Abstract
Purpose:
To investigate the role of fibroblast growth factors (FGFs) in the production of neural retina (NR) and retinal pigmented epithelium (RPE) in a human pluripotent stem cell model of early retinal development.
Methods:
Human induced pluripotent stem cell (hiPSC) lines from an individual with microphthalmia caused by a functional null mutation (R200Q) in Visual systems homeobox 2 (VSX2), a transcription factor involved in early NR progenitor cell (NRPC) production, and a normal sibling were differentiated along the retinal and forebrain lineages using an established protocol. Quantitative and global gene expression analyses (microarray and RNAseq) were used to investigate endogenous FGF expression profiles in these cultures over time. Based on these results, mutant and control hiPSC cultures were treated exogenously with selected FGFs and subjected to gene and protein expression analyses to determine their effects on RPE and NR production.
Results:
We found that FGF9 and 19 were selectively increased in early hiPSC-derived optic vesicles (OVs) when compared to isogenic cultures of hiPSC-derived forebrain neurospheres. Furthermore, these same FGFs were downregulated over time in (R200Q)VSX2 hiPSC-OVs relative to sibling control hiPSC-OVs. Interestingly, long-term supplementation with FGF9, but not FGF19, partially rescued the mutant retinal phenotype of the (R200Q)VSX2 hiPSC-OV model. However, antagonizing FGF9 in wildtype control hiPSCs did not alter OV development.
Conclusions:
Our results show that FGF9 acts in concert with VSX2 to promote NR differentiation in hiPSC-OVs and has potential to be used to manipulate early retinogenesis and mitigate ocular defects caused by functional loss of VSX2 activity.
INTRODUCTION
The purpose of this thesis is to ascertain the role and impact of specific fibroblast growth factors (FGFs) in the production of neural retina (NR) and retinal pigment epithelium (RPE) from human pluripotent stem cells (hPSCs). In recent years, multiple hPSC-based clinical trials have been initiated that seek to replace RPE in age-related macular degeneration or Stargardt disease1–6. Furthermore, improvements in hPSC-NR differentiation protocols, most notably those that incorporate three dimensional culture techniques, portend future clinical trials aimed at photoreceptor replacement in late stage retinal degenerative diseases7–23 In addition to its in vivo applications, hPSC biology has been employed to create in vitro retinal cell and tissue “disease-in-a-dish” models17,24–41, which have in turn been used to establish preclinical efficacy for gene therapy trials that are underway (choroideremia) or pending (CEP290 mutation in Leber congenital amaurosis type 10)42–45. As such, hPSCs now occupy a position at the intersection of developmental biology, vision science, and ophthalmology, with rapidly increasing clinical relevance despite many gaps that remain in our understanding of this young technology7, 8, 10, 46. One such gap pertains to our limited knowledge regarding molecular cues that govern production of specific cell types and lineages in differentiating hPSCs.
One of the earliest and most important steps in vertebrate retinogenesis occurs during the optic vesicle (OV) stage, when primitive cells face the seminal decision to develop either as a neural retinal progenitor cell (NRPC; the anlage of all NR cell types) or an RPE cell47. This decision occurs shortly after the OV evaginates from the anterior neural tube, with the distal portion destined to become the NR domain, whereas the proximal portion becomes RPE48, 49. However, for an unknown period of time, the presumptive NR remains competent to develop into RPE and vice-versa50–52. The forces influencing the adoption and maintenance of these two broad retinal cell fates are not fully understood, but likely require the coordinated efforts of multiple extrinsic and intrinsic factors. One such postulated relationship involves the Fgf family of signaling molecules53–58 and the pleiotropic homeodomain transcription factor Visual system homeobox 2 (Vsx2, formerly called Chx10)53, 59–62.
Fibroblast growth factor (Fgf) signaling plays a critical role in the development of numerous tissues, including those of the eye47, 55, 63, 64. During vertebrate retinogenesis, spatiotemporal expression patterns of specific Fgfs overlap that of Vsx2, which is the earliest marker of NRPCs and is found in all of these cells throughout retinal development58–61. Early perturbations in Fgf and Vsx2 expression in vertebrate animal models caused similar ocular phenotypes, including microphthalmia and ectopic production of RPE at the expense of NR 65–67,68,69–71. Notably, NR did form to some extent when Vsx2 function or Fgf signaling was perturbed, suggesting that these factors were not strictly or solely necessary for NR specification72, 73. Using a broad and powerful, multi-ligand inhibitor of FGF signaling (SU5402), we later extended these findings to humans by showing that FGFs played a similarly prominent role in NR vs. RPE differentiation from human embryonic stem cells (hESCs)74, 75. However, the expression profiles and roles of individual FGF ligands within the 22 member family76, and their cellular influences relative to VSX2, had not been examined in differentiating hPSC retinal cultures prior to the present study.
Beginning in 200975, we and others demonstrated that both types of hPSCs, human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), could differentiate along the retinal lineage in a manner that closely paralleled normal retinogenesis16, 33, 46, 74, 77–91. Within the first weeks of differentiation, near uniform expression of VSX2 was found in a subpopulation of cell aggregates that possessed numerous characteristics of the OV46, 74, 78. These hPSC-OVs adopted a vesicular structure in suspension culture that allowed them to be visibly distinguished and manually separated from forebrain neurospheres (FBNs), which arose concurrently in culture in keeping with the co-development of these tissues during embryogenesis74. Additional investigation revealed that VSX2+ hPSC-OVs were highly proliferative and gave rise to all NR cell types in a developmentally appropriate sequence and time frame, which further identified them as multipotent NRPC cultures74, 82. Moreover, when allowed to remain as adherent cultures, cells immediately surrounding OV colonies invariably gave rise to RPE92. These and other reports established the capacity of hPSC culture systems to provide the first and still only window into the earliest stages of human retinal development. Importantly, this process was governed largely by cell and/or tissue autonomous mechanisms, since it occurred in isolated hPSC-OV cultures grown under fully defined conditions in the absence of exogenous, retina-inducing morphogens74, 75, 78. Thus, hPSC-OVs offered a unique opportunity to examine the discrete roles and relationships of endogenous developmental factors in a deconstructed model of early human retinogenesis.
To test the extent to which hPSC-OVs rely on the same developmental mechanisms as their in vivo counterparts, we embarked on a series of studies that examined the roles of specific transcription factors and signaling cues in early retinal differentiation33, 72, 92. Similar to the Vsx2 mutant mouse model, hiPSC-OVs derived from a microphthalmic patient with a functional knockout mutation (R200Q) in the VSX2 homeodomain (i.e., DNA binding) region demonstrated an NR-to-RPE shift in differentiation and delayed photoreceptor maturation, among other findings33, 93. Furthermore, RNAseq analysis of (R200Q)VSX2 hiPSC-OVs showed significant changes in the expression of key transcription factor and signaling pathway genes, including numerous FGFs33. Given that hiPSC-OVs mimic the spatiotemporal sequence of human retinal development when grown in isolation33, these cultures provide an ideal system to interrogate the roles of endogenous FGFs without confounding influences from other tissues. hiPSC-OVs also provide the only human experimental platform to corroborate or contrast findings from other species, which is particularly important for the present study given that species-specific differences in Fgf-mediated regulation are known to exist47, 64, 69, 94, 95.
Herein, we used mutant (R200Q)VSX2 and wild-type control hiPSC-OV cultures to probe the relationship between FGF signaling and VSX2 in NR production and maintenance. We hypothesized, based on prior published studies by our group72, 74, 75, 78, 96, and others53, 62, 66, that specific, endogenously expressed FGF ligands act in concert with VSX2 to establish and/or maintain NR identity in hiPSC-OVs. Quantitative RT-PCR and global gene expression analyses (microarray and RNAseq) in (R200Q)VSX2 and control hiPSC-derived cultures showed increased expression of FGF3, FGF8, and FGF9 at time points associated with eye field and OV formation. In addition, when compared to FBNs derived from the same cultures, hiPSC-OVs displayed increased expression of FGF8, FGF9, and FGF19 (equivalent to Fgf15 in mouse), three FGFs that have been specifically implicated in vertebrate retinogenesis58, 97, 98. FGF9 and FGF19 expression levels were also significantly lower in (R200Q)VSX2 vs. wildtype control hiPSC-OVs. These and other findings pointed most strongly toward FGF9, and perhaps FGF19, as having particular importance in the differentiation of NR from hiPSCs. However, treatment with FGF19 failed to reverse the NR-to-RPE conversion phenotype of (R200Q)VSX2 retinal cultures, which prompted us to focus our investigations on FGF9.
The role of FGF9 in NR vs. RPE differentiation in (R200Q)VSX2 hiPSC-OV cultures was further investigated by timed administration of exogenous FGF9, which stimulated a major downstream effector, extracellular signal-regulated kinase 1/2 (ERK1/2), and succeeded in partially rescuing the mutant phenotype. Specifically, FGF9 supplementation blunted RPE production and enhanced NR marker expression in (R200Q)VSX2 hiPSC-OVs. In contrast, directly antagonizing FGF9 signaling in wildtype control hiPSC-OVs did not affect the relative production of NR vs. RPE cells despite a reduction in ERK1/2 activation. Together, these data supported our hypothesis that FGF9 acts in concert with VSX2 to maintain NR identity in differentiating hiPSC-OVs, but also indicated that multiple, redundant mechanisms exist that support normal NR:RPE patterning. In addition, our collective results suggest that FGF9 plays a predominantly pro-NR role during early retinal development, whereas VSX2 acts in large part to suppress RPE formation. In this way, FGF9 and VSX2 exert distinct but complementary influences on NR production. This knowledge, combined with earlier published reports, could lead to more efficient methods for retinal differentiation in vitro and perhaps contribute to future strategies to combat developmental disorders of the eye and retina.
METHODS
hiPSC generation, culture, and differentiation along the retinal and forebrain lineages.
hiPSCs utilized in this study were derived from activated T-cells of a patient with a homozygous R200Q mutation in VSX2 (designated (R200Q)VSX2) and an unaffected sibling control33, 78, 93. In brief, whole blood samples were collected from both individuals and shipped to Cellular Dynamics International for reprogramming (Madison, WI). T cells within the peripheral blood mononuclear cell population were activated with OKT3 mAb (10 ng/mL, eBioscience, San Diego, CA) and recombinant human IL-2 (300 U/mL, Peprotech, Rocky Hill, NJ). Two days later, Moloney murine leukemia virus (MMLV) bicistronic constructs were used to deliver the reprogramming genes OCT4 , SOX2, c-MYC, KLF4, NANOG, and LIN28. Colonies with distinctive hiPSC morphology were visible between day 17 and day 20 after transduction, confirmed with live-cell Tra-1–60 antibody (MAB4770, R&D Systems, Minneapolis, MN), and manually picked for subsequent propagation. The control and R200Q(VSX2) hiPSC lines utilized in this study were previously characterized for expression of pluripotency markers (NANOG, OCT4, SSEA4, TRA-1–60, TRA-1–81), presence of normal karyotype and ability to form teratomas in vivo33, 78.
The control and patient samples were obtained in accordance with an approved IRB protocol at University of Wisconsin-Madison and the Helsinki declaration. After reprogramming and characterization, up to three distinct hiPSC clonal lines from both the control and R200Q(VSX2) individuals were cultured and maintained in an undifferentiated state in mTeSR1 medium99) on Matrigel® (BD Biosciences, San Jose, CA) or on irradiated mouse embryonic fibroblast feeder layers (WiCell, Madison, WI) in hiPSC culture medium (DMEM/F12, 20% knockout serum replacement or KOSR, 1% MEM nonessential amino acids, 1 mM L-glutamine, 0.1 mM β-mercaptoethanol, and basic FGF, 100 ng/ml). To differentiate hiPSCs towards the retinal lineage, we utilized our original protocol that does not employ exogenous growth factors or undefined elements such as serum33, 78. Specifically, hiPSC colonies were enzymatically lifted with dispase (1 mg/ml) and grown as three dimensional aggregate embryoid bodies (EBs) in EB medium (DMEM/F12, 20% KOSR, 1% MEM nonessential amino acids, 1 mM L-glutamine, 0.1 mM β-mercaptoethanol). On day 4 after EB generation, the culture medium was replaced with neural induction medium (NIM, DMEM/F12, 1% N2 supplement, MEM nonessential amino acids, and 2 μg/mL heparin to stabilize endogenously secreted growth factors). Two days after switching to NIM medium, EBs were plated onto laminin-coated 6-well plates and grown in NIM as an adherent culture for an additional 10 days. Subsequently, at day 16, neural clusters were mechanically lifted from the tissue culture plate and grown as free-floating suspension culture in retinal differentiation medium (RDM, DMEM/F12 [3:1], 2% B27 supplement (without retinoic acid), MEM nonessential amino acids, 1% penicillin-streptomycin). Four days later, at day 20, hiPSC-OVs and hiPSC-FBNs were manually isolated based on their distinctive appearance by light microscopy. Subsequently, hiPSC-OVs and hiPSC-FBNs were maintained in a suspension culture in RDM for up to 90 or 30 days in culture, respectively.
Recombinant FGF and FGF neutralizing antibody treatments.
Adherent retinal cultures from at least two (R200Q)VSX2 hiPSC differentiation runs were divided into at least four separate wells of a 24-well plate. Starting on day 20, cells in each well were either cultured in 500 ml RDM alone or 500 ml RDM supplemented with recombinant FGF9 or FGF19 (100 ng/ml, Pepro Tech, Rocky Hill, NJ) for the duration of the experiment. Similarly, wildtype control hiPSC retinal cultures were cultured in 500 μl RDM alone or 500 μl RDM plus neutralizing antibody against FGF9 (anti-FGF9; 500 ng/ml, R&D systems, Minneapolis, MN). RDM with or without FGF9, FGF19, or anti-FGF9 was replaced daily for the duration of the experiment. At the end of the experiments, cells were collected and processed for analysis by quantitative real time PCR (qRT-PCR), Western blot, or immunocytochemistry.
Microarray and RNAseq analysis.
Microarray74 and RNASeq33 data from wildtype control and/or R200Q(VSX2) hiPSC OV cultures at day 20 and/or day 30 were analyzed with GeneSifter software (Perkin Elmer, Waltham, MA). Of note, the raw microarray and RNASeq data utilized in this study has been previously published33, 74, 75.
Quantitative real time PCR (qRT-PCR).
Total RNA extraction was carried out using either RNeasy Mini Plus Kit (Qiagen, Germantown, MD) or ARCTURUS® PicoPure® RNA Isolation Kit (ThermoFisher Scientific, Waltham, MA) in accordance with the manufacturer’s instructions. Of note, any residual genomic DNA contamination was removed by DNase I treatment (Qiagen, Venlo, Netherlands). Subsequently, the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA) was utilized to synthesize cDNA from total RNA. Next, our previously published protocol for qRT-PCR33 was employed using a Bio-Rad CFX Thermal cycler (40 cycles), gene-specific primers (Supplementary Table 1), and the Sso Advanced SYBR Green Supermix (Bio-Rad). Data was analyzed using Bio-Rad CFX software (Bio-Rad) and Microsoft Excel.
Immunocytochemical analyses.
Immunocytochemistry was performed in accordance with our previously published protocol78. Briefly, free-floating hiPSC-OVs were fixed in 4% paraformaldehyde for 30 minutes and cryosectioned. Next, fixed hiPSC-OV cryosections were incubated in blocking solution (10% normal donkey or goat serum and 0.5% triton-X100 in PBS) for 1 hour followed by overnight incubation at 4°C in blocking buffer containing mouse primary antibody directed against Ki67 (1:500, BD Pharmingen, San Jose, CA). The next day, samples were washed 2 times in 0.05% Triton-X100 in 1X PBS and incubated for 1 hour at room temperature in blocking buffer containing host-specific
Alexa-Fluor conjugated secondary antibody (1: 500, ThermoFisher Scientific). Samples were then washed twice in 0.05% Triton-X100 in 1X PBS, incubated with the nuclear staining dye, DAPI (ThermoFisher Scientific), for 15 minutes in PBS and treated with Prolong gold (ThermoFisher Scientific) prior to placing the cover slip. Image acquisition was carried out on a Nikon 80i laser scanning confocal microscope (Nikon Corporation, Tokyo, Japan).
Western blot analysis.
hiPSC-derived retinal cultures (OVs in suspension or adherent cultures) were lysed in protein extraction buffer containing RIPA (Pierce, Rockford, IL) and protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Of note, in experiments evaluating ERK phosphorylation, a phosphatase inhibitor cocktail (Sigma-Aldrich) was also added to the protein extraction buffer. Total protein was quantified using the Bio-Rad DC protein assay (Bio-Rad) in accordance with the manufacturer’s instructions. Subsequently, protein samples were mixed with 1X Laemelli buffer containing 5% β-mercaptoethanol buffer, resolved on 4–20% Tris-HCl gradient gels (Bio-Rad), and transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad) as previously described33. The PVDF membranes were then incubated in blocking buffer (Licor Biosciences, Lincoln, NE, USA) for 1 hour at room temperature followed by overnight incubation in blocking buffer containing one of the following primary antibodies: TYR (1: 500, mouse, Abcam), ACTN (1:500, goat, Santa Cruz), Phospho-p44/42 Erk1/2 (1:1000, rabbit, Cell Signaling Technology, Danvers, MA), ERK (1: 1000, Cell Signaling Technology), RCVRN (1:2000, rabbit, Abcam), or RPE65 (1:500, mouse, EMD Millipore, Burlington, MA). The next day, PVDF membranes were washed 5 times in 0.1% Tween in 1X PBS and incubated for 1 hour at room temperature in blocking buffer solution containing host-specific infrared secondary antibodies (1:10,000, Licor Biosciences). Subsequently, blots were washed 5 times in 0.1% Tween in 1X PBS and imaged on an Odyssey Infrared Imager (Licor Biosciences).
Measurement of secreted FGF9.
hiPSC-OV cultures in 24 well plates were fed with fresh RDM and 24 hour later the conditioned medium was collected and the amount of FGF9 in the media was determined using a commercially available FGF9 ELISA kit (Abcam, Cambridge, MA) in accordance with the manufacturer’s instructions.
Statistics.
Data throughout the manuscript are expressed as mean ± SEM and compared using two tailed student’s t-test with Welch’s correction. A P value less than 0.05 was used as a cut off for significance. Specific P values approaching (but not reaching) significance were also provided where appropriate.
RESULTS
Increased expression of FGF9 and FGF19 in hiPSC-derived optic vesicles (OVs) vs. early forebrain progenitor neurosphere (FBN) cultures.
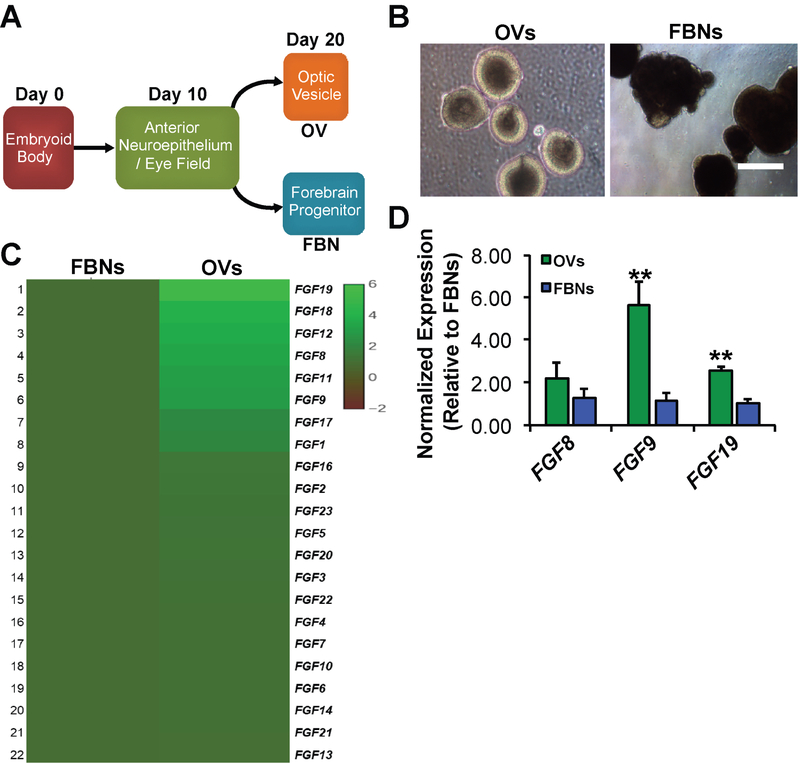
Fgf signaling is known to be involved in the development of the anterior neuroectoderm and its primary derivatives, the forebrain and retina, with certain Fgfs demonstrating differential expression between these tissues47, 55, 63, 64, 100, 101. Using an established serum-free “minimal media” hPSC differentiation protocol, which generates distinct OVs and FBNs from embryoid bodies without the need for exogenous FGFs, we sought to determine the expression levels of endogenous FGF genes in these two culture populations102, 103 (Figure 1A and 1B). Retrospective analysis of a previously published microarray data set74 comparing day 20 (D20) gene expression in isolated hiPSC-OV vs. hiPSC-FBN cultures showed differential expression of specific FGF family members, most notably FGF8, FGF9, and FGF19 (Figure 1C). Quantitative real-time PCR (qRT-PCR) analysis confirmed the increased expression of FGF9 and FGF19 in D30 hiPSC-OVs relative to hiPSC-FBNs, but not FGF8 (Figure 1D). In addition, both microarray and qRT-PCR analyses demonstrated expression of the major FGF receptors, FGFR1, FGFR2, and FGFR3, in both hiPSC-OV and hiPSC-FNB cultures (data not shown). Together, these results pointed most strongly toward FGF9 and FGF19 as potentially having selective roles in early human hiPSC-OV development.
Figure 1. Gene expression of FGF ligands in early optic vesicles (OVs) and forebrain neurospheres (FBNs) derived from hiPSCs.
A) Schematic showing the timeline of early hiPSC differentiation to OVs and FBNs. At day 0 (D0), pluripotent hiPSCs are subjected to a well-established, fully defined differentiation protocol that generates anterior neuroepithelium/eye field cells by D10, followed 10 days later by the appearance of OVs and FBNs74, 75, 78. B) hiPSC-OVs and hiPSC-FBNs can be easily distinguished in live cultures by their light microscopic appearances, manually picked, and cultured separately. C) Comparative microarray analysis showing the relative expression (see accompanying heat map legend) of FGF ligands in wildtype hiPSC-OVs vs. hiPSC-FBNs isolated from the same cultures at D2074. D) Quantitative real-time PCR analysis at D30 revealed a significantly sustained increase in the expression of FGF9 and FGF19 in hiPSC-OVs relative to hiPSC-FBNs, but not FGF8 (**P<0.01).
Expression levels of FGF9 and FGF19 are decreased in (R200Q)VSX2 vs. sibling control hiPSC-OVs.
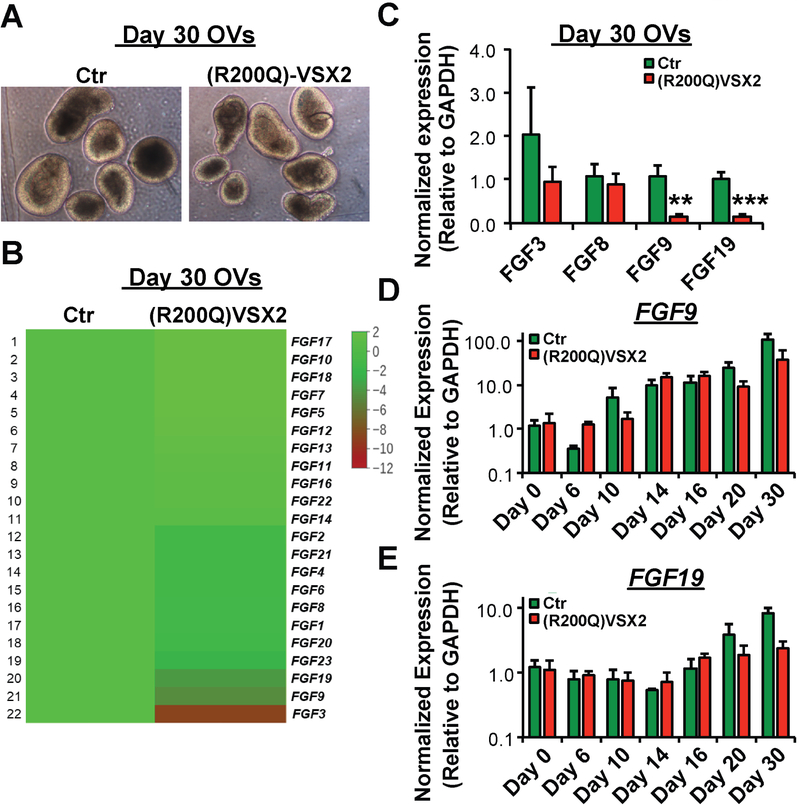
The role of FGF signaling during early vertebrate OV development has been associated with the activity of the homeodomain transcription factor VSX2, most notably in conjunction with the segregation of the NR and RPE domains62, 104. In differentiating wildtype hiPSCs, we previously demonstrated that broad inhibition of endogenous FGF signaling reduced VSX2 expression and NR cell production and concurrently increased RPE generation and expression of the RPE-specific gene Microphthalmia-associated transcription factor (MITF)33. In keeping with this finding, hiPSC-OVs derived from a patient with a functional null mutation in the homeodomain region of VSX2 (R200Q) showed increased production of MITF+ RPE at the expense of NR33, 93. RNAseq data comparing D30 OV cultures from (R200Q)VSX2 and wildtype sibling control hiPSCs revealed decreased expression of a subset of FGFs, including FGF3, FGF9, and FGF19 (and to a lesser extent FGF8), all of which have been previously linked to NR development33 (Figure 2A and 2B). However, subsequent qRT-PCR analysis across multiple cultures (n=3) narrowed the list of FGFs with significantly and consistently reduced expression in D30 (R200Q)VSX2 vs. control hiPSC-OVs to FGF9 and FGF19 , with FGF3 showing only a non-significant trend (Figure 2C). Given that FGF signaling can exert important and disparate effects at different stages of retinal development53, we next examined the expression of FGF9 and FGF19 in differentiating (R200Q)VSX2 and wildtype control hiPSC cultures over time, starting with embryoid body formation (D0), followed by production of anterior neuroectoderm/eye field (D6–D10), and finally early differentiation of OVs (D14-D30) (Figure 2D,E). From D0 to D10 (i.e., prior to VSX2 expression), FGF9 expression levels increased significantly in both (R200Q)VSX2 and wildtype control hiPSC-OVs (Figure 2D), whereas FGF19, whose expression is restricted predominantly to the developing retina33, 98, was almost nonexistent in both cultures over this time period (Figure 2E). A second rise in FGF9 expression, along with an initial increase in FGF19 expression, was seen in (R200Q)VSX2 and wildtype control hiPSC-OVs between D16-D30 (Figure 2D,E). However, consistent with results presented above (Figure 2C), FGF9 and FGF19 expression at D30 was higher in wildtype control vs. (R200Q)VSX2 hiPSC-OVs (Figure 2D,E). Therefore, FGF9 is expressed endogenously in differentiating hiPSCs at time points corresponding to anterior neuroectoderm/eye field development, whereas both FGF9 and FGF19 are in a temporal position to affect early NR development. Furthermore, our results using (R200Q)VSX2 hiPSCs suggested that the expression of both FGF9 and FGF19 in early OVs is influenced by the presence or absence of functional VSX2.
Figure 2. Comparative gene expression of FGF ligands in early wildtype control vs. (R200Q)VSX2 hiPSC-OVs.
No difference in light microscopic appearance at day 20 (D20) was observed between wildtype control and (R200Q)VSX2 hiPSC-OVs, as expected since VSX2 is first expressed around this time. B) Comparative RNAseq analysis showing the relative expression (see accompanying heat map legend) of FGF ligands in D30 (R200Q)VSX2 hiPSC-OVs relative to parallel D30 cultures of wildtype control hiPSC-OVs. C) Confirmatory qRT-PCR analysis of selected FGF ligands revealed significantly decreased expression of FGF9 and FGF19, but not FGF3 or FGF8, in (R200Q)VSX2 hiPSC-OV cultures relative to wildtype control hiPSC-OVs (**P<0.01, ***P<0.001). D,E) Quantitative RT-PCR analyses of wildtype and (R200Q)VSX2 hiPSC-OVs showing FGF9 (D) and FGF19 (E) expression levels at multiple differentiation time points between D0 and D30 (relative to D0 wildtype hiPSC-OVs; note the logarithmic y axis scale). FGF9 demonstrated a biphasic rise in expression levels between D10-D14 and again between D20-D30, whereas FGF19 expression increased between D16-D30.
Exogenous administration of FGF9, but not FGF19, antagonizes RPE cell production in differentiating (R200Q)VSX2 hiPSC retinal cultures.
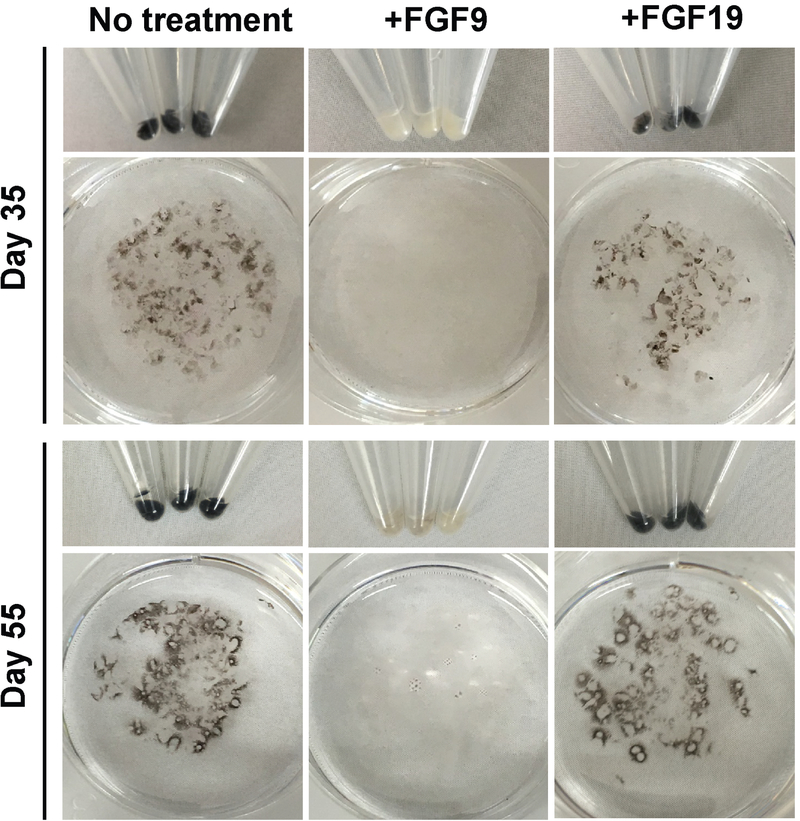
Previous in vivo mouse studies have shown that localized ectopic expression of Fgf9 or Fgf15 (equivalent to human FGF19) led to formation of excess NR tissue at the expense of RPE62. However, it is unclear to what extent these two FGFs can exert this effect in the absence of functional VSX2 and whether the effect is limited to a particular developmental time window. To address the former question using our system, we treated adherent cultures of differentiating (R200Q)VSX2 hiPSC-OVs daily with 100 ng/ml FGF9 or FGF19 starting at D20 and extending to D35–D55. Consistent with previously published data33, untreated cultures gave rise to numerous patches of deeply pigmented RPE (Figure 3). Treatment with FGF19 resulted in no phenotypic change relative to untreated control cultures, but FGF9 treatment drastically reduced production of pigmented RPE patches at D35 and D55 (Figure 3). Of note, given the comparative RNAseq results shown in Figure 2B, we also treated cultures with 100 ng/ml FGF3, which, like FGF19, had no phenotypic effect on mutant cultures (data not shown).
Figure 3. Treatment with exogenous FGF9, but not FGF19, substantially reduced production of pigmented RPE in (R200Q)VSX2 hiPSC-OV cultures.
Culture wells containing equal amounts of adherent (R200Q)VSX2 hiPSC-OVs were treated with or without 100 ng/ml FGF9 or FGF19 beginning at day 20 (D20) of differentiation. Wells were photographed on D35 and again on D55 to qualitatively assess the relative production of pigmented RPE.
Continuous FGF9 treatment within an early developmental time window is required for long-term antagonism of RPE production in differentiating (R200Q)VSX2 hiPSC retinal cultures.
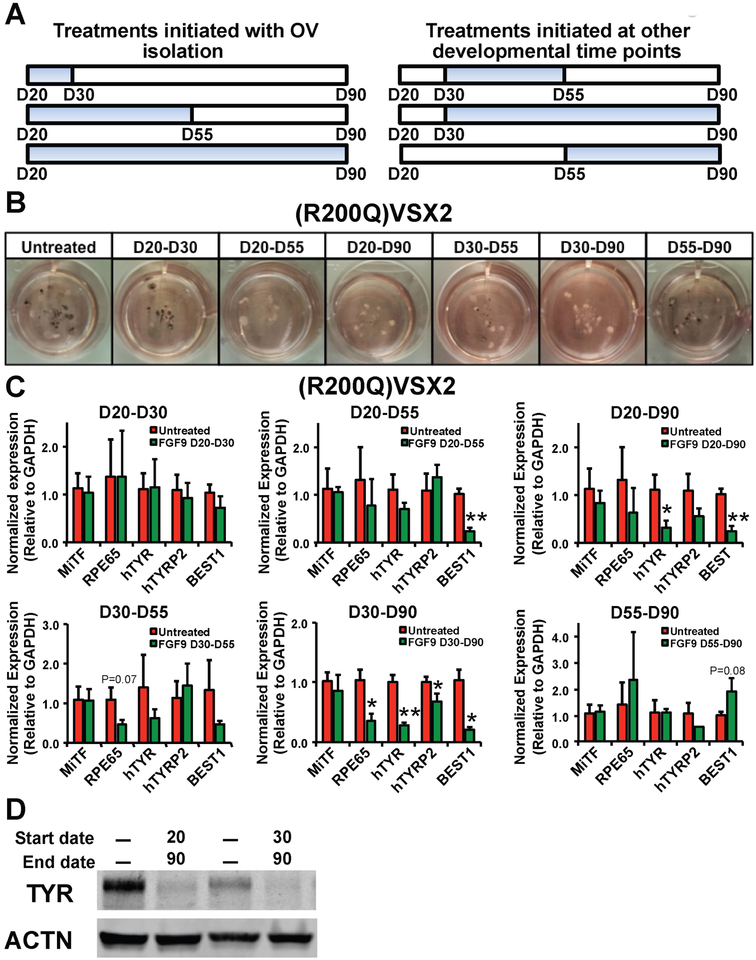
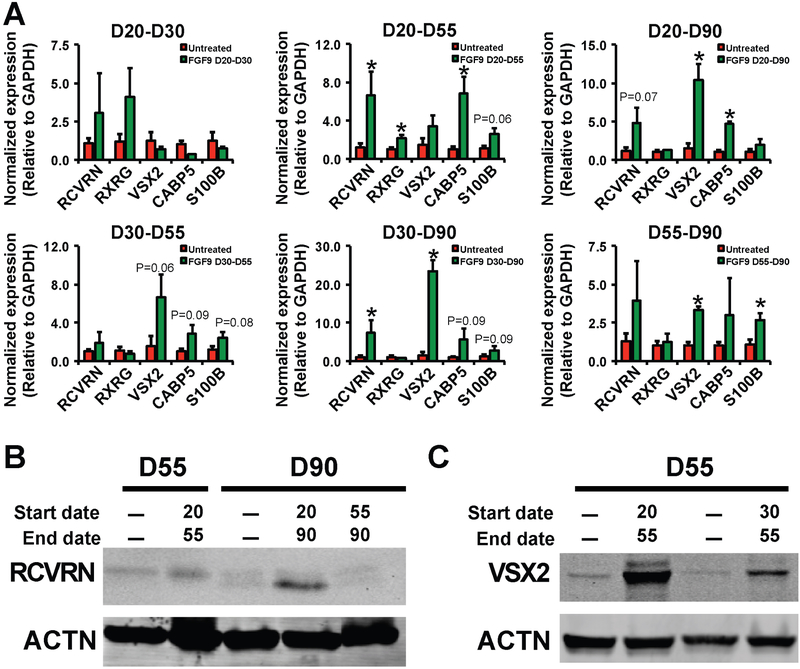
We next sought to delineate the developmental time window within which FGF9 could ameliorate the functional null VSX2 phenotype in (R200Q)VSX2 hiPSC-derived retinal cultures. For end points of FGF9 administration, we chose D30, D55, or D90, which correspond to peaks of NRPC, RPE, and photoreceptor precursor production, respectively33, 46. We also varied the day that FGF9 treatment was initiated (D20, D30, or D55). 100 ng/ml FGF9 was added to cultures daily during the prescribed window of treatment (with control cultures receiving no exogenous FGF9) and all cultures were carried to D90 (Figure 4A). Of note, ELISA confirmed that exogenous FGF9 administration led to a sustained increase in the level of FGF9 in culture media 24 hours after treatment (Supplementary Figure 1). Visual examination of cultures at D90 revealed decreased RPE-associated pigmentation in (R200Q)VSX2 hiPSC-OVs treated with FGF9 from D20–D55, D20–D90, and D30–D90, with lesser or no effects seen with treatments administered between D20–D30, D30–D55, or D55–D90 (Figure 4B). Subsequent qRT-PCR analysis confirmed significantly decreased levels of one or more RPE signature genes at D90 following FGF9 treatment from D20–D55, D20–D90, and D30–D90, but not following shorter treatments within this time window (Figure 4C). To further examine the effect of FGF9 supplementation on RPE cell differentiation in the presence or absence of functional VSX2, we compared the protein expression of the RPE marker Tyrosinase (TYR) in control vs. (R200Q)VSX2 hiPSC-OVs. Consistent with our phenotypic observations and qRT-PCR analyses, Western blot analysis showed that prolonged, daily supplementation of FGF9 (D20–D90 or D30–D90) reduced the protein expression of TYR (Figure 4D). Of note, expression of MITF, a major RPE gene directly repressed by VSX292, was not significantly altered in (R200Q)VSX2 hiPSC-OVs after FGF9 treatment from D20-D90 despite a concurrent upregulation of the functionally inert mutant VSX2 gene (Supplementary Figure 2). These findings revealed that FGF9 effects on MITF expression, unlike other RPE genes, are wholly VSX2-dependent. The persistence of MITF in treated (R200Q)VSX2 hiPSC-OVs also indicates that exposure to FGF9, while capable of antagonizing the mutant phenotype, cannot fully override the molecular consequences of loss of VSX2 function. Collectively, results from these experiments show that early, prolonged, and selective exposure to FGF9 can partially overcome the pro-RPE phenotype brought about by the functional loss of VSX2. However, the time window for achieving this effect in (R200Q)VSX2 hiPSC cultures is limited, since initiation of FGF9 treatment past D55 failed to affect RPE differentiation.
Figure 4. Early and prolonged exposure to exogenous FGF9 is required for long-term maintenance of RPE antagonism in (R200Q)VSX2 hiPSC-OV cultures.
A) Schematic depicting the time periods of FGF9 treatment tested in panels B and C (blue bars). Treatments were initiated at day 20 (D20; the day OVs are isolated), D30, or D55 and carried to D30, D55, or D90. Time points were chosen to coincide with peaks of NRPC, RPE, and photoreceptor precursor production in wildtype cultures. B) Photographs of culture wells containing equal amounts of adherent (R200Q)VSX2 hiPSC-OVs treated for the time periods shown in A. C) Quantitative RT-PCR showing expression levels of selected RPE genes relative to GAPDH in adherent cultures of (R200Q)VSX2 hiPSC-OVs treated with FGF9 for the indicated time periods (*P<0.05, **P<0.01, or otherwise indicated). D) Western blot of Tyrosinase (TYR) protein expression in untreated adherent (R200Q)VSX2 hiPSC-OV cultures (1st and 3rd lanes) and the same cultures treated with FGF9 from D20-D90 (2nd lane) or D30-D90 (4th lane). Expression of Actin (ACTN) protein was used as a control.
FGF9 supplementation promotes NR differentiation in (R200Q)VSX2 hiPSC-OVs.
After examining its impact on RPE differentiation, we wished to determine whether exogenous FGF9 also influenced NR differentiation in (R200Q)VSX2 hiPSC retinal cultures. We previously observed that (R200Q)VSX2 hiPSC-OVs exhibited delayed photoreceptor marker expression and attenuated bipolar cell marker expression33. Comparative qRT-PCR analysis of several NR genes (photoreceptors: RCVRN; retinal ganglion cells: RXRG; NRPCs and/or bipolar cells: VSX2, CABP5; Müller glia: S100B) revealed that early and prolonged FGF9 treatment significantly increased expression of RCVRN, CABP5, and VSX2 (Figure 5A). Furthermore, similar to experiments examining the effect of FGF9 on RPE cell differentiation, supplementation of FGF9 from D20–D55, D20–D90, and D30–D90 had the most profound effect on NR gene expression, although later administration of FGF9 led to an increase in S100B, a marker of proliferating Müller glia (Figure 5A). To further interrogate the effect of FGF9 supplementation on photoreceptor marker expression, we performed Western blot analysis for RCVRN. Once again, early and prolonged (D20–D90) treatment of (R200Q)VSX2 hiPSC-OVs with FGF9 increased RCVRN expression at D90, but later and shorter treatment (D55–D90) had no such effect (Figure 5B). Western blot analysis at D55 also revealed higher expression of VSX2 protein in (R200Q)VSX2 hiPSC-OVs treated with FGF9 from D20–55, with a more modest effect seen when FGF9 was introduced later (D30–D55) (Figure 5C). Altogether, our data demonstrate that early and persistent exposure to exogenous FGF9 can limit the NR-to-RPE shift in hiPSC-OVs caused by the functional absence of VSX2.
Figure 5. Early and prolonged exposure to FGF9 also led to upregulation of NR genes in (R200Q)VSX2 hiPSC-OV cultures.
A) Quantitative RT-PCR showing expression levels of selected NR genes relative to GAPDH in adherent cultures of (R200Q)VSX2 hiPSC-OVs treated with FGF9 for the same time periods investigated in Figure 4 (*P<0.05 or otherwise indicated). B) Western blot of Recoverin (RCVRN) protein expression in untreated adherent (R200Q)VSX2 hiPSC-OV cultures (1st and 3rd lanes) and the same cultures treated with FGF9 from D20-D55 (2nd lane), D20-D90 (4th lane), or D55–90 (5th lane). C) Western blot of VSX2 protein expression in untreated adherent (R200Q)VSX2 hiPSC-OV cultures (1st and 3rd lanes) and the same cultures treated with FGF9 from D20-D55 (2nd lane) or D30-D55 (4th lane). Expression of Actin (ACTN) protein was used as a control in panels B and C.
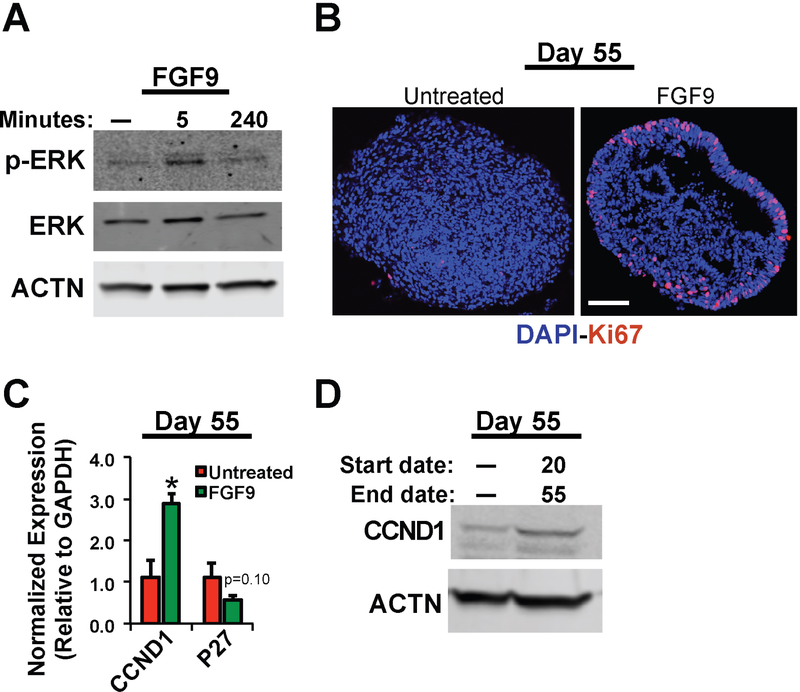
FGF9 treatment leads to activation of its downstream effector, ERK1/2, and promotes cell proliferation in (R200Q)VSX2 hiPSC-OVs.
FGF signaling is mediated by a variety of intracellular signaling pathways, including phospholipase Cγ, protein kinase C, and the ERK/mitogen-activated/protein kinase (MAPK) pathways105. Of these potential mediators, prior studies have suggested that FGF signaling utilizes the ERK/MAPK pathway to influence ocular development106,68,69–71. We found that acute administration of FGF9 to (R200Q)VSX2 hiPSC-OVs transiently activated the ERK/MAPK pathway, as shown by an increase in the amount of phosphorylated ERK1/2 (p-ERK) at five minutes post-exposure (Figure 6A). Among numerous other consequences, ERK/MAPK pathway activation can promote cell proliferation, which is deficient in differentiating (R200Q)VSX2 hiPSC-OVs33. Daily treatment with FGF9 from D20-D55 resulted in an increase in cell proliferation at D55 as determined by Ki67 immunostaining (Figure 6B), and also maintained organized, neuroepithelial structure longer than in untreated (R200Q)VSX2 hiPSC-OVs (Figure 6B and Supplementary Figure 3). Furthermore, qRT-PCR and Western blot analyses showed increased expression of the cell cycle regulator CCND1/CCND1 and decreased expression of the cell cycle inhibitor P27 in (R200Q)VSX2 hiPSC-OV cultures after daily FGF9 supplementation from D20-D55 (Figure 6C and 6D). Therefore, at least some of the effects of FGF9 administration on (R200Q)VSX2 hiPSC-OVs likely involve ERK/MAPK pathway activation and cell cycle regulation.
Figure 6. FGF9 treatment increased ERK phosphorylation and cell proliferation in (R200Q)VSX2 hiPSC-OV cultures.
A) Western blot showing the temporal effects of FGF9 treatment on levels of phosphorylated ERK (p-ERK) in (R200Q)VSX2 hiPSC-OV cultures. Expression of unphosphorylated ERK and Actin (ACTN) is also shown. B) Immunocytochemical analysis on fixed cryosections showing increased nuclear expression of the cell proliferation marker Ki67 in FGF9-treated vs. untreated (R200Q)VSX2 hiPSC-OV (Scale bar = 50 mm). C,D) Quantitative RT-PCR (C) and Western Blot (D) analyses showing increased gene and protein expression of the pro-proliferative marker CCND1/CCND1 (C,D) and decreased expression of the cell cycle inhibitor P27 (C) at day 55 (D55) in (R200Q)VSX2 hiPSC-OVs treated with or without FGF9 beginning at D20.
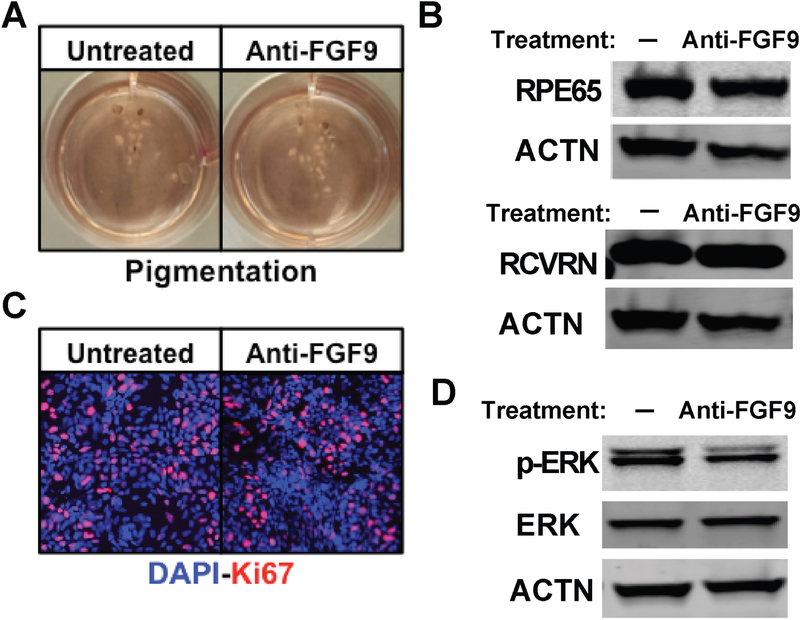
Inhibition of endogenous FGF9 does not impact differentiation of wildtype hiPSC-OVs.
Given that FGF9 supplementation partially rescued the phenotype of (R200Q)VSX2 hiPSC-OVs, we next investigated whether suppressing endogenous FGF9-mediated signaling would induce an (R200Q)VSX2 mutant-like phenotype in wildtype sibling control hiPSC-OVs. No effect of prolonged daily (D20-D90) anti-FGF9 neutralizing antibody treatment (500 ng/ml) was seen on cellular pigmentation (Figure 7A) or expression of the RPE-specific protein RPE65 in control hiPSC-OV cultures (Figure 7B). Similarly, there was no difference between treated or untreated hiPSC-OVs in the expression of the photoreceptor protein RCVRN as measured by Western blot (Figure 7B), or in cellular proliferation as determined by Ki67 immunostaining following OV dissociation and plating (Figure 7C). To confirm the activity of the FGF9 neutralizing antibody, we performed FGF9 ELISA and found that antibody treatment resulted in a decrease in endogenously secreted FGF9 to less than 10% of untreated levels (data not shown). Intracellular p-ERK levels were also transiently reduced after administration of FGF9 neutralizing antibodies in control hiPSC-OVs (Figure 7D), further confirming its anti-FGF9 activity. These results demonstrate that suppression of FGF9 alone is not sufficient to mimic the (R200Q)VSX2 phenotype in wildtype control hiPSC-OVs, which in turn suggests that NR production is likely supported by multiple signaling molecules and/or pathways with at least partially redundant activities.
Figure 7. Inhibition of FGF9 activity in wildtype hiPSC-OV cultures did not alter RPE or NR gene expression, ERK phosphorylation, or cellular proliferation.
A) Photographs taken at day 90 (D90) of culture wells containing equal amounts of adherent wildtype hiPSC-OVs treated with or without 500 ng/ml FGF9 neutralizing antibody (anti-FGF9) beginning at D20. No qualitative difference in the production of pigmented RPE was observed. B) Western blots showing similar protein expression levels of the RPE marker RPE65 and the NR (and photoreceptor) marker RCVRN with or without treatment with FGF9 neutralizing antibody from D20-D90. ACTN expression was used as a control. C) Immunocytochemical analysis also showed no difference in nuclear Ki67 expression in hiPSC-OV cultures with or without treatment with FGF9 neutralizing antibody from D20-D90. D) Western blot demonstrating a reduction in the level of phosphorylated ERK (p-ERK) protein in wildtype hiPSC-OVs 5 min after treatment with or without FGF9 neutralizing antibody (demonstrating activity of the anti-FGF9 antibody). Expression levels of ACTN and unphosphorylated ERK are also shown.
DISCUSSION
Deciphering the roles of developmental signaling factors is a challenging task, particularly when multiple factors are present that can exert competitive, redundant, and/or synergistic effects on a target cell or tissue. Adding to this complexity is the existence of large signaling factor families whose individual members may have unique or tissue-specific activities76. Such variables are brought to bear during vertebrate retinogenesis, a process that is influenced by a host of factors elaborated by the developing retina and surrounding tissues53–56. While gain and loss of function experiments in nonhuman organisms has yielded significant insight into the effects of secreted factors53, 62, 66, 107–109, hPSC model systems have the exclusive ability to test effects of molecules in isolated human cells and tissues without confounding influences from surrounding non-target tissues.
Within the retinal lineage, the choice to become either RPE or NR is of significant importance to both stem cell biology and ophthalmology given current and future therapeutic applications of these cell types or their derivatives (e.g., photoreceptors). Pharmacological and gene therapy testing has also successfully employed hPSC-derived retinal cells and tissues as model systems to support investigational new drug (IND) submissions. Therefore, it stands to reason that increased knowledge of the intrinsic and extrinsic factors governing production of specific retinal cell populations from hPSCs will enhance the clinical utility of – and confidence in – this promising technology.
In addition to its scientific and clinical significance, the RPE/NR decision fork in retinal development is particularly suitable for studies seeking to de-convolute the effects of multiple signaling factors. This step is one of the earliest in retinogenesis, and it occurs during a transient period of relatively minimal retinal tissue complexity. In addition, RPE and NRPCs show marked differences in pigmentation and morphology and are discernible based on their distinct gene and protein expression profiles. Foremost among the early RPE-and NRPC-specific genes are the transcription factors Mitf and Vsx2. In mammals, Mitf is expressed earlier than Vsx2 and is initially present throughout the early optic vesicle. Soon thereafter, upregulation of Vsx2 and downregulation of Mitf in the distal OV establishes the NR domain, whereas the proximal OV retains Mitf expression and becomes RPE53, 57, 62, 66, 97, 110–119.
The importance of Mitf/MITF and Vsx2/VSX2 during retinal development is further underscored by the phenotypes of mice and humans who lack normal function of either protein. Patients with homozygous MITF mutations exhibit anophthalmia120, while those with homozygous mutations in VSX2 display microphthalmia and retinal dysgenesis93, 121. In addition, mice with loss of function mutations in Mitf or Vsx2 exhibit profound shifts in RPE:NRPC production, with mutations in Mitf leading to excess NR tissue at the expense of RPE and mutations in Vsx2 eliciting the contrary phenotype62, 65–67, 107–109. These effects led to the speculation that Mitf and Vsx2 directly or indirectly suppressed each other’s expression or activity. Indeed, Vsx2 was shown to directly inhibit Mitf expression in mice through binding and repression of specific Mitf isoform promoter sites66, 122 and via protein-protein interactions67. However, prior to the advent of hPSC technology, the activities of these and other developmental signaling factors in differentiating human cells and tissues remained uninvestigated due to the absence of source material for such studies.
Using hiPSCs derived from a microphthalmic patient with a homozygous R200Q mutation in VSX2 that eliminates its DNA binding capacity (thus rendering it a functional “null” protein), we previously showed that mutant hiPSC-OVs grew considerably slower than wildtype sibling control hiPSC-OVs, consistent with the patient’s clinical phenotype33. In addition, (R200Q)VSX2 hiPSC-OVs demonstrated increased production of RPE at the expense of NR, as had been observed in Vsx2−/− animal models65–67, 107. Lentivirus-mediated expression of wildtype VSX2 in mutant hiPSC cultures restored production of NR while simultaneously reducing RPE generation33. To directly test MITF function during early human retinal development, we also engineered a genetic MITF−/− knockout in a hESC line92. (Of note, no patients with homozygous MITF mutations were known to exist at the time, although we later collaborated with a team from the National Eye Institute that described two such patients who exhibited anophthalmia and deafness120.) Compared to isogenic control hESCs, the MITF−/− hESC line showed defects in cell proliferation and RPE production, also mimicking effects seen in mammalian model systems92. A similar phenotype could be obtained by directly downregulating MITF expression using short hairpin RNAs (shRNAs) directed against MITF92. We then employed chromatin immunoprecipitation (ChIP) analyses to show that VSX2 bound directly to a subset of MITF isoform promoters and downregulated its expression92. These two studies established for the first time the roles of VSX2 and MITF in the establishment of the NR and RPE domains in a human developmental model system.
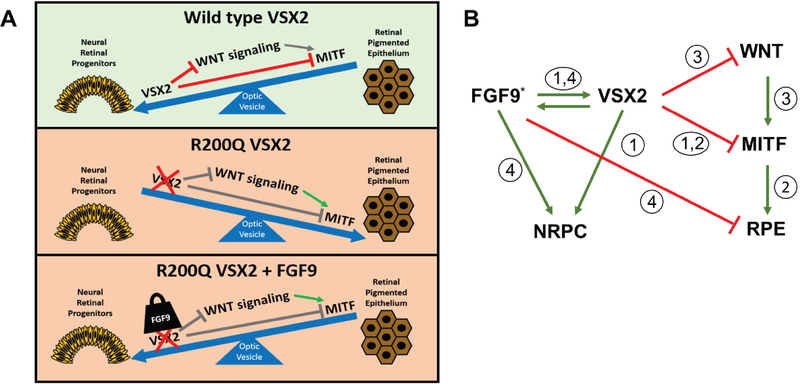
In the course of examining the functions of VSX2 and MITF in differentiating hPSC cultures, we also found relationships between developmental signaling pathways and the targeted production of NR or RPE from hPSCs33, 72, 92. RNAseq signaling pathway analysis of (R200Q)VSX2 hiPSC-OVs revealed upregulation of multiple canonical Wingless/Integrated (Wnt) pathway genes and downregulation of specific FGF family members compared to sibling wildtype control hiPSC-OVs33. Wnt agonists are similar to FGFs in that they are secreted and act on the same or nearby cells to regulate gene transcription; however, activation of the Wnt or FGF pathways yields opposing results, with Wnt stimulation favoring formation of RPE over NR. We found that pharmacological inhibition of Wnt signaling in (R200Q)VSX2 hiPSC-OVs rescued the NR-to-RPE mutant phenotype, while augmentation of Wnt signaling in wildtype hiPSC-OVs induced a NR-to-RPE production, mimicking the (R200Q)VSX2 hiPSC-OV mutant phenotype72. ChIPseq assays subsequently uncovered multiple Wnt pathway genes that, like MITF, are direct regulatory targets of VSX272. These experiments uncovered a role for VSX2 as a direct transcriptional repressor of Wnt pathway constituents and suggested a means in addition to MITF repression whereby VSX2 promoted NR production at the expense of RPE (Figure 8A, top and middle panels).
Figure 8. Model generated by our study series depicting the inter-related effects of VSX2, MITF, FGF9, and Wnt signaling on RPE and NR production in hPSCs.
Top panel: In normal hiPSC-OVs, VSX2 binds directly to and inhibits MITF and Wnt pathways genes, leading to the generation of NRPCs over RPE72, 92 in the early optic vesicle. Middle panel: In the absence of functional VSX2, inhibition of pro-RPE genes is lifted and RPE production is favored over NRPCs33. Lower panel: Application of exogenous FGF9 can tip balance back towards NRPC production and at least partially override the phenotypic consequences of the functional loss of VSX2 activity in hiPSC-OVs (present study). B) Simplified schematic showing the relative impact of VSX2, MITF, FGF9, and Wnt signaling on RPE and NRPC production in hPSCs. Note that FGF9 works in parallel with VSX2 but is not strictly required for NR production and maintenance in wildtype cultures, likely due to the redundant activity of other pro-NR factors. The asterisk denotes the existence of additional pro-NR influences from factors other than FGF9. Circled numbers demarcate the following individual studies and the aspect(s) of RPE and NR production on which the indicated study focused: 1 = Phillips and colleagues (2014)33; 2 = Capowski and colleagues (2014)92; 3 = Capowski and colleagues (2016)72; 4 = Gamm and colleagues (present study).
Unlike WNT pathway genes, FGF family member genes were not found to be direct targets of transcriptional repression by VSX272, in keeping with the synergistic effects of VSX2 and FGFs during early mammalian NR development. The overall importance of FGF signaling in the formation of NR from hPSCs was evident from our earlier study using the FGF receptor-1 inhibitor SU5402, which caused a profound reduction in VSX2 expression and a reciprocal increase in MITF expression75. This finding spurred our interest in examining the relationship between VSX2 and specific FGFs in the maintenance of NR vs. RPE cell identity in hPSC-OVs. In accordance with previously published studies, we found that FGF3, FGF8, FGF9, and FGF19 were robustly expressed in wildtype hPSC-derived OVs33. Among these FGFs, FGF9 had a peak in gene expression at time points corresponding to both neuroectoderm/eye field specification (D10) as well as OV formation (D20-D30), whereas FGF19 expression peaked only during the latter time period. Most strikingly, we discovered that supplementation with FGF9, but not FGF19, was sufficient to partially overcome the NR-to-RPE fate switch associated with the (R200Q)VSX2 hiPSC-OV mutant phenotype (Figure 8A, lower panel). However, we did not see any discernible effect of anti-FGF9 treatment in control hiPSC-derived retinal cultures. The fact that FGF9 suppression alone did not adversely affect NR production in wildtype hiPSC-OVs is likely due to redundancy in FGF signaling (or other pro-NR morphogen pathways) during retinal development.
Together, these findings suggest that FGF9 and VSX2 act in parallel to promote NR production and antagonize RPE production. This conclusion is a departure from previously held theories based on non-human, whole organism model systems that hypothesized that FGFs and VSX2 worked in series to achieve this effect53, 54, 56–58, 97, 117, 123. Our combined data further revealed that FGF9 and VSX2 are part of a redundant quality control system that assures proper NR and RPE production during retinal development. Other FGFs or signaling factors controlled by VSX2, including Wnt pathway components, are prime or known candidates in this heavily orchestrated event (Figure 8B). The requirement for continuous and prolonged FGF9 treatment in our studies also points toward a significant degree of plasticity in NR and RPE development in early OVs. This information not only sheds light on mechanisms of human retinogenesis, but may also be helpful in efforts to manipulate hPSC differentiation for applications in cell replacement therapies and disease modeling. Furthermore, although treatments for genetic defects that affect early ocular development face a high barrier for implementation in humans, our results show that such therapies are at least theoretically possible via spatiotemporally targeted application of specific developmental signaling factors.
Supplementary Material
Supplementary Figure 1. Quantification of FGF9 protein levels in (R200Q)VSX2 hiPSC-OVs with and without addition of exogenous FGF9.
The concentration of FGF9 in (R200Q)VSX2 hiPSC-OV culture media was determined by ELISA 24 hours after full media change with or without 100 ng/ml FGF9.
Supplementary Figure 2. MITF expression was unaffected by exogenous FGF9 treatment in adherent cultures of (R200Q)VSX2 hiPSC-OVs despite a substantial increase in mutant VSX2 expression.
(R200Q)VSX2 hiPSC-OV cultures were treated with exogenous FGF9 from day 20 (D20) to D90 and the levels of VSX2 and MITF gene expression were determined by qRT-PCR. FGF9 caused an increase in VSX2 expression, consistent with an increase in production of NRPCs (*P<0.05). However, there was no concurrent effect on MITF expression since mutant VSX2 protein lacks the capacity to bind MITF regulatory sequences and inhibit its expression. This result demonstrates that FGF9-induced effects on MITF expression during retinal differentiation in normal hPSCs must be VSX2-dependent.
Supplementary Figure 3. Exogenous FGF9 enhanced retention of neuroepithelial structure in (R200Q)VSX2 hiPSC-OVs.
Parallel cultures of (R200Q)VSX2 hiPSC-OVs underwent daily media changes with or without FGF9 from day 20 (D20) to D55 and the fold difference in retained, phase bright neuroepithelial structure in treated vs. untreated OVs (Figure 1B and Figure 2A) was determined by light microscopic analysis at days 20, 30, 40, and 55 (*P<0.05).
ACKNOWLEDGMENTS
Funding Support: Funded by a grant from the National Eye Institute (R01 EY021218), the Retina Research Foundation Emmett Humble Distinguished Directorship of the McPherson Eye Research Institute, Research to Prevent Blindness, and the Sandra Lemke Trout Chair in Eye Research. Drs. Gamm and Capowski are full-time employees of the University of Wisconsin-Madison, Mr. Clark was a research specialist when he assisted in this study and is currently a graduate student at the Medical College of Wisconsin, and Dr. Singh is currently employed full-time at the University of Rochester.
Other Acknowledgments: Kyle Wallace, Rasa Valiauga, David Kuai, Michael Miller, and Sarah Dickerson were research specialists and undergraduate researchers who provided assistance with hiPSC culture maintenance at times during the course of this study.
Financial Disclosures: Dr. Gamm is co-founder of Opsis Therapeutics.
Abbreviations:
- ChIP
Chromatin immunoprecipitation
- EB
Embryoid body
- ELISA
Enzyme-linked immunosorbent assay
- ERK 1/2
Extracellular signal-regulated kinase 1/2
- FBN
Forebrain neurospheres
- FGF
Fibroblast growth factor
- hPSC
Human pluripotent stem cell
- hESC
Human embryonic stem cell
- hiPSC
Human induced pluripotent stem cell
- IND
Investigational new drug
- MAPK
Mitogen-activated/protein kinase
- Mitf/MITF
Microphthalmia associated transcription factor
- NR
Neural retina
- NRPC
Neural retina progenitor cell
- OV
Optic vesicle
- PVDF
Polyvinylidene difluoride
- RPE
Retinal pigmented epithelium
- qRT-PCR
Quantitative real time polymerase chain reaction
- Vsx2/VSX2
Visual system homeobox 2
- Wnt
Wingless/Integrated
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hussain RM, Ciulla TA, Berrocal AM, Gregori NZ, Flynn HW, Lam BL. Stargardt macular dystrophy and evolving therapies. Expert Opin Biol Ther 2018;18:1049–1059. [DOI] [PubMed] [Google Scholar]
- 2.Zarbin M Cell-Based Therapy for Retinal Disease: The New Frontier. Methods Mol Biol 2019;1834:367–381. [DOI] [PubMed] [Google Scholar]
- 3.da Cruz L, Fynes K, Georgiadis O, et al. Phase 1 clinical study of an embryonic stem cell-derived retinal pigment epithelium patch in age-related macular degeneration. Nat Biotechnol 2018;36:328–337. [DOI] [PubMed] [Google Scholar]
- 4.Kashani AH, Lebkowski JS, Rahhal FM, et al. A bioengineered retinal pigment epithelial monolayer for advanced, dry age-related macular degeneration. Sci Transl Med 2018;10. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz SD, Regillo CD, Lam BL, et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet 2015;385:509–16. [DOI] [PubMed] [Google Scholar]
- 6.Mehat MS, Sundaram V, Ripamonti C, et al. Transplantation of Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cells in Macular Degeneration. Ophthalmology 2018;125:1765–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasparini SJ, Llonch S, Borsch O, Ader M. Transplantation of photoreceptors into the degenerative retina: Current state and future perspectives. Prog Retin Eye Res 2018. [DOI] [PubMed] [Google Scholar]
- 8.Jin ZB, Gao ML, Deng WL, et al. Stemming retinal regeneration with pluripotent stem cells. Prog Retin Eye Res 2018. [DOI] [PubMed] [Google Scholar]
- 9.Jung YH, Phillips MJ, Lee J, et al. 3D Microstructured Scaffolds to Support Photoreceptor Polarization and Maturation. Adv Mater 2018;30:e1803550. [DOI] [PubMed] [Google Scholar]
- 10.Gamm DM, Wong R, and working group panelists. Report on the National Eye Institute Audacious Goals Initiative: Photoreceptor Regeneration and Integration Workshop. Transl Vis Sci Technol 2015;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamba DA, Gust J, Reh TA. Transplantation of human embryonic stem cell-derived photoreceptors restores some visual function in Crx-deficient mice. Cell Stem Cell 2009;4:73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamba DA, McUsic A, Hirata RK, Wang PR, Russell D, Reh TA. Generation, purification and transplantation of photoreceptors derived from human induced pluripotent stem cells. PLoS One 2010;5:e8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hambright D, Park KY, Brooks M, McKay R, Swaroop A, Nasonkin IO. Long-term survival and differentiation of retinal neurons derived from human embryonic stem cell lines in un-immunosuppressed mouse retina. Mol Vis 2012;18:920–36. [PMC free article] [PubMed] [Google Scholar]
- 14.Barnea-Cramer AO, Wang W, Lu SJ, et al. Function of human pluripotent stem cell-derived photoreceptor progenitors in blind mice. Sci Rep 2016;6:29784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao JR, Lamba DA, Klesert TR, et al. Transplantation of Human Embryonic Stem Cell-Derived Retinal Cells into the Subretinal Space of a Non-Human Primate. Transl Vis Sci Technol 2017;6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Cordero A, Kruczek K, Naeem A, et al. Recapitulation of Human Retinal Development from Human Pluripotent Stem Cells Generates Transplantable Populations of Cone Photoreceptors. Stem Cell Reports 2017;9:820–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirai H, Mandai M, Matsushita K, et al. Transplantation of human embryonic stem cell-derived retinal tissue in two primate models of retinal degeneration. Proc Natl Acad Sci U S A 2016;113:E81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandai M, Fujii M, Hashiguchi T, et al. iPSC-Derived Retina Transplants Improve Vision in rd1 End-Stage Retinal-Degeneration Mice. Stem Cell Reports 2017;8:69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J, Cifuentes H, Reynolds J, Lamba DA. Immunosuppression via Loss of IL2rγ Enhances Long-Term Functional Integration of hESC-Derived Photoreceptors in the Mouse Retina. Cell Stem Cell 2017;20:374–384.e5. [DOI] [PubMed] [Google Scholar]
- 20.Iraha S, Tu HY, Yamasaki S, et al. Establishment of Immunodeficient Retinal Degeneration Model Mice and Functional Maturation of Human ESC-Derived Retinal Sheets after Transplantation. Stem Cell Reports 2018;10:1059–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakowski J, Welby E, Budinger D, et al. Isolation of Human Photoreceptor Precursors via a Cell Surface Marker Panel from Stem Cell-Derived Retinal Organoids and Fetal Retinae. Stem Cells 2018;36:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLelland BT, Lin B, Mathur A, et al. Transplanted hESC-Derived Retina Organoid Sheets Differentiate, Integrate, and Improve Visual Function in Retinal Degenerate Rats. Invest Ophthalmol Vis Sci 2018;59:2586–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagliardi G, Ben M’Barek K, Chaffiol A, et al. Characterization and Transplantation of CD73-Positive Photoreceptors Isolated from Human iPSC-Derived Retinal Organoids. Stem Cell Reports 2018;11:665–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Artero Castro A, Lukovic D, Jendelova P, Erceg S. Concise Review: Human Induced Pluripotent Stem Cell Models of Retinitis Pigmentosa. Stem Cells 2018;36:474–481. [DOI] [PubMed] [Google Scholar]
- 25.Galloway CA, Dalvi S, Hung SSC, et al. Drusen in patient-derived hiPSC-RPE models of macular dystrophies. Proc Natl Acad Sci U S A 2017;114:E8214–E8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha D, Phillips J, Joseph Phillips M, Gamm DM. Mimicking Retinal Development and Disease With Human Pluripotent Stem Cells. Invest Ophthalmol Vis Sci 2016;57:ORSFf1–9. [DOI] [PubMed] [Google Scholar]
- 27.Singh R, Shen W, Kuai D, et al. iPS cell modeling of Best disease: insights into the pathophysiology of an inherited macular degeneration. Hum Mol Genet 2013;22:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R, Kuai D, Guziewicz KE, et al. Pharmacological modulation of photoreceptor outer segment degradation in a human iPS cell model of inherited macular degeneration. Mol Ther 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gamm DM, Phillips MJ, Singh R. Modeling retinal degenerative diseases with human iPS-derived cells: current status and future implications. Expert Rev Ophthalmol 2013;8:213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tucker BA, Scheetz TE, Mullins RF, et al. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci U S A 2011;108:E569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucker BA, Mullins RF, Streb LM, et al. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. Elife 2013;2:e00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin ZB, Okamoto S, Xiang P, Takahashi M. Integration-free induced pluripotent stem cells derived from retinitis pigmentosa patient for disease modeling. Stem Cells Transl Med 2012;1:503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips MJ, Perez ET, Martin JM, et al. Modeling human retinal development with patient-specific induced pluripotent stem cells reveals multiple roles for visual system homeobox 2. Stem cells (Dayton, Ohio) 2014;32:1480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida T, Ozawa Y, Suzuki K, et al. The use of induced pluripotent stem cells to reveal pathogenic gene mutations and explore treatments for retinitis pigmentosa. Mol Brain 2014;7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arno G, Agrawal SA, Eblimit A, et al. Mutations in REEP6 Cause Autosomal-Recessive Retinitis Pigmentosa. Am J Hum Genet 2016;99:1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parfitt DA, Lane A, Ramsden CM, et al. Identification and Correction of Mechanisms Underlying Inherited Blindness in Human iPSC-Derived Optic Cups. Cell Stem Cell 2016;18:769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Megaw R, Abu-Arafeh H, Jungnickel M, et al. Gelsolin dysfunction causes photoreceptor loss in induced pluripotent cell and animal retinitis pigmentosa models. Nat Commun 2017;8:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz N, Lane A, Jovanovic K, et al. Arl3 and RP2 regulate the trafficking of ciliary tip kinesins. Hum Mol Genet 2017;26:2480–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma TP, Wiley LA, Whitmore SS, et al. Patient-specific induced pluripotent stem cells to evaluate the pathophysiology of TRNT1-associated Retinitis pigmentosa. Stem Cell Res 2017;21:58–70. [DOI] [PubMed] [Google Scholar]
- 40.Shimada H, Lu Q, Insinna-Kettenhofen C, et al. In Vitro Modeling Using Ciliopathy-Patient-Derived Cells Reveals Distinct Cilia Dysfunctions Caused by CEP290 Mutations. Cell Rep 2017;20:384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng WL, Gao ML, Lei XL, et al. Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients. Stem Cell Reports 2018;10:2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patrício MI, Barnard AR, Xue K, MacLaren RE. Choroideremia: molecular mechanisms and development of AAV gene therapy. Expert Opin Biol Ther 2018;18:807–820. [DOI] [PubMed] [Google Scholar]
- 43.Ruan GX, Barry E, Yu D, Lukason M, Cheng SH, Scaria A. CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10. Mol Ther 2017;25:331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiCarlo JE, Mahajan VB, Tsang SH. Gene therapy and genome surgery in the retina. J Clin Invest 2018;128:2177–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramlogan-Steel CA, Murali A, Andrzejewski S, Dhungel B, Steel JC, Layton CJ. Gene therapy and the adeno associated virus in the treatment of genetic and acquired ophthalmic diseases in humans: trials, future directions and safety considerations. Clin Exp Ophthalmol 2018. [DOI] [PubMed] [Google Scholar]
- 46.Capowski EE, Samimi K, Mayerl SJ, et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuhrmann S Eye morphogenesis and patterning of the optic vesicle. Curr Top Dev Biol 2010;93:61–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorval KM, Bobechko BP, Fujieda H, Chen S, Zack DJ, Bremner R. CHX10 targets a subset of photoreceptor genes. J Biol Chem 2006;281:744–51. [DOI] [PubMed] [Google Scholar]
- 49.Livne-Bar I, Pacal M, Cheung MC, et al. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc Natl Acad Sci U S A 2006;103:4988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sridhar A, Steward MM, Meyer JS. Nonxenogeneic growth and retinal differentiation of human induced pluripotent stem cells. Stem Cells Transl Med 2013;2:255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujimura N, Taketo MM, Mori M, Korinek V, Kozmik Z. Spatial and temporal regulation of Wnt/beta-catenin signaling is essential for development of the retinal pigment epithelium. Dev Biol 2009;334:31–45. [DOI] [PubMed] [Google Scholar]
- 52.Westenskow P, Piccolo S, Fuhrmann S. Beta-catenin controls differentiation of the retinal pigment epithelium in the mouse optic cup by regulating Mitf and Otx2 expression. Development 2009;136:2505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen M, Arnheiter H. Signaling and transcriptional regulation in early mammalian eye development: a link between FGF and MITF. Development 2000;127:3581–91. [DOI] [PubMed] [Google Scholar]
- 54.Pittack C, Grunwald GB, Reh TA. Fibroblast growth factors are necessary for neural retina but not pigmented epithelium differentiation in chick embryos. Development 1997;124:805–16. [DOI] [PubMed] [Google Scholar]
- 55.Esteve P, Bovolenta P. Secreted inducers in vertebrate eye development: more functions for old morphogens. Curr Opin Neurobiol 2006;16:13–9. [DOI] [PubMed] [Google Scholar]
- 56.Yang XJ. Roles of cell-extrinsic growth factors in vertebrate eye pattern formation and retinogenesis. Semin Cell Dev Biol 2004;15:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fuhrmann S, Levine EM, Reh TA. Extraocular mesenchyme patterns the optic vesicle during early eye development in the embryonic chick. Development 2000;127:4599–609. [DOI] [PubMed] [Google Scholar]
- 58.Zhao S, Hung FC, Colvin JS, et al. Patterning the optic neuroepithelium by FGF signaling and Ras activation. Development 2001;128:5051–60. [DOI] [PubMed] [Google Scholar]
- 59.Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci 2001;2:333–42. [DOI] [PubMed] [Google Scholar]
- 60.Liu IS, Chen JD, Ploder L, et al. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron 1994;13:377–93. [DOI] [PubMed] [Google Scholar]
- 61.Liang L, Sandell JH. Focus on molecules: homeobox protein Chx10. Exp Eye Res 2008;86:541–2. [DOI] [PubMed] [Google Scholar]
- 62.Horsford DJ, Nguyen MT, Sellar GC, Kothary R, Arnheiter H, McInnes RR. Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development 2005;132:177–87. [DOI] [PubMed] [Google Scholar]
- 63.Spence JR, Madhavan M, Aycinena JC, Del Rio-Tsonis K. Retina regeneration in the chick embryo is not induced by spontaneous Mitf downregulation but requires FGF/FGFR/MEK/Erk dependent upregulation of Pax6. Mol Vis 2007;13:57–65. [PMC free article] [PubMed] [Google Scholar]
- 64.Vergara MN, Del Rio-Tsonis K. Retinal regeneration in the Xenopus laevis tadpole: a new model system. Mol Vis 2009;15:1000–13. [PMC free article] [PubMed] [Google Scholar]
- 65.Burmeister M, Novak J, Liang MY, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet 1996;12:376–84. [DOI] [PubMed] [Google Scholar]
- 66.Rowan S, Chen CM, Young TL, Fisher DE, Cepko CL. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development (Cambridge, England) 2004;131:5139–52. [DOI] [PubMed] [Google Scholar]
- 67.Zou C, Levine EM. Vsx2 controls eye organogenesis and retinal progenitor identity via homeodomain and non-homeodomain residues required for high affinity DNA binding. PLoS Genet 2012;8:e1002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai Z, Feng GS, Zhang X. Temporal requirement of the protein tyrosine phosphatase Shp2 in establishing the neuronal fate in early retinal development. J Neurosci 2010;30:4110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinez-Morales JR, Del Bene F, Nica G, Hammerschmidt M, Bovolenta P, Wittbrodt J. Differentiation of the vertebrate retina is coordinated by an FGF signaling center. Dev Cell 2005;8:565–74. [DOI] [PubMed] [Google Scholar]
- 70.McCabe KL, Gunther EC, Reh TA. The development of the pattern of retinal ganglion cells in the chick retina: mechanisms that control differentiation. Development 1999;126:5713–24. [DOI] [PubMed] [Google Scholar]
- 71.McFarlane S, Zuber ME, Holt CE. A role for the fibroblast growth factor receptor in cell fate decisions in the developing vertebrate retina. Development 1998;125:3967–75. [DOI] [PubMed] [Google Scholar]
- 72.Capowski EE, Wright LS, Liang K, et al. Regulation of WNT Signaling by VSX2 During Optic Vesicle Patterning in Human Induced Pluripotent Stem Cells. Stem cells (Dayton, Ohio) 2016;34:2625–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stavridis MP, Collins BJ, Storey KG. Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation. Development 2010;137:881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyer JS, Howden SE, Wallace KA, et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells 2011;29:1206–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci U S A 2009;106:16698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem 2006;281:15694–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakano T, Ando S, Takata N, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012;10:771–785. [DOI] [PubMed] [Google Scholar]
- 78.Phillips MJ, Wallace KA, Dickerson SJ, et al. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest Ophthalmol Vis Sci 2012;53:2007–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phillips MJ, Jiang P, Howden S, et al. A Novel Approach to Single Cell RNA-Sequence Analysis Facilitates In Silico Gene Reporting of Human Pluripotent Stem Cell-Derived Retinal Cell Types. Stem Cells 2018;36:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sridhar A, Ohlemacher SK, Langer KB, Meyer JS. Robust Differentiation of mRNA-Reprogrammed Human Induced Pluripotent Stem Cells Toward a Retinal Lineage. Stem Cells Transl Med 2016;5:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reichman S, Terray A, Slembrouck A, et al. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc Natl Acad Sci U S A 2014;111:8518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhong X, Gutierrez C, Xue T, et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun 2014;5:4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuwahara A, Ozone C, Nakano T, Saito K, Eiraku M, Sasai Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat Commun 2015;6:6286. [DOI] [PubMed] [Google Scholar]
- 84.Mellough CB, Collin J, Khazim M, et al. IGF-1 Signaling Plays an Important Role in the Formation of Three-Dimensional Laminated Neural Retina and Other Ocular Structures From Human Embryonic Stem Cells. Stem Cells 2015;33:2416–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh RK, Mallela RK, Cornuet PK, et al. Characterization of Three-Dimensional Retinal Tissue Derived from Human Embryonic Stem Cells in Adherent Monolayer Cultures. Stem Cells Dev 2015;24:2778–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lowe A, Harris R, Bhansali P, Cvekl A, Liu W. Intercellular Adhesion-Dependent Cell Survival and ROCK-Regulated Actomyosin-Driven Forces Mediate Self-Formation of a Retinal Organoid. Stem Cell Reports 2016;6:743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wiley LA, Burnight ER, DeLuca AP, et al. cGMP production of patient-specific iPSCs and photoreceptor precursor cells to treat retinal degenerative blindness. Sci Rep 2016;6:30742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wahlin KJ, Maruotti JA, Sripathi SR, et al. Photoreceptor Outer Segment-like Structures in Long-Term 3D Retinas from Human Pluripotent Stem Cells. Sci Rep 2017;7:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ovando-Roche P, West EL, Branch MJ, et al. Use of bioreactors for culturing human retinal organoids improves photoreceptor yields. Stem Cell Res Ther 2018;9:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hallam D, Hilgen G, Dorgau B, et al. Human-Induced Pluripotent Stem Cells Generate Light Responsive Retinal Organoids with Variable and Nutrient-Dependent Efficiency. Stem Cells 2018;36:1535–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo Z, Zhong X, Li K, et al. An Optimized System for Effective Derivation of Three-Dimensional Retinal Tissue via Wnt Signaling Regulation. Stem Cells 2018;36:1709–1722. [DOI] [PubMed] [Google Scholar]
- 92.Capowski EE, Simonett JM, Clark EM, et al. Loss of MITF expression during human embryonic stem cell differentiation disrupts retinal pigment epithelium development and optic vesicle cell proliferation. Hum Mol Genet 2014;23:6332–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ferda Percin E, Ploder LA, Yu JJ, et al. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet 2000;25:397–401. [DOI] [PubMed] [Google Scholar]
- 94.Dias da Silva MR, Tiffin N, Mima T, Mikawa T, Hyer J. FGF-mediated induction of ciliary body tissue in the chick eye. Dev Biol 2007;304:272–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Willardsen M, Hutcheson DA, Moore KB, Vetter ML. The ETS transcription factor Etv1 mediates FGF signaling to initiate proneural gene expression during Xenopus laevis retinal development. Mech Dev 2014;131:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wright LS, Pinilla I, Saha J, et al. VSX2 and ASCL1 Are Indicators of Neurogenic Competence in Human Retinal Progenitor Cultures. PLoS One 2015;10:e0135830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vogel-Höpker A, Momose T, Rohrer H, Yasuda K, Ishihara L, Rapaport DH. Multiple functions of fibroblast growth factor-8 (FGF-8) in chick eye development. Mech Dev 2000;94:25–36. [DOI] [PubMed] [Google Scholar]
- 98.Kurose H, Bito T, Adachi T, Shimizu M, Noji S, Ohuchi H. Expression of Fibroblast growth factor 19 (Fgf19) during chicken embryogenesis and eye development, compared with Fgf15 expression in the mouse. Gene Expr Patterns 2004;4:687–93. [DOI] [PubMed] [Google Scholar]
- 99.Ludwig T A Thomson J. Defined, feeder-independent medium for human embryonic stem cell culture. Curr Protoc Stem Cell Biol 2007;Chapter 1:Unit 1C.2. [DOI] [PubMed] [Google Scholar]
- 100.LaVaute TM, Yoo YD, Pankratz MT, Weick JP, Gerstner JR, Zhang SC. Regulation of neural specification from human embryonic stem cells by BMP and FGF. Stem Cells 2009;27:1741–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev Dyn 1999;216:72–88. [DOI] [PubMed] [Google Scholar]
- 102.Plaza Reyes A, Petrus-Reurer S, Antonsson L, et al. Xeno-Free and Defined Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cells Functionally Integrate in a Large-Eyed Preclinical Model. Stem Cell Reports 2016;6:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buchholz DE, Pennington BO, Croze RH, Hinman CR, Coffey PJ, Clegg DO. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl Med 2013;2:384–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dyer MA. Regulation of proliferation, cell fate specification and differentiation by the homeodomain proteins Prox1, Six3, and Chx10 in the developing retina. Cell Cycle 2003;2:350–7. [PubMed] [Google Scholar]
- 105.LaVallee TM, Prudovsky IA, McMahon GA, Hu X, Maciag T. Activation of the MAP kinase pathway by FGF-1 correlates with cell proliferation induction while activation of the Src pathway correlates with migration. J Cell Biol 1998;141:1647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Galy A, Neron B, Planque N, Saule S, Eychene A. Activated MAPK/ERK kinase (MEK-1) induces transdifferentiation of pigmented epithelium into neural retina. Dev Biol 2002;248:251–64. [DOI] [PubMed] [Google Scholar]
- 107.Green ES, Stubbs JL, Levine EM. Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development 2003;130:539–52. [DOI] [PubMed] [Google Scholar]
- 108.Nakayama A, Nguyen MT, Chen CC, Opdecamp K, Hodgkinson CA, Arnheiter H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech Dev 1998;70:155–66. [DOI] [PubMed] [Google Scholar]
- 109.Scholtz CL, Chan KK. Complicated colobomatous microphthalmia in the microphthalmic (mi/mi) mouse. Development 1987;99:501–8. [DOI] [PubMed] [Google Scholar]
- 110.Bora N, Conway SJ, Liang H, Smith SB. Transient overexpression of the Microphthalmia gene in the eyes of Microphthalmia vitiligo mutant mice. Dev Dyn 1998;213:283–92. [DOI] [PubMed] [Google Scholar]
- 111.Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development 1997;124:2377–86. [DOI] [PubMed] [Google Scholar]
- 112.Hodgkinson CA, Moore KJ, Nakayama A, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 1993;74:395–404. [DOI] [PubMed] [Google Scholar]
- 113.Steingrímsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 2004;38:365–411. [DOI] [PubMed] [Google Scholar]
- 114.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 2006;12:406–14. [DOI] [PubMed] [Google Scholar]
- 115.Hemesath TJ, Steingrímsson E, McGill G, et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 1994;8:2770–80. [DOI] [PubMed] [Google Scholar]
- 116.Hyer J, Mima T, Mikawa T. FGF1 patterns the optic vesicle by directing the placement of the neural retina domain. Development 1998;125:869–77. [DOI] [PubMed] [Google Scholar]
- 117.Müller F, Rohrer H, Vogel-Höpker A. Bone morphogenetic proteins specify the retinal pigment epithelium in the chick embryo. Development 2007;134:3483–93. [DOI] [PubMed] [Google Scholar]
- 118.Bharti K, Nguyen MT, Skuntz S, Bertuzzi S, Arnheiter H. The other pigment cell: specification and development of the pigmented epithelium of the vertebrate eye. Pigment Cell Res 2006;19:380–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martínez-Morales JR, Rodrigo I, Bovolenta P. Eye development: a view from the retina pigmented epithelium. Bioessays 2004;26:766–77. [DOI] [PubMed] [Google Scholar]
- 120.George A, Zand DJ, Hufnagel RB, et al. Biallelic Mutations in MITF Cause Coloboma, Osteopetrosis, Microphthalmia, Macrocephaly, Albinism, and Deafness. Am J Hum Genet 2016;99:1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reis LM, Khan A, Kariminejad A, Ebadi F, Tyler RC, Semina EV. VSX2 mutations in autosomal recessive microphthalmia. Mol Vis 2011;17:2527–32. [PMC free article] [PubMed] [Google Scholar]
- 122.Bharti K, Gasper M, Ou J, et al. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet 2012;8:e1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ohkubo Y, Chiang C, Rubenstein JL. Coordinate regulation and synergistic actions of BMP4, SHH and FGF8 in the rostral prosencephalon regulate morphogenesis of the telencephalic and optic vesicles. Neuroscience 2002;111:1–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Quantification of FGF9 protein levels in (R200Q)VSX2 hiPSC-OVs with and without addition of exogenous FGF9.
The concentration of FGF9 in (R200Q)VSX2 hiPSC-OV culture media was determined by ELISA 24 hours after full media change with or without 100 ng/ml FGF9.
Supplementary Figure 2. MITF expression was unaffected by exogenous FGF9 treatment in adherent cultures of (R200Q)VSX2 hiPSC-OVs despite a substantial increase in mutant VSX2 expression.
(R200Q)VSX2 hiPSC-OV cultures were treated with exogenous FGF9 from day 20 (D20) to D90 and the levels of VSX2 and MITF gene expression were determined by qRT-PCR. FGF9 caused an increase in VSX2 expression, consistent with an increase in production of NRPCs (*P<0.05). However, there was no concurrent effect on MITF expression since mutant VSX2 protein lacks the capacity to bind MITF regulatory sequences and inhibit its expression. This result demonstrates that FGF9-induced effects on MITF expression during retinal differentiation in normal hPSCs must be VSX2-dependent.
Supplementary Figure 3. Exogenous FGF9 enhanced retention of neuroepithelial structure in (R200Q)VSX2 hiPSC-OVs.
Parallel cultures of (R200Q)VSX2 hiPSC-OVs underwent daily media changes with or without FGF9 from day 20 (D20) to D55 and the fold difference in retained, phase bright neuroepithelial structure in treated vs. untreated OVs (Figure 1B and Figure 2A) was determined by light microscopic analysis at days 20, 30, 40, and 55 (*P<0.05).