Significance
The mechanisms that lead to establishment of pregnancy and formation of a functional placenta during early human development are poorly understood due to ethical and technical constraints limiting research, yet this period is when many pregnancies fail and placental pathologies are most likely initiated. We cultured donated human embryos to days 8, 10, and 12 postfertilization and, for each stage, performed single-cell RNA sequencing on the trophoblast cells that compose the early placenta and enclose the embryo proper as it implants. Over the 5 d of culture, 2 cell lineages emerged from a progenitor stem cell population. One was syncytial and produced placental hormones. The second became motile and expressed genes associated with the innate immune system and invasion.
Keywords: single-cell RNA sequencing, transcriptome, interferon, human endogenous retroviruses
Abstract
Single-cell RNA sequencing of cells from cultured human blastocysts has enabled us to define the transcriptomic landscape of placental trophoblast (TB) that surrounds the epiblast and associated embryonic tissues during the enigmatic day 8 (D8) to D12 peri-implantation period before the villous placenta forms. We analyzed the transcriptomes of 3 early placental cell types, cytoTB (CTB), syncytioTB (STB), and migratoryTB (MTB), picked manually from cultured embryos dissociated with trypsin and were able to follow sublineages that emerged from proliferating CTB at the periphery of the conceptus. A unique form of CTB with some features of STB was detectable at D8, while mature STB was at its zenith at D10. A form of MTB with a mixed MTB/CTB phenotype arose around D10. By D12, STB generation was in decline, CTB had entered a new phase of proliferation, and mature MTB cells had begun to move from the main body of the conceptus. Notably, the MTB transcriptome at D12 indicated enrichment of transcripts associated with IFN signaling, migration, and invasion and up-regulation of HLA-C, HLA-E, and HLA-G. The STB, which is distinct from the STB of later villous STB, had a phenotype consistent with intense protein export and placental hormone production, as well as migration and invasion. The studies show that TB associated with human embryos is in rapid developmental flux during peri-implantation period when it must invade, signal robustly to the mother to ensure that the pregnancy continues, and make first contact with the maternal immune system.
Implantation of the human embryo into the uterine wall is poorly understood and not closely paralleled in model organisms, such as the mouse. It is estimated that 40 to 60% of human conceptions fail, with the majority of the losses occurring just prior to or during implantation (1, 2), a process that is initiated soon after the blastocyst attaches to the uterine wall at about day 6 (D6) postconception. What little is known of early human embryo development and emergence of the earliest form of the placenta (3, 4) has come from histological studies performed more than 50 y ago (5–7), archived material (8), and parallels with nonhuman primates (8–10). The generally accepted view is that soon after the polar trophectoderm of the D6 to D7 blastocyst adheres to the uterine wall, a zone of invasive syncytiotrophoblast (STB) forms at the implantation site where it locally displaces uterine epithelial cells, thereby allowing the conceptus to penetrate the underlying stroma. The STB soon surrounds the conceptus and, as it advances into the decidualized endometrium, hollows out lacunae, which become filled with blood and uterine secretions that likely provision the conceptus (11–14). The trophoblasts of this tiny conceptus must also release sufficient human chorionic gonadotropin (hCG) to rescue the corpus luteum; otherwise, the pregnancy fails. By D12, columns of cytotrophoblast (CTB) begin to penetrate the STB to form primary villi, which will eventually branch, acquire cores of blood vessels and connective tissue, and form the emerging villous placenta, while the fate of the original STB layer remains unclear. All these events occur prior to the time that a new menstrual cycle would normally begin in a nonpregnant woman.
The above histological studies have only provided a glimpse of events that occur during the second week of pregnancy and have been unable to provide insights into the dynamic process of implantation and what can go wrong. Some models for understanding placental trophoblast emergence have shown promise. For example, human pluripotent stem cells exposed to BMP4 can be driven efficiently to what has been proposed to be trophoblast analogous to that encountered around the periphery of an implanting conceptus (15, 16). Trophoblast stem cells derived from either preimplantation blastocysts or first trimester villi (17) and from trophoblast organoids (18, 19) also can be induced to differentiate along trophoblast sublineages. Recently, a system has been described that allows human embryos to be cultured in vitro for up to 14 d (20, 21). Here we have used such a system to compare trophoblast lineages from embryos at increasing days in culture, primarily through use of single-cell RNA sequencing (RNA-seq) to gain insights into events driving early placental emergence. It should be noted that there have been 3 previous papers that explored the nature of trophoblast in cultured human embryos by using the single-cell RNA-seq approach (22–24), but these studies were confined to the preimplantation period.
Results
Embryo Extended Culture and Single-Cell Collection.
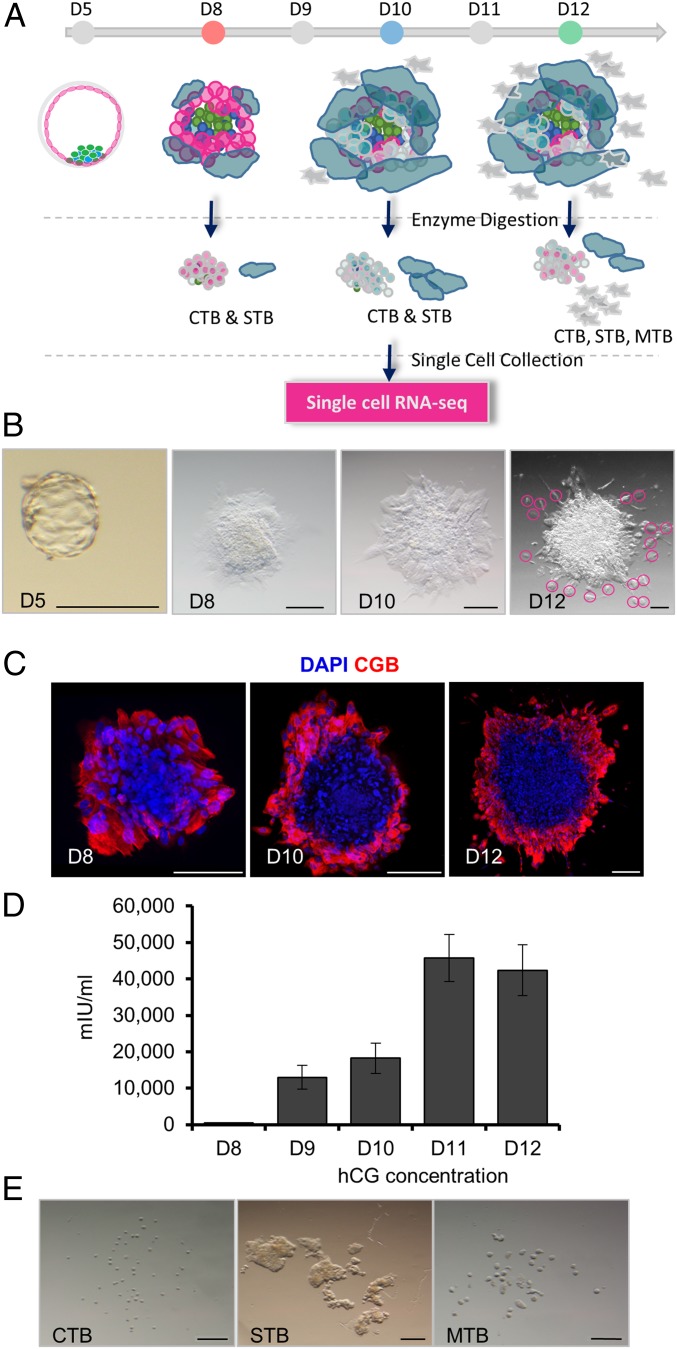
Vitrified and warmed D5 donated human blastocysts were cultured to embryonic D8, D10, and D12 according to a published protocol (20, 21) (Fig. 1 A and B and SI Appendix, Table S1). By D8, most embryos attached to the bottom of the dish and possessed a peripherally located, apparently syncytial region that was positive for CG subunit beta (CGB) (Fig. 1C), a definitive STB marker, and had initiated secretion of hCG into the medium (Fig. 1D). Syncytial areas expanded over time and stained more intensely for CGB at D10 and D12 than at D8 (Fig. 1C and SI Appendix, Fig. S1A). Daily release of hCG increased 41-fold by D10 compared to D8 (Fig. 1D). The expression of epiblast marker, POU5F1, was confined to a central area that represents the formation of the embryonic disk (SI Appendix, Fig. S1 A and B). The commonly used trophoblast markers, KRT7 and GATA3, were also confirmed in these outer regions by immunofluorescence staining (SI Appendix, Fig. S1B). In some D10 cultures, it was possible to observe a few cells migrating away from the embryo. By D12, these migrating cells had become more numerous. A time-lapse video that followed development between D8 and D12 was consistent with these patterns of cell fusion and migration during extended culture (Movie S1).
Fig. 1.
Isolation of single cells from extended cultured human embryos. (A) Workflow for culturing human embryos and isolating cytotrophoblast (CTB), syncytiotrophoblast (STB), and migratory trophoblast (MTB) for single-cell RNA-seq. (B) Morphologies of a D5 human blastocyst after zona pellucida removal and embryos at embryonic D8, D10, and D12. Pink circles outline MTB seen at D12 but rarely earlier. (Scale bar, 200 µm.) (C) Beta-human CG (CGB) immunofluorescence at embryonic D8, D10, and D12 (n = 3). (Scale bar, 200 µm.) (D) Levels of hCG in cell culture medium at embryonic D8 to D12 (mean ± SEM) (n = 13). (E) Morphologies of CTB, STB, and MTB after single-cell enzyme digestion. (Scale bar, 200 µm.)
Finally, after treating D8, D10, and D12 embryos with trypsin, 3 different classes of placental cells were selected and individually picked, based on their size and location, for RNA-seq. The smallest, round cells (Fig. 1E) were inferred to be mononucleated CTB and possibly also similarly sized cells from other lineages. The multinucleated STB (SI Appendix, Fig. S1A) were easily identified as irregular shaped structures significantly larger than the small cells (Fig. 1E). The migratory trophoblast cells (MTB) recognized as seemingly moving away from the main body of the embryo were collected before the embryos had been fully dissociated by the trypsin (Fig. 1 B and E). Although we were able to pick populations of CTB and STB at D8, D10, and D12, MTB cells were solely from D12 because of their scarcity at earlier times (Fig. 1 A and B). RNA was extracted from 139 samples derived from 11 embryos (SI Appendix, Table S2) and, upon sequencing, generated ∼1.4 billion reads (∼10 million reads per individual cell). To ensure we were only analyzing the transcriptomes of trophoblast cells and not other cell types, cell lineages were first predicted based on reported expression of lineage markers derived from human blastocysts (24) by Spearman correlation algorithm. Initially, 7 single-cell samples were considered to be nontrophoblast by these criteria. However, when additional known lineage marker genes (POU5F1, GATA6, KRT7, GATA3, SOX2, NANOG, and CD24) (20) were used to check these 7 cells, 5 were considered to be trophoblast because of their high expression of KRT7 and GATA3. The remaining 2 were confirmed as epiblast (high expression of POU5F1, SOX2, and NANOG and low in KRT7 and GATA3) and were excluded from the extended analysis (SI Appendix, Table S3). Principal component analysis (PCA) and clustering analyses identified 9 cells as outliers, while 3 cells had low read counts (<8,000 genes detected). These were also discarded. In the final analysis of the remaining 125 cells (78 CTB, 31 STB, and 16 MTB), there were 14,105 genes with a maximum FPKM ≥ 1 and 15,420 genes with a maximum FPKM ≥ 0.3 in at least 4 cells (SI Appendix, Fig. S2A). These genes remained in the analysis, while all others were excluded. In general, the CTB expressed the highest number of expressed genes per cell, but there was no significant difference in number within any grouping between D8, D10, and D12 (SI Appendix, Fig. S2B).
Cell Type Specific Marker Expression and Pathway Analysis.
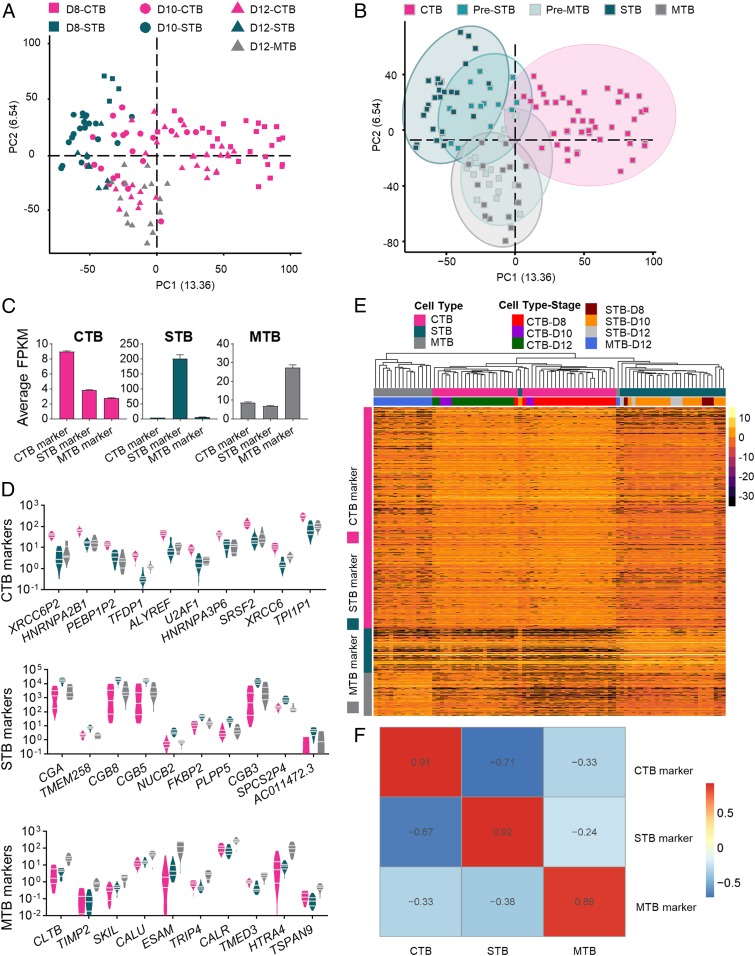
PCA was performed to show transcriptional profiles of all 3 cell populations at the different collection time points (Fig. 2A). The transcriptomic profiles clustered better according to cell type (CTB, STB, and MTB) than by day, i.e., developmental stage (SI Appendix, Fig. S3 A and B). Unsupervised clustering analysis (K-means) further revealed 17 small cells (CTB) had a partial MTB signature and were classified as Pre-MTB, while 15 small cells had a partial STB signature and were designated Pre-STB (Fig. 2B). We infer that these intermediate-stage cells were in the process of differentiating from CTB to either the MTB or STB sublineages. Cells considered to be in these 2 intermediate classes were removed from the remainder of the CTB group in order to identify the unique transcriptome signature for CTB. We also identified genes that were enriched in each cell type. Specifically, 2932, 592, and 569 genes were identified as enriched marker genes for CTB, STB, and MTB, respectively (Fig. 2 C and D and Dataset S1). Based on such gene panels, each main cell type could be clearly separated by hierarchical clustering analysis (Fig. 2E), which indicated strong correlations to their corresponding cell type specific genes (Fig. 2F) and demonstrated the robustness of each panel to identify each respective cell lineage.
Fig. 2.
Identification of cell type specific markers for CTB, STB, and MTB. (A) PCA of trophoblast cells showing discrete clusters based on cell type. (B) PCA analysis reveals subsets of CTB that had partial STB and MTB signatures, classified as Pre-STB and Pre-MTB, respectively. (C) Average FPKM for CTB, STB, and MTB marker gene expression in each cell type. (D) Expression, based on least likelihood of false discovery of the top 10 marker genes for CTB, STB, and MTB in each cell type. (E) Hierarchical clustering analysis of cell type marker genes in trophoblast cells. These 3 panels of marker genes clearly partitioned each major TE cell type. Three panels of marker genes specifically for CTB, STB, and MTB are presented in the y axis, and cell types with different developmental stages are presented in the x axis. The color spectrum, ranging from yellow to black, indicates high to low normalized levels of gene expression. (F) Spearman correlation map indicating strong correlations between each cell type and their marker genes. The color spectrum, ranging from red to blue, indicates the correlation from high to low.
Next, gene ontology (GO) and pathway analysis was performed to identify pathways most strongly associated with each cell type. The top GO terms for CTB were ones linked to cell proliferation, transcription, and energy metabolism (SI Appendix, Fig. S4), consistent with the inferred role of CTB in supplying cells to the other trophoblast lineages and ultimately the placenta as a whole. For STB, the top terms were protein folding, transport, and hormone processing (SI Appendix, Fig. S4), which are again consistent with known functions of syncytial trophoblast, whether in the villous placenta or the primitive placenta encountered in very early pregnancy (16). Pathways involved in protein export and uptake, steroidogenesis, and migration were also up-regulated. The latter term may reflect the suspected invasive as well as endocrine nature of early STB during the implantation phase of pregnancy (25). As anticipated, MTB-enriched pathways underscored roles in cell migration and invasion into the extracellular matrix. More surprising was the presence of terms and pathways associated with type I and type II IFN signaling, innate immune responses, and antigen processing (SI Appendix, Fig. S4).
Dynamics of Trophoblast Differentiation.
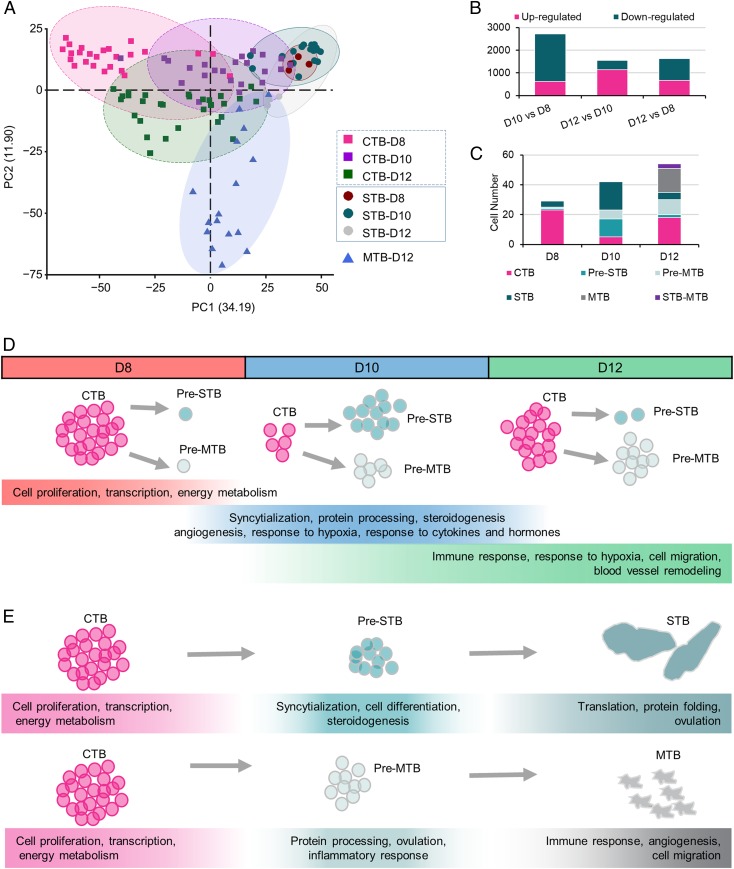
While the transcriptomic profiles of the STB remained relatively stable between D8 and D12, those of the CTB showed considerable change, possibly reflecting the developmental plasticity of the CTB as the cells prepare for differentiation (Fig. 3 A and B). A comparison of the transcriptomes of CTB between D8 and D10, for example, reflected a switch from a proliferative phenotype at D8 to one with a greater emphasis on syncytialization, protein processing, and steroidogenesis at D10, suggesting that differentiation toward STB had become an emphasis. Additionally, several MTB-related GO pathways, including angiogenesis, response to hypoxia, and response to cytokines and hormones, had also become active at D10 (Fig. 3D and SI Appendix, Fig. S5). These data suggest that there are 2 distinct differentiation routes ongoing for CTB at D10, one to STB and the other to MTB. This inference is supported by the fact that the majority of small cells collected at D10 were either Pre-STB (12 out of 23) or Pre-MTB (6 out of 23), and only 5 (21.7%) possessed the phenotype of bona fide undifferentiated CTB (Fig. 3C). By D12, there was evidence of a more emphatic shift toward a Pre-MTB phenotype, as evidenced by an up-regulation of pathways related to IFN signaling, hypoxia, cell migration, and vascular remodeling, while genes associated with syncytium formation had become less dominant than in D10 CTB (Fig. 3D and SI Appendix, Fig. S5). Additionally, more Pre-MTB (10 out of 30 small cells) and fewer Pre-STB (2 out of 30 small cells) were present in the population of CTB collected at D12 (Fig. 3C). There also appeared to be a partial restoration of undifferentiated CTB numbers at D12, as well as the appearance of a possible new phenotype in the form of 2 cells with a mixed STB/MTB signature (Fig. 3C).
Fig. 3.
Dynamics of CTB differentiation. (A) PCA of trophoblast cells clustered by cell type and developmental stages. Data suggest that CTB cells are considerably more dynamic than STB in different developmental stages. (B) Numbers of differentially expressed genes in CTB from different developmental stages. (C) The number of cells in each cell type collected at different developmental stages. (D) Diagram to illustrate the switch of cell functions of CTB at different developmental stages as revealed by GO terms and pathway analysis. (E) Diagram to illustrate the switch of cell functions of CTB as they undergo differentiation to STB (Upper) and MTB (Lower) as revealed by GO terms and pathway analysis.
We then compared the transcriptomes of Pre-STB and Pre-MTB, with undifferentiated CTB and differentiated STB and MTB regardless of their collection day. While Pre-STB had up-regulated genes associated with syncytium formation (Fig. 3E and SI Appendix, Fig. S6A), they also had retained high mitotic activity as suggested by their expression levels of the cell proliferation markers PCNA and MCM genes (26, 27) (SI Appendix, Fig. S7). By contrast, the differentiated STB showed evidence for departure from the cell cycle and possessed gene networks linked to protein transport and hormone production that were largely absent from Pre-STB (Fig. 3E and SI Appendix, Fig. S6A). We conclude that the Pre-STB represent a population of dividing CTB primed to fuse to form syncytium but that had not yet up-regulated the machinery required for hormone production and cell fusion (Fig. 3E and SI Appendix, Fig. S6A). The Pre-MTB existed in an analogous state to the Pre-STB, i.e., committed to differentiation but lacking the full-blown phenotype of MTB. For example, they appeared to be continuing to proliferate, although the pathways related to RNA processing had begun to decline (Fig. 3E and SI Appendix, Fig. S6B). GO terms related to IFN signaling and antigen processing that had been strongly represented in mature MTB had apparently not been acquired by the Pre-MTB.
Gene Networks Linked to IFN Responses.
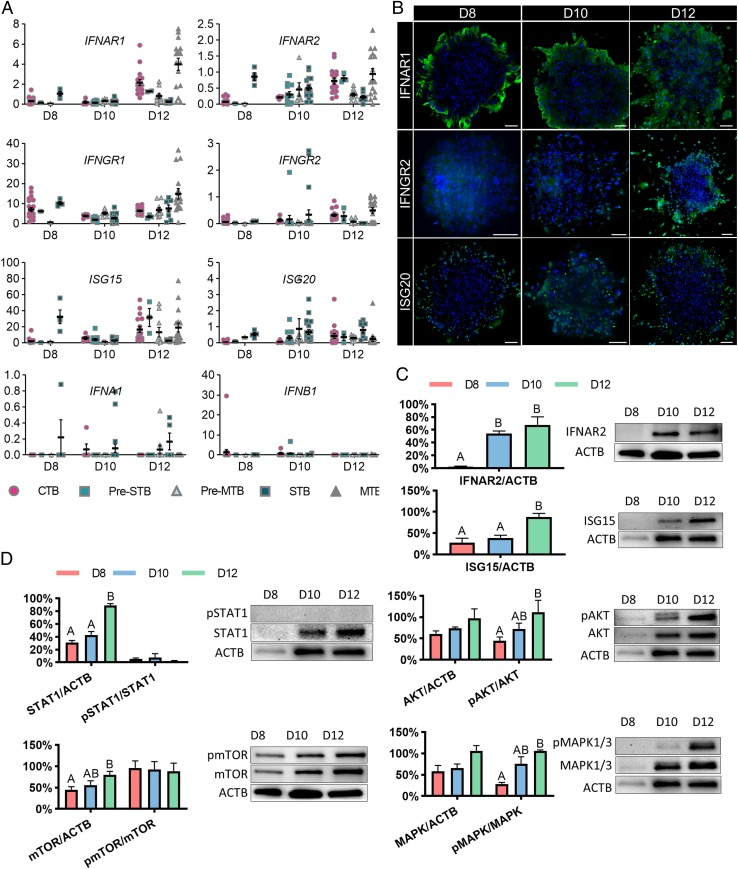
Closer analysis of the RNA-seq data revealed that several IFN response genes were expressed as early as D8 in trophoblast cells and increased in expression as development proceeded (Fig. 4A). GO pathway analysis indicated that D12 CTB (SI Appendix, Fig. S5) and MTB (SI Appendix, Fig. S4) had several terms associated with type I and type II IFN signaling pathways. To confirm these findings, we immunostained embryos for type I IFN receptor subunits 1 (IFNAR1), type II IFN receptor subunit 2 (IFNGR2), and ISG20 (Fig. 4B). We found expression of INFAR1 and ISG20 as early as D8, with IFNAR1 and ISG20 mainly located to the periphery of the colonies where differentiation occurs (Fig. 4B). All of the IFN-related markers became positive by D12. The increase in expression of IFNAR2 and ISG15 were also confirmed by Western blotting (Fig. 4C).
Fig. 4.
IFN response pathways in early pregnancy. (A) FPKM values of the IFN-related genes: type I and type II IFN receptors (IFNAR1, IFNAR2, IFNGR1, and IFNGR2), type I IFN (IFNA1 and IFNB1), and IFN-stimulated genes (ISG15 and ISG20). (B) Immunofluorescent localization of proteins implicated in potential IFN responses in human embryos between D8 and D12 (n = 3). (Scale bar, 100 µm.) (C) Western blot analysis of IFNAR2 and ISG15 (n = 3) and (D) phosphorylation of STAT1 (n = 3), mTOR (n = 3), AKT (n = 3), and MAPK1/3 (n = 3) in human embryos between D8 and D12. The expression of protein levels was normalized to ACTB (actin beta). The ratio of phosphorylated (p) and total protein abundance was used to determine the phosphorylation level of target proteins. All data are presented as the mean ± SEM.
An IFN response triggered by type I and type II IFN is mediated by the JAK-STAT pathway (28). Although total STAT1 increased over time, we found no evidence for the presence of its phosphorylated form, indicating the JAK-STAT pathway was probably not activated (Fig. 4D). In addition, mTOR signaling activity, a mechanism that has been considered to mediate IFN-induced mRNA translation (28), remained unchanged over time, also suggesting that the classical JAK-STAT mediated IFN signaling pathway was inactive (Fig. 4D). Although increased concentrations of phosphorylated AKT and ERK1/2 were detected, these factors play more general roles in metabolism, rather than up-regulating type I and type II IFN responses. We also found no evidence for either bacterial contamination or endotoxin in the medium that might have triggered a type II IFN response, nor were IFNG or IFNA/B detectable in the medium by immunoassay. Finally, FPKM values for IFNA subtypes and IFNB were extremely low (Fig. 4A), while IFNG transcripts were undetectable. These data suggest that the up-regulation of IFN receptors and downstream IFN response genes were not triggered by exogenous factors but were components of a constitutive developmental process associated with normal development.
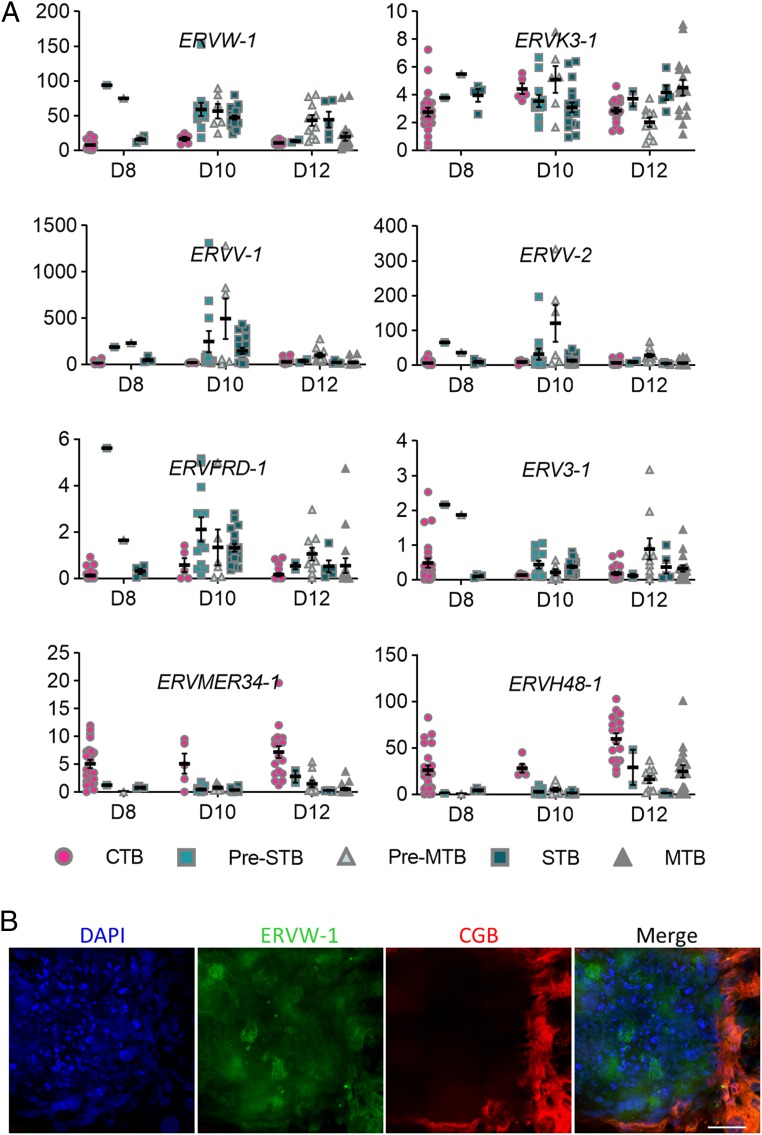
About 8% of the human genome (29) consists of human endogenous retroviruses (HERV) capable of producing virus-like particles competent to induce an IFN response. The placenta expresses several HERV that have been implicated in trophoblast differentiation and syncytialization (30, 31). Eight HERV were expressed in cultured human embryos (Fig. 5A). Of these, 5 (ERV3-1, ERVFRD-1, ERVV-1, ERVV-2, and ERVW-1) were more abundantly expressed in differentiated trophoblast cells (Pre-STB, Pre-MTB, STB, and MTB) than in CTB. ERVV-1, ERV-2, and ERVW-1 had the highest transcript levels, with the highest expression at D10 when the presence of ERVW-1 could be confirmed by immunofluorescence (Fig. 5B). By contrast, expression of ERVH48-1 and ERVMER34-1 was higher in undifferentiated CTB than in the more differentiated cell populations. ERVH48-1 is known to inhibit rather than promote cell fusion, which it accomplishes by competing with ERVW-1 for binding to its cell surface receptor (32). ERVMER34-1 has barely been studied, but its presence in CTB rather than STB suggests that it, too, might be an inhibitor of cell fusion.
Fig. 5.
Human endogenous retroviruses in peri-implantation embryos. (A) FPKM values (mean ± SEM) of several HERV in human embryos between D8 and D12. (B) Immunofluorescence of ERVW-1 in D10 human embryos (n = 3). (Scale bar, 100 µm.)
Discussion
Here we have employed an extended culture system for human embryos, combined it with single-cell RNA-seq, and successfully captured transcriptome dynamics in trophoblast cells occurring within the primitive placenta between D8 and D12 postfertilization, a time that in vivo corresponds to the first 5 d after the embryo begins to implant into the uterine wall. It is important to note that during this period, the familiar villous placenta, with its characteristic thin outer syncytialized epithelium overlaying a mitotic population of CTB stem cells has not yet emerged. Instead, the embryo proper is surrounded by a mass of trophoblast, with STB and, as demonstrated here, a population of migratory cells (here called MTB) toward the exterior. Our data are consistent with the hypothesis that STB and MTB are replenished from below by a progenitor population of CTB. However, several facts should be born in mind about this early placental structure. First, unlike villous STB (33), it has invasive/migratory features that are probably responsible for its ability to burrow into the endometrium and place the conceptus within a hollowed-out niche where it can gain nutritional support from close proximity with surrounding maternal decidual cells, capillaries, and glands. Second, the conceptus must produce sufficient hCG and possibly other supporting factors to prevent a decline in progesterone resulting from regression of the corpus luteum as would occur in a nonfertile cycle. Third, this early placenta, although functional, is short lived. Columns of CTB begin to penetrate the STB layer beginning around D12 and ultimately give rise to the villous placenta while the fate of the initial STB remains unclear. It is mistaken to believe, as others have done, that the STB associated with the D8 to 12 placenta is equivalent to villous STB, which it clearly is not, although it may have an analogous function in supplying the embryo proper with nutrient support. Additionally, the migratory cells (called MTB here) should probably not be referred to as extravillous TB, since there are no villous structures at this stage from which MTB could arise. In terms of marker gene expression, the transcriptome profiles of the MTB, including the expression of HLA-G, do appear to resemble those of first trimester EVT arising from the tips of anchoring villi, but whether the latter are of the same lineage as MTB is not clear.
At D8, when implantation in vivo would have just begun, the majority of TB cells in the embryos were small and apparently mitotically active and had a predominantly CTB phenotype, although some STB and 1 or 2 Pre-STB and Pre-MTB cells were also present in the CTB population (Fig. 3C) and were probably responsible for the small amount of hCG produced at this time (Fig. 1D). Trophectoderm, a simple epithelium, may provide the progenitors for trophoblast outgrowth from cells at the polar end of the blastocyst (24) but is clearly functionally distinct from the trophoblast associated with implantation. Additionally, our transcriptomic data reveal that the small cell population, while appearing morphologically homogeneous, contained a subset of mitotically active intermediate cytotrophoblast cells already transcriptionally preparing either to fuse to form syncytium, particularly apparent at D10, or to differentiate toward MTB (Figs. 2B and 3C). This lineage bifurcation seemed to initiate around D8 but was most apparent at D10, when commitment to STB was at a maximum and production of hCG was rising sharply (Figs. 1D and 3C). By D12, STB differentiation was in decline, but MTB production was on the upswing (Fig. 3C). At D12, there was also an indication of an upsurge in the proportion of undifferentiated, mitotically active CTB (Fig. 3C), possibly reflecting the beginning of villous formation and the demise of the early placenta. Together these data suggest that the early placenta is distinct from trophectoderm and, although short-lived, is able to prioritize highly specialized cell functions at very specific time points throughout the implantation period.
The transcriptomic data also provided some surprises. One was the identification of several genes and GO pathways associated with IFN signaling and partial activation of antiviral responses in the MTB. Although the production of type I and type II IFN by trophoblast has been demonstrated in some ungulates (34), and is especially important in ruminants where IFN-tau (IFNT) plays a critical role in maternal recognition of pregnancy (35, 36), no homologous role for IFN was evident here for the human, where neither type I nor type II IFN genes were being transcribed (Fig. 4A). Rather, the MTB seemed poised to respond to IFN, having both the receptors and some component effectors already in place. Conceivably, IFN released by cells resident in the endometrium would be able to trigger an IFN response in trophoblast and provide some survival advantage to the embryo in addition to serving as a first line of defense against viral and bacterial infections. For example, in vitro fertilization (IVF) patients with recurrent implantation failure have significantly lower concentrations of intrauterine IFNG than controls (37). Of course, overstimulation of IFN may desensitize IFN pathways, a mechanism that contributes to unresponsiveness of type I IFN in HIV disease (38). In human embryos, a high level of IFN exposure as a result of uterine infection or inflammation could lead to desensitization of IFN pathways and blockage of trophoblast differentiation, resulting in embryo loss, thereby sparing maternal costs associated with continuation of a blighted pregnancy. Thus, IFN response pathways may act as a safety switch that determines the success of implantation in vivo.
As anticipated, expression of transcripts for classical MHC class I molecules, HLA-A and HLA-B (39), as well as MHC class II molecules (40), illustrated here by HLA-DOB and HLA-DRB1 (SI Appendix, Fig. S9C), were poorly expressed in each class of trophoblast cell at each developmental time point. By contrast, HLA-C, and the nonclassical HLA-E, and HLA-G genes were robustly expressed by D12 (SI Appendix, Fig. S9 A and B), especially in MTB. These human versions of the major histocompatibility complex (MHC) molecules have been proposed to be responsible for creating an in vivo immunomodulatory environment that favors extravillous trophoblast invasion (41). Such features of the MHC system, like the IFN signaling pathway discussed previously, may be critical for early conceptus survival prior to formation of the villous placenta at a perilous time in pregnancy when embryonic wastage is high. A deeper investigation of these transcriptomic data will likely elucidate additional insights into mechanisms that facilitate human implantation.
It should be recognized that the embryos used in our experiments were cultured in a medium in absence of maternal decidual cells and on a substratum, namely, fibronectin, that would only be a single component of the mixed matrix encountered in the endometrium during implantation. For example, both substrate composition and stiffness can influence transcriptomes of stem cells (42, 43). Thus, the developmental trajectory observed here in vitro might be somewhat different from that occurring in vivo in a normal pregnancy. As demonstrated in Fig. 5, however, major metabolism pathways remained active at D12, and there was an apparent increase of mitotically active CTB and a rise of MTB. Nonetheless, it would be overly optimistic to believe that the embryos were not losing viability by this stage. As the technology for extended embryo culture improves, it is likely that a system more reflective of endometrium–conceptus interactions as they occur in vivo will emerge. Therefore, even though the data provided in the present paper can only be an incomplete indication of what goes on in vivo, we suspect that they still provide insight into the functioning of the early placenta, a structure essential for pregnancy maintenance prior to the time that the villous placenta emerges. Our results are consistent with the inference that initial implantation events and endocrine support of ovarian progesterone production by the mother are a function of the STB surrounding the conceptus as it invades, while the motile MTB is responsible for initiating additional colonization of the uterine endometrium prior to the outgrowth of placental villi.
Materials and Methods
Ethics Statement.
This research was approved by the Western Institutional Review Board (study no. 1179872) and followed international guidelines for extended embryo culture. Embryos were donated for research with patients’ informed consent. Once a patient has completed treatment and has no further medical need for their frozen embryos and no longer wants to retain them, they have several options: discard and thus destroy their embryos, donate their embryos to another couple for embryo adoption, or donate them to research. This choice is certified by a signed and notarized disposition document. This process is completed by nonresearch personnel. Once a patient has chosen and finalized “donation to research” and their decision has been confirmed, those embryos become available for research.
Human Embryo Thawing and Zona Pellucida Removal.
Human embryos (see SI Appendix for further details) were voluntarily donated by patients at the Colorado Center for Reproductive Medicine. D5 and D6 vitrified human blastocysts were thawed in prewarmed solution for 1 min and transferred to modified Kitazato dilution solution (Kitazato) for 3 min prior to washing solution (WS) for 5 min (44). Individual blastocysts were placed in individual culture drops of IVC1 medium (Cell Guidance Systems, M11 25) overlaid in mineral oil and left for 2 h at 37 °C under 20% O2 and 7.5% CO2 to adjust for the high elevation where these studies have been performed. After 2 h, embryos were moved to 3-(N-morpholino)propanesulfonic acid (Mops) buffered medium before removal of the zona pellucida by 3 sequential washes in Acidic Tyrode’s solution (Millipore Sigma, T1788) followed by 3 sequential washes in Mops.
Extended Embryo Culture.
The extended embryo culture procedure was performed as previously described (20, 21). Briefly, blastocysts immediately after zona removal were washed in a 60-mm center-well organ culture dish (Corning, 353037) containing warmed IVC1 medium before transfer into 8-well chamber dishes (Ibidi, 80841). Each chamber had been coated with sterile fibronectin (Millipore Sigma, F0895) diluted in PBS at a 3:100 dilution. Plates were coated overnight, and remaining soluble fibronectin were then removed before addition of 300 µL equilibrated IVC1 medium. After at least 1 h, embryos lacking zonas were placed in the medium and cultured for a further 48 h. On extended embryo culture day 2, embryo attachment was assessed for all embryos. For embryos that had attached, half the IVC1 medium was removed and replaced with fresh IVC2 medium (Cell Guidance Systems, M12-25). After day 2, a 50% media change with IVC2 media was performed daily on all attached embryos.
Isolation of Single Cells.
Single cells were isolated from embryos at embryonic D8, D10, and D12; incubated with TrypLE Express reagent for 10 min; and dissociated into single cells. For D12 embryos, migratory TB (MTB) that were located outside the main colony were collected before the main colony had been dissociated. After gentle pipetting, rounded, single mononucleated cytoTB (CTB) and multinucleated syncytioTB (STB) were collected. Each cell was transferred into a 0.2-mL PCR tube (Eppendorf) containing cell lysis buffer and kept at −80 °C until library preparation.
Single-Cell RNA-Seq Library Preparation.
The RNA-seq libraries were generated from individual cells by using the Smart-seq2 v4 kit with minor modification from manufacturer’s instructions. Briefly, individual cells were lysed, and mRNA was captured and amplified with the Smart-seq2 v4 kit (Clontech). After AMPure XP beads purification, amplified RNAs were quality checked by using Agilent High Sensitivity D5000 kit (Agilent Technologies). High-quality amplified RNAs were subject to library preparation (Nextera XT DNA Library Preparation Kit; Illumina) and multiplexed by Nextera XT Indexes (Illumina). The concentration of sequencing libraries was determined by using Qubit dsDNA HS Assay Kit (Life Technologies) and KAPA Library Quantification Kits (KAPA Biosystems). The size of sequencing libraries was determined by means of High Sensitivity D5000 Assay in at Tapestation 4200 system (Agilent). Pooled indexed libraries were then sequenced on the Illumina HiSeq X platform with 100-bp pair-end reads.
Single-Cell RNA-Seq Data Filtration and Processing.
Multiplexed sequencing reads that passed filters were trimmed to remove low-quality reads and adaptors by TrimGalore-0.4.3. The quality of reads after filtering was assessed by fastQC, followed by alignment to the human genome (hg38) by STAR (2.5.3a) with default parameters. Approximately 10 million reads per individual cell were generated. Individual mapped reads were adjusted to provide FPKM (fragments per kilobase of exon model per million mapped fragments) values with RefSeq genes as reference. The raw FASTQ files and normalized gene expression profiles (FPKM) are available at Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE 130289. Differential gene expression analysis was performed by a Partek Flow GSA algorithm with default parameters. The genes were deemed differentially expressed if they provided a false discovery rate of <0.05 and fold change >2. DAVID (https://david.ncifcrf.gov) and IPA (Ingenuity Pathway Analysis) were used to reveal the Gene Ontology (GO) and pathways, respectively. Cells with more than 8,000 detected genes, each with FPKM values >0.3, were used for downstream analysis for lineage specification. Lineage specification was performed by Spearman correlation. PCA and cluster analysis were performed by using R. Identification of cell type and stage-specific genes was performed as previously described (45). Briefly, to identify cell type and stage-specific genes, a unit vector was constructed for each cell type (stage), followed by the calculation of the Pearson correlation between the vector and individual genes. This unit vector was designed to represent cell type (stage)-specific gene expression patterns. More specifically, the entries of the vector corresponding to samples of a cell type (or at a stage) were set to 1, with all others being 0. Genes with P value (after Bonferroni correction for multiple testing) less than 0.05 were labeled as cell type (stage) specific.
Immunofluorescence and Image Acquisition.
Embryos were washed 3 times in warmed, filtered PBS and fixed in 4% paraformaldehyde (PFA) for 20 min. They were then washed in 0.1% Tween20 (Millipore Sigma, P1379) in PBS (PBS-T) 3 times and permeabilized by 0.5% Triton X-100 (Millipore Sigma, X100) in PBS for 30 min. Embryos were washed again in PBS-T and exposed to 10% vol/vol FBS and 3% wt/vol BSA in PBS-T overnight at 4 °C. Primary antibodies (SI Appendix, Table S4) were added at a 1:200 dilution in blocking buffer (10% FBS/3% BSA/PBS-T) and incubated at 4 °C overnight. Samples were then washed in PBS-T 3 times and incubated with secondary antibodies overnight at 4 °C. After incubation, embryos were washed again in PBS-T and then mounted in ProLong Gold antifade reagent with DAPI (ThermoFisher Scientific, P36931) mounting medium. Embryos were imaged on a 3I Marianas inverted spinning disk microscope. Z-stack images were captured by using a 40x/1.52 numerical aperture oil-immersion objective. The lasers 405, 488, and 561 were used to capture DAPI, Alexa Fluor 488, and Alexa Fluor 594, respectively. Laser intensity and exposure time was manually adjusted for each sample to avoid oversaturation or photobleaching of samples. After image capture, images were immediately deconvoluted by means of Slidebook software (3i) set to the “Nearest Neighbors” setting. Images were rendered and processed with Imaris microscopy image analysis software (Bitplane) to generate maximum intensity projections.
Other methods including Western blotting, ELISA and ECLIA analyses, and statistical analysis are summarized in SI Appendix. Antibodies used for immunofluorescence and Western blotting are listed in SI Appendix, Tables S4 and S5.
Supplementary Material
Acknowledgments
We thank Karen Maruniak and the Colorado Center for Reproductive Medicine (CCRM) Clinical Laboratory for processing hCG samples and Sue McCormick and the CCRM IVF Laboratory for embryo collection. We thank Dr. Radu Moldovan and Dr. Dominik Stich at the University of Colorado Advanced Light Microscopy Core for their help with the confocal image acquisition and image processing. We also thank Dr. Thomas Hansen, Dr. Gerrit Bouma at Colorado State University, Dr. Danny Schust, Dr. Laura Schultz, and Dr. Jie Zhou at University of Missouri for their critical inputs. This project was funded by internal research funds provided by the CCRM. The microscopy and imaging services provided by the Advanced Light Microscopy Core Facility at the University of Colorado Anschutz School of Medicine to the CCRM were billed at the nonacademic (non-NIH funded user) rate.
Footnotes
The authors declare no competing interest.
Data deposition: Data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE 130289).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911362116/-/DCSupplemental.
References
- 1.Jarvis G. E., Early embryo mortality in natural human reproduction: What the data say. F1000 Res. 5, 2765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macklon N. S., Geraedts J. P., Fauser B. C., Conception to ongoing pregnancy: The ‘black box’ of early pregnancy loss. Hum. Reprod. Update 8, 333–343 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Carter A. M., Animal models of human placentation–A review. Placenta 28 (suppl. A), S41–S47 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Enders A. C., Trophoblast-uterine interactions in the first days of implantation: Models for the study of implantation events in the human. Semin. Reprod. Med. 18, 255–263 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Hertig A. T., Rock J., Adams E. C., A description of 34 human ova within the first 17 days of development. Am. J. Anat. 98, 435–493 (1956). [DOI] [PubMed] [Google Scholar]
- 6.Boyd J. D., Hamilton W. J., The Human Placenta (Heffer & Sons, Cambridge, 1970). [Google Scholar]
- 7.Amoroso E., “Placentation” in Marshall’s Physiology of Reproduction, Parkes A., Ed. (Little Brown & Co., Boston, 1952), vol. 2, pp. 127–311. [Google Scholar]
- 8.Enders A. C., Trophoblast differentiation during the transition from trophoblastic plate to lacunar stage of implantation in the rhesus monkey and human. Am. J. Anat. 186, 85–98 (1989). [DOI] [PubMed] [Google Scholar]
- 9.Enders A. C., King B. F., Early stages of trophoblastic invasion of the maternal vascular system during implantation in the macaque and baboon. Am. J. Anat. 192, 329–346 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Enders A. C., Lantz K. C., Peterson P. E., Hendrickx A. G., From blastocyst to placenta: The morphology of implantation in the baboon. Hum. Reprod. Update 3, 561–573 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Hustin J., Schaaps J. P., Echographic [corrected] and anatomic studies of the maternotrophoblastic border during the first trimester of pregnancy. Am. J. Obstet. Gynecol. 157, 162–168 (1987). [DOI] [PubMed] [Google Scholar]
- 12.Foidart J. M., Hustin J., Dubois M., Schaaps J. P., The human placenta becomes haemochorial at the 13th week of pregnancy. Int. J. Dev. Biol. 36, 451–453 (1992). [PubMed] [Google Scholar]
- 13.Burton G. J., Jauniaux E., Watson A. L., Maternal arterial connections to the placental intervillous space during the first trimester of human pregnancy: The Boyd collection revisited. Am. J. Obstet. Gynecol. 181, 718–724 (1999). [DOI] [PubMed] [Google Scholar]
- 14.James J. L., Carter A. M., Chamley L. W., Human placentation from nidation to 5 weeks of gestation. Part I: What do we know about formative placental development following implantation? Placenta 33, 327–334 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Xu R. H., et al. , BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Yabe S., et al. , Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc. Natl. Acad. Sci. U.S.A. 113, E2598–E2607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okae H., et al. , Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Haider S., et al. , Self-renewing trophoblast organoids recapitulate the developmental program of the early human placenta. Stem Cell Reports 11, 537–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turco M. Y., et al. , Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 564, 263–267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deglincerti A., et al. , Self-organization of the in vitro attached human embryo. Nature 533, 251–254 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Shahbazi M. N., et al. , Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 18, 700–708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan L., et al. , Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Blakeley P., et al. , Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3613 (2015). Erratum in: Development 142, 3151–3165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petropoulos S., et al. , Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knöfler M., Pollheimer J., Human placental trophoblast invasion and differentiation: A particular focus on Wnt signaling. Front. Genet. 4, 190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tye B. K., MCM proteins in DNA replication. Annu. Rev. Biochem. 68, 649–686 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Kelman Z., PCNA: Structure, functions and interactions. Oncogene 14, 629–640 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Platanias L. C., Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005). [DOI] [PubMed] [Google Scholar]
- 29.de Parseval N., Lazar V., Casella J. F., Benit L., Heidmann T., Survey of human genes of retroviral origin: Identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 77, 10414–10422 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson P. M., Lyden T. W., Mwenda J. M., Endogenous retroviral expression in the human placenta. Am. J. Reprod. Immunol. 23, 115–120 (1990). [DOI] [PubMed] [Google Scholar]
- 31.Rote N. S., Chakrabarti S., Stetzer B. P., The role of human endogenous retroviruses in trophoblast differentiation and placental development. Placenta 25, 673–683 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto J., Sugimoto M., Bernstein H., Jinno Y., Schust D., A novel human endogenous retroviral protein inhibits cell-cell fusion. Sci. Rep. 3, 1462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain A., Ezashi T., Roberts R. M., Tuteja G., Deciphering transcriptional regulation in human embryonic stem cells specified towards a trophoblast fate. Sci. Rep. 7, 17257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Bonnardière C., et al. , Production of two species of interferon by Large White and Meishan pig conceptuses during the peri-attachment period. J. Reprod. Fertil. 91, 469–478 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Imakawa K., et al. , Interferon-like sequence of ovine trophoblast protein secreted by embryonic trophectoderm. Nature 330, 377–379 (1987). [DOI] [PubMed] [Google Scholar]
- 36.Roberts R. M., Chen Y., Ezashi T., Walker A. M., Interferons and the maternal-conceptus dialog in mammals. Semin. Cell Dev. Biol. 19, 170–177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inagaki N., et al. , Analysis of intra-uterine cytokine concentration and matrix-metalloproteinase activity in women with recurrent failed embryo transfer. Hum. Reprod. 18, 608–615 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Hardy G. A., et al. , Desensitization to type I interferon in HIV-1 infection correlates with markers of immune activation and disease progression. Blood 113, 5497–5505 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apps R., et al. , Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology 127, 26–39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy S. P., Choi J. C., Holtz R., Regulation of major histocompatibility complex class II gene expression in trophoblast cells. Reprod. Biol. Endocrinol. 2, 52 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holtan S. G., Creedon D. J., Haluska P., Markovic S. N., Cancer and pregnancy: Parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin. Proc. 84, 985–1000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B., et al. , Effect of substrate topography and chemistry on human mesenchymal stem cell markers: a transcriptome study. Int. J. Stem Cells 12, 84–94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darnell M., Gu L., Mooney D., RNA-seq reveals diverse effects of substrate stiffness on mesenchymal stem cells. Biomaterials. 181, 182–188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fasano G., et al. , A randomized controlled trial comparing two vitrification methods versus slow-freezing for cryopreservation of human cleavage stage embryos. J. Assist. Reprod. Genet. 31, 241–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Z., et al. , Transcriptional profiles of bovine in vivo pre-implantation development. BMC Genomics 15, 756 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.