Significance
The development of novel, effective treatments for patients with advanced/recurrent cervical cancer remains an unmet medical need. Using whole-exome sequencing, we identified multiple genes with recurrent alterations in cervical cancer, including the ERBB2/PI3K/AKT/mTOR, apoptosis, chromatin remodeling, and cell cycle pathways. Using fully sequenced cell lines and xenografts, we found pan-HER (afatinib/neratinib) and PIK3CA (copanlisib) inhibitors to be active, but only transiently effective in controlling the in vivo growth of PIK3CA-mutated cervical tumor xenografts. In contrast, the combination of irreversible pan-HER kinase and PIK3CA inhibitors was highly synergistic and able to induce durable regression of xenografts harboring derangements in the ERBB2/PI3K/AKT/mTOR pathway in vivo. These findings suggest a large subset of cervical tumors (>70%) might benefit from existing ERBB2/PIK3CA/AKT/mTOR-targeted drugs.
Keywords: cervical cancer, PIK3CA, HER2/neu, neratinib, copanlisib
Abstract
The prognosis of advanced/recurrent cervical cancer patients remains poor. We analyzed 54 fresh-frozen and 15 primary cervical cancer cell lines, along with matched-normal DNA, by whole-exome sequencing (WES), most of which harboring Human-Papillomavirus-type-16/18. We found recurrent somatic missense mutations in 22 genes (including PIK3CA, ERBB2, and GNAS) and a widespread APOBEC cytidine deaminase mutagenesis pattern (TCW motif) in both adenocarcinoma (ACC) and squamous cell carcinomas (SCCs). Somatic copy number variants (CNVs) identified 12 copy number gains and 40 losses, occurring more often than expected by chance, with the most frequent events in pathways similar to those found from analysis of single nucleotide variants (SNVs), including the ERBB2/PI3K/AKT/mTOR, apoptosis, chromatin remodeling, and cell cycle. To validate specific SNVs as targets, we took advantage of primary cervical tumor cell lines and xenografts to preclinically evaluate the activity of pan-HER (afatinib and neratinib) and PIK3CA (copanlisib) inhibitors, alone and in combination, against tumors harboring alterations in the ERBB2/PI3K/AKT/mTOR pathway (71%). Tumors harboring ERBB2 (5.8%) domain mutations were significantly more sensitive to single agents afatinib or neratinib when compared to wild-type tumors in preclinical in vitro and in vivo models (P = 0.001). In contrast, pan-HER and PIK3CA inhibitors demonstrated limited in vitro activity and were only transiently effective in controlling in vivo growth of PIK3CA-mutated cervical cancer xenografts. Importantly, combinations of copanlisib and neratinib were highly synergistic, inducing long-lasting regression of tumors harboring alterations in the ERBB2/PI3K/AKT/mTOR pathway. These findings define the genetic landscape of cervical cancer, suggesting that a large subset of cervical tumors might benefit from existing ERBB2/PIK3CA/AKT/mTOR-targeted drugs.
Cervical cancer is the second leading cause of cancer death in women aged 20 to 39 y (1). While most of cervical cancer-related deaths take place in women living in low-income countries, it is estimated that over 13,170 women will be diagnosed with cervical cancer in the United States in 2019, and over 4,250 women will die of the disease (1). Persistent infection with sexually transmitted, oncogenic types of human papillomavirus (HPV) represents the most important risk factor for the development of cervical cancer (2). Although more than 100 distinct HPV genotypes have been described, and at least 20 are associated with squamous cell carcinoma (SCC) and adenocarcinoma (ACC) of the uterine cervix, HPV types 16 and 18 are the most frequently detected, regardless of the geographical origin of the patients (2). While cervical cancer is a largely preventable disease through early detection with regular screening tests (i.e., Papanicolaou smear) and ablation of dysplastic tissues, disseminated carcinoma of the cervix remains a discouraging clinical entity, with a 1-y survival rate between 10% and 15% (3). A deeper understanding of the molecular basis of cervical cancer and the development of novel more effective treatment modalities remain an unmet medical need.
In this study, we have sequenced the exomes of a large cohort of fresh-frozen tumors, as well as 15 primary cervical cancer cell lines with limited passages (≤15), and identified genes with recurrent mutations and increased numbers of somatic single nucleotide variants (SNVs) and copy number variants (CNVs). The results demonstrate high APOBEC3B-mediated mutagenesis and alterations in multiple genes of the ERBB2/PI3K/AKT/mTOR, cell cycle, chromatin remodeling, and apoptosis pathways. The establishment of primary fully sequenced cervical tumor cell lines provided us with the opportunity for in vitro and in vivo assessment of whether their mutation profiles are predictive of drug response. Accordingly, primary tumors and xenografts harboring HER2 extracellular domain mutations demonstrated tumor regression when treated with irreversible pan-HER tyrosine kinase inhibitors afatinib or neratinib, in both in vitro and in vivo experiments, while cervical cancer xenografts harboring alterations of at least 1 of the PI3K/AKT/mTOR components demonstrated durable regression only when exposed to combinations of pan-HER and PIK3CA inhibitors. These data help define the genetic landscape of cervical cancer and provide a strong preclinical rationale for clinical trials targeting HER2/PIK3CA/AKT-activating mutations with drug combinations in a large subset of metastatic or recurrent cervical cancer patients.
Results
Whole-Exome Sequencing.
Sixty-nine patients with cervical carcinoma were studied. The clinical features of these patients are presented in SI Appendix, Table S1. Upon biopsy or surgical removal of tumors, primary cell lines were established (15 tumors), or tumors were frozen (54 tumors). Whole-exome sequencing was performed on all tumor specimens. Normal tissue-matched DNA was available for 56 of the tumor samples and sequenced. Whole-exome sequencing was performed using the NimbleGen/Roche capture reagent, followed by 74 base paired-end DNA sequencing on the Illumina HiSeq platform (4). By design, tumor samples were sequenced to a greater depth of coverage to allow detection of somatic mutations in tumors, despite a mixture of normal and tumor cells in these samples. Among tumor samples, each targeted base was sequenced by a mean of 200 independent reads while matched normal DNA had a mean coverage of 173 independent reads per targeted base (SI Appendix, Table S2). Segments of loss of heterozygosity (LOH) were called from the difference in B-allele frequency between tumor-normal pairs, allowing estimates of tumor purity, which was above 60% for frozen tumors and higher for primary cell lines. Somatic mutations were identified in tumors by finding variant reads that were significantly more frequent than expected by chance. Variants in genes implicated in the pathogenesis of cervical cancer were verified by direct Sanger sequencing and RT-PCR, confirming that these genes were expressed in the cervical cancer cell lines (5).
Analysis of Single Nucleotide Variants.
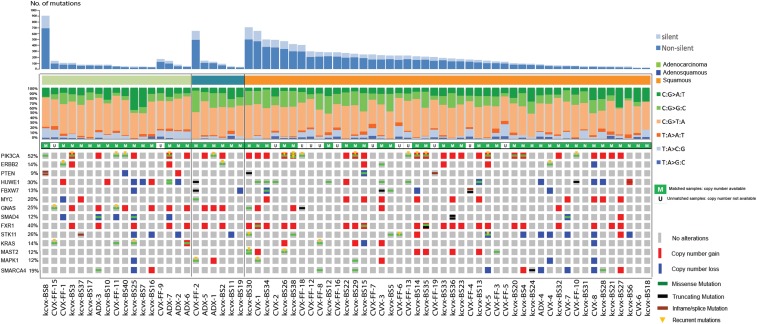
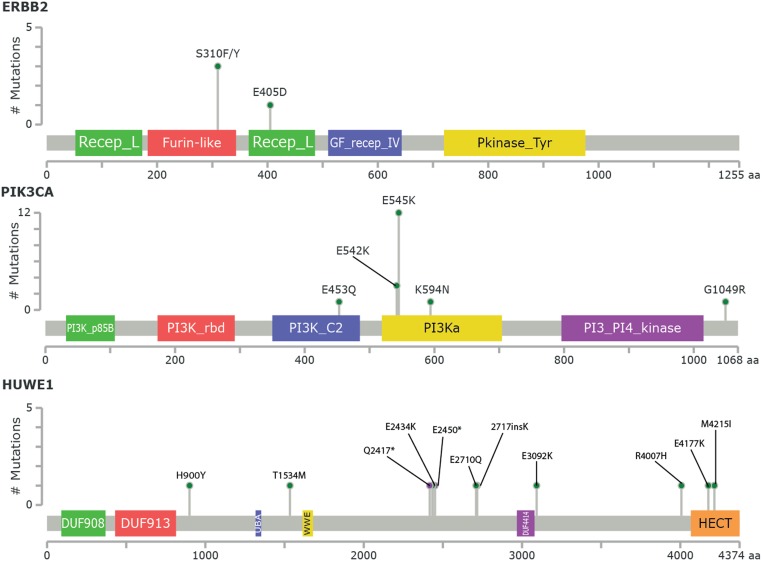
We analyzed somatic mutations in a total of 69 cervical tumors. Results are shown in Fig. 1 and SI Appendix, Tables S3–S5. Recurrent somatic mutations were looked for in 56 different tumor-normal pairs and 13 unmatched samples. The aggregate nonsilent mutation rate across squamous tumors (SCCs) was 5.26 mutations per Mb, significantly higher than the rate of nonsilent mutations found in adenocarcinoma (ACC) (2.04 mutations per Mb) (P = 0.003). Hierarchical clustering of all tumors based on the mutational context revealed that cervical tumor samples are characterized by enrichment in APOBEC cytidine deaminase mutagenesis patterns, with predominantly TCW and WGA mutations (SI Appendix, Figs. S1 and S2). Bayesian nonnegative matrix factorization analysis confirmed the APOBEC signature (SI Appendix, Fig. S3 and Table S6). The mean percentage of APOBEC-related mutations out of the total mutations in SCC was 36.1% while, for ACC, was 24.3% (P = 0.002). Accounting for the rate of protein-altering somatic mutations in these tumors and the size of the exome, the likelihood of seeing the same protein-altering mutation twice by chance at any position among these tumors is <0.002. We also adjusted for the level of expression of each gene from expression data in normal human cervical keratinocytes (6) since we found a higher somatic mutation rate among genes with lower expression, consistent with the effects of transcription-coupled DNA repair to reduce the mutation rate among expressed genes (7). We identified 22 genes with recurrent somatic missense mutations (Fig. 1 and SI Appendix, Table S3). Among these, we found activating mutations E542K and E545K in PIK3CA, the catalytic subunit of phosphoinositide-3 kinase (3 and 12 tumors, respectively), and extracellular domain mutations at S310F/S310Y of the receptor tyrosine kinase gene ERBB2 (3 tumors) (Fig. 2). Furthermore, we found mutations at S216F and S216Y in STK11, a highly conserved serine-threonine kinase known to phosphorylate a threonine residue in the activation loop of 14 kinases in the AMPK family (3 tumors); mutations in GNAS, a gene encoding the guanine nucleotide-binding protein (G protein) alpha subunit (Gsα) in 2 tumors; and mutations at R135T and R135K in MAPK1, a serine/threonine kinase in the MAP kinase signal transduction pathway in 2 tumors. Recurrent mutations also occurred in BAZ2B, ZNF385B, and ZNF493 (SI Appendix, Table S3), the 2 latter genes encoding for zinc finger proteins involved in various aspects of transcriptional regulation (8). Of interest, we also found D297V and D297Y mutations in the receptor tyrosine kinase gene ERBB3 (2 tumors). To prioritize genes with a distinct pattern of somatic mutations from the background, we used the IntOGen web resource (https://www.intogen.org/search) (9). Oncogenes and tumor suppressor genes were considered significant when the q-values for OncodriveCLUST or OncodriveFM, respectively, were smaller than or equal to 0.1. (SI Appendix, Tables S4 and S5, respectively). These genes include but are not limited to PIK3CA (17 tumors, 24.6%), STK11/LKB1, a tumor suppressor gene previously found inactivated in cervical cancer (10, 11) in 6 (8.7%) tumors, and MAPK1 in 3 (4.3%) of the tumors (Fig. 1 and SI Appendix, Tables S4 and S5). Mutations in HUWE1, an E3 ubiquitin that targets c-MYC, Cdc6, histones, and the proapoptotic molecule p53 for degradation (12), were detected in 11 (15.9%) of the tumors. Mutations R4007H, E4177K, and M4215I all occur in the N-lobe of the ubiquitin transferase HECT domain of HUWE1 (Fig. 2). Residue R4007H is on the surface of the N-terminal α1-helix: This helix is conserved over HECT domains, stabilizes the fold, and its deletion increases HECT domain activity and HUWE1 auto-ubiquitination (13), and mutations in this helix of UBE3A are associated with autism (14). Residue E4177K is part of the β5-β6 hairpin that aids recognition of the E2 (15, 16), raising the potential that a charge reversal at this residue could alter recognition of specific E2 partner(s). M4215 is completely buried in a hydrophobic region within the N-lobe. This position is conserved as a hydrophobic residue in other HECT domains (e.g., Ile652 in RSP5) (SI Appendix, Fig. S4) (17). Additional genes found to harbor an increased number of somatic mutations include, but are not limited to, SMAD4, a gene previously correlated to the loss of the tumor suppression function of transforming growth factor type β (TGF-β) in multiple tumors, including cervical cancer (18) (mutated 5 times at different positions in 5.8% of the samples) (SI Appendix, Table S5 and Fig. 1). Additional mutated genes included the fragile X related gene (FXR1), reported to down-regulate tumor necrosis factor α (TNFα) production (i.e., a key mediator of inflammation) at the posttranscriptional level by TGF-β (19, 20). Mutations were also found in the FBXW7 gene (Fig. 1 and SI Appendix, Table S5), a ubiquitin ligase known to regulate a network of proteins with central roles in cell division, cell growth, and differentiation including c-MYC (21, 22), and in multiple zinc finger genes (SI Appendix, Table S5), a family of genes involved in various aspects of gene regulation and previously linked to HPV-induced carcinogenesis (2). Interestingly, 5.8% of samples harbored nonsynonymous mutations in the receptor tyrosine kinase gene ERBB2. We found 3 patients harboring recurrent mutations S310F and/or S310Y (1 of the 3 patients harbored both mutations). S310F and S310Y were previously demonstrated to be driver mutations and potential drug targets in breast, lung, and colorectal cancers (23). Moreover, we found a patient harboring an ERBB2 mutation (E405D) (Fig. 2).
Fig. 1.
Somatic variation pattern underlying cervical cancer. Distribution of number of protein-altering somatic mutations and copy number variations in 69 cervical cancers. Representative significantly mutated genes are listed vertically.
Fig. 2.
Schematic representation of ERBB2, PIK3CA, and HUWE1 functional domains and mutation conservation analysis. Somatic mutations observed are shown. Green dots represent each case of the indicated mutation, and purple and black dots represent truncating alterations.
Analysis of Copy Number Variants.
For the 56 matched tumors, copy number variants (CNVs) were identified. Twelve chromosome segments with more frequent gains of copy number and 40 chromosomes segments with more frequent deletions than expected by chance were found (SI Appendix, Fig. S5 and Tables S7 and S8), including large duplications on chromosome 3 that contains the PIK3CA and FXR-1 genes in over 40% of the samples. There was also amplification of a segment of chromosome 8 containing c-MYC in 12 (17.4%) tumors. Among deletions in chromosome 4, FBXW7 was deleted in 7.2% of tumors (21, 22). Frequent somatic deletions were also detected on chromosome 18, including SMAD4 in 6 tumors (8.7%) (18) and on chromosome 19, including the STK11 gene.
Taken together, CNV and SNV results identified the ERBB2/PIK3CA/AKT/mTOR pathway (71%) (SI Appendix, Figs. S6 and S10), the cell cycle pathway (57.9%) (SI Appendix, Figs. S7 and S10), the apoptosis pathways (69.6%) (SI Appendix, Figs. S8 and S10), and the chromatin remodeling pathway (75.3%) (SI Appendix, Figs. S9 and S10) as major altered pathways in cervical cancer. Finally, alterations in genes involved in the antigen processing and presentation pathway (i.e., KEGG hsa04612), such as HSPA8, CREB1, CTSB, HLA-A, and HLA-B Class I, TAP1 and TAP2, and B2M, were identified in 19%, 14%, 13%, 13%, 13%, 12%, 10%, and 4% of the samples, respectively (SI Appendix, Figs. S11 and S12).
Inhibition of Tumor Cell Growth by Afatinib/Neratinib in HER2/neu-Mutated Cervical Cancer Cell Lines and Xenografts.
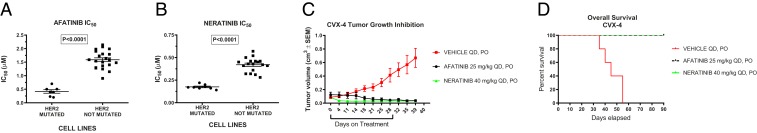
A small subset of lung and breast cancer cell lines (about 1%) harbor mutations in the ERBB2 extracellular and kinase domain. These mutations may confer sensitivity to treatment with irreversible ErbB family inhibitors in some, but not other, human tumor types (23–25). Since 5.8% of our cervical tumors (including 2 primary cell lines) (Fig. 1) harbored ERBB2 extracellular domain mutations, we evaluated the effect of afatinib (Boehringer Ingelheim GmbH, Ingelheim, Germany) and neratinib (Puma Biotechnologies, Los Angeles, CA) on cell growth, cell cycle distribution and signaling in 8 fully sequenced primary cell lines. We found the IC50 (concentration that inhibits response by 50%) values of afatinib and neratinib to be significantly lower in the group of mutated cell lines (i.e., CVX-3 and CVX-4, harboring the E405D mutation and the previously reported S310F extracellular ERBB2 gene mutation, respectively) than in the nonERBB2-mutated control group of tumors (Fig. 3 A and B and SI Appendix, Fig. S13) (mean ± SEM = 0.418 ± 0.065 vs. 1.589 ± 0.071 and 0.175 ± 0.008 vs. 0.420 ± 0.018 μM, P < 0.0001, for afatinib and neratinib, respectively). In ERBB2-mutated cell lines, afatinib and neratinib growth inhibition was associated with a significant and dose-dependent increase in the percentage of cells arrested in the G1 cell cycle phase, as well as a significant and dose-dependent dephosphorylation of HER2 and S6 (SI Appendix, Fig. S14 A and B). Finally, we found afatinib and neratinib to be highly active against human HER2 mutant cervical cancer xenografts. Indeed, as shown in Fig. 3C, daily oral administration of afatinib at 25 mg/kg or neratinib at 40 mg/kg showed a significant tumor growth inhibition in the treated group after 30 d of treatment (P = 0.001 and P = 0.0002, respectively) and significantly improved the overall survival when compared to the control group (P = 0.0001 and P = 0.0001, respectively) (Fig. 3D).
Fig. 3.
ERBB2 extracellular domain mutant cervical cancers are sensitive to ERBB2 inhibition by afatinib or neratinib in vitro and in vivo. (A and B) Mean IC50 values of ERBB2-mutated cervical carcinoma (CC) compared to ERBB2 wild-type CC cell lines (P < 0.0001) in response to afatinib (A) and to neratinib (B). (C) Tumor growth inhibition and (D) overall survival (OS) in response to afatinib and neratinib in a representative Her2/neu-mutated xenograft (CVX-4). The control group received vehicle per os (PO), the afatinib group 25 mg/kg afatinib, and the neratinib group 40 mg/kg neratinib (tumor volume [TV] control vs. afatinib P = 0.001 and control vs. neratinib P = 0.0002, OS control vs. afatinib P = 0.0001 and OS control vs. neratinib P = 0.0001).
Combination of pan-HER and PIK3CA Inhibitors Induces Long-Lasting Regression of Primary Cervical Cancer in Xenografts.
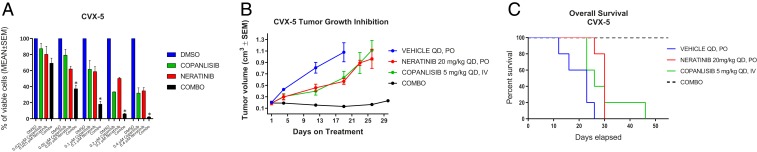
Since 52% of the cervical tumors sequenced, including multiple primary cell lines (Fig. 1), harbored “hot spot” mutations and/or amplifications in the PIK3CA gene and derangements in the PIK3CA pathway, known to play a central role in tumor cell proliferation and survival in multiple human cancers, we evaluated the effect of copanlisib (Bayer Pharmaceuticals Inc.), an intravenous, selective pan-Class I PI3K inhibitor highly active in PIK3CA mutant tumors recently approved by the Food and Drug Administration (FDA) for the treatment of patients with relapsed follicular lymphoma, as single agent or in combination with a pan-HER inhibitor (neratinib) on cell growth of multiple cervical cancer cell lines. Tumor-harboring alterations in the PI3K/AKT/mTOR pathway demonstrated variable but overall limited in vitro sensitivity to single agent copanlisib or neratinib (Fig. 4A and SI Appendix, Fig. S15). In contrast, the combination of the 2 inhibitors induced a potent and highly synergistic (evaluated by CompuSyn software) (SI Appendix, Table S9) cell growth inhibition in vitro, as well as decreased pS6, when compared to single agent therapy in all primary tumor models available (Fig. 4A and SI Appendix, Figs. S15 and S16). Consistent with the in vitro data, when the activity of copanlisib and neratinib was evaluated in vivo against CVX-5, a PIK3CA-mutated cervical tumor, we found single agent copanlisib and neratinib to be active but only transiently effective in controlling tumor growth (Fig. 4B). Remarkably, the combination of the 2 inhibitors induced a durable tumor growth inhibition when compared to single agent copanlisib and neratinib (P = 0.02 and P = 0.006, respectively), with no evidence of increased acute or chronic toxicity (SI Appendix, Fig. S17). This effect lasted for the entire treatment period (i.e., 4 wk) and significantly prolonged the life of the CVX-5–xenografted animals when compared to all of the other treatment groups (Fig. 4 B and C, P = 0.0001).
Fig. 4.
Combinations of pan-HER and PIK3CA inhibitors induce synergistic in vitro and in vivo responses in tumors with derangements in the ERBB2/PI3K/AKT/mTOR pathway. (A) Cell viability of the PIK3CA-mutated and amplified CVX-5 primary cell line treated with neratinib, copanlisib, and their combination for 72 h. Cell viability was analyzed by flow cytometry and was normalized to the mean of the untreated control (Materials and Methods) and expressed as mean ± SEM; 3 independent experiments were performed (*P < 0.05 when compared to the control, to neratinib, and to copanlisib). (B and C) In vivo treatment of CVX-5 xenografts. Mice were treated with vehicle, neratinib (20 mg/kg), copanlisib (5 mg/kg), or the combination of the two for up to 30 d. Tumor volumes are reported as mean ± SEM. Tumor growth is significantly different between control (CTRL) and the treated groups (CTRL vs. copanlisib, neratinib, and combination, P = 0.001, P = 0.0002, and P < 0.0001, respectively). Similarly, the OS shows significant difference (P = 0.0001, P = 0.0001, and P < 0.0001, respectively). DMSO, dimethyl sulfoxide; IV, intravenous.
Discussion
We report the whole-exome sequencing (WES) of a large cervical cancer cohort of fresh-frozen tumors and primary cervical cancer cell lines. Importantly, the cell lines establishment and comprehensive WES characterization of 15 primary cervical tumors gave us the opportunity to preclinically evaluate whether specific mutation profiles, such as recurrent extracellular ERBB2 mutations and PIK3CA hot spot mutations, may be “druggable” in cervical cancer patients.
We found 22 genes to harbor recurrent missense mutations in cervical cancer and a strong APOBEC-mediated mutagenesis pattern in both SCC as well as ACC. These results are in keeping with the high APOBEC3B mRNA transcript levels we found in HPV-infected tumors when compared to normal HPV-negative cervical keratinocytes (SI Appendix, Fig. S2) (6), the analysis of the landscape of 115 early stages cervical cancer samples by Ojesina et al. (26), and The Cancer Genome Atlas (TCGA) comprehensive genetic analysis on 226 cervical cancers (27). The APOBEC mutagenesis signatures (28) were more common in cervical tumors with squamous and adenosquamous histology when compared to adenocarcinomas (SI Appendix, Fig. S3 and Table S6) and, importantly, extended into a subset of genes considered by multiple criteria to be cancer drivers (Fig. 1 and SI Appendix, Tables S3–S5). A comparison of the similarities and differences between the WES results of our study and those reported in previous studies by Ojesina et al. (26) and the TCGA Network (27) are depicted in SI Appendix, Tables S10–S12).
We also identified 12 somatic CNV gains and 40 CNV losses that occurred more often than expected by chance, with the most frequent events occurring in pathways similar to those found from analysis of SNVs, including the ERBB2/PI3K/AKT/mTOR, apoptosis, chromatin remodeling, and cell cycle pathway. Of interest, mutations in HUWE1, an E3 ubiquitin ligase that targets multiple substrates including c-MYC, were detected in 11 (15.9%) tumors (Figs. 1 and 2) (12, 29). Although the function(s) of these mutations in cervical cancer are uncertain, alteration of HUWE1 has been previously reported in multiple human tumors, including lung, breast, and colorectal carcinomas (12, 29). Inoue et al. have recently reported that selective HUWE1 deletion in the skin epithelium augmented the formation of highly proliferative skin tumors (29). This phenotype was largely rescued by genetic deletion of c-MYC, an oncogene known to promote cell cycle progression and proliferation and frequently amplified in multiple human tumors, suggesting that HUWE1-mediated tumor suppression is dependent on its regulation of c-MYC (29). Notably, in this regard, 12 additional cervical tumors had amplification of the segment of chromosome 8 containing c-MYC (Fig. 1). Taken together, our results suggest a potential role for HUWE1 mutations in cervical carcinoma and identify the HUWE1/c-MYC pathway as one of the frequent altered pathways in this disease (Fig. 1 and SI Appendix, Figs. S7 and S10).
Recurrent mutations at a single codon (R201) and gene amplifications in the GNAS were identified in 13 cervical tumors. Of interest, GNAS amplifications were significantly more common in adenocarcinoma when compared to squamous tumors (ACC 8 of 17 vs. SCC 3 of 46, P = 0.0001). GNAS mutations have previously been identified in mucinous endocervical adenocarcinomas and pituitary and intraductal mucinous neoplasms of the pancreas and found to constitutively activate GNAS function, causing high adenyl cyclase activity and increased adenosine 3′,5′-monophosphate (cAMP) levels (30–32). Because the guanine-binding protein-couple receptor (GPCR) pathway is a known major target for drug development and drugs targeting GPCR have already been shown to suppress malignant phenotypes of various human cancer cell lines, our results suggest potential treatment options in cervical cancer patients, particularly in patients with cervical adenocarcinomas.
We also identified genetic alterations in several genes encoding for proteins known to play crucial roles during viral and tumor–host immune system interactions. For example, mutations in SMAD4, an intermediate of TGF-β signaling, were identified in 5.8% of the samples, with an additional 8.7% of the tumors found to have deletion of the SMAD4 gene by CNV analysis. Loss of SMAD4 function has been previously shown to cause alterations in the TGF-β1 signal transduction route, and lack of sensitivity of the SMAD4-mutated cells to the inhibitory signal mediated by TGF-β (i.e., inhibition in epithelial cell proliferation) may cause increased angiogenesis due to increased TGF-β secretion (18). Since TGF-β is also a powerful immunosuppressive cytokine, it is tempting to speculate that cervical tumors with SMAD4 mutations secreting high TGF-β may also potentially suppress or down-regulate antitumor immune response against HPV-infected cervical tumor cells (18, 33). Alterations in multiple genes playing crucial roles in the antigen-presenting capability of cervical tumor cells (i.e., HLA, TAP1/2), as well as in the regulation of apoptosis (CASP8, BCL2L1, and MCL-1), were also detected, further supporting the notion that HPV-infected carcinomas may undergo host immune system selective pressure and, consequently, develop escape mechanisms to avoid immune system recognition/destruction. In agreement with this hypothesis, novel immunotherapeutic strategies based on immune checkpoint inhibitors (i.e., nivolumab/pembrolizumab) have demonstrated limited clinical responses in HPV-infected cervical cancer patients with advanced/recurrent disease (34).
The ERBB2 oncogene, a member of the EGF family of receptor tyrosine kinases (RTKs) encoding for the HER2 receptor, leads to phosphorylation and regulation of the activity of a variety of intracellular signaling molecules and transcription factors that modulate various biological responses, such as proliferation, survival, migration, and differentiation (23–25). Moreover, ERBB2 functions as an upstream regulator of the PIK3CA/AKT/mTOR signaling pathway. In our study, we identified mutations in the extracellular domain of ERBB2 in 5.8% of the carcinomas. Because of the establishment and sequencing of multiple primary cancer cell lines (2 of which harboring the HER2/neu extracellular domain mutations), we assessed in preclinical studies whether the HER2/neu mutation profile may be predictive of drug response in cervical cancer patients. We found both ERBB2 mutations to confer significant cervical tumor sensitivity to the exposure to afatinib or neratinib (i.e., 2 potent, irreversible pan-HER inhibitors). Taken together, our in vitro and in vivo experimental data in cervical cancer xenografts support and expand previous findings showing that the S310F extracellular ERBB2 gene mutation identified in breast, lung, and colorectal cancers and the E405D mutation may act as druggable mutations in cervical tumors. Overall, these data suggest that afatinib and neratinib, 2 FDA-approved drugs in lung and breast cancer patients, may represent a potentially valid therapeutic option for patients harboring ERBB2-mutated advanced/recurrent cervical cancers.
The phosphatidylinositol 3-kinase/AKT (PI3K/AKT)-mammalian target of rapamycin (mTOR) signaling cascade plays a central role in diverse cellular responses, such as proliferation, survival, mobility, metabolism, and control of malignant cellular growth (35). WES results demonstrated 24.6% of the tumor samples to harbor activating recurrent missense mutations in exon 9 of PIK3CA and an additional 42% to harbor amplification of the PIK3CA gene (Fig. 1 and SI Appendix, Figs. S6 and S10). These findings suggest the common involvement of this pathway in both squamous and adenocarcinoma of the uterine cervix. Accordingly, we preclinically evaluated the efficacy of copanlisib and neratinib as single agents and in combination against multiple genetically characterized primary cervical cancer cell lines harboring derangements in the ERBB2/PI3K/AKT/mTOR pathway. We found single agent copanlisib and neratinib to have limited activity in vitro against PIK3CA-mutated cell lines, and, accordingly, both single agents were only transiently effective in vivo in controlling the growth of CVX-5 xenografts harboring oncogenic PIK3CA mutations. These results support the view of a weak driver oncogenic activity of PIK3CA in cervical tumors and/or the rapid feedback up-regulation of compensatory mechanisms when PI3K is blocked, which may both confer resistance to single agent treatment (35). The combination treatment, however, showed a remarkable synergistic effect against CVX-5 and all other primary tumor model available harboring derangements in the PI3K/AKT/mTOR pathway. Importantly, this effect was also consistently detected in PIK3CA-mutated xenograft models. Taken together, these results suggest that a large subset of cervical tumors might greatly benefit from existing ERBB2/PIK3CA/AKT/mTOR-targeted drugs.
In our analysis, other potentially druggable targets include, but are not limited to, the antiapoptosis genes BCL2L1 and MCL1 (Fig. 1 and SI Appendix, Figs. S8 and S10), and the chromatin-remodeling genes ARID1A, MLL2, and EZH2 (SI Appendix, Figs. S9 and S10) were identified in cervical tumors. Further efforts will be necessary to preclinically validate the impact of these mutations on the sensitivity of primary cervical tumor cell lines to antiapoptosis and antimethyltransferase inhibitors that are currently in clinical trials (i.e., navitoclax, S63845, and tazemetostat).
In conclusion, our results define the genetic landscape of squamous cell carcinoma and adenocarcinoma of the uterine cervix and identify multiple genes/pathways that are frequently mutated in these tumors. While cervical cancer patients with early vs. advanced stage disease have different treatment options and survival outcomes, preclinical in vitro and in vivo data suggest that ERBB2/PI3K/AKT/mTOR pathway-deranged tumors may be “targetable” in a large subset of cervical cancer patients by using pan-HER and PIK3CA inhibitors. These findings will help guide further research and targeted therapies against this highly prevalent cancer worldwide.
Materials and Methods
Patients and Specimens.
Briefly, DNA and RNA fractions from consented patients were isolated from biological samples (i.e., fresh tumor specimens, primary cell lines, venous blood, primary fibroblasts, or frozen myometrium) using an AllPrep DNA/RNA Mini Kit (Qiagen) following the manufacturer’s procedure and prepared as described (6) (see SI Appendix, Materials and Methods for details). The study protocol was approved by the Yale Human Investigation Committee and was conducted in accordance with the Declaration of Helsinki. DNA was extracted from 54 primary tumors and 15 primary cell lines with limited passages.
WES.
Genomic DNA was captured and analyzed as described (4, 36–38). Identified variants were annotated based on novelty, impact on the encoded protein, conservation, and expression using an automated pipeline. See SI Appendix, Materials and Methods for details.
Somatic Copy Number Variation.
Copy number variants were identified by means of Excavator (version 2.1) (39) and GISTIC (version 2.0.16) (40) to calculate significantly amplified or deleted regions as previously described (39, 40). See SI Appendix, Materials and Methods for details.
Real-Time Reverse Transcription-PCR (qRT-PCR).
RNA isolation and quantitative RT-PCR were performed using standard protocols on the AB 7500 Real-Time PCR instrument. Primer sequences are described in SI Appendix.
Drug Preparation and In Vitro Chemosensitivity Assay.
Cells were exposed to afatinib (Boehringer) or neratinib (Puma) or copanlisib (Bayer), dissolved per the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO) as single agents or in combination, at concentrations ranging from 0.1 to 10 mM. After 72 h of incubation, cells were harvested for flow cytometric count. Detailed information is provided in SI Appendix, Materials and Methods.
In Vivo Experiments in Tumors Harboring ERBB2/PI3K/AKT/mTOR Pathway Alterations.
Briefly, HER2/neu-mutated (CVX-4) and PIK3CA-mutated (CVX-5) cell lines were injected into the s.c. region of 5- to 6-wk-old SCID mice (Envigo Dublin VA, Horst, Netherlands). The mice were divided into multiple treatment groups (i.e., controls, afatinib, neratinib, copanlisib, single agents, or combinations) and received vehicle (0.5% methylcellulose-0.4% Tween 80) or the drugs, as described in Figs. 3 and 4, and in SI Appendix, Materials and Methods. All of the mice were housed and treated in accordance with the policies set forth by the Yale Institutional Animal Care and Use Committee (IACUC).
Statistical Analysis.
All statistical analyses were performed using Prism 6 software (GraphPad Prism Software Inc., San Diego, CA). A P value of <0.05 was considered as the level of statistical significance. See SI Appendix, Materials and Methods for details.
Data Availability.
All data discussed in the paper are available in Dataset S1.
Supplementary Material
Acknowledgments
This work was supported in part by Gilead Sciences Inc. (Foster City, CA), Puma Biotechnology, Inc. (Los Angeles, CA), NIH Grants R01 CA154460-01 and U01 CA176067-01A1, the Deborah Bunn Alley Foundation, the Tina Brozman Foundation, the Discovery to Cure Foundation, and the Guido Berlucchi Foundation (to A.D.S.). This investigation was also supported by NIH Research Grant CA-16359 from the National Cancer Institute and by Stand-up-to Cancer (SU2C) Convergence Grant 2.0 (to A.D.S.). We thank Katerina Politi (Department of Pathology, Yale University) for generous support with reagents. Preliminary results for this work were presented at the 50th Annual Meeting of the Society of Gynecologic Oncology, March 16–19, 2019, Honolulu, HI (Poster ID 1215: “Whole exome sequencing (WES) reveals novel therapeutic targets in cervical cancer”, by Lopez S. et al.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911385116/-/DCSupplemental.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Bosch F. X., Lorincz A., Muñoz N., Meijer C. J., Shah K. V., The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55, 244–265 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tewari K. S., Monk B. J., “Invasive cervical cancer” in Clinical Gynecologic Oncology, DiSaia Philip J., Creasman William T., Eds. (Elsevier Inc., ed. 8, 2012), pp. 51–119. [Google Scholar]
- 4.Choi M., et al. , Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 106, 19096–19101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao S., et al. , Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc. Natl. Acad. Sci. U.S.A. 110, 2916–2921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santin A. D., et al. , Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: Identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology 331, 269–291 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Pleasance E. D., et al. , A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishna S. S., Majumdar I., Grishin N. V., Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 31, 532–550 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Perez A., et al. , IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods 10, 1081–1082 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alessi D. R., Sakamoto K., Bayascas J. R., LKB1-dependent signaling pathways. Annu. Rev. Biochem. 75, 137–163 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Wingo S. N., et al. , Somatic LKB1 mutations promote cervical cancer progression. PLoS One 4, e5137 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen D., et al. , ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121, 1071–1083 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Pandya R. K., Partridge J. R., Love K. R., Schwartz T. U., Ploegh H. L., A structural element within the HUWE1 HECT domain modulates self-ubiquitination and substrate ubiquitination activities. J. Biol. Chem. 285, 5664–5673 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi J. J., et al. , An autism-linked mutation disables phosphorylation control of UBE3A. Cell 162, 795–807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L., et al. , Structure of an E6AP-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321–1326 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Kamadurai H. B., et al. , Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamadurai H. B., et al. , Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. Elife 2, e00828 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloth J. N., et al. , Expression of Smad2 and Smad4 in cervical cancer: Absent nuclear Smad4 expression correlates with poor survival. Mod. Pathol. 21, 866–875 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Garnon J., et al. , Fragile X-related protein FXR1P regulates proinflammatory cytokine tumor necrosis factor expression at the post-transcriptional level. J. Biol. Chem. 280, 5750–5763 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Khera T. K., Dick A. D., Nicholson L. B., Fragile X-related protein FXR1 controls post-transcriptional suppression of lipopolysaccharide-induced tumour necrosis factor-alpha production by transforming growth factor-beta1. FEBS J. 277, 2754–2765 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P., Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Welcker M., Clurman B. E., FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Cocco E., Lopez S., Santin A. D., Scaltriti M., Prevalence and role of HER2 mutations in cancer. Pharmacol. Ther. 199, 188–196 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose R., et al. , Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 3, 224–237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greulich H., et al. , Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc. Natl. Acad. Sci. U.S.A. 109, 14476–14481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojesina A. I., et al. , Landscape of genomic alterations in cervical carcinomas. Nature 506, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network et al. , Integrated genomic and molecular characterization of cervical cancer. Nature 543, 378–384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexandrov L. B., et al. , Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue S., et al. , Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev. 27, 1101–1114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freda P. U., et al. , Analysis of GNAS mutations in 60 growth hormone secreting pituitary tumors: Correlation with clinical and pathological characteristics and surgical outcome based on highly sensitive GH and IGF-I criteria for remission. Pituitary 10, 275–282 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Wu J., et al. , Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci. Transl. Med. 3, 92ra66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsubara A., et al. , Lobular endocervical glandular hyperplasia is a neoplastic entity with frequent activating GNAS mutations. Am. J. Surg. Pathol. 38, 370–376 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Malkoski S. P., Wang X. J., Two sides of the story? Smad4 loss in pancreatic cancer versus head-and-neck cancer. FEBS Lett. 586, 1984–1992 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frenel J. S., et al. , Safety and efficacy of pembrolizumab in advanced, programmed death ligand 1-positive cervical cancer: Results from the phase Ib KEYNOTE-028 trial. J. Clin. Oncol. 35, 4035–4041 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Hanker A. B., Kaklamani V., Arteaga C. L., Challenges for the clinical development of PI3K inhibitors: Strategies to improve their impact in solid tumors. Cancer Discov. 9, 482–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cibulskis K., et al. , Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magi A., et al. , EXCAVATOR: Detecting copy number variants from whole-exome sequencing data. Genome Biol. 14, R120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beroukhim R., et al. , Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc. Natl. Acad. Sci. U.S.A. 104, 20007–20012 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper are available in Dataset S1.