SUMMARY
Many stem cells utilize asymmetric cell division (ACD) to produce a self-renewed stem cell and a differentiating daughter cell. How non-genic information could be inherited differentially to establish distinct cell fates is not well understood. Here, we report a series of spatiotemporally regulated asymmetric components, which ensure biased sister chromatid attachment and segregation during ACD of Drosophila male germline stem cells (GSCs). First, sister centromeres are differentially enriched with proteins involved in centromere specification and kinetochore function. Second, temporally asymmetric microtubule activities and polarized nuclear envelope breakdown allow for the preferential recognition and attachment of microtubules to asymmetric sister kinetochores and sister centromeres. Abolishment of either the asymmetric sister centromeres or the asymmetric microtubule activities results in randomized sister chromatid segregation. Together, these results provide the cellular basis for partitioning epigenetically distinct sister chromatids during stem cell ACDs, which opens new directions to study these mechanisms in other biological contexts.
Graphical Abstract

eTOC paragraph:
Drosophila male germline stem cell undergoes asymmetric cell division to produce two different cells. Ranjan et al. show that a temporal asymmetry of microtubules tightly coordinates with quantitative asymmetries in sister centromeres and sister kinetochores to ensure non-random sister chromatids segregation and asymmetric epigenetic inheritance, which regulate distinct cell fates.
INTRODUCTION
Epigenetic mechanisms play important roles in regulating cell identity and function. Mis-regulation of epigenetic information may lead to abnormalities in cellular behaviors that underlie many diseases, such as developmental defects, cancer, and tissue degeneration (Allis and Jenuwein, 2016). Histone proteins, which play important roles in DNA packaging and chromosomal structure, represent a major group of epigenetic information carriers (Sitbon et al., 2017).
Many types of adult stem cells undergo asymmetric cell divisions (ACDs) to generate both a self-renewed stem cell and a daughter cell, which will subsequently differentiate (Clevers, 2005; Kahney et al., 2017; Knoblich, 2010; Morrison and Kimble, 2006; Venkei and Yamashita, 2018. Since stem cells have unique gene expression [e.g. (Amcheslavsky et al., 2014; Blanpain and Fuchs, 2006; Kai et al., 2005; Seale et al., 2004; Terry et al., 2006; Terskikh et al., 2003; Young, 2011)], the field has long sought to understand how stem cells could maintain their epigenetic information and gene expression through many cell divisions.
During the asymmetric division of Drosophila male germline stem cells (GSCs) (Figure 1A), we showed previously that the preexisting (old) histone H3 is selectively segregated to the GSC, whereas the newly synthesized (new) H3 is enriched in the differentiating daughter cell known as a gonialblast (GB) (Tran et al., 2012). This asymmetry is unique to stem cells, as symmetrically dividing spermatogonial cells (SGs) show symmetric patterns of histone H3 inheritance. We also identified that differential phosphorylation at Threonine 3 of histone H3 (H3T3P) distinguishes old versus new H3 in the asymmetrically dividing GSCs. Mis-regulation of this phosphorylation leads to randomized segregation of old H3 versus new H3, as well as stem cell loss and early-stage germline tumors (Xie et al., 2015). We hypothesize that prior to mitosis, old H3 and new H3 are differentially distributed at the two sets of sister chromatids (Wooten et al., 2019). During the subsequent mitosis, the two sets of epigenetically distinct sister chromatids are asymmetrically segregated (Tran et al., 2013; Xie et al., 2017). According to this model, sister chromatids carrying distinct epigenetic information need to communicate with the mitotic machinery to achieve differential attachment, followed by asymmetric segregation. It has been shown previously that centrosomes display asymmetry in Drosophila male GSCs, wherein the mother centrosome is retained by the GSC while the daughter centrosome is inherited by the GB (Yamashita et al., 2007). However, it remains unclear whether this asymmetry in centrosome inheritance is related to the phenomenon of asymmetric histone inheritance.
Figure 1: Asymmetric CID inheritance in Drosophila male GSCs.
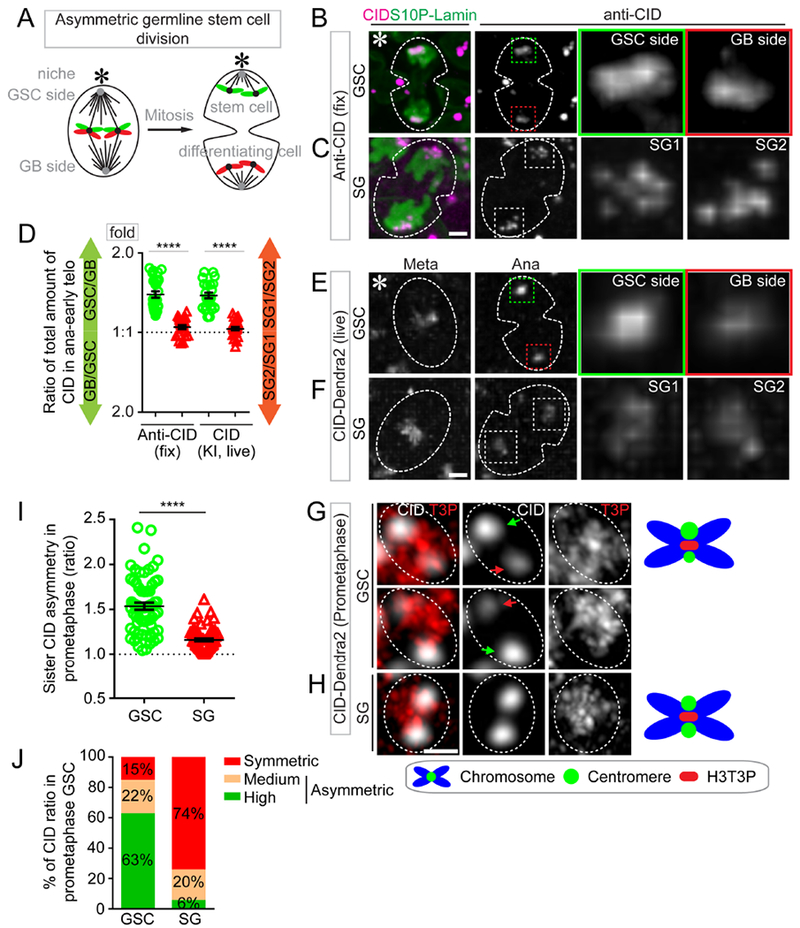
(A) A cartoon depicting asymmetric Drosophila male GSC division and asymmetric histone H3 inheritance (old H3 - green, new H3 - red). (B-F) Asymmetric CID segregation in asymmetrically dividing GSCs with more CID toward the GSC side, using both immunostaining of fixed cells (B) and live cell imaging of a CID-Dendra2 knock-in line (E). In symmetric SG cell division, CID is symmetrically distributed, as determined using both immunostaining (C) and live cell imaging (F). Quantification of all four data sets in (D): 1.41± 0.04-fold for GSC/GB (n= 27), 1.06± 0.02-fold for SG1/SG2 (n= 23), as determined by anti-CID, Table S1; 1.39± 0.04-fold for GSC/GB (n= 23), 1.04± 0.02-fold for SG1/SG2 (n= 21), as determined by live cell imaging using CID-Dendra2, Table S2. (G-H) At prometaphase, using Airyscan resolved individual sister centromeres show more asymmetry in GSCs [green arrow-stronger centromere, red arrow-weak centromere, (G)], compared with resolved sister centromeres in SGs (H). (I) Quantification of individual pairs of resolved sister centromeres in GSCs (1.54± 0.04-fold, n= 65) and SGs (1.16± 0.02-fold, n= 66) at prometaphase, Table S4. (J) Percentage of different categories of symmetric (< 1.20-fold), medium asymmetric (1.20- to 1.40-fold) and highly asymmetric sister centromeres (> 1.40-fold) for GSCs (n= 65) and SGs (n= 66) in prometaphase, examples of resolved individual sister centromeres are shown in GSCs (Figure S1H) and SGs (Figure S1I). All ratios= Avg± SE. P-value: paired t test. ****: P< 10−4. Asterisk: hub. Scale bars: 2μm (B-C, E-F), 0.5μm (G-H).
During mitosis, microtubules emanate from centrosomes and attach to sister chromatids at the centromeric region through the kinetochore protein complex (Cheeseman, 2014; Fukagawa and Earnshaw, 2014; Rieder, 2005). Centromeres are specialized chromosomal regions which are often defined by epigenetic characteristics (Bodor et al., 2014; Dunleavy et al., 2005; Karpen and Allshire, 1997; Schueler and Sullivan, 2006; Vos et al., 2006; Westhorpe and Straight, 2014). In most eukaryotic cells, the critical epigenetic feature of centromeres is the centromere-specific histone H3 variant, known as the Centromere identifier (CID) in flies (Henikoff et al., 2000) and Centromere protein A (CENP-A) in mammals (Allshire and Karpen, 2008; Mendiburo et al., 2011; Palmer et al., 1987). Both are structurally different from the canonical H3 (McKinley and Cheeseman, 2016; Melters et al., 2015; Miell et al., 2013). Based on the unique features and essential roles of centromeres in coordinating sister chromatid segregation during mitosis, it has been proposed that epigenetic differences between sister centromeres could ensure that stem cells retain their unique epigenetic information and gene expression after many cell divisions (Lansdorp, 2007).
RESULTS
Asymmetric Sister Centromere Inheritance in Male GSCs
During Drosophila male GSC asymmetric division at anaphase or early telophase, the level of endogenous CID showed a 1.41-fold enrichment at sister chromatids segregating toward the future GSC side compared to the future GB side, stained by anti-CID antibodies (Figures 1B and 1D, Figures S1A and S1C, Table S1). Conversely, symmetrically dividing SGs showed nearly equal CID distribution at anaphase or early telophase (Figures 1C and 1D, Figures S1B and S1D, Table S1). To confirm this result, live cell imaging was performed using a knock-in fly strain with the endogenous cid gene tagged with a fluorescent protein-encoding Dendra2 (Chudakov et al., 2007), which was generated by CRISPR/Cas9-mediated genome editing (Horvath and Barrangou, 2010; Wright et al., 2016). Consistent with the immunostaining results using anti-CID, the CID-Dendra2 fusion displayed a 1.39-fold overall enrichment at sister chromatids segregating toward the future GSC side compared to the future GB side (Figures 1D and 1E, Movie S1, Table S2). Additionally, CID-Dendra2 showed largely symmetric distribution patterns in SGs (Figures 1D and 1F, Movie S2). Finally, a CID-GFP genomic transgene used in previous studies (Henikoff et al., 2000) showed a 1.73-fold enrichment toward the future GSC side compared to the future GB side in asymmetrically dividing GSCs (Figures S1E and S1G, Movie S8, Table S3). By contrast, a nearly equal distribution of CID-GFP was found in symmetrically dividing SGs (Figures S1F and S1G, Movie S9, Table S3). In summary, both endogenous and transgenic CID showed enrichment toward the future stem cell side during GSC asymmetric division, using both immunostaining on fixed samples and live cell imaging.
We hypothesize that the asymmetries in CID segregation observed in anaphase and early telophase GSCs are due to the asymmetry in CID levels between individual pairs of sister centromeres. To test this hypothesis, we examined sister centromeres at prophase or prometaphase. Using Airyscan high spatial resolution microscopy, sister centromeres could be resolved, which displayed quantitative differences in prophase or prometaphase GSCs with a 1.52-fold average asymmetry in CID levels (Figures 1G and 1I–J, Figures S1H, Table S4). Conversely, the vast majority of sister centromeres resolved in SGs showed no obvious level differences (Figures 1H and 1I–J, Figures S1I, Table S4). Even though a range of patterns could be detected in both GSCs and SGs (Figure 1J, Figures S1H–I), the overall percentage of asymmetric sister centromeres in GSCs was significantly higher than that in SGs (Figure 1I–J). These results also suggest that the establishment of asymmetric CID in GSCs likely occurred prior to mitosis, when this asymmetry could be detected at resolved sister centromeres during mitosis.
The Temporal and Spatial Distribution of the CAL1 Chaperone in GSCs
In Drosophila, it has been demonstrated that Chromosome alignment defect 1 (CAL1) is both necessary (Erhardt et al., 2008; Goshima et al., 2007) and sufficient (Chen et al., 2014) to interact with CID and assemble CID-containing nucleosomes. Using CRISPR/Cas9 technology, we generated a Dendra2 knock-in line at the endogenous cal1 gene. In non-replicative cells such as hub cells at the apical tip of testis, Dendra2-CAL1 was undetectable (Figure S2A). In GSCs, Dendra2-CAL1 fusion protein was found to localize at the centromeric region in G2 phase as well as in early M phase (from prophase to prometaphase, Figure 2A). However, its level decreased as the cell progressed from G2 to early M phase, and kept decreasing from prophase to prometaphase (Figures 2A and 2B). Furthermore, at prophase or prometaphase when sister centromeres could be resolved in GSCs using Airyscan, asymmetric distribution of CAL1 could be detected along with the asymmetric CID (Figures 2C–E, Figure S2B, Table S5). The percentages of asymmetric CAL1 (Figure 2E and S2B) were comparable with the percentages of asymmetric CID distribution at sister centromeres (Figures 1J and S1H).
Figure 2: The temporal and spatial distribution of the CAL1 chaperone in male GSCs.

(A) Two representative GSCs from one testis, one in G2 phase and the other in prometaphase, show higher CAL1 levels in G2 phase compared to prometaphase, visualized within the same testis using the same parameters. (B) Quantification of total amount of CAL1 in GSCs at different cell cycle stages: the highest CAL1 levels are detected in G2 phase, which gradually decreases in mitosis. The CAL1 signals in hub cells are used as a control. (C) In prometaphase GSCs, using Airyscan resolved individual sister centromeres show asymmetric distribution of CAL1 (green arrow-stronger centromere with more CID and CAL1, red arrow-weaker centromere with less CID and CAL1). (D) Quantification of individual pairs of resolved sister centromeres in prometaphase GSCs show asymmetric CAL1 distribution (1.66± 0.09-fold, n= 28), Table S5. (E) Quantification of percentages of different categories in GSCs (n= 28). Images shown in Figure S2B. (F) Photoconversion experiments at the apical tip of testis (circled area), which convert CID-Dendra2 protein from green fluorescent protein to red fluorescent protein. Insets show the very tip of the testis where GSCs are located. (G) After photoconversion, testes were cultured for 5hrs before fixation. New CID (green Dendra2) incorporation is detectable in early prophase GSCs. All ratios= Avg± SE, Asterisk: hub. Scale bars: 10μm (A), 0.5μm (C), 25μm (F), 5μm (G).
Unlike the canonical histones such as H3, deposition of the histone variant CID is independent on DNA replication, and the timing could be cell type-specific (Dunleavy et al., 2012; Garcia Del Arco et al., 2018; Mellone et al., 2011; Schuh et al., 2007). To study the timing of new CID incorporation in Drosophila male GSCs, we utilized the knock-in cid gene that produces the CID-Dendra2 fusion protein. Dendra2 is a photoconvertable protein that can switch irreversibly from green fluorescence to red fluorescence. Following photoconversion any newly synthesized CID will be green fluorescent. Therefore, old CID would be labeled in red while new CID in green (Chudakov et al., 2007). We first checked that CID-Dendra2 had approximately 90% photoconversion efficiency in cells at the apical tip of testis (Figure 2F, STAR Methods). To analyze the timing of new CID incorporation in GSCs, we photoconverted cells located at the testis tip and then captured mitotic GSCs five hours later (Figure 2G). Considering that the elongated G2 phase in GSCs lasts for approximately 10-12 hours (Sheng and Matunis, 2011; Tran et al., 2012; Yadlapalli et al., 2011; Yadlapalli and Yamashita, 2013), the significantly increased green fluorescence detected in prophase GSCs indicates that the new CID has been incorporated prior to mitosis, likely in mid- to late G2 phase or in early mitosis (Figures 2G and S2C). As a control, the non-replicative hub cells showed no new CID incorporation during the same period of time (Figure S2C), consistent with the lack of CAL1 expression in hub cells (Figure S2A). Furthermore, in asymmetrically dividing GSCs at anaphase or early telophase, both old CID and new CID displayed asymmetric patterns with significantly more CID toward the GSC side, suggesting that new CID is deposited in a sister chromatid-biased manner. In contrast, no such bias could be detected in symmetrically dividing SGs (Figure S2D).
Asymmetric Sister Kinetochores Correlate with the Asymmetry between Sister Centromeres in GSCs
The recognition of centromeres by microtubules is mediated by the kinetochore, a highly organized multiprotein complex that nucleates at the surface of centromere and coordinates the attachment of sister chromatids by the mitotic spindle (Cheeseman, 2014; Cleveland et al., 2003). Next, we examined a key kinetochore protein, NDC80 (Schittenhelm et al., 2007), using a Dendra2-Ndc80 knock-in fly strain. Consistent with the CID asymmetry, the kinetochore component NDC80 displayed a 1.49-fold overall enrichment at sister chromatids segregated toward the future GSC side compared to the future GB side in anaphase or early telophase GSCs (Figures 3A and 3C, Table S6). In contrast, the symmetrically dividing SGs showed a nearly symmetric NDC80 segregation pattern (Figures 3B and 3C, Table S6).
Figure 3: Asymmetric sister kinetochore correlates with the asymmetry of sister centromeres in GSCs.

(A) Asymmetric kinetochore component NDC80 in asymmetrically dividing GSCs with more NDC80 toward the GSC side, using immunostaining of a Dendra2-NDC80 knock-in line. (B) In symmetric SG cell division, NDC80 is symmetrically distributed. (C) Quantification of both data sets: GSC/GB= 1.49± 0.07 (n= 19), SG1/SG2= 1.03± 0.03 (n= 28), Table S6. (D-E) In prometaphase, using Airyscan resolved sister kinetochores show more asymmetry in GSCs (NDC80 in D, with the same polarity with CID enrichment), compared to resolved sister kinetochores in SGs (E). (F) Quantification of individual pairs of resolved sister kinetochores in prometaphase: 1.76± 0.08-fold in GSCs (n= 46); 1.19± 0.02-fold in SGs (n= 34), Table S7. (G) Percentage of different categories of symmetric (< 1.20-fold), medium asymmetric (1.20- to 1.40-fold) and highly asymmetric sister kinetochores (> 1.40-fold) for GSCs (n= 46) and SG (n= 34) in prometaphase, examples of resolved individual sister kinetochores are shown in GSCs (Figure S3A) and SGs (Figure S3B). (H) In prometaphase GSCs, asymmetric NDC80 is positively correlated (correlation coefficient, r = 0.2) with asymmetric CID. The degree of asymmetry for sister kinetochore is more than that for sister centromere [CID = 1.49-fold, NDC80 = 1.76-fold (n= 43), Table S8]. All ratios= Avg± SE. P-value: paired t test: ****: P< 10−4, Asterisk: hub. Scale bars: 2μm (A-B) and 0.5μm (D-E).
Furthermore, differences between individual pairs of sister kinetochores were already detectable using Airyscan in prophase or prometaphase GSCs (Figure 3D), but not in SGs (Figure 3E). Individual pairs of sister kinetochores showed, on average, a 1.76-fold difference in GSCs (Figure 3F–G, Figures S3A, Table S7). In addition, although a range of distinct patterns among sister kinetochores could be detected in SGs, the percentage of asymmetric sister kinetochores was considerably less compared to that in GSCs (Figure 3F–G, Figures S3B).
Noticeably, sister kinetochore showed a similar asymmetric pattern compared to sister centromere (i.e. the side with more CID also had more NDC80, Figure 3D), but quantitatively sister kinetochores showed a greater degree of asymmetry than sister centromeres (Figures 3D and 3H, Table S8). Collectively, these results demonstrate that sister kinetochores are also quantitatively different in mitotic GSCs (Figure S3C).
Temporally Asymmetric Microtubule Dynamics in GSCs
Since NDC80 is a key microtubule interacting protein at the kinetochore (Cheeseman et al., 2006; DeLuca et al., 2006), we next sought to understand how asymmetry on sister kinetochores could communicate with the mitotic spindle. We tracked microtubules using a GFP-tagged α-Tubulin under the control of an early-stage germline-specific nanos-Gal4 driver (Van Doren et al., 1998), as used previously in this system (Yamashita et al., 2003). Using high temporal resolution movies, we tracked microtubules in real-time throughout the cell cycle of germ cells at distinct stages, including both GSCs and SGs. Due to the unique morphology of the mitotic spindle at metaphase, we used metaphase as a landmark to define time point zero; other cell cycle phases prior to metaphase were then labeled accordingly. In male GSCs, the mother centrosome is retained in proximal to the GSC-niche interface while the daughter centrosome migrates to the distal side after duplication in early G2 phase (Yamashita et al., 2003; Yamashita et al., 2007). From −250 minutes (−250’) at approximately mid-G2 phase to −150’ at late-G2 phase, more microtubules emanated from the mother centrosome than microtubules from the daughter centrosome (Figure 4A–B and S4A, Movie S3 and S5, Table S9). Subsequently, this dominance of mother centrosome-emanating microtubules declined at the G2-to-M phase transition from −50’ to −35’, when the daughter centrosome-emanating microtubules increased (Figures 4A and 4D, Movie S3). The decrease of microtubules from the mother centrosome and the increase of microtubules from the daughter centrosome persisted from −30’ to −10’ in prophase GSCs, as well as at −5’ in prometaphase GSCs (Figures 4A and 4D, Movie S3, Table S9). Subsequently, at metaphase, microtubules from both centrosomes formed the mitotic spindle perpendicular to the GSC-niche interface (Figures 4A and 4D, Movie S3), consistent with previous reports (Chen et al., 2018; Inaba et al., 2010; Yamashita et al., 2003).
Figure 4: Temporally asymmetric microtubules in GSCs.

3D reconstructed montage made from live cell imaging using a nanos-Gal4; UAS-α-tubulin-GFP line in GSCs (A) and quantified in (D, n= 22, Table S8), as well as in SGs (C) and quantified in (E, n= 24, Table S9). Metaphase was set as time 0, and other time points prior to metaphase were labeled as minus minutes accordingly. (A) In GSCs the mother centrosome is activated in mid G2 (− 250 min) prior to the daughter centrosome at G2/M transition (− 50 min). (B) Using Airyscan, more microtubules emanating from mother centrosome could be detected from mid- to late-G2 phase GSCs. (C) In SGs, both centrosomes are active at the G2/M transition (−35 min) simultaneously. (D-E) All ratios= Avg± SE, see Table S8. Magenta arrow: microtubules poke-in. Asterisk: hub. Scale bars: 2μm (A, C), 5μm (B).
In comparison, microtubules from both centrosomes became almost simultaneously detectable at −35’ at the G2-to-M phase transition in SGs. After that, no obvious difference in the amount of microtubules could be detected between the two centrosomes throughout mitosis in SGs (Figures 4C and 4E, Movie S4, Table S9). In summary, these results demonstrate that microtubules, the key component of the mitotic machinery, display a temporal asymmetry in male GSCs.
Polarized Nuclear Envelope Breakdown Coincides and Colocalizes with Temporally Asymmetric Microtubules
Next, using antibodies against Drosophila Lamin B (Chen et al., 2013), we found that invagination of the nuclear lamina first appeared exclusively at the future stem cell side in early prophase GSCs (Figure 5A and 5C, Movie S8A–C). Local disassembly of nuclear lamina was also first detected at the future stem cell side (green arrows in Figure 5A and 5C, Movie S6, S10–S11), after which invagination of nuclear lamina was detected from the GB side, leading to nuclear envelope breakdown (NEBD, red arrowheads in Figure 5B and Movie S7, S12). By contrast, such a temporal polarity in NEBD was not observed in symmetrically dividing SGs (yellow arrowheads in Figure 5D and yellow arrows in Figure 5E).
Figure 5: Polarized disassembly of nuclear lamina in GSCs.
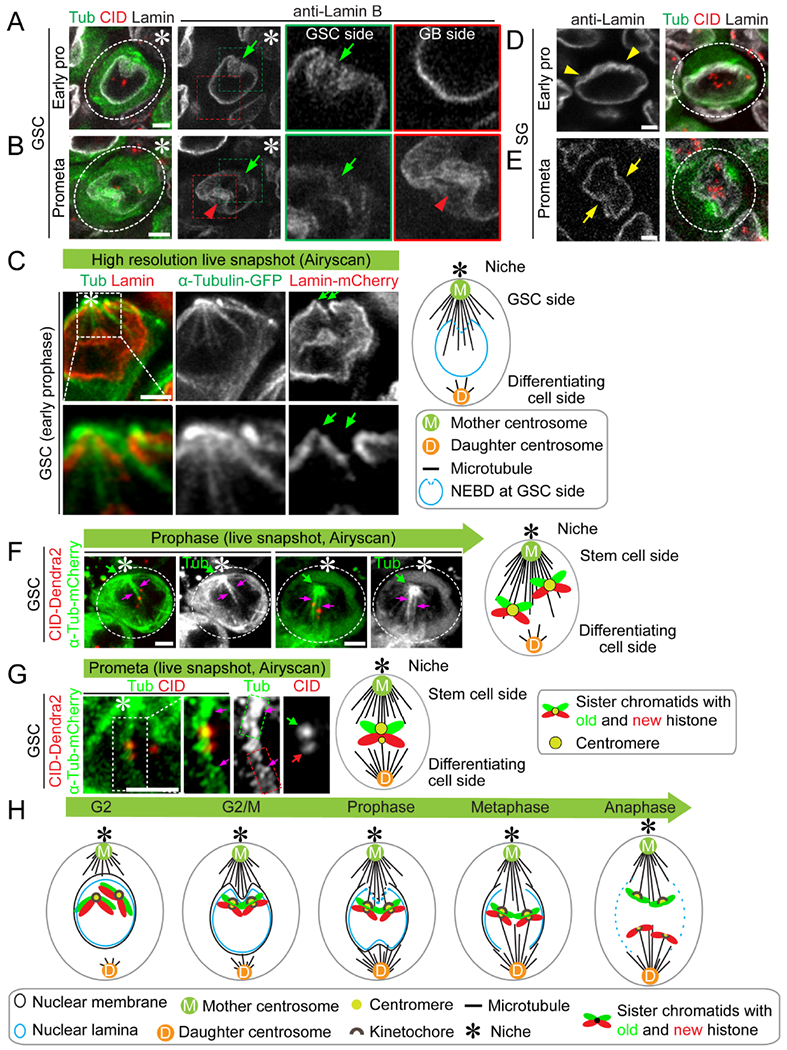
(A-B) Airyscan images showing morphology of nuclear lamina in GSCs at early prophase (A) and at prometaphase (B), visualized by immunostaining using anti-Lamin B (white), co-stained with anti-CID (red) using nanos-Gal4; UAS-α-tubulin-GFP line (α-Tub in green). Early active mother centrosome-nucleating microtubules correspond with the initial disassembly of nuclear lamina toward the hub (green arrows in A and B), whereas later daughter centrosome-nucleating microtubules lead to disassembly of nuclear lamina toward the other side (red arrowhead in B). (C) Airyscan images showing morphology of nuclear lamina and microtubules in GSCs at early prophase, using nanos-Gal4; UAS-α-tubulin-GFP; UAS-lamin-mCherry line (α-Tub in green and Lamin in red). Mother centrosome-emanating microtubules break through the nuclear lamina toward the stem cell side, leading to polarized disassembly of nuclear lamina. (D-E) Airyscan images showing morphology of nuclear lamina in SGs at early prophase (D) and at prometaphase (E), visualized by immunostaining using anti-Lamin B (white), co-stained with anti-CID (red) using nanos-Gal4; UAS-α-tubulin-GFP line (α-Tub in green). In comparison with GSCs (A-C), SGs show no polarized disassembly of nuclear lamina (yellow arrowheads label invagination sites in D, while yellow arrows point to symmetric disassembly sites of nuclear lamina in E). (F) Airyscan images showing CID and microtubules in GSCs at early prophase (two left panels) and late prophase (two right panels), using nanos-Gal4; UAS-α-tubulin-mCherry; cid-Dendra2 line (α-Tub in green and CID in red). Microtubules emanating from the mother centrosome anchor the centromere in early prophase and maintain the attachment to late prophase. (G) Airyscan images showing CID and microtubules in GSCs at prometaphase, using nanos-Gal4; UAS-α-tubulin-mCherry; cid-Dendra2 line (α-Tub in green and CID in red). Stronger centromere is attached by more microtubules (green dotted box) emanating from the mother centrosome whereas the weaker centromere is attached by less microtubules (red dotted box) emanating from the daughter centrosome. (H) A cartoon depicting the sequential events in Drosophila male GSCs. Magenta arrow: K-fiber. Asterisk: hub. Scale bars: 2 μm.
Similar results were also obtained using wheat germ agglutinin (WGA), which binds to the cytoplasmic part of nuclear pore, allowing for visualization of nuclear membrane morphology in GSCs (Figures S5A–B) and SGs (Figures S5C–D). Together these data showed that in GSCs a polarized nuclear lamina disassembly was detected, first at the GSC side and then at the GB side.
We next asked whether the asymmetric microtubule activity and the polarized nuclear lamina disassembly coordinate with chromosomes at the centromeric region, visualized by CID signals. Noticeably, in late G2 phase GSCs, close proximity between chromosomes and microtubules could be detected: When the mother centrosome-emanating microtubules dynamically interacted with the nuclear envelope (α-Tubulin in Figures S4B and S5E), centromeres from all chromosomes clustered preferentially toward the future GSC side (CID in Figures S4B, S4C and S5E). Given the higher microtubule dynamics from the mother centrosome, this close proximity of all centromeres toward the future GSC side might initiate crosstalk between centromeres and mother centrosome-emanating microtubules. A series of immunostaining images to label microtubules (α-Tubulin), nuclear lamina (Lamin), the mitotic chromosomes (H3S10P or S10P), and centromeres (CID) showed coordinated microtubule activity and nuclear lamina disassembly with chromosome and centromere movement in GSCs at different cell cycle stages (Figures S5E–J). High-resolution imaging showed that microtubules from mother centrosome attached to centromeres in prophase GSCs (Figure 5F). In addition, the stronger centromere was attached by more microtubules emanating from the mother centrosome whereas the weaker centromere by less microtubules emanating from the daughter centrosome in prometaphase GSCs (Figure 5G). In comparison, these events were also examined in symmetrically dividing SGs: Even though microtubules emanating from both centrosomes still coincided with chromosomal condensation and centromere movement, no obvious sequential order was detected (Figures S5K–O).
Together, these immunostaining images with markers for both microtubules and nuclear lamina, as well as the centromere marker CID, delineate a series of events at sister centromeres, likely allowing for differential segregation of sister chromatids (Figure S5P). In addition, this series of interactions has cellular specificity to asymmetrically dividing GSCs, which were not detectable in symmetrically dividing SGs.
Compromising the CAL1 Chaperone Abolishes the CID Asymmetry between Sister Centromeres
Based on these results, we propose a two-step model for the establishment and recognition of CID asymmetry in GSCs (Figure 6A): Step 1 involves the establishment of asymmetric CID at individual pairs of sister centromeres, likely from late G2 phase to early M phase; Step 2 requires asymmetric microtubule activity to recognize asymmetric sister centromeres to allow for their differential attachment and segregation during ACD of GSCs.
Figure 6: Compromising the CAL1 chaperone disrupts the CID asymmetry and results in GSC defects.
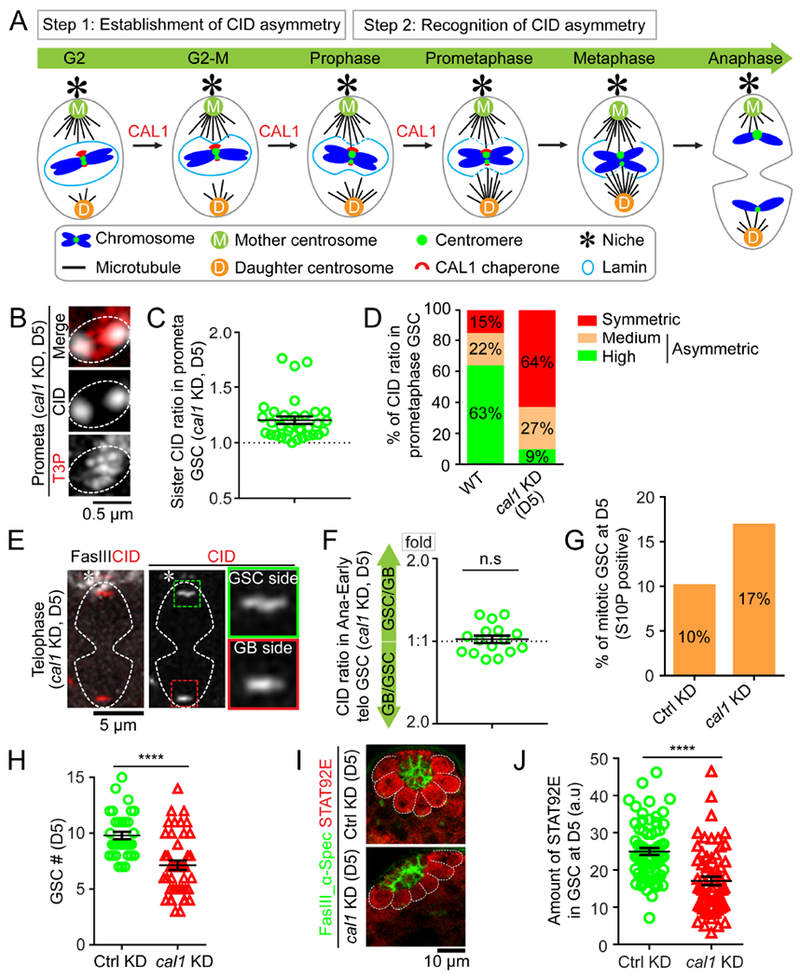
(A) A cartoon depicting a two-step model for asymmetric CID inheritance: Step 1 involves establishment of CID asymmetry at individual sister centromeres; Step 2 requires recognition of asymmetric CID by the mitotic machinery. CAL1 is likely contributing to Step 1. (B) In prometaphase GSCs after knocking down cal1 for five days (D5), resolved individual sister centromeres show symmetry, quantified in (C): 1.20± 0.03-fold, n= 33, Table S10. (D) Percentage of different categories of symmetric (< 1.20-fold), medium asymmetric (1.20- to 1.40-fold) and highly asymmetric sister centromeres (> 1.40-fold) for wild-type (WT) GSCs (n= 65) and cal1 KD GSCs (D5, n=33) at prometaphase. (E) Symmetric CID segregation in anaphase or early telophase GSCs after knocking down cal1 for five days (D5), quantified in (F): 1.03± 0.03-fold, n= 16, Table S11. (G) Mitotic index of Ctrl mCherry KD GSCs (H3S10P-positive GSCs%= 10%, n= 396) vs. cal1 KD GSCs (H3S10P-positive GSCs%= 17%, n= 480). (H) Quantification of GSCs in cal1 KD versus Ctrl testes. (I) Immunostaining using anti-Stat92E (red) in Ctrl GSCs vs. cal1 KD GSCs at D5, signals were quantified in (J). Testes were costained with a hub marker Fas III (green) and a spectrosome/fusome marker α-Spectrin (α-Spec) (green). All ratios = Avg± SE. P-value: paired t test. ****: P< 10−4; n.s: not significant. Asterisk: hub. Scale bars: 0.5μm (B), 5μm (E), 10μm (I).
Next, to investigate how these two steps act together in GSCs, we designed experiments attempting to disrupt either Step 1 or Step 2. First, because cal1 is an essential gene required for CID incorporation, we compromised cal1 in a spatiotemporally controlled manner in adult flies. The temperature-sensitive Gal80 controlled by the ubiquitous tubulin promoter (tub-Gal80ts) was used in combination with the early germline-specific nos-Gal4 driver, in order to temporally control the expression of call shRNA (short hairpin RNA) in early-stage germline in adult flies [tub-Gal80ts, nos-Gal4>cal1 shRNA (cal1 KD)]. We found that the CID asymmetry in anaphase or early telophase GSCs was still detectable two days after shifting from the permissive to the restrictive temperature as adult flies (D2). However, at five days (D5) this asymmetry became very much compromised (Figure S6A). In addition, the CAL1 protein was almost undetectable at D5 (Figure S6B), suggesting an efficient knocking down at this time point.
We then examined whether cal1 KD disrupts the CID asymmetry at individual pair of sister centromeres. In D5 prometaphase GSCs, approximately 63.6% of sister centromeres displayed symmetric CID distribution compared to 15.4% in WT GSCs (Figures 6B–D and Figure S6C, Table S10), suggesting that the establishment of asymmetric CID at sister centromeres in Step 1 was indeed compromised in cal1 KD GSCs at D5. Furthermore, the asymmetric segregation of CID was greatly reduced in anaphase or early telophase GSCs at D5 (Figures 6E–F, Table S11), consistent with the prediction that disruption of Step 1 would lead to symmetric segregation patterns (Figure 6A).
Next, we asked if compromising CAL1 would lead to any GSC defects. Because cal1 is an essential gene, we are mainly interested in the cellular defects after disrupting CID asymmetry but before detecting any potential secondary defects, such as cell cycle arrest. As shown above, at D5 in cal1 KD GSCs, CID asymmetry was compromised (Figures 6B–D), but measurement of the mitotic index did not show decreased mitotic index in cal1 KD GSCs, compared with Ctrl KD GSCs (tub-Gal80ts, nos-Gal4>mcherry shRNA) at D5 (Figure 6G, Table S12). However, the average GSC number per testis had a reduction at D5 (Figure 6H, Table S12), raising a possibility for GSC self-renewal failure. Next, we examined expression of a key transcription factor required for GSC maintenance, Stat92E (Feng et al., 2018; Kiger et al., 2001; Leatherman and Dinardo, 2008, 2010; Tarayrah et al., 2015; Tulina and Matunis, 2001). Indeed, Stat92E showed an overall decreased level in cal1 KD GSCs compared to the Ctrl GSCs (Figure 6I–J).
Consistent with decreased Stat92E in cal1 KD GSCs, there was a significant loss of GSCs in cal1 KD testes at D10, compared to the Ctrl testes under the same condition (Figures S6E–G, Table S13). At D10, the mitotic index also decreased in cal1 KD GSCs compared to the Ctrl GSCs, suggesting that prolonged cal1 KD could lead to defective mitosis (Figure S6D). Finally, an increase of the hub area was detected in cal1 KD testes compared to the Ctrl testes (hub regions in Ctrl versus cal1 KD in Figure S6F and S6G, Figure S6H, Table S14), likely as a secondary defect due to GSC loss, as reported previously (Dinardo et al., 2011; Gonczy and DiNardo, 1996; Monk et al., 2010; Tarayrah et al., 2015; Tazuke et al., 2002; Xie et al., 2015). In summary, these results demonstrate that knockdown of the CAL1 chaperone in adult testes leads to symmetric CID distribution at individual sister centromeres followed by an overall symmetric CID segregation pattern in GSCs. Under this condition, we also detected decreased immunostaining signals of Stat92E, whose activity is required for GSC maintenance (Figure S6I).
Depolymerizing Microtubules Disrupts the Temporal Asymmetry of Microtubules and Results in Randomized Sister Chromatid Segregation
In order to investigate how temporal asymmetries in microtubules may regulate asymmetric sister chromatid segregation in GSCs, we utilized a microtubule depolymerizing drug Nocodazole (NZ) to acutely destabilize microtubules in a temporally controlled manner. We applied NZ on adult testes ex vivo followed by washing out and immunostaining (Figure 7A, STAR Methods). Here, GSCs were arrested at the G2/M transition when treated with NZ, but were released and progressed through mitosis in a time-dependent manner following washing out (Figures 7A and S7A).
Figure 7: Depolymerizing microtubules disrupts the temporal microtubule asymmetry and results in randomized sister chromatid segregation.
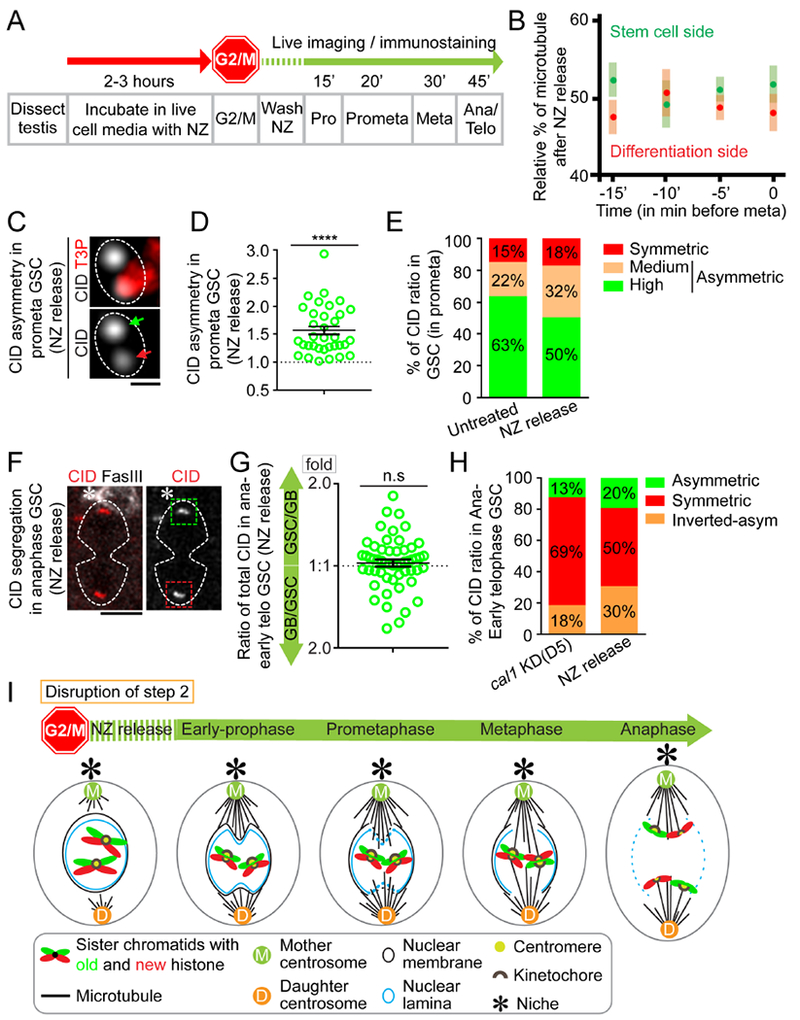
(A) A regime of Nocodazole (NZ) treatment experiment whereby testes were treated with NZ for two to three hours to arrest GSCs primarily at the G2/M transition. Immediately after washing out NZ, GSCs progress into different stages of mitosis in a time-dependent manner. (B) Immediately after releasing from NZ treatment, microtubules nucleate from both mother centrosome and daughter centrosome almost simultaneously. Percentage of microtubules emanated from mother centrosome versus daughter centrosome were quantified in GSCs (n= 10, Table S14), using α-Tubulin-GFP signals with 3D reconstructed montage made from live cell imaging, similar to quantification in Figure 4D–E. (C) In prometaphase GSCs immediately after releasing from NZ treatment, using Airyscan resolved sister centromeres display asymmetric distribution (green arrow: stronger centromere, red arrow: weaker centromere). (D) Quantification of individual pairs of resolved sister centromeres in prometaphase GSCs immediately after releasing from NZ treatment: 1.56± 0.08-fold (n= 34), Table S15. (E) Percentages of CID ratios at resolved individual sister centromeres in prometaphase with different categories in untreated GSCs (n= 65) and GSCs immediately after releasing from NZ arrest (n= 34). (F) Randomized CID segregation pattern in anaphase or early telophase GSCs immediately after releasing from NZ treatment, quantified in (G): 1.05± 0.03-fold, n= 54, Table S16. (H) Percentage of GSCs showing conventional asymmetric (see Supplemental Information for classification, GSC side/GB side≥ 1.2), symmetric (0.9< GSC side/GB side < 1.2), and inverted asymmetric (GSC side/GB side≤ 0.9, i.e. more toward GB side than to the GSC side) patterns of CID segregation at anaphase or early telophase, in cal1 KD (disrupts Step 1) and immediate release from NZ treatment (NZ release, disrupts Step 2). More symmetric segregation patterns are seen in cal1 KD compared to NZ release; both conventional asymmetric and inverted asymmetric patterns are detected with NZ release, suggesting the segregation pattern is randomized. (I) A cartoon depicting randomized sister chromatids segregation upon disrupting Step 2 without affecting Step 1. Asterisk: hub. All ratios= Avg± SE. P-value: paired t test. ****: P< 10−4; n.s: not significant. Scale bars: 0.5μm (C), 5μm (F).
Immediately after washing out NZ, GSCs entered mitosis with compromised temporal asymmetry of microtubules, as comparable microtubules emanated from both the mother centrosome and the daughter centrosome (Figures 7B, Movie S13, Table S15). Furthermore, the sequential disassembly of nuclear lamina detected in untreated GSCs (Figures 5A and 5C, Figure S5F–I) was now undetectable in treated GSCs after releasing from NZ-induced arrest (α-Tubulin and Lamin in Figures S7B–C). These results indicate that NZ treatment disrupted temporal asymmetry of microtubules and likely led to loss of sequential disassembly of nuclear lamina. Noticeably, in treated GSCs, the centrosomes were duplicated and both centrosomes were oriented perpendicular to the GSC-niche interface at early prophase (α-Tubulin in Figure S7B), whereas in SGs duplicated centrosomes were in proximity with each other at early prophase (α-Tubulin in Figure S7D) and were at the opposite sides only later during mitosis (α-Tubulin signals in Figure S7E). This observation is consistent with a previous report showing that centrosomes in GSCs duplicate and migrate during the G2 phase and prior to mitosis (Yamashita et al., 2003). This observation also confirmed that the NZ treatment followed by washing out specifically changed the temporal asymmetry of microtubules emanated from mother versus daughter centrosomes, but not the centrosome duplication and migration processes.
Next, we examined CID distribution at individual sister centromeres in prometaphase GSCs and found that this asymmetry was largely undisrupted (Figures 7C–E and S7F, Table S16). These results demonstrate that NZ treatment did not perturb the establishment of asymmetric sister centromeres (Step 1 in Figure 6A). However, asymmetric CID segregation in anaphase and early telophase GSCs was largely compromised (Figures 7F–G, Table S17). Overall, treated GSCs showed a high incidence of symmetric CID segregation pattern. Both asymmetric (more CID toward GSC side than to GB side) and inverted asymmetric (more CID toward GB side than to GSC side) patterns could also be detected but at lower ratios compared to the symmetric pattern (Figures 7G–H). However, in NZ treated GSCs both asymmetric ratios were greater than those ratios in cal1 KD GSCs (Figure 7H). These results suggest that Step 2 was likely disrupted with little to no effect on Step 1 (Figure 6A) using NZ treatment. Furthermore, when Step 1 was disrupted in cal1 KD GSCs, most GSCs displayed symmetric CID segregation patterns. In contrast, when Step 2 was disrupted without affecting Step 1, randomized CID segregation patterns were detectable including both asymmetric and inverted asymmetric patterns (Figure 7H). Moreover, asymmetric individual pairs of sister centromeres with a randomized orientation could be observed in metaphase GSCs soon after NZ release (Figures S7G–H). Together, these results suggest that the temporal asymmetry of microtubules is required for asymmetric segregation of sister chromatids (Figure 7I).
Consistent with the hypothesis that NZ treatment randomizes sister chromatid segregation, both old and new histone H3 displayed randomized patterns at anaphase or telophase GSCs immediately after NZ release (Figures S7I–J, Table S18). Taken together, these results suggest that the temporal asymmetries from the mitotic machinery are required for segregating the asymmetric sister centromeres and histones during the process of asymmetric sister chromatid inheritance (Figure S7K).
DISCUSSION
Here, we show that asymmetrically dividing GSCs utilize a cohort of cellular mechanisms to segregate epigenetically distinct sister chromatids. The previously identified asymmetrically inherited centrosomes (Yamashita et al., 2007), the herein reported temporally asymmetric microtubules, polarized NEBD, and asymmetric sister kinetochores serve as an axis of asymmetric factors of the mitotic machinery. Meanwhile, asymmetric H3 (Tran et al., 2012) and H4 (Wooten et al., 2019), differential phosphorylation at Thr3 of H3 (Xie et al., 2015), as well as the herein reported asymmetric sister centromeres are a series of epigenetic asymmetries between sister chromatids. Together, temporally asymmetric microtubules coordinate with asymmetric sister centromeres to ensure asymmetric attachment and segregation of sister chromatids, in order to generate two epigenetically distinct daughter cells, a phenomenon consistent with what has been previously hypothesized (Kahney et al., 2017; Lansdorp, 2007; Malik, 2009; Yamashita, 2013). We propose this series of interactions represent either a stem cell-specific or an ACD-specific phenomenon during the asymmetric divisions of Drosophila male GSCs (Figure 5H).
Here, both asymmetric microtubules and epigenetically different sister centromeres act together to ensure asymmetric sister chromatid segregation (Figure 6A). In order to understand how key aspects of this process are regulated, we designed several experiments trying to investigate Step 1 and Step 2 in this model. First, using temporally controlled knockdown experiments we demonstrated that the CID asymmetry in GSCs requires the CAL1 chaperone. When Step 1 is disrupted when cal1 is knocked down, individual pairs of sister centromeres show symmetry and the symmetric segregation pattern of overall CID is prevalent in anaphase or early telophase GSCs (Figures 6B–F). We then developed a drug arrest and release procedure to test whether recognition of the CID asymmetry depends on temporally asymmetric microtubules. Our results demonstrate when temporally asymmetric microtubule activity is compromised using NZ, asymmetry between each individual pairs of sister centromeres is preserved (Figures 7D–F and S7F). However, loss of temporally asymmetric microtubules severely affected asymmetric attachment and segregation of sister centromeres, leading to a range of CID segregation patterns from asymmetric, inverted asymmetric to symmetric patterns (Figure 7H–I). Because male flies have two sex chromosomes (X and Y chromosomes) and two major autosomes (2nd and 3rd chromosomes), even randomized segregation of sister chromatids could lead to an asymmetric pattern, albeit at a low percentage as shown previously (Xie et al., 2015). Accordingly, the low occurrence of the asymmetric patterns should have the opposite polarities with comparable percentages, which was indeed what we observed in anaphase or early telophase GSCs immediately after releasing from NZ treatment (Figure 7I). These results demonstrate that these two steps could be uncoupled; disruption of either step could lead to largely symmetric segregation pattern of sister centromeres at anaphase or early telophase, but distinct patterns at resolved individual pairs of sister centromeres in prophase or prometaphase GSCs.
In general, ACD gives rise to two daughter cells with distinct cell fates, which could occur during development, as well as tissue homeostasis and regeneration of multicellular organisms (Clevers, 2005; Kahney et al., 2017; Knoblich, 2010; Morrison and Kimble, 2006; Venkei and Yamashita, 2018). Many studies have shed light on how these different cell fates could be achieved in stem cells: While extrinsic cues could contribute to this asymmetric outcome after ACD, intrinsic factors that are asymmetrically segregated during ACD could also be critical for distinct cell fate decisions. More than four decades ago, an “immortal strand” hypothesis proposed that stem cells could retain “immortal” DNA strands (Cairns, 1975) to minimize the incidence of mutations, which may arise during pathological progression or aging. However, later in vivo studies using more rigorous methods and precise labeling have challenged this hypothesis (Klar and Bonaduce, 2013; Sauer et al., 2013; Schepers et al., 2011; Yadlapalli and Yamashita, 2013). On the other hand, examples of biased chromosome segregation have been reported in multiple stem cell systems, including both invertebrates and vertebrates (Rocheteau et al., 2012; Yadlapalli and Yamashita, 2013). It has been proposed that epigenetic differences between sister chromatids, especially at the centromeric region, could direct the asymmetric outcome during ACD (Klar, 1994, 2007; Lansdorp, 2007). It is conceivable that the centromere and kinetochore asymmetry may provide cellular mechanisms for selective sister chromatid segregation during ACD (Yadlapalli and Yamashita, 2013). Interestingly, asymmetric kinetochores were reported previously as a mechanism to define a specific lineage in yeast Saccharomyces cerevisiae (Thorpe et al., 2009). Yet, it remains unclear whether a similar phenomenon could occur in multicellular organisms. Here, our findings not only provide in vivo evidence in support of these hypotheses, but also reveal the cellular and molecular specificities of such selective chromatid segregation patterns in a well characterized adult stem cell model.
The “meiotic drive” hypothesis has proposed that the allele with a stronger centromere is retained in the oocyte while the allele with a weaker centromere is segregated to polar bodies for degeneration during female meiosis (Henikoff et al., 2001; Malik and Henikoff, 2009; Pardo-Manuel de Villena and Sapienza, 2001a, b). It has been shown that in female mice, the stronger kinetochore often associates with the longer, “selfish” centromere and has more affinity to the meiotic spindle. Moreover, the meiotic spindle itself has been asymmetrically modified due to polarized signaling (Akera et al., 2017; Chmatal et al., 2014; Iwata-Otsubo et al., 2017; Kursel and Malik, 2018). Recently, it has also been shown that a microtubule motor protein regulates the “meiotic drive” in maize, indicating a role for microtubules in selective attachment to centromeres in this process (Dawe et al., 2018; Schroeder and Malik, 2018).
Up to date, it remains unclear whether these asymmetric functions of the kinetochore and centromere found in meiosis could act in mitosis, in order to regulate sister chromatid segregation. Here, our results show both commonalities and distinctions in mitotic stem cells, compared to the “meiotic drive”. First, in meiosis, the centromere difference occurs between specific homologous chromosomes, whereas in mitosis, it occurs between genetically identical sister chromatids. Second, in meiosis, the microtubule itself does not have temporal asymmetries, but has different modifications instead. In contrast, in mitosis, temporal differences in microtubule nucleation activity from mother versus daughter centrosomes could be detected, which bias their attachment to epigenetically distinct centromeres. Third, the biological outcomes are different: Meiotic drive leads to retention of a particular allele with a stronger kinetochore and a “selfish” centromere to the haploid oocyte, whereas these events in a mitotic cell leads to two diploid cells, each with distinct epigenetic information in preparation for potentially differential gene expression and distinct cell fates. We speculate that this series of spatiotemporally regulated events in the mitotic stem cells a ‘mitotic drive’ phenomenon. Although the meiotic drive and the herein reported ‘mitotic drive’ have distinct features, they also share certain commonalities at the cellular level. For example, in both processes, the centromere and the microtubule differences serve as key mechanisms to ensure differential inheritance of chromosomes.
In summary, our results demonstrate that the asymmetry between sister chromatids tightly coincides and coordinates with the asymmetric components of mitotic machinery. Together, this spatiotemporally regulated axis of asymmetries allows for differential segregation of epigenetically distinct sister chromatids. In summary, our discovery helps further understanding of a fundamental biological question regarding how the mitotic machinery could distinguish and ensure asymmetric partitioning of epigenetic information. This work also opens a new avenue to study whether and how similar mechanisms might be used in other contexts of multicellular organisms, such as other asymmetrically dividing cells or other stem cells, in order to generate cells with distinct identities.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xin Chen (xchen32@jhu.edu).
Materials Availability Statement: Fly lines generated in this study will be available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Transgenic Drosophila melanogaster (fly) strains
Fly stocks were raised using standard Bloomington medium at 18°C, 25°C, or 29°C as noted. The following fly stocks were used: hs-flp on the X chromosome (Bloomington Stock Center BL-26902), nos-Gal4 on the 2nd chromosome (Van Doren et al., 1998), UASp-FRT-H3-GFP-PolyA-FRT-H3-mKO on the 3rd chromosome as reported previously (Tran et al., 2012), UASp-FRT-H3-mCherry -PolyA-FRT-H3-eGFP on the 3rd chromosome, UAS-α-Tubulin-GFP (Bloomington Stock Center BL-7373) on the 3rd chromosome, GFP-cid (Bloomington Stock Center BL-25047) on the 2nd chromosome, UAS-cal1-shRNA (Bloomington Stock Center BL-41716) on the 3rd chromosome, UAS-cal1-shRNA (Bloomington Stock Center BL-55730) on the 2nd chromosome, and nos-Gal4 (without VP16)/Cyo; tub-Gal80ts/TM6B (from Yukiko Yamashita, University of Michigan, Ann Arbor, Michigan, USA).
Generating knock-in Drosophila melanogaster (fly) strains
In collaboration with Fungene Inc. (Beijing, China), the following fly lines were generated using the CRISPR-Cas9 technology: CG613329 (cid) with Dendra2 tag at the internal site (between 118th - 119th codon), in order to generate the following fusion protein: CID N term-Dendra2-CID C term; CG5148 (cal1) with Dendra2 tag at the 5’ immediately downstream of the START codon, in order to generate the following fusion protein: Dendra2-CAL1; CG9938 (Ndc80) with Dendra2 tag at the 5’ immediately downstream of the START codon, in order to generate the following fusion protein: Dendra2-NDC80; CG6944 (Lamin B) with mCherry tag at the 3’ immediately before the STOP codon, in order to generate the following fusion protein: Lamin-mCherry. Related to Figure 1, Figure 2, Figure 3, Figure 5, Figure7, Figure S1, Figure S2, Figure S3, Figure S4, Figure S6, and Figure S7.
METHOD DETAILS
Immunostaining
Immunofluorescence staining was performed using standard procedures (Hime et al., 1996; Tran et al., 2012). For immunofluorescence staining, testes from young (0-2 day old) flies were dissected in PBS buffer. Samples were then fixed in 4% formaldehyde in phosphate-buffered saline (PBS) for 10 minutes at room temperature, washed thrice for 15 minutes per wash in PBST (PBS with 0.1% Triton X-100) for at least 10 minutes and blocked for at least 30 minutes in PBST with 3% bovine serum albumin at room temperature. Samples were incubated for 16 hours at 4°C in primary antibodies. Then sample were washed thrice for 15 minutes per wash in PBST and incubated in a 1:1000 dilution of Alexa Fluor-conjugated secondary antibody (from Molecular Probes) in PBST for 2 hours at room temperature. Sample were washed quickly three times with PBST and then four times 10 minutes with PBST. Samples were mounted for microscopy in Vectashield antifade mounting medium (Vector Laboratories, Cat# H-1400) with/without DAPI, and examined using Zeiss LSM 780 confocal microscope, or Zeiss LSM 800 confocal microscope with Airyscan with 63x oil immersion objectives. Images of separate fluorochromes from multiply stained tissues were collected individually and combined using Fiji and Adobe illustrator (Adobe).
Chromosome spreading
Chromosome spreading was performed to better visualize sister centromeres and sister kinetochores at prophase and prometaphase. Adult Drosophila testes were dissected in Schneider’s medium and incubated in a hypotonic solution (0.5% sodium citrate) for 5 minutes (min) at room temperature (RT). Testes were then fixed in freshly prepared 4% paraformaldehyde in PBS for 5-7 min at RT on a Superfrost plus slide, a cover slip was placed on top of fixed tissue, and squashed as hard as possible with a thumb. Testes were then frozen by immersing them in liquid nitrogen, the cover slip was popped off using a razor blade, followed by immediate incubation in chilled 95% ethanol (−20°C) for 10 min. The immobilized testes were washed three times with 1x PBST, 10 min each time, followed by overnight incubation with primary antibodies in blocking solution (1x PBST with 3% BSA) at 4°C. The testes were then washed three times with 1x PBST, 15 min each time, followed by incubation with secondary antibodies for two hours at RT, washed three times with 1x PBST, 15 min each time, and mounted in Vectashield without DAPI (Vector, Cat# H-1400). Related to Figure 1, Figure 2, Figure 3, Figure 6, Figure 7, Figure S1, Figure S2, Figure S3, Figure S6, and Figure S7.
Heat shock scheme
Flies with UASp-dual color histone transgenes were paired with the nos-Gal4 driver. Flies were raised at 18°C throughout development until adulthood to avoid pre-flip (Tran et al., 2012). Before heat shock, 1-3 day old males were transferred to vials that had been air dried for 24 hours. Vials were submerged underneath water up to the plug in a circulating 37°C water bath for 90 minutes and recovered in a 29°C incubator for indicated time before dissection for immunostaining experiments. Related to Figure S7.
Photoconversion of Dendra2 protein
Drosophila testes from CID-Dendra2 knock-in strain were dissected and mounted in the live cell medium in a fluorodish, and put under the LSM 780 microscope. The 40X objective was used, the region of interest at the apical tip of the testis was selected; the 405 laser at 10-15% laser power for 30sec/pulse and ~100 iterations were used to photoconvert Dendra2 protein from green fluorescent protein to red fluorescent protein. The photoconversion efficiency was measured by quantifying amount of green Dendra2 before photoconversion (Db) and the amount of green Dendra2 immediately after photoconversion (Da) using the following formula: Photoconversion efficiency= (Db-Da)/Db X 100 %. Related to Figure 2 and Figure S2.
Live cell imaging
To examine the temporal dynamics of cellular processes during asymmetric GSC divisions, we conducted live cell imaging with high temporal resolution (e.g. 30sec, 60sec or 5min interval as mention in the figure legend and supplemental movie legend). To perform live cell imaging, adult Drosophila testes were dissected in a medium containing Schneider’s insect medium with 200 μg/ml insulin, 15% (vol/vol) FBS, 0.6x pen/strep, with pH value at approximately 7.0, which we called “live cell medium”. Testes were then placed on a Poly-D-lysine coated FluoroDish (World Precision Instrument, Inc.), which contains the live cell medium as described.
All movies were taken using spinning disc confocal microscope (Zeiss) equipped with an evolve−™ camera (Photometrics), using a 63x Zeiss objective (1.4 NA) at 29°C. The ZEN 2 software (Zeiss) was used for acquisition with 2×2 binning. Mitotic cells were used to reconstruct 3-D movies using Imaris software (Bitplane). All videos for live cells are shown in Movie S1 – Movie S7 and Movie S13.
High-resolution live snapshot
To examine the temporal dynamics of cellular processes and the interaction between cellular components during asymmetric GSC divisions, we conducted live snapshot cell imaging with a high spatial resolution (Airyscan). To perform live snapshot, adult Drosophila testes, either expressing GFP-α-tubulin (Figure 4B) or GFP-α-tubulin and Lamin-mCherry (Figure 5C), were dissected in live cell medium (containing Schneider’s insect medium with 200 μg/ml insulin, 15% (vol/vol) FBS, 0.6x pen/strep, with pH value at approximately 7.0). Testes were then placed on a Poly-D-lysine coated FluoroDish (World Precision Instrument, Inc.), which contains the live cell medium as described. Testes were incubated at room temperature for ~ 2-3 hrs (incubate longer if needed) before imaging to stabilize the tissue in ex-vivo condition and to minimize the interference due to movement. To see kinetochores are attached to microtubules (K-fiber) adult’s testes expressing mCherry-α-tubulin and CID-Dendra2 were dissected and placed into ice (cold) briefly for ~5 min. Brief cold treatment depolymerizes most of the unattached microtubules but not attached microtubules; this would allow better visualization of K-fibers (Figure 5F–5G). All live snapshot was taken using Zeiss LSM 800 confocal microscope with AiryScan superresolution module equipped with highly sensitive GaAsP (Gallium Arsenide Phosphide) detectors using a 63x Zeiss objective (1.4 NA) at ~20°C. Related to Figure 4, Figure 5 and Figure S4.
Fix cell imaging
To examine the spatial dynamics of cellular processes and spatial localization of cellular component during asymmetric GSC divisions, we conducted fix cell imaging with high spatial resolution. After immunostaining, images were taken using Zeiss LSM 780 confocal microscope, or Zeiss LSM 800 confocal microscope with Airyscan with 63x oil immersion objectives. Airyscan high resolution imaging was performed to resolve sister centromeres (Figures 1G–H, 2C, 6B, 7C, S1H–I, S2B, S5G, S5L, S6C, and 7F–H), sister centromeres and sister kinetochores (Figure 3D–E and S3A–B), to detect microtubules (Figures 4B, 5C, and S4A), NEBD (Figures 5A–E, S5) and to detect microtubules-centromeres attachment (Figure 5 F–G) in normal cells, as well as to resolve sister centromeres (Figures 6B, S6C, S7C and S7F–H) in cal1-KD and NZ release, to detect microtubules-centromeres attachment (Figures S7G), and loss of polarized NEBD (Figures S7B–E) in NZ-treated cells. Images were processed using Imaris software (3D image reconstruction) and Fiji software (to quantify the total amount of protein “RawIntDen” or to generate maximum intensity projection).
Knockdown of cal1 in the germline of adult testes
The UAS-call-shRNA flies were crossed with nanos-Gal4 (without VP16); tub-Gal80ts flies to generate tub-Gal80ts, nos-Gal4 (Ctrl), tub-Gal80ts, nos-Gal4>mCherry shRNA (Ctrl KD), and tub-Gal80ts, nos-Gal4>cal1 shRNA (cal1 KD) male flies. Crosses were maintained at 18°C (permissive temperature for Gal80ts) to keep the Gal4 repressed therefore no CAL1 knockdown during development. After eclosion flies were shifted to the 29°C (restrictive temperature for Gal80ts) for 2-10 days. Related to Figure 6 and Figure S6.
Disruption of microtubule asymmetry
To disrupt the temporal asymmetry in microtubules activity at mother and daughter centrosomes, testes were briefly (2-3 hours) treated with microtubule depolymerizing drug nocodazole (NZ), and then NZ was washed out to allow cells to resume the cell cycle. We considered that #1. This would disrupt pre-established microtubule asymmetry in GSCs, and #2. When cell resume cell cycle after NZ wash, microtubules activity at mother and daughter centrosome would be nearly symmetric.
Prior to dissection, Nocodazole (NZ) solution was freshly prepared by adding 1 μl of 2 mg/ml stock solution of NZ in DMSO per 200 μl of “live cell media” for a final NZ concentration at 10μg/ml. This solution was left in the dark at room temperature (RT) until needed. Testes were dissected in Schneider’s insect media as quickly as possible and transferred to tubes where the excess media was carefully removed. After removing the Schneider’s media, 50 μl of NZ solution was added to each tube, which was left open in darkness at RT. Every hour, the old NZ solution was removed and 50 μl of new NZ solution was added for a total of three exchanges or four total hours in NZ solution. At the end of the four hours, the NZ solution was removed and 1 ml of Schneider’s insect media was added to each tube except the batch for immediate fixation after NZ, for which 4% formaldehyde was added to the tube instead. For the tubes where NZ was washed out, the media was removed and fresh media was added, repetitively for a total of at least five washes within 15 minutes. Testes were then fixed immediately after washout to catch prophase cells (15 min after release), prometaphase or metaphase cells (30 min after release), metaphase or anaphase cells (45 min after release), and anaphase or telophase cells (60 to 75 min after release), see Figures 7A and S7A. Related to Figure 7 and Figure S7.
QUANTIFICATION AND STATISTICAL ANALYSIS
The 3D quantification of movies and fix images
To quantify the amount of proteins segregated during asymmetric GSC divisions and symmetric SG divisions, we conducted a 3D quantification by measuring the fluorescence signal in each plane from the Z-stack (e.g. Figure S1A). The 3D quantification was done at different cell cycle stages (as labeled in the corresponding figures) in GSCs and SGs (e.g. 8-cell cyst), using time lapse movie with cid-Dendra2, Dendra2-cal1, Dendra2-Ndc80 knock-in lines, CID-GFP, α-tubulin-GFP, and UASp-FRT-H3-mCherry -PolyA-FRT-H3-eGFP transgenic lines, respectively. No antibody was added to enhance Dendra2, GFP or mCherry signals for quantification.
The fixed immunostaining images used fluorescence signal of Dendra2 (CID, CAL1, and NDC80) or antibodies recognizing endogenous CID. The 3D quantification of the fluorescence signal was done manually. Un-deconvolved raw images as 2D Z-stacks were saved as un-scaled 16-bit TIF images, and the sum of the gray values of pixels in the image (“RawIntDen”) was determined using Fiji (Image J). A circle was drawn to include all fluorescence signal (marked by Dendra2, GFP or mCherry), and an identical circle was drawn in the hub region as the background. The gray values of the fluorescence signal pixels for each Z-stack (Foreground signal, Fs) was calculated by subtracting the gray values of the background signal pixels (Background signal, Bs) from the gray values of the raw signal pixels (Raw signal, Rs). The total amount of the fluorescence signal in the nuclei was calculated by adding the gray values of the fluorescence signal from all Z-stacks. The total amount of the fluorescence signal (Fs) in the nuclei with Z-stacks (Z1+…+Zn): Fs (Z1+…+Zn) = [(Rs-Bs)1+…+(Rs-Bs)n]. All quantification of fluorescence signal was done using this method.
Define different categories of asymmetric
To define different categories (highly asymmetric, medium asymmetric, and symmetric) for sister centromeres in prometaphase, we used those ratios in symmetrically dividing SGs (SG1/SG2) to define the symmetric range. For example, in prometaphase, ratios above {mean + SE [1.161 + 0.015 = 1.176 (~1.2)]} are called ‘asymmetric’ and below are called ‘symmetric’. Among the asymmetric group, we further classified them into two categories, medium asymmetric (between 1.2- to 1.4-fold difference) and highly asymmetric (> 1.4-fold difference). To define different categories (highly asymmetric, medium asymmetric, and symmetric) for sister kinetochores in prometaphase, we used those ratios in symmetrically dividing SGs (SG1/SG2) to define the symmetric range. For example, in prometaphase, ratios above {mean + SE [1.19 + 0.02 = 1.21 (~ 1.2)]} are called ‘asymmetric’ and below are called ‘symmetric’. Among the asymmetric group, we further classified them into two categories, medium asymmetric (between 1.2- to 1.4-fold difference) and highly asymmetric (> 1.4-fold difference). Related to Figure 1, Figure 2, Figure 3, Figure 6, Figure 7, Figure S1, Figure S2, Figure S3, Figure S6, and Figure S7.
Define conventional and inverted asymmetries
For CID inheritance patterns in anaphase-to-early telophase, ratios above {mean + SE [1.056 + 0.017 = 1.073 (~1.1)]} are called ‘asymmetric’ and below are called ‘symmetry.
To define different categories of symmetric, conventional asymmetric and inverted asymmetric for CID inheritance in anaphase or early telophase GSCs, we used those ratios in symmetrically dividing SGs (SG1/SG2) to define the symmetric range. For example, in anaphase or early telophase, ratios above {mean + SE [1.056 + 0.017 = 1.073 (~1.1)]} are called ‘conventional asymmetry’, ratios below {SG2/SG1 mean − SE [0.952 − 0.017 = 0.935 (~0.9)]} are called ‘inverted asymmetry’, and ratio between 0.9 – 1.1 are called ‘symmetry’. Related to Figure 7.
Statistics
Statistics analysis (paired t test, with a 95% confidence interval) was performed in Prism 6 (GraphPad). Data are presented as Average ± SE and significant difference between two groups were noted by asterisks (**** P < 10−4). Correlation between the centromere and the kinetochore pairs were analyzed using Pearson correlation method in Prism 6 (Figure 3H). Number represent number of centromeres/chromosomes in Figure 1G–J, Figure 2C–E, Figure 3D–H, Figure 6B–D, Figure 7C–E, Figure S1H–I, Figure S2B, Figure S3A–B, Figure S6C, and Figure S7F. Number represent number of cells in Figure 1B–F, Figure 2B, Figure 3A–C, Figure 4D–E, Figure 6F–H, Figure 6J, Figure 7G–H, Figure S1G, Figure S2D, Figure S6A, Figure S6D–E, Figure S6H and Figure S7J.
DATA AND CODE AVAILABILITY
The published article includes all datasets generated or analyzed during this study. Original data for Supplemental movies and tables in the paper is deposited to Mendeley Data and available at: https://data.mendeley.com/datasets/vytyg7sdxx/3 (DOI: 10.17632/vytyg7sdxx.3).
Supplementary Material
Movie S1. Related to Figure 1: Asymmetric CID-Dendra2 segregation in GSC. Live cell imaging of the CID-Dendra2 in a Drosophila male GSC: This video is a 3D reconstruction of individual images (8 × 1-μm interval optical sections per frame), showing asymmetric CID-Dendra2 segregation during asymmetric GSC division. The video was acquired at 30sec intervals for 5-6 minutes from prophase to anaphase. Metaphase is used as a landmark to define time point zero, and other time points are labeled as minus minutes prior to metaphase. The quantification is shown in Figure 1D. Asterisk: hub. Scale bar: 2μm.
Movie S2. Related to Figure 1: Symmetric CID-Dendra2 segregation in SG. Live cell imaging of the CID-Dendra2 in a Drosophila male SG: This video is a 3D reconstruction of individual images (8 × 1-μm interval optical sections per frame), showing symmetric CID-Dendra2 segregation during symmetric SG division. The video was acquired at 30sec intervals for 5-6 minutes from prophase to anaphase. Metaphase is used as a landmark to define time point zero, and other time points are labeled as minus minutes prior to metaphase. The quantification is shown in Figure 1D. Scale bar: 2μm.
Movie S3. Related to Figure 4: Temporal asymmetry of α-Tubulin in GSC. Live cell imaging of α-Tubulin-GFP in a Drosophila male GSC: This video is a 3D reconstruction of individual images (14 × 1-μm interval optical sections per frame), showing temporal asymmetry in α-Tubulin-GFP activity at mother centrosome (labeled by green arrow) and daughter centrosome (labeled by red arrow). The mother centrosome is actively nucleating microtubules from mid- to late G2 phase (−240min to −50min), whereas the daughter centrosome nucleates microtubules upon mitotic entry (~ −50 min). Also, in prophase, the dominance of mother centrosome activity becomes less, while microtubules from the daughter centrosome start to increase (−35min to −5min). However, at metaphase microtubules from both sides become equal (0 min). The video was acquired at 5-min interval for 5-6 hrs from mid G2 to G1. Metaphase is used as a landmark to define time point zero, and other time points are labeled as minus minutes prior to metaphase. The quantification is shown in Figure 4D. Asterisk: hub. Scale bar: 5μm.
Movie S4. Related to Figure 4: Symmetric α-tubulin in SG. Live cell imaging of the α-tubulin-GFP in a Drosophila male SG: This video is a 3D reconstruction of individual images (8 × 1-μm interval optical sections per frame), showing neither the mother nor the daughter centrosome nucleates microtubules throughout G2 phase. However, both centrosomes start nucleating microtubules simultaneously upon mitotic entry (~ −35min). The video was acquired at 5-min interval for 5-6 minutes from G2/M to G1. Metaphase is used as a landmark to define time point zero, and other time points are labeled as minus minutes prior to metaphase. The quantification is shown in Figure 4E. Scale bar: 5 μm.
Movie S5. Related to Figure 4: A high-resolution time lapse movie of GSC to visualize asymmetric microtubule ‘poking in’ phenomenon. Live cell imaging of the α-tubulin-GFP in a Drosophila male GSC using LSM800 confocal microscopy equipped with Airyscan: This video is a 3D reconstruction of individual images (6 × 0.15μm interval optical sections per frame), showing microtubules spike “poking in” (labeled by green arrow) at GSC side at G2/M transition. The video was acquired at 5-min interval for 30 minutes from G2/M transition to prophase. The G2/M is time point zero; snapshots are shown in Figure S4A. Asterisk: hub. Scale bar: 5μm.
Movie S6. Related to Figure 5: The 3D reconstructed GSC in early-prophase showing NEBD at the GSC side. This video is a 3D reconstruction of a high-resolution airyscan individual fixed images (5 × 0.15μm interval optical sections per frame) of α-tubulin-GFP-expressing Drosophila male GSC at early prophase, immunostained with anti-Lamin B and anti-CID. A 3D reconstruction with α-tubulin-GFP (green), anti-Lamin B (magenta) and anti-CID (red) signals. This 3D reconstruction shows asymmetric nuclear envelope breakdown at the GSC side in early prophase. The maximum projection of the image is shown in Figure 5A. Asterisk: hub. Scale bar: 5μm.
Movie S7. Related to Figure 5: The 3D reconstructed GSC in late-prophase showing NEBD at the GSC side and “poking in” phenomenon at the GB side. This video is a 3D reconstruction of individual fixed images (10 × 0.15μm interval optical sections per frame) of α-tubulin-GFP-expressing Drosophila male GSC at late-prophase, immunostained with anti-Lamin B and anti-CID. A 3D reconstruction with α-tubulin-GFP (green), anti-Lamin B (magenta) and anti-CID (red) signals. This image shows that the nuclear envelope at stem cell side is broken, and the nuclear envelope at GB side is “poking in” at late-prophase. The maximum projection of the image is shown in Figure 5B. Asterisk: hub. Scale bar: 5μm.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Fascilin III (Fas III) | Developemental Studies Hybridoma Bank (DSHB) | RRID: AB_528238 |
| Mouse monoclonal anti-alpha Spectrin | Developemental Studies Hybridoma Bank (DSHB) | RRID: AB_528473 |
| Mouse monoclonal anti-Armadillo | Developemental Studies Hybridoma Bank (DSHB) | RRID: AB_528089 |
| Rat monoclonal anti-DE-cadherin | Developemental Studies Hybridoma Bank (DSHB) | RRID: AB_528120 |
| Mouse monoclonal anti-lamin | Developemental Studies Hybridoma Bank (DSHB) | RRID: AB_528336 |
| Rabbit polyclonal anti-CID | Active motif | RRID:AB_2793320 |
| Rat monoclonal anti-CID | Active motif | RRID:AB_2793749 |
| Rabbit monoclonal anti-H3T3P | Millipore | 05-746R |
| Rabbit anti-Vasa | Santa Cruz | SC-30210 |
| Mouse monoclonal anti-H3S10P | Abcam | AB-14995 |
| Guinea pig anti-Traffic jam | Kindly provided by Mark Van Doren, Johns Hopkins University, MD, USA | N/A |
| Rabbit anti-Stat92E | Kindly provided by Denise Montell, University of Santa Barbara, CA,USA | N/A |
| Chicken anti-CID | Kindly provided by Sylvia Erhardt, ZMBH, Heidelberg University, Heidelberg, Germany | N/A |
| Rabbit anti-WGA | Kindly provided by Joe Gall, Carnegie Institute, MD, USA | N/A |
| Experimental Models: Transgenic Organisms/Strains | ||
| D. melanogaster. UAS-GFP-CID | Bloomington Drosophila Stock Center | RRID: BDSC_25047 |
| D. melanogaster. UAS-α-tubulin-GFP | Bloomington Drosophila Stock Center | RRID: BDSC_7373 |
| D. melanogaster. hs-flp | Bloomington Drosophila Stock Center | RRID: BDSC_26902 |
| D. melanogaster. UAS-CAL1RNAi | Bloomington Drosophila Stock Center | RRID: BDSC_55730 |
| D. melanogaster. UAS-CAL1RNAi | Bloomington Drosophila Stock Center | RRID: BDSC_41716 |
| D. melanogaster. Nos-Gal4 | Kindly provided by Mark Van Doren, Johns Hopkins University, MD, USA | N/A |
| D. melanogaster. Nos-Gal4; tubulin-Gal80ts | Kindly provided by Yukiko Yamashita, University of Michigan, Ann Arbor, Michigan, USA | N/A |
| D. melanogaster. UAS-H3-mCherry-H3-GFP | Our lab has developed | N/A |
| Experimental Models: Knock-in Organisms/Strains | ||
| D. melanogaster. Dendra2-CAL1 | Our lab has developed | N/A |
| D. melanogaster. CID-Dendra2 | Our lab has developed | N/A |
| D. melanogaster. Dendra2-NDC80 | Our lab has developed | N/A |
| D. melanogaster. Lamin B-mCherry | Our lab has developed | N/A |
| Chemicals | ||
| FBS | Sigma | Cat#F2442 |
| Penicillin/Streptomycin | Sigma | Cat#P0781 |
| Nocodazole | Sigma | Cat#M1404 |
| DMSO | Sigma | Cat#D2650 |
| Mounting Medium | Vector | Cat# H-1400 |
| BSA | Cell Signaling Technology | Cat#9998 |
| Formaldehyde | Fisher Scientific | Cat#F79-500 |
| Schneider Medium | ThermoFisher Scientific | Cat# 21720024 |
| Insulin | ThermoFisher Scientific | Cat#12585014 |
| FluoroDish | World Precision Instrument, Inc. | Cat#FD35PDL |
| Software and algorithms | ||
| Adobe Illustrator CS6 | Adobe | N/A |
| Prism 6 | Graphpad | N/A |
| Fiji | NIH | N/A |
| EndNote | Clarivate Analytics | N/A |
| IMARIS | Bitplane | N/A |
| Data and Code Availability | ||
| Supplemental Data | Original/source data for [supplemental movies and tables] | Mendeley Data DOI: 10.17632/vytyg7sdxx.3 |
Highlights:
Sister chromatids contain asymmetric levels of CID in male Drosophila GSCs
Mother and daughter centrosomes sequentially nucleate microtubules
Asymmetric centrosomes preferentially attach to microtubules (MTs)
Centromeres and MTs coordinate to mediate non-random sister chromatid segregation
Acknowledgements:
We thank Y. Yamashita, S. Erhardt, M. Van Doren, R. Johnston and X.C. lab members for insightful suggestions. We thank J. Gall for WGA, S. Erhardt for anti-CID, R. Quinn for technical assistance, Johns Hopkins Integrated Imaging Center for confocal imaging. Supported by NIGMS/NIH T32GM007231 (J.S.), NIGMS/NIH R01GM112008 and R35GM127075, the Howard Hughes Medical Institute, the David and Lucile Packard Foundation, and Johns Hopkins University startup funds (X.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
The supplemental movies and tables have been deposited at Mendeley data and are available at: https://data.mendeley.com/datasets/vytyg7sdxx/3 (DOI: 10.17632/vytyg7sdxx.3).
Declaration of interests: The authors declare no competing interests.
References:
- Akera T, Chmatal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, and Lampson MA (2017). Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358, 668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, and Jenuwein T (2016). The molecular hallmarks of epigenetic control. Nat Rev Genet 17, 487–500. [DOI] [PubMed] [Google Scholar]
- Allshire RC, and Karpen GH (2008). Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9, 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amcheslavsky A, Nie Y, Li Q, He F, Tsuda L, Markstein M, and Ip YT (2014). Gene expression profiling identifies the zinc-finger protein Charlatan as a regulator of intestinal stem cells in Drosophila. Development 141, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, and Fuchs E (2006). Epidermal stem cells of the skin. Annu Rev Cell Dev Biol 22, 339–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, and Jansen LE (2014). The quantitative architecture of centromeric chromatin. Elife 3, e02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J (1975). Mutation selection and the natural history of cancer. Nature 255, 197–200. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM (2014). The kinetochore. Cold Spring Harb Perspect Biol 6, a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, and Desai A (2006). The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997. [DOI] [PubMed] [Google Scholar]
- Chen C, Cummings R, Mordovanakis A, Hunt AJ, Mayer M, Sept D, and Yamashita YM (2018). Cytokine receptor-Eb1 interaction couples cell polarity and fate during asymmetric cell division. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, and Mellone BG (2014). CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol 204, 313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen X, and Zheng Y (2013). The Nuclear Lamina Regulates Germline Stem Cell Niche Organization via Modulation of EGFR Signaling. Cell Stem Cell 13, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmatal L, Gabriel SI, Mitsainas GP, Martinez-Vargas J, Ventura J, Searle JB, Schultz RM, and Lampson MA (2014). Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol 24, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudakov DM, Lukyanov S, and Lukyanov KA (2007). Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc 2, 2024–2032. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, and Sullivan KF (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421. [DOI] [PubMed] [Google Scholar]
- Clevers H (2005). Stem cells, asymmetric division and cancer. Nat Genet 37, 1027–1028. [DOI] [PubMed] [Google Scholar]
- Dawe RK, Lowry EG, Gent JI, Stitzer MC, Swentowsky KW, Higgins DM, Ross-Ibarra J, Wallace JG, Kanizay LB, Alabady M, et al. (2018). A Kinesin-14 Motor Activates Neocentromeres to Promote Meiotic Drive in Maize. Cell 173, 839–850 e818. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, and Salmon ED (2006). Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, 969–982. [DOI] [PubMed] [Google Scholar]
- Dinardo S, Okegbe T, Wingert L, Freilich S, and Terry N (2011). lines and bowl affect the specification of cyst stem cells and niche cells in the Drosophila testis. Development 138, 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E, Pidoux A, and Allshire R (2005). Centromeric chromatin makes its mark. Trends in biochemical sciences 30, 172–175. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Beier NL, Gorgescu W, Tang J, Costes SV, and Karpen GH (2012). The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol 10, e1001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, and Straight AF (2008). Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol 183, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Shi Z, Xie J, Ma B, and Chen X (2018). Enhancer of polycomb maintains germline activity and genome integrity in Drosophila testis. Cell Death Differ 25, 1486–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, and Earnshaw WC (2014). The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30, 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Del Arco A, Edgar BA, and Erhardt S (2018). In Vivo Analysis of Centromeric Proteins Reveals a Stem Cell-Specific Asymmetry and an Essential Role in Differentiated, Nonproliferating Cells. Cell Rep 22, 1982–1993. [DOI] [PubMed] [Google Scholar]
- Gonczy P, and DiNardo S (1996). The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development 122, 2437–2447. [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, and Stuurman N (2007). Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, and Malik HS (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Platero JS, and van Steensel B (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci U S A 97, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime GR, Brill JA, and Fuller MT (1996). Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci 109 (Pt 12), 2779–2788. [DOI] [PubMed] [Google Scholar]
- Horvath P, and Barrangou R (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170. [DOI] [PubMed] [Google Scholar]
- Inaba M, Yuan H, Salzmann V, Fuller MT, and Yamashita YM (2010). E-cadherin is required for centrosome and spindle orientation in Drosophila male germline stem cells. PLoS One 5, e12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmatal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, and Black BE (2017). Expanded Satellite Repeats Amplify a Discrete CENP-A Nucleosome Assembly Site on Chromosomes that Drive in Female Meiosis. Curr Biol 27, 2365–2373 e2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahney EW, Ranjan R, Gleason RJ, and Chen X (2017). Symmetry from Asymmetry or Asymmetry from Symmetry? Cold Spring Harb Symp Quant Biol 82, 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai T, Williams D, and Spradling AC (2005). The expression profile of purified Drosophila germline stem cells. Dev Biol 283, 486–502. [DOI] [PubMed] [Google Scholar]
- Karpen GH, and Allshire RC (1997). The case for epigenetic effects on centromere identity and function. Trends Genet 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, and Fuller MT (2001). Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 294, 2542–2545. [DOI] [PubMed] [Google Scholar]
- Klar AJ (1994). A model for specification of the left-right axis in vertebrates. Trends Genet 10, 392–396. [DOI] [PubMed] [Google Scholar]
- Klar AJ (2007). Lessons learned from studies of fission yeast mating-type switching and silencing. Annu Rev Genet 41, 213–236. [DOI] [PubMed] [Google Scholar]
- Klar AJ, and Bonaduce MJ (2013). Unbiased segregation of fission yeast chromosome 2 strands to daughter cells. Chromosome Res 21, 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol 11, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel LE, and Malik HS (2018). The cellular mechanisms and consequences of centromere drive. Curr Opin Cell Biol 52, 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp PM (2007). Immortal strands? Give me a break. Cell 129, 1244–1247. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, and Dinardo S (2008). Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell 3, 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman JL, and Dinardo S (2010). Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol 12, 806–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS (2009). The centromere-drive hypothesis: a simple basis for centromere complexity. Prog Mol Subcell Biol 48, 33–52. [DOI] [PubMed] [Google Scholar]
- Malik HS, and Henikoff S (2009). Major evolutionary transitions in centromere complexity. Cell 138, 1067–1082. [DOI] [PubMed] [Google Scholar]
- McKinley KL, and Cheeseman IM (2016). The molecular basis for centromere identity and function. Nature reviews Molecular cell biology 17, 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, and Karpen GH (2011). Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet 7, e1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Nye J, Zhao H, and Dalal Y (2015). Chromatin Dynamics in Vivo: A Game of Musical Chairs. Genes (Basel) 6, 751–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, and Heun P (2011). Drosophila CENH3 is sufficient for centromere formation. Science 334, 686–690. [DOI] [PubMed] [Google Scholar]
- Miell MD, Fuller CJ, Guse A, Barysz HM, Downes A, Owen-Hughes T, Rappsilber J, Straight AF, and Allshire RC (2013). CENP-A confers a reduction in height on octameric nucleosomes. Nature structural & molecular biology 20, 763–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk AC, Siddall NA, Volk T, Fraser B, Quinn LM, McLaughlin EA, and Hime GR (2010). HOW is required for stem cell maintenance in the Drosophila testis and for the onset of transit-amplifying divisions. Cell Stem Cell 6, 348–360. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, and Kimble J (2006). Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068–1074. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, Andrews BS, and Margolis RL (1987). A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol 104, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, and Sapienza C (2001a). Female meiosis drives karyotypic evolution in mammals. Genetics 159, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, and Sapienza C (2001b). Nonrandom segregation during meiosis: the unfairness of females. Mamm Genome 12, 331–339. [DOI] [PubMed] [Google Scholar]
- Rieder CL (2005). Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to a draw. Chromosoma 114, 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, and Tajbakhsh S (2012). A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148, 112–125. [DOI] [PubMed] [Google Scholar]
- Sauer S, Burkett SS, Lewandoski M, and Klar AJ (2013). A CO-FISH assay to assess sister chromatid segregation patterns in mitosis of mouse embryonic stem cells. Chromosome Res 21, 311–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Vries R, van den Born M, van de Wetering M, and Clevers H (2011). Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J 30, 1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, and Lehner CF (2007). Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116, 385–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder CM, and Malik HS (2018). Kindr Motors Drive in Meiosis. Cell 173, 813–815 [DOI] [PubMed] [Google Scholar]
- Schueler MG, and Sullivan BA (2006). Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet 7, 301–313. [DOI] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, and Heidmann S (2007). Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17, 237–243. [DOI] [PubMed] [Google Scholar]
- Seale P, Ishibashi J, Holterman C, and Rudnicki MA (2004). Muscle satellite cell-specific genes identified by genetic profiling of MyoD-deficient myogenic cell. Dev Biol 275, 287–300 [DOI] [PubMed] [Google Scholar]
- Sheng XR, and Matunis E (2011). Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development 138, 3367–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon D, Podsypanina K, Yadav T, and Almouzni G (2017). Shaping Chromatin in the Nucleus: The Bricks and the Architects. Cold Spring Harb Symp Quant Biol 82, 1–14. [DOI] [PubMed] [Google Scholar]
- Tarayrah L, Li Y, Gan Q, and Chen X (2015). Epigenetic regulator Lid maintains germline stem cells through regulating JAK-STAT signaling pathway activity. Biol Open 4, 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazuke SI, Schulz C, Gilboa L, Fogarty M, Mahowald AP, Guichet A, Ephrussi A, Wood CG, Lehmann R, and Fuller MT (2002). A germline-specific gap junction protein required for survival of differentiating early germ cells. Development 129, 2529–2539. [DOI] [PubMed] [Google Scholar]
- Terry NA, Tulina N, Matunis E, and DiNardo S (2006). Novel regulators revealed by profiling Drosophila testis stem cells within their niche. Dev Biol 294, 246–257. [DOI] [PubMed] [Google Scholar]
- Terskikh AV, Miyamoto T, Chang C, Diatchenko L, and Weissman IL (2003). Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood 102, 94–101. [DOI] [PubMed] [Google Scholar]
- Thorpe PH, Bruno J, and Rothstein R (2009). Kinetochore asymmetry defines a single yeast lineage. Proc Natl Acad Sci U S A 106, 6673–6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Feng L, and Chen X (2013). Asymmetric distribution of histones during Drosophila male germline stem cell asymmetric divisions. Chromosome Res 21, 255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Lim C, Xie J, and Chen X (2012). Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science 338, 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulina N, and Matunis E (2001). Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 294, 2546–2549. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, and Lehmann R (1998). Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol 8, 243–246. [DOI] [PubMed] [Google Scholar]
- Venkei ZG, and Yamashita YM (2018). Emerging mechanisms of asymmetric stem cell division. J Cell Biol 217, 3785–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos LJ, Famulski JK, and Chan GK (2006). How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem Cell Biol 84, 619–639 [DOI] [PubMed] [Google Scholar]
- Westhorpe FG, and Straight AF (2014). The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb Perspect Biol 7, a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten M, Snedeker J, Nizami ZF, Yang X, Ranjan R, Urban E, Kim JM, Gall J, Xiao J, and Chen X (2019). Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nat Struct Mol Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nunez JK, and Doudna JA (2016). Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 164, 29–44. [DOI] [PubMed] [Google Scholar]
- Xie J, Wooten M, Tran V, Chen BC, Pozmanter C, Simbolon C, Betzig E, and Chen X (2015). Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline. Cell 163, 920–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Wooten M, Tran V, and Chen X (2017). Breaking Symmetry - Asymmetric Histone Inheritance in Stem Cells. Trends Cell Biol 27, 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadlapalli S, Cheng J, and Yamashita YM (2011). Drosophila male germline stem cells do not asymmetrically segregate chromosome strands. J Cell Sci 124, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadlapalli S, and Yamashita YM (2013). Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature 498, 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM (2013). Nonrandom sister chromatid segregation of sex chromosomes in Drosophila male germline stem cells. Chromosome Res 21, 243–254. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, and Fuller MT (2003). Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, and Fuller MT (2007). Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315, 518–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA (2011). Control of the embryonic stem cell state. Cell 144, 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Related to Figure 1: Asymmetric CID-Dendra2 segregation in GSC. Live cell imaging of the CID-Dendra2 in a Drosophila male GSC: This video is a 3D reconstruction of individual images (8 × 1-μm interval optical sections per frame), showing asymmetric CID-Dendra2 segregation during asymmetric GSC division. The video was acquired at 30sec intervals for 5-6 minutes from prophase to anaphase. Metaphase is used as a landmark to define time point zero, and other time points are labeled as minus minutes prior to metaphase. The quantification is shown in Figure 1D. Asterisk: hub. Scale bar: 2μm.
Movie S2. Related to Figure 1: Symmetric CID-Dendra2 segregation in SG. Live cell imaging of the CID-Dendra2 in a Drosophila male SG: This video is a 3D reconstruction of individual images (8 × 1-μm interval optical sections per frame), showing symmetric CID-Dendra2 segregation during symmetric SG division. The video was acquired at 30sec intervals for 5-6 minutes from prophase to anaphase. Metaphase is used as a landmark to define time point zero, and other time points are labeled as minus minutes prior to metaphase. The quantification is shown in Figure 1D. Scale bar: 2μm.
Movie S3. Related to Figure 4: Temporal asymmetry of α-Tubulin in GSC. Live cell imaging of α-Tubulin-GFP in a Drosophila male GSC: This video is a 3D reconstruction of individual images (14 × 1-μm interval optical sections per frame), showing temporal asymmetry in α-Tubulin-GFP activity at mother centrosome (labeled by green arrow) and daughter centrosome (labeled by red arrow). The mother centrosome is actively nucleating microtubules from mid- to late G2 phase (−240min to −50min), whereas the daughter centrosome nucleates microtubules upon mitotic entry (~ −50 min). Also, in prophase, the dominance of mother centrosome activity becomes less, while microtubules from the daughter centrosome start to increase (−35min to −5min). However, at metaphase microtubules from both sides become equal (0 min). The video was acquired at 5-min interval for 5-6 hrs from mid G2 to G1. Metaphase is used as a landmark to define time point zero, and other time points are labeled as minus minutes prior to metaphase. The quantification is shown in Figure 4D. Asterisk: hub. Scale bar: 5μm.
Movie S4. Related to Figure 4: Symmetric α-tubulin in SG. Live cell imaging of the α-tubulin-GFP in a Drosophila male SG: This video is a 3D reconstruction of individual images (8 × 1-μm interval optical sections per frame), showing neither the mother nor the daughter centrosome nucleates microtubules throughout G2 phase. However, both centrosomes start nucleating microtubules simultaneously upon mitotic entry (~ −35min). The video was acquired at 5-min interval for 5-6 minutes from G2/M to G1. Metaphase is used as a landmark to define time point zero, and other time points are labeled as minus minutes prior to metaphase. The quantification is shown in Figure 4E. Scale bar: 5 μm.
Movie S5. Related to Figure 4: A high-resolution time lapse movie of GSC to visualize asymmetric microtubule ‘poking in’ phenomenon. Live cell imaging of the α-tubulin-GFP in a Drosophila male GSC using LSM800 confocal microscopy equipped with Airyscan: This video is a 3D reconstruction of individual images (6 × 0.15μm interval optical sections per frame), showing microtubules spike “poking in” (labeled by green arrow) at GSC side at G2/M transition. The video was acquired at 5-min interval for 30 minutes from G2/M transition to prophase. The G2/M is time point zero; snapshots are shown in Figure S4A. Asterisk: hub. Scale bar: 5μm.
Movie S6. Related to Figure 5: The 3D reconstructed GSC in early-prophase showing NEBD at the GSC side. This video is a 3D reconstruction of a high-resolution airyscan individual fixed images (5 × 0.15μm interval optical sections per frame) of α-tubulin-GFP-expressing Drosophila male GSC at early prophase, immunostained with anti-Lamin B and anti-CID. A 3D reconstruction with α-tubulin-GFP (green), anti-Lamin B (magenta) and anti-CID (red) signals. This 3D reconstruction shows asymmetric nuclear envelope breakdown at the GSC side in early prophase. The maximum projection of the image is shown in Figure 5A. Asterisk: hub. Scale bar: 5μm.
Movie S7. Related to Figure 5: The 3D reconstructed GSC in late-prophase showing NEBD at the GSC side and “poking in” phenomenon at the GB side. This video is a 3D reconstruction of individual fixed images (10 × 0.15μm interval optical sections per frame) of α-tubulin-GFP-expressing Drosophila male GSC at late-prophase, immunostained with anti-Lamin B and anti-CID. A 3D reconstruction with α-tubulin-GFP (green), anti-Lamin B (magenta) and anti-CID (red) signals. This image shows that the nuclear envelope at stem cell side is broken, and the nuclear envelope at GB side is “poking in” at late-prophase. The maximum projection of the image is shown in Figure 5B. Asterisk: hub. Scale bar: 5μm.
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. Original data for Supplemental movies and tables in the paper is deposited to Mendeley Data and available at: https://data.mendeley.com/datasets/vytyg7sdxx/3 (DOI: 10.17632/vytyg7sdxx.3).


