Significance
The parasite Neospora caninum has a worldwide distribution and causes fetal loss in livestock. It is transmitted in cows vertically (from mother to fetus), and in some herds nearly all cattle are infected. We genotyped Neospora strains isolated from an array of infected hosts (dogs, cattle, deer, horse, rhinoceros) and observed a single, highly inbred genome that has swept globally. At first glance, data support vertical transmission as a primary mode for the expansion of the parasite genome; however, genome-level analyses identified large introgression blocks of a distinct ancestry that has recombined into the genome, consistent with a unisexual expansion model. The successful genome appears to have piggy-backed on the expansion of European cattle breeds that have been exported globally.
Keywords: Neospora caninum, protozoan, population genetics, genome, unisexual
Abstract
Neospora caninum, a cyst-forming apicomplexan parasite, is a leading cause of neuromuscular diseases in dogs as well as fetal abortion in cattle worldwide. The importance of the domestic and sylvatic life cycles of Neospora, and the role of vertical transmission in the expansion and transmission of infection in cattle, is not sufficiently understood. To elucidate the population genomics of Neospora, we genotyped 50 isolates collected worldwide from a wide range of hosts using 19 linked and unlinked genetic markers. Phylogenetic analysis and genetic distance indices resolved a single genotype of N. caninum. Whole-genome sequencing of 7 isolates from 2 different continents identified high linkage disequilibrium, significant structural variation, but only limited polymorphism genome-wide, with only 5,766 biallelic single nucleotide polymorphisms (SNPs) total. Greater than half of these SNPs (∼3,000) clustered into 6 distinct haploblocks and each block possessed limited allelic diversity (with only 4 to 6 haplotypes resolved at each cluster). Importantly, the alleles at each haploblock had independently segregated across the strains sequenced, supporting a unisexual expansion model that is mosaic at 6 genomic blocks. Integrating seroprevalence data from African cattle, our data support a global selective sweep of a highly inbred livestock pathogen that originated within European dairy stock and expanded transcontinentally via unisexual mating and vertical transmission very recently, likely the result of human activities, including recurrent migration, domestication, and breed development of bovid and canid hosts within similar proximities.
Eukaryotic pathogens, including protozoan parasites and fungi, are the causative agents of devastating sequelae in both humans and livestock. Most of these eukaryotic pathogens selectively utilize 2 modes of reproduction, sexual and asexual, to evolve and fix their population genetic structures. Sexual reproduction is ubiquitous throughout the eukaryotic kingdom and genetic recombination is known to shape population genetic structures, allowing these pathogens to evolve nonclonal population structures, as has been demonstrated for several fungal pathogens (1). However, recent reports have documented the potential for both sexual macroevolution and asexual microevolution among clonal outbreak clusters of Cryptococcus gattii (2). In contrast, the expansion of a clonal Plasmodium falciparum population structure, the parasite that causes human malaria and is responsible for the death of 1 million African children annually (3, 4), was largely shaped by a rapid spread of drug-resistant parasites across continents through selective sweeps that act on alleles in their chromosomes (5, 6). In contrast to Plasmodium, Toxoplasma gondii, another widespread apicomplexan parasite, utilizes both sexual and asexual transmission to shape its global population structure (7–9).
Neospora caninum is a spore-forming, single-celled obligate intracellular parasite belonging to the apicomplexan phylum (10), which includes many medically important parasites, such as Plasmodium spp., Cryptosporidium spp., and T. gondii. After its discovery in 1984, infecting a dog in Norway (11), neosporosis has since become recognized as a major cause of bovine abortions, causing significant economic loss in the dairy and beef cattle industries worldwide (12, 13). Like other closely related apicomplexan parasites, N. caninum possesses a relatively limited host range, including—but not limited to—dogs, cattle, and sheep. Its success in nature is thought to be the result of its highly flexible life cycle with multiple routes of transmission possible. In cattle, it is transmitted vertically (transplacental infection) either by recrudescence of a preexisting maternal infection (endogenous) or by ingestion of oocysts, products of the parasite’s sexual cycle, that sporulate in the environment after being shed by its definitive canid host (exogenous). It can also be transmitted horizontally (acquired postnatally) by ingestion of sporulated oocysts (10, 14, 15). No evidence currently exists documenting the transmission of infectious tissue cysts by carnivory between intermediate hosts, as is seen for T. gondii (7, 16). Surprisingly, no studies to date have investigated whether the parasite’s sexual cycle has impacted the population genetics and transmission of strains in nature, nor whether meiosis genes or other genes associated with sexual reproduction are conserved within the genome. Sexual transmission, which has been demonstrated to occur only in canid species (17), is thought to occur at low levels, based on previous modeling studies and published data (14), and this may explain the paucity of studies reported.
Central to understanding whether genetic diversity within a species influences either their pathogenicity or host range, only a few molecular epidemiological studies have been systematically applied against Neospora, and those that have been done were of typically limited resolution (18). This lack of knowledge is due primarily to a dearth of suitable population genetic tools applied against only limited isolates that do not capture the parasite’s extensive host range or wide geographic distribution. These few studies used the 18S and ITS1 regions within the small subunit RNA, and found no nucleotide differences (19, 20). Recently, single gene sequences from polymorphic surface antigens (SAG1, SAG4, and SRS2) and secreted dense granule proteins (GRA6 and GRA7) were developed; however, they have not been widely applied and were only used to genotype very few bovine and canine Neospora isolates from a restricted geography (21–23). Unlike the polymorphic gene-sequence markers, multilocus microsatellite (MS) markers have been utilized extensively for molecular characterization of N. caninum isolates, and they show extensive genetic diversity that imply a recent genetic diversification from a common ancestor (24, 25). Interestingly, the identification of Neospora hughesi, which is considered a separate species because it has only been isolated from horses (26), did show significant sequence polymorphism at the polymorphic genes compared to N. caninum (21–23). Taken together, these data suggest that Neospora population genetics is composed of at least 2 separate species, N. caninum and N. hughesi, that have evolved from a common ancestor and expanded independently to adapt to their various vertebrate hosts. However, the drawback of these previous analyses is that they were based on only a few genetic and MS markers that lack sufficient resolution to be informative genome-wide; the polymorphic markers are too few to capture polymorphism throughout the genome, and the MS markers used are prone to misclassify variants due to homoplasy (27). Hence, the primary objective of this study was to conduct a systematic population genetic analysis to compare existing, and newly developed, polymorphic markers alongside MS markers to prioritize isolates for whole-genome sequencing (WGS) and produce a population genetic model to identify the extent to which Neospora is expanding in nature by outcrossing or inbreeding.
The impact of host migrations on the population structure of a pathogen can be difficult to determine with many factors contributing to the overall success and expansion of a pathogen when it occupies a new ecological niche. However, several examples have shown how ancient traditions, such as nomadic migrations, as well as the purchase of infected livestock have contributed extensively toward the introduction of pathogens, such as Mycobacterium avium and the virus that causes foot and mouth disease into previously uninfected farms (28). Neospora could prove to be another such interesting example as it can be maintained as a chronic infection in livestock, which would likely aid in its transmission, not only locally but also across continents. Such global movement of the bovine host may contribute extensively to fixing the population genetic structure, but this question remains largely unexplored.
Here, we sought to improve the understanding of Neospora’s population genetic structure by analyzing genome-wide genetic diversity using next-generation sequencing of Neospora strains isolated from different hosts and geographic locations. To this end, we genotyped 47 N. caninum and 3 N. hughesi strains from a wide host range, using 12 (9 nuclear and 3 mitochondrial) sequenced markers to define the population genetic structure. Eight of the 50 strains were subjected to further characterization at whole-genome resolution using next-generation sequencing. Genetic characterization using sequenced markers and WGS supports a model whereby a highly inbred genome of N. caninum has evolved recently and swept, the result of a genetic bottleneck. In this study we used seroprevalence data from African cattle to gain insight into the profound role the migration of European breeds of domesticated cattle have played in this massive genetic “selective sweep” of a highly inbred Neospora genome that has piggy-backed on global livestock trade to become distributed globally.
Results and Discussion
Global Population Genetic Structure of Neospora Using Linked and Unlinked Genetic Markers.
To understand the true extent of genetic diversity and to determine the population genetic structure of N. caninum, 9 linked and unlinked sequence markers (SM) (SI Appendix, Table S2) were developed based on genetic markers that have been successfully applied to genotype T. gondii strains. This robust set of genetic markers comprises 5,647 bp and was used to characterize 47 Neospora isolates collected globally (SI Appendix, Table S1). These strains were isolated from various geographic locations in North and South America, Europe, Asia, and Australia (SI Appendix, Fig. S1) and from a wide host range, including cattle, dogs, horses, wolves, rhinoceros, and deer (SI Appendix, Table S1). To perform a cross-species comparison of genetic diversity, we also included 3 N. hughesi strains. After sequencing, we conducted maximum-likelihood analysis using single nucleotide polymorphisms (SNPs) from concatenated sequences of SM markers. Strikingly, all 47 N. caninum strains clustered tightly into a single monophyletic node (Fig. 1A). Four independent SNPs separated 4 strains from the remaining 43, and those 43 were identical based on maximum-likelihood analysis. Population structure derived from the program STRUCTURE (29), which utilizes a Bayesian statistical approach for linkage analysis, closely resembled the maximum-likelihood analysis, and revealed only 2 populations with no admixture between the 2 (Fig. 1B and SI Appendix, Fig. S2), which is in close agreement with the population structure derived from principle component analysis (SI Appendix, Fig. S3). Interestingly, the 3 N. hughesi strains were markedly different from the N. caninum strains and formed a distinct cluster not only in the phylogenetic analysis, but also in STRUCTURE, supporting their distinction as a separate species (26). However, an alternative explanation could be that N. hughesi exists as a strain of N. caninum but is polymorphic and less successful. Examples of isolates, those that are highly polymorphic but belong to the same species based on sexual recombination, are well documented to occur in T. gondii, for example haplogroups 2 and 10. Thus, the ability of cross-species mating between N. caninum and N. hughesi remains to be tested or inferred by comparative phylogenomic sequencing studies. Collectively, our analyses identified a single genotype of N. caninum that has expanded globally.
Fig. 1.
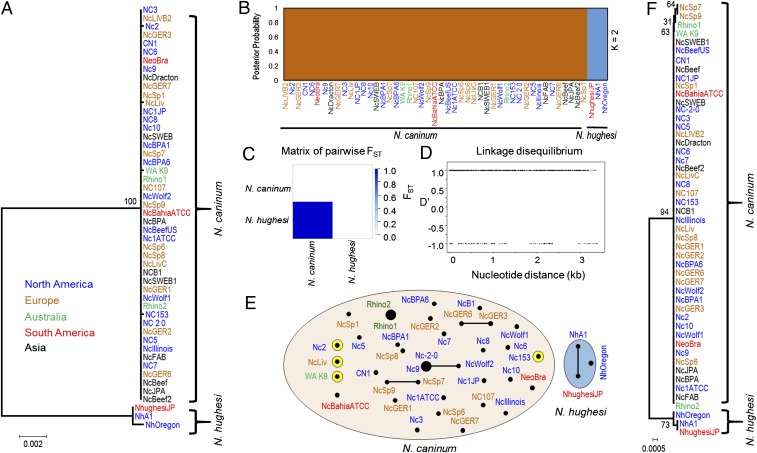
Expansion of a single clonal lineage of N. caninum throughout the world. (A) Concatenated phylogenetic tree using 9 sequenced markers comprised of linked and unlinked introns, antigens, and housekeeping genes (5,647 bp) (SI Appendix, Table S2) from 50 globally distributed strains (SI Appendix, Fig. S1 and Table S1). Two populations of Neospora are identified: N. caninum and N. hughesi. All N. caninum strains are tightly clustered in a single node (with only 4 private SNPs), demonstrating a global selective sweep. An unrooted maximum-likelihood tree was constructed using MEGA (73) with 1,000 bootstrap replicates. Bootstrap values are indicated at each node as a percentage. Strains are colored based on their continent of origin. (Scale = number of SNPs per site.) (B) Population STRUCTURE analysis using an admixture model with a Bayesian clustering algorithm (ancestral population K = 2) shows 2 expanded populations of Neospora (SI Appendix, Fig. S2) with no admixture blocks identified between the populations based on sequenced markers (5,647 bp). Strains are colored based on their continent of origin. (C) Pairwise FST calculation identified a close genetic relatedness among the globally distributed isolates of N. caninum (FST ≤ 0.2 supports the minimal genetic differentiation). (D) The LD (D′) plot demonstrated complete linkage disequilibrium among N. caninum strains. Sequenced markers were concatenated to generate the plot. (E) Modified eBURST analysis with an n − 1 rule (where n = no. of MS markers) using intron and antigen markers as well as 7 sequenced MS makers. The intron and antigen markers from N. caninum formed a monophyletic group (red circle) that were separated from the N. hughesi isolates (blue circle). However, the MS markers were sufficiently resolved and readily demonstrated a fine separation among the N. caninum lineage isolates. Circle size is proportional to the number of strains present within each circle. Yellow circles highlight strains that were separated using the DNA sequence markers from A. (F) Although phylogenetic construction using 3 mitochondrial markers (COX1, COX3, CYTB) demonstrated a uniparental maternal inheritance among the majority of the N. caninum isolates, there are signatures for different sources of maternal inheritance in some strains including NcSp7, NcSp9, WA K9, and Rhino1 could be differentiated from Rhino2. The maximum-likelihood tree was constructed using MEGA with 1,000 bootstrap replicates. Bootstrap values are indicated at each node as a percentage. Strains are colored based on their continent of origin. (Scale = no. of SNPs per site.)
A Global Selective Sweep of a Single Genotype of N. caninum.
The genetic makeup of N. caninum isolates was very closely related (FST = 0.0, P < 0.0001) and highly divergent from that of N. hughesi (FST = 0.7293, P < 0.0001) based on a pairwise fixation index FST calculation (Fig. 1C). Pairwise haplotype distance matrix also indicated that the genetic distance among N. caninum strains was very low (SI Appendix, Fig. S4), which supports a nonsubdivided population. We also calculated linkage disequilibrium (LD) to understand the influences of sexual recombination on the population genetic structure of N. caninum, as a high cross-over rate can introduce rapid LD breakdown. The N. caninum strains possessed a high LD by the standard test of D′ among different unlinked markers (Fig. 1D). The low genetic diversity observed led to an effectively clonal population structure and high LD among the N. caninum isolates. Collectively, the phylogenetic analyses, calculation of genetic distance indexes including FST, haplotype distance matrix (SI Appendix, Fig. S4), average number of pairwise differences (SI Appendix, Fig. S4), and LD all identified a global expansion of a single N. caninum genotype. To our knowledge, this global population structure is completely unprecedented among closely related apicomplexan parasites and exits as a pandemic outbreak clone.
Drug-selection sweeps of a single chromosome for pathogens like Plasmodium have been shown to occur previously in endemic regions (30), and it has been likewise postulated that endogenous vertical transmission or congenital transmission could conceivably facilitate the penetrance of a single Neospora genome. In cattle, sheep, and small ruminants, for example, a preexisting maternal infection with Neospora is thought to be the primary mode of N. caninum transmission, allowing the parasite to effectively bypass its sexual cycle within its definitive canid host (15). In contrast, unisexual transmission of certain pathogenic fungi, as well as the protozoan parasite Toxoplasma, can generate clonal population structures by inbreeding among highly related strains or by self-mating (also referred to as uniparental sexual transmission) in which a single parasite clone can generate both male and female gametes that fuse within its definitive host (8). To determine which of these 2 transmission models, endogenous or unisexual, best explain the origin and global expansion of a single N. caninum genome, we performed both microsatellite and WGS analyses
Substructure Exists within the Single N. caninum Genotype.
A limited number of previous studies utilizing MS markers suggested extensive genetic diversity within the N. caninum population genetic structure (24, 25, 31–33). Our results using the SM markers did not support this model and rather suggested a pandemic expansion of a single genome. We next applied 7 MS markers to assess intratypic variation among our global collection of isolates to formally test whether the genetic relationship between the isolates is consistent with an outbreak clone of a single N. caninum genotype. Three of the 7 MS markers applied were used in previous studies (33), giving us the opportunity to compare our results with those found in other studies (24, 25, 31–33). To visualize the genetic diversity identified, we conducted a modified eBURST analysis in 2 ways: First with nuclear markers, which resolved the isolates into just 2 clonal complexes, N. caninum and N. hughesi (Fig. 1E); and second, using the MS markers. Interestingly, eBURST analysis of the MS markers showed a collapse in the clonal complexes and a high degree of divergence among the N. caninum isolates investigated (Fig. 1E). Of the 43 strains analyzed, only 2 clonal complexes (Rhino1 and Rhino2; NC-2-0 and NC9) were observed (Fig. 1E), which is consistent with previous studies (24, 25, 31–33). Thus, the microsatellite analysis showed a clear incongruence among strains genotyped using the SM markers and did support intratypic subdivision of the single genotype identified. Combining the 2 datasets from eBURST, our results support a model in which N. caninum has evolved recently from a common ancestor, and the high degree of divergence among the strains using MS markers is consistent with the fact that microsatellite mutations evolve at a much faster rate than the rest of the genome (34, 35).
Signature of Sexual Recombination.
The existence of a monomorphic genome suggests that N. caninum has expanded either asexually as a clone or by uniparental mating after sexual inbreeding. Mitochondrial genomes have been used in other organisms to differentiate between asexual versus sexual reproduction because mitochondria are exclusively inherited maternally, and are less prone to genetic exchange (36). To evaluate genetic relationships in Neospora, SNPs from 3 mitochondrial gene markers, comprising 2,211 bp, were used in a maximum-likelihood analysis (Fig. 1F). This analysis was used to determine whether mitochondrial genomes were genetically diverse within the N. caninum clonal population and likely inherited from various sources, which would be a signature for sexual recombination. The phylogenetic analysis showed that 2 strains (NcSp7 and NcSp9) had inherited mitochondrial genomes from a different source, resolved by 2 SNPs not shared with the other N. caninum strains (Fig. 1F). NcSp7 and NcSp9, isolated from 2 different cattle on the same farm in Spain, were not only identical to each other at the SM markers, they also formed a clonal complex using the MS markers. Two other strains, Rhino1 and Rhino2, isolated from a single host, also differed by a single SNP in their mitochondrial markers but not at any of the SM markers (Fig. 1F). These results may indicate that these 2 sets of strains evolved from separate recombination events. Alternatively, it is also conceivable that they have simply accumulated SNPs in their mitochondrial genomes, the result of genetic drift. However, the fact that the 4 strains were identical to all N. caninum isolates at the nuclear genome level, and the specific accumulation of shared SNPs was present only within the maternally inherited mitochondrial genome, rather supports a scenario in which unisexual expansion is occurring among circulating strains of N. caninum.
Population Structure at Whole-Genome Resolution.
To produce a genetic ancestry model for Neospora and test whether unisexual reproduction can explain the origin of its population genetic structure, we prioritized WGS on the following 7 Neospora isolates, consisting of 6 N. caninum (4 from the United States and 2 from Europe) and 1 N. hughesi (from the United States) strain using Illumina HiSeq (Illumina) (SI Appendix, Table S3). These N. caninum strains were selected because they were isolated as single clones and could be expanded by in vitro culture to generate sufficient genomic DNA for WGS. One isolate (NcSp7) possessed a divergent mitochondrial genome. The small reads obtained were reference-mapped against the N. caninum reference genome, NcLiv (https://.toxodb.org/toxo). A total of 375,560 high-quality biallelic SNPs were identified across all 8 strains, including reference NcLiv (Fig. 2A, SI Appendix, Table S3, and Dataset S1, SNPs found in Neospora strains). More than 372,000 biallelic SNPs (∼0.5% polymorphism) were identified between N. caninum and N. hughesi. In contrast, only 5,766 SNPs (SI Appendix, Table S3) differentiated the 6 N. caninum strains, representing a polymorphism rate of 0.0005 to 0.0059%. This range is significantly lower than the 1 to 2% polymorphism observed among globally distributed strains of the closely related apicomplexan parasite T. gondii (16), indicating that intraspecific genetic diversity is substantially lower among the N. caninum isolates (SI Appendix, Figs. S5 and S6 and Table S3).
Fig. 2.
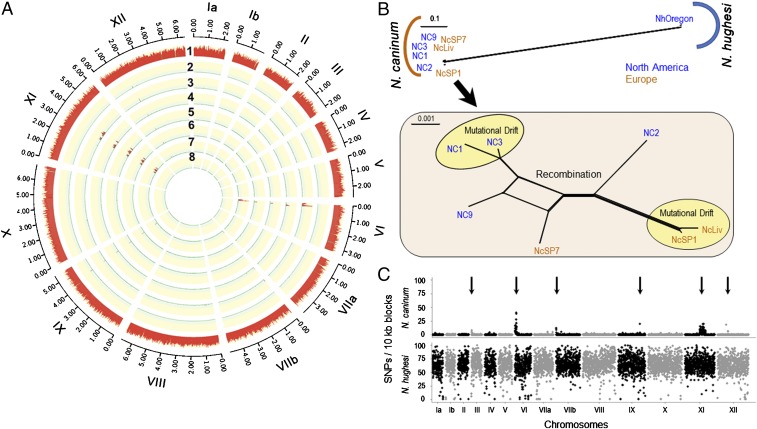
Phylogenomic analysis of Neospora based on WGS. (A) N. caninum: Circos plot depicting SNP density using NcLiv as the reference strain (SI Appendix, Fig. S6 and Dataset S1, SNPs found in Neospora strains). Chromosomes are numbered by Roman numerals and size intervals are plotted using Arabic numerals. The red track with the yellow background indicates the number of SNPs per 10-kb sliding window. Strain names: 1) NhOregon, 2) NcSP1, 3) NC2, 4) NC3, 5) NcSP7, 6) NC9, 7) NC1, and 8) NcLiv. (B) Neighbor-net analysis of 8 Neospora strains using genome-wide SNPs (375,560) (SI Appendix, Table S3) indicates 2 distinct populations of Neospora (N. caninum and N. hughesi) with significantly high genetic distance. Zooming-in on the N. caninum population (Inset) shows very low genetic diversity (total = 5,766 SNPs). The yellow circles denote strains that are in genome-wide LD and are separated by mutational drift, whereas the other strains possess a reticulated structure, indicating that gene flow has occurred between these remaining isolates (red). Strains are colored based on their continent of origin. (Scale = no. of SNPs per site.) (C) SNP density plots using the average number of SNPs present within 10-kb sliding windows when compared with the reference strain NcLiv. The y axis shows the number of SNPs per 10-kb sliding window and identified 6 distinct sequence blocks that possessed elevated SNPs (arrows). The x axis indicates the relative size of the chromosome. Chromosomes are separated by black and gray colors.
Using genome-wide SNPs, a neighbor-net analysis (37) similarly identified the existence of 2 distinct genetic clusters: 1) N. caninum and 2) N. hughesi (Fig. 2B). Seven of the 8 strains grouped very tightly within the N. caninum cluster, which was consistent with our findings using the SM markers. Importantly, the increased resolution within the neighbor-net analysis using genome-wide SNPs identified substructure and supported recombination between different N. caninum strains, which was largely separated by geographic location. To identify the precise locations of genetic admixture detected in Fig. 2B, we developed SNP plots across Neospora’s 14 chromosomes by binning total number of SNPs per 10-kb window per strain (Fig. 2C and SI Appendix, Fig. S6). SNP plots showed that the N. caninum genome is monomorphic, except at 6 large, distinct sequence blocks. These haploblocks each possessed elevated SNP rate signatures, some of which were shared and appeared to have independently segregated across the 7 isolates sequenced. These signature blocks were found specifically within the 5′ regions of chromosomes III, VI, and VIIb; the 3′ region of chromosome IX; and the middle portion of chromosome XI (between 3 and 4 Mb) and XII (Fig. 2C and SI Appendix, Figs. S6 and S7). Strikingly, more than half of the total N. caninum SNPs (n = ∼3,000 of 5,766) clustered within these 6 distinct haploblocks. An unrooted phylogenetic tree utilizing SNPs derived from the 6 nonconserved, haploblock-only regions, was incongruent with the rest of the genome, and did not share the same overall topology (SI Appendix, Fig. S9). The SNP rate was ∼5-fold higher compared to the monomorphic or conserved regions of the genome (which had fewer than than 1 SNP/kb). To confirm that the nonconserved regions were statistically different, and consistent with a distinct ancestry, a Robinson–Foulds (RF) distance metric (38) was calculated. The RF distance metric statistic indicated that the 2 regions (conserved vs. nonconserved) were genetically distinct. In addition, each haploblock possessed limited allelic diversity (with only 4 to 6 haplotypes resolved at each cluster), consistent with genetic hybridization. The haploblocks on chromosomes VI and XI appeared to possess signatures of introgression with shared haplotypes that had independently assorted across the strains examined, supporting a unisexual expansion model (SI Appendix, Fig. S7).
In contrast, the N. hughesi strain, which represents a separate species of Neospora (21, 26), showed extensive genetic diversity compared to N. caninum strains (∼6 to 9 SNPs/kb) (Fig. 2 B and C and SI Appendix, Figs. S5 and S6). Although the 0.5% rate of polymorphism between the 2 Neospora species is lower than that observed among circulating T. gondii strains known to undergo genetic hybridization, the Neospora interspecies SNP density plot (SI Appendix, Fig. S5) failed to detect any genetic admixture between N. caninum and N. hughesi. In addition to genetic diversity, the maternally inherited apicoplast genome of N. hughesi had a different ancestry than that of N. caninum (Fig. 3A). The available data appear to support their designation as separate species, but additional comparative phylogenomic sequencing studies utilizing more isolates or experimental crosses should be attempted to confirm whether or not these 2 species are mating compatible, as many interspecies crosses have now been performed successfully among different Leishmania species, for example (39).
Fig. 3.
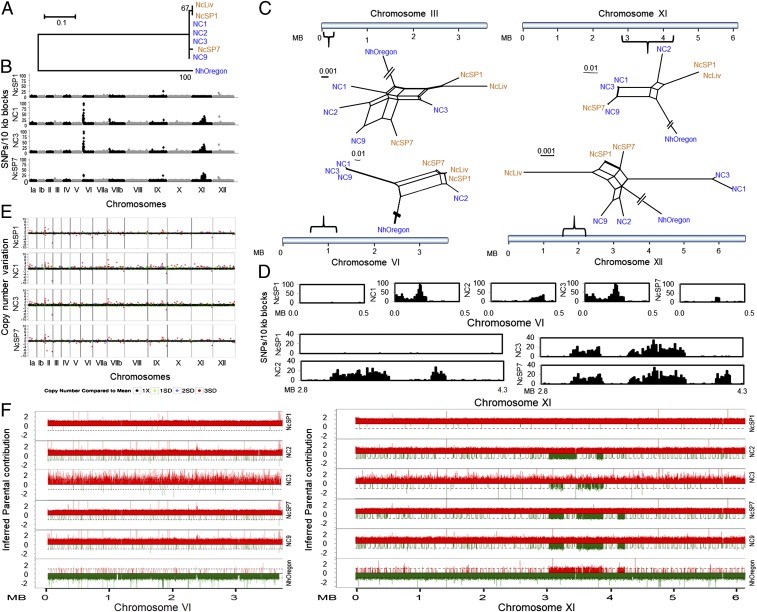
Structural variation and introgression of haploblocks by sexual recombination in the genomes of N. caninum. (A) Different sources of maternal inheritance in Neospora were depicted by a phylogenetic tree using SNPs from the apicoplast genome (total = 105). A maximum-likelihood tree was constructed using MEGA with 1,000 bootstrap replicates. Bootstrap values are indicated at each node as a percentage. (Scale = no. of SNPs per site.) (B) Graphical representation of SNP density plots. SNPs were calculated by comparing isolates against the reference strain NcLiv, and the total number of SNPs present within 10-kb blocks were plotted. Alternating black and gray highlighted sections represent different chromosomes. Low SNP density indicates a high degree of similarity with the NcLiv strain. The y axis indicates SNP density per 10-kb windows and the x axis represents the position on the chromosomes. (C) Neighbor-net networks were developed using SNPs present within 4 variable regions of the N. caninum genomes. (Scale = no. of SNPs per site.) Colors are based on the continent of origin of these strains as depicted in Fig. 2B. Brackets represent the relative positions of the variable regions in the chromosomes. The blue horizontal line indicates the relative size of each chromosome. (D) Detailed comparison of SNP density plots of 10-kb sliding windows indicates patterns of introgressed haploblocks in chromosomes VI and XI. NcLiv was used as the reference strain for comparison. The y axis indicates total number of SNPs per 10-kb blocks and the x axis represents the position on the chromosomes. (E) Genome-wide structural variation was revealed by analysis of the CNV in 10-kb tiling windows. The 10-kb blocks with no CNV are plotted as black dots (1X). Green, blue, and red dots indicate 1, 2, and 3 SDs from the mean (1X), respectively. y axis indicates the CNV. (F) Comparison of parental SNPs across chromosomes VI and XI of N. caninum using bottle-brush plots indicated shared ancestry with N. hughesi on chromosome XI, but not chromosome VI. SNPs mapping to N. caninum are shown in red, whereas N. hughesi SNPs are shown in green. The y axis indicates the inferred allelic depth normalized across the entire genome. The x axis represents the position of the chromosomes.
Expansion of a Highly Successful Inbred Population.
The accumulated evidence indicates that a single genome of N. caninum has expanded globally, but the relative contribution of endogenous vertical transmission versus unisexual reproduction in the expansion of this highly successful genome is enigmatic. To understand the effective forces shaping the transmission and population genetic structure of N. caninum, we performed a comparative WGS phylogenomic analysis using the maternally inherited 35-kb circular apicoplast genome (Fig. 3A). Surprisingly, the maximum-likelihood analysis identified only 3 SNPs, and these separated the mitochondrial sequences from the 7 N. caninum isolates into 3 distinct matrilineages. However, the paucity of genetic diversity within the apicoplast genomes failed to resolve whether the single genome of N. caninum was the product of multiple recombination events or if the few SNPs identified in the mitochondrial sequences were due to genetic drift.
To investigate whether genetic hybridization can explain the origin of the elevated SNP regions within each of the 6 distinct haploblocks (Fig. 3B and SI Appendix, Figs. S6, S7, S11A, S14A, S15A, and S16A), a pairwise FST analysis was performed. The genome-wide SNP-based FST measurement identified a breakdown of the FST in each of the 6 distinct haploblock regions (SI Appendix, Fig. S12), indicating significant genetic differentiation. Collectively, the RF distance metric and FST plots support a model whereby the haploblocks possess distinct ancestries that have introgressed into the most recent common ancestor (RCA) of N. caninum by genetic recombination.
To differentiate between genetic hybridization from that of selective retention or somatic recombination, 2 alternative scenarios capable of generating and maintaining elevated SNP regions within an otherwise monomorphic genome, we examined all genes contained in the 6 haploblocks. We did not observe any obvious signatures in the functional categories of the genes present that could explain the high genetic diversity observed (Dataset S2, Neospora gene list in elevated SNP regions); however, published RNA-sequencing datasets showed that several genes, particularly in the region on chromosome VI, are highly overexpressed in NcLiv compared to NC3, for example (SI Appendix, Fig. S10A). Pairwise comparison of genome-wide nucleotide substitution rates (dN/dS) also indicated that several of these genes are under purifying selection, supporting a model of adaptive evolution (SI Appendix, Fig. S10B).
To determine the origin and the extent of haploblock diversity, we reconstructed the genetic ancestry of each haploblock individually using a neighbor-net network analysis (Fig. 3C). All 6 trees were reticulated, and the topology of shared ancestry was incongruent between the 7 isolates, depending on the genomic region analyzed. A maximum of 7 haplotypes was resolved at chromosome III, whereas only 4 haplotypes were identified at chromosomes VI, VIIb, and XII, indicating shared ancestry (Fig. 3 C and D and SI Appendix, Figs. S11A, S14A, S15A, and S16A). To determine if the genetic relationship within the 6 haploblocks was consistent with that of recombination, as opposed to genetic drift, the inheritance of each haploblock was assessed across the 7 isolates. NC9 shared essentially the same haploblock sequence with NC1 and NC3 at chromosomes VI and VIIb, but not at chromosome XI, where it shared haploblock sequence identity with NcSp7 (Fig. 3 C and D and SI Appendix, Fig. S16), thus supporting recombination. In addition, the NC1 haploblock was distinct from NC3 on chromosome III, indicating that these 2 isolates were not in LD. NcLiv shared 5 of 6 haploblocks with NcSP1, except at chromosome XII, whereas NC2 possessed a distinct haplotype at each of the 6 haploblocks. The best illustration of genetic hybridization occurred within the 3- to 4-Mb region on chromosome XI, which possessed ostensibly 3 tandemly arrayed introgression blocks. An SNP density plot clearly showed that NC1, NC3, NC9, and NcSP7 possessed 2 identical haploblocks, but that the third introgressed block present in NcSP7 and NC9 was not present in NC1 and NC3 (Fig. 3F and SI Appendix, Fig. S11A). Similarly, NC2 shared identity with the first haploblock, but only part of the second haploblock, and not the third. NC2 also possessed a distinct recombination haploblock not present in all other isolates (SI Appendix, Fig. S11).
This overall pattern of introgressed blocks in chromosome XI was also present in the N. hughesi isolate NcOregon. To show that each punctate recombination block was genetically distinct, the ancestry of each SNP position was enumerated using a bottle brush plot (40), which clearly identified sequence blocks that were neither N. caninum nor N. hughesi, but shared ancestry with both species (SI Appendix, Fig. S11B). Distinct recombination blocks harboring this divergent genetic ancestry were also observed in the 5′ region of chromosome VIIb (SI Appendix, Fig. S16). The most parsimonious explanation for the existence of these admixture patterns is that the genome of N. caninum has previously undergone genetic hybridization in nature with an ancestry distinct from N. caninum. However, with only limited recombination blocks identified genome-wide, the relationship among the isolates sequenced is consistent with a genetic model whereby sexual recombination between sister clones has since purified much of this distinct genetic ancestry from the genome by unisexual mating, except within 6 haploblocks (8, 41, 42).
Another hallmark of unisexual mating is increased structural variation among isolates that undergo sexual expansion, as opposed to exclusive asexual replication, as would be seen in vertically transmitted clones of Neospora. We therefore investigated structural variation in N. caninum by determining copy number variation (CNV) in 10-kb sliding windows across the genome. Significant structural variation was detected among the monomorphic N. caninum genomes (Fig. 3E and SI Appendix, Fig. S8). For example, despite NC1, NC3, and NC9 having similar SNP density plots (Fig. 3B and SI Appendix, Fig. S6) and the same maternally inherited mitochondrial genome (Fig. 3A), they each possessed different patterns of genome-wide CNVs (Fig. 3E and SI Appendix, Fig. S8). Alternatively, the NcSP1 and NcSP7 isolates had essentially identical CNV plots (Fig. 3E and SI Appendix, Fig. S8) but different mitochondrial genomes and were incongruent at the 6 haploblocks, suggesting that they were recent recombinants (Fig. 3 A and B and SI Appendix, Fig. S6). De novo generation of aneuploidy and localized expansion of copy number are commonly associated with meiotic reproduction in somatic cells (43, 44). Meiotic reproduction and aneuploidy or expansion of copy number in the yeast Saccharomyces cerevisiae or in Leishmania is known to confer genotypic and phenotypic plasticity to better adapt to new environments (43, 44). Although somatic replication can also generate structural variants in both yeast and kinetoplastid parasites, and mitotic recombination is known to produce chimeric variants within the Plasmodium surface-associated Var genes, such somatic recombination has not been identified to occur within gene families from other well-studied, closely related apicomplexan parasites, such as T. gondii. Furthermore, this type of structural variation is typically localized and is not likely to account for the significant, genome-scale CNV differences we found in N. caninum. Bottle-brush plots (45) generated to map parental SNPs across N. caninum and N. hughesi identified 2 large sequence haploblocks on chromosomes VIIb and XI that encoded a distinct genetic ancestry (i.e., it was neither N. caninum nor N. hughesi) (Fig. 3F and SI Appendix, Figs. S11B and S16B). Moreover, no gene families (e.g., the Srs or Ropk gene families analogous to the Var genes in Plasmodium) or those associated with meiosis (there are no mating types in Neospora) are observed in these regions. Rather, these haploblocks possess a diverse array of different genes. Collectively, the identification of large, introgressed sequence haploblocks bearing different genetic ancestry coupled with the evidence of localized CNVs is more consistent with sexual recombination having impacted the population genetics of N. caninum.
Recent Expansion of N. caninum Worldwide.
As discussed above, apart from the 6 variable haploblocks, our genome-wide pairwise SNP comparison identified nearly identical genomes among the N. caninum strains sequenced. Based on this finding, we hypothesized that the current N. caninum population was recently derived, and is presumably the result of recombination events and a genome-selective sweep that has effectively expanded a highly inbred and successful genome throughout the world. Because no fossil record or mutation rate calculation has been performed for Neospora (46), we next estimated the time to most RCA by comparative genome analysis using 2 closely related apicomplexan parasites; T. gondii and P. falciparum. Neospora and T. gondii separated from a common ancestor ∼28 million y ago and have since evolved independently (47). Taking advantage of WGS data for 36 T. gondii strains obtained by a community white paper (https://www.jcvi.org/gcid/project/toxoplasma-gondii) (16), short DNA sequencing reads were mapped against the reference strain ME49 and 1,065,685 SNPs were identified. Network analysis using genome-wide SNP data revealed a complex population genetic structure for T. gondii, which divided into ∼15 distinct haplogroup subpopulations with deep branch lengths (48–50) (Fig. 4A). Thus, T. gondii, a close relative of N. caninum, possessed a completely different population genetic structure and showed a high degree of genetic diversity compared to the unprecedented finding of a single genome of N. caninum. Previous studies estimated that the North American and European T. gondii clonal lineages (types I, II, and III) expanded ∼500 to 5,000 y ago, based on Plasmodium mutation rates (48, 51). More recently, a pairwise comparison of genome-wide SNPs between 2 type I strains identified only 1,394 SNPs and insertions/deletions (INDELs) that separated RH from GT1 (52), which is higher than the number of SNPs/INDELs separating any pair-wise combination within our sequenced N. caninum isolates (having a range of 21 to ∼1,000 SNPs), supporting a recent global expansion of this species.
Fig. 4.
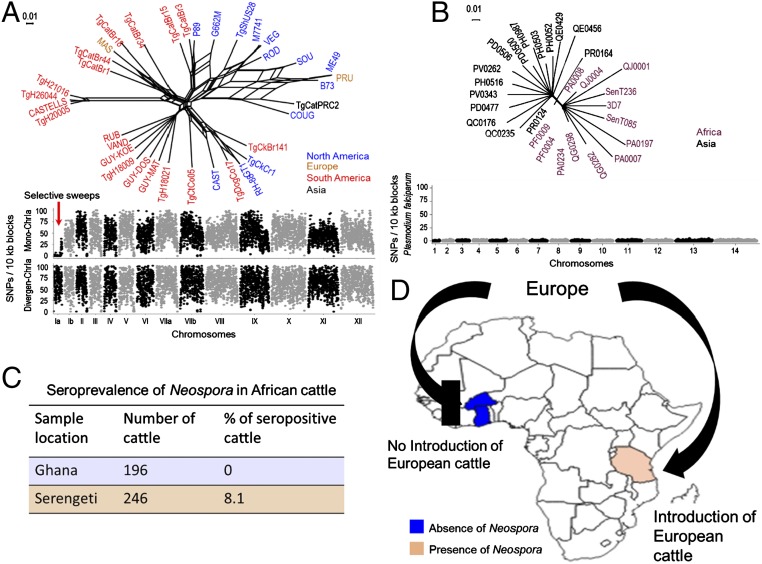
Recent expansion of N. caninum worldwide through European cattle movement. (A) Population genetic structure of T. gondii based on neighbor-net analysis using genome wide SNPs (total = 1,065,685) from 36 strains (SI Appendix, Table S4). Strains are colored based on their continent of origin. (Scale = no. of SNPs per site.) The SNP density plot shows the monomorphic distribution of SNPs on chromosome Ia for most of the T. gondii strains (red arrow). Each dot indicates the average number of SNPs present within 10-kb sliding windows compared with the reference strain Me49. y axis = no. of SNPs per 10-kb window, x axis = relative size of chromosome. (B) Neighbor-net analysis indicates that recent drug-resistance selective sweeps that have shaped the population genetic structure of P. falciparum. Neighbor-net analysis was conducted based on 8,810 SNPs among 28 strains (SI Appendix, Table S5). Strains are colored based on their continent of origin. (Scale = no. of SNPs per site.) SNP density plots indicate the monomorphic distribution of SNPs in the P. falciparum genome. Each dot indicates the average number of SNPs present within 10-kb sliding windows comparing with the reference strain 3D7. y axis = no. of SNPs per 10-kb window; x axis = relative size of chromosome. (C) Blood samples collected from West and East African cattle (SI Appendix, Table S6) were subjected to serological testing for the presence of Neospora antibodies. Orange and blue regions indicate the presence and absence of Neospora antibodies, respectively. (D) Schematic representation and map of Africa with the location of the cohort enrollment sites and direction of European cattle movement (C) (SI Appendix, Table S6).
To better understand the rapid emergence and evolution of the highly inbred N. caninum genome, we generated SNP density plots of T. gondii using 2 sets of strains: 1) those that have selectively retained a monomorphic chromosome Ia (ChrIa), versus 2) those that possess a divergent ChrIa. It has been postulated that genes unique to the monomorphic version of ChrIa have facilitated the recent sweep of highly successful lineages and improved strain fitness in the population (51, 53). An age calculation for ChrIa showed that the fixation of the monomorphic ChrIa took place very recently, within 10,000 y. Strikingly, SNP density plots showed that the genetic architecture and pairwise SNP rate of the monomorphic ChrIa of T. gondii was higher than the pairwise SNP rate calculated for N. caninum (fewer than 1 SNP in 10,000 bp genome-wide, and ∼2-fold lower when the 6 recombination blocks are excluded) (Fig. 4A), further supporting the recent emergence of N. caninum worldwide.
We then compared the genetic architecture of N. caninum with P. falciparum. The population genetic structure of P. falciparum has been shaped by numerous population genetic bottlenecks, principally the result of several recent drug-resistance sweeps throughout the world (54, 55). Taking advantage of 28 sequenced strains part of the Pf3K project (Pf3K Project [2014]: pilot data release 2. https://www.malariagen.net/data/pf3k-2) (56, 57), our network analysis identified only 8,810 biallelic SNPs after reference-mapping with sequence 3D7, and the SNP density plot showed that the population genetic structure of P. falciparum highly resembles the population structure of N. caninum; it likewise has 2 distinct subpopulations: Asian and African (Fig. 4B). Previous reports documented that the current P. falciparum population evolved within the past few thousand years and has recently (within 60 to 70 y) experienced several drug-selection sweeps (58). Given the vagaries of genetic clock calculations that are influenced by the biology and the calculated mutation rate for each species, the 5,766 biallelic SNPs across the 7 sequenced isolates would suggest that the current, highly inbred genome of N. caninum likely evolved as a single lineage that expanded throughout the world as recently as 100 to as late as 10,000 y ago.
Seroprevalence of Neospora Antibodies in East and West African Cattle.
During the past 125 y, the global cattle trade has been instrumental in the distribution and establishment of several transboundary breeds, including Holstein Friesian cattle (Bos taurus) that have been exported from Europe to 126 countries. During this time, European cattle populations were first exported to America, followed by United Kingdom cattle exports to America as well as United Kingdom, French, and Portuguese cattle to Africa (59). Because Neospora is considered a long-term persistent infection within the cattle industry, we hypothesized that N. caninum expanded relatively recently within Europe’s intensive dairy cattle systems and has been subsequently exported with livestock and farm dogs on a large scale sometime during colonization, between the 15th to the 19th centuries, and that the intensive global movement of domesticated cattle breeds in the last 125 y has facilitated parasite expansion and dissemination.
To test our hypothesis, we compared the seroprevalence rate for Neospora in 196 West African cattle, including mixed breeds of B. taurus and Bos indicus from a wide geographical region that has not experienced importation of cattle breeds from Europe, against a cohort of 246 East African cattle, including introduced cattle breeds from Europe (Fig. 4 C and D and SI Appendix, Table S6). Neosporosis seroprevalence rates for West and East African cattle were determined using a highly sensitive and specific multiple antigen-based ELISA that utilizes a water-soluble fraction of sonicated NC1 strain tachyzoites as antigen (60). The ELISA was also controlled for no cross-reactivity with sera from cattle infected with T. gondii or Sarcocystis cruzi. In support of our hypothesis, strikingly no West African cattle were infected with Neospora, whereas 8.1% of East African cattle were seropositive (P = 0.00017) (Fig. 4 C and D and SI Appendix, Table S6). Historically, West Africa and East Africa were not colonized by the Europeans in the same manner with respect to the introduction of European-based agricultural systems. There are several accounts of attempts to introduce European cattle into West Africa, all meeting with complete failure (61). Beal, the first British veterinarian to visit the country in 1911, concluded that both disease and environment were incompatible with European cattle breeds (62). As a result, relatively few White settlers brought farm systems and livestock into the West African region, and in conjunction with the ravages of disease, fewer European cattle were introduced to the area. Thus, the absence of Neospora in regions such as West Africa, where European cattle were not imported, supports a model whereby increased colonization of regions of Africa with intensive European dairy cattle systems may be responsible for the global radiation of Neospora to the region of East Africa, and to native East African cattle. Although deciphering the exact mechanism of Neospora’s expansion remains complex, the difference in seroprevalence between East and West Africa is noteworthy and may indicate that recent movement of cattle industries from Europe have placed a strong selective pressure that resulted in the radiation of a single Neospora lineage throughout the world.
While our data certainly support a recent radiation of a single lineage worldwide, the origin and evolution of this highly inbred genome harboring distinct haploblocks of genetic hybridization is less clear. The most parsimonious explanation is that a heterologous genome recombined with the RCA of N. caninum sometime within the last 10,000 y and that subsequent unisexual mating with N. caninum, in concert with the parasite’s endogenous vertical transmission cycle (59, 60), has purified out the other ancestry, except in 6 sequence haploblocks that have been selectively retained, presumably for their evolutionary advantage. Several aspects of cattle breed evolution and the biology of the Neospora transmission cycle may have contributed to this unique outcome. Specifically, the generation and eventual purification of a hybrid N. caninum genome was presumably selected for by several distinct genetic bottlenecks associated with the domestication of dogs (63, 64) and cattle in the Near East (65–68), followed by an extensive movement of cattle and dogs during human migration (69) which expanded a single, highly successful N. caninum genome. Indeed, archeozoological and genetic data have shown that modern taurine cattle breeds are descendants of local wild ox (aurochs) domestication events that occurred in the Near East prior to extensive cross-breeding of domestic and wild-cattle populations throughout Europe (70) during the Neolithic transition ∼8,000 to 10,000 y ago, which corresponds to one of the timelines for generation of the RCA of N. caninum (65, 67).
In conclusion, by utilizing a wide variety of genetic markers from Neospora’s nuclear and mitochondrial genomes, coupled with WGS of Neospora isolates from a wide host range, we have demonstrated a global expansion of a single lineage of N. caninum. This single lineage evolved recently from a common ancestor since the domestication of livestock and has expanded worldwide alongside the movement of cattle industries from Europe. Furthermore, the presence of sequence haploblocks introgressed from a distinct ancestry, the existence of several organellar matrilineages, and significant structural variation among largely identical, but independent sister lines supports a model whereby unisexual inbreeding has significantly contributed to and fixed the population genetic structure of N. caninum. Unisexual inbreeding, or a cryptic sexual cycle, has emerged in eukaryotic pathogens, including fungi and parasites as an adaptive mechanism to preserve a well-adapted genomic architecture while simultaneously allowing for rapid expansion and diversification of existing clones (71). The advantage of such a cryptic sexual cycle over an asexual cycle is the ability to reshuffle the genome to purge deleterious mutations and alter gene dosage (71), which could be experimentally tested using dogs as the animal model for sexual recombination (72). Hence, our genome sequencing results do not appear to support an asexual expansion model via vertical transmission as the only mode of transmission of Neospora in nature. Rather, our data support a model whereby the fixation of a highly inbred genome in conjunction with the domestication, expansion, and movement of a limited number of cattle and dog breeds worldwide has led to a global selective sweep of a single lineage of N. caninum.
Data Availability.
All sequences related to this work are published (see footnote) and data files related to gene lists, and SNPs for Toxoplasma and Plasmodium are available for download as SI files.
Supplementary Material
Acknowledgments
A.K., A.W.F., J.S.S., K.S., A.J.O., M.Q., and M.E.G. are supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health. Spanish contribution was supported by the Spanish Ministry of Economy and Competitiveness (AGL2013-44694-R and AGL2016-75935-C2-1-R). We thank Patricia Sikorski and Beth Gregg at NIAID for careful reading of the manuscript and helpful comments.
Footnotes
The authors declare no competing interest.
Data deposition: Sequence reads from the Illumina sequences were deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov, under BioProject number PRJNA534109 (accession nos. SRX5723126 [NC1], SRX5723132 [NC2], SRX5723128 [NC3], SRX5723130 [NC9], SRX5723131 [NcSP1], SRX5723129 [NcSP7], and SRX5723127 [NhOregon]).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913531116/-/DCSupplemental.
References
- 1.Heitman J., Sexual reproduction and the evolution of microbial pathogens. Curr. Biol. 16, R711–R725 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Billmyre R. B., et al. , Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. MBio 5, e01494-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenwood B., Mutabingwa T., Malaria in 2002. Nature 415, 670–672 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Breman J. G., The ears of the hippopotamus: Manifestations, determinants, and estimates of the malaria burden. Am. J. Trop. Med. Hyg. 64(1–2 suppl.), 1–11 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Hayton K., Su X. Z., Drug resistance and genetic mapping in Plasmodium falciparum. Curr. Genet. 54, 223–239 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Dondorp A. M., et al. , Artemisinin resistance: Current status and scenarios for containment. Nat. Rev. Microbiol. 8, 272–280 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Su C., et al. , Recent expansion of Toxoplasma through enhanced oral transmission. Science 299, 414–416 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Grigg M. E., Sundar N., Sexual recombination punctuated by outbreaks and clonal expansions predicts Toxoplasma gondii population genetics. Int. J. Parasitol. 39, 925–933 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sibley L. D., Khan A., Ajioka J. W., Rosenthal B. M., Genetic diversity of Toxoplasma gondii in animals and humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2749–2761 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubey J. P., Schares G., Ortega-Mora L. M., Epidemiology and control of neosporosis and Neospora caninum. Clin. Microbiol. Rev. 20, 323–367 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjerkås I., Mohn S. F., Presthus J., Unidentified cyst-forming sporozoon causing encephalomyelitis and myositis in dogs. Z. Parasitenkd. 70, 271–274 (1984). [DOI] [PubMed] [Google Scholar]
- 12.Haddad J. P., Dohoo I. R., VanLeewen J. A., A review of Neospora caninum in dairy and beef cattle—A Canadian perspective. Can. Vet. J. 46, 230–243 (2005). [PMC free article] [PubMed] [Google Scholar]
- 13.Dubey J. P., Neosporosis in cattle: Biology and economic impact. J. Am. Vet. Med. Assoc. 214, 1160–1163 (1999). [PubMed] [Google Scholar]
- 14.Williams D. J., Hartley C. S., Björkman C., Trees A. J., Endogenous and exogenous transplacental transmission of Neospora caninum—How the route of transmission impacts on epidemiology and control of disease. Parasitology 136, 1895–1900 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Dubey J. P., Hemphill A., Calero-Bernal R., Schares G., Neosporosis in Animals (CRC Press, 2017). [Google Scholar]
- 16.Lorenzi H., et al. , Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat. Commun. 7, 10147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubey J. P., Hattel A. L., Lindsay D. S., Topper M. J., Neonatal Neospora caninum infection in dogs: Isolation of the causative agent and experimental transmission. J. Am. Vet. Med. Assoc. 193, 1259–1263 (1988). [PubMed] [Google Scholar]
- 18.Calarco L., Barratt J., Ellis J., Genome wide identification of mutational hotspots in the apicomplexan parasite Neospora caninum and the implications for virulence. Genome Biol. Evol. 10, 2417–2431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh A. E., et al. , Sequence analysis and comparison of ribosomal DNA from bovine Neospora to similar coccidial parasites. J. Parasitol. 81, 530–535 (1995). [PubMed] [Google Scholar]
- 20.Stenlund S., Björkman C., Holmdahl O. J., Kindahl H., Uggla A., Characterization of a Swedish bovine isolate of Neospora caninum. Parasitol. Res. 83, 214–219 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Marsh A. E., et al. , Differentiation of Neospora hughesi from Neospora caninum based on their immunodominant surface antigen, SAG1 and SRS2. Int. J. Parasitol. 29, 1575–1582 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Fernández-García A., Risco-Castillo V., Zaballos A., Alvarez-García G., Ortega-Mora L. M., Identification and molecular cloning of the Neospora caninum SAG4 gene specifically expressed at bradyzoite stage. Mol. Biochem. Parasitol. 146, 89–97 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Walsh C. P., et al. , Molecular comparison of the dense granule proteins GRA6 and GRA7 of Neospora hughesi and Neospora caninum. Int. J. Parasitol. 31, 253–258 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Regidor-Cerrillo J., Pedraza-Díaz S., Gómez-Bautista M., Ortega-Mora L. M., Multilocus microsatellite analysis reveals extensive genetic diversity in Neospora caninum. J. Parasitol. 92, 517–524 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Basso W., et al. , Molecular comparison of Neospora caninum oocyst isolates from naturally infected dogs with cell culture-derived tachyzoites of the same isolates using nested polymerase chain reaction to amplify microsatellite markers. Vet. Parasitol. 160, 43–50 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Marsh A. E., Barr B. C., Packham A. E., Conrad P. A., Description of a new Neospora species (Protozoa: Apicomplexa: Sarcocystidae). J. Parasitol. 84, 983–991 (1998). [PubMed] [Google Scholar]
- 27.Adams R. I., Brown K. M., Hamilton M. B., The impact of microsatellite electromorph size homoplasy on multilocus population structure estimates in a tropical tree (Corythophora alta) and an anadromous fish (Morone saxatilis). Mol. Ecol. 13, 2579–2588 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Maradei E., et al. , Characterization of foot-and-mouth disease virus from outbreaks in Ecuador during 2009-2010 and cross-protection studies with the vaccine strain in use in the region. Vaccine 29, 8230–8240 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Falush D., Stephens M., Pritchard J. K., Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldberg D. E., Siliciano R. F., Jacobs W. R. Jr, Outwitting evolution: Fighting drug-resistant TB, malaria, and HIV. Cell 148, 1271–1283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regidor-Cerrillo J., et al. , Genetic diversity and geographic population structure of bovine Neospora caninum determined by microsatellite genotyping analysis. PLoS One 8, e72678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Qassab S., Reichel M. P., Ellis J., A second generation multiplex PCR for typing strains of Neospora caninum using six DNA targets. Mol. Cell. Probes 24, 20–26 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Pedraza-Díaz S., et al. , Microsatellite markers for the molecular characterization of Neospora caninum: Application to clinical samples. Vet. Parasitol. 166, 38–46 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Schlötterer C., Evolutionary dynamics of microsatellite DNA. Chromosoma 109, 365–371 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Ellegren H., Microsatellite mutations in the germline: Implications for evolutionary inference. Trends Genet. 16, 551–558 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Hutchison C. A. 3rd, Newbold J. E., Potter S. S., Edgell M. H., Maternal inheritance of mammalian mitochondrial DNA. Nature 251, 536–538 (1974). [DOI] [PubMed] [Google Scholar]
- 37.Huson D. H., Bryant D., Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Robinson D. R., Foulds L. R., Comparison of phylogenetic trees. Math. Biosci. 53, 131–147 (1981). [Google Scholar]
- 39.Romano A., et al. , Cross-species genetic exchange between visceral and cutaneous strains of Leishmania in the sand fly vector. Proc. Natl. Acad. Sci. U.S.A. 111, 16808–16813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inbar E., et al. , Whole genome sequencing of experimental hybrids supports meiosis-like sexual recombination in Leishmania. PLoS Genet. 15, e1008042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wendte J. M., et al. , Self-mating in the definitive host potentiates clonal outbreaks of the apicomplexan parasites Sarcocystis neurona and Toxoplasma gondii. PLoS Genet. 6, e1001261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heitman J., Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8, 86–99 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni M., et al. , Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol. 11, e1001653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akopyants N. S., et al. , Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324, 265–268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inbar E., et al. , The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet. 9, e1003672 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Escalante A. A., Ayala F. J., Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc. Natl. Acad. Sci. U.S.A. 92, 5793–5797 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reid A. J., et al. , Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: Coccidia differing in host range and transmission strategy. PLoS Pathog. 8, e1002567 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan A., et al. , Recent transcontinental sweep of Toxoplasma gondii driven by a single monomorphic chromosome. Proc. Natl. Acad. Sci. U.S.A. 104, 14872–14877 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehmann T., Marcet P. L., Graham D. H., Dahl E. R., Dubey J. P., Globalization and the population structure of Toxoplasma gondii. Proc. Natl. Acad. Sci. U.S.A. 103, 11423–11428 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su C., et al. , Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc. Natl. Acad. Sci. U.S.A. 109, 5844–5849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan A., et al. , A monomorphic haplotype of chromosome Ia is associated with widespread success in clonal and nonclonal populations of Toxoplasma gondii. MBio 2, e00228-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang N., et al. , Genetic basis for phenotypic differences between different Toxoplasma gondii type I strains. BMC Genomics 14, 467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khan A., et al. , Common inheritance of chromosome Ia associated with clonal expansion of Toxoplasma gondii. Genome Res. 16, 1119–1125 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su X. Z., Mu J., Joy D. A., The “Malaria’s Eve” hypothesis and the debate concerning the origin of the human malaria parasite Plasmodium falciparum. Microbes Infect. 5, 891–896 (2003). [DOI] [PubMed] [Google Scholar]
- 55.White N. J., et al. , Malaria. Lancet 383, 723–735 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Miotto O., et al. , Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 45, 648–655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miotto O., et al. , Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 47, 226–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mu J., et al. , Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat. Genet. 42, 268–271 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Groeneveld L. F., et al. ; GLOBALDIV Consortium , Genetic diversity in farm animals—A review. Anim. Genet. 41 (suppl. 1), 6–31 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Osawa T., Wastling J., Maley S., Buxton D., Innes E. A., A multiple antigen ELISA to detect Neospora-specific antibodies in bovine sera, bovine foetal fluids, ovine and caprine sera. Vet. Parasitol. 79, 19–34 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Oppong E. N. W., Veterinary Medicine in the Service of Mankind (Ghana Academy of Arts and Sciences, 1999). [Google Scholar]
- 62.Beal W. P. B., (1973) “Gold Coast (Ghana)” in A History of the Overseas Veterinary Services, Ware F., West G. P., Eds. (British Veterinary Association; ), pp. 107–111. [Google Scholar]
- 63.Botigué L. R., et al. , Ancient European dog genomes reveal continuity since the Early Neolithic. Nat. Commun. 8, 16082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freedman A. H., Wayne R. K., Deciphering the origin of dogs: From fossils to genomes. Annu. Rev. Anim. Biosci. 5, 281–307 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Elsik C. G., et al. ; Bovine Genome Sequencing and Analysis Consortium , The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science 324, 522–528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibbs R. A., et al. ; Bovine HapMap Consortium , Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324, 528–532 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bollongino R., et al. , Modern taurine cattle descended from small number of near-eastern founders. Mol. Biol. Evol. 29, 2101–2104 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Loftus R. T., MacHugh D. E., Bradley D. G., Sharp P. M., Cunningham P., Evidence for two independent domestications of cattle. Proc. Natl. Acad. Sci. U.S.A. 91, 2757–2761 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McTavish E. J., Decker J. E., Schnabel R. D., Taylor J. F., Hillis D. M., New World cattle show ancestry from multiple independent domestication events. Proc. Natl. Acad. Sci. U.S.A. 110, E1398–E1406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Upadhyay M. R., et al. ; European Cattle Genetic Diversity Consortium ; RPMA Crooijmans , Genetic origin, admixture and population history of aurochs (Bos primigenius) and primitive European cattle. Heredity 119, 469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feretzaki M., Heitman J., Unisexual reproduction drives evolution of eukaryotic microbial pathogens. PLoS Pathog. 9, e1003674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lindsay D. S., Dubey J. P., Duncan R. B., Confirmation that the dog is a definitive host for Neospora caninum. Vet. Parasitol. 82, 327–333 (1999). [DOI] [PubMed] [Google Scholar]
- 73.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences related to this work are published (see footnote) and data files related to gene lists, and SNPs for Toxoplasma and Plasmodium are available for download as SI files.