Summary
Transcriptional regulators, including the cohesin complex member STAG2, are recurrently mutated in cancer. The role of STAG2 in gene regulation, hematopoiesis, and tumor suppression remains unresolved. We show Stag2 deletion in hematopoietic 5 stem/progenitor cells (HSPC) results in altered hematopoietic function, increased self-renewal, and impaired differentiation. ChIP-sequencing revealed that while Stag2 and Stag1 bind a shared set of genomic loci, a component of Stag2 binding sites are unoccupied by Stag1 even in Stag2-deficient HSPCs. While concurrent loss of Stag2 and Stag1 abrogated hematopoiesis, Stag2 loss alone decreased chromatin accessibility and transcription of lineage-specification genes, including Ebf1 and Pax5, leading to increased self-renewal and reduced HSPC commitment to the B-cell lineage. Our data illustrate a role for Stag2 in transformation and transcriptional dysregulation distinct from its shared role with Stag1 in chromosomal segregation.
eTOC Blurb
In murine hematopoietic Stag2 deletion, Stag1 rescues topologically associated domains in the absence of Stag2 but cannot restore chromatin architecture required for hematopoietic lineage commitment. PU.1 target genes lose accessibility and expression. Induced target gene expression but not PU.1 overexpression is sufficient to restore differentiation in the altered chromatin state.
Graphical Abstract
Introduction
Cell-type specific transcriptional programs are regulated in part by the activity of tissue-specific transcription factors and enhancer elements within structurally defined topologically associating domains (TADs) (Dixon et al., 2012; Nora et al., 2012; Rao et al., 2017). The genes which contribute to transcriptional regulation, including members of the cohesin complex, are frequently mutated in human cancers, including leukemias (Bailey et al., 2014; Balbas-Martinez et al., 2013; Cancer Genome Atlas Research et al., 2013; Kon et al., 2013; Solomon et al., 2011; Thota et al., 2014). Cohesin is a tripartite ring comprised of three structural proteins SMC1A, SMC3, and RAD21; this core complex binds to either STAG1 or STAG2. STAG2, which is X-linked, is the most commonly mutated cohesin complex member in human cancer, with recurrent nonsense, frameshift, and missense mutations in Ewing’s Sarcoma (40–60%)(Solomon et al., 2011), bladder cancer (20–35%)(Bailey et al., 2014; Balbas-Martinez et al., 2013), glioblastoma (4–5%)(Bailey et al., 2014), myelodysplastic syndrome (MDS; 5–20%)(Kon et al., 2013; Thota et al., 2014), and acute myeloid leukemia (AML; 2–12%)(Cancer Genome Atlas Research et al., 2013; Kon et al., 2013; Thota et al., 2014). This is in contrast to SMC1A, SMC3, and RAD21, which are more commonly mutated in myeloid malignancies than in the broader spectrum of human cancers. The cohesin complex is essential in pleiotropic cellular and gene regulatory functions, including chromosome segregation (Losada et al., 1998; Toth et al., 1999) and in the formation of DNA loops (Rao et al., 2014; Rao et al., 2017; Schwarzer et al., 2017; Seitan et al., 2013) which regulate gene expression (Cuartero et al., 2018; Kagey et al., 2010; Seitan et al., 2013). However, the high mutational frequency and unique mutational spectrum of STAG2 in human cancers (Bailey et al., 2014; Kon et al., 2013; Thota et al., 2014) suggests a distinct role for STAG2 in homeostasis and tumor suppression (Mazumdar et al., 2015; Mullenders et al., 2015) which has not been delineated.
Results
Loss of Stag2 alters hematopoietic stem cell function
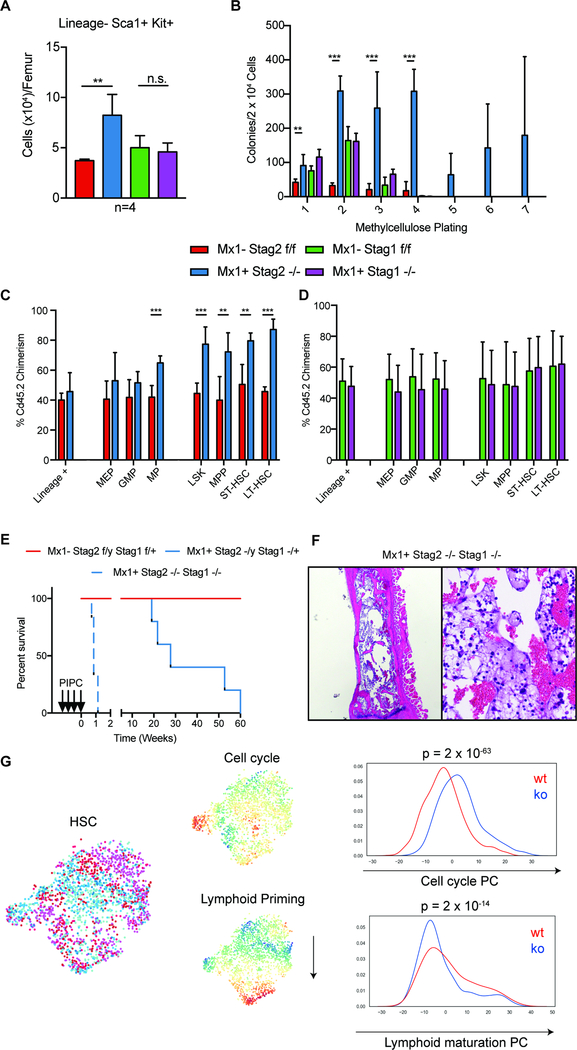
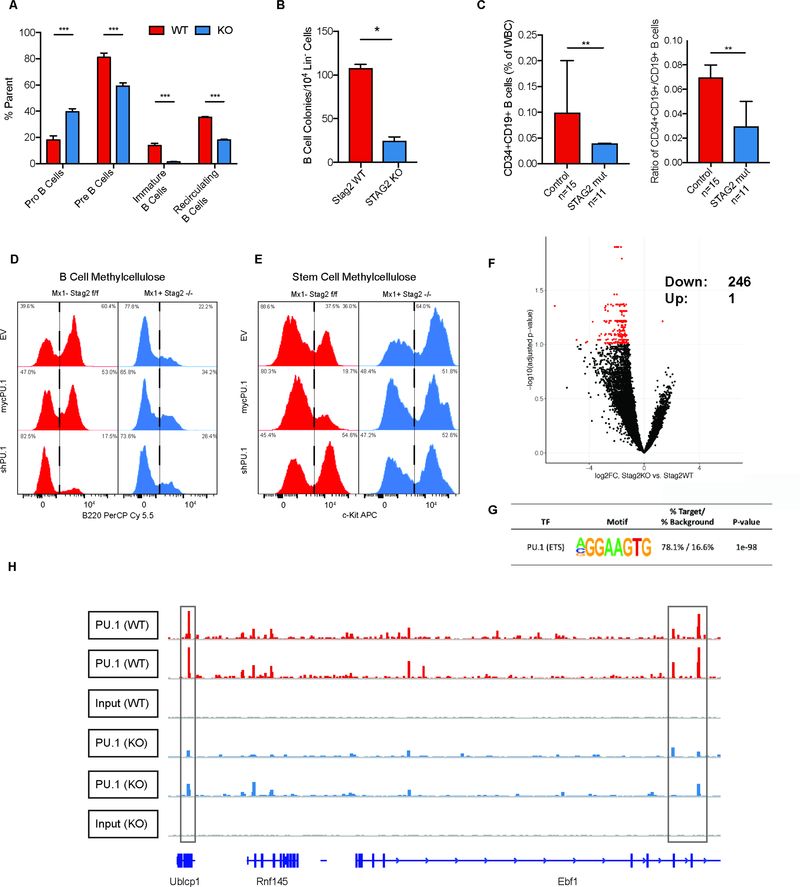
In order to elucidate the role of Stag1 and Stag2 in hematopoietic function/transformation, we generated Stag1 and Stag2 conditional knockout alleles. We induced somatic deletion of Stag1 or Stag2 in hematopoietic cells through Mx1-Cre mediated excision (Figure S1A–H). Following polyinosinic:polycytadinic (PIPC)-mediated deletion, Mx1-cre Stag2 −/− mice compared to cre-negative Stag2fl/fl (referred henceforth as KO and WT respectively) manifested an increase in hematopoietic stem cells (HSC), including a significant increase (2.2-fold; p<0.01) in the proportion of LSK (Lineage−Sca1+cKit+) cells (Figure 1A). Stag2 loss, but not Stag1 loss, induced an expansion of multipotent progenitors (MPP: LSK+Cd150−Cd48+Cd127−; p<0.04), short term HSCs (ST-HSC: LSK+Cd150−Cd48−; p<0.001), and long-term HSCs (LT-HSC: LSK+Cd150+Cd48−; p<0.02) (Figure S1I). We also observed a significant increase in granulocyte macrophage progenitors (GMP: Lineage−cKit+Sca1−Cd34+Fcγ+; p<0.001; Figure S1J–K) in the Stag2-deficient hematopoietic compartment.
Fig 1. Hematopoietic specific loss of Stag2, but not Stag1, results in altered stem cell function.
A) Stag2 KO, but not Stag1 KO mice have increased LSK (Lin−Kit+Sca1+) hematopoietic stem cells (Log2FC=2.2; p<0.01) B) Whole bone marrow plated in cytokine-enriched methylcellulose. Stag2 KO marrow, but not WT and Stag1 KO, has increased self-renewal capacity and can serially replate (>7 platings). Competitive bone marrow transplantation of C) Stag2 WT (Red)/KO (Blue) or D) Stag1 WT (Green)/KO (Purple) bone marrow mixed 1:1 with Cd45.1 normal marrow. Stag2 KO shows increased chimerism in stem and progenitor populations in the bone marrow at 16 weeks. E) Kaplan-Meier curve shows Stag2−/y/Stag1−/− is lethal with a median survival of 0.7 weeks (p=0.01). Stag2−/y/Stag1−/+ has a lethal phenotype with a median survival of 27.7 weeks (p=0.05). F) Hematoxylin and eosin (H&E) staining of Stag2/Stag1 KO bone marrow reveals marked aplasia. G) Left: t-SNE projection of library-size normalized and log transformed data for inferred HSC subset (2025 cells). Each dot represents a single cell colored by genetic condition (Stag2 KO (shades of blue), Stag2 WT (shades of red). Middle: t-SNE projection colored by the second and third principal components most correlated with lymphoid priming and cell cycle, respectively. Right: Distribution of cells along lymphoid and cell cycle components; p-values for Mann-Whitney U test are shown.
We next investigated the role of Stag2 and Stag1 in hematopoietic self-renewal. Stag2-deficient hematopoietic cells displayed increased serial replating capacity in methylcellulose culture assays (≥7 platings), whereas WT and Stag1 KO cells were not able to form colonies beyond 3 platings (Figure 1B). We next performed competitive transplantation assays to assess the impact of Stag2/Stag1 loss on self-renewal in vivo. We observed reduced Stag2-deficient derived chimerism in the peripheral blood of mice reconstituted with Stag2 KO and WT competitor cells (p<0.001; Figure S1L–N). However, within the bone marrow of transplant recipient mice, we found a significant increase in Stag2 KO derived chimerism, which was most significant in the LSK compartment (p<0.001) including in LT-HSCs (p<0.001; Figure 1C) but not in lineage positive cells (p≤0.37). Stag1 loss did not impact in vivo self-renewal or differentiation output (Figure 1D; S1O–Q). These data suggest that Stag2 loss induces both an increase in self-renewal and reduced differentiation capacity in HSCs, which are critical features of hematopoietic transformation.
To determine if Stag2 loss was associated with the expansion of hematopoietic cells with clonal cytogenetic alterations, we performed karyotype analysis in Stag2 and Stag1 WT and KO bone marrow cells. Chromosomal aberrations by metaphase karyotyping were not detected in either Stag2- or Stag1-deficient hematopoietic cells (n=7; Figure S2A–B). We did observe an increase in the proportion of cells with premature sister chromatid separation in Stag2 KO bone marrow (Figure S2C); however, consistent with metaphase cytogenetics, we did not observe clonal copy number alterations on low-pass whole genome sequencing (Figure S2D–E). This data suggested that the increase in hematopoietic self-renewal induced by Stag2 loss is not due to genomic instability and resultant clonal evolution.
Stag2 and Stag1 concurrent deletion is synthetic lethal
The observation that Stag2-deficient HSCs displayed increased self-renewal is in contrast to the lethality observed with loss of the core cohesin component Smc3 in hematopoietic cells (Viny et al., 2015). We next sought to evaluate potential compensatory function among the Stag paralogs. We found Stag1 transcript and protein expression were significantly increased in Stag2 KO hematopoietic cells (p<0.05), whereas Stag2 expression was not altered in Stag1 KO hematopoietic cells (p<0.65; Figure S2F–I). Analysis of the AML TCGA dataset showed that AML patients with STAG2 mutations expressed higher levels of STAG1 (p<0.006; Figure S2J), suggesting compensatory regulation. We hypothesized that Stag1 and Stag2 may have redundant roles with respect to chromatid segregation. In vitro studies have recently nominated STAG1 as a synthetic lethal target in STAG2 mutant cell lines (Benedetti et al., 2017; van der Lelij et al., 2017); however, this has not been assessed in vivo. We generated mice with hematopoietic cells retaining one or zero copies of Stag1 on the background of a Stag2 (hemizygous male) KO genotype. Stag2−/y/Stag1+/− mice developed rapid, progressive thrombocytopenia (p<0.001) and impaired survival (p<0.001) due to bone marrow failure (median survival of 27.7 weeks). Complete loss of Stag1 and Stag2 in hematopoietic cells induced rapid lethality (median survival 0.7 weeks; Figure 1E; Figure S2K). Stag1/Stag2 deficient mice were characterized by pancytopenia and bone marrow aplasia, consistent with redundancy in hematopoietic function and in alignment with the absolute requirement for core cohesin complex members (Viny et al., 2015) (Figure 1F). Similar to mice with bi-allelic deletion of Smc3, we observed marked chromosomal alterations on metaphase spreads of the Stag1/Stag2 deleted bone marrow (Figure S2L). Collectively, these data suggest a role for Stag2 in gene regulation and tumor suppression that is not shared with Stag1, whereas they have a shared, redundant role in chromosomal segregation.
Stag2 alters transcriptional lineage commitment
We hypothesized that Stag2 loss induced alterations in transcriptional output which dysregulate hematopoietic function and promote transformation. To investigate transcriptional mechanisms regulating self-renewal and differentiation in the context of Stag2 loss, we performed RNA-sequencing (RNAseq) on Stag2 KO and WT LSK cells. We identified 186 genes which were significantly downregulated and 42 genes which were significantly upregulated in Stag2 KO compared to WT cells (Figure S3A). We observed reduced expression of Ebf1, Vbrep3, Cd19, Bank1, and Pax5, suggesting a loss of B-cell lineage commitment (Herman et al., 2018; Zandi et al., 2008). We observed reduced expression of Vwf (Grover et al., 2016), Hba-a1, Hba-a2, Hbb-b1(Kingsley et al., 2013), Fcgr2b (Ferrari et al., 2007), Ccr2 (Reid et al., 1999), Spib (Su et al., 1997), S100a8, and S100a9 (Lagasse and Weissman, 1992), genes associated with myeloid/erythroid lineage commitment. Gene set enrichment analysis (GSEA) revealed that Stag2 deficient LSKs showed negative enrichment of genes associated with cell lineage specification (Figure S3B). We further found increased expression of the Hoxb4-target Hemgn (Jiang et al., 2010), as well as of Itgb3 (Umemoto et al., 2006), a Periostin-binding integrin shown to mediate HSC self-renewal. Coupled with downregulation of Flt3 (Adolfsson et al., 2001) and Selp (P-selectin) (Sullivan et al., 2011), these findings indicate an LSK pool that is transcriptionally skewed towards increased self-renewal.
Given the heterogeneity of the LSK compartment, we sought to determine if the transcriptional alterations observed in bulk LSKs were due to a subpopulation wide shift or a result of cell intrinsic differential expression. We performed single cell RNA-seq (scRNAseq) of ~24,000 Lin− hematopoietic stem/progenitor cells (HSPC) from Stag2 KO (n=3) and WT (n=3) mice (cohort annotations in Figure S3C–E). We found that within the HSC population, representing ~8% of cells, that the compartment distribution was skewed with KO HSCs having enriched expression of cell cycle gene signatures (p<10−63; Figure 1G top panel; S3F) and decreased expression of early lymphoid commitment signatures (p<10−14; Figure 1G bottom panel; S3G) consistent with negative enrichment for cell specification. We expanded upon these findings by comparing bulk LSK differentially expressed genes to gene expression data from dormant and active HSCs separately isolated through genetic lineage tracing studies (Cabezas-Wallscheid et al., 2017). We observed that genes that were decreased in expression in Stag2-deficient mice were most highly expressed in slow-cycling quiescent HSCs, while the upregulated genes were most highly expressed in the proliferative MPP1 fraction (LSK, Cd34+, Flt3−) (Figure S3H). Similar findings were observed when comparing our data to single cell RNA-seq data, such that the most downregulated genes in Stag2-deficient LSKs were most highly expressed in the most quiescent HSCs (Figure S3I). Collectively, these analyses highlight a transcriptional signature associated with decreased lineage commitment, decreased quiescence, and enhanced self-renewal in the setting of STAG2 loss.
Stag2 loss impairs differentiation and induces myeloid dysplasia
To determine the consequences of these transcriptional alterations in vivo, we assessed the phenotype of mice with Stag2-deficient hematopoiesis over time. Hematopoietic-specific Stag2 loss resulted in progressive leukopenia (p<0.02) and thrombocytopenia (p<0.01; Figure S4A). We observed an expansion in myeloid cells and a significant reduction in B lymphocytes (p<0.001; Figure 2A) in the peripheral blood, which was not seen with Stag1 deletion. Stag2 loss, but not Stag1 loss, induced morphologic alterations including an increase in immature myeloid cells, an expansion of small hypolobated megakaryocytes, and nuclear:cytoplasmic dyssynchrony in the erythroid lineage (Figure 2B). These abnormalities are features of human myelodysplasia—consistent with the observation that 5–15% of MDS patients have loss-of-function mutations in STAG2 (Kon et al., 2013; Thota et al., 2014).
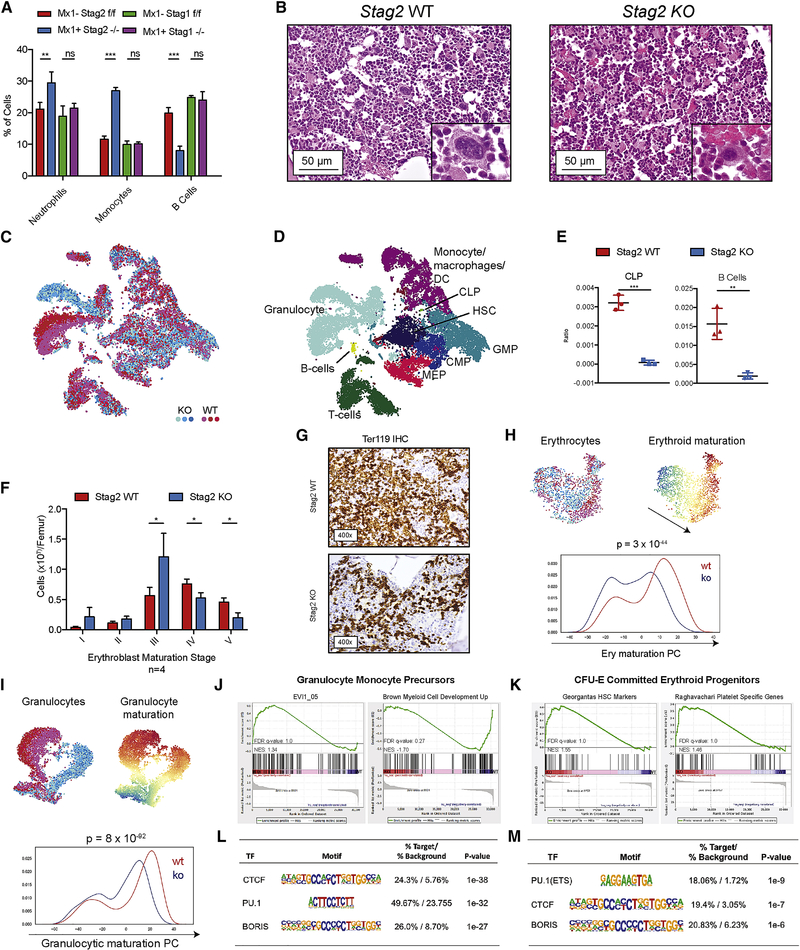
Fig 2. Stag2 alters transcriptional lineage commitment in stem and progenitor cells.
A) Peripheral blood with increase in Neutrophils (Gr1+Mac1hi), Monocytes (Gr1+Mac1lo), and decrease in B cells (B220+) in Stag2 KO mice. No statistical differences were observed in Stag1 KO mice. B) Hematoxylin and eosin (H&E) staining of sternal bone marrow (400x) from Stag2 KO mice. Increased immature myeloid cells, nuclear-to-cytoplasmic dyssynchrony in erythropoiesis, and small, hypolobated megakaryocytes (Inset image, 1000x). C) Single cell RNA sequencing of Stag2 WT (n=3; shades of red) and Stag2 KO (n=3; shades of blue) Lin− HSPC. (C) t-SNE projection of library-size normalized and log transformed data for complete collection (24,153 cells). Each dot represents a single cell colored by genetic condition. D) t-SNE map colored by inferred cell type, as detailed in Figure S4. E) Frequency of CLPs (left) and B-cells (right) in Stag2 WT (red) and KO (blue) samples; asterisks indicate statistical significance (student’s t test, **p<0.01, ***p<0.001) F) Flow cytometry of erythroblast stage in Stag2 WT and KO bone marrow reveals increased erythroblast stage III (p<0.05), reduction of mature erythroblast stage IV (p<0.02) and stage V (p<0.01). G) Immunohistochemical analysis of Stag2 WT and KO bone marrow reveals a marked reduction in Ter-119 expression in Stag2 KO. H) Top left: t-SNE projection of library-size normalized and log transformed data for inferred MEP subset (1787 cells). Each dot represents a single cell colored by genetic condition (Stag2 KO: shades of blue, Stag2 WT: shades of red). Top right: t-SNE projection colored by the second principal component most correlated with erythroid maturation. Bottom: Distribution of cells along erythroid maturation component. I) Top left: t-SNE projection of library-size normalized and log transformed data for inferred granulocytic subset (6316 cells). Each dot represents a single cell colored by genetic condition (Stag2 KO: blue, Stag2 WT: red). Top right: t-SNE projection colored by the first principal component most correlated with granulocyte maturation. Bottom: Distribution of cells along granulocyte maturation component. J) Gene-set enrichment analysis of GMP RNAseq shows increased expression of genes in the Evi1 gene set and decreased expression of myeloid development genes. K) CFU-E RNAseq shows enrichment for the Georgantas HSC markers gene set and retention of platelet specific genes. Integration of differentially expressed genes in L) GMP and M) CFU-E populations with ATAC-sequencing commonly have genes with CTCF/CTCFL (BORIS) motif signatures and PU.1 (ETS) motif signatures with decreased expression and decreased accessibility in both populations.
To determine at which level of differentiation multi-lineage dysplasia emerges, we analyzed lineage composition through analysis of scRNAseq data (Figure 2C–D). Supporting the reduction in B cell output observed in the peripheral blood, analysis of scRNAseq data showed a significant decrease in B cells and in B cell progenitors (Figure 2E). Although the paucity of lymphoid cells precluded further transcriptional tracing, we observed significant population shifts in erythroid and granulocyte development consistent with observed histologic myelodysplasia. Flow cytometric analysis of erythroblast maturation revealed attenuation at Stage III of erythropoiesis (Chen et al., 2009) (Figure 2F, S4B–C), a stage that is demarcated by condensed cell size by forward scatter (FSC) and decreased CD44 expression relative to prior stages of erythroid differentiation. Stag2-deficient erythroid cells were also characterized by morphologic dysplasia; immunohistochemical analysis demonstrated a marked decrease in Ter-119, a glycophorin-associated murine erythroid marker (Kina et al., 2000) (Figure 2G). By comparison, Stag1 KO mice did not manifest hematopoietic alterations, morphologic abnormalities, or defects in erythroid differentiation (Figure S4A–C). Transcriptional analysis of the erythroid lineage in Stag2 deficient cells further supported impaired terminal maturation (p<10−44; Figure 2H). Genes defining this principle component define the progression from common myeloid progenitor (CMP) to megakaryocyte erythroid progenitor to erythroid A (Figure S4D–E). Overall, the most significant transcriptional difference identified in the scRNAseq was impaired granulocytic terminal maturation (p<10−92; Figure 2I). Genes defining this principle component define the progression from CMP to GMP to granulocyte (Figure S4F–G).
Given the alterations in transcriptional output and differentiation potential, we hypothesized that Stag2 loss led to altered chromatin accessibility at differentially expressed loci. We therefore performed RNAseq and ATAC-sequencing (ATACseq) on sorted Stag2 WT and KO GMP and erythroid progenitor (CFU-E: Lineage−cKit+Sca1−Cd34−Fcγ−Cd105+) cells. Compared to WT GMPs, Stag2-deficient GMPs had enrichment for a stem-cell gene expression signature associated with increased Evi1 expression and negative enrichment of myeloid cell development signatures by GSEA (Figure 2J). Stag2-deficient CFU-Es had increased expression of genes normally expressed in HSCs and in the megakaryocyte lineage, consistent with the increased self-renewal and impaired differentiation seen with Stag2 loss (Figure 2K). Integrated analysis of RNA-seq and ATAC-seq revealed that a significant subset of the downregulated genes had reduced chromatin accessibility. Both GMPs and CFU-E shared loss of accessibility and expression for genes with either CTCF/CTCFL (BORIS) or PU.1(ETS) motifs (Figure 2L–M). CTCF is a well-described cohesin binding partner (Wendt et al., 2008) that cooperates with cohesin to mediate enhancer/promoter associations (Rao et al., 2014). Notably, PU.1 and its target genes have a critical role in determining hematopoietic lineage commitment (Antony-Debre et al., 2017; Will et al., 2015).
As we had previously reported defective lineage commitment and dynamic effects on accessibility in the heterozygous loss of Smc3, we compared the differentially expressed and differentially accessible genes between the two models, with the caveat that our transcriptional/epigenomic studies of Smc3 heterozygous cells used c-kit enrichment. We identified 36 common differentially expressed genes, which were decreased in expression in both datasets. Integration of ATAC-seq data from the 2 models allowed us to identify 236 loci with altered accessibility by ATACseq in Stag2 KO and Smc3 heterozygous HSPC. This included 116 sites with loss of accessibility, with HOMER analysis identifying these as being highly enriched for PU.1(ETS) motifs (p<10−15; Figure S4H). This suggests that although Smc3 haploinsufficiency and Stag2 loss may have non-overlapping effects on gene expression, we were able to identify alterations in gene regulation including at PU.1 target genes, consistent with an important role for the cohesin complex in hematopoietic lineage commitment.
Stag1/2 possess shared and independent chromatin binding
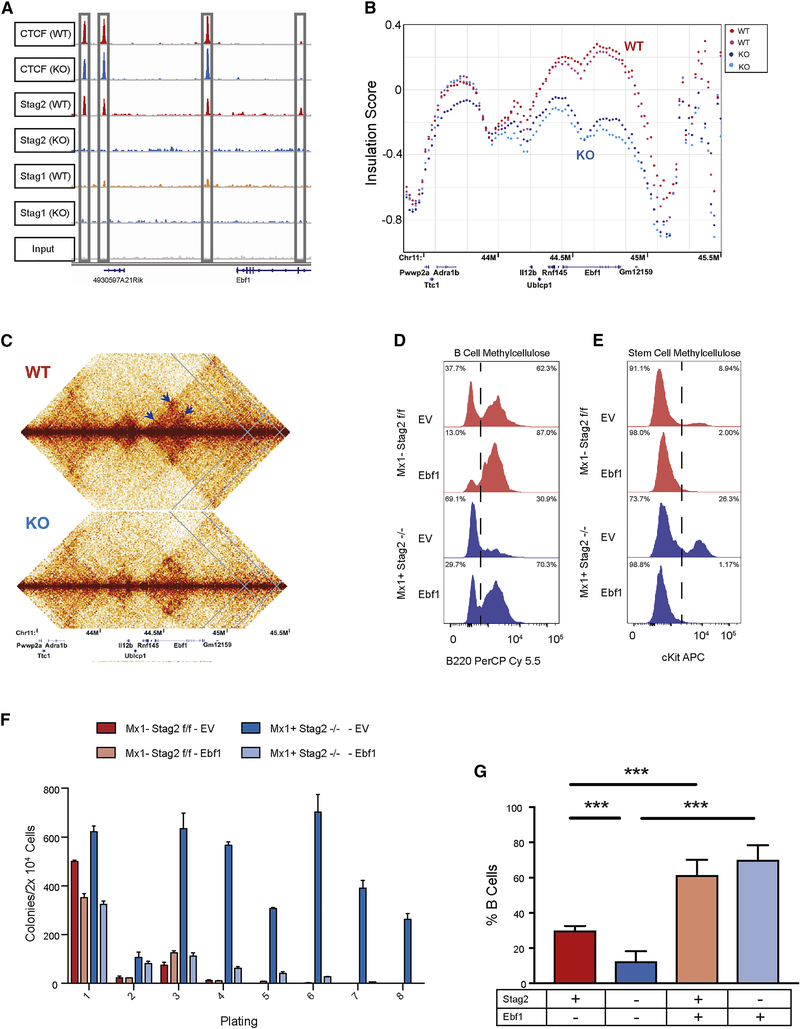
It has been shown using in vitro model systems that cohesin is essential for genomic cisinteractions and that rapid depletion of cohesin ring members result in complete loss of interphase architecture (Haarhuis et al., 2017; Rao et al., 2017; Schwarzer et al., 2017). However, these model systems do not reflect the genomic alterations observed in either human cancers or in developmental syndromes with germline cohesin mutations, such as Cornelia de Lange syndrome (Liu and Krantz, 2009). As the complete loss of cohesin structural loop components may not be compatible with a viable cell division event, the absence of transcriptional changes observed using in vitro systems of cohesin degradation (Schwarzer et al., 2017) may not reflect the pathophysiology of Stag2 loss of function. We next sought to determine how Stag2-loss affected chromatin topology in primary cells using Hi-C chromosome conformation capture in HSPCs. Stag2 KO cells did not show alterations in topologically associating domain (TAD) boundaries, in global insulation, and did not show differences in the spatially segregated genomic regions known as A/B compartments compared to WT HSPCs (Lieberman-Aiden et al., 2009) (Figure 3A–C).
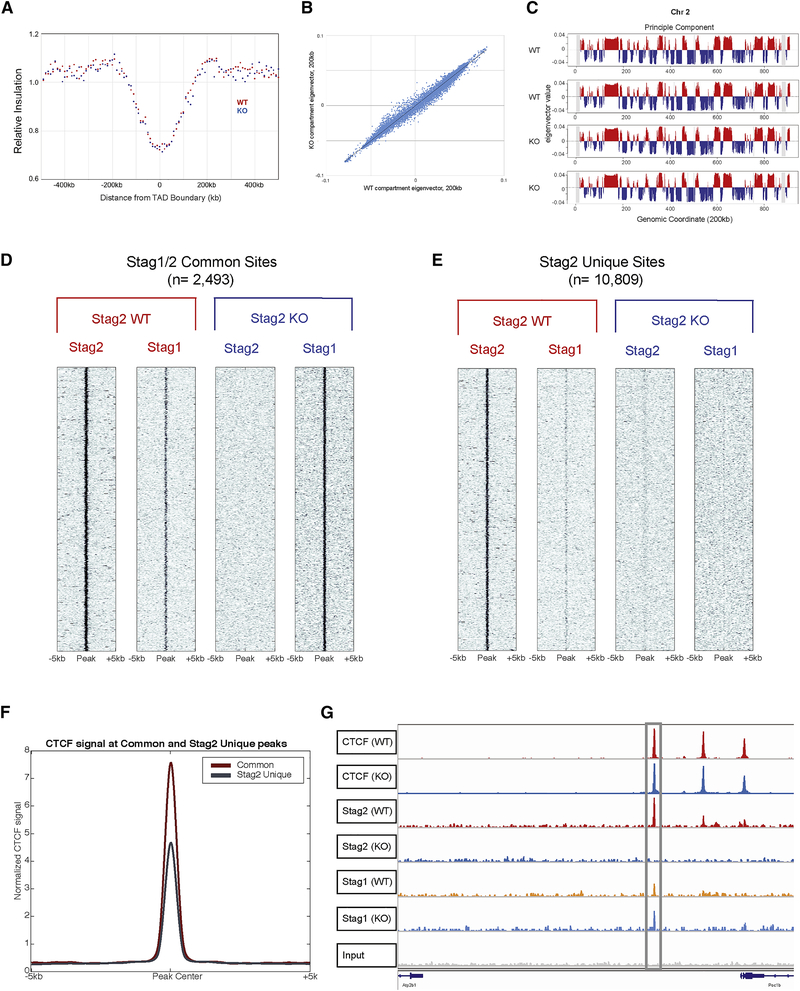
Fig 3. Stag2 and Stag1 possess shared and independent chromatin binding.
A) Insulation pileup plot from each dot shows the median of the interactions for each 200kb bins located 500kb upstream or downstream of a TAD border, normalized by the median interactions for WT and KO, respectively. B) Hi-C insulation scores were normalized by the median interactions for WT and KO, respectively. The diagonal bins were excluded for calculating the median. C) A/B compartments of chr2 for each biological replicate. The A compartments, positive PC1 signals, are highlighted in red, while the B compartments, negative PC1 signals, are highlighted in blue. D) Chromatin immunoprecipitation and sequencing for Stag1 and Stag2 in Stag2 WT and KO HSPC show a discrete subset of the genome where Stag1 is able to bind Stag2 loci in its absence (common sites) and E) discrete loci where Stag1 is unable to bind in the absence of Stag2 (Stag2 unique sites). F) Enrichment of Stag1/2-common sites and Stag2-unique sites for CTCF shows strong enrichment in both sets. G) IGV track at TAD boundary as measured by Hi-C insulation on chromosome 10 showing CTCF occupancy as well as Stag2 in WT with Stag1 occupancy increased in Stag2 KO HSPC.
Given the lack of changes in global structure, we sought to determine if Stag2 loss would affect the ability of either CTCF or the structural cohesin components Smc1a and Smc3 to localize to canonical cohesin/CTCF binding sites (Nora et al., 2017). Chromatin immunoprecipitation sequencing revealed that the occupancy of these three proteins was largely unaffected by the presence or absence of Stag2 in HSPCs (Figure S5A–C). We hypothesized that complete loss of Stag2 might not alter higher order DNA topology due to redundancy with Stag1, particularly at CTCF bound sites. We therefore performed ChIP-seq to delineate Stag2 and Stag1 occupancy in HSPCs from Stag2 WT and KO mice. We identified a set of peaks where Stag2 and Stag1 were bound in WT mice and where Stag1 occupancy increased in the absence of Stag2 (n=2,493; Figure 3D, S5D–E). Conversely, we identified a second set of peaks where Stag2, but not Stag1, was bound in WT HSPCs and lost in Stag2 KO cells without compensatory Stag1 occupancy (n=10,809; Figure 3E, S5F–G). Previous work has demonstrated CTCF-dependent and independent localization of the cohesin complex (Parelho et al., 2008; Rubio et al., 2008) which in epithelial cell lines have preferential binding sites for Stag1 and Stag2(Kojic et al., 2018). We hypothesized that altered regulation of Stag2-bound loci in WT cells which are not bound by Stag1 in WT or KO cells, were key to the altered differentiation and self-renewal seen in Stag2 KO mice. We found that loci with both Stag2 and Stag1 binding had the highest enrichment for CTCF binding (Figure 3F).
Loss of chromatin insulation impairs transcriptional output in Stag2 deficient hematopoietic progenitors
Stag1 was able to bind a subset of loci previously occupied by Stag2 or both Stag1 and Stag2 in WT cells; this included TAD boundaries determined by Hi-C (Figure 3G). By contrast, when we focused on Stag2-only bound sites, we observed locus-specific alterations in short-range interactions as calculated by the insulation metric (Crane et al., 2015). We quantified these alterations through measurement of cumulative local contacts and insulation score changes (ISC) that reflect local differences in chromosomal organization. To examine whether loss of Stag2 binding affects local chromosomal organization, we compared ISC in WT and Stag2KO HSPCs and identified 71,815 ISC bins that showed local chromosomal contact changes (10kb bins with insulation score change less than −0.1 or greater than 0.1, Figure S6A, left panel). We then overlapped ISC bins with Stag1/2 co-bound and Stag2-unique peaks. Annotation analysis of both Stag1/Stag2 common and Stag2-unique peaks showed enrichment of Stag2-unique ISC peaks within promoter regions compared to Stag1/Stag2 common peaks (fisher’s exact test p<3.03 ×10−26 for ISC up; p<1.22 ×10−10 for ISC down; Figure S6A, right panel), suggesting that the ISC seen in Stag2-deficient cells may contribute to altered gene expression.
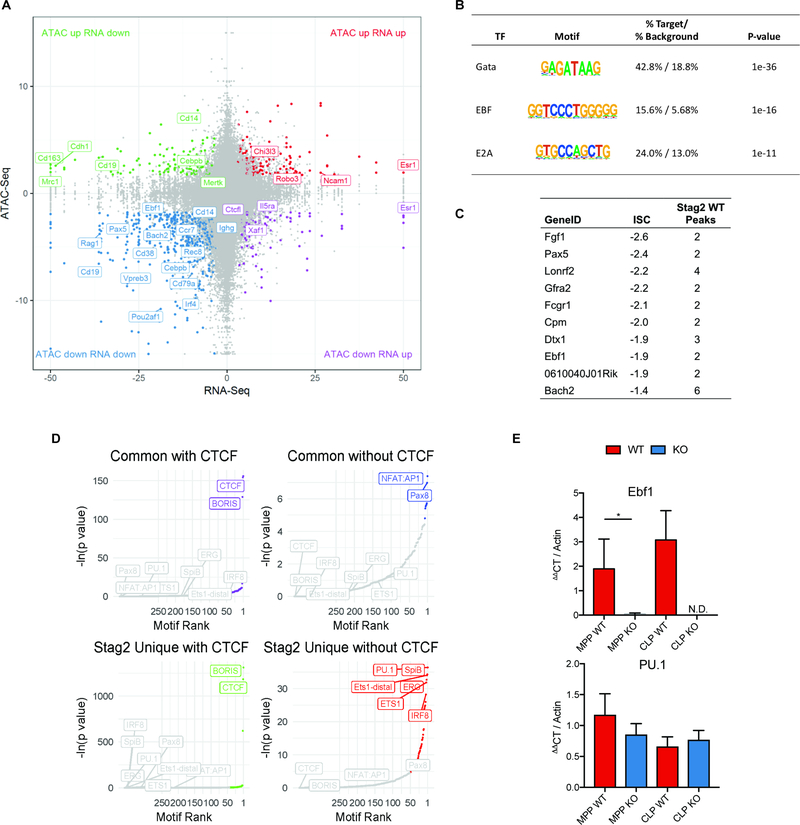
These data suggested that the role of Stag2 in gene regulation and in hematopoiesis might be due to regulation of loci bound by factors other than CTCF. To determine the impact of Stag2 loss on chromatin structure and transcriptional output we performed RNA-seq and ATAC-seq in WT and Stag2 KO HSPCs. Similar to gene expression/ATAC-seq data in sorted hematopoietic subsets, the majority of genes with significantly altered chromatin accessibility and gene expression in the absence of Stag2 had decreased accessibility and reduced transcription (Figure 4A). Several of the ATAC peaks with reduced accessibility were in proximity to critical B cell and myeloid lineage defining transcription factors, including Ebf1, Pax5, and Cebpb. To uncover transcription factor networks underlying these changes, we performed motif enrichment on peaks with concomitant reduction in accessibility/gene expression. We observed enrichment for Gata, Ebf, and E2A targets within the loci with decreased expression and accessibility (Figure 4B), but not for Cebpb motifs. The genes associated with loss of insulation by ISC significantly overlapped with those loci with decreased accessibility and decreased expression, with Ebf1 and Pax5 among the genes with the greatest magnitude of insulation change (Figure 4C, S6B–D). Notably, this effect was not confounded by sequencing depth as there were not significant differences in read counts across replicates (Figure S6E) and Hi-C contact frequencies were corrected and scaled to exclude sequencing depth bias (Crane et al., 2015). We therefore performed motif analysis on Stag1/2 common and Stag2-unique sites by dichotomizing those that were bound and not bound by CTCF. As expected, loci with shared Stag1/Stag2 occupancy and CTCF binding sites were enriched for CTCF and CTCFL (BORIS) binding motifs. By contrast, Stag2-unique loci (not bound by Stag1) were enriched for key lineage-specific transcription factor motifs including ERG, IRF8, and PU.1 (Figure 4D). These transcription factors are essential for myeloid and lymphoid development (McKercher et al., 1996; Wang et al., 2014) and attenuated function of these transcription factors has transformative potential in murine (Will et al., 2015) and human hematopoietic cells (Mueller et al., 2006; Pogosova-Agadjanyan et al., 2013; Vangala et al., 2003).
Fig 4. Stag2 loss decreases chromatin insulation and results in impaired transcriptional output.
A) Intersection of RNAseq and ATACseq in Lin− bone marrow plotted as −log10(pvalue) *sign of the Log2 fold change of Stag2 KO compared to WT and each point representing a one ATAC peak. The majority of differentially expressed genes lose accessibility, including distinct B-cell regulators (e.g. Ebf1, Pax5, Cd19). B) HOMER motif analysis of genes in lower left quadrant of Figure 4A from intersection of RNAseq and ATACseq in Lin− bone marrow. Genes that are downregulated and lose accessibility are targets of Gata (p=10−36), EBF (p=10−16), and E2A (p=10−11). C) Top 10 genes with greatest magnitude of ISC and number of WT Stag2 peaks in the gene. D) Motif analyses of common and Stag2-specific sites with and without CTCF show common enrichment for CTCF and CTCFL (BORIS). Stag2-unique sites bind targets of key lineage priming factors PU.1, SpiB, ERG, ETS1, and IRF8, which Stag1 is unable to bind. E) Putative expressers of PU.1 and Ebf1 from Stag2 WT and KO bone marrow were sorted and Ebf1 and PU.1 expression was measured by RT-PCR in multipotent progenitors (MPP) and common lymphoid progenitors (CLP). Ebf1 expression was decreased in both populations (MPP p=0.04; CLP not detectable). PU.1 expression was not statistically different in either population (MPP p=0.13; CLP p=0.32).
Altered Ebf1 chromatin structure results in attenuated B-cell development in Stag2-deficient hematopoietic progenitors
We next sought to determine if the altered expression of hematopoietic transcription factors were due to alterations in the population distribution in HSPC or was due to altered expression of these factors in purified hematopoietic populations, including in MPPs antecedent to full lineage commitment. We measured PU.1 and Ebf1 expression and B cell differentiation in MPPs and in common lymphoid progenitors (CLP: LSK+Cd150−Cd48+Cd127+) from Stag2 KO and WT mice. We found that Ebf1 expression was reduced/absent in purified Stag2 KO MPPs (p<0.04, Log2FC=−1.8) and CLPs (not detectible in KO), while there was no significant difference in PU.1 expression in either cell type (Figure 4E). Consistent with these data, B cell maturation was blocked from the pro-B cell to early pre-B cell stage (Pucella et al., 2015) in Stag2 KO mice, with almost no late pre-B cells (Figure 5A). These findings were further bolstered by in vitro colony assays in B cell cytokine enriched methylcellulose where Stag2 KO HSPCs failed to generate B cell colonies (p<0.003; Figure 5B). We next investigated if the same was true in Stag2-mutant MDS patients by enumerating B cells and B cell progenitors in STAG2-mutant MDS patient bone marrow samples compared to a cohort of non-MDS individuals evaluated for refractory cytopenias (Figure 5C, S6F). Immature B cells (CD34+CD19+) were decreased by 1.7-fold (p<0.01) in STAG2-mutant MDS patients, demonstrating that altered B-cell lineage commitment is observed in STAG2-mutant MDS.
Fig 5. Altered Ebf1 chromatin structure results in a blockade of B cell development.
A) Enumeration of B lymphocyte maturation in Stag2 WT and KO mice. Bone marrow was analyzed using flow cytometry for pro- (Cd43+) and pre- (Cd43−) B-cells in parent gate B220loIgM− showing B-cell development block in the pro-B to pre-B transition (p<0.003). Immature (B220loIgM+) and recirculating B cells (B220hiIgM+) were analyzed as a percentage of live singlets, which were both markedly reduced in Stag2 KO mice (asterisks indicate statistical significance (student’s t test, **p<0.01, ***p<0.001). B) Methylcellulose colony enriched with IL-7, SCF, and FLT3-L shows reduction in the number of B cell colonies in Stag2 KO bone marrow compared to WT (p<0.003). C) Enumeration of immature B cells (CD34+CD19+) and ratio compared to mature B cells (CD34-CD19+) show reduced immature B cells (p<0.010) and immature:mature ratio in STAG2 mutant MDS patients (p<0.008; n=11) compared to controls (n=15). D-E) Stag2 WT and KO HSPC were infected with lentivirus containing GFP-tagged empty vector, GFP-mycPU.1, or GFP-shPU.1; GFP+ cells were plated in either B cell colony methylcellulose or stem cell methylcellulose. Cells were harvested after 7 days and analyzed by flow cytometry for (D) the B marker B220 or (E) stem cell marker cKit. F) Volcano plot for differentially PU.1-occupied loci by chromatin immunoprecipitation sequencing in HSPCs of Stag2 WT and Stag2 KO (data points in red indicate adjusted p<0.1). Loci with decreased in PU.1 occupancy in Stag2 KO to the left (n=246) and loci with increased in PU.1 occupancy in Stag2 KO to the right (n=1). G) HOMER analysis of 246 loci with decreased PU.1 binding shows enrichment for PU.1(ETS) motif (p<1098). H) IGV track of the Ebf1 locus for Stag2 WT (n=2) and KO (n=2) shows decreased PU.1 binding at 3 loci (gray boxes).
Given that PU.1 can regulate myeloid and lymphoid lineage commitment (Scott et al., 1997) and the enrichment for PU.1 motif/targets in our epigenomic/transcriptional analysis, we investigated the role of PU.1 in Stag2-deficient alterations in B-cell lineage commitment and self-renewal. PU.1 silencing reduced B-lineage output derived from WT hematopoietic cells, similar to the effects of Stag2 deletion (Figure 5D, top row). However, the reduced B-cell lineage output of Stag2 KO cells was not further attenuated by PU.1 silencing as assessed by flow cytometry of B220 expression (Mansson et al., 2008) and B-lineage colony formation (Figure 5D, bottom row; S6G). By contrast, overexpression of PU.1 was unable induce Ebf1 expression in Stag2 KO cells (Figure S6H) and unable to restore B220 expression in B cell methylcellulose cultures (Figure 5D, middle row). Moreover, increased PU.1 expression did not attenuate the increased expression of the stem/progenitor marker cKit seen in Stag2-deficient hematopoietic cells (Figure 5E) or attenuate the capacity of Stag2-deficient hematopoietic cells to serially replate (Figure S6I). Given these results, we suspected that PU.1 occupancy would be affected by Stag2 loss and we performed ChIPseq for PU.1 in Stag2 WT and KO HSPC (Figure 5F). We identified 246 loci with reduced PU.1 occupancy in Stag2 KO bone marrow. HOMER analysis confirmed that these sites contain the PU.1(ETS) motif (p<10−98; Figure 5F–G). These data suggest that PU.1 occupancy is reduced in the absence of Stag2, but restored PU.1 expression is not sufficient to reverse the alterations in hematopoietic differentiation and self-renewal seen in Stag2 KO cells.
This led us to hypothesize that key transcription factors downstream of PU.1 which are bound by Stag2 have locus specific alterations in conformation in the absence of Stag2 that lead to alterations in target gene expression with functional importance which cannot be rescued by re-expression of upstream transcription factors. Ebf1 has three PU.1 bound sites which are decreased in Stag2 KO (Figure 5H) and four Stag2 bound sites in hematopoietic progenitors, each of which is not bound by Stag1 in WT or KO cells (Figure 6A). As noted above, we observed near-complete abrogation of Ebf1 transcription including in MPPs preceeding Blineage commitment (Figure 4E). Although Ebf1 expression increased in PU.1 overexpressing Stag2 WT cells (p<0.014), we did not observe induction of Ebf1 expression when PU.1 was overexpressed in Stag2 KO cells (Figure S6H). We observed a significant reduction in local insulation throughout the Ebf1 locus (Figure 6B) including loss of cis-interactions at three Stag2 binding sites (Figure 6C). Consistent with this hypothesis, expression of Ebf1 in Stag2 KO progenitors (Figure S6J) restored B-lineage marker expression and B-cell colony formation in vitro (Figure 6D). Ebf1-expressing Stag2 KO progenitors had decreased cKit expression (Figure 6E) and Ebf1 restoration abrogated the serial replating capacity of Stag2-deficient cells (Figure 6F). Stag2 WT and KO HSPC infected with GFP-tagged Ebf1 were then transplanted into lethally irradiated recipients, which led to restored B cell output in vivo (p<0.001, Figure 6G). Taken together, these data demonstrate a key role for Stag2-mediated regulation of Ebf1 expression in modulating the balance between hematopoietic differentiation and self-renewal.
Fig 6. Induced Ebf1 expression rescues B cell development.
A) IGV track of the Ebf1 locus with Stag2 and Ctcf binding at 4 distinct sites lost in Stag2 KO and not bound by Stag1 either in WT or KO. B) ISC plotted across Ebf1 for Stag2 WT (n=2; shades of red) and KO (n=2; shades of blue) shows marked loss of insulation. C) Contact map of Ebf1 shows loss of cis-interaction at three loci (arrows). D-E) Lin− Stag2 WT and KO marrow were infected with retrovirus containing GFP-tagged empty vector or GFP-pCMV-Ebf1. GFP+ cells were plated in either B-cell colony methylcellulose or stem cell methylcellulose. Cells were harvested after 7 days and analyzed by flow cytometry for mature cell markers including (D) the B marker B220 or (E) stem cell markers including cKit. F) Cells plated in stem cell 10 methylcellulose were serially replated with exhaustion of Stag2 KO cell overexpressing Ebf1. G) GFP-tagged empty vector or GFP-pCMV-Ebf1 transduced Lin− cells were injected into lethally irradiated CD45.1 recipient mice. Leukocyte populations from the parent gate (GFP+, Cd45.2+, Cd45.1−) were analyzed by flow cytometry for percentage of mature B cells. Induced Ebf1 expression restored B cell population frequency from the Stag2 KO HSPC (p<0.001). Asterisks 15 indicate statistical significance (student’s t test, **p<0.01, ***p<0.001)
Discussion
The cohesin complex is essential in pleiotropic cellular and gene regulatory functions, including in chromosome segregation (Losada et al., 1998; Toth et al., 1999) and in maintaining DNA interactions (Rao et al., 2017; Schwarzer et al., 2017) which regulate gene expression (Cuartero et al., 2018; Kagey et al., 2010; Seitan et al., 2013). Here we demonstrate a non-redundant role of Stag2 and Stag1 in hematopoiesis, with a specific role for Stag2 in regulating the balance between hematopoietic differentiation and self-renewal. In addition, we show that Stag2 bound loci that are not bound by Stag1 undergo locus specific changes in local interactions that, in the absence of Stag2, leads to altered expression of key loci governing hematopoietic function. Taken together, these data illustrate a critical link between Stag2, DNA interactions, and gene regulation that contributes to hematopoietic transformation.
Stag2 and Stag1 have both shared and distinct roles (Kojic et al., 2018). These paralogs have a redundant role in chromatid separation such the presence of either protein within the cohesin complex is adequate for intact sister chromatid alignment. In contrast to deletion of core cohesin ring components, which abrogates the survival of hematopoietic cells (Viny et al., 2015), somatic deletion of either Stag1 or Stag2 was compatible with hematopoietic stem cell viability, whereas loss of both genes led to hematopoietic failure and defective chromosomal segregation. Our in vivo studies underscore previous in vitro dependency studies (van der Lelij et al., 2017) and suggest that therapies which directly or indirectly alter Stag1 function will induce a specific dependency in Stag2 mutant cancer cells. However, increased self-renewal, impaired lineage commitment, and progressive hematopoietic alterations were observed with Stag2 deletion but not with Stag1 loss. These studies illustrate that Stag2 has a key role in hematopoiesis and in hematopoietic transformation that is not shared by Stag1. Future studies will illustrate how Stag2 loss of function cooperates with other alleles to promote leukemic transformation and how co-occurring disease alleles cooperate with the chromatin architecture alterations induced by Stag2 loss.
Our data suggest that the role of Stag2 in regulating hematopoiesis and in hematopoietic transformation is Stag1-independent. Key hematopoietic regulators which show differential expression, accessibility, and insulation in the absence of Stag2 are bound by Stag2 and not by Stag1. This includes several key genes essential for lineage commitment to lymphoid fate, including the PU.1 targets Ebf1 and Pax5. Previous studies demonstrated that PU.1-mediated regulation of Ebf1 is a key event in B-cell lineage commitment (Medina et al., 2004; Scott et al., 1997), and PU.1 has been defined to be a pioneer factor with nucleosome remodeling capacity. Given these data and the known role of Ebf1 in B cell commitment/differentiation (Zandi et al., 2008), we show that PU.1 restoration cannot rescue the impact of Stag2 loss on hematopoietic self-renewal and lineage commitment and point to non-hierarchical roles for transcription factor expression and the permissive chromatin context regulated by Stag2. By contrast, restoration of Ebf1 expression restores differentiation and abrogates the aberrant self-renewal of Stag2 deficient cells.
Our data illustrate a key role for Stag2 in the balance between self-renewal and differentiation, a critical feature of transformation across different tumor types. Moreover, the observation that reactivation of the Stag2-target Ebf1 can reverse these alterations suggests a key role for Stag2-regulated transcription factor networks in tumor suppression, which is reversible when key networks are restored in Stag2 mutant cells. We hypothesize that Stag2 loss-of-function induces lineage specific alterations in transcription factor function in a spectrum of malignant contexts. Most importantly, our studies identify a key pathophysiologic role for mutations in the factors that govern gene regulatory architecture in malignant transformation. Our data point to the reversibility of transcriptional dysregulation which mediates aberrant self-renewal and differentiation in cancer cells and suggest that therapies that reverse these alterations may have therapeutic benefit in cohesin-mutant cancers.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ross L. Levine (leviner@mskcc.org).
Mouse lines generated in this study have been deposited to Jackson Laboratory (Jax), Stag2flox (JAX#030902); Stag1flox (JAX#030904).
Experimental Models and Subject Details
Animals
All animals were housed at Memorial Sloan Kettering Cancer Center. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at Memorial Sloan Kettering Cancer Center.
Generation of Stag1/2-deficient mice
The Stag2 and Stag1 conditional allele were each deleted by targeting exon 7. Two LoxP sites flanking exon 7 and an Frt-flanked neomycin selection cassette were inserted in the upstream intron (Figure S1A). 10 μg of the targeting vector was linearized by NotI and then transfected by electroporation of BAC-BA1 (C57BL/6 × 129/SvEv) hybrid ES cells. After selection with G418 antibiotic, surviving clones were expanded for PCR analysis to identify recombinant ES clones. Secondary confirmation of positive clones identified by PCR was performed by Southern blotting analysis. DNA was digested with BamHI and electrophoretically separated on a 0.8% agarose gel. After transfer to a nylon membrane, the digested DNA was hybridized with a probe targeted against the 3′ or 5′ external region. DNA from C57BL/6 (B6), 129/SvEv (129), and BA1 (C57BL/6 × 129/SvEv; Hybrid) mouse strains was used as WT controls. Positive ES clones were expanded and injected into blastocysts. The generated mice (Stag2fl/fl and Stag1fl/fl) were initially crossed to a germline Flp-deleter (The Jackson Laboratory), to eliminate the neomycin cassette, and subsequently to the IFN-α–inducible Mx1-cre (The Jackson Laboratory) (Kuhn et al., 1995; Lakso et al., 1996). Mice were backcrossed for six generations to C57BL/6 mice. Stag2fl/fl or y, Stag2fl/+ or y, and Stag2+/+ or y littermate mice were genotyped by PCR with primers Stag2-NF (5′-CACTCATGCTGGCAAGTATTGTAC-3′) and Stag2-NR (5′-AACAGCCTGAGCAAAGAATCCAAAG-3′) and Stag2–3’ (5′-TGTGTGCCTCTTTGAACAATGCCC-3′) using the following parameters: 94°C for 3 min, followed by 35 cycles of 94°C for 15 s, 64°C for 30 s, and 72°C for 90 s, and then 72°C for 5 min. The WT allele was detected as a band at 385 bp, whereas the floxed allele was detected as a band of 554 bp. Excision after Cre recombination was confirmed by PCR with primers to detect a band at 294 bp. Stag1fl/fl, Stag1fl/+, and Stag1+/+ littermate mice were genotyped by PCR with primers Stag1-CommonF (5′-GACTGGTATCTGACGGCTTATACC-3′) and Stag1-CommonR (5′-CACTGAGGACCAGGCATTGTAAGG-3′) and Stag1-FloxR (5′-TGAACTGATGGCGAGCTCAGACC-3′) using the following parameters: 94°C for 3 min, followed by 35 cycles of 94°C for 15 s, 62°C for 30 s, and 72°C for 90 s, and then 72°C for 5 min. The WT allele was detected as a band at 1104 bp, whereas the floxed allele was detected as a band of 745 bp. Excision after Cre recombination was confirmed by PCR with primers to detect a band at 619 bp.
Patient Samples
Diagnostic bone marrow flow cytometric data was accessed from patients seen at Memorial Sloan Kettering Cancer Center with STAG2-mutant myelodysplastic syndrome or non-STAG2 mutant patients with refractory cytopenias. Patient characteristics are depicted in Table S1. This study was approved by Institutional Review Board at the MSKCC (protocol #16–1591).
Method Details
In vivo experiments
Mx1-cre+ Stag2fl/fl conditional KO and Cre− Stag2fl/fl control WT mice or Mx1-cre+ Stag1fl/fl conditional KO and Cre− Stag1fl/fl control WT mice received four intraperitoneal injections of polyinositric:polycytidinic acid (PIpC) every other day at a dose of 20 mg/kg of body weight starting at 6 weeks after birth. Deletion/recombination was confirmed 2 weeks after PIPC injection and mice were used for experiments at least 8 weeks after PIPC injection. All mice were analyzed between 8 and 60 weeks of age. BM, spleen, and peripheral blood were analyzed by flow cytometry. Formalin-fixed paraffin-embedded tissue sections were stained with hematoxylin and eosin (H&E). Peripheral blood was smeared on a slide and stained using the Wright-Giemsa staining method. Tissue sections and blood smears were evaluated by a hematopathologist (B. Durham). Deletion of the Stag1 and Stag2 allele and transcript was measured by genomic PCR, qRT-PCR, and Western blot analysis (described below). All experiments were repeated and results confirmed in male Mx1-cre+ Stag2fl/y conditional KO and Cre− Stag2fl/y control WT mice.
BM transplantation
Freshly dissected femurs and tibias were isolated from Stag1fl/fl and Stag2fl/fl CD45.2+ or Mx1-cre+ Stag1fl/fl and Stag2fl/fl CD45.2+ mice. Bones were transected at the epiphyses and were centrifuged at 4°C to extract whole bone marrow int o PBS and RBCs were lysed in ammonium chloride-potassium bicarbonate lysis buffer for 10 min. After centrifugation, cells were resuspended in PBS plus 3% FBS, passed through a cell strainer, and counted. Finally, 0.5 × 106 total BM cells from Stag1fl/fl and Stag2fl/fl CD45.2+ or Mx1-cre+ Stag1fl/fl and Stag2fl/fl CD45.2+ mice were mixed with 0.5 × 106 WT CD45.1+ support BM and transplanted via tail vein injection into lethally irradiated (two times 450 cGy) CD45.1+host mice. Recipient mice were B6.SJL-Ptprca/BoyAiTac female mice between 6–12 weeks of age. Mice were purchased from Taconic. Chimerism was measured by FACS in peripheral blood at 4 weeks after transplant (week 0, pre-PIpC). Chimerism was followed via FACS in the peripheral blood every 4 weeks (week 0, 4, 6, 8, 12, and 16 after PIpC injection). Additionally, for each bleeding, whole blood cell counts were measured on a blood analyzer, and peripheral blood smears were scored. Chimerism in the BM and spleen was evaluated at 16 weeks via animal sacrifice and subsequent FACS analysis.
In vitro colony-forming assays
BM of Stag1fl/fl and Stag2fl/fl and littermate Mx1-cre Stag1fl/fl and Stag2fl/fl mice were extracted and seeded at a density of 20,000 cells/replicate into cytokine-supplemented methylcellulose medium (Methocult M3434; STEMCELL Technologies) or methylcellulose medium for mouse pre-B cells (M3630) supplemented with FLT3L (20 ng/mL), SCF (100 ng/mL), and IL7 (10 ng/mL)(de Boer et al., 2011). Colonies propagated in culture were scored at day 7. Representative colonies were isolated from the plate for cytospins and flow cytometry. Remaining cells were resuspended and counted, and a portion was taken for replating (20,000 cells/replicate) for a total of eight platings. Cytospins were performed by resuspending in warm PBS and spun onto the slides at 350 g for 5 min. Slides were air-dried and stained using the Giemsa-Wright method.
Antibodies, FACS, and Western blot analysis
Antibody staining and FACS analysis was performed as previously described (Abdel-Wahab et al., 2013). BM or spleen mononuclear cells were stained with a lineage cocktail comprised of antibodies targeting CD4, CD8, B220, NK1.1, Gr-1, CD11b, and Ter119. Cells were also stained with antibodies against c-Kit, Sca-1, IL-7Rα, FcγRII/III, and CD34. Cell populations were analyzed using a Fortessa Flow Cytometer (BD) and sorted with a FACS-SH800 instrument (Sony). All FACS antibodies were purchased from BD or eBioscience. We used the following antibodies: c-Kit (2B8), Sca-1 (D7), Mac-1/CD11b (M1/70), Gr-1 (RB6–8C5), NK1.1 (PK136), Ter-119, IL7-Rα (A7R34), IgD (11–26c.2a), IgM (RMM-1), CD34 (RAM34), FcγRII/III (2.4G2), CD4 (RM4–5), CD8 (53–6.7), CD19 (HIB19), CD43 (1B11), CD44 (IM7), CD45.1 (A20), CD45.2 (104), CD45R/B220 (RA3–6B2), CD71 (R17217), CD105 (MJ7/18), CD150 (9D1), and CD48 (HM48–1). The following antibodies were used for Chromatin Immunoprecipitation and Western blot analysis: Stag2 (Bethyl, A302–580A), Stag1 (Bethyl, A302–579A), Smc3(Bethyl, A300–060A), Smc1a (Active Motif, 61067), Ctcf (Cell Signaling, 3418S), PU.1 (Cell Signaling, #2266S), and Actin (CalBiochem, JLA-20).
Multiparameter flow cytometry was performed on bone marrow aspirates at diagnosis for patients with STAG2 mutant MDS and matched controls patients with nonmalignant cytopenias. Briefly, up to 1.5 million cells from freshly drawn bone marrow aspirate were stained with a 10-”color” panel (CD15-FITC, CD33-PE, CD117-PC5, CD13-PE-Cy7, CD34-APC, CD71-APC-A700, CD38-APC-A750, HLA-DR-PAC Blue, CD45-V500C, and CD19-BV605), washed, and acquired on a Canto-10 cytometer (BD Biosciences, San Jose, CA). The results were analyzed with custom Woodlist software (generous gift of Wood BL, University of Washington). In order to enumerate the B cells, CD19+CD15-CD33-CD13- cells were gated. Plasma cells were excluded based on the bright CD38 expression. CD34+/CD19+ cells were considered as immature B cells.
Histological analyses
Mice were sacrificed and autopsied, and dissected tissue samples were fixed for 24 h in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 4 μm and stained with H&E. Images were acquired using an Axio Observer A1 microscope (Carl Zeiss).
Peripheral blood analysis
Blood was collected by submandibular bleeding using a 5mm lancet (MEDipoint Inc). Automated peripheral blood counts were obtained using a ProCyte Dx (IDEXX Laboratories) according to standard manufacturer’s instruction. Differential blood counts were realized on blood smears stained using Wright-Giemsa staining and visualized using an Axio Observer A1 microscope.
Cytogenetic Analysis and Metaphase Karyotyping
Bone marrow acquired at necropsy was resuspended in cytokine enriched media containing 5mL RPMI 10% FCS supplemented with 2mM L-glutamine and after documented excision or 8 weeks after PIpC. Harvested cells were cultured in T25 tissue culture flasks with 25 uL of Colcemid (10mg/mL) (Gibco Life Technologies, Inc) for 45 minutes and 4 hours respectively, resuspended in 0.075 mol/L KCl for 10 minutes at 37°C and fixed in methanol-acetic acid (3:1). Metaphases were scored and counted. Chromosome analysis was performed on a minimum of 20 DAPI-banded metaphases and all metaphases were fully karyotyped. Low depth whole genome sequencing was performed to assess genome wide copy number. One microgram of genomic DNA was sheared using an E220 Sonicator (Covaris) to +/− 300bps. Fragmented DNA was end-repaired, A-tailed, and ligated to Illumina TruSeq dual indexed adaptors using standard methods. Indexed libraries were enriched by PCR amplified, quantified, pooled and sequenced in multiplex fashion on an Illumina HiSeq instrument to obtain roughly 1 million sequencing reads per sample, sufficient to call copy number variation at a bin resolution of 150kb) (Baslan et al., 2015). Data was processed for copy number analysis as described previously (Baslan et al., 2012).
PU.1/EBF1 overexpression and knockdown
Lentiviral constructs expressing PU.1-IRES-GFP, PU.1 shRNA-IRES-GFP, and GFP alone were generously provided by the laboratory of Dr. Ulrich Steidl (Steidl et al., 2006; Will et al., 2015). Whole BM was negatively selected for lineage markers using antibodies conjugated to magnetic beads and separated using EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit (STEMCELL Technologies). Lineage depleted BM cells were transduced at a cell density of 5 ×105 using virus concentrated through ultracentrifugation. After 48h cells were sorted for GFP expression. Overexpression/knockdown was confirmed using qRT-PCR. Retroviral constructs expressing EBF1-IRES-GFP and GFP alone were generously provided by the laboratory of Dr. Charles Mulligan. Lineage depleted BM cells were transduced at a cell density of 5 ×105 using virus concentrated using Retro-X concentrator (Clontech). After 48h cells were sorted for GFP expression and either used for in vitro assays or resuspended in PBS and tail-vein injected into lethally irradiated recipient mice. Blood and BM of transplanted mice were analyzed by flow cytometry as above. Overexpression was confirmed using qRT-PCR.
RNA-Seq and quantitative real-time PCR analysis
For qRT-PCR experiments, all samples were prepared in biologic triplicate. Whole BM was negatively selected for lineage markers using antibodies conjugated to magnetic beads and separated using EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit (STEMCELL Technologies). Total RNA was isolated using the Trizol (Invitrogen), and cDNA was synthesized using the Verso cDNA Synthesis kit (Fisher). Quantitative PCR was performed using Taqman reagents and probes (Thermo Fisher) for ActinB (Mm02619580_g1), Spi1, (Mm00488140_m1) and Ebf1 (Mm00432948_m1) and FastStart Universal SYBR Green (Sigma) and primers for Stag2 (F: 5’-TGCTATGCAGTCGGTGGTAG-3’) and (R: 5’-AGGACCAGCCATGGTAAGTG-3’), Stag1 (F: 5’-CTACAAGCATGACCGGGACAT-3’) and (R: 5’-AGGGTACTTGTATGCCTAAAAGC-3’), and ActinB (F: 5’-GGCTGTATTCCCCTCCATCG-3’) and (R: 5’-CCAGTTGGTAACAATGCCATGT-3’). For mRNA-Seq analysis, samples were prepared and analyzed in biologic triplicate. For RNA-sequencing, RNA was isolated by TriZOL extraction from sorted-cell population or lineage negative bone marrow as indicated. RNA-sequencing libraries were generated by 3’ sequencing and SMART-Seq2 amplification and sequenced on an Illumina NextSeq 500.
Single cell RNA-sequencing and data analysis
Lineage−bone marrow cells from 3 Stag2 WT and 3 Stag2 KO mice were sorted for viability (4′,6-diamidino-2-phenylindole (DAPI)-negative). Individual samples were loaded on 10X Genomics Chromium System aiming to generate 7,000 Gel Beads in Emulsion (GEMs) per sample. scRNA-seq libraries were prepared following 10X Genomics protocols (Chromium Single Cell 3’ Reagent Kits User Guide v2 Chemistry). Libraries were sequenced on NovaSeq 6000 (Illumina) system (S2 flow cell, paired-end) recovering a median of 239,350,330 reads/sample.
ATAC-sequencing
Chromatin accessibility assays utilizing the bacterial Tn5 transposase were performed as described (Corces et al., 2016) with minor modifications. Cells (1.0 × 104) were lysed and incubated with transposition reaction mix containing PBS, 1% Digitonin, Tween-20, and Transposase (llumina). Samples were incubated for 30 minutes at 37°C in a thermomixer at 1000rpm. Prior to amplification, samples were concentrated with the DNA Clean and Concentrator Kit-5 (Zymo). Samples were eluted in 20uL of elution buffer and PCR-amplified using the NEBNext 2X Master Mix (NEB) for 10 cycles and sequenced on a NextSeq 500 (Illumina).
Chromatin Immunoprecipitation-sequencing
Cells (~0.5–1 million cells were used for one ChIP reaction) were cross-linked with 1% formaldehyde (Sigma, F1635) at 37°C for 12 min, inv erting occasionally, and quenched with 1.25 M glycine and Tris-HCl and placed on ice. Cells pellets were washed with cold PBS and transferred to Eppendorf tubes. Cell pellets were resuspended in 1ml SDS lysis buffer (1% SDS, 10mM EDTA, 50mM Tris-HCl pH8) containing proteinase inhibitor and centrifuged at maximum speed for 10 minutes at room temperature. After aspirating the supernatant, the pellet was resuspended in 130uL ChIP buffer supplemented with protease inhibitor and transferred to a AFA microtube (Covaris). Sonication was performed on a E220 Sonicator (Covaris) for 360 seconds using manufacturer settings (Peak Power 105, 200 Cycles/burst, Duty Factor 3). ChIP inputs were decrosslinked by adding 5M NaCl and 0.1X TE and incubated overnight at 65C followed by bead purification with AMPure XP beads (Beckman Coulter). Sonicated chromatin was incubated with primary antibody 1:50 total volume of chromatin and incubated overnight, rotating at 4C. Dyna beads prewashed in ChIP buffer were added at 5X volume of antibody and incubated for 3 hours rotating at 4C and then washed with salt buffers mixed micelle, B500, LiCl, and TBS. The ChIP product was then eluted from the Dynabeads in 52uL elution buffer and incubating for 15 minutes. ChIP product was decrosslinked with 2uL 5M NaCl and incubated overnight at 65C. Libraries were prepared using the NEBNext® ChIP-seq Library Prep Master Mix Set for Illumina ® (NEB, E6240L) and Swift Accel-NGS® 2S Plus and Swift Accel-NGS® 2S MID Indexed adapters. Samples were QC’d using a Bioanalyzer TapeStation (Agilent Technologies 2200) to determine fragment size. Samples were pooled and submitted for SE50 sequencing using an Illumina NextSeq500.
Hi-C
Hi-C was performed as previously described (Belaghzal et al., 2017). Briefly, Lin− bone marrow cells (5×106) were cross-linked in 1% formaldehyde for 10 minutes and quenched in 125mM glycine. Cross-linked cells were lysed in Hi-C lysis buffer (10mM Tris-HCl pH8.0, 10mM NaCl, 0.2% Igepal CA-630 and Halt protease inhibitors Thermo Fisher 78429). After disruption, chromatin was solubilized and digested using 400Units of DpnII at 37°C overnight. DNA overhangs were then filled in with biotin-14-dATPs and ligated with T4 ligases at 16°C for 4 hours. Cross-links of ligated DNAs were reversed with proteinase K (Life Technologies, 25530–031) at 65°C overnight and purified using phenol:ch olorform. After removal of biotin from unligated ends, DNA was fragmented to 150–350 bps using an E220 sonicator (Covaris). After end repair, biotinylated DNA was collected using streptavidin beads (MyOne C1 beads, Life Technologies, 650.01) to prepare Hi-C libraries using the Illumina TruSeq Nano DNA kit. Hi-C libraries were sequenced on an Illumina NextSeq500 and raw sequencing data in the Fastq format were obtained.
Experimental Design
No specific methods were used for randomization and investigators were not blinded to the identity of samples. No statistical methods were utilized to determine sample size. The experiments described in this study were designed to use the minimum number of animals required. Each experiment was performed using separate cohorts for males and females, both in triplicate, to ensure reproducibility.
Quantification and Statistical Analysis
In vivo experiments
The number of animals, cells and experimental replication can be found in the respective figure legend. The unpaired Student’s t test was used to compare the mean of two groups. Data were analyzed and plotted using GraphPad Prism 6 software. Data shown in graphs indicate mean ± S.D. Kaplan-Meier survival analysis and log-rank test were used to compare survival outcomes.
RNA-Seq data analysis
Fastq files were mapped to the mouse genome (mm9) and reads counts per gene were quantified using STAR(Dobin et al., 2013) with default parameters and genecode (vM1) annotation file. Differentially expressed genes (DEGs) were identified with DESeq2 (Love et al., 2014), with a fold change cutoff of +/− 2 and a FDR of 10%. Motif enrichment on DEGs was performed with HOMER using a window of −1kb to +100bp around the transcription start site. Gene ontology analysis was also performed using HOMER.
Single cell RNA-sequencing and data analysis
FASTQ files were processed using the Sequence Quality Control (SEQC) pipeline (Azizi et al., 2018) and reads were aligned to the mouse genome mm38, resulting in a median of 4226 cells/samples with a median of 5036 molecules/cell. Cells from the lower molecule counts, determined by lower mode of molecule counts distribution (5.8% of cells), were additionally filtered out to remove putative empty droplets, resulting in a final collection of 24,153 cells. The resulting count matrices from all samples were then combined and normalized to median library-size and log transformed shown in Figures 1G, 2C–D and S4C. For individual subsets of granulocytes, MEP, HSCs, cells were normalized within the subset. Ribosomal genes were excluded in downstream analyses. In the global cohort, t-SNE was performed on 20 principal components, explaining ~83.6% of data variance, with a perplexity of 150. For subpopulations analyses, the number of PCs considered was determined based on the knee point of explained variance (granulocytes: 8, MEP: 14, HSC: 7) and perplexity was fixed at 150.
In order to annotate principal components (Figure 1G, 2H, 2I), the Pearson correlation between each principal component and expression of gene signatures was computed. Gene lists were sorted by correlation and ranked Gene Set Enrichment Analysis (GSEA) was performed. In addition, expression of most correlated and anticorrelated was assessed in bulk RNA-sequencing data of sorted populations to identity components defined by lineage maturation.
To annotate subsets of cells, we performed clustering of normalized and log transformed data using Phenograph (Levine et al., 2015). Then, Pearson correlation between the centroid of each cluster and standardized bulk RNA-seq data on sorted subsets (from Haemopedia-Mouse RNAseq,(de Graaf et al., 2016)) was computed (Figure S3D). The subset with highest correlation to each cluster was used to annotate the cluster, as shown in Figures 2D, S3D. The cluster annotations were confirmed through studying differentially expressed genes in each cluster. Differential expression analysis was performed using the Wasserstein distance (Santambrogio and SpringerLink (2015)) between normalized and log-transformed expression in cells from each inferred lineage and all other cells (Figure S3E).
Epigenomic data analysis
Reads were trimmed for both quality and Illumina adapter sequences using ‘trim_galore’ then aligned to mouse assembly mm9 with bowtie2 using the default parameters. Aligned reads with the same start site and orientation were removed using the Picard tool MarkDuplicates (http://broadinstitute.github.io/picard). ChIP density profiles were created by extending each read to the average library fragment size and then computing density using the BEDTools suite (http://bedtools.readthedocs.io). Reads were not extended when generating ATAC-seq read density. Enriched regions were discovered using MACS2 and scored against matched input libraries (fold change > 2 and p-value < 0.005). Peaks were then filtered against genomic ‘blacklisted’ regions (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm9-mouse/mm9-blacklist.bed.gz) and filtered peaks within 500 bp were merged to create a full peak atlas. Raw read counts were tabulated over this peak atlas using featureCounts (http://subread.sourceforge.net). All genome browser tracks and read density tables were normalized to a sequencing depth of ten million mapped reads. Peaks were annotated using linear genomic distance, with a gene assigned to a peak if it was within 50 kb upstream or downstream of the gene start or end, respectively. Motif signatures were obtained using the ‘de novo’ approach in Homer v4.5 (http://homer.ucsd.edu). Rescued peaks were defined as Stag2 regions in which Stag1 was enriched above input in the Stag2 KO, whereas non-rescued peaks showed no evidence of Stag1 binding in the Stag2 KO background.
Hi-C
Hi-C data was processed based on the previous method(Belton et al., 2012). Fastq files were mapped binned using c-World pipeline from the Dekker lab, which is available at a GitHub repository(Lajoie et al., 2015), https://github.com/dekkerlab/cMapping; https://github.com/dekkerlab/balance; https://github.com/dekkerlab/cworld-dekker). Briefly, 50bp paired end reads were truncated to 25bp starting at the 5-prime end and then were iteratively mapped onto mm9. Uniquely mapped, paired end reads were collected and assigned to DpnII restriction fragments based on their 5-prime locations. Mapped reads with same fragment ends and uniqueness were kept, and PCR duplicates were removed. Interaction heat-maps, insulation scores and loop pile-up were generated using scripts included in c-world pipeline. The loop coordinates used for loop pileup analysis were obtained from the mouse CH12.LX line(Rao et al., 2014). To increase resolution of Hi-C interaction profiles, intermediate validpairs files from the same genotype mice were pooled together using cooltools from the Mirny Lab (https://github.com/mirnylab). The insulation score change (ISC) between Stag2−/y and Stag2fl/y was calculated using 10kb binned Hi-C data and the ISC bins containing Stag2 rescued or non-rescued peaks were identified with Bedtools and annotated using ChIPseeker (Yu et al., 2015).
Data and Code Availabilities
The ATACseq, RNAseq, scRNAseq, ChIPseq, and Hi-C datasets generated during this study are available and deposited on Gene Expression Common under accession number GEO:134583
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-mouse CD11b | BioLegend | Cat#101205; RRID: AB_312788 |
| PE anti-mouse CD11b | BioLegend | Cat#101207; RRID: AB_312790 |
| APC/Cy7 anti-mouse CD11b | BioLegend | Cat#101225; RRID: AB_830641 |
| PE anti-mouse Gr-1 | BioLegend | Cat#108407; RRID: AB_313372 |
| APC/Cy7 anti-mouse Gr-1 | BioLegend | Cat#108423; RRID: AB_2137486 |
| PE anti-mouse CD71 | BioLegend | Cat#113807; RRID: AB_313568 |
| APC anti-mouse Ter119 | BioLegend | Cat#116211; RRID: AB_313712 |
| APC/Cy7 anti-mouse Ter119 | BioLegend | Cat#116223; RRID: AB_2137788 |
| APC/Cy7 anti-mouse NK-1.1 | BioLegend | Cat#108723; RRID: AB_830870 |
| APC/Cy7 anti-mouse B220 | BioLegend | Cat#103223; RRID: AB_313006 |
| APC/Cy7 anti-mouse CD3 | BioLegend | Cat#100221; RRID: AB_2057374 |
| APC/Cy7 anti-mouse CD19 | BioLegend | Cat#115529; RRID: AB_830706 |
| APC/Cy7 anti-mouse CD4 | BioLegend | Cat#100413; RRID: AB_312698 |
| PE anti-mouse CD117/cKit | BioLegend | Cat#105807; RRID: AB_313216 |
| APC anti-mouse CD117/cKit | BioLegend | Cat#105811; RRID: AB_313220 |
| PE/Cy7 anti-mouse Sca-1 | BioLegend | Cat#108113; RRID: AB_493597 |
| APC anti-mouse Sca-1 | BioLegend | Cat#108111; RRID: AB_313348 |
| PE anti-mouse CD150 | eBioscience | Cat#12-1502-82; RRID: AB_1548765 |
| PerCP/Cy5.5 anti-mouse CD48 | BioLegend | Cat#103421; RRID: AB_1575045 |
| eFluor 450 anti-mouse CD16/32 | eBioscience | Cat#48-0161-82; RRID: AB_1272191 |
| eFluor 450 anti-mouse CD45.1 | eBioscience | Cat#48-0453-82; RRID: AB_1272189 |
| PE/Cy7 anti-mouse CD45.2 | BioLegend | Cat#109829; RRID: AB_1186103 |
| APC/Cy7 anti-mouse CD45.2 | BioLegend | Cat#109823; RRID: AB_830788 |
| PE anti-mouse CD34 | BD Biosciences | Cat#551387 RRID: AB_394176 |
| AlexaFluor 700 anti-mouse CD45.2 | Biolegend | Cat#109822 RRID: AB_493731 |
| PerCPCy5.5 anti-mouse CD16/32 | Biolegend | Cat#101323 RRID: AB_1877268 |
| PE Cy7 anti-mouse CD71 | Biolegend | Cat# 113811 RRID:AB_2203383 |
| APC anti-mouse CD44 | Biolegend | Cat# 103011 RRID:AB_312962 |
| PE anti-mouse CD41 | Biolegend | Cat#133906 RRID:AB_2129745 |
| FITC anti-mouse CD45.1 | Biolegend | Cat#110706 RRID:AB_313495 |
| cKIT (CD117) anti-mouse BV785 | Biolegend | Cat#:105841 RRID:AB_2629799 |
| PE anti-mouse IgD | Biolegend | Cat#: 405705 RRID:AB_315027 |
| AlexaFluor700 anti-mouse CD19 | Biolegend | Cat#: 302226 RRID:AB_493751 |
| APC Cy7 anti-mouse CD43 | Biolegend | Cat# 121220 RRID:AB_2194192 |
| PerCP Cy5.5 anti-mouse CD45.2 | Biolegend | Cat# 109828 RRID:AB_893350 |
| PE Cy7 Anti-mouse CD45R/B220 antibody | BioLegend | Cat#103222 RRID: AB_313005 |
| APC Anti-mouse Cd11b | BioLegend | Cat# 101212 RRID: AB_312795 |
| PerCP Cy5.5 Anti-mouse CD3 | BioLegend | Cat# 100218 RRID: AB_1595492 |
| Brilliant Violet 421™ anti-mouse CD45.1 Antibody | BioLegend | Cat#110732 RRID: AB_2562563 |
| Alexa 700 anti-mouse CD45.2 | BioLegend | Cat# 109822 RRID: AB_493731 |
| FITC anti-mouse CD45.1 | BioLegend | Cat#110706 RRID: AB_313495 |
| Alexa 700 anti-mouse CD45.2 | BioLegend | Cat# 109822 RRID: AB_493731 |
| APC Anti-mouse Cd11b | BioLegend | Cat# 101212 RRID: AB_312795 |
| APC/Cy7 Anti-mouse CD117/cKIT | BioLegend | Cat# 105826 RRID: AB_1626278 |
| Rabbit anti-SA2 | Bethyl | Cat#A302-580A RRID: AB_2034860 |
| Rabbit anti-SA1 | Bethyl | Cat#A302-579A RRID: AB_2034857 |
| Rabbit anti-SMC1a | Active Motif | Cat#61067 RRID: AB_2688006 |
| Rabbit anti-PU.1 | Cell Signaling | Cat#2266S RRID: AB_10692379 |
| Rabbit anti-CTCF | Cell Signaling | Cat#3418S RRID: AB_2086791 |
| Mouse anti-Actin | CalBiochem | Cat#JLA-20 RRID:AB_528068 |
| Peroxidase Goat anti-Mouse IgM | CalBiochem | Cat#JA-1200 |
| Goat anti-Rabbit IgG, HRP conjugate | Millipore | Cat#12-348 RRID:AB_390191 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Patient samples | Memorial Sloan Kettering Cancer Center | Table S1 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Polyinosinic:polycytidylic acid | Amersham | Cat#27-4732-01 |
| Bovine serum albumin | Fisher | Cat#BP1600-1 |
| Phosphate Buffered Saline | MSKCC Media Core | N/A |
| RPMI medium | MSKCC Media Core | N/A |
| IMDM medium | MSKCC Media Core | N/A |
| Fetal bovine serum | MSKCC Media Core | N/A |
| Fetal calf serum | MSKCC Media Core | N/A |
| Penicillin-Streptomycin | Fisher | Cat#15140122 |
| L-Glutamine | Thermofisher | Cat#21051040 |
| Sodium Pyruvate | Thermofisher | Cat# 11360070 |
| Colcemid | Gibco | Cat#15212012 |
| MethoCult GF M3434 | StemCell Technologies | Cat#03434 |
| MethoCult M3630 | StemCell Technologies | Cat#03630 |
| Recombinant mouse GM-CSF | Peprotech | Cat#315-03 |
| Recombinant mouse IL-3 | Peprotech | Cat#213-13 |
| Recombinant mouse SCF | Peprotech | Cat#250-03 |
| Recombinant mouse IL-7 | Biolegend | Cat# 577802 |
| Recombinant mouse FLT3-L | Peprotech | Cat# 250-31 |
| Dimethyl sulfoxide | Fisher | Cat#D128-500 |
| 16% Paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat#15710 |
| DAPI | Sigma | Cat#D9542 |
| RNase-free DNase set (50) | Qiagen | Cat#79254 |
| TaqMan Master Mix (2X) | Thermofisher | Cat#4304437 |
| Polybrene | American Bio | Cat#AB01643 |
| HEPES buffer | Fisher | Cat#BP299 |
| Retro-X concentrator | Takara | Cat# 631455 |
| 5M NaCl | Thermofisher | Cat#AM9760G |
| Tris-EDTA | Fisher | Cat#BP2473-500 |
| 0.5M EDTA, pH 8.0 | Life Technologies | Cat#15575-020 |
| SDS Solution, 20% Sodium Dodecyl Sulfate Solution | Fisher | Cat#BP1311-200 |
| Sucrose | Sigma | Cat#84097-250G |
| 1M MgCl2 | Ambion(Thermo) | Cat#AM9530G |
| Tween 20 | Sigma Aldrich | Cat#P7949-500mL |
| N,N-Dimethylformamide | Sigma Aldrich | Cat#D4551-250mL |
| Transposase,TAGMENT DNA Enzyme | Illumina | Cat# 15027865 |
| Dynabeads Protein A | Invitrogen | Cat#10002D |
| Dynabeads Protein G | Invitrogen | Cat#10004D |
| Lithium Chloride | Acros Organics | Cat#413271000 |
| TBS | Licor | Cat#927-50000 |
| D1000 HS Buffer | Agilent | Cat#5190-6504 |
| NP40 | Fisher | Cat#FNN021 |
| Deoxycholic Acid | Fisher | Cat#BP349-100 |
| NaHCO3 | Fisher | Cat#BP329-500 |
| NaN3 | USB | Cat#21610 |
| Triton X-100 | Sigma | Cat#0992-93-1 |
| Igepal CA-630 | Sigma | Cat#I8896 |
| Halt protease inhibitors | Thermofisher | Cat#78429 |
| Streptavidin beads (MyOne C1 beads) | Life Technologies | Cat#650.01 |
| DpnII | NEB | Cat# R0543L |
| 10X NEB 3.1 Buffer | NEB | Cat#B7203 |
| Halt Protease Inhibitor Cocktail (100X) | Thermo | Cat#78429 |
| Adenosine 5’-triphosphate magnesium salt from bacterial source (ATP) - 1G | Sigma | Cat#A9187-1G |
| Biotin-14-dATP | Invitrogen | Cat#19524-016 |
| Biotin-14-dCTP | Invitrogen | Cat#19518-018 |
| Proteinase K (Fungal) | Invitrogen | Cat#25530-031 |
| DNA polymerase I, large (Klenow) fragment | NEB | Cat#M0210S |
| T4 DNA ligase 1U/ μl | Invitrogen | Cat#15224090 |
| T4 DNA polymerase | NEB | Cat#M0203L |
| 5X ligation buffer | Invitrogen | Cat#46300-018 |
| T4 polynucleotide kinase | NEB | Cat#M0201 |
| 5X T4 DNA ligase buffer | Invitrogen | Cat#P/N y90001 |
| Klenow fragment (3’ → 5’ exo−) | NEB | Cat#M0212L |
| Agencourt AMPure XP | Beckman Coulter | Cat#A63881 |
| Critical Commercial Assays | ||
| Verso cDNA Synthesis Kit | Fisher | Cat#AB1453A |
| EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit | STEMCELL Technologies | Cat# 19856 |
| TruSeq Nano DNA kit | Illumina | Cat#20015965 |
| Accel-NGS 2S Plus DNA Library Kit | Swift | Cat#21096 |
| Accel NGS 2S MID Indexing Kit | Swift | Cat#27148 |
| NEBNext® ChIP-Seq Library Prep Master Mix Set for Illumina | NEB | Cat# E6240L |
| DNA Clean and Concentrator - 5 | Zymo | Cat# D4013 |
| Deposited Data | ||
| Gene Expression Omnibus (GEO:GSE134583) | N/A | RRID:SCR_005012 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Stag2fl/fl | The Jackson Laboratory | Cat#030902: RRID: IMSR_JAX:030902 |
| Mouse: Stag1fl/fl | The Jackson Laboratory | Cat#030904: RRID: IMSR_JAX:030904 |
| Mouse: Mx1-Cre | The Jackson Laboratory | Cat#003556: RRID: IMSR_JAX:003556 |
| C57BL/6J | The Jackson Laboratory | Cat#000664: RRID: IMSR_JAX:000664 |
| B6.SJL-Ptprca/BoyAiTac | Taconic | Cat#4007: RRID: IMSR_TAC:4007 |
| Oligonucleotides | ||
| Stag2 NF 5′- CACTCATGCTGGCAAGTATTGTAC-3′ | Invitrogen | N/A |
| Stag2-NR 5′-AACAGCCTGAGCAAAGAATCCAAAG-3′ | Invitrogen | N/A |
| Stag2-3’ 5′-TGTGTGCCTCTTTGAACAATGCCC-3′ | Invitrogen | N/A |
| Stag1-CommonF 5′-GACTGGTATCTGACGGCTTATACC-3′ | Invitrogen | N/A |
| Stag1-CommonR 5′- CACTGAGGACCAGGCATTGTAAGG-3′ | Invitrogen | N/A |
| Stag1-FloxR 5′- TGAACTGATGGCGAGCTCAGACC-3′ | Invitrogen | N/A |
| Mouse Stag2 RT-PCR For 5’-TGCTATGCAGTCGGTGGTAG-3’ | Invitrogen | N/A |
| Mouse Stag2 RT-PCR Rev 5’-AGGACCAGCCATGGTAAGTG-3’ | Invitrogen | N/A |
| Mouse Stag1 RT-PCR For 5’-GAGTCCTGATTCGACAGTGTCA-3’ | Invitrogen | N/A |
| Mouse Stag1 RT-PCR Rev 5’-AGGGTACTTGTATGCCTAAAAGC-3’ | Invitrogen | N/A |
| Mouse Actin RT-PCR For 5’- GGCTGTATTCCCCTCCATCG-3’ | Invitrogen | N/A |
| Mouse Actin RT-PCR Rev 5’- CCAGTTGGTAACAATGCCATGT-3’ | Invitrogen | N/A |
| Taqman Spi1 (PU.1) Probe | Thermofisher | Mm00488140_m1 |
| Taqman Ebf1 Probe | Thermofisher | Mm00432948_m1 |
| Taqman Actin Probe | Thermofisher | Mm02619580_g1 |
| Recombinant DNA | ||
| pMSCV-IRES-GFP | Mulligan Lab | N/A |
| pMSCV-IRES-GFP-Ebf1 | Mulligan Lab | N/A |
| pCAD-IRES-GFP | Will et al., 2015 | N/A |
| pCAD-IRES-GFP-PU1 | Will et al., 2015 | N/A |
| pRRLsin-PGK-eGFP-WPRE | Will et al., 2015 | N/A |
| pRRLsin-PGK-eGFP-WPRE-PU1 | Will et al., 2015 | N/A |
| EcoPack plasmids | Will et al., 2015 | N/A |
| pRRLsin-PGK-eGFP-WPRE | Will et al., 2015 | N/A |
| psPAX2 | Will et al., 2015 | N/A |
| pMD2.G | Will et al., 2015 | N/A |
| Software and Algorithms | ||
| Prism 6 | GraphPad | https://www.graphpad.com |
| FlowJo V9.9.6 | Tree Star | https://www.flowjo.com/ |
| Other | ||
Highlights.
Hematopoietic Stag2 loss enhances stem cell self-renewal and impairs differentiation
Stag1 can maintain TAD boundary integrity in the absence of Stag2
Stag2 is required for intra-TAD interactions at lineage genes (e.g. PU.1 targets)
Stag2 target expression but not PU.1 overexpression restores B cell differentiation
Acknowledgments
We acknowledge the use of the Integrated Genomics Operation Core, funded by the MSKCC Support Grant NIH P30 CA008748, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. We would like to thank Dr. Gouri Nandakumar and the staff of the Cytogenetics Core Facility for their assistance in imaging and processing of 5 cytogenetic data.
Funding: This work was supported by National Cancer Institute awards R35 CA197594–01A1 (RLL), R01 CA216421 (RLL), and PS-OC U54 CA143869–05 (RLL and JD) and a grant from Leukemia & Lymphoma Society Translational Research Foundation 6499–17 (RLL). ADV is supported by a National Cancer Institute career development grant K08 CA215317, the William Raveis Charitable Fund Fellowship of the Damon Runyon Cancer Research Foundation (DRG 117–15), and an EvansMDS Young Investigator grant from the Edward P. Evans Foundation. RLB is supported by the Sohn Foundation Fellowship of the Damon Runyon Cancer Research Foundation (DRG 22–17). TB is supported by the William C. and Joyce C. O’Neil Charitable Trust, MSKCC Single Cell Sequencing Initiative. J.D. is an investigator of the Howard Hughes Medical Institute.
Declaration of interests: ADV received travel support from Mission Bio and is on the Editorial Advisory Board of Hematology News. JD is on the Scientific Advisory Board of Arima Genomics, and a consultant for Omega Therapeutics. RLL is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics and Isoplexis. He receives research support from and consulted for Celgene and Roche and has consulted for Janssen, Astellas, Morphosys and Novartis. He has received honoraria from Roche, Lilly and Amgen for invited lectures and from Gilead for grant reviews. RLB, YL, WX, BHD, SEE, AN, ISC, MW, EA, TB, CJO, and RK have nothing to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, Kuscu C, Hricik T, Ndiaye-Lobry D, Lafave LM, et al. (2013). Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. The Journal of experimental medicine 210, 2641–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, Sasaki Y, and Jacobsen SE (2001). Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 15, 659–669. [DOI] [PubMed] [Google Scholar]
- Antony-Debre I, Paul A, Leite J, Mitchell K, Kim HM, Carvajal LA, Todorova TI, Huang K, Kumar A, Farahat AA, et al. (2017). Pharmacological inhibition of the transcription factor PU.1 in leukemia. J Clin Invest 127, 4297–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, et al. (2018). Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308 e1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey ML, O’Neil NJ, van Pel DM, Solomon DA, Waldman T, and Hieter P (2014). Glioblastoma cells containing mutations in the cohesin component STAG2 are sensitive to PARP inhibition. Mol Cancer Ther 13, 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Marquez M, Vazquez M, Lapi E, Castro-Giner F, Beltran S, Bayes M, et al. (2013). Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet 45, 1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, Troge J, Ravi K, Esposito D, Lakshmi B, et al. (2012). Genome-wide copy number analysis of single cells. Nat Protoc 7, 1024–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslan T., Kendall J., Ward B., Cox H., Leotta A., Rodgers L., Riggs M., D’Italia S., Sun G., Yong M., et al. (2015). Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res 25, 714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaghzal H, Dekker J, and Gibcus JH (2017). Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, and Dekker J (2012). Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti L, Cereda M, Monteverde L, Desai N, and Ciccarelli FD (2017). Synthetic lethal interaction between the tumour suppressor STAG2 and its paralog STAG1. Oncotarget 8, 37619–37632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N, Buettner F, Sommerkamp P, Klimmeck D, Ladel L, Thalheimer FB, Pastor-Flores D, Roma LP, Renders S, Zeisberger P, et al. (2017). Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell 169, 807–823 e819. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N., Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ Jr., Laird, PW, et al. (2013). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368, 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Liu J, Heck S, Chasis JA, An X, and Mohandas N (2009). Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci U S A 106, 17413–17418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu BJ, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Gkeenleaf WJ, et al. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nature Genetics 48, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, and Meyer BJ (2015). Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, Robles-Rebollo I, Xiao X, Wang YF, Barozzi I, et al. (2018). Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Yeung J, Ellu J, Ramanujachar R, Bornhauser B, Solarska O, Hubank M, Williams O, and Brady HJ (2011). The E2A-HLF oncogenic fusion protein acts through Lmo2 and Bcl-2 to immortalize hematopoietic progenitors. Leukemia 25, 321–330. [DOI] [PubMed] [Google Scholar]
- de Graaf CA, Choi J, Baldwin TM, Bolden JE, Fairfax KA, Robinson AJ, Biben C, Morgan C, Ramsay K, Ng AP, et al. (2016). Haemopedia: An Expression Atlas of Murine Hematopoietic Cells. Stem Cell Reports 7, 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F, Bortoluzzi S, Coppe A, Basso D, Bicciato S, Zini R, Gemelli C, Danieli GA, and Ferrari S (2007). Genomic expression during human myelopoiesis. BMC Genomics 8, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A, Sanjuan-Pla A, Thongjuea S, Carrelha J, Giustacchini A, Gambardella A, Macaulay I, Mancini E, Luis TC, Mead A, et al. (2016). Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat Commun 7, 11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yanez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. (2017). The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169, 693–707 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JS, Sagar, and Grun D (2018). FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat Methods 15, 379–386. [DOI] [PubMed] [Google Scholar]
- Jiang J, Yu H, Shou Y, Neale G, Zhou S, Lu T, and Sorrentino BP (2010). Hemgn is a direct transcriptional target of HOXB4 and induces expansion of murine myeloid progenitor cells. Blood 116, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kina T, Ikuta K, Takayama E, Wada K, Majumdar AS, Weissman IL, and Katsura Y (2000). The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br J Haematol 109, 280–287. [DOI] [PubMed] [Google Scholar]
- Kingsley PD, Greenfest-Allen E, Frame JM, Bushnell TP, Malik J, McGrath KE, Stoeckert CJ, and Palis J (2013). Ontogeny of erythroid gene expression. Blood 121, e5–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic A, Cuadrado A, De Koninck M, Gimenez-Llorente D, Rodriguez-Corsino M, Gomez-Lopez G, Le Dily F, Marti-Renom MA, and Losada A (2018). Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol 25, 496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. (2013). Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 45, 1232–1237. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, and Rajewsky K (1995). Inducible gene targeting in mice. Science 269, 1427–1429. [DOI] [PubMed] [Google Scholar]
- Lagasse E, and Weissman IL (1992). Mouse MRP8 and MRP14, two intracellular calcium-binding proteins associated with the development of the myeloid lineage. Blood 79, 1907–1915. [PubMed] [Google Scholar]
- Lajoie BR, Dekker J, and Kaplan N (2015). The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods 72, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, and Westphal H (1996). Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, and Krantz ID (2009). Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet 76, 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, and Hirano T (1998). Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12, 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson R, Zandi S, Anderson K, Martensson IL, Jacobsen SE, Bryder D, and Sigvardsson M (2008). B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood 112, 1048–1055. [DOI] [PubMed] [Google Scholar]
- Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, Li R, Corces MR, Flynn RA, Buenrostro JD, Chan SM, et al. (2015). Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell 17, 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, et al. (1996). Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 15, 5647–5658. [PMC free article] [PubMed] [Google Scholar]
- Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, and Singh H (2004). Assembling a gene regulatory network for specification of the B cell fate. Dev Cell 7, 607–617. [DOI] [PubMed] [Google Scholar]
- Mueller BU, Pabst T, Fos J, Petkovic V, Fey MF, Asou N, Buergi U, and Tenen DG (2006). ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood 107, 3330–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K, Kayembe C, Rocha PP, Raviram R, Gong Y, et al. (2015). Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med 212, 1833–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, and Bruneau BG (2017). Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930–944 e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. (2008). Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433. [DOI] [PubMed] [Google Scholar]
- Pogosova-Agadjanyan EL, Kopecky KJ, Ostronoff F, Appelbaum FR, Godwin J, Lee H, List AF, May JJ, Oehler VG, Petersdorf S, et al. (2013). The prognostic significance of IRF8 transcripts in adult patients with acute myeloid leukemia. PLoS One 8, e70812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucella JN, Yen WF, Kim MV, van der Veeken J, Luo CT, Socci ND, Naito Y, Li MO, Iwai N, and Chaudhuri J (2015). miR-182 is largely dispensable for adaptive immunity: lack of correlation between expression and function. J Immunol 194, 2635–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. (2017). Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S, Ritchie A, Boring L, Gosling J, Cooper S, Hangoc G, Charo IF, and Broxmeyer HE (1999). Enhanced myeloid progenitor cell cycling and apoptosis in mice lacking the chemokine receptor, CCR2. Blood 93, 1524–1533. [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, and Krumm A (2008). CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A 105, 8309–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santambrogio F, and SpringerLink (Online service) Optimal Transport for Applied Mathematicians : Calculus of Variations, PDEs, and Modeling, 1st edn.
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, C HH, Mirny L, et al. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Fisher RC, Olson MC, Kehrli EW, Simon MC, and Singh H (1997). PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity 6, 437–447. [DOI] [PubMed] [Google Scholar]
- Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al. (2013). Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res 23, 2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, et al. (2011). Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333, 1039–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl U, Rosenbauer F, Verhaak RG, Gu X, Ebralidze A, Otu HH, Klippel S, Steidl C, Bruns I, Costa DB, et al. (2006). Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet 38, 1269–1277. [DOI] [PubMed] [Google Scholar]
- Su GH, Chen HM, Muthusamy N, Garrett-Sinha LA, Baunoch D, Tenen DG, and Simon MC (1997). Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J 16, 7118–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan C, Chen Y, Shan Y, Hu Y, Peng C, Zhang H, Kong L, and Li S (2011). Functional ramifications for the loss of P-selectin expression on hematopoietic and leukemic stem cells. PLoS One 6, e26246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thota S, Viny AD, Makishima H, Spitzer B, Radivoyevitch T, Przychodzen B, Sekeres MA, Levine RL, and Maciejewski JP (2014). Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood 124, 1790–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, and Nasmyth K (1999). Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 13, 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto T, Yamato M, Shiratsuchi Y, Terasawa M, Yang J, Nishida K, Kobayashi Y, and Okano T. (2006). Expression of Integrin beta3 is correlated to the properties of quiescent hemopoietic stem cells possessing the side population phenotype. J Immunol 177, 7733–7739. [DOI] [PubMed] [Google Scholar]
- van der Lelij P, Lieb S, Jude J, Wutz G, Santos CP, Falkenberg K, Schlattl A, Ban J, Schwentner R, Hoffmann T, et al. (2017). Synthetic lethality between the cohesin subunits STAG1 and STAG2 in diverse cancer contexts. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangala RK, Heiss-Neumann MS, Rangatia JS, Singh SM, Schoch C, Tenen DG, Hiddemann W, and Behre G (2003). The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood 101, 270–277. [DOI] [PubMed] [Google Scholar]
- Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, Papalexi E, Yelin D, Shank K, Reyes J, Chiu A, et al. (2015). Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J Exp Med 212, 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yan M, Sun J, Jain S, Yoshimi R, Abolfath SM, Ozato K, Coleman WG Jr., Ng AP, Metcalf D, et al. (2014). A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. J Immunol 193, 1766–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. (2008). Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801. [DOI] [PubMed] [Google Scholar]
- Will B, Vogler TO, Narayanagari S, Bartholdy B, Todorova TI, da Silva Ferreira M, Chen J, Yu Y, Mayer J, Barreyro L, et al. (2015). Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med 21, 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, and He QY (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. [DOI] [PubMed] [Google Scholar]
- Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, and Sigvardsson M (2008). EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol 181, 3364–3372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.