independent experiments are shown. Even though the transcription of the three genes started 6 hours post-infection, only the induction of ompF was significant (*, p<0.02 in a two-way analysis of variance with Dunnett’s correction for multiple comparisons).
OmpR-EnvZ maximizes colonization of the proventriculus but not colonization of the midgut.
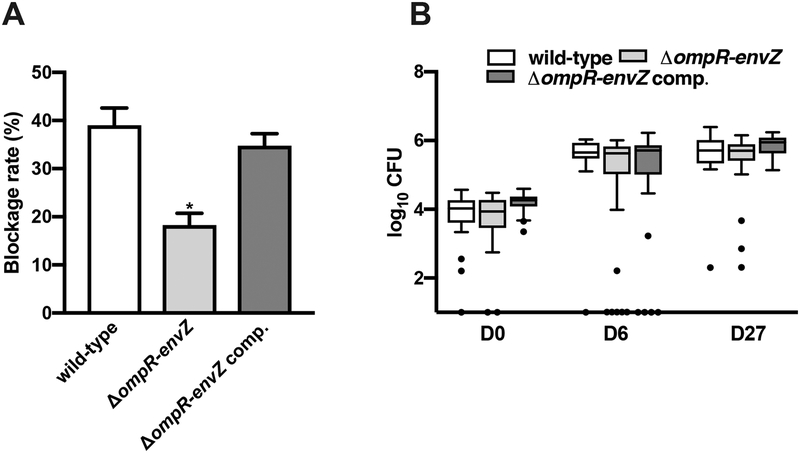
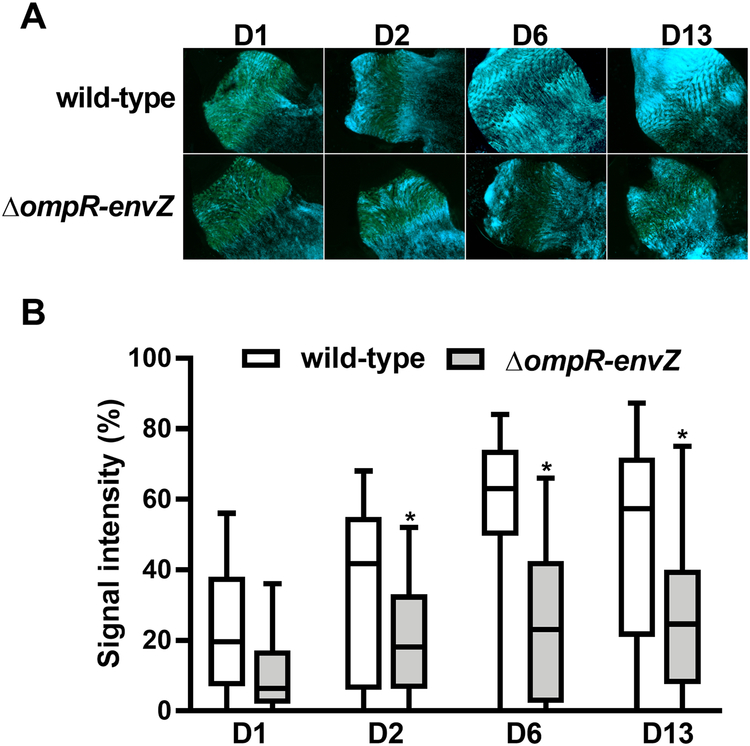
Since OmpR-EnvZ is activated in fleas, we sought to determine the 2CS’s role in the production of a transmissible infection in the flea. To this end, we generated a ΔompR-envZ mutant and monitored the latter’s ability, relative to the wild-type (WT) strain, to block fleas over the 4-week period following an infected blood meal. We found that Y. pestis lacking ompR-envZ blocked half the number of fleas blocked by the WT strain. However, the mutant could be rescued by transformation with a high-copy-number plasmid bearing the ompR-envZ operon under the control of its own promoter (Fig. 3A). The impaired blockage by the ΔompR-envZ strain does not mean that it was unable to colonize the flea gut. Indeed, the numbers of ΔompR-envZ and WT Y. pestis per flea were similar at both 6 and 27 days post-infection (Fig. 3B). Our data suggest that OmpR-EnvZ is specifically needed to obstruct the proventriculus. To confirm this hypothesis, we used GFP-expressing Y. pestis strains to quantify the relative amount of ΔompR-envZ mutant and WT bacteria in the proventriculus on the first day and up to 13 days post-infection. Like the WT strain, the ΔompR-envZ mutant colonized most of the proventriculus, regardless of the day of infection (Fig. 4). However, by the second day after infection, the density of the mutant bacteria in the proventriculus was always significantly lower than that of the WT strain. In fact, the density of ΔompR-envZ bacteria remained stable over time. Overall, our data show that OmpR-EnvZ is essential for producing a dense bacterial mass that blocks the foregut and not for colonizing the gut.
Figure 3. The Y. pestis OmpR-EnvZ system is needed for optimal flea blockage but not flea infectivity.
Comparison of the ability of the Y. pestis WT strain (white bars), the ΔompR-envZ mutant strain (light grey bars), and the same ΔompR-envZ mutant strain expressing a functional ompR-envZ operon under the control of its own promoter on a high copy plasmid (dark grey bars) to block (A) and colonize (B) fleas over a 4-week period. (A) The bars correspond to the mean ± SEM of three independent experiments with: (i) the WT strain, ΔompR-envZ::aphA, and its complemented strain; (ii) the WT strain, ΔompR-envZ::dfrB, and its complemented strain; and (iii) the WT, the two independent mutants and their complemented strains. The ΔompR-envZ mutant strain (but not its derived complemented strain) blocked significantly fewer fleas than the WT strain (*, p<0.01 in a one way analysis of variance with Holm-Sidak’s correction for multiple comparisons). (B) Box-and-whiskers (Tukey) plots representing the bacterial loads determined in 17 to 20 fleas collected immediately after feeding, and then 6 and 27 days after. The symbols indicate outliers. All three strains colonized the fleas to a similar extent (p>0.05 in a two-analysis of variance with Tukey’s correction for multiple comparisons).
Figure 4. The Y. pestis OmpR-EnvZ system is required for massive colonization of the flea’s proventriculus.
(A) Representative fluorescence photos of the proventriculus colonized by the WT and the ΔompR-envZ mutant strain on different days (D) post-infection. The photos came from two independent experiments in which 17–20 proventriculi per strain were analyzed at each time point. (B) Box-and-whiskers (Tukey) represent the surface area occupied by bacteria in the proventriculus of 37 X. cheopis fleas that had fed on blood infected with the Y. pestis WT (white bars) and the ΔompR-envZ mutant strain (grey bars) and collected on different days (D) post-infection. The data represent the sum of 2 independent experiments in which 17–20 fleas were dissected (i.e. n= 37 per time point). The symbols indicate outliers. The signal intensities of the proventriculus infected with the WT vs. the ΔompR-envZ mutant strain differed significantly from the second day post-infection onwards (*, p<0.01 in a two-way analysis of variance with Sidak’s correction for multiple comparisons).
EnvZ-OmpR is required for biofilm formation under conditions mimicking a nutrient-limited environment.
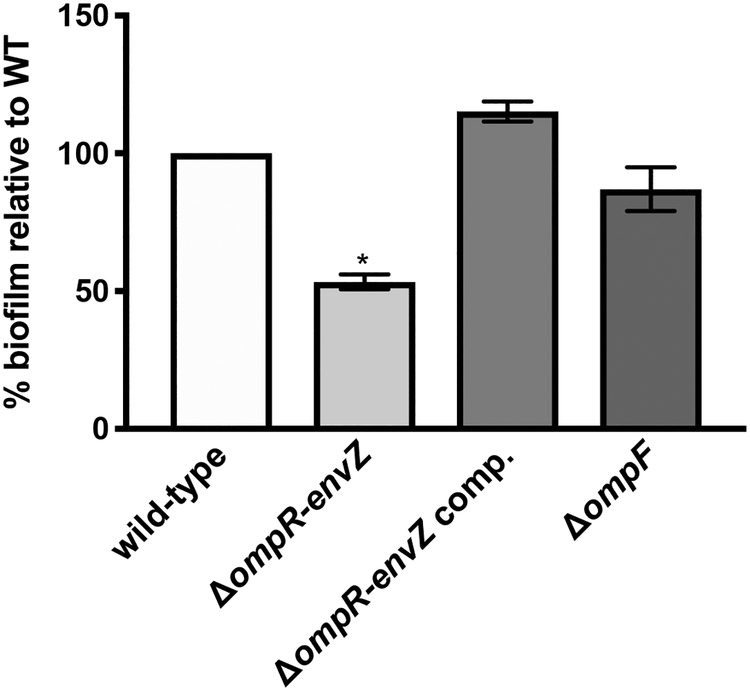
The data on flea infection highlighted the role of OmpR-EnvZ in the blockage of the proventriculus (Fig. 4). Our assessment of the flea’s physiology and transcription suggests that the flea gut is predominantly a nutrient-depleted environment where Y. pestis uses EnvZ-OmpR to sense and respond to starvation (Fig. 1 and 2). Hence, we hypothesized that OmpR-EnvZ is required for proventriculus blockage because it is needed for bacterial growth and/or biofilm formation in a nutrient-limited environment. To test this hypothesis, we compared the growth and biofilm formation of the WT and ΔompR-envZ strains in minimal medium (used here as surrogate for a nutrient-depleted environment). We found that the lack of OmpR-EnvZ did not impact bacterial growth (Fig. S1); this observation agrees with the fact that the mutant colonized the flea to the same extent as the WT (Fig. 3B). In contrast, the lack of ompR-envZ partially reduced the mutant’s ability to form a biofilm (Fig. 5), which is reminiscent of its partial loss of the ability to block fleas (Fig. 3A).
Figure 5. The Y. pestis OmpR-EnvZ system is a positive regulator of biofilm formation in vitro.
In vitro biofilm formation by the Y. pestis WT strain (white bars), the ΔompR-envZ mutant strain (light grey bars), the ΔompR-envZ mutant strain expressing a functional ompR-envZ operon under the control its own promoter on a high copy plasmid (dark grey bars), and the ΔompF mutant strain (very dark grey bars) in minimal media. The mean ± SEM values from three independent experiments are shown. Only the ΔompR-envZ mutant displayed a significant impairment in biofilm formation (*, p<0.0005 in a one-way analysis of variance with Holm-Sidak’s correction for multiple comparisons).
OmpF (but neither OmpC nor OmpX) is required for flea blockage but has no obvious role in biofilm formation in vitro.
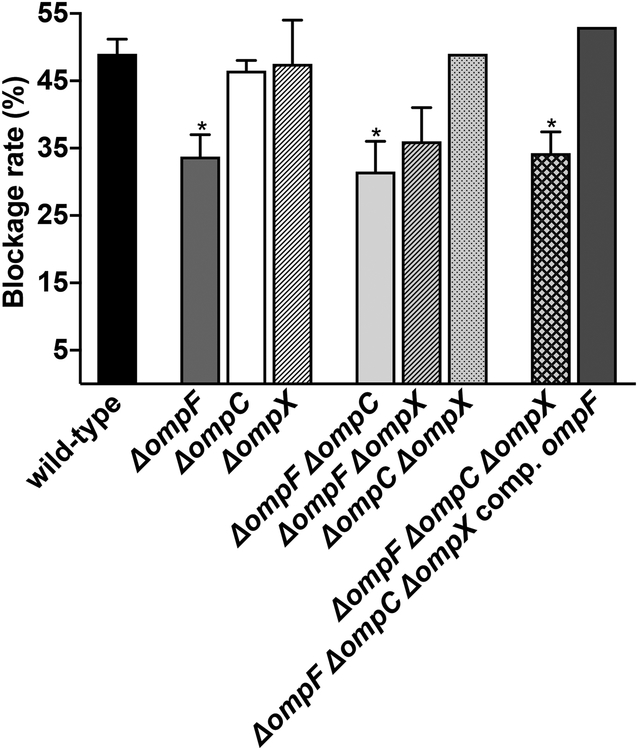
To further study the role of OmpR-EnvZ in flea blockage, we sought to identify the genes in its regulon that were needed to block fleas. To this end, we assessed the ability of Y. pestis strains lacking one, two or all three of the porin genes activated by the system (i.e. ompF, ompC and ompX) in flea infection (Fig. 6). We found that ompC and ompX were dispensable for flea blockage, even when the Y. pestis strain lacked both genes (i.e. a ΔompC ΔompX mutant). In contrast, the mutation of ompF alone was associated with a significantly lower blockage rate. This reduction was not accentuated by the absence of ompC and/or ompX (i.e. in a ΔompF ΔompC or ΔompF ΔompX double mutant or the ΔompF ΔompC ΔompX triple mutant). Lastly, the triple ΔompF ΔompC ΔompX mutant blocked fleas normally when it expressed a WT copy of ompF. Taken as a whole, these data suggest that OmpF is involved in flea blockage. However, OmpF alone does not reflect the entire role of OmpR-EnvZ in vivo. Indeed, strains lacking ompF had a lower blockage rate (25% of the WT value) than the ΔompR-envZ mutant (50% of the WT value). Lastly, we found that deletion of ompF did not impact biofilm formation in vitro (Fig. 5), which indicates that OmpR-EnvZ regulates several genes involved in biofilm formation but also that ompF is probably more important for biofilm formation in vivo than it appeared to be in our (imperfect) in vitro conditions.
Figure 6. OmpF (but not OmpC and OmpX) is needed for effective blockage of the flea’s digestive tract.
The figure shows the percentages of fleas that developed proventricular blockage after feeding on blood contaminated with Y. pestis lacking one, two or three of the ompF, ompC and ompX genes. The mean ± SEM values from three independent experiments are shown. *, p<0.05, in a one-way analysis of variance with Dunnett’s correction for multiple comparisons.
DISCUSSION
Arthropod-borne pathogens are often perceived as replicating in a stable environment, relative to those replicating outside a host. However, this does not prevent pathogens from experiencing sudden and sometimes challenging changes in their environment. This is the case for Y. pestis, which is quickly ingested when a flea takes a blood meal on a bacteremic host, and which then remains confined in the luminal part of the flea’s digestive tract until it is quickly transmitted to a new host (Bacot & Martin, 1914). Following ingestion, Y. pestis in slightly alkaline blood (pH 7.4) becomes suddenly exposed to a slightly acidic medium (pH 6.8) in the gut and remains so until the pathogen is transmitted to a new host. Furthermore, the pH oscillates, with an amplitude of 0.5 units during blood digestion (Fig. 1A). Like the pH, the luminal osmolarity slightly oscillates (from ~300 to 500 mOsM) but this variation is driven by the cycles of meal ingestion and digestion (Fig. 1B). It is interesting to note that pH and osmolarity variations occur successively and within the first 24 hours post-feeding (Fig. 1). This indicates that X. cheopis (the flea species used in this study) digests the mouse blood very quickly in two distinct steps. This concept is consistent with a report that X. cheopis digests mouse blood in less than 12 hours (Vashchenok et al., 1976). Hence, given that X. cheopis feeds every 2 to 4 days, it appears that Y. pestis would perceive the flea’s digestive tract to be a nutrient-limited environment for most of the time. Overall, the data indicate that Y. pestis replicates in a weakly acidic, nutrient-poor environment in the flea gut, where the osmolarity oscillates slightly with the ingestion and digestion of a food bolus.
It is noteworthy that the osmolarity profile encountered by Y. pestis in the flea gut is similar to that encountered by Borrelia burgdorferi in the tick gut (Bontemps-Gallo et al., 2016). In contrast, the gut pH is respectively acidic and alkaline (Yang et al., 2000). It is also noteworthy that as in the tick, alkalinization of the digestive tract after a blood meal has been reported in the Anopheles mosquito (Nepomuceno et al., 2017, Billker et al., 2000). Hence, it appears that nutrient depletion and variations in pH and osmolarity are common environmental variations experienced by arthropod-borne pathogens during colonization of their vector. It is probable that most of these pathogens sense and respond to these variations so that the infection is successfully transmitted by the vector. In the case of Y. pestis, we found that the bacterium uses its OmpR-EnvZ regulatory system to infect fleas; this bacterial 2CS is known to be involved in sensing and adapting to acidic pH, osmolarity, and nutrient starvation (Chakraborty & Kenney, 2018, Chakraborty et al., 2017, Gao et al., 2011b, Gao et al., 2011c, Liu et al., 2015, Mizuno & Mizushima, 1990). Our comparative analysis showed that the activation of OmpR-EnvZ system is not associated with variations in acidity or osmolarity in the gut – indicating that the third known stimulus of OmpR-EnvZ (nutrient limitation) triggers the system in vivo. Accordingly, OmpR-EnvZ is activated when X. cheopis has digested >99.5% of the ingested red blood cells (Vaughan & Azad, 1993). Although the evidence presented here suggests that EnvZ senses nutrient depletion in the flea gut, we cannot rule out the possibility that osmolarity or pH oscillations are still involved indirectly in regulation - for example, by initiating a cascade of reactions that result in gene expression profiles that differ from those regulated directly by OmpR-EnvZ. Alternatively, other environmental signals might also contribute to the stimulation of OmpR-EnvZ. For instance, at least one unidentified but cytotoxic blood plasma digestion product is generated in the flea gut during blood digestion (Hinnebusch et al., 2002). This toxic compound would presumably trigger the system by disturbing the bacterial membrane, since the EnvZ is considered to sense the latter’s physical (Clarke et al., 2011). Lastly, one can hypothesize that the stress caused by the presence of a particularly important blood-derived nutrient can also trigger this system.
The bacteriologic and fluorescence microscopy analyses of infected fleas suggest that OmpR-EnvZ is required to produce the biofilm obstructing the flea gut, rather than bacterial growth per se. Consistently, our in vitro experiments showed that a ΔompR-envZ mutant produced less biofilm than the WT strain, while its growth rate is unaffected (Fig. 5 and S1). It is noteworthy that even though Y. pestis needs OmpR-EnvZ to form a biofilm in vivo, the ΔompR-envZ mutant could still block a fair proportion of fleas. This observation suggest that OmpR-EnvZ is a key player that strengthens the structure of the biofilm so that the biofilm resists the incoming blood flow. This conclusion is reminiscent of a study using Yersinia enterocolitica (one of the three human pathogens in the Yersinia genus); the absence of OmpR-EnvZ in this species affected the biofilm’s general architecture and the thickness (Raczkowska et al., 2011). Although the data suggest that OmpR-EnvZ is needed to produce the flea-blocking biofilm in vitro and in vivo, we found that ompF (which is upregulated in fleas, known to be activated by OmpR, and needed to block fleas (biofilm formation in vivo) (Fig. 2,6)) is not required for biofilm formation in vitro under conditions that still require OmpR-EnvZ (Fig. 5). One possible explanation for this discrepancy is that several of the 224 putative targets regulated by the system (Gao et al., 2011a) might be involved in biofilm formation in vitro, and that the genes required for biofilm formation in vitro might not be the same as those required in vivo. In other words, the loss of only one of the regulated genes might induce a phenotype in vivo but not in vitro. If this hypothesis is true, it suggests that OmpF imports a nutrient that is essential for biofilm formation but only in the flea gut. However, we cannot rule out another role for OmpF in biofilm formation (e.g. the export of a molecule that is essential for the biofilm’s extracellular matrix (Prehna et al. 2012)), whereas OmpR-EnvZ might control several different processes leading to flea blockage. For instance, it has been suggested that in order to block fleas, the OmpR-regulated operon phoPQ modulates physiological adaptation to the low pH in the gut (Fukuto et al., 2018, Rebeil et al., 2013, Vadyvaloo et al., 2015). However, a phoP mutant produces biofilms (albeit less adherent ones) in vitro and in the flea gut (Rebeil et al., 2013, Sun et al., 2009) - suggesting that PhoPQ has a subtle role in the biofilm’s architecture.
One may suggest that it is possible that the rovM transcriptional regulator is part of the OmpR-regulon and is involved in the formation of the gut-blocking biofilm. Indeed, rovM is known to be upregulated in the flea gut, and it has been suggested that rovM senses nutritional cues and thus facilitates the bacterium’s metabolic adaptation to the insect environment (Vadyvaloo et al., 2010, Vadyvaloo et al., 2015). However, there are several lines of evidence to suggest that rovM is not part of the OmpR regulon in Y. pestis and is not involved in biofilm formation in fleas. Firstly, in silico analyses have not found any OmpR consensus-like sequences within the region upstream of rovM. Secondly, Gao et al.’s comparative transcriptomic analysis found that that rovM expression was not affected in an ompR-negative mutant (Gao et al. 2011a). Thirdly, the urease locus in Yersinia pseudotuberculosis (the recent ancestor of Y. pestis) is regulated by OmpR and the carbon storage regulator (Csr)-RovM cascade because both OmpR and RovM bind to the urease locus promoter and not because OmpR regulates rovM (Achtman et al., 1999, Hu et al., 2009, Dai et al., 2018). Even though it has been suggested that RovM is involved in Y. pestis’s metabolic adaptation to the flea gut (because it provides a fitness advantage), it has been clearly stated that RovM does not contribute to biofilm blockage or Y. pestis’s growth or survival in the flea gut (Vadyvaloo et al., 2015). We therefore hypothesize that during flea infection, nutrient cues activate the Csr-RovM regulatory cascade and the OmpR-EnvZ regulatory system independently. Activation of the Csr-RovM cascade would increase the bacterium’s fitness in the flea gut, whereas the stimulation of the OmpR-EnvZ system would lead to biofilm production and, ultimately, gut blockage.
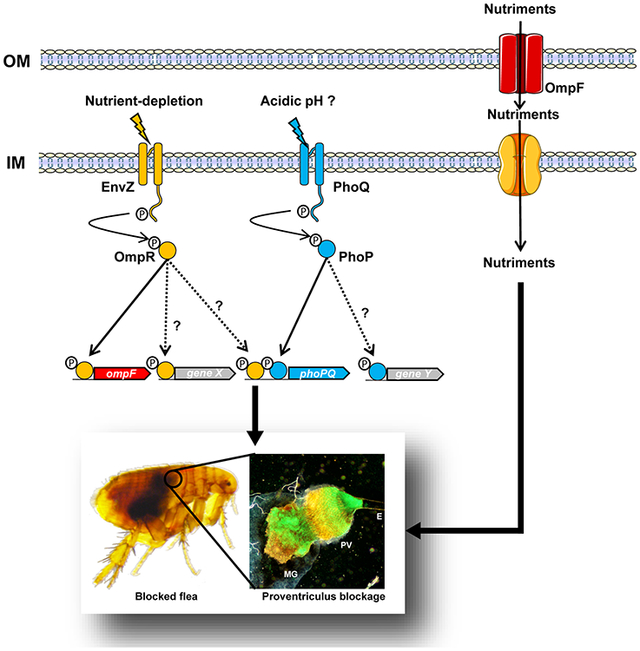
Although some pieces of the puzzle are still missing, our present results and the literature data prompted us to draw up a model of the flea environment in which Y. pestis senses and responds to physiological challenges to the establishment of a successful infection (Fig. 7). In this model, the flea gut is a slightly acidic environment with average osmolarity. The sudden entry and then the digestion of a blood meal causes harmonic oscillations in the pH, osmolarity, and nutrient concentrations. Hence, from Y. pestis’ point of view, an acid shock is the first stress encountered immediately upon ingestion in a blood meal. Subsequently, the bacterium undergoes cyclical variations in osmolarity and rapid nutrient depletion. In this complex, challenging, suddenly fluctuating environment, Y. pestis senses the depletion in nutrient content and (possibly) acidity through the EnvZ-OmpR and PhoPQ 2CSs, respectively. The stimulated EnvZ and PhoQ activate their cognate regulators (OmpR and PhoQ) and thus set in motion a molecular program (including OmpF, at least) that facilitates the production of the biofilm required for efficient plague transmission. We hypothesize that OmpF increases the flow of nutrients required for the formation of a biofilm matrix that is strong enough to block the blood drawn by the flea.
Figure 7. The environment encountered by Y. pestis during its life cycle, and the response that produce an infection transmitted by the flea.
(A) Y. pestis experiences environment fluctuations in pH and osmolarity when it passes from the mammal’s blood into the arthropod vector and vice versa, and (according to our present data) also while it colonizes the flea gut. (B) In response to nutrient depletion and (perhaps) acidic pH, the EnvZ and PhoQ sensors self-phosphorylate and then transfer the phosphate to their cognate regulators OmpR and PhoQ, which sets in motion a molecular program needed to block the flea (e.g. ompF, and a gene X that has yet to be identified) (this study, and Rebeil et al., 2013 Vadyvaloo et al., 2015). The molecular program (unknown gene Y) activated by PhoQ to block fleas is unknown, while OmpR’s molecular program includes ompF and other loci (perhaps including phoPQ; this study and Gao et al., 2011b). The photo on the left represents a blocked flea, which is characterized by the presence of fresh red blood in the foregut (at the right of the circle) but not in the midgut (at the left of the cycle). The photo on the right shows the infected flea gut, which is blocked by a mass containing a myriad of Y. pestis (green). The mass extends from the esophagus (E) to the proventriculus (PV) and the midgut (MG). Genes X and Y have yet to be identified.
Lastly, from an evolutionary point of view, it is interesting to note that at least one OmpR-regulated gene (the urease locus) was inactivated during the emergence of Y. pestis (Hu et al., 2009). This silencing was a key factor in increasing the potential transmission of the bacteria by fleas (Chouikha & Hinnebusch, 2014, Sebbane et al., 2001). In other words, Y. pestis had to modify its regulatory network during its emergence in order to take full advantage of OmpR-EnvZ’s role in flea-mediated transmission. Taken as a whole, our present results and the literature data provide a fine example of how bacteria have evolved to spread effectively.
EXPERIMENTAL PROCEDURES
Strains, mutants, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1. Mutant Y. pestis strains were generated using the lambda red recombinase-based-method (Datsenko & Wanner, 2000) and the previously described primer sets and plasmids (Table S2) (Pradel et al., 2014). Each mutation was confirmed by PCR assays using (i) primer sets that were complementary to the sequence to be deleted, and (ii) primer sets that targeted a flanking sequence not involved in the allelic exchange, and the antibiotic resistance gene (Table S2). Mutant strains were complemented with the PCR Zero blunt plasmid (Life technologies) or using the mini-Tn7 (Choi et al., 2005) containing a WT (WT) copy of the gene of interest under the control of its own promoter (Table S1). The strains were grown in lysogeny broth, brain heart infusion (BHI) medium and/or minimal medium (M9; 33.7 mM Na2HPO4, 22 mM KH2PO4, 58.55 mM NaCl, 9.35 mM NH4Cl, 1 mM thiamine hydrochloride, 0.2% casamino acids, 2 mM MgSO4, 0.1 mM CaCl2) at 21, 28 and/or 37°C. When needed, agar (15 g.L−1), kanamycin (50 μg.mL−1), trimethoprim (25 μg.mL−1), zeocin (25 μg.mL−1), spectinomycin (100 μg.mL−1) or ampicillin (100 μg.mL−1) was added to obtain solid and/or selective media. All the products used for bacterial culture were purchased from Becton Dickinson (France) or Sigma-Aldrich (France).
Measurement of osmolarity and pH in the flea gut content and in mouse blood.
Intact digestive tracts lacking their Malpighian tubule system were collected from fleas under a dissecting microscope, transferred to a microfuge tube, and then gently pressed with a grinder pestle (Kimble Chase, Vineland, NJ, USA) in order to carefully extract the gut contents. For the measurement of osmolarity, it was sometimes been necessary to pool the contents of up to four guts. Mouse blood was collected in a heparinized tube (Becton Dickinson) after cardiac puncture. The osmolarity and pH of the flea gut contents and mouse blood were measured with a vapor pressure osmometer (Model 3320, Advanced Instruments) and a pH Micro Orion electrode (Thermo) connected to a FiveEasy pH Meter (Mettler Toledo).
Flea infection.
A flea population confined in the same jar and composed of enough individuals to test all the Y. pestis strains of interest on the day of infection had been starved for five days prior to infection. The fleas were infected using an artificial system described previously (Quintard et al., 2015). Around 500 Xenopsylla cheopis fleas were allowed to feed for 1 h on an artificial feeding system containing heparinized OF-1 mouse blood contaminated with 3 × 108 bacteria/ml. After feeding, a cohort composed of 50 fed males and 50 fed females was collected, maintained at 21°C and 75% humidity, and allowed to feed twice a week for a 4-week period (Quintard et al., 2015). Immediately after each feed (except at 2 days post-infection), blocked fleas were counted under a binocular microscope (i.e., presence of fresh red blood in the flea’s foregut but not in the midgut). A cohort of females was also collected, in order to measure the ingested bacterial load and/or the bacteriak load present at different days post-infection, as previously described (Quintard et al., 2015). In particular, 20 insects were sterilized (by successive washes with hydrogen peroxide, ethanol and PBS) and then individually triturated in PBS using lysing matrix H beads and a FastPrep homogenizer (MP Biomedicals, Illkirch-Graffenstaden, France). After trituration, samples were plated on BHI agar supplemented with 1 μg/ml Irgasan and 10 μg/ml hemin. The CFUs were counted after a 48-hour incubation at 28°C.
To monitor the strains’ ability to colonize the proventriculus, female fleas were infected with Y. pestis expressing the green fluorescent protein (GFP) from the pAcGFP plasmid (Addgene). When required, 17 to 20 fleas were selected at random, and dissected under a binocular microscope to remove the digestive tract. Immediately after dissection, fluorescence photos of the digestive tract were captured using the fluorescence Eclipse CiS microscope (Nikon) and the Sight DS-F1c camera (Nikon). All the photos had the same magnification, exposure time, and sensor sensitivity. The photos were then processed (using ImageJ software) to measure the surface area occupied by bacteria within the mass anchored to the proventriculus. To do this, the blue channel of each of the proventriculus images was extracted, and an intensity threshold was applied to remove the autofluorescence in the proventriculus while maintaining the signal emitted by fluorescent bacteria. Next, the surface area of selected part of the image area was measured and compared with the surface area of the whole image. Lastly, the photos shown in the manuscript were processed using the curve adjustment tool in Adobe Photoshop CS4, in order to highlight the GFP-expressing bacteria and facilitate their identification within the proventriculus (which autofluoresces in green).
Transcription study.
We studied gene transcription in infected female fleas collected at different time points post-infection in two independent experiments and in the contaminated bloods on which they fed. More specifically, groups of 50 infected fleas were rapidly dissected in a bath of bacteria RNAprotect reagent under a binocular microscope, with removal of the gut. Immediately after dissection, each gut was immerged and disrupted in a bath of bacteria RNAprotect reagent (Qiagen). RNA from the pooled guts and the contaminated bloods were purified using RNeasy kit (Qiagen), treated with DNAse (using a DNA-free kit (Ambion)), assessed for quality and quantity (with a Bioanalyzer [Agilent Technologies] and a Nano-Drop [GE Healthcare], respectively), and reverse-transcribed using Superscript IV First-Strand Synthesis (Invitrogen). Quantitative PCR was performed using the MX3005P thermocycler (Agilent Technologies), the SYBR green PCR master mix (Applied Biosystems), and the primer sets listed in Table S2. Data were normalized against levels of the proC transcript, and the relative fold change was calculated using the ΔCT method (Pfaffl, 2001, Koch et al., 2019).
Assessment of biofilm formation in vitro.
Bacteria grown overnight in M9 with supplements at 21°C in 24-well plates were diluted in 1 mL of fresh medium per well to give a density of 2 × 106 bacteria/ml. Twenty-four hours later (after incubation at 21°C with shaking), the medium and planktonic bacteria were removed, the wells were washed three times with sterile water. A crystal violet dye solution (0.01%) was added to stain the attached bacterial biofilm. After a 15 min incubation at room temperature, the wells were rinsed four times with 2 ml sterile water. Next, 1.5 mL of an ethanol-acetone (80:20) solution was added to solubilize the dye attached to the biofilm. Lastly, the solution was transferred to a 96-well plate, and the absorbance of each well was quantified at 542 nm using an ELx800 microplate reader (BioTek). The absorbance ratio for each strain relative to the parental strain was calculated.
Ethics statement.
The animal experiments were carried out in accordance with the European directive 2010/63/EU and the French governmental decree 2013–118, and all protocols had been approved by the French Ministry of Higher Education, Research and Innovation, and the Ethics Committee in Animal Experimentation No 75. All animal husbandry adhered to the current European and French regulations.
Data availability.
All data that support the findings of this study are available from the corresponding authors upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr B.J. Hinnebush (RML/NIH) for his help in collecting the flea gut contents. This work was funded by the Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Institut Pasteur de Lille, Université Lille, and an Agence National de la Recherche grant (reference ANR-15-CE39–0017 to F. S.). F.C.G was funded by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. S.B-G was funded by a grant from the Agence National de la Recherche. F.M. was funded by a PhD fellowship from the University of Lille. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare that no competing interests exist.
References
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E (1999) Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 96: 14043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacot AW, and Martin CJ (1914) Observations on the mechanism of the transmission of plague by fleas. J. Hyg Plague 13 (Plague Suppl. 3): 423–439. [PMC free article] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, and Miller SI (2005) Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122: 461–472. [DOI] [PubMed] [Google Scholar]
- Billker O, Miller AJ, and Sinden RE (2000) Determination of mosquito bloodmeal pH in situ by ion-selective microelectrode measurement: implications for the regulation of malarial gametogenesis. Parasitology 120 (Pt 6): 547–551. [DOI] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Vadyvaloo V, Koestler BJ, Hinz AK, Mack D, Waters CM, and Perry RD (2015) The Y. pestis HmsCDE regulatory system is essential for blockage of the oriental rat flea (Xenopsylla cheopis), a classic plague vector. Environ Microbiol 17: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontemps-Gallo S, Lawrence K, and Gherardini FC (2016) Two Different Virulence-Related Regulatory Pathways in Borrelia burgdorferi Are Directly Affected by Osmotic Fluxes in the Blood Meal of Feeding Ixodes Ticks. PLoS pathogens 12: e1005791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler T (1983) Plague and Other Yersinia Infections. Plenum Medical Book Company, New and London. [Google Scholar]
- Chakraborty S, and Kenney LJ (2018) A New Role of OmpR in Acid and Osmotic Stress in Salmonella and E. coli. Front Microbiol 9: 2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Winardhi RS, Morgan LK, Yan J, and Kenney LJ (2017) Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nat Commun 8: 1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, and Schweizer HP (2005) A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2: 443–448. [DOI] [PubMed] [Google Scholar]
- Chopra I, and Eccles SJ (1978) Diffusion of tetracycline across the outer membrane of Escherichia coli K-12: involvement of protein Ia. Biochem Biophys Res Commun 83: 550–557. [DOI] [PubMed] [Google Scholar]
- Chouikha I, and Hinnebusch BJ (2014) Silencing urease: a key evolutionary step that facilitated the adaptation of Y. pestis to the flea-borne transmission route. Proc Natl Acad Sci U S A 111: 18709–18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke EJ, and Voigt CA (2011) Characterization of combinatorial patterns generated by multiple two-component sensors in E. coli that respond to many stimuli. Biotechnol Bioeng 108: 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, and Rosenbusch JP (1992) Crystal structures explain functional properties of two E. coli porins. Nature 358: 727–733. [DOI] [PubMed] [Google Scholar]
- Dai Q, Xu L, Xiao L, Zhu K, Song Y, Li C, Zhu L, Shen X, Wang Y (2018) RovM and CsrA negatively regulate urease expression in Yersinia pseudotuberculosis. Front Microbiol 9: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ, and Gage KL (2012). Transmission of flea-borne zoonotic agents. Annu Rev Entomol 57: 61–82. [DOI] [PubMed] [Google Scholar]
- Forst S, Delgado J, Ramakrishnan G, and Inouye M (1988) Regulation of ompC and ompF expression in Escherichia coli in the absence of envZ. J Bacteriol 170: 5080–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JW, and Hall HK (1990) Adaptive acidification tolerance response of Salmonella typhimurium. J Bacteriol 172: 771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuto HS, Vadyvaloo V, McPhee JB, Poinar HN, Holmes EC, and Bliska JB (2018) A Single Amino Acid Change in the Response Regulator PhoP, Acquired during Y. pestis Evolution, Affects PhoP Target Gene Transcription and Polymyxin B Susceptibility. J Bacteriol 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Zhang Y, Han Y, Yang L, Liu X, Guo Z, Tan Y, Huang X, Zhou D, and Yang R (2011a) Phenotypic and transcriptional analysis of the osmotic regulator OmpR in Y. pestis. BMC Microbiol 11: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Zhang Y, Tan Y, Wang L, Xiao X, Guo Z, Zhou D, and Yang R (2011b) Transcriptional regulation of ompF2, an ompF paralogue, in Y. pestis. Can J Microbiol 57: 468–475. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhang Y, Yang L, Liu X, Guo Z, Tan Y, Han Y, Huang X, Zhou D, and Yang R (2011c) Regulatory effects of cAMP receptor protein (CRP) on porin genes and its own gene in Y. pestis. BMC Microbiol 11: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Vescovi E, Soncini FC, and Groisman EA (1996) Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84: 165–174. [DOI] [PubMed] [Google Scholar]
- Grandjean O (1983) Blood digestion in Ornithodorus moubata Murray sensu stricto Walton females (Ixodoidea: Argasidae) II. Modifications of midgut cells related to the digestive cycle and to the triggering action of mating. Ann Parasitol Hum Comp 58: 493–514. [DOI] [PubMed] [Google Scholar]
- Hall MN, and Silhavy TJ (1981) Genetic analysis of the ompB locus in Escherichia coli K-12. J Mol Biol 151: 1–15. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ (2004) The evolution of flea-borne transmission in Y. pestis In Yersinia molecular and cellular biology. Horizon bioscience; Norfolk, UK: 49–74. [Google Scholar]
- Hinnebusch BJ, Perry RD, and Schwan TG (1996) Role of the Y. pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273: 367–370. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, and Forsberg A (2002) Role of Yersinia murine toxin in survival of Y. pestis in the midgut of the flea vector. Science 296: 733–735. [DOI] [PubMed] [Google Scholar]
- Hu Y, Lu P, Wang Y, Ding L, Atkinson S, and Chen S (2009) OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 155: 2522–2531. [DOI] [PubMed] [Google Scholar]
- Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, Kobayashi SD, DeLeo FR, and Hinnebusch BJ (2004) Transmission of Y. pestis from an infectious biofilm in the flea vector. J. Infect. Dis 190: 783–792. [DOI] [PubMed] [Google Scholar]
- Jung K, Fried L, Behr S, and Heermann R (2012) Histidine kinases and response regulators in networks. Curr Opin Microbiol 15: 118–124. [DOI] [PubMed] [Google Scholar]
- Koch L, Poyot T, Schnetterle M, Guillier S, Soule E, Nolent F, Gorge O, Neulat-Ripoll F, Valade E, Sebbane F, and Biot F (2019) Transcriptomic studies and assessment of Y. pestis reference genes in various conditions. Sci Rep 9: 2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, and Ramos JL (2010) Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol 64: 539–559. [DOI] [PubMed] [Google Scholar]
- Liu X, and Ferenci T (1998) Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. J Bacteriol 180: 3917–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang H, Wang H, Wang J, Bi Y, Wang X, Yang R, and Han Y (2015) Intrinsic plasmids influence MicF-mediated translational repression of ompF in Y. pestis. Front Microbiol 6: 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau M (2005) Transcriptional regulation in Yersinia: an update. Curr Issues Mol Biol 7: 151–177. [PubMed] [Google Scholar]
- Mizuno T, and Mizushima S (1990) Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol Microbiol 4: 1077–1082. [DOI] [PubMed] [Google Scholar]
- Mortimer PG, and Piddock LJ (1993) The accumulation of five antibacterial agents in porin-deficient mutants of Escherichia coli. J Antimicrob Chemother 32: 195–213. [DOI] [PubMed] [Google Scholar]
- Nepomuceno DB, Santos VC, Araujo RN, Pereira MH, Sant’Anna MR, Moreira LA, and Gontijo NF (2017) pH control in the midgut of Aedesaegypti under different nutritional conditions. J Exp Biol 220: 3355–3362. [DOI] [PubMed] [Google Scholar]
- Nikaido H (1994) Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem 269: 3905–3908. [PubMed] [Google Scholar]
- Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67: 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M (1897) Über die Pestepidemie in Formosa. Zentralbl Bakte-riol Parasitenkd Infektionskr 21: 769–777. [Google Scholar]
- Oyston PC, Dorrell N, Williams K, Li SR, Green M, Titball RW, and Wren BW (2000) The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Y. pestis. Infect Immun 68: 3419–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Lemaitre N, Merchez M, Ricard I, Reboul A, Dewitte A, and Sebbane F (2014) New insights into how Y. pestis adapts to its mammalian host during bubonic plague. PLoS pathogens 10: e1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LA, Hsing W, Gibson KE, and Silhavy TJ (1996) From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol 20: 911–917. [DOI] [PubMed] [Google Scholar]
- Prehna G, Zhang G, Gong X, Duszyk M, Okon M, McIntosh LP, Weiner JH, Strynadka NC (2012) A protein export pathway involving Escherichia coli porins. Structure 20: 1154–66. [DOI] [PubMed] [Google Scholar]
- Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, and Miller SI (2007) Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell 26: 165–174. [DOI] [PubMed] [Google Scholar]
- Quintard K, Dewitte A, Reboul A, Madec E, Bontemps-Gallo S, Dondeyne J, Marceau M, Simonet M, Lacroix JM, and Sebbane F (2015) Evaluation of the Role of the opgGH Operon in Yersinia pseudotuberculosis and Its Deletion during the Emergence of Y. pestis. Infect Immun 83: 3638–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczkowska A, Skorek K, Brzostkowska M, Lasinska A, and Brzostek K (2011) Pleiotropic effects of a Yersinia enterocolitica ompR mutation on adherent-invasive abilities and biofilm formation. FEMS Microbiol Lett 321: 43–49. [DOI] [PubMed] [Google Scholar]
- Ramsay JA (1950) The determination of sodium in small volumes of fluid by flame photometry. J Exp Biol 27: 407–419. [DOI] [PubMed] [Google Scholar]
- Rebeil R, Jarrett CO, Driver JD, Ernst RK, Oyston PC, and Hinnebusch BJ (2013) Induction of the Y. pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J Bacteriol 195: 1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul A, Lemaitre N, Titecat M, Merchez M, Deloison G, Ricard I, Pradel E, Marceau M, and Sebbane F (2014) Y. pestis requires the 2-component regulatory system OmpR-EnvZ to resist innate immunity during the early and late stages of plague. J Infect Dis 210: 1367–1375. [DOI] [PubMed] [Google Scholar]
- Rempe KA, Hinz AK, and Vadyvaloo V (2012) Hfq regulates biofilm gut blockage that facilitates flea-borne transmission of Y. pestis. J Bacteriol 194: 2036–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos VC, Nunes CA, Pereira MH, and Gontijo NF (2011) Mechanisms of pH control in the midgut of Lutzomyia longipalpis: roles for ingested molecules and hormones. J Exp Biol 214: 1411–1418. [DOI] [PubMed] [Google Scholar]
- Sebbane F, Devalckenaere A, Foulon J, Carniel E, and Simonet M (2001) Silencing and reactivation of urease in Yersinia pestis is determined by one G residue at a specific position in the ureD gene. Infect Immun 69: 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simond P (1898) La propagation de la peste. Ann Inst Pasteur, 12: 626–686. [Google Scholar]
- Sun YC, Koumoutsi A, and Darby C (2009) The response regulator PhoP negatively regulates Yersinia pseudotuberculosis and Y. pestis biofilms. FEMS Microbiol Lett 290: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YC, Koumoutsi A, Jarrett C, Lawrence K, Gherardini FC, Darby C, and Hinnebusch BJ (2011) Differential control of Y. pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One 6: e19267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam C, Demke O, Hermanas T, Mitchell A, Hendrickx AP, and Schneewind O (2014) YfbA, a Yersinia pestis regulator required for colonization and biofilm formation in the gut of cat fleas. J Bacteriol 196: 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra WR, and Ferreira C (1994) Insect digestive enzymes: properties, compartimetalization and function. Comp. Biochem. Physiol 109B: 1–62. [Google Scholar]
- Vadyvaloo V, Jarrett C, Sturdevant D, Sebbane F, and Hinnebusch BJ (2007) Analysis of Y. pestis gene expression in the flea vector. Advances in experimental medicine and biology 603: 192–200. [DOI] [PubMed] [Google Scholar]
- Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, and Hinnebusch BJ (2010) Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Y. pestis. PLoS Pathogens 6: e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadyvaloo V, and Hinz AK (2015) A LysR-type transcriptional regulator, RovM, senses nutritional cues suggesting that it is involved in metabolic adaptation of Yersinia pestis to the flea Gut. PLos One 10: e0137508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadyvaloo V, Viall AK, Jarrett CO, Hinz AK, Sturdevant DE, and Joseph Hinnebusch B (2015) Role of the PhoP-PhoQ gene regulatory system in adaptation of Y. pestis to environmental stress in the flea digestive tract. Microbiology 161: 1198–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashchenok VS, Solina LT, and Zhirnov AE (1976) [Characteristics of digestion of blood of different animals by the flea Xenopsylla cheopis]. Parazitologiia 10: 544–549. [PubMed] [Google Scholar]
- Vaughan JA, and Azad AF (1993) Patterns of erythrocyte digestion by bloodsucking insects: constraints on vector competence. J Med Entomol 30: 214–216. [DOI] [PubMed] [Google Scholar]
- Vescovi EG, Ayala YM, Di Cera E, and Groisman EA (1997) Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+. J Biol Chem 272: 1440–1443. [DOI] [PubMed] [Google Scholar]
- Wigglesworth VB (1972) The principles of insect physiology. Chapman and Hall; London. [Google Scholar]
- Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, and Norgard MV (2000) Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol Microbiol 37: 1470–1479. [DOI] [PubMed] [Google Scholar]
- Yersin A (1894) La peste bubonique à Hong-Kong. Annales de l’Institut Pasteur 8: 662–667. [Google Scholar]
- Yoshimura F, and Nikaido H (1985) Diffusion of beta-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob Agents Chemother 27: 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Jin F, Glatter T, and Sourjik V (2017) Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc Natl Acad Sci U S A 114: E10792–E10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Russell CW, Johnson KL, Mortensen RD, and Erickson DL (2012) Gene expression analysis of Xenopsylla cheopis (Siphonaptera: Pulicidae) suggests a role for reactive oxygen species in response to Y. pestis infection. J Med Entomol 49: 364–370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available from the corresponding authors upon reasonable request.