Significance
Archaea synthesize distinctive membrane-spanning lipids (GDGTs) that are readily preserved in ancient sediments and utilized as paleotemperature proxies to reconstruct sea surface temperatures deep in Earth’s past. However, properly interpreting GDGT-based biomarker proxies requires an accurate assessment of the archaea that contribute to GDGT pools in modern environments and of the proteins necessary for synthesizing GDGTs. In this study, we identify 2 radical SAM proteins in Sulfolobus acidocaldarius that are required to produce these molecules. Bioinformatics analyses of these GDGT ring synthesis proteins reveal that Thaumarchaeota are the dominant source of cyclized GDGTs in the open ocean, allowing us to constrain one factor of uncertainty in the application of GDGT-based paleotemperature proxies.
Keywords: GDGT, radical SAM, Sulfolobus, paleotemperature proxies
Abstract
Glycerol dibiphytanyl glycerol tetraethers (GDGTs) are distinctive archaeal membrane-spanning lipids with up to eight cyclopentane rings and/or one cyclohexane ring. The number of rings added to the GDGT core structure can vary as a function of environmental conditions, such as changes in growth temperature. This physiological response enables cyclic GDGTs preserved in sediments to be employed as proxies for reconstructing past global and regional temperatures and to provide fundamental insights into ancient climate variability. Yet, confidence in GDGT-based paleotemperature proxies is hindered by uncertainty concerning the archaeal communities contributing to GDGT pools in modern environments and ambiguity in the environmental and physiological factors that affect GDGT cyclization in extant archaea. To properly constrain these uncertainties, a comprehensive understanding of GDGT biosynthesis is required. Here, we identify 2 GDGT ring synthases, GrsA and GrsB, essential for GDGT ring formation in Sulfolobus acidocaldarius. Both proteins are radical S-adenosylmethionine proteins, indicating that GDGT cyclization occurs through a free radical mechanism. In addition, we demonstrate that GrsA introduces rings specifically at the C-7 position of the core GDGT lipid, while GrsB cyclizes at the C-3 position, suggesting that cyclization patterns are differentially controlled by 2 separate enzymes and potentially influenced by distinct environmental factors. Finally, phylogenetic analyses of the Grs proteins reveal that marine Thaumarchaeota, and not Euryarchaeota, are the dominant source of cyclized GDGTs in open ocean settings, addressing a major source of uncertainty in GDGT-based paleotemperature proxy applications.
Archaea are distinct from bacteria and eukaryotes in many respects, including, most prominently, the chemistry of their membrane lipids. While bacterial and eukaryotic lipids are composed of fatty acid bilayers, archaeal membranes comprise isoprenoidal bilayers that may be fused to form unique 86 carbon membrane-spanning structures known as glycerol dibiphytanyl glycerol tetraethers (GDGTs) (1). GDGTs are further modified through the insertion of up to 8 cyclopentyl moieties at the C-7 and C-3 positions of each of the biphytanyl chains (Fig. 1), cross-linking of the 2 biphytanyl chains, and inclusion of a cyclohexane ring at C-11 believed to be specific to taxa within the Thaumarchaeota phylum (2). It is proposed that archaea modify their GDGTs as a way to adjust membrane rigidity in response to changes in environmental temperature, pH, and/or salinity and that these modifications provide a protective effect under fluctuating environmental conditions (3). Several studies have specifically documented a relationship between growth temperature and GDGT cyclization (4–6), and this association is the basis for a variety of GDGT-based paleoenvironmental proxies, such as the TEX86 (Tetraether Index of 86 carbons) sea surface temperature proxy. This index, and other related parameters such as the Ring Index, allow the occurrence of cyclic GDGTs and their isomers in marine surface sediments to be correlated with overlying sea surface temperatures (7, 8). Given the ubiquity of GDGTs in marine sediments and their resistance to diagenetic alterations, these proxies are potentially powerful paleoclimatology tools that allow for reconstruction of past temperature changes, providing insight into Earth system dynamics deep in geologic time (9).
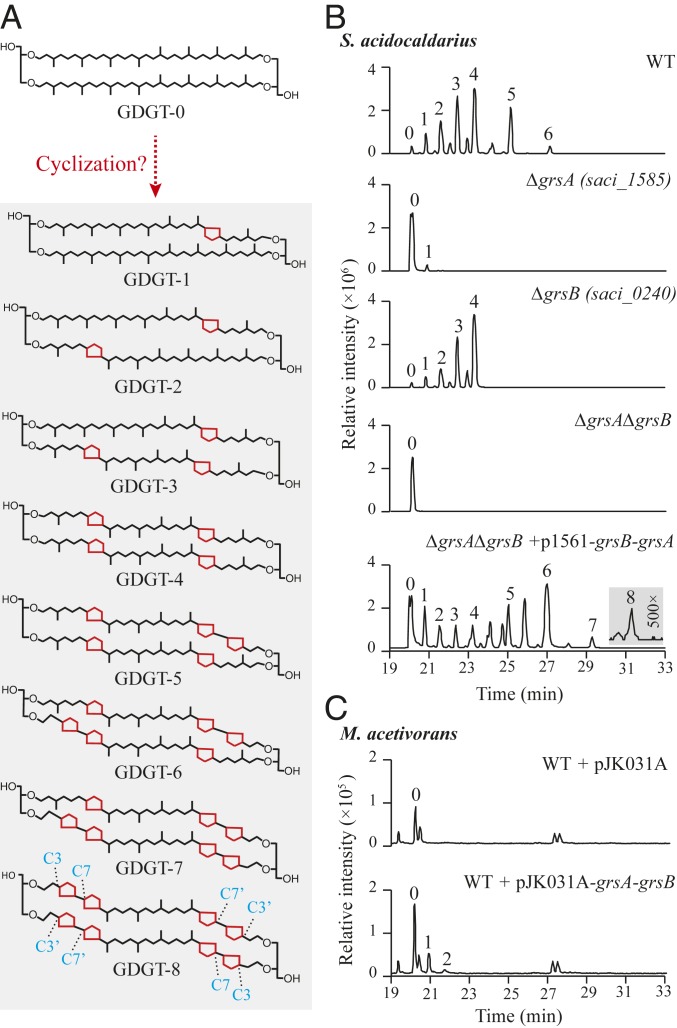
Fig. 1.
GrsA (Saci_1585) and GrsB (Saci_0240) are required for GDGT ring formation. (A) Various GDGT structures produced by S. acidocaldarius. Acyclic GDGT-0 is hypothesized to be the biosynthetic precursor to cyclic GDGTs. The C-7 and C-3 positions are indicated on GDGT-8. (B) LC-MS merged extracted ion chromatograms (EICs) of acid-hydrolyzed lipid extracts of S. acidocaldarius wild type (WT), grs single and double mutants, and the complemented double deletion strain. Grey boxes indicate amplification of y axis to highlight smaller peaks. (C) LC-MS merged EICs of acid-hydrolyzed lipid extracts of M. acetivorans WT with empty plasmid pJK031A showing only a minor production of GDGT-0 and M. acetivorans WT with S. acidocaldarius grsA and grsB heterologously expressed producing GDGTs with up to 2 rings. The numbers above each chromatographic peak indicate the number of cyclopentane rings in the GDGT structure. Multiple isomers of ring-containing GDGTs and their mass spectra are shown in SI Appendix, Fig. S1.
However, a robust assessment of GDGT-based proxies is hampered by a variety of unresolved factors, including the influence of environmental factors other than temperature on increased tetraether cyclization (9–13) and ambiguity in the archaeal taxa contributing to GDGT pools in marine ecosystems (14, 15). Specifically, applications of TEX86 assume that mesophilic Marine Group I (MG-I) Thaumarchaeota are the dominant source of cyclic GDGTs in the pelagic zone (9). However, studies have shown that MG-II Euryarchaeota are abundant in near-surface waters (14, 15), and some have suggested that marine Euryarchaeota are significant contributors to ocean cyclic GDGT pools at shallower depths (13, 16). A mixed archaeal population producing cyclized GDGTs in the marine water column could significantly affect TEX86-based temperature estimates, especially if GDGT distribution patterns reflect changes in archaeal community structure rather than changes in temperature. Therefore, constraining the archaeal sources of GDGTs in the open oceans is critical but has been difficult, as there are currently no MG-II isolates in culture to directly test for GDGT production and the proteins required for tetraether biosynthesis are still unknown (17–20). Identification of GDGT ring formation proteins would allow for a direct assessment through bioinformatics analyses of the archaeal taxa potentially producing cyclized GDGTs in marine environments. This would also enable transcriptomic and biochemical studies that directly assess how changes in temperature affect the expression and biochemical activity of these proteins. These types of studies would provide significant insight into current GDGT-based proxy ambiguities and vastly improve the development and understanding of these paleotemperature proxies.
In this study, we utilized a comparative genomics approach coupled to gene deletion and lipid analyses to identify 2 GDGT ring synthase proteins, GrsA and GrsB, in Sulfolobus acidocaldarius. Although S. acidocaldarius is a thermoacidophile not detected in marine systems that would be applicable to TEX86 proxies, it does produce GDGTs with up to 8 cyclopentyl rings (Fig. 1A), and it is one of a few genetically tractable archaeal model systems (21). Our studies revealed that deletion of grsA and grsB in S. acidocaldarius results in complete loss of GDGT cyclization. In addition, we demonstrate that each tetraether cyclization protein inserts cyclopentane rings at specific locations on the GDGT core lipid and provide insight into the overall biochemical mechanism of GDGT synthesis. Finally, bioinformatics analyses show that Grs homologs in marine systems are dominated by MG-I Thaumarchaeota species and validate the underlying TEX86 assumption that Thaumarchaeota, and not Euryarchaeota, are the dominant contributors to cyclic GDGT pools in the surface ocean.
Results
Identification of 2 GDGT Cyclization Proteins in S. acidocaldarius.
To identify potential GDGT cyclization candidate proteins, we first hypothesized that the introduction of cyclopentane rings into the GDGT core structure would require a free radical mechanism, most likely carried out by a radical S-adenosylmethionine (SAM) protein. The radical SAM protein family encompasses a wide class of enzymes that generate a deoxyadenosyl radical to initiate a diverse set of otherwise difficult chemical reactions (22). We have previously shown that radical SAM proteins are critical for fine-tuning of various lipid structures, including formation of the nonhydrolyzable calditol head group in the membrane lipids of S. acidocaldarius and modifications to hopanoid lipids in bacteria (23–26). Both modifications involve formation of new C−C bonds at otherwise unreactive carbon atoms. The S. acidocaldarius genome contains 18 annotated radical SAMs (Pfam: 04055), and we searched for homologs of each of these proteins that were present in the genomes of archaea that produce cyclic GDGTs and absent from those that produce only GDGT-0 or which do not produce any GDGTs at all. This analysis resulted in 3 GDGT ring synthase (grs) candidate genes: saci_0240, saci_1585, and saci_1785.
Deletion of saci_1785 in S. acidocaldarius did not alter tetraether lipid production, nor did we observe any other phenotypic consequences of this deletion. However, deletion of saci_0240 and saci_1585 resulted in a dramatic reduction in the number of GDGT rings (Fig. 1B and SI Appendix, Fig. S1). Mutants lacking saci_1585 (grsA) produced primarily GDGT-0 with a small amount of GDGT-1, while saci_0240 (grsB) deletion mutants produced GDGTs with up to a maximum of 4 rings. Deleting both grsA and grsB together resulted in the complete loss of GDGT cyclization, indicating that ring formation is not essential in S. acidocaldarius, although we do observe a reduced growth rate (SI Appendix, Fig. S2). Complementation of the double deletion strain by expressing grsA and grsB together restored the ability to produce GDGTs with up to 8 cyclopentyl rings (Fig. 1B).
We next wanted to determine whether any other proteins are required for inserting cyclopentyl rings into the core GDGT structure. To do so, we heterologously expressed each gene in Methanosarcina acetivorans, a methanogen that produces minor amounts of GDGT-0 but does not produce any cyclized GDGTs. Expression of grsA and grsB together in M. acetivorans produced GDGTs with 1 or 2 cyclopentyl rings (Fig. 1C), indicating that these 2 proteins are sufficient for ring formation. However, GrsA and GrsB did not produce GDGTs with more than 2 rings in M. acetivorans, and expression of grsA alone resulted in the production of GDGT-1, while expression of grsB alone did not produce any cyclized GDGTs (SI Appendix, Fig. S3). This partial complementation is most likely due to a combination of factors, including inefficient expression of thermophilic proteins in a mesophilic strain and the small amount of endogenous GDGT-0, a potential Grs substrate, available in the heterologous host. Nonetheless, these expression results in M. acetivorans together with the gene deletions analyses in S. acidocaldarius confirm that GrsA and GrsB are necessary for GDGT ring formation.
Grs Proteins Add Cyclopentane Rings at Distinct Locations.
Having established that GrsA and GrsB are required for GDGT cyclization, we subsequently wanted to assess how these 2 proteins together generate the various ring patterns observed in S. acidocaldarius membrane-spanning lipids. Deletion of grsB indicated that GrsA adds 4 rings to the core structure (Fig. 1B), and this was confirmed by complementing the double deletion mutant with grsA alone (Fig. 2A). In contrast, deletion of grsA suggested that GrsB only introduces one ring (Fig. 1B). But complementation of the double deletion mutant with only grsB resulted in the production of GDGT-1 and a small amount GDGT-2 (Fig. 2A), demonstrating that GrsB can add multiple rings but is less efficient than GrsA. Because the ∆grsA∆grsB strain only produces GDGT-0, this inefficiency may be a result of GDGT-0 not being the preferred GrsB substrate. We hypothesized that GrsB might favor a cyclized GDGT substrate with rings already introduced at a specific position and that each protein is potentially cyclizing the isoprenoid structure at distinct locations (Fig. 2B).
Fig. 2.
GrsA and GrsB insert rings at distinct locations on the core GDGT structure. (A) LC-MS merged EICs of acid-hydrolyzed extracts from the complemented ΔgrsAΔgrsB deletion strains showing that GrsA can generate GDGT-1 to GDGT-4 (Top) and GrsB only produces GDGT-1 and GDGT-2 (Bottom). (B) GrsA is proposed to utilize GDGT-0 as a substrate and introduces 4 rings at the C-7 position, while GrsB prefers a substrate with rings at C-7 and introduces rings at the C-3 position. GrsB is also able to introduce rings at the C-3 position of GDGT-0, but this is thought to be an unfavorable side reaction in the complementation experiments. (C) GC-MS total ion chromatograms of biphytanes released by ether cleavage of lipid extracts from S. acidocaldarius WT, ΔgrsAΔgrsB complemented with grsA alone, grsB alone, and both genes together, showing that GrsA synthesizes rings exclusively at C-7 and GrsB at C-3. Grey boxes indicate amplification of y axis to highlight smaller peaks. bp, biphytane. Mass spectra of biphytanes are shown in SI Appendix, Fig. S4.
If our hypothesis is correct, then the GDGT-1 and GDGT-2 produced by grsB complementation and the GDGT-1, GDGT-2, GDGT-3, and GDGT-4 generated through grsA complementation should have rings at different positions (Fig. 2A). To test this, biphytane structures were generated from complemented and wild-type lipid extracts through ether bond cleavage and analyzed by gas chromatography−mass spectrometry (GC-MS) (27). In the extracts of the grsA-complemented mutant, biphytanes with no rings or with 1 or 2 rings at C-7 were detected, indicating that GrsA only adds rings at the C-7 position (Fig. 2C and SI Appendix, Fig. S4). In the lipid extracts of the grsB-complemented mutant, only a small amount of biphytane with one ring could be detected, and this ring was located exclusively at the C-3 position. Further, we only observed biphytanes with rings at both the C-3 and C-7 positions when grsA and grsB are expressed together and did not detect any biphytanes with rings only at C-3 in this strain. All told, these data demonstrate that GrsA and GrsB introduce rings at C-7 and C-3, respectively, and that GrsB prefers a substrate with existing C-7 rings (Fig. 2B). This suggests that cyclization at C-7 by GrsA is occurring prior to cyclization by GrsB at C-3, and, therefore, ring formation progresses from the center of the tetraether lipid outward toward the glycerol moieties.
Redundant Grs Copies Identified in Archaeal Genomes Are Functional.
The discovery that specific tetraether rings are introduced by distinct proteins indicates that we may be able to predict the cyclized GDGT structures produced by different archaea based on the Grs homologs present in their genomes. To determine whether this is possible, we searched for GrsA and GrsB homologs in a subset of Crenarchaeota, Thaumarchaeota, and Euryarchaeota species for which GDGT profiles have been determined (12). We predicted that archaea that produce GDGTs with 1 to 4 rings at C-7 would have one GrsA homolog in their genome, while archaea that produce more-complex structures such as GDGT-5 through GDGT-8 or crenarchaeol, with an additional cyclohexane ring at C-11, would have a second synthase in their genomes. This pattern was observed for several of the archaea in our search (SI Appendix, Table S1). However, we also found that some archaea have multiple GrsA or GrsB copies, which was unexpected given their lipid profiles. For example, Saccharolobus solfataricus, a Sulfolobales species phylogenetically related to S. acidocaldarius that also produces GDGTs with up to 8 cyclopentane rings (18), has 1 GrsA (SSO0604) and 2 potential GrsB homologs (SSO0477 and SSO2881). Based on sequence homology alone, it is unclear whether these multiple GrsB copies are functional and whether they are both catalyzing the insertion of cyclopentane rings at C-3.
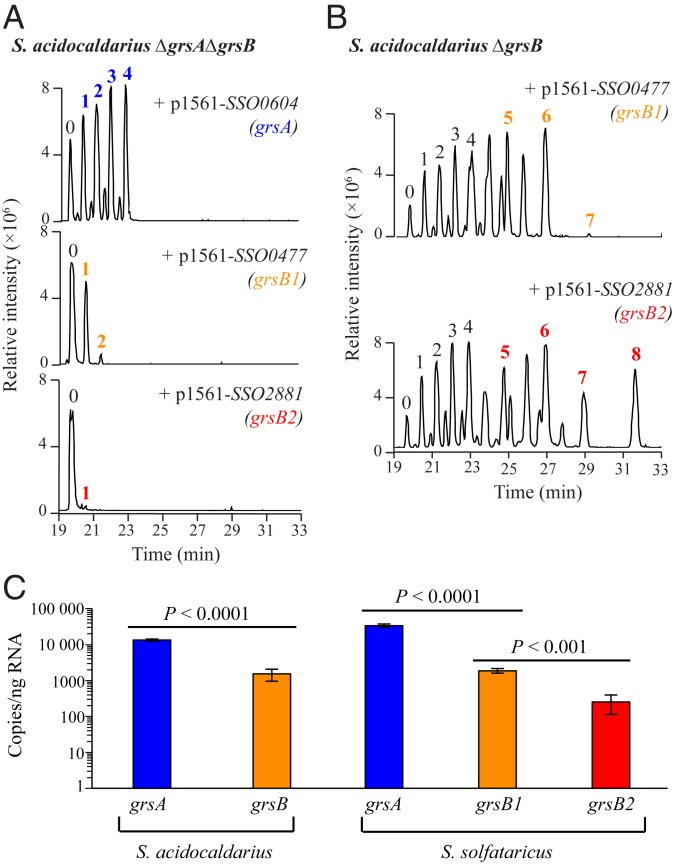
To test the functionality of these potentially redundant Grs homologs, we expressed the S. solfataricus grs genes in the S. acidocaldarius ∆grsA∆grsB mutant. The S. solfataricus GrsA (SSO0604) homolog produced GDGT-1 through GDGT-4, indicating that this protein has the same function as the S. acidocaldarius GrsA (Fig. 3A). Expression of grsB1 (SSO0477) produced GDGT-1 and a minor amount of GDGT-2, while grsB2 (SSO2881) expression resulted in a small amount of GDGT-1 (Fig. 3A). This minor production of cyclized GDGTs in the double deletion mutant by the S. solfataricus GrsB homologs indicates that both GrsB homologs are functional but also suggests that, similar to what we observe with GrsB in S. acidocaldarius, their preferred substrate might be a cyclized GDGT. To test this, we expressed the grsB homologs individually in the S. acidocaldarius grsB mutant which produces GDGT-1 through GDGT-4. Complementation of this strain demonstrated that GrsB1 produces significant amounts of GDGT-5 through GDGT-7 and GrsB2 produces GDGT-5 through GDGT-8 (Fig. 3B). Thus, both of these S. solfataricus proteins are functional GrsB homologs that prefer a cyclized GDGT substrate and are cyclizing at the C-3 position.
Fig. 3.
S. solfataricus Grs homologs are functional. (A) LC-MS merged EICs of acid-hydrolyzed lipid extracts of S. acidocaldarius ∆grsA∆grsB expressing S. solfataricus grs homologs. Expression of SSO0604 results in the production of GDGT-1 through GDGT-4, expression of SSO0477 produces GDGT-1 and GDGT-2, and expression of SSO2881 produces only GDGT-1. (B) LC-MS merged EICs of acid-hydrolyzed lipid extracts of S. acidocaldarius ∆grsB expressing the 2 S. solfataricus GrsB homologs (SSO0477 and SSO2881). In this strain, the S. solfataricus GrsB1 (SSO0477) produces GDGT-5 through GDGT-7, while GrsB2 (SSO2881) produces GDGT-5 through GDGT-8 (bold numbers). Because the S. acidocaldarius ∆grsB strain produces GDGT-1 through GDGT-4, these results suggest that S. solfataricus GrsB1 and GrsB2 prefer cyclized GDGT substrates. (C) Quantification of grsA and grsB transcripts in S. acidocaldarius and S. solfataricus. In S. acidocaldarius, grsA is expressed 9-fold higher than grsB. In S. solfataricus, grsA is expressed 18-fold higher than grsB1 and 134-fold higher than grsB2, while grsB1 is expressed 7-fold higher than grsB2.
The occurrence of multiple Grs homologs in a variety of archaea raises the question as to what purpose these redundant copies serve. It is possible these multiple grs gene copies may be differentially expressed under distinct environmental conditions or that there are differences in the enzymatic properties of the various Grs proteins that provide a physiological advantage. To begin to distinguish between these 2 possibilities, we measured transcript levels of the grs genes in the wild-type strains of S. acidocaldarius and S. solfataricus under standard growth conditions. We found that grsB1 transcript levels in S. solfataricus were 7-fold higher than the transcript levels of grsB2 (Fig. 3C), demonstrating that these redundant proteins are expressed at different levels. In addition, both the S. acidocaldarius and S. solfataricus grsA genes are expressed 1 to 2 orders of magnitude higher than the grsB genes (Fig. 3C). Thus, there does appear to be some differential transcriptional control of grsA and grsB.
Grs Homologs in Open Oceans Are Restricted to the Thaumarchaeota.
Our identification of the Grs proteins now allows us to more directly address the potential contributions of Thaumarchaeota and Euryarchaeota to cyclic GDGT pools in marine settings. As previously noted, TEX86 paleotemperature proxies are based on the assumption that mesophilic MG-I Thaumarchaeota are the dominant source of cyclic GDGTs in the pelagic zone (9). However, it has been suggested that MG-II Euryarchaeota are also significant contributors to ocean cyclic GDGT pools at shallower depths (13, 16) but this has been difficult to assess, as MG-II organisms have not been cultured and shown to produce cyclic GDGTs. One study attempted to correlate cyclic GDGT distribution in the North Pacific Subtropical Gyre (NPSG) with the occurrence of Euryarchaeota and Thaumarchaeota 16S ribosomal RNA (rRNA) and metagenomic gene sequence counts (16). Although the correlation between GDGTs detected and the occurrence of MG-II Euryarchaeota above 150 m was robust, these findings were contentious, because detection of a specific archaeal group with genetic markers unrelated to GDGT biosynthesis does not necessarily indicate that this group is producing the observed GDGTs (28, 29).
To better clarify whether MG-II Euryarchaeota are a potential source of cyclic GDGTs in the NPSG, we searched for GrsA and GrsB homologs in 38 publicly available NPSG metagenomes (SI Appendix, Table S2) in the Joint Genome Institute (JGI) Integrated Microbial Genomes (IMG) database (30). We retrieved 709 Grs homologs (>400 amino acids, e value < 1e−20, >20% identity) from metagenomes collected at 125 m and deeper. Above this depth, only one Grs homolog was detected from a metagenome collected at 25 m. The majority of Grs homologs were present in the metagenome samples collected from deeper in the water column, with the most homologs retrieved from 500 m (291) and 770 m (235). However, only 8 (1%) of the Grs sequences retrieved were unique when redundant sequences were removed from the dataset, indicating that the diversity of Grs sequences in the NPSG water column is limited.
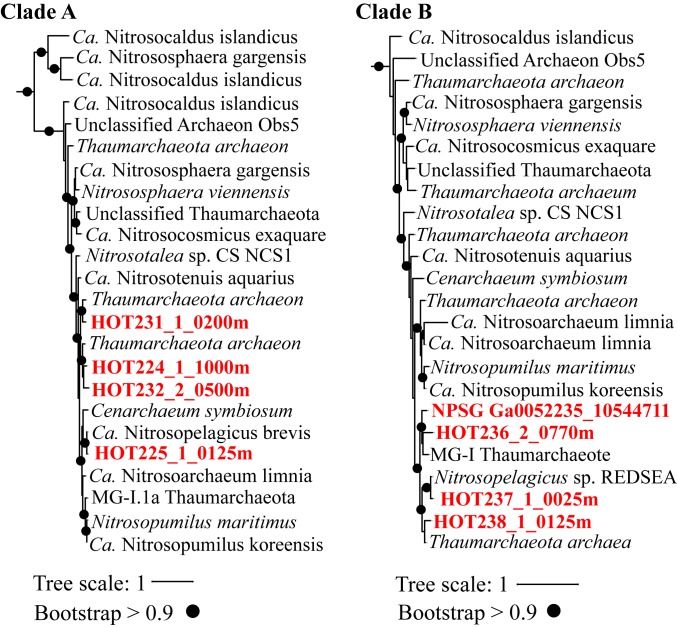
To determine which archaeal group the NPSG homologs are derived from, a maximum-likelihood phylogenetic tree was generated with 329 unique Grs homologs (e value < 1e−20, >20% identity) retrieved from the JGI IMG archaeal isolates genomes database (SI Appendix, Fig. S5). Due to short sequences, present transcriptome data were insufficient to include in our phylogenetic trees. The clustering of the NPSG metagenome sequences revealed that all 8 homologs are closely related to MG-I Thaumarchaeota Grs sequences (Fig. 4). This indicates that, even though MG-II Euryarchaeota dominate the surface waters of the NPSG based on 16S rRNA gene and metagenomic analyses (16), the MG-I Thaumarchaeota are the only source of cyclic GDGTs in both the shallow and deep waters of the NPSG. The lack of Grs homologs at shallower depths also suggests that MG-II Euryarchaeota, more broadly, may not produce any cyclic GDGTs. To test this, we searched for Grs homologs in 228 MG-II metagenome acquired genomes (MAGs; taxon ID: 133814) recently assembled from a variety of marine environments (31) and available in the National Center for Biotechnology Information database. We did not detect any Grs homologs in these searches (e value < 1e−20, >20% identity), suggesting that MG-II Euryarchaeota do not have the genetic potential to produce any cyclized GDGTs. Thus, our analyses support the hypothesis that mesophilic marine Thaumarchaeota are the dominant source of cyclic GDGTs in the open ocean.
Fig. 4.
NPSG metagenomic Grs homologs cluster only with Thaumarchaeota Grs homologs. Two clades from a rooted maximum-likelihood phylogenetic tree of Grs homologs (e value, <1e−20; protein identity, >20%) identified in cultured archaeal genomes, single-cell acquired genomes (SAGs), MAGs, or NPSG metagenomes. Grs homologs from the NPSG metagenomes are shown in red. Full phylogenetic maximum likelihood (PhyML) tree is shown in SI Appendix, Fig. S5. Black circles indicate branches that have bootstrap values greater than 0.9, and the scale bar represents one change per nucleotide site.
Discussion
The discovery of isoprenoidal tetraether lipids in archaea over 40 y ago (17) has generated significant interest in the biosynthesis and biomarker applications of these unique molecules. From a biosynthesis perspective, the proteins and catalytic mechanisms behind the potential condensation of 2 diether lipids to form GDGT-0 and the reactions that insert the cyclopentane or cyclohexane rings into the core GDGT have been difficult to elucidate (1, 19, 20). Although a few studies have suggested potential mechanisms for the diether condensation (32–36), no potential mechanism has been proposed for the formation of the cyclopentane rings. Our finding that 2 radical SAM proteins are necessary for GDGT cyclization establishes that a free radical mechanism is involved in their formation—most likely generating the radical necessary to initiate the internal cyclopentane ring formation from the isoprenoid methyl groups (37). Our analyses also demonstrate that GrsA and GrsB introduce rings specifically at the C-7 or C-3 position, respectively, and suggest that the preferred substrate for each protein differs, with GrsA appearing to prefer the acyclic GDGT-0 and GrsB favoring C-7 cyclized GDGTs. This is significant, as it shows that there is an ordered process to cyclization, with the C-7 rings being introduced prior to the C-3 rings. Further, it suggests that GDGTs with rings only at the C-3 position are unlikely to occur naturally, thereby constraining the number of possible GDGT isomers in nature. These findings also confirm that cyclization occurs after the formation of the ether bonds to glycerol-1-phosphate (SI Appendix, Fig. S6) and after the condensation of 2 diether lipids, as has been suggested by previous studies (32, 38, 39). However, it is still unclear whether the GDGT substrate must be unsaturated for cyclization and whether different phospholipid and hexose head groups influence ring formation (SI Appendix, Fig. S6). Additional biochemical and structural characterizations of GrsA and GrsB, as well as identification of the proteins required for diether condensation and head group modification, are necessary to resolve these open questions.
The finding that GrsA and GrsB introduce rings at distinct locations is also significant from a geochemical perspective. The TEX86 paleotemperature proxy is a ratio of a subset of cyclized GDGTs that correlates the degree of cyclization with sea surface temperature (7), despite surface waters not being the primary habitat of these organisms. The direct relationships between GDGT ring distributions and thermal properties therefore remain unclear. One cyclized GDGT structure that features prominently in the TEX86 paleotemperature proxy is an isomer of crenarchaeol, a GDGT that appears to be restricted to Thaumarchaeota species and contains 4 cyclopentane rings at C-7 and a unique cyclohexane ring at C-11 (7, 9). Given our findings, we would predict that Thaumarchaeota would harbor a GrsA homolog as well as a unique Grs responsible for generating the C-11 cyclohexane ring. Our bioinformatics analyses did reveal several potential Grs homologs in Thaumarchaeota genomes (SI Appendix, Table S1), and future analyses of these homologs may reveal the synthase responsible for the cyclization at C-11. We also demonstrate that the grsA and grsB genes are differentially expressed under standard conditions in both S. acidocaldarius and S. solfataricus. This implies that the regulatory mechanisms that control cyclization at C-7 and C-3 (and perhaps C-11 of crenarchaeol) may differ and be influenced by distinct environmental factors. Because paleotemperature proxies are derived from ratios of a subset of cyclized GDGTs, the robustness of TEX86 and other indices can be improved by determining which cyclization sites (C-3 vs. C-7 vs. C-11), if any, respond to environmental or physiological fluctuations, through in vivo and in vitro analyses of the respective Grs proteins.
Finally, this work demonstrates the usefulness of identifying GDGT ring synthase proteins in constraining the environmental sources of cyclized GDGTs. Through bioinformatics analyses of metagenomic samples from the North Pacific, we show that MG-I Thaumarchaeota are indeed the dominant source of cyclic GDGTs in the open ocean. Further, we show that genomes of MG-II Euryarchaeota acquired from metagenomic data do not harbor any Grs homologs, suggesting that these archaea do not contribute to cyclic GDGT pools in any marine environment. However, it is possible that MG-II Euryarchaeota can cyclize their GDGTs through distinct proteins not related to Grs, and, therefore, cultivation and lipid analyses of marine Euryarchaeota directly should be a priority to confirm these results. Additionally, coanalyses of pelagic marine GDGTs and Grs homologs in pelagic metagenomes are still needed to determine whether these patterns are widespread and whether they are influenced by seasonal changes. Nonetheless, the results of this study demonstrate the importance of understanding the molecular details of GDGT biosynthesis in extant organisms to better constrain paleotemperature proxies which, in turn, will allow for more accurate reconstruction of ancient global temperatures and impact our understanding of past temperature variability and Earth system dynamics.
Methods
Microbial Strains, Media, and Growth Conditions.
Strains used in this study are listed in SI Appendix, Table S3. S. acidocaldarius MW001 and S. solfataricus DSM1617 were cultured at 75 °C in Brock medium supplemented with 0.1% NZ-Amine and 0.2% sucrose (40). The pH of the growth medium was adjusted with sulfuric acid to pH 3.5. S. acidocaldarius MW001 and mutant strains are uracil auxotroph and required supplementation with 10 μg/mL uracil. For growth on solid medium, Brock medium was solidified with 0.6% gelrite.
M. acetivorans C2A strains were grown in single-cell morphology (41) at 37 °C in bicarbonate-buffered high-salt liquid medium containing 50 mM trimethylamine in Balch tubes with a N2/CO2 (80/20) headspace. Growth on medium solidified with 1.6% agar was performed as previously described by Sowers et al. (41), except cells were spread directly on plates without the use of top agar. Solid media plates were incubated in 3.5-L anaerobic jars under an N2/CO2/H2S (80/20/1,000 ppm) atmosphere contained inside an anaerobic glove chamber. All M. acetivorans manipulations were carried out under anaerobic conditions in an anaerobic glove chamber unless otherwise stated. Puromycin dihydrochloride (Research Products International) was added to a final concentration of 2 μg/mL from a sterile, anaerobic stock solution. Sterile, anaerobic tetracycline hydrochloride solutions were prepared fresh and added 1.6 μg/mL to induce protein expression.
Escherichia coli strains were grown in lysogeny broth (LB) at 37 °C with shaking at 225 rpm. LB media was solidified with 1.5% agar. The E. coli strain WM4489, a DH10B derivative designed to control oriV-based plasmid copy number, was used for M. acetivorans plasmid construction and maintenance (42). LB media was supplemented, if necessary, with 100 μg/mL ampicillin, 50 μg/mL kanamycin, and 20 μg/mL chloramphenicol for E. coli growth, and 10 mM rhamnose to increase plasmid copy number.
Molecular Cloning.
All plasmids and oligonucleotides used in this study are described in SI Appendix, Tables S4 and S5. Details of plasmid construction are described in SI Appendix.
Construction of S. acidocaldarius Mutants and Complementation.
S. acidocaldarius markerless gene deletions were constructed through homologous recombination mediated allelic replacement as previously described by Wagner et al. (21) with minor modifications as outlined in SI Appendix and in Zeng et al. (23). For complementation experiments, plasmids were transformed into the appropriate strains as described in SI Appendix, and all complemented strains (containing pSVA1561 plasmids) were supplemented with 0.4% maltose to induce protein expression.
Lipid Extraction and Analyses.
Cultures of S. acidocaldarius (45 mL) or M. acetivorans (250 mL) were harvested at stationary phase of growth by centrifugation at 11,000 × g for 10 min, and pellets were frozen at −20 °C prior to extraction. A modified ultrasonic extraction method (43) was used to extract both intact polar and core GDGTs as described in SI Appendix. Lipids analysis by liquid chromatography–mass spectrometry (LC-MS) was performed on an Agilent 1290 series UHPLC (ultrahigh-performance liquid chromatography) system coupled to an Agilent 6530 quadrupole time-of-flight (Q-tof) mass spectrometer through an Agilent jet stream dual electrospray ionization (AJS-ESI) interface as described in SI Appendix.
Ether Cleavage and Biphytane Analyses.
To elucidate the ring configuration on biphytanes, GDGTs were subjected to ether cleavage. Briefly, aliquots of total lipid extracts were transferred into a 2-mL vial and dried with N2 prior to the addition of boron tribromide (BBr3) (0.5 mL, 1 M in DCM; Sigma-Aldrich). The vial was sealed and heated at 60 °C for 2 h. After removing the solvent and residual BBr3 with N2, the bromides were reduced to hydrocarbons by the addition of superhydride (0.5 mL, lithium triethylborohydride [LiEt3BH], in DCM, Sigma-Aldrich), and the mixture was maintained sealed at 60 °C for 2 h. A few drops of water were added to quench the reaction, and the hydrocarbon products were extracted (3×) with n-hexane. The combined extracts were dried, redissolved in n-hexane, and analyzed by GC-MS. GC-MS was performed using an Agilent 5977B Inert Plus Mass Selective Detector (MSD) system equipped with an Agilent DB-5MS+DG column (60 m × 250 µm × 0.25 µm; Agilent) with helium as carrier gas (1.2 mL⋅min−1). Separation was achieved using an oven temperature ramp program of 60 °C held for 2 min and then raised to 320 °C at 4 °C min−1 and held for 63 min. In addition to the ring modification on biphytanes, the arrangement of 2 glycerols in a GDGT molecule results in 2 regioisomers, the parallel and antiparallel glycerol configured GDGT (44, 45). Structures of isomers caused by both glycerol and ring configurations will be discussed, separately, in another work. The present study focuses on the ring modification, with all GDGT molecules illustrated in the figures in the antiparallel glycerol configuration, for simplicity.
Quantification of Gene Expression.
Quantitative PCR was carried out as described by Tolar et al. (46), with minor modifications as described in SI Appendix.
Bioinformatics Analyses.
Comparative genomics analyses, Basic Local Alignment Search Tool (BLAST) homology searches, protein alignments, and phylogenetic tree construction of Grs homologs were carried out as described in SI Appendix.
Supplementary Material
Acknowledgments
We thank 2 anonymous reviewers for their critical review of our manuscript. Support for this study was provided by the Simons Collaboration on the Origins of Life to P.V.W. (Grant 511568) and R.E.S. (Grant 290361) and by the National Science Foundation to P.V.W. under Grant 1752564. K.R.F. and W.W.M. were supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through Grant DE-FG02-02ER15296.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See Commentary on page 22423.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909306116/-/DCSupplemental.
References
- 1.Koga Y., Morii H., Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev. 71, 97–120 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Damsté J. S., Schouten S., Hopmans E. C., van Duin A. C., Geenevasen J. A., Crenarchaeol: The characteristic core glycerol dibiphytanyl glycerol tetraether membrane lipid of cosmopolitan pelagic crenarchaeota. J. Lipid Res. 43, 1641–1651 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Valentine D. L., Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 5, 316–323 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Boyd E. S., et al. , Temperature and pH controls on glycerol dibiphytanyl glycerol tetraether lipid composition in the hyperthermophilic crenarchaeon Acidilobus sulfurireducens. Extremophiles 15, 59–65 (2011). [DOI] [PubMed] [Google Scholar]
- 5.De Rosa M., Esposito E., Gambacorta A., Nicolaus B., Bulock J. D., Effects of temperature on ether lipid composition of Caldariella acidophila. Phytochemistry 19, 827–831 (1980). [Google Scholar]
- 6.Lai D., Springstead J. R., Monbouquette H. G., Effect of growth temperature on ether lipid biochemistry in Archaeoglobus fulgidus. Extremophiles 12, 271–278 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Schouten S., Hopmans E. C., Schefuss E., Sinninghe Damste J. S., Distributional variations in marine crenarchaeotal membrane lipids: A new tool for reconstructing ancient sea water temperatures? Earth Planet. Sci. Lett. 204, 265–274 (2002). [Google Scholar]
- 8.Zhang Y. G., Pagani M., Wang Z., Ring Index: A new strategy to evaluate the integrity of TEX86 paleothermometry. Paleoceanography 31, 220–232 (2016). [Google Scholar]
- 9.Tierney J. E., “Biomarker-based inferences of past climate: The TEX86 paleotemperature proxy” in Treatise on Geochemistry, Holland H. D., Turekian K. K., Eds. (Organic Geochemistry, Elsevier Science, Oxford, ed. 2, 2014), vol. 12, pp. 379–393. [Google Scholar]
- 10.Qin W., et al. , Confounding effects of oxygen and temperature on the TEX86 signature of marine Thaumarchaeota. Proc. Natl. Acad. Sci. U.S.A. 112, 10979–10984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurley S. J., et al. , Influence of ammonia oxidation rate on thaumarchaeal lipid composition and the TEX86 temperature proxy. Proc. Natl. Acad. Sci. U.S.A. 113, 7762–7767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schouten S., Hopmans E. C., Sinninghe Damste J. S., The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: A review. Org. Geochem. 54, 19–61 (2013). [Google Scholar]
- 13.Turich C., et al. , Lipids of marine Archaea: Patterns and provenance in the water-column and sediments. Geochim. Cosmochim. Acta 71, 3272–3291 (2007). [Google Scholar]
- 14.DeLong E. F., et al. , Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311, 496–503 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Zhang C. L., Xie W., Martin-Cuadrado A. B., Rodriguez-Valera F., Marine Group II Archaea, potentially important players in the global ocean carbon cycle. Front. Microbiol. 6, 1108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lincoln S. A., et al. , Planktonic Euryarchaeota are a significant source of archaeal tetraether lipids in the ocean. Proc. Natl. Acad. Sci. U.S.A. 111, 9858–9863 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Rosa M., De Rosa S., Gambacorta A., Minale L., Bu’lock J. D., Chemical structure of the ether lipids of thermophilic acidophilic bacteria of the Caldariella group. Phytochemsitry 16, 1961–1965 (1977). [Google Scholar]
- 18.De Rosa M., Gambacorta A., The lipids of archaebacteria. Prog. Lipid Res. 27, 153–175 (1988). [DOI] [PubMed] [Google Scholar]
- 19.Jain S., Caforio A., Driessen A. J., Biosynthesis of archaeal membrane ether lipids. Front. Microbiol. 5, 641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villanueva L., Damsté J. S., Schouten S., A re-evaluation of the archaeal membrane lipid biosynthetic pathway. Nat. Rev. Microbiol. 12, 438–448 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Wagner M., et al. , Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front. Microbiol. 3, 214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broderick J. B., Duffus B. R., Duschene K. S., Shepard E. M., Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng Z., Liu X. L., Wei J. H., Summons R. E., Welander P. V., Calditol-linked membrane lipids are required for acid tolerance in Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. U.S.A. 115, 12932–12937 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welander P. V., Coleman M. L., Sessions A. L., Summons R. E., Newman D. K., Identification of a methylase required for 2-methylhopanoid production and implications for the interpretation of sedimentary hopanes. Proc. Natl. Acad. Sci. U.S.A. 107, 8537–8542 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welander P. V., et al. , Identification and characterization of Rhodopseudomonas palustris TIE-1 hopanoid biosynthesis mutants. Geobiology 10, 163–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmerk C. L., et al. , Elucidation of the Burkholderia cenocepacia hopanoid biosynthesis pathway uncovers functions for conserved proteins in hopanoid-producing bacteria. Environ. Microbiol. 17, 735–750 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Liu X. L., De Santiago Torio A., Bosak T., Summons R. E., Novel archaeal tetraether lipids with a cyclohexyl ring identified in Fayetteville Green Lake, NY, and other sulfidic lacustrine settings. Rapid Commun. Mass Spectrom. 30, 1197–1205 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Lincoln S. A., et al. , Reply to Schouten et al.: Marine Group II planktonic Euryarchaeota are significant contributors to tetraether lipids in the ocean. Proc. Natl. Acad. Sci. U.S.A. 111, E4286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schouten S., Villanueva L., Hopmans E. C., van der Meer M. T., Sinninghe Damsté J. S., Are Marine Group II Euryarchaeota significant contributors to tetraether lipids in the ocean? Proc. Natl. Acad. Sci. U.S.A. 111, E4285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen I. A., et al. , IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinke C., et al. , A phylogenomic and ecological analysis of the globally abundant Marine Group II archaea (Ca. Poseidoniales ord. nov.). ISME J. 13, 663–675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eguchi T., Nishimura Y., Kakinuma K., Importance of the isopropylidene terminal of geranylgeranyl group for the formation of tetraether lipid in methanogenic archaea. Tetrahedron Lett. 44, 3275–3279 (2003). [Google Scholar]
- 33.Fitz W., Arigoni D., Biosynthesis of 15,16-dimethyltriacontanedioic acid (diabolic acid) from [16-(H-2)3]- and [14-(H-2)2]-palmitic acids. J. Chem. Soc. Chem. Commun., 20, 1533–1534 (1992). [Google Scholar]
- 34.Galliker P., Gräther O., Rümmler M., Fitz W., Arigoni D., “New structural and biosynthetic aspects of the unusual core lipids from archaebacteria” in Vitamin B12, and B12-Proteins, Kräutler B., Arigoni D., Golding B. T., Eds. (Wiley-VCH, Weinheim, Germany, 1998), pp. 447–458. [Google Scholar]
- 35.Nemoto N., Shida Y., Shimada H., Oshima T., Yamagishi A., Characterization of the precursor of tetraether lipid biosynthesis in the thermoacidophilic archaeon Thermoplasma acidophilum. Extremophiles 7, 235–243 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Poulter C. D., Aoki T., Daniels L., Biosynthesis of isoprenoid membranes in the methanogenic archaebacterium Methanospirillum hungatei. J. Am. Chem. Soc. 110, 2620–2624 (1988). [Google Scholar]
- 37.De Rosa M., De Rosa S., Gambacorta A., 13C-NMR assignments and biosynthetic data for the ether lipids of Caldariella. Phytochemistry 16, 1909–1912 (1977). [Google Scholar]
- 38.Murakami M., et al. , Geranylgeranyl reductase involved in the biosynthesis of archaeal membrane lipids in the hyperthermophilic archaeon Archaeoglobus fulgidus. FEBS J. 274, 805–814 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Pearson A., “Lipidomics for geochemistry” in Treatise on Geochemistry, Holland H. D., Turekian K. K., Eds. (Organic Geochemistry, Elsevier Science, Oxford, UK, ed. 2, 2014), vol. 12, pp. 291–336. [Google Scholar]
- 40.Brock T. D., Brock K. M., Belly R. T., Weiss R. L., Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84, 54–68 (1972). [DOI] [PubMed] [Google Scholar]
- 41.Sowers K. R., Boone J. E., Gunsalus R. P., Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl. Environ. Microbiol. 59, 3832–3839 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S. Y., et al. , Different biosynthetic pathways to fosfomycin in Pseudomonas syringae and Streptomyces species. Antimicrob. Agents Chemother. 56, 4175–4183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huguet C., Martens-Habbena W., Urakawa H., Stahl D. A., Ingalls A. E., Comparison of extraction methods for quantitative analysis of core and intact polar glycerol dialkyl glycerol tetraethers (GDGTs) in environmental samples. Limnol. Oceanogr. Methods 8, 127–145 (2010). [Google Scholar]
- 44.Liu X. L., Lipp J. S., Birgel D., Summons R. E., Hinrichs K. U., Predominance of parallel glycerol arrangement in archaeal tetraethers from marine sediments: Structural features revealed from degradation products. Org. Geochem. 115, 12–23 (2018). [Google Scholar]
- 45.Liu X. L., Russell D. A., Bonfio C., Summons R. E., Glycerol configurations of environmental GDGTs investigated using a selective sn2 ether cleavage protocol. Org. Geochem. 128, 57–62 (2019). [Google Scholar]
- 46.Tolar B. B., King G. M., Hollibaugh J. T., An analysis of Thaumarchaeota populations from the northern Gulf of Mexico. Front. Microbiol. 4, 72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.