One of the most powerful molecular motors discovered, to date, belongs to a phage, a type of virus that infects bacteria (1). The motor serves to package stiff, double-stranded DNA (dsDNA) into a preassembled, proteinaceous polyhedral container called the procapsid. The DNA is about 50 times longer than the size of the capsid and gets packaged and compressed by the motor at near-crystalline densities. The capsid holds up its end of the bargain, withstanding positive pressure differentials of up to 40 atm (2). The level of current understanding of this fascinating biological process was made possible, in great measure, by the development of in singulo approaches to packaging dynamics, adapted to phages. However, what works for one virus may hardly apply to another. Thus, in a great number of single-stranded RNA (ssRNA) viruses—the largest virus group on Earth—nucleic acid packaging occurs concurrently with, and indeed is part of, spontaneous assembly. No ATP-powered motor is involved here. Instead of great positive pressures, the final pressure exerted by the RNA, at equilibrium, is modest and negative (3). Disentangling the interplay between the 2 types of concurrent driving interactions—those among coat proteins and those between coat proteins and RNA—is far from trivial. Adding to the challenge, intermediates have a fleeting existence (milliseconds), while the sizes of ssRNA viruses that can assemble in vitro are small (tens of nanometers). This is why, up to now, the principal features of ssRNA virus assembly have been mainly supplied by theoretical models, structural arguments from static data, and ensemble-averaged dynamic experiments. In singulo methods that would offer the same level of real-time dynamics detail and direct insight as those applied to dsDNA phages 2 decades ago have been scarce. In PNAS, Garmann et al. (4) provide a compelling demonstration of the potential held by a wide-field optical microscopy method for tightly controlled, real-time single-particle studies of the ssRNA bacteriophage MS2 assembly with near-molecular accuracy and broad temporal dynamic range.
To measure assembly kinetics in real time, Garmann et al. (4) adapt an imaging technique called interferometric scattering microscopy (5). In their approach, sparsely distributed single molecules of viral RNA are tethered via flexible linkers to the surface of a microscope coverslip, in solution. Upon injection of virus coat proteins, RNA−protein association results in an increase of the effective local protein density at tethered RNA locations. Since the optical polarizability of proteins is different from that of water, there is increased light scattering by the growing nucleoprotein complex. The scattered light is phase-shifted with respect to the incident light. This phase shift provides a way to boost contrast at the detector plane, where the coherent scattered light is made to interfere with light reflected by the coverslip. Thus, multiple growing virus particles can be imaged in parallel via a charge-coupled device in real time, as diffraction-limited spots of an intensity proportional to the local mass accumulation. The reported detection limit is 6 coat-protein dimers at 1-Hz bandwidth.
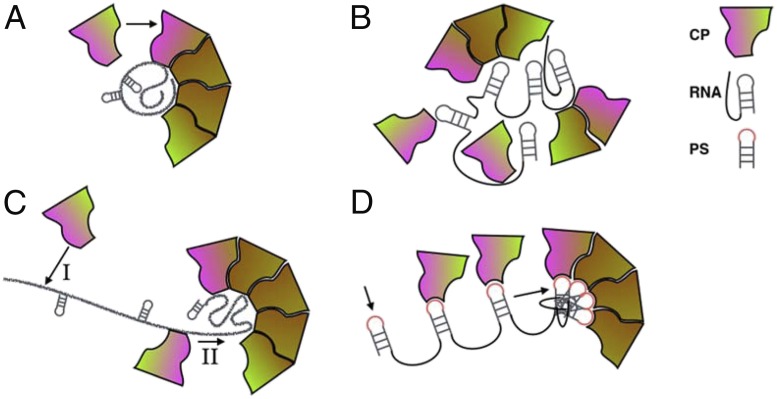
MS2 has a capsid formed of 180 identical proteins organized with icosahedral symmetry as 12 pentamers and 20 hexamers. A single genomic RNA molecule, 3,569 nucleotides long, is encapsulated inside the capsid. Like other ssRNA viruses, the genome does not occupy uniformly the entire available space. Instead, it forms 2 concentric layers adjacent to the luminal capsid surface and has relatively low density near center. However, while the genome of other simple T = 3 ssRNA viruses, such as the well-studied cowpea chlorotic mottle virus (CCMV), is smeared nearly isotropically across the lumenal surface, genome distribution in MS2 is anisotropic and correlates with capsid geometry (6). This difference in genome organization helped formulate the hypothesis that genome packaging during MS2 assembly may be guided by different constraints and therefore may follow qualitatively different pathways from those proposed for other simple viruses. For instance, 3 possible models exist for the broad class of icosahedral viruses represented by CCMV (Fig. 1 A−C). The coat proteins of this class will readily assemble and form capsids in vitro around heterologous nucleic acids, other polyanions, and even nanoparticles, and thus the first driving interactions taken into consideration in building these models were nonspecific.
Fig. 1.
Cartoon representation of intermediates specific to 4 proposed mechanisms for icosahedral ssRNA virus growth. (A) Nucleation−elongation model (7–9). (B) Micellar condensation (“en masse”) (10). (C) RNA antenna model (11). (D) PS-mediated sequential assembly model (14). Arrows indicate impending binding events, vivid hues indicate interfaces activated for binding, and subdued hues represent subunits stabilized in postassembly conformations. CP, coat protein.
The first model was adapted from the kinetic nucleation−growth approach of Oosawa and Asakura (7), successfully applied to empty capsid assembly first (8), then later extended to the study of assembly kinetics of coat proteins around genomic cargo (9). This model is predicated on strong interactions between coat proteins, relative to RNA−coat-protein interactions. Here, the nucleation step is rate-limiting. Growth occurs through rapid addition of subunits at the growing edge once a critical size oligomer has formed on a precompacted RNA molecule. Intermediates along the elongation leg of the assembly pathway are very short-lived (Fig. 1A).
The second model (“en masse” or “micelle/crystallization” model) corresponds to the limit where interactions between RNA and coat protein are dominating. Here, after rapid initial formation of a disordered nucleoprotein complex, the high effective protein concentration increases the chances of multiple weak protein interactions acting cooperatively to stabilize the precursors of a symmetric capsid along the growth pathway (10) (Fig. 1B). Interactions among coat proteins are necessarily weak in this case, which leads to a fluid previrion surface, open to structural transitions, and able to find, in the end, the free-energy minimum, that is, the icosahedral cage structure.
Since the magnitude of interactions between coat proteins, and coat protein and RNA, can be modulated by pH and ionic strength, different assembly scenarios are expected to be favored by the same system. Indeed, coarse-grained simulations and in vitro small-angle X-ray scattering experiments with CCMV both support this expectation (11, 12). This suggests that virus assembly is a resilient process, which can adapt to changes in the chemical environment by switching between mechanisms.
The third model (Fig. 1C) reserves yet another role for RNA, whereby the nucleic acid not only locally concentrates the coat protein, but its strand acts like an extended antenna that guides the RNA-associated coat proteins, through one-dimensional diffusion, toward the live edge of a nucleus from which the virus grows (13). The emphasis on the multifaceted, central role of RNA sets this model as a precursor of the current model for the mechanism of MS2 assembly.
The defining feature of the mechanistic model for MS2 assembly is the pivotal role it assigns to specific interactions between RNA and coat protein throughout the virus growth process (14). Thus, increased coat-protein binding affinity sequences called packaging signals (PS) have been identified on the MS2 RNA, and PS−protein binding events at low (nanomolar) protein concentration were found to lead to RNA condensation. In this process, RNA-bound proteins are believed to be brought together in ways that avoid potential kinetic traps. Orderly binding and folding of the viral RNA thus orchestrates the growth of the capsid. It would be a fascinating experience to be able to see this process at work in real time, from A to Z. This is what Garmann et al. (4) set out to do.
In PNAS, Garmann et al. provide a compelling demonstration of the potential held by a wide-field optical microscopy method for tightly controlled, real-time single-particle studies of the ssRNA bacteriophage MS2 assembly with near-molecular accuracy and broad temporal dynamic range.
Their single particle assembly kinetics data (at micromolar dimer concentrations) show that there is a broad distribution of start times for the elongation step, which strongly suggests the existence of a critical nucleus; that is, assembly has to overcome a free-energy barrier before rapid, cooperative growth can take place (4). Interestingly, during the rapid growth phase, the time to capsid completion is 4 orders of magnitude longer than the timescale required for 90 dimers to cross the RNA strand by diffusion. Therefore, only a small fraction of protein−RNA collisions are productive at this protein concentration, with the system having the ability of sampling a large number of pathway steps before proceeding to add new subunits. No major disassembly events could be observed.
The size of the nucleus could not be estimated, since it appears to be smaller than the detection limit, that is, about 6 dimers (dimers, not monomers, are the more stable coat-protein state in solution, prior to assembly) (4). The findings are intriguing because the number of putative PS sites should be greater than that (14), yet it appears that the initiation step corresponding to proteins binding at PS sites could not be detected by the interferometric scattering approach. This could be due to a number of reasons: drift in the background intensity, and/or differences in the buffer ionic strength and protein concentration with respect to previous work (15). In those previous studies, protein concentration was in the nanomolar range, and total assembly times exceeded 103 s, which would be challenging to achieve with interferometric scattering, due to drift. In addition, lower ionic strength in ref. 15 favored stronger RNA−coat-protein interactions. Could it be that, like other viruses, MS2 is able to switch between mechanisms, for example, depending on the stage of infection? Might it be that PS activity depends on the kinetics of the nucleoprotein complex folding with PS sites fading in and out of existence through a slow ramping of protein concentration, as Dykeman et al. (16) have suggested? What would in singulo kinetics look like if the RNA of a different virus were used instead of MS2 RNA? What instrumentation improvements could lower the detection limit of interferometric scattering microscopy to the point where the nature of the critical nucleus could be determined? The path ahead is wide open and promises to add to an already fascinating story.
Acknowledgments
My current research is supported by the Army Research Laboratory-Army Research Office, Grant W911NF-17-1-0329, and by the National Science Foundation, under Awards CBET 1803440 and CHE 1808027.
Footnotes
The author declares no competing interest.
See companion article on page 22485.
References
- 1.Smith D. E., et al. , The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 413, 748–752 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Gelbart W. M., Knobler C. M., The physics of phages. Phys. Today 61, 42–47 (2008). [Google Scholar]
- 3.Siber A., Božič A. L., Podgornik R., Energies and pressures in viruses: contribution of nonspecific electrostatic interactions. Phys. Chem. Chem. Phys. 14, 3746–3765 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Garmann R. F., Goldfain A. M., Manoharan V. N., Measurements of the self-assembly kinetics of individual viral capsids around their RNA genome. Proc. Nat. Acad. Sci. U.S.A. 116, 22485–22490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortega Arroyo J., Cole D., Kukura P., Interferometric scattering microscopy and its combination with single-molecule fluorescence imaging. Nat. Protoc. 11, 617–633 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Toropova K., Basnak G., Twarock R., Stockley P. G., Ranson N. A., The three-dimensional structure of genomic RNA in bacteriophage MS2: Implications for assembly. J. Mol. Biol. 375, 824–836 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Oosawa F., Asakura S., Thermodynamics of the Polymerization of Protein (Academic, 1975). [Google Scholar]
- 8.Zlotnick A., To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J. Mol. Biol. 241, 59–67 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Kler S., et al. , RNA encapsidation by SV40-derived nanoparticles follows a rapid two-state mechanism. J. Am. Chem. Soc. 134, 8823–8830 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McPherson A., Micelle formation and crystallization as paradigms for virus assembly. BioEssays 27, 447–458 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Perlmutter J. D., Perkett M. R., Hagan M. F., Pathways for virus assembly around nucleic acids. J. Mol. Biol. 426, 3148–3165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevreuil M., et al. , Nonequilibrium self-assembly dynamics of icosahedral viral capsids packaging genome or polyelectrolyte. Nat. Commun. 9, 3071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu T., Shklovskii B. I., Kinetics of viral self-assembly: Role of the single-stranded RNA antenna. Phys. Rev. E 75, 051901 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Rolfsson Ó., et al. , Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J. Mol. Biol. 428, 431–448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borodavka A., Tuma R., Stockley P. G., Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc. Natl. Acad. Sci. U.S.A. 109, 15769–15774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dykeman E. C., Stockley P. G., Twarock R., Solving a Levinthal’s paradox for virus assembly identifies a unique antiviral strategy. Proc. Natl. Acad. Sci. U.S.A. 111, 5361–5366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]