Abstract
High osmolarity, bound water, and hydrostatic pressure contribute to notochord mechanics and its morphogenesis into the nucleus pulposus (NP) compartment of the intervertebral disc. Indeed, the osmoadaptive transcription factor, nuclear factor of activated T-cells 5 (NFAT5 aka TonEBP), is robustly expressed by resident cells of the notochord and NP. Nevertheless, the molecular mechanisms that drive notochord osmoregulation and the functions of NFAT5 in disc embryogenesis remain largely unexplored. In this study, we show that deletion of NFAT5 in mice results in delayed vertebral column development and a reduced NP aspect ratio in the caudal spine. This phenotype is associated with lower levels of the T-box transcription factor, Brachyury, delayed expression of notochord phenotypic markers, and decreased collagen II deposition in the perinotochordal sheath and condensing mesenchyme. In addition, NFAT5 mutants showed a stage-dependent dysregulation of sonic hedgehog (Shh) signaling with non-classical expression of Gli1. Generation of mice with notochord-specific deletion of IFT88 (ShhcreERT2;Ift88f/f) supported this mode of Gli1 regulation. Using isolated primary NP cells and bioinformatics approaches, we further show that Ptch1 and Smo expression is controlled by NFAT5 in a cell autonomous manner. Altogether, our results demonstrate that NFAT5 contributes to notochord and disc embryogenesis through its regulation of hallmark notochord phenotypic markers, extracellular matrix, and Shh signaling2.
Keywords: Intervertebral disc, notochord, NFAT5 transcription factor, sonic hedgehog, brachyury, extracellular matrix
INTRODUCTION
The notochord is an evolutionarily conserved, rod-like structure encased by a sheath of extracellular matrix. It arises from the node during gastrulation by convergent extension and elongates at the midline, around which the rest of the body plan is oriented (Stemple, 2005). The fundamental roles of the notochord are to coordinate rostro-caudal elongation, provide the embryo with mechanical support, and produce morphogenic factors, such as sonic hedgehog (Shh), which pattern adjacent embryonic tissues (Corallo et al., 2015). To achieve elongation, vertebrate notochord cells exert an outward force against the sheath by increasing their osmotic activity and inflating large lysosome-related cytosolic vacuoles (Adams et al., 1990; Ellis et al., 2013). This restricted osmotic swelling increases the internal pressure of the notochord, forming a stiffened hydrostatic system. As vacuolated notochord cells enlarge and vertebral bodies gradually develop, the physical constraint of the sheath forces notochord cells into regions between developing vertebrae where they settle to form nucleus pulposus (NP) tissue of the intervertebral disc (Choi et al., 2008; Walmsley, 1953). The molecular mechanisms that drive osmoregulation in the notochord and early disc formation, however, remain largely unknown.
In situ RNA hybridization and recent RNA sequencing studies have revealed that the osmoadaptive transcription factor, nuclear factor of activated T-cells 5 (NFAT5 aka TonEBP), is expressed with high specificity in the Ciona Intestinalis notochord (José-Edwards et al., 2011; Reeves et al., 2017). Previous studies have also shown that NFAT5 is abundantly expressed in hyperosmotic tissues such as the kidneys and intervertebral discs, where it serves an osmoadaptive role (López-Rodríguez et al., 2004; Tsai et al., 2006). The notochord-derived NP is a hyperosmotic tissue due to an influx of cations drawn by high concentrations of negatively charged proteoglycans (Ghosh et al., 1975; Ishihara et al., 1997). In NP cells, NFAT5 maintains intracellular osmotic balance by enhancing the expression of key osmoadaptive genes, including tuarine transporter (Slc6A6), sodium-myo-inositol co-transporter (Slc5A3), betaine-GABA transporter (Slc6A12), and aldose reductase (AKR1B1) (Johnson et al., 2014; Tsai et al., 2006). These genes encode proteins involved in the import and synthesis of organic, non-ionic osmolytes (Burg and Ferraris, 2008), and are of interest with respect to notochord inflation and elongation. Notably, in addition to maintaining intracellular osmotic balance, NFAT5 also controls the extracellular osmotic environment by regulating the expression of key matrix-related genes, including aggrecan, β1,3-glucoronosyltransferase, and collagen II (Hiyama et al., 2009; Tsai et al., 2006; van der Windt et al., 2010). Consequently, NP cell homeostasis is largely maintained by the collective activity of NFAT5-regulated genes.
By employing NFAT5 knockout mice, we have characterized its contribution in the morphogenesis of notochord into NP. Our results do not support the view that NFAT5 is indispensable for notochord inflation and intervertebral disc development. Instead, we show that lack of NFAT5 results in delayed vertebral column development and changes in the aspect ratio of the caudal NP compartment. This phenotype is associated with lower levels of the T-box transcription factor, Brachyury, delayed expression of notochord phenotypic markers, altered collagen deposition, and dysregulated Shh signaling. In vitro experiments using primary NP cells also demonstrate that NFAT5 controls Shh signaling in a cell autonomous manner, lending mechanistic and broader insight into the important role of NFAT5 during notochord and intervertebral disc embryogenesis.
METHODS
Mice
C57BL/6 mice heterozygous for deletion of exons 6 and 7 in the Nfat5 gene were kindly provided by H. Moo Kwan, Ulsan National Institute of Science and Technology (Go et al., 2004). Nfat5 mice and embryos were genotyped by PCR using primers that span the site of deletion as previously described (Go et al., 2004). Ift88f/f mice were kindly provided by Bradley Yoder, University of Alabama, maintained on a mixed genetic background, and genotyped as previously described (Haycraft et al., 2007). ShhcreERT2 mice were obtained from The Jackson Laboratory (Stock # 005623) and crossed with Ift88f/f mice to generate ShhcreERT2; Ift88f/+ mice. To perform timed pregnancies, mice were paired, allowed to mate overnight, and immediately separated the following morning. Pregnancies were confirmed by copulation plugs and increase in body weight. To generate conditional Ift88 knockout embryos, pregnant Ift88f/f dams crossed with ShhcreERT2; Ift88f/+ males were given three consecutive i.p. injections of tamoxifen (50 mg/kg, Sigma-Aldrich, St. Louis, MO, USA) dissolved in corn oil (Sigma-Aldrich) starting at E12.5. Time of tamoxifen injection was chosen such that loss of canonical Shh sensing in the notochord would not affect sheath formation and therefore disc development (Choi and Harfe, 2011). Housing, breeding, and embryo collection at E12.5, E13.5, and E17.5 were performed under the guidelines of the Institutional Animal Care and Use Committee of Thomas Jefferson University. Aseptic technique and barrier conditions were used for social housing. Mice were given Lab Diet 5010 Laboratory Autoclavable Rodent ad libitum. Experiments were performed with two or more pairs of control and mutant littermates.
Histological Analysis
E17.5 embryos were decalcified in 20% EDTA at 4°C for 3 days, while E12.5 and E13.5 embryos did not require decalcification. Embryos were fixed in 4% paraformaldehyde (PFA) for 48 hours and embedded in paraffin for sectioning in the sagittal plane. 7μm sections were stained with 1% Safranin-O, 0.05% Fast Green, and 1% Hematoxylin, and then visualized by light microscopy (Axio Imager 2, Carl Zeiss) using 20×/0,5 EC Plan-Neofluar (Carl Zeiss) or 63×/1,4 Plan-Apochromat (Carl Zeiss) objectives. Imaging of sections was conducted with the Axiocam 105 color camera (Carl Zeiss) using Zen2™ software (Carl Zeiss). Morphological analysis was performed on eight E12.5, two E13.5, and ten E17.5 null and wild-type embryos. Aspect ratio was measured using ImageJ 1.52a (http://rsb.info.nih.gov/ij/), where the major axis was divided by the minor axis determined by Fit Ellipse measurements. Aspect ratio data were collected from five E17.5 embryos per genotype with three discs per embryo (15 discs).
Whole Mount Skeletal Preparation
Collected embryos were washed and scalded in hot tap water for 30 seconds at 65°C to facilitate removal of skin. Embryos were eviscerated, soft connective tissues were removed, and then embryos were fixed in 4% PFA for 48 hours. After fixation, embryos were submerged in Alcian blue (0.03% w/v, 80% EtOH, 20% glacial acetic acid) overnight at room temperature to stain for cartilage. Next, embryos were washed in 70% EtOH followed by 95% EtOH overnight. Alcian blue-stained embryos were then pre-cleared with 1% KOH for 1 hour. The KOH solution was replaced with Alizarin red (0.05% w/v in 95% EtOH) solution, stained for 4 hours at room temperature, and then overnight at 4°C to control the rate of staining. To clear embryos, Alizarin red was replaced with a 50% glycerol: 50% (1%) KOH solution and incubated at room temperature until samples were transparent (Rigueur and Lyons, 2014). Morphological analysis was performed on four E17.5 null and three wild-type skeletal preparations.
TUNEL Assay
The in situ Cell Death Detection Kit, TMR Red (MilliporeSigma), was used to measure cell death of E12.5 and E17.5 embryonic sections. Slides were heated at 60°C for 3 hours, de-paraffinized, rehydrated through a graded series of ethanol solutions, and permeabilized with boiling citrate-based unmasking solution (pH 6) (Vector Laboratories, H-3301) for 30 minutes at room temperature. The TUNEL assay was then performed as per manufacturer’s protocol. The positive control was generated by treatment with RNase Free DNase I (QIAGEN) (Suppl. Figure 1). Sections were washed with PBS before mounting with ProLong® Gold Antifade Mountant containing DAPI (Thermo Fisher Scientific, P36934), and visualized by fluorescence microscopy (Axio Imager 2, Carl Zeiss) using the 20x/0,5 EC Plan-Neofluar (Carl Zeiss) objective. The stained sections were imaged using X-Cite® 120Q Excitation Light Source (Excelitas), the AxioCam MRm camera (Carl Zeiss), and Zen2™ software (Carl Zeiss). Staining was performed on four embryonic notochords at E12.5 and five embryos at E17.5 with three representative discs per embryo (15 discs).
Immunohistochemistry
Sagittal sections were de-paraffinized in histoclear and rehydrated in a series of ethanol solutions (100%-70%). De-paraffinized sections were incubated in boiled citrate-based unmasking solution (Vector Laboratories, H-3301) for 20 minutes and then cooled to room temperature for 30 minutes. Next, the sections were incubated for 1 hour in the appropriate blocking solution (either 5-10% Normal Goat Serum, 10% Fetal Bovine Serum, or reagent from M.O.M.™ Immunodetection Kit; vector Laboratories, BMK-2202) and subsequently incubated overnight at 4°C with primary antibody against Brachyury (1:20; R&D, AF2085), CA3 (1:150; Santa Cruz, sc-50715), vimentin (1:200; Cell Signaling, D21H3), β-actin (1:100; Cell Signaling, 13E5), aggrecan (1:50, MilliporeSigma, AB1031), chondroitin sulfate (1:300; Abcam, ab11570), collagen II (1:400, Fitzgerald, 70R-CR008), collagen I (1:100; Abcam, ab34710), Shh (1: 300, Novus, NBP2-22139), Ptch1 (1:200; R&D, MAB41051), Smo (1:50; Abcam, ab72130), Gli1 (1:250; Abcam, ab151796), and IFT88 (1:250; MilliporeSigma, ABC932). After washing with PBS, sections were incubated for 1 hour at room temperature with either Alexa Fluor®-594 or Alexa Fluor®-647 secondary antibodies (1:700, Jackson ImmunoResearch Lab, Inc.). The sections were washed with PBS before mounting with ProLong® Gold Antifade Mountant containing DAPI (Thermo Fisher Scientific, P36934), and visualized by fluorescence microscopy (Axio Imager 2, Carl Zeiss) using the 20x/0,5 EC Plan-Neofluar (Carl Zeiss) objective. The stained sections were imaged using X-Cite® 120Q Excitation Light Source (Excelitas), the AxioCam MRm camera (Carl Zeiss), and Zen2™ software (Carl Zeiss). Staining of NFAT5 null embryos was performed on four embryos at E12.5, two littermates at E13.5, and five embryos at E17.5 with three representative discs per embryo (n=15 discs). Staining of Ift88 conditional knockout embryos was performed on two littermates with four representative discs per embryo (8 discs).
Digital Image Analysis
All imaged sections stained by immunohistochemistry were analyzed using ImageJ 1.52a (http://rsb.info.nih.gov/ij/) in the grayscale. The boundaries of the notochord, perinotochordal sheath, and NP were digitally traced using the Freehand Tool. The perinotochordal mesenchyme and condensing mesenchyme were delineated by a rectangular fixed area (300×1000 pixels) and circular fixed area (200×215 pixels), respectively. These images were then thresholded to create binary images. Designated regions of interest (ROI) were analyzed using the Area Fraction measurement for each section. Area Fraction represents the percentage of positive pixels normalized to ROI, therefore representing protein expression within a given area. Analysis of the notochord, perinotochordal sheath (bilateral average), and perinotochordal mesenchyme (bilateral average) was conducted on four E12.5 embryos per genotype. Analysis of NP tissue was conducted on two E13.5 at four levels per genotype (8 discs) and five E17.5 embryos per genotype with three discs per embryo (15 discs). Fractions of Brachyury-positive nuclei were manually quantified, and fluorescence intensity signals along the line were measured using Zen2™ software (Carl Zeiss). The number of IFT88-positive cell was also manually quantified.
NP cell isolation and treatment
NP cells were isolated from Sprague Dawley rats (Charles River) as previously described (Risbud et al., 2006). Collection of animal tissues for cell isolation was approved by Thomas Jefferson University IACUC. Cells were maintained in DMEM containing 1g /L glucose with 10% FBS and 1% penicillin-streptomycin, and cultured in a hypoxia work station (Invivo2, Ruskinn, UK) with a mixture of 1% O2, 5% CO2 and 94% N2 to mimic the natural physiological environment of the avascular NP compartment. Either 100/500 nM of SAG (ab142160, abcam) or 5/10 μM of CyA (C-8700, LC Laboratories) was added to medium for 24 hour treatment.
Lentiviral particle production and viral transduction
HEK 293 T cells (ATCC, CRL-3216) were plated in 10 cm plates (5 × 106cells/plate) in DMEM with 10% heat-inactivated FBS a day before their transfection. Cells were transfected with ShCtr or ShTonEBP plasmids along with psPAX2 and pMD2.G using Lipofectamine 2000 (MilliporeSigma). Six hours after transfection, medium was replaced with DMEM with 10% heat-inactivated FBS and penicillin-streptomycin. Lentiviral particles were harvested at 48 to 60 hours post-transfection of HEK cells, and precipitated using 7% PEG 6000 (Sigma Aldrich, 81253) solution. NP cells plated in DMEM with 10% heat-inactivated FBS were transduced with lentiviral particles along with 8 μg/ml polybrene (Sigma Aldrich, H9268). Cells were incubated for 16 hours and the medium with remaining particle was replaced. Four-five days after transduction, cells were harvested for protein extraction to ensure maximum knockdown efficiency without affecting cell viability. At least 3 independent experiments were performed.
Real-Time qRT-PCR
Total RNA was extracted from primary rat NP cells using RNeasy mini columns (Qiagen) and RNase free DNase I (Qiagen). The eluted DNA-free RNA was made into cDNA using EcoDry premix (Clontech). PCR reactions using gene-specific primers (IDT, IA) and SYBR Green master mix (Applied Biosystems) were measured by the Step-One Plus System (Applied Biosystems).
Protein Extraction and Western Blotting
Following experimental treatments, notochordal-NP cells were placed on ice and washed with ice-cold PBS. All buffers included 1X-protease inhibitor cocktail (Roche), NaF (4mM) and Na3VO4 (20mM), NaCl (150mM), β-glycerophosphate (50mM), and DTT (0.2mM). Protein was resolved on 8-10% SDS-polyacrylamide gels and transferred by electroblotting to activated PVDF membranes (Bio-Rad, CA). Membranes were blocked with 5% non-fat dry milk in TBST (50 mM Tris, pH 7.6, 150 mM NaCl, 0.1% tween 20) and incubated overnight at 4°C in 3% non-fat dry milk in TBST with anti-Shh (1:1000, Novus, NBP2-22139), Ptch1 (1:500; R&D, MAB41051), Smo (1:1000; Abcam, ab72130), and Gli1 (1:500; Abcam, 151796). Immunolabeling was detected using ECL reagent (Amersham Biosciences). Relative expression levels were determined by quantitative densitometric analysis using 1D image analysis software (Quantity One, BIO-RAD).
Bioinformatics Analysis
The nucleotide sequence of the 3 kb proximal promoter of Shh, Ptch1, and Smo genes were identified using the UCSC Table Browser data retrieval tool (Karolchik et al., 2004). Putative TonE consensus sequences were determined using MatInspector (Genomatix Software Suite) with a matrix-similarity threshold of 0.80 and TF family p-value of ~0.05 or below. Analysis of species conservation of putative TonE motifs in rat, mouse, and human was performed by multiz alignment using the UCSC Genome Browser (Kent et al., 2002).
Statistical Analysis
Data is presented as mean ± SD. Differences between genotypes were analyzed using the Student’s t test when only two groups presented on graph, or one-way ANOVA with a Sidak’s multiple comparison test between groups. All statistical analyses were done using Prism7 (GraphPad Software). P ≤ 0.05 is considered statistically significant change.
A Key Resources Table (KRT) is included to help identify key materials used in this study.
KEY RESOURCES TABLE
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Brachyury (goat polyclonal) | R&D Systems, Inc., Minneapolis, MN | AF2085 |
| Carbonic anhydrase 3 (goat polyclonal) | Santa Cruz Biotechnology, Inc., Dallas, TX | sc-50715 |
| Vimentin (rabbit monoclonal) | Cell Signaling Technology, Inc., Danvers, MA | D21H3 |
| β-actin (rabbit monoclonal) | Cell Signaling Technology, Inc., Danvers, MA | 13E5 |
| Aggrecan (rabbit polyclonal) | MilliporeSigma, Bedford, MA | AB1031 |
| Chondroitin sulfate (mouse monoclonal) | Abcam, Cambridge, MA | ab34710 |
| Collagen II (rabbit polyclonal) | Fitzgerald Industries International, Acton, MA | 70R-CR008 |
| Collagen I (rabbit polyclonal) | Abcam, Cambridge, MA | ab34710 |
| Sonic Hedgehog (rabbit polyclonal) | Novus Biologicals, Centennial, CO | NBP2-22139 |
| Patched 1 (rat monoclonal) | R&D Systems, Inc., Minneapolis, MN | MAB41051 |
| Smoothened (rabbit polyclonal) | Abcam, Cambridge, MA | ab72130 |
| Gli1 (rabbit polyclonal) | Abcam, Cambridge, MA | ab151796 |
| IFT88 (rabbit polyclonal) | MilliporeSigma, Bedford, MA | ABC932 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Smoothened agonist (SAG) | Abcam, Cambridge, MA | Ab142160 |
| Cylcopamine (CyA) | LC Laboratories, Woburn, MA | C-8700 |
| Critical Commercial Assays | ||
| In situ Cell Death Detection Kit, TMR Red | MilliporeSigma, Bedford, MA | 12156792910 |
| Experimental Models: Cell Lines | ||
| HEK 293 T cells | ATCC, Manassas, VA | CRL-3216 |
| Experimental Models: Organisms/Strains | ||
| Nfat5+/Δ mice | Provided by H. Moo Kwan, Ulsan National Institute of Science and Technology | |
| Ift88f/f mice | Provided by Bradley Yoder, University of Alabama | |
| ShhcreERT2 mice | The Jackson Laboratory | 005623 |
| Sprague Dawley Rats | Charles River, Wilmington, MA | Code: 400 |
| RT-PCR Oligonucleotides | ||
| Nfat5 mouse genotyping WT (F): 5’-AAGGGCTTCTTCCAGAATGG-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Nfat5 mouse genotyping NFAT5+/Δ (F): 5’-GTGTTTCTCAGCCAAGTAAGGTC-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Nfat5 mouse genotyping (R): 5’-GGGCTATAGACATGCACCACCACACAG-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Ift88f/f mouse genotyping (common 5’ primer): GCCTCCTGTTTCTTGACAACAGTG | Integrated DNA Technologies, Inc. Skokie, IL | |
| Ift88f/f mouse genotyping (3’ flox and wild type primer): GGTCCTAACAAGTAAGCCCAGTGTT | Integrated DNA Technologies, Inc. Skokie, IL | |
| Ift88f/f mouse genotyping (3’ delta allele primer): CTGCACCAGCCATTTCCTCTAAGTCATGTA | Integrated DNA Technologies, Inc. Skokie, IL | |
| ShhcreERT2 mouse genotyping (transgene forward): 5’-TGGCTACCCGTGATATTGCT-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| ShhcreERT2 mouse genotyping (transgene reverse): 5’-GAGCGGCGATACCGTAAAG-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| ShhcreERT2 mouse genotyping (internal positive control forward): 5’-CACGTGGGCTCCAGCATT-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| ShhcreERT2 mouse genotyping (internal positive control reverse): 5’-TCACCAGTCATTTCTGCCTTTG-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| qRT-PCR Oligonucleotides | ||
| TonEBP (F) 5’-AAACGAAATCCAAAGCAGAGGCCG-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| TonEBP (R) 5’-TTGCCTTTGTCCGTGGTAAGC-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Shh (F) 5’-AGCCTACAAGCAGTTTATCCC-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Shh (R) 5’-GTTCCTTAAATCGTTCGGAGTTTC-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Ptch1 (F) 5’-AGACAAGCCCATCGACATTAG-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Ptch1 (R) 5’-GCGGTCAGGTAGATGTAGAAAG-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Smo (F) 5’-TGAGTGGCATCTGCTTTGT-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Smo (R) 5’-GGAAGTAGCCTCCCATAAG-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Gli1 (F) 5’-ATGGATACTAGAGGGCTACAGG-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Gli1 (R) 5’-CCAGAGTGTCAGCAGAAGAAA-3’ | Integrated DNA Technologies, Inc. Skokie, IL | |
| Plasmids | ||
| shNFAT5 | MilliporeSigma, Bedford, MA | TRCN0000020019 |
RESULTS
Loss of NFAT5 delays the development of the vertebral column without inhibiting the formation of intervertebral discs
We investigated intervertebral disc embryogenesis in mice harboring a deletion of exons 6 and 7 of the Nfat5 gene. Exons 6 and 7 encode a crucial region of the NFAT5 DNA binding domain (Figure 1A) (Go et al., 2004). Genotypes confirming exon deletion were validated by PCR as described by Go et al. (Figure 1B) (Go et al., 2004). On gross examination, embryos with homozygous deletions evidenced an overall decrease in body size or were found resorbed due to gestational lethality (Figure 1C). This decrease in body size was further evidenced by whole mount skeletal preparations stained with alcian blue (cartilage) and alizarin red (bone) (Figure 1D). Comparison of isolated spines revealed that the thoracic, lumbar, and caudal regions of the vertebral column were notably smaller than controls, likely accounting for the shorter stature of mutants (Figure 1E). Likewise, individual vertebrae from lumbar (L1-L6), sacral (S1-S4), and caudal (Cd1-Cd-5) levels were undersized compared to level-matched controls (Figure 1F). While the morphology of the vertebral bodies was generally maintained, the vertebrae showed a marked reduction of primary centers of ossification throughout the lumbar region and complete absence in the sacrocaudal region starting at S4 (Figure 1F). Since development of the vertebral column progresses from the rostral to the caudal end, this phenotype was consistent with a delay in vertebral body ossification and spinal development. Similar to the axial skeleton, other skeletal structures appeared smaller and the sternum showed lack of ossification. Moreover, lengths of the humerus and femur in mutant embryos were smaller compared to the wild-type littermates (Suppl. Figure 2).
Figure 1:

Deletion of NFAT5 results in delayed development of the vertebral column. (A) Diagram showing targeting of exons 6 and 7 of the Nfat5 gene and resultant mRNA product. (B) Agarose gel of PCR-amplified genomic DNA from wild-type (+/+), heterozygous (+/Δ) and null (Δ/Δ) mice. (C) Gross comparison of E17.5 embryos showing shorter stature in nulls. (D) Whole mount skeletal preparations of NFAT5 mutant embryos at E17.5 stained with alcian blue (cartilage) and alizarin red (bone). (E) Comparison of isolated spines showed a shorter vertebral column in mutants than controls. (F) Individual lumbar (L1-L6), sacral (S1-S4), and caudal (Cd1-Cd-5) vertebrae of E.17.5 mutant mice compared to level-matched controls showed absence of primary ossification centers in the sacrocaudal levels starting at S4 (Green box).
To determine tissue localization of NFAT5 expression during intervertebral disc development, we performed immunohistochemistry on embryos at E12.5, E13.5, and E17.5. Staining revealed that NFAT5 is strongly expressed by cells of the notochord, condensing perinotochordal mesenchyme, early NP, and the hypertrophic zone of the developing vertebral body (Figure 2A). Mining of deposited RNA-seq data (GSE100934) confirmed that NFAT5 is expressed by notochord cells at E12.5 and NP cells at P0 (Figure 2B) (Peck et al., 2017). To our surprise, gross histological analysis of the NFAT5 null notochord revealed an intact structure marked by vacuolated notochord cells and strong Safranin-O staining of the perinotochordal sheath (Figure 2C). However, notochord cell intercalation appeared disorganized in a few embryos, which was evident observing DAPI stained nuclei (Suppl. Figure 3). Further, removal of the notochord at the level of the developing vertebral bodies appeared to be slightly delayed in mutants compared to control mice, in conjunction with a delay in enlargement of prevertebral cells (Figure 2D). Similar to NFAT5-depleted notochord cells at E12.5, mutant notochordal-NP cells at E13.5 did not show a deficiency in vacuole inflation (Figure 2D). Likewise, at E17.5, the thoracic and lumbar intervertebral discs developed fully and were populated by vacuolated NP cells (Figure 2E). These results suggested that while NFAT5 appears to be important for the enlargement of prevertebral cells, it is dispensable for notochord inflation and vacuolation, as well as intervertebral disc formation. Notably, however, E17.5 mutant embryos showed a lower aspect ratio of the caudal NP compartment, while the aspect ratios of the thoracic and lumbar NP were comparable to controls (Figure 2F). This decrease in aspect ratio was not associated with a decrease in the number of cells per area (Figure 2G) or increase in cell death as measured by TUNEL assay (Figure 2H). Indeed, the decrease in aspect ratio of the caudal NP compartment suggests further that NFAT5-deficient embryos experienced developmental delay of the axial skeleton.
Figure 2:
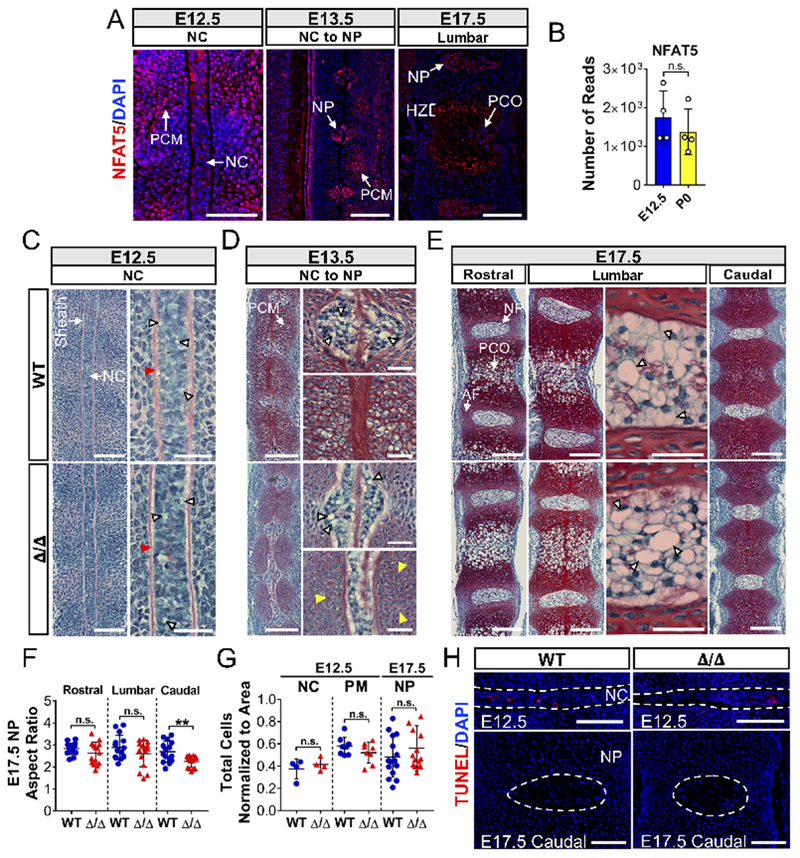
Loss of NFAT5 results in caudal discs with decreased aspect ratios. (A) Localization and expression of NFAT5 at E12.5, E13.5, and E17.5 by immunostaining. Scale bar = 100 μm. PCM: Perinotochordal condensing mesenchyme; NC: Notochord; NP: Nucleus pulposus; PCO: primary center of ossification; HZ: Hypertrophic zone. (B) NFAT5 mRNA expression by notochord cells at E12.5 and NP cells at P0 from deposited RNA-seq data (GSE100934). (C-E) Sagittal sections of E12.5, E13.5, and E17.5 embryos stained by Safranin-O/Fast Green/Hematoxylin showing notochord morphogenesis into NP at the thoracic, lumbar, and caudal levels. Scale bar = 50 μm. High magnification images showing intact perinotochordal sheath (red arrowheads), delayed enlargement of prevertebral cells at E13.5 (yellow arrowheads), and large intracellular vacuoles at all stages (white arrowheads) (Scale bar = 20 μm).(F) Aspect ratios of the caudal but not lumbar and rostral NP of NFAT5 null embryos were significantly smaller than WT embryos. (G, H) Changes in aspect ratio were not associated with decreased cell number per area or increased TUNEL positive cells. Scale bar = 100 μm. Quantitative measurements represent mean ± SD. n.s. = not significant; **, p ≤ 0.01.
NFAT5 is required for notochord cells to acquire their molecular signature and maintain cytoskeletal integrity
Notochord and developing NP cells have a unique molecular phenotype characterized by expression of several specific markers, including the T-box transcription factor, Brachyury/T, CA3, and the notochord-enriched intermediate filament, vimentin (Risbud et al., 2015; Risbud and Shapiro, 2011; Sagstad et al., 2011; Silagi et al., 2018a). Analysis of RNA-Seq data (GSE100934) showed that expression of these markers increased from E12.5 to P0, suggesting that their expression coincides with NP cell maturation and/or differentiation (Peck et al., 2017) (Suppl. Figure 4). Interestingly, the expression levels of Brachyury, as measured by quantitative immunohistochemistry, showed a significant decrease at both E12.5 and E17.5 (Figure 3A, A’, C). Importantly, NFAT5 mutants also showed reduced Brachyury nuclear localization, evident from a decrease in the fraction of Brachyury-positive nuclei (Figure 3B, B’, D), as well as out-of-phase fluorescence intensity signals of Brachyury and DAPI (Figure 3D’, D”). In addition, E12.5 mutants showed nearly undetectable levels of carbonic anhydrase 3 (CA3) (Figure 4A, D) and vimentin (Figure 4B, E), suggesting delayed notochord maturation. Unlike Brachyury, CA3 and vimentin levels were restored at E17.5, with a small but statistically significant increase in vimentin over wild type embryos (Figure 4A’, B’, D, E). These changes in vimentin led us to further examine the cytoskeleton by staining against β-actin, which showed no evidence of altered levels in the notochord, but perturbed and diffused localization at the cortical surface of notochord cells in three of four null embryos (Figure 4C, F). Similar to vimentin, we also observed that β-actin expression in NP cells was moderately elevated at E17.5 (Figure 4C’, F). Thus, taken altogether, these results reveal that the NFAT5 null notochord exhibits delayed acquisition of its molecular phenotype and shows evidence of a disorganized cytoskeleton.
Figure 3:
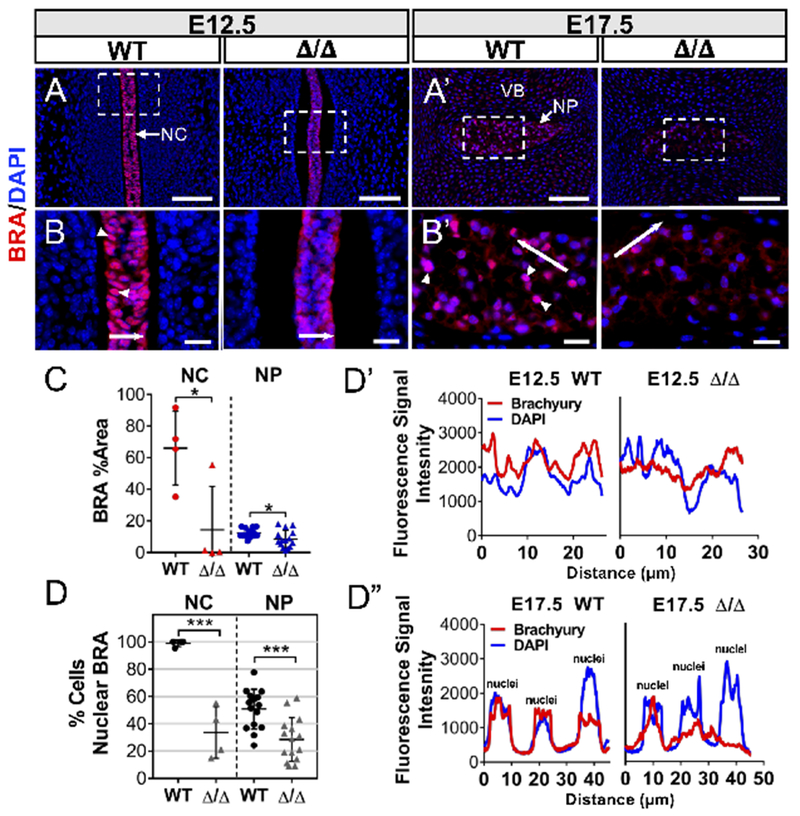
NFAT5 controls expression and nuclear localization of Brachyury in the notochord and early NP. (A, A’) Immunofluorescence staining of Brachyury (BRA) showed decreased expression in both E12.5 notochord (NC) and E17.5 nucleus pulposus (NP). VB: presumptive vertebral body. Scale bar = 100 μm. (B, B’) Corresponding high magnification images showed decreased Brachyury nuclear localization (white arrowheads). Scale bar = 20 μm. (C) Staining quantified by Area Fraction (% Area) showed lower Brachyury levels in mutants. (D) Quantification of Brachyury positive nuclei showed marked reduction in NFAT5 mutants at E12.5 and E17.5 compared to WT embryos. (D’, D”) Fluorescence intensity along the line in B and B’ is plotted in D’ for E12.5 and D” for E17.5 embryos, which showed out-of-phase Brachyury and DAPI signals in mutants. Quantitative measurements represent mean ± SD. n.s. = not significant; *, p ≤ 0.05; ***, p ≤ 0.001.
Figure 4:
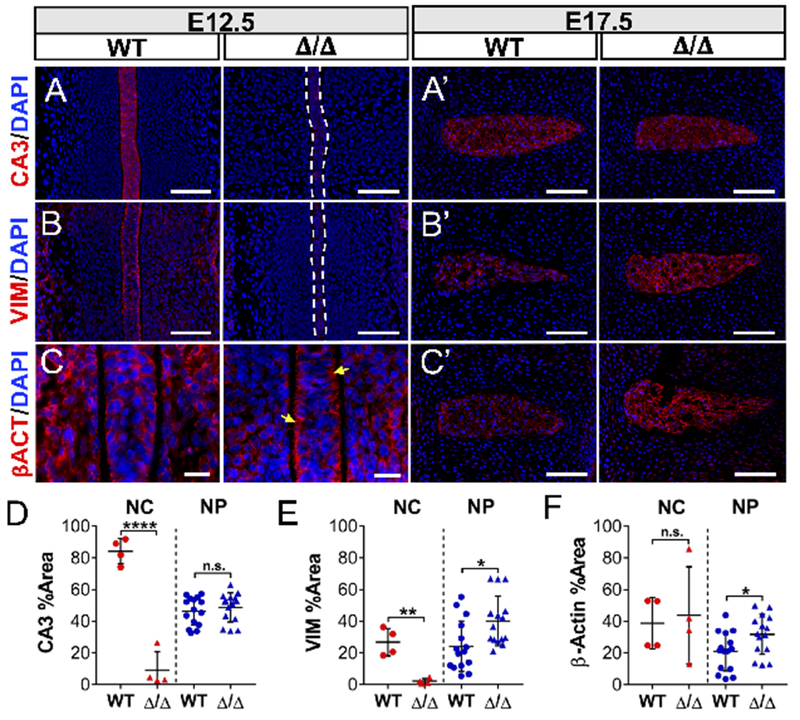
The NFAT5 null notochord exhibits delayed acquisition of its molecular phenotype and shows evidence of a disorganized cytoskeleton. (A, A’) Deletion of NFAT5 significantly reduced notochordal expression levels of carbonic anhydrase 3 (CA3), whereas expression was restored in the NP at E17.5. (B, B’) Mutant notochord showed nearly undetectable expression levels of vimentin (VIM), with increased levels in the NP at E17.5. Scale bar = 100 μm (C) High magnification images show reduced β-actin localization to the submembranous cortex in null notochord cells (yellow arrows). Scale bar = 20 μm. (C, C’) There were no changes in β-actin expression at E12.5 between genotypes, however, E17.5 embryos showed moderately increased levels. Scale bar = 100 μm. Staining against CA3 (D), vimentin (E), and β-actin (F) was quantified by Area Fraction (% Area). Quantitative measurements represent mean ± SD. n.s. = not significant; *, p ≤ 0.05; **, p ≤ 0.01; ****, p ≤ 0.0001.
Loss of NFAT5 alters deposition of major collagen subtypes during intervertebral disc embryogenesis
We next examined critical extracellular matrix components of the perinotochordal sheath and intervertebral disc in NFAT5 null embryos. Surprisingly, aggrecan (ACAN) and chondroitin sulfate (CS), a glycosaminoglycan that substitutes proteoglycans including aggrecan and versican (Silagi et al., 2018b), showed no differences between null and control embryos at both E12.5 (Figure 5A, B, E, F) and E17.5 (Figure 5A’, B’, E, F), consistent with our Safranin-O staining results. However, collagen II was markedly decreased in the sheath and perinotochordal condensing mesenchyme (PCM) (Figure 5C, G), while collagen I staining was primarily localized to the sheath and unaffected in E12.5 mutants (Figure 5D, H). By E17.5, null embryos showed a small increase in levels of both collagen II and I in the NP compartment, with a concomitant decrease in collagen I in the outer annulus fibrosis, possibly reflecting a tissue-specific response to loss of NFAT5 (Figure 5C’, D’, G, H). Changes in collagen content were in accord with previous studies using human articular chondrocytes and disc cells, which showed a role of NFAT5 and hyperosmolarity in regulating collagen I and II expression (van der Windt et al., 2010; Wuertz et al., 2007).
Figure 5:

NFAT5 mutants show altered levels of collagen I and II. Immunofluorescence staining of (A, A’) aggrecan (ACAN) and (B, B’) chondroitin sulfate (CS) showed no discernable differences between genotypes at both E12.5 and E17.5. Decreased levels of (C) collagen II (COLII) in the sheath (arrowhead) and perinotochordal condensing mesenchyme (PCM) were observed at E12.5. (C’) At E17.5, collagen II levels were restored and moderately increased in the NP. (D, D’) While collagen I (COLI) was unaffected by NFAT5 deletion at E12.5 (D), expression levels were increased in the NP, with lower levels in the outer AF (D’). Scale bar =100 μm. Inset scale bar = 20 μm. (E-H) Immunofluorescence staining of the notochord, perinotochordal sheath, PCM, and NP were quantified by Area Fraction (% Area). Quantitative measurements represent mean ± SD. n.s. = not significant; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
Deletion of NFAT5 results in stage-dependent dysregulation of sonic hedgehog signaling in the notochord and early NP
Sonic hedgehog (Shh) secretion and signaling from the notochord is required for perinotochordal sheath formation and thus patterning of the intervertebral discs (Choi et al.,2012; Choi and Harfe, 2011). Interestingly, the notochord and perinotochordal mesenchyme (PM) showed robust staining of Shh in NFAT5 mutants, demonstrating that NFAT5-deficient notochord cells secreted increased levels of Shh (Figure 6A, E). On the other hand, patched-1 (Ptch1) expression did not mirror increased Shh levels in both notochord and PM (Figure 6B, F). Expression of smoothened (Smo) in the notochord of mutants was also similar to wild-type embryos, but with reduced expression in the PM (Figure 6C, G). Importantly, the downstream effector of Shh signaling glioma-associated oncogene homologue 1 (Gli1) showed elevated expression in notochord and PM, indicating elevated Shh activity (Figure 6D, H). Interestingly, by E13.5, Shh levels in mutants were reduced and comparable to wild-type controls, with a concomitant decrease in Ptch1 (Figure 6A’, B’, E, F). Smo and Gli1 levels were unaffected by this shift (Figure 6C’, D’, G, H). By E17.5, Shh and Ptch1 levels in NFAT5 nulls were further reduced, falling significantly below wild type levels (Figure 6A”, B”, E, F). Smo, and interestingly Gli1, were unaffected in the NP at this stage despite a precipitous decrease in Shh, suggesting that Gli1 expression was maintained by non-classical mechanisms (Figure 6C”, D”,G, H). These results present an interesting model in NFAT5 null embryos wherein the temporal pattern of Shh signaling is exaggerated with induction at E12.5 and suppression by E17.5. To test the hypothesis that Gli1 expression in notochordal-NP cells at E17.5 is maintained in a non-classical fashion, we generated mice with conditional deletion of Ift88 in notochord cells using ShhcreERT2 driver (Figure 6I–K) (Haycraft et al., 2007). Deletion of IFT88 disrupts the formation of functional primary cilia which are critical for canonical Shh sensing (Huangfu et al., 2003). Targeting of Ift88 in Shh expressing notochord cells did not prevent the formation of intervertebral discs or alter NP cell morphology, as shown by Safranin-O staining (Figure 6L). Immunostaining and quantification of IFT88-positive NP cells confirmed that IFT88 was knocked out with high efficiency (Figure 6M, M’). Importantly, Gli1 levels were unaffected in ShhcreERT2; Ift88f/f embryos (Figure 6N, N’), suggesting that primary cilia-mediated Shh signaling was not required for maintenance of Gli1 expression in NP cells at this stage of development.
Figure 6:
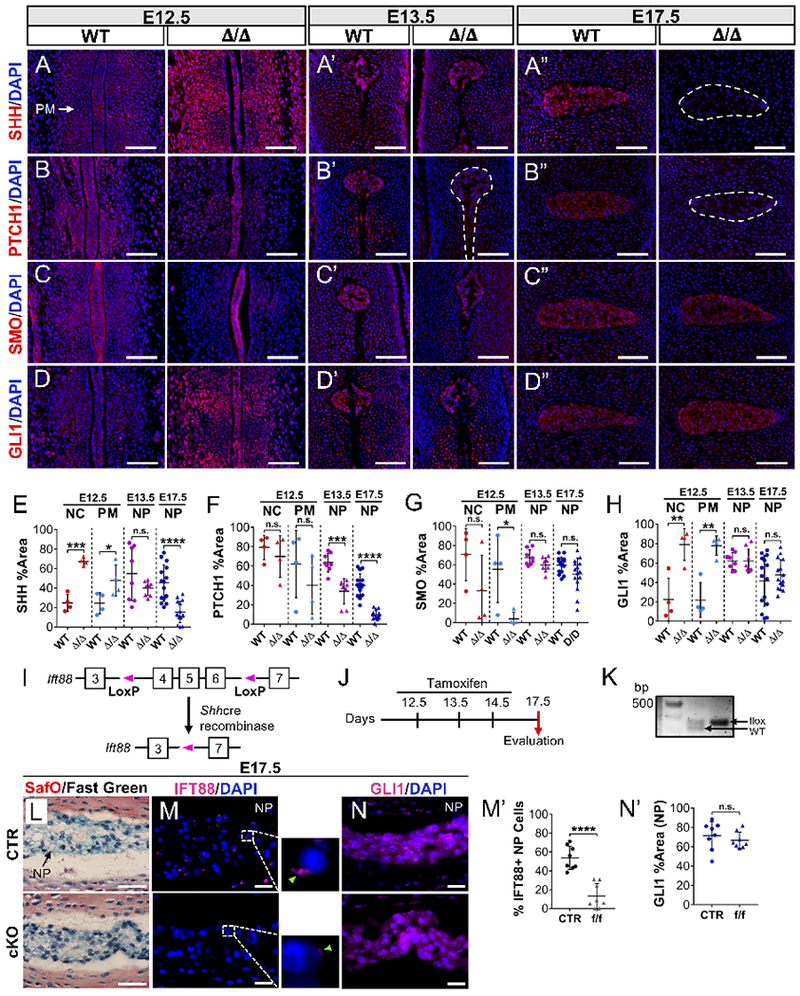
NFAT5 null embryos show stage-dependent dysregulation of sonic hedgehog signaling involving nonclassical expression of Gli1. (A) Sonic hedgehog (SHH) in NFAT5 mutants showed elevated expression levels in both the notochord and perinotochordal mesenchyme (PM) at E12.5. (A’, A”) SHH Levels in early NP were similar to wild-type at E13.5 and then markedly reduced at E17.5. (B) Patched-1 (PTCH1) levels in mutants did not mirror elevated SHH in the notochord and PM. (B’, B”) Minimal PTCH1 expression was observed in the mutant NP at E13.5 and E17.5. (C) NFAT5 nulls showed normal levels of smoothened (SMO) in the notochord with decreased levels in the PM. (C’, C”) At E13.5 and E17.5, no differences were seen between groups. (D, D’) In mutants GLI1 levels increased in the notochord and PM at E12.5, and mirrored SHH levels in the early NP at E13.5. (D”) GLI1 levels were refractory to NFAT5 deletion at E17.5. Scale bar = 100 μm. (E-H) Staining of the notochord, PM, and NP was quantified by Area Fraction (% Area). (I) Schematic representing the generation of Ift88 null allele in Shh expressing cells. (J) Timeline showing tamoxifen injection strategy to conditionally knockout Ift88 without disrupting disc development. (K) Agarose gel of PCR-amplified genomic DNA confirming Ift88 floxed alleles. (L) Representative images of sagittal sections of intervertebral discs from E17.5 ShhcreERT2;Ift88f/f (cKO) animals stained by Safranin-O/Fast Green/Hematoxylin compared to controls. (M, M’) Immunostaining and quantification of IFT88-positive NP cells confirmed that IFT88 was knocked out with high efficiency. (N, N’) Gli1 levels were unaffected in ShhcreERT2; Ift88f/f embryos. Sale bars = 20 μm. Quantitative measurements represent mean ± SD. n.s. = not significant; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
NFAT5-mediated regulation of sonic hedgehog signaling is cell autonomous in NP cells
We aimed to establish a mechanistic relationship between NFAT5 and Shh signaling. In particular, we investigated whether NFAT5 controlled Shh or whether fluctuations in Shh were compensatory due to loss of NFAT5. To this effect, we stably knocked down NFAT5 in primary NP cells and measured expression of Shh and its signaling components. Knockdown resulted in increased Shh mRNA levels, whereas a robust decrease in Ptch1 and Smo transcripts were seen without any concomitant change in Gli1 (Figure 7A). Western blot analysis confirmed a significant decrease in protein levels of Ptch1 and to a smaller extent Smo without changes in Gli1. In contrast to Shh transcript, however, Shh protein levels remained unaffected in NFAT5 knockdown cells, suggesting the possibility of further translational or post-translational regulation (Figure 7B). These results also raised a possibility that NFAT5 may control expression of Ptch1 and Smo independent of Shh. It is known that NFAT5 transactivates downstream targets by binding to tonicity-response elements (TonE) in gene promoters; therefore, we investigated whether Shh, Ptch1 and Smo contain potential TonE sites within their proximal promoters. Analysis of 3 kb proximal promoter sequences using MatInspector (Genomatix Software Suite) identified putative TonE binding sites in the rat (4 sites) and mouse (3 sites) Ptch1 promoter (Figure 7C). Multiz alignment showed that two of these TonE binding sites were highly conserved among rat, mouse, and human, suggesting that these sites are physiologically relevant. Analysis also identified four putative TonE binding sites in the rat and mouse and six sites in human Smo promoter, with one site conserved between rat and mouse (Figure 7C). Interestingly, however, we did not find predicted TonE binding sites in the Shh promoter, suggesting the possibility that its transcriptional induction in NFAT5 knocked down cells was due to a compensatory response to reduced Ptch1 and Smo levels. To investigate if Shh signaling modulates NFAT5 expression, we measured NFAT5 mRNA and protein levels in NP cells following treatment with smoothened agonist, SAG, and smoothened inhibitor, cyclopamine (CyA). NFAT5 mRNA and protein levels were refractory to either treatment whereas Glil mRNA was responsive to both treatments (Figure 7D–F’). Altogether, these results suggest that NFAT5 controls Shh signaling by functioning as a positive regulator of Ptch1 and possibly Smo expression in an NP cell autonomous manner.
Figure 7:
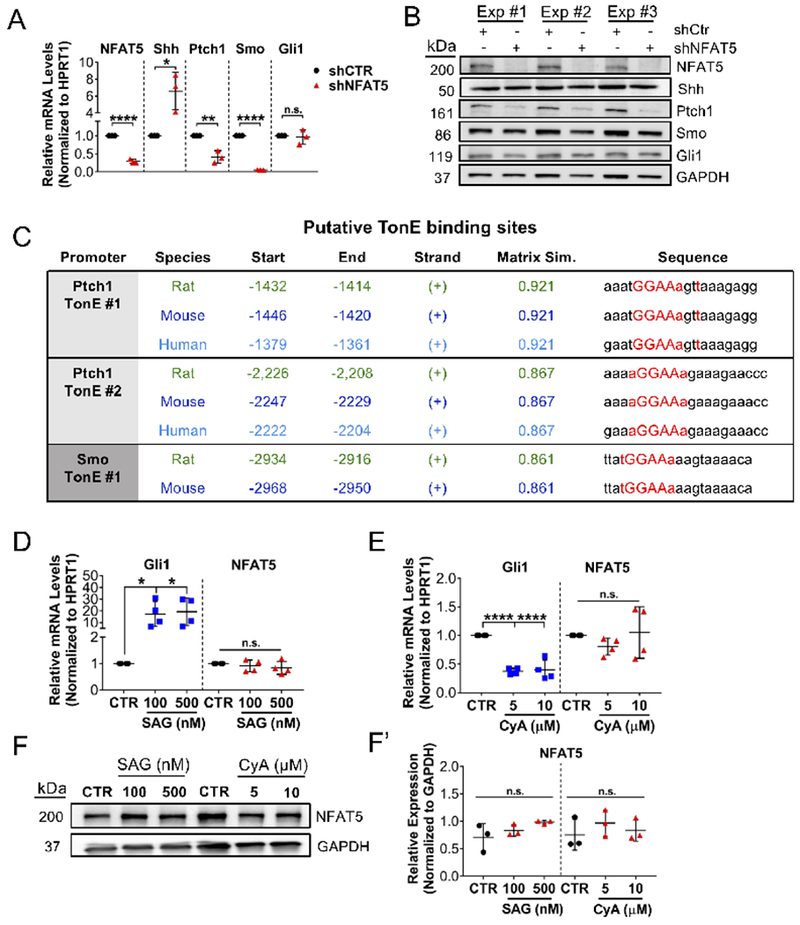
NFAT5-mediated regulation of Shh signaling is cell-autonomous in notochordal-NP cells. (A) mRNA levels of Nfat5, Ptch1, and Smo were significantly suppressed in notochordal-NP cells transduced with Nfat5 shRNA (shNFAT5) compared to control shRNA (shCtr). mRNA levels of Shh were markedly increased in shNFAT5 cells, whereas Gli1 mRNA levels were not affected. (n = 3) (B) Western blot showing three independent experiments showed significantly decreased protein levels of PTCH1 with a downward trend in SMO and no change in GLI1 following Nfat5 knockdown. In contrast to Shh transcript, SHH protein levels remained unaffected in knockdown cells. (C) Predicted TonE binding sites identified by MatInspector (Genomatix Software Suite) in the proximal promoters of Ptch1 and Smo in rat, mouse, and human show high conservation. (E) Activation of canonical SHH signaling in notochordal-NP cells with SMO agonist (SAG) induced Gli1 mRNA but not Nfat5 mRNA levels. (n = 4) (F) Treatment with the SMO inhibitor cyclopamine (CyA) reduced levels of Gli1 mRNA but not Nfat5 mRNA (n = 4). Protein levels of NFAT5 measured by Western blot (G) and corresponding densitometry from three independent experiments (G’) show no effect on expression by both SAG and CyA treatment (n = 3). In G, one representative Western blot is shown. Quantitative measurements represent mean ± SD. n.s. = not significant; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
DISCUSSION
NFAT5 is robustly expressed by resident cells of the Ciona notochord and, as shown here, by cells of the mouse notochord, and plays a critical role in maintaining osmoadaptation and matrix homeostasis in NP cells (Johnson et al., 2014; José-Edwards et al., 2011; Reeves et al., 2017). However, there are no studies that have directly addressed the role of NFAT5 in notochord biology and its morphogenesis into NP, or in the formation of the intervertebral discs. Using genetic mouse models, and in vitro and bioinformatics approaches, we provide the first insights into the contribution of NFAT5 during these early morphogenic events.
Our analysis suggests that NFAT5 deletion results in developmental delay of the vertebral column. First, at E17.5, mutant spines were shorter in length with undersized vertebral bodies and a significant delay in the formation of primary ossification centers, most noticeably in the sacrocaudal region. Since the caudal-most vertebral column is the last to develop during spinal skeletogenesis, it is not unreasonable to assume that a delay in its development would be more apparent in the caudal region. This idea is further supported by the observation that null embryos showed decreased aspect ratios of the NP compartment at the caudal levels, but not at the thoracic and lumbar levels. Second, at E13.5, we observed what appeared to be a delay in the enlargement or maturation of prevertebral cells, lending the possibility that NFAT5 may play an important role in chondrocyte development. A delay in chondrocyte development might also explain why the condensing mesenchyme of E12.5 embryos showed lower levels of collagen II. Third, the notochord phenotypic markers Brachyury, carbonic anhydrase 3 (CA3), and vimentin, all of which were seen to be associated with notochord cell maturation and NP cell differentiation, were under expressed at E12.5. Lastly, as Shh is most strongly expressed by notochord cells at early stages of development, increased Shh levels at E12.5 in both NFAT5 null notochord and adjacent mesenchyme is consistent with a delay in development. Altogether, these results suggest that NFAT5 is required for early notochord cell maturation and acquisition of their molecular signature, and for controlling the temporal progression of vertebral column development.
It is plausible that the observed caudal abnormalities were in part due to compromised Brachyury expression and activity. Indeed, it is known that adult mice heterozygous for Brachyury show a short-tail phenotype—hence the ancient Greek name “Brachy (short) -ury (tail)”, and several examples of sacral/caudal defects associated with dysregulated Brachyury have been examined (Chiba et al., 2009; Ghebranious et al., 2008; Korzh and Grunwald, 2001; Pennimpede et al., 2012; Postma et al., 2014; Stott et al., 1993). While our data suggests that NFAT5 maintains Brachyury expression in both the mouse notochord and newly formed NP, Jose-Edwards and colleagues have also demonstrated that notochord expression of the Ciona NFAT5 ortholog is markedly decreased in a large percentage of Brachyury null embryos (José-Edwards et al., 2011). These results lend the possibility that a bidirectional regulatory circuit exists between NFAT5 and Brachyury. Interestingly, it has been shown that attenuation of Brachyury by overexpression of dominant-negative NFAT5 can be rescued by stabilizing β-catenin (Adachi et al., 2012), suggesting that this relationship might be mediated by canonical Wnt signaling.
In addition to changes in notochord phenotypic marker expression, NFAT5 null embryos showed a stage-dependent dysregulation of Shh signaling, in which Shh levels were significantly elevated at E12.5 and decreased by E17.5. Shh signaling is required for the formation of the matrix-rich perinotochordal sheath, which is necessary for containing notochord cells during early disc patterning, but not for maintaining the embryonic nuclei pulposi once they are formed (Choi and Harfe, 2011). It is therefore possible that Shh was elevated in NFAT5 mutants at E12.5 to ensure that the collagen II-depleted sheath was formed sufficiently for disc patterning to occur. Indeed, Shh is most strongly expressed by notochord cells at early stages of mouse development and fall precipitously by 88 fold from E12.5 to P0 (Peck et al., 2017). Notably, while we observed elevated Shh and Gli1 levels at E12.5, Ptch1 levels did not mirror this increase, and were later decreased at E13.5 and E17.5. These results, along with our in vitro findings that Ptch1 transcript and protein levels were suppressed by knockdown of NFAT5, raises an intriguing possibility that NFAT5 may regulate Ptch1 expression independent of Shh. In corroboration of this hypothesis, analysis of the proximal Ptch1 promoter revealed two highly conserved TonE binding sites, suggesting that Ptch1 is a target of NFAT5. Interestingly, analysis of the Smo promoter also predicted a conserved TonE binding site, which might explain why Smo levels showed a downward trend in vivo and decreased with NFAT5 knockdown in vitro. Another notable observation in mutant embryos was that Gli1 levels did not decrease with declining Shh and Ptch1 levels at E17.5, consistent with our observation that Gli1 transcript and protein levels were refractory to NFAT5 knockdown in vitro. Furthermore, notochord-specific conditional deletion of IFT88, an intraflagellar transport protein which maintains functional primary cilia and thus canonical Shh sensing (Huangfu et al., 2003), did not decrease Gli1 levels at E17.5. These results clearly suggest that Gli1 expression by embryonic NP cells can be maintained by non-classical mechanisms. Indeed, non-classical activation of Gli1 can occur through numerous pathways, including TGF-β and Notch signaling, which are active in embryonic and post-natal NP cells (Aberger et al., 2012; Chan et al., 2014; Hiyama et al., 2011; Tran et al., 2010).
While NFAT5 null embryos showed interesting phenotypic and molecular changes, loss of NFAT5 had modest effects on the formation and morphology of the notochord and intervertebral discs, implying that NFAT5 was dispensable for notochord inflation and notochordal-NP cell vacuolation. This result is quite surprising because NFAT5 is the only known tonicity-responsive transcription factor in mammals (López-Rodríguez et al., 2001). Second, notochord inflation is widely understood to utilize transmembrane channels, transporters, and pumps to build an osmotic gradient and draw osmotically obliged water into the notochord (Adams et al., 1990; Deng et al., 2013; Reeves et al., 2017). Accordingly, while NFAT5 is primarily involved in regulating organic, non-ionic osmolytes, perhaps ionic gradients independent of NFAT5 activity are of greater importance in driving notochord inflation and notochordal-NP cell vacuolation. In addition, because NP cells begin to deposit negatively-charged glycosaminoglycan-rich matrix during the later stages of disc development, it is possible that the osmoprotective role of NFAT5 is not apparent until greater amounts of matrix are accumulated. Consistent with this speculation, NFAT5-deficient NP cells at E17.5 upregulated their production of vimentin and β-actin, perhaps to resist the osmotic challenge presented by continued extracellular matrix deposition. Yet still, it is possible that osmoprotection is more important in Ciona, given that, unlike vertebrates, the Ciona notochord forms extracellular lumens of water-imbibing matrix (Lu et al., 2018). It is also important to note that the modest phenotype in mutants is unlikely due to compensation of NFAT5 function by other members of the NFAT family, NFAT1-4. While these family members can regulate expression of NFAT5 target genes associated with inflammatory responses, they cannot compensate osmoadaptive functions that require binding to TonE sites, where cooperative binding with AP1 and p65 is not involved (Johnson et al., 2017, 2016; Lopez-Rodríguez et al., 1999; Macián et al., 2001; Miyakawa et al., 1999; Stroud et al., 2002).
In summary, our results do not support the view that NFAT5 is indispensable for notochord inflation and intervertebral disc development in vertebrates. Instead, we reveal that deletion of NFAT5 results in delayed vertebral column development and delayed expression of notochord phenotypic markers. Further, we show that NFAT5 controls Shh signaling by regulation of Ptch1 and Smo, and that Gli1 expression in embryonic NP cells can be maintained despite loss of Shh signaling. For the first time, these results thus offer mechanistic and broader insight into the important role of NFAT5 during notochord and intervertebral disc embryogenesis.
Supplementary Material
Supplementary Figure 1: Wild-type E17.5 disc treated with DNase served as a positive control for TUNEL assay and shows labeled cells. Scale bar = 100 μm.
Supplementary Figure 2: NFAT5 mutant embryos show smaller skeletal structures. (A) Comparison of alizarin red and alcian blue stained cranium, ribcage, forelimb, and hindlimb of mutant and wild-type E17.5 embryos. Black arrows indicate lack of ossification of the sternum. h: Humerus; f: Femur. (B) Length measurements of the humerus and femur. Data is represented as mean ± SD. *, p ≤ 0.05.
Supplementary Figure 3: Disorganized intercalation and clustering of DAPI stained nuclei in notochord cells was observed in a few mutant embryos. Scale bar = 20 μm.
Supplementary Figure 4: Analysis of RNA-Seq data (GSE100934) showing expression of Brachyury, CA3, and vimentin by notochord cells at E12.5 and NP cells at P0. Quantitative measurements represent mean ± SD. *, p ≤ 0.05.
Figure 8:
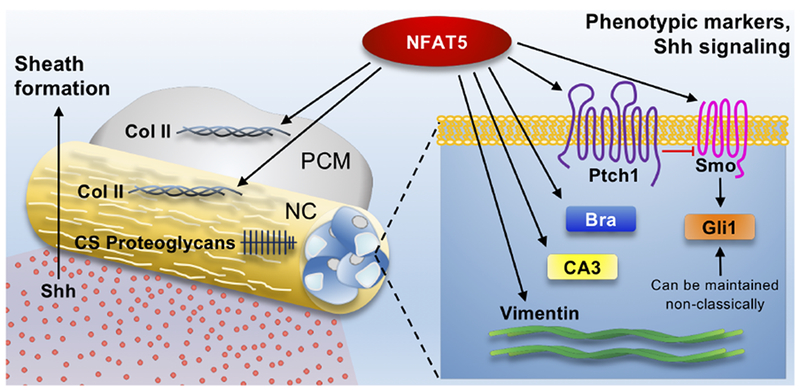
Model showing the role of NFAT5 in notochord morphogenesis. NFAT5 regulates expression levels of notochordal phenotypic markers Brachyury, CA3, and vimentin, while also controlling Shh signaling in notochord cells and collagen II deposition in the perinotochordal sheath and condensing mesenchyme. NC: Notochord; PCM: Perinotochordal condensing mesenchyme.
Highlights.
Loss of NFAT5 delays the development of the vertebral column.
NFAT5 is required for notochord cells to acquire their molecular signature.
NFAT5 regulates collagen deposition during intervertebral disc embryogenesis.
NFAT5 controls Shh signaling in an NP cell-autonomous manner.
Acknowledgements
We kindly thank Emanuel Novais for performing tamoxifen injections and Elizabeth Silagi for the bioinformatics analysis.
Funding: This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health under [grant numbers R01AR064733, R01AR055655, R01AR074813, T32AR052273].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: Declarations of interest: None.
Supplemental material has been included with this submission.
References
- Aberger F, Kern D, Greil R, Hartmann TN, 2012. Canonical and noncanonical Hedgehog/GLI signaling in hematological malignancies. Vitam Horm 88, 25–54. doi: 10.1016/B978-0-12-394622-5.00002-X [DOI] [PubMed] [Google Scholar]
- Adachi A, Takahashi T, Ogata T, Imoto-Tsubakimoto H, Nakanishi N, Ueyama T, Matsubara H, 2012. NFAT5 regulates the canonical Wnt pathway and is required for cardiomyogenic differentiation. Biochem Biophys Res Commun 426, 317–323. doi: 10.1016/j.bbrc.2012.08.069 [DOI] [PubMed] [Google Scholar]
- Adams DS, Keller R, Koehl MA, 1990. The mechanics of notochord elongation, straightening and stiffening in the embryo of Xenopus laevis. Development 110, 115–130. [DOI] [PubMed] [Google Scholar]
- Burg MB, Ferraris JD, 2008. Intracellular organic osmolytes: function and regulation. J Biol Chem 283, 7309–7313. doi: 10.1074/jbc.R700042200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WCW, Au TYK, Tam V, Cheah KSE, Chan D, 2014. Coming together is a beginning: the making of an intervertebral disc. Birth Defects Res C Embryo Today 102, 83–100. doi: 10.1002/bdrc.21061 [DOI] [PubMed] [Google Scholar]
- Chiba S, Jiang D, Satoh N, Smith WC, 2009. Brachyury null mutant-induced defects in juvenile ascidian endodermal organs. Development 136, 35–39. doi: 10.1242/dev.030981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-S, Cohn MJ, Harfe BD, 2008. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn 237, 3953–3958. doi: 10.1002/dvdy.21805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-S, Harfe BD, 2011. Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proc Natl Acad Sci U S A 108, 9484–9489. doi: 10.1073/pnas.1007566108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-S, Lee C, Harfe BD, 2012. Sonic hedgehog in the notochord is sufficient for patterning of the intervertebral discs. Mech Dev 129, 255–262. doi: 10.1016/j.mod.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corallo D, Trapani V, Bonaldo P, 2015. The notochord: structure and functions. Cell Mol Life Sci 72, 2989–3008. doi: 10.1007/s00018-015-1897-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Nies F, Feuer A, Bocina I, Oliver D, Jiang D, 2013. Anion translocation through an Slc26 transporter mediates lumen expansion during tubulogenesis. Proc Natl Acad Sci U S A 110, 14972–14977. doi: 10.1073/pnas.1220884110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis K, Bagwell J, Bagnat M, 2013. Notochord vacuoles are lysosome-related organelles that function in axis and spine morphogenesis. J Cell Biol 200, 667–679. doi: 10.1083/jcb.201212095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebranious N, Blank RD, Raggio CL, Staubli J, McPherson E, Ivacic L, Rasmussen K, Jacobsen FS, Faciszewski T, Burmester JK, Pauli RM, Boachie-Adjei O, Glurich I, Giampietro PF, 2008. A missense T (Brachyury) mutation contributes to vertebral malformations. J Bone Miner Res 23, 1576–1583. doi: 10.1359/jbmr.080503 [DOI] [PubMed] [Google Scholar]
- Ghosh P, Taylor TK, Horsburgh BA, 1975. The composition and protein metabolism in the immature rabbit intervertebral disc. Cell Tissue Res 163, 223–238. doi: 10.1007/BF00221729 [DOI] [PubMed] [Google Scholar]
- Go WY, Liu X, Roti MA, Liu F, Ho SN, 2004. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc Natl Acad Sci U S A 101, 10673–10678. doi: 10.1073/pnas.0403139101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft CJ, Zhang Q, Song B, Jackson WS, Detloff PJ, Serra R, Yoder BK, 2007. Intraflagellar transport is essential for endochondral bone formation. Development 134, 307–316. doi: 10.1242/dev.02732 [DOI] [PubMed] [Google Scholar]
- Hiyama A, Gajghate S, Sakai D, Mochida J, Shapiro IM, Risbud MV, 2009. Activation of TonEBP by calcium controls {beta}1,3-glucuronosyltransferase-I expression, a key regulator of glycosaminoglycan synthesis in cells of the intervertebral disc. J Biol Chem 284, 9824–9834. doi: 10.1074/jbc.M807081200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama A, Skubutyte R, Markova D, Anderson DG, Yadla S, Sakai D, Mochida J, Albert TJ, Shapiro IM, Risbud MV, 2011. Hypoxia activates the notch signaling pathway in cells of the intervertebral disc: implications in degenerative disc disease. Arthritis Rheum 63, 1355–1364. doi: 10.1002/art.30246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV, 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87. doi: 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Ishihara H, Warensjo K, Roberts S, Urban JP, 1997. Proteoglycan synthesis in the intervertebral disk nucleus: the role of extracellular osmolality. Am J Physiol 272, C1499–506. doi: 10.1152/ajpcell.1997.272.5.C1499 [DOI] [PubMed] [Google Scholar]
- Johnson ZI, Doolittle AC, Snuggs JW, Shapiro IM, Le Maitre CL, Risbud MV, 2017. TNF-α promotes nuclear enrichment of the transcription factor TonEBP/NFAT5 to selectively control inflammatory but not osmoregulatory responses in nucleus pulposus cells. J Biol Chem 292, 17561–17575. doi: 10.1074/jbc.M117.790378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, Shapiro IM, Risbud MV, 2014. Extracellular osmolarity regulates matrix homeostasis in the intervertebral disc and articular cartilage: evolving role of TonEBP. Matrix Biol 40, 10–16. doi: 10.1016/j.matbio.2014.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, Shapiro IM, Risbud MV, 2016. RNA Sequencing Reveals a Role of TonEBP Transcription Factor in Regulation of Pro-inflammatory Genes in Response to Hyperosmolarity in Healthy Nucleus Pulposus Cells: A HOMEOSTATIC RESPONSE? J Biol Chem 291, 26686–26697. doi: 10.1074/jbc.M116.757732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- José-Edwards DS, Kerner P, Kugler JE, Deng W, Jiang D, Di Gregorio A, 2011. The identification of transcription factors expressed in the notochord of Ciona intestinalis adds new potential players to the brachyury gene regulatory network. Dev Dyn 240, 1793–1805. doi: 10.1002/dvdy.22656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ, 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 1;32(Database issue):D493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D, 2002. The human genome browser at UCSC. Genome Res 12(6):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzh V, Grunwald D, 2001. Nadine Dobrovolskaïa-Zavadskaïa and the dawn of developmental genetics. Bioessays 23, 365–371. doi: 10.1002/bies.1052 [DOI] [PubMed] [Google Scholar]
- López-Rodríguez C, Antos CL, Shelton JM, Richardson JA, Lin F, Novobrantseva TI, Bronson RT, Igarashi P, Rao A, Olson EN, 2004. Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression. Proc Natl Acad Sci U S A 101, 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Rodríguez C, Aramburu J, Jin L, Rakeman AS, Michino M, Rao A, 2001. Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity 15, 47–58. [DOI] [PubMed] [Google Scholar]
- Lopez-Rodríguez C, Aramburu J, Rakeman AS, Rao A, 1999. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci U S A 96, 7214–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Bhattachan P, Dong B, 2018. Ascidian notochord elongation. Dev Biol. doi: 10.1016/j.ydbio.2018.11.009 [DOI] [PubMed] [Google Scholar]
- Macián F, López-Rodríguez C, Rao A, 2001. Partners in transcription: NFAT and AP-1. Oncogene 20, 2476–2489. doi: 10.1038/sj.onc.1204386 [DOI] [PubMed] [Google Scholar]
- Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM, 1999. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci U S A 96, 2538–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck SH, McKee KK, Tobias JW, Malhotra NR, Harfe BD, Smith LJ, 2017. Whole Transcriptome Analysis of Notochord-Derived Cells during Embryonic Formation of the Nucleus Pulposus. Sci Rep 7, 10504. doi: 10.1038/s41598-017-10692-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennimpede T, Proske J, König A, Vidigal JA, Morkel M, Bramsen JB, Herrmann BG, Wittler L, 2012. In vivo knockdown of Brachyury results in skeletal defects and urorectal malformations resembling caudal regression syndrome. Dev Biol 372, 55–67. doi: 10.1016/j.ydbio.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Postma AV, Alders M, Sylva M, Bilardo CM, Pajkrt E, van Rijn RR, Schulte-Merker S, Bulk S, Stefanovic S, Ilgun A, Barnett P, Mannens MMAM, Moorman AFM, Oostra RJ, van Maarle MC, 2014. Mutations in the T (brachyury) gene cause a novel syndrome consisting of sacral agenesis, abnormal ossification of the vertebral bodies and a persistent notochordal canal. J Med Genet 51, 90–97. doi: 10.1136/jmedgenet-2013-102001 [DOI] [PubMed] [Google Scholar]
- Reeves WM, Wu Y, Harder MJ, Veeman MT, 2017. Functional and evolutionary insights from the Ciona notochord transcriptome. Development 144, 3375–3387. doi: 10.1242/dev.156174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigueur D, Lyons KM, 2014. Whole-mount skeletal staining. Methods Mol Biol 1130, 113–121. doi: 10.1007/978-1-62703-989-5_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud MV, Guttapalli A, Stokes DG, Hawkins D, Danielson KG, Schaer TP, Albert TJ, Shapiro IM, 2006. Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J Cell Biochem 98, 152–159. doi: 10.1002/jcb.20765 [DOI] [PubMed] [Google Scholar]
- Risbud MV, Schoepflin ZR, Mwale F, Kandel RA, Grad S, Iatridis JC, Sakai D, Hoyland JA, 2015. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J Orthop Res 33, 283–293. doi: 10.1002/jor.22789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud MV, Shapiro IM, 2011. Notochordal cells in the adult intervertebral disc: new perspective on an old question. Crit Rev Eukaryot Gene Expr 21, 29–41. doi: 10.1615/CritRevEukarGeneExpr.v21.i1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagstad A, Grotmol S, Kryvi H, Krossøy C, Totland GK, Malde K, Wang S, Hansen T, Wargelius A, 2011. Identification of vimentin- and elastin-like transcripts specifically expressed in developing notochord of Atlantic salmon (Salmo salar L.). Cell Tissue Res 346, 191–202. doi: 10.1007/s00441-011-1262-y [DOI] [PubMed] [Google Scholar]
- Silagi ES, Batista P, Shapiro IM, Risbud MV, 2018a. Expression of Carbonic Anhydrase III, a Nucleus Pulposus Phenotypic Marker, is Hypoxia-responsive and Confers Protection from Oxidative Stress-induced Cell Death. Sci Rep 8, 4856. doi: 10.1038/s41598-018-23196-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silagi ES, Shapiro IM, Risbud MV, 2018b. Glycosaminoglycan synthesis in the nucleus pulposus: Dysregulation and the pathogenesis of disc degeneration. Matrix Biol 368–379. doi: 10.1016/j.matbio.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemple DL, 2005. Structure and function of the notochord: an essential organ for chordate development. Development 132, 2503–2512. doi: 10.1242/dev.01812 [DOI] [PubMed] [Google Scholar]
- Stott D, Kispert A, Herrmann BG, 1993. Rescue of the tail defect of Brachyury mice. Genes Dev 7, 197–203. [DOI] [PubMed] [Google Scholar]
- Stroud JC, Lopez-Rodriguez C, Rao A, Chen L, 2002. Structure of a TonEBP-DNA complex reveals DNA encircled by a transcription factor. Nat Struct Biol 9, 90–94. doi: 10.1038/nsb749 [DOI] [PubMed] [Google Scholar]
- Tran CM, Markova D, Smith HE, Susarla B, Ponnappan RK, Anderson DG, Symes A, Shapiro IM, Risbud MV, 2010. Regulation of CCN2/connective tissue growth factor expression in the nucleus pulposus of the intervertebral disc: role of Smad and activator protein 1 signaling. Arthritis Rheum 62, 1983–1992. doi: 10.1002/art.27445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai T-T, Danielson KG, Guttapalli A, Oguz E, Albert TJ, Shapiro IM, Risbud MV, 2006. TonEBP/OREBP is a regulator of nucleus pulposus cell function and survival in the intervertebral disc. J Biol Chem 281, 25416–25424. doi: 10.1074/jbc.M601969200 [DOI] [PubMed] [Google Scholar]
- van der Windt AE, Haak E, Das RHJ, Kops N, Welting TJM, Caron MMJ, van Til NP, Verhaar JAN, Weinans H, Jahr H, 2010. Physiological tonicity improves human chondrogenic marker expression through nuclear factor of activated T-cells 5 in vitro. Arthritis Res Ther 12, R100. doi: 10.1186/ar3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley R, 1953. The development and growth of the intervertebral disc. Edinb Med J 60, 341–364. [PMC free article] [PubMed] [Google Scholar]
- Wuertz K, Urban JPG, Klasen J, Ignatius A, Wilke HJ, Claes L, Neidlinger-Wilke C, 2007. Influence of extracellular osmolarity and mechanical stimulation on gene expression of intervertebral disc cells. J Orthop Res 25, 1513–1522. doi: 10.1002/jor.20436 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Wild-type E17.5 disc treated with DNase served as a positive control for TUNEL assay and shows labeled cells. Scale bar = 100 μm.
Supplementary Figure 2: NFAT5 mutant embryos show smaller skeletal structures. (A) Comparison of alizarin red and alcian blue stained cranium, ribcage, forelimb, and hindlimb of mutant and wild-type E17.5 embryos. Black arrows indicate lack of ossification of the sternum. h: Humerus; f: Femur. (B) Length measurements of the humerus and femur. Data is represented as mean ± SD. *, p ≤ 0.05.
Supplementary Figure 3: Disorganized intercalation and clustering of DAPI stained nuclei in notochord cells was observed in a few mutant embryos. Scale bar = 20 μm.
Supplementary Figure 4: Analysis of RNA-Seq data (GSE100934) showing expression of Brachyury, CA3, and vimentin by notochord cells at E12.5 and NP cells at P0. Quantitative measurements represent mean ± SD. *, p ≤ 0.05.


