Significance
Ribosomes—the universal biosynthesis machinery central to all cellular life—are composed of 4 ribosomal RNA (rRNA) and up to 80 ribosomal proteins (RPs), yet detecting fluctuations in their RP composition with possible functional consequences remains a challenge. Using quantitative proteomics we show an invariant RP composition of ribosomes across 3 murine brain and liver tissues. The RP pool does not change with age and remains constant in tissues of juvenile, adult, and middle-aged mice groups. In a broader context, the high robustness in the overall protein architecture of the ribosomes across tissues and ages implies that ribosomal heterogeneity, if any, may result from other processes, including postsynthesis modifications of rRNA and RPs.
Keywords: ribosome, translation, mass spectrometry, neuronal tissues, aging
Abstract
Across phyla, the ribosomes—the central molecular machines for translation of genetic information—exhibit an overall preserved architecture and a conserved functional core. The natural heterogeneity of the ribosome periodically phases a debate on their functional specialization and the tissue-specific variations of the ribosomal protein (RP) pool. Using sensitive differential proteomics, we performed a thorough quantitative inventory of the protein composition of ribosomes from 3 different mouse brain tissues, i.e., hippocampus, cortex, and cerebellum, across various ages, i.e., juvenile, adult, and middle-aged mouse groups. In all 3 brain tissues, in both monosomal and polysomal ribosome fractions, we detected an invariant set of 72 of 79 core RPs, RACK1 and 2 of the 8 RP paralogs, the stoichiometry of which remained constant across different ages. The amount of a few RPs punctually varied in either one tissue or one age group, but these fluctuations were within the tight bounds of the measurement noise. Further comparison with the ribosomes from a high-metabolic-rate organ, e.g., the liver, revealed protein composition identical to that of the ribosomes from the 3 brain tissues. Together, our data show an invariant protein composition of ribosomes from 4 tissues across different ages of mice and support the idea that functional heterogeneity may arise from factors other than simply ribosomal protein stoichiometry.
The molecular composition of ribosomes—a large complex composed of ribosomal RNA (rRNA) and ribosomal proteins (RPs)—varies from bacteria to eukaryotes and among organelles; however, the overall architecture is preserved and the functional unit of decoding and peptide bond formation are both conserved (1). Along the phylogenetic tree, the ribosomes have evolved by idiosyncratic expansion of the RPs and rRNAs without perturbing the conserved functional core (2).
Mammalian cells typically contain ∼106 cytoplasmic ribosomes, and the question arises as to whether various types of cells maintain homogenous pools of ribosomes with the exact same stoichiometry of the individual RPs. Expression of functionally related genes consisting of functional modules, e.g., RPs constituting the ribosome, is conserved at great evolutionary distances by maintaining coregulation while the regulatory mechanisms controlling single genes can diverge (3). Across human tissues, the messenger RNA (mRNA) levels of RPs span several orders of magnitude (4), and the RP mRNA levels poorly correlate with the protein levels (5), implying additional regulation at the level of translation of the RP transcripts. In addition to their canonical role in translation, many RPs exhibit extraribosomal function (reviewed in ref. 6), which also suggests disproportional quantities of RPs. The half-life of single RPs substantially differs for each individual RP (7), but also shows variation among tissues (8). On average, the protein lifetime is 3 times shorter in liver than in brain (8). Various tissues also exhibit different transcript dynamics over age. In brain tissues of Caenorhabditis elegans or the short-lived fish Nothobranchius fuzeri, the RP mRNA levels increase with age, while in other tissues they decrease or remain constant (9–11). In addition, individual RPs are exchanged at different rates with the cytoplasmic pool of their free counterparts (12, 13). Collectively, these variations could potentially give a rise to ribosomes with different RP pools across tissues. Do ribosomes differ in their protein composition across tissues, and does this composition change over the lifetime of the cell?
In yeast, RP genes are generally duplicated, and incorporation of RP paralogs results in compositional heterogeneity of the ribosomes (14). Genomes of humans and mammals encode multiple copies of the 4 rRNAs (15, 16), and unlike yeast, most of the 79 RPs are typically encoded by a single gene. Of the 8 total human paralogs (4, 17), few exhibit tissue-specific patterns of transcription; e.g., uL3-like is expressed in striated muscle (18) and uL16-like and eL39-like in testis (19), supporting the hypothesis of tissue-specific variations in ribosomal composition. However, the depletion or even complete deletion of some paralogs does not cause phenotypic effects, challenging the role of the paralogs as a unique component of the ribosomes in those tissues (20, 21). Compositional heterogeneity of ribosomes could have message-specific effects; i.e., subpools of ribosomes might be specialized in translation of specific mRNAs, a hypothesis referred to as “specialized ribosomes.” Ribosomes deficient in a single RP, such as eL38 or uS2 (22, 23), posttranslational modifications (24, 25), or detachment of ribosome-associated factors (26) provide examples for tissue- and message-specific expression, prompting the debate on whether a specialized ribosome pool may shape tissue development (27). This argument is challenged by recent observations in C. elegans that tissue diversification is efficiently maintained during embryonic development by maternal ribosomes and synthesis of a new distinct pool of ribosomes is not required (28). The argument for heterogeneous ribosomal pools conferring message-specific expression is used to explain the tissue specificity of haploinsufficiencies associated with germline mutations disrupting one copy of RP genes in a type of ribosomopathies (14, 27). Opposing views suggest instead that tissue-specific dosage is shaped by affecting auxiliary factors or ribosome concentration relative to mRNA levels, which can alter translation of poorly initiated messages (29–31). Using sensitive differential mass spectrometric proteomics, we set out to systematically quantify the RP constituents of ribosomes in mice across various tissues and ages and address the following questions: 1) Do ribosomes from murine brain tissues exhibit compositional differences in their RPs compared to ribosomes from a high-metabolic-rate organ (e.g., liver)? 2) Does RP stoichiometry change with age?
Results
Quantitative Proteomics Shows Stable Stoichiometry of RPs Across Various Brain Tissues.
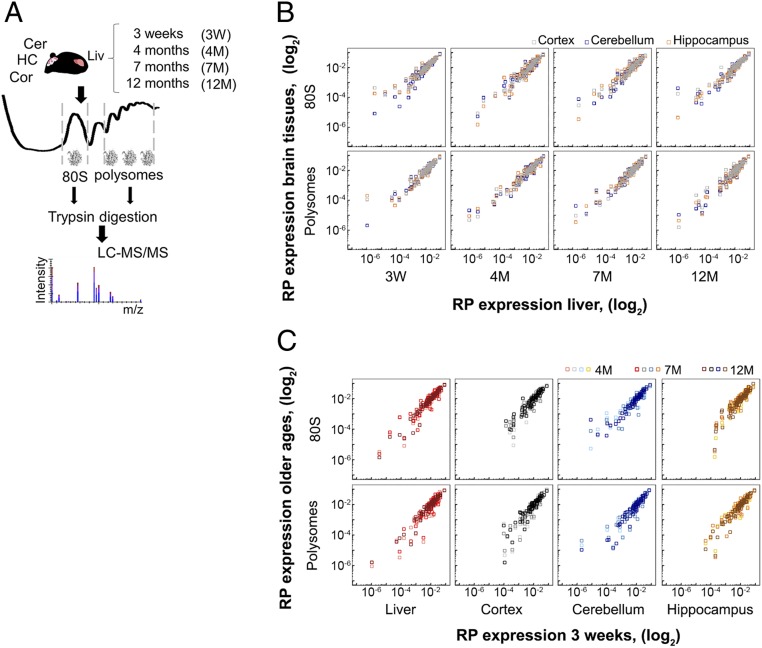
To determine the RP stoichiometry in brain tissues, we isolated translating ribosomes from 3 different brain tissues—cerebellum, hippocampus, and cortex—using velocity sedimentation in sucrose gradients and quantified the RPs by differential proteomics using a liquid chromatography system coupled to a tandem mass spectrometer (LC-MS/MS) (Fig. 1A). We compared them to the RPs constituting the ribosomes of a high-metabolic-rate organ, e.g., the liver (Fig. 1A). We used the outbred Swiss RjOrl (CD-1) mouse leveraging the higher genetic diversity of an outbred or wild-derived mouse that captures population variations (32, 33). Measurements of RP composition of mouse embryonic stem cells and yeast suggest differential stoichiometry between monoribosomes (80S) and ribosomes engaged in polysomes (34); hence we separated the 80S monoribosomes from the polysomes (SI Appendix, Fig. S1) and separately quantified their RPs by tandem mass spectrometry (Fig. 1A). In total, 1,163 proteins met the requirements for quantitative comparison between samples at a false discovery rate (FDR) of < 0.01. The numbers of ribosomes vary by an order of magnitude across tissues (17). Thus, it is important to note that our analysis does not provide any absolute quantitative measures on the ribosome number or protein copy number in tissues. Normalization of the level of each RP to the total amount of detected RPs provides a means for assessing the relative amount or fraction of each RP (34) and thus enables exploring quantitative variations among RPs in different tissues (for more details see Materials and Methods).
Fig. 1.
RP expression over tissues and ages. (A) Overview of the approach. Cer, cerebellum; HC, hippocampus; Cor, cortex; Liv, liver. (B and C) Expression levels of RPs in polysomes and 80S of brain tissues compared to liver for each age group (B) and within each tissue across ages compared to mice at 3 wk of age (C). Pearson correlation coefficients are summarized in SI Appendix, Table S3. W, week; M, month.
The mass spectrometric proteome analysis achieved high coverage of the RPs: 72 of 79 core RPs, RACK1, and 2 of the 8 RP paralogs (eL22-like and eS27-like) were faithfully quantified in all samples (SI Appendix, Table S1) with very good sequence coverage for the majority of the RPs (SI Appendix, Table S2). Missing RPs (uS19, eS27, eL29, eL34, eL37, eL40) either were localized on the ribosomal surface (hence they might have been lost during isolation) or following trypsinolysis they delivered peptides that were too short (eL41) and thus escaped detection. Strikingly, we detected the identical set of the same 72 core RPs, RACK1, and 2 paralogs (in total 75 proteins) in the polysomal fraction across all tested tissues; e.g., 3 brain tissues and liver, within each age group, and the RP expression correlated well (Fig. 1B). Furthermore, the ribosomes of the 80S and polysomal fractions were remarkably similar, and across all tissues we identified the same set of 75 proteins in both the 80S fractions and in the polysomal fractions (SI Appendix, Fig. S2). The RP levels spanned an order of magnitude with overall excellent reproducibility between independent biological replicates (R2 ≥ 0.84; SI Appendix, Figs. S3 and S4 and Table S4), despite the higher intrinsic heterogeneity of an outbred mouse strain. Low-abundance RPs (the lower one-third of abundance levels) contributed more to the scattering (SI Appendix, Table S4).
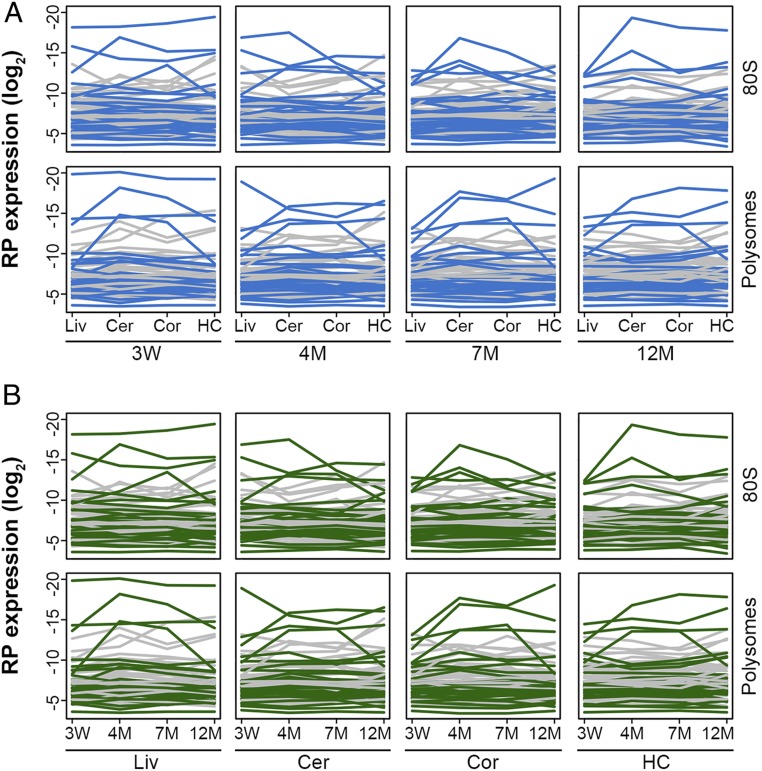
Although globally the RP levels among tissues correlated very well (Fig. 1B), single proteins may oppose this trend. Thus, we plotted the amount of each single RP across tissues (Fig. 2A). Some RPs saw much higher fluctuations across tissues, and, using Student’s t test, we selected 11 RPs in the 80S and 14 in the polysomal ribosomes with marked changes in at least one tissue. However, all these changes in RP levels scored “insignificant” with an FDR < 0.1 (SI Appendix, Fig. S5). Together, our data suggest that across the 4 different tissues that we analyzed, the RP set was identical in both the polysomal and monoribosomal fractions. The RP amounts were remarkably similar among tissues, and the fluctuations remained within tight bounds of the measurement noise.
Fig. 2.
Changes in expression of single RPs across tissue and age are not significant. (A and B) Comparison of the RP levels in tissues of the same age (A) or across ages within the same tissues (B). Gray, proteins of 40S; green or blue, proteins of 60S. Despite fluctuating expression, none of the proteins show significant changes in their amounts (FDR < 0.1) in the respective sample or population. Cer, cerebellum; HC, hippocampus; Cor, cortex; Liv, liver; W, week; M, month.
The RP Stoichiometry Remains Unchanged with Aging.
Earlier reports suggest that aging is accompanied by progressive decline in concentration of polysomes in skeletal muscles (35) and that translation activity of ribosomes from liver (36) and brain (37) decreases in an age-dependent manner. Also, genetic experiments support the view that loss of particular RPs affects the rate of protein synthesis (reviewed in ref. 38). To assess changes in the RP amount with aging, we compared 4 age groups with different age characteristics, i.e., juvenile mice of 3 wk which are still in development, young adult mice of 4 mo, and 2 groups of middle-aged adults of 7 and 12 mo, respectively (Fig. 1A). Each of these age groups in mice corresponds to different ages in humans, e.g., < 10, ∼23, 33, and 43 y, respectively (39). In our analysis we used female mice; under same calorie restrictions, the female murine brains undergo age-related changes much earlier (6 to 9 mo of age) than male brains (9 to 12 mo of age) (40). Because of the marked drop-off in survivorship and enhanced disease susceptibility of the wild-derived mice, we used the oldest mice (12 mo old).
Across different age groups we detected the same 72 of 79 core RPs, 2 paralogs, and RACK1 in both 80S and polysomal fractions (Fig. 1C and SI Appendix, Table S1), which we detected across tissues (Fig. 1B), implying that the core RP composition of the ribosomes does not change with aging. Globally, the RP levels in 80S and polysomal fractions of the tissues from the adult age groups correlated well with those of the 3-wk-old mice (Fig. 1C). After plotting the level of each single protein at different ages (Fig. 2B), in some age groups we observed notable fluctuations for some proteins. Using Student’s t test, we selected 2 RPs in the 80S and 3 in the polysomal ribosomes, none of which, however, scored significant even at an FDR < 0.1 (SI Appendix, Fig. S6), implying that the fluctuations were within the measurement noise. Importantly, we realized that the amount of the RPs that we selected with notable changes only punctually fluctuated in a single age group, but did not follow a consistent pattern of decline or increase with age (SI Appendix, Fig. S6). We also tested the differential RP expression in the 80S and polysomal fraction in liver with an independent method, immunoblot analysis (SI Appendix, Fig. S7). Consistent with the LC-MS-MS data, RACK1, uS3, and eL22 showed stable amounts across ages (SI Appendix, Fig. S7).
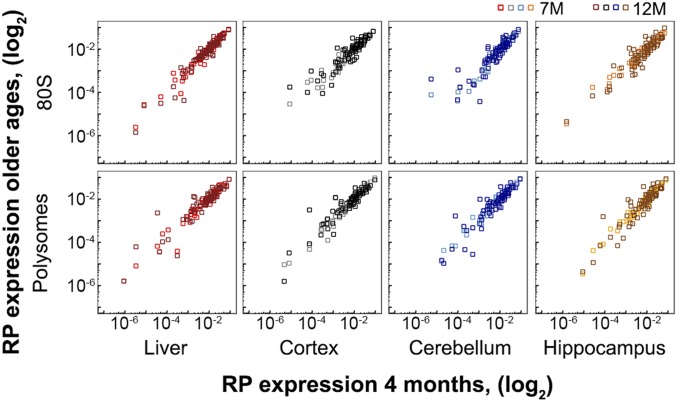
In our analysis, we compared the RP levels in adult mice samples to that of juvenile mice of 3 wk of age (Fig. 1C). Although the brain development in mice is accomplished at 29.7 d after conception (or 12 d after birth), the 3-wk-old mice are within the juvenile age group and still undergoing developmental and growth changes (41). Hence, we next compared the RP pattern in 7- and 12-mo-old mice to that in 4-mo-old mice which belongs to the mature adult group past development, yet unaffected by senescence. Gross comparison of the quantitative levels of RPs in the middle-aged groups to that of young, 4-mo-old mice also showed invariant RP stoichiometry with age (Fig. 3). Again, t test-based selection of single RPs from the 80S or polysomal fraction with marked fluctuations in abundance at least at one age (9 and 22 proteins, respectively) did not confirm those changes to be significant at an FDR < 0.1.
Fig. 3.
RP stoichiometry does not significantly change in adult ages. Expression levels of RPs in polysomes and 80S for each tissue compared to 4 mo of age. M, month. Pearson correlation coefficients are summarized in SI Appendix, Table S5.
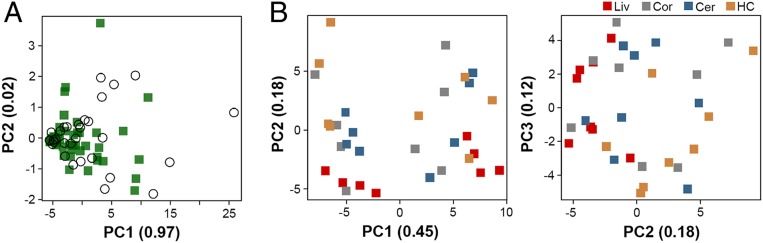
To reduce the dimensions of the dataset and to extract directions of the highest variance, we applied principal component analysis (PCA). PCA of the RP fractions in the coordinates of tissues and ages revealed no apparent clusters; the uS3 protein—the protein with the highest detection levels (SI Appendix, Fig. S3)—exhibited the largest difference from the rest of the RPs (Fig. 4A). PCA of the tissues and age groups in the coordinates of RPs showed some separation of 2 groups along principal component 1 (PC1), with no distinct clustering of a single tissue or age group (Fig. 4B). The liver and brain tissue samples were unevenly spread in the PC2 direction, but no clear tissue groups could be selected (Fig. 4B). PC3 direction reflected mostly the difference between the monoribosomal and polysomal fractions. Together, PCA detected no clear clustering of the RPs, tissues, or ages.
Fig. 4.
RP expression values in tissues or ages do not separate in the principal component space. (A) PCA of RP sets of 80S and polysomes from cortex, cerebellum, hippocampus, and liver (n = 2 biological replicates) across all age groups. Green squares, RPs from 60S; black open circles, RPs from 40S. (B) The studied tissues and age groups were heterogeneously spread within PC1, PC2, and PC3 directions. Transposed matrix of PC1 of tissues and age groups in the coordinates of RPs. Cer, cerebellum; HC, hippocampus; Cor, cortex; Liv, liver.
In sum, our quantitative proteomic analysis shows that in mice the protein composition of the ribosomes is equal across 3 brain tissues and liver and that RP stoichiometry remains stable with aging in both monosomal and polysomal fractions. The quantity of some proteins may punctually vary, but those fluctuations remain within tight bounds of the measurement noise.
Discussion
The levels of mRNAs encoding RPs reveal high variation across tissues (11). Together with the identification of tissue-specific RP paralogs (19), tissues with developmental specialization of ribosomes became an appealing place to look for tissue-specific heterogeneity in the ribosomal composition (42). Here, we report a systematic quantification by differential mass spectrometric proteomics of RPs in 3 murine brain tissues and liver across 4 age groups in mice—juvenile (3 wk), young (4 mo), and middle-aged (7 and 12 mo). Across tissues we detect the same RP sets with constant stoichiometry within the monoribosomes and polysomes which remains stable with aging. Our results align well with observations in other systems. Quantitative measurements of RPs show constant composition of ribosomes across other rodent tissues (19) during human hematopoietic lineage commitment (29) and among various cancer cell lines (43).
The role of RP paralogs in higher eukaryotes might be far more subtle than originally proposed (20, 21), and the compensatory drift model may explain the preserved expression of functionally redundant paralogs (44). In this model, a dosage-dependent balance constrains the total expression of a protein from redundant functional duplicates (paralogs) (44). In support of this is that we detected ubiquitous expression of 2 RP paralogs (eL22-like and eS27-like) along the major gene product (eL22 and eS27) in all tested tissues. An earlier study, which analyzed RP composition in rodent liver, mammary gland, and testis, also identified a ubiquitously expressed paralog, eL22-like, in all 3 tissues (19). Even paralogs with a distinct tissue-specific expression pattern, e.g., uL3-like in striated muscle (18) and uL16-like and eL39-like in testis (19), are always accompanied by their major RP counterpart (18, 21), and changes in one amount of paralog compensate for the other (20, 21). The sequence similarity of the paralogs led to a suggestion of a similar location in the ribosome (18). uL3-like is an example for a physical association with ribosomes; specialized functions of the uL3-like–containing ribosomes have not yet been reported. The role of the paralogs as a unique component of the ribosome is challenged by the lack of evidence for a distinct function in translation. The examples support functional redundance among the paralogs (18, 21) and align with a dosage-dependent balance model of their total expression (44) rather than supporting a specialized function in translation.
Our steady-state measurements are not predictive of synthesis and degradation rates, which are regulated in a tissue- and cell-type–dependent manner; proteins with similar abundances exhibit a wide range of turnover rates (8). Difference in abundance of RP mRNAs across tissues (11) cannot be extrapolated to their protein levels, since posttranscriptional processing actively buffers protein concentration (5). Protein dynamics is shaped by mRNA and protein half-lives, both of which differ among individual transcripts and proteins but also exhibit a tissue-specific pattern of stability with somewhat higher stability in neuronal tissues (8–11). A recent study in a genetically well-tractable system, C. elegans (28), and earlier observations in Xenopus embryos (45) suggests that tissue diversification may not rely on ribosomes with a heterogeneous RP pool. Along that line, another study shows that ribosome levels determine the human hematopoietic lineage commitment, while the RP sets remain unchanged (29). Together, these examples, combined with our observations on the remarkable invariability of the RP pool in translating ribosomes, suggest that ribosomes with uniform RP pools may be a common feature across tissues and species. Thus, ribosome heterogeneity that can potentially reflect functional differences or “specialization” may rather stem from other processes than from changes in RP stoichiometry. Among these processes could be rRNA variations and modifications (16, 46, 47), alteration of ribosome-associated auxiliary (26, 48) or signaling factors (49), or posttranslational modifications of ribosomal proteins (24, 25, 50, 51).
Materials and Methods
Animals and Tissue Isolation.
Mouse colonies were maintained in accordance with the European Union’s (directive 2010/63) and German animal welfare guidelines and approved by the State and Institutional review board (GZ G21305/591–00.33, Behörde für Gesundheit und Verbraucherschutz, Hamburg, Germany). Wild-type female mice (SWISS RjOrl [Crl:CD1 (ICR)]) of 3 or 18 wk of age were purchased from Janvier Labs. The cohort of the 18-wk-old mice was housed in the in-house animal facility until they reached the desired ages. Animals that developed tumors and showed health aberrancies as they aged were excluded. Animals at different ages were collected in groups, so that tissues from litters of 3 wk and 12, 4, and 7 mo were collected together, respectively.
The animals were killed at the same time of the day to minimize the effect of daily fluctuation in ribosome number (52). Anesthesia was induced with isofluran, and termination took place by beheading the mice. Tissues of interest [cortex (Cor), cerebellum (Cer), hippocampus (HC), and liver] were harvested simultaneously. Blood was washed away using ice-cold PBS. All tissues were flash-frozen in liquid nitrogen and stored at −80 °C for further analysis. The tissues of 3 littermates of each age group were mixed together and represent one biological replicate.
Polysome Profiling and Isolation of Ribosome Fractions.
In order to consider animal heterogeneity, tissues from 3 littermates were pooled, treated as one biological replicate, and used to isolate the 80S and polysomal fractions. Tissues were homogenized on ice for 90 s in 400 µL precooled polysome lysis buffer [20 mM Hepes–KOH, pH 7.4, containing 5 mM MgCl2, 100 mM KCl, 2 mM dithiothreitol (DTT), 100 µg/mL cycloheximide (CHX), 1× complete protease inhibitor (Roth), 1% Nonidet P-40, 2% sodium deoxycholate] and centrifuged at 12,000 × g at 4 °C for 10 min to pellet the cell debris. Supernatants were transferred into fresh RNase-free tubes, and the RNA concentration was measured at 260 nm. Equal amounts of supernatants from each tissue were loaded on the sucrose gradient, consisting of 8, 16, 24, 32, 40, and 48% (wt/vol) layers dissolved in RNase-free sucrose in buffer (20 mM Hepes–KOH, pH 7.4, containing 5 mM MgCl2, 100 mM KCl, 100 µg/mL CHX, and 2 mM DTT in H2ODEPC) and centrifuged at 35,000 × g at 4 °C for 2.5 h (SW40Ti rotor, Beckman Coulter). Fractions of 80S, disomes, and polysomes were collected with piston gradient fractionator (Biocomp) and stored frozen at −80 °C. The disomes were separated and discarded from analysis to achieve a clear, baseline separation between 80S and polysomes.
RP Digestion and Preparation for LC-MS/MS.
Since the monosome and polysome fractions have different sucrose concentrations (e.g., ∼32 and 40%, respectively), all samples were diluted with keratin-free water to equalize the amount of sucrose between them to ∼20%. The 80S and polysome fractions were centrifuged at 54,000 × g at 4 °C for 4 h (SW55Ti; Beckman Coulter). Ribosome-containing pellets were resuspended in 8 M urea and loaded onto 15% sodium dodecyl sulfate/polyacrylamide gel electrophoresis and run to concentrate the samples to a single protein band at the top of the separating gel. Gels were stained in NEUHOFF brilliant blue [0.08% (wt/vol) CBB-G250, 1.2% (vol/vol) of 85% H3PO4, 10% (wt/vol) ammonium sulfate], and the single band, containing the protein mixture, was cut out. In-gel digestion was performed with trypsin (Promega) at 37 °C for 16 h. Digestion was stopped by adding 65% acetonitrile and 5% formic acid (vol/vol). The digested peptides were air-dried and stored at −80 °C for LC-MS/MS analysis.
LC-MS/MS in Data Analysis.
LC-MS/MS experiments were performed on the same day for all age groups of both biological replicates; hence some samples (e.g., from 4- and 7-mo-old mice) were stored longer than those from 3-wk- and 12-mo-old mice. Samples were resuspended in 0.1% formic acid (FA) and transferred into a full recovery autosampler vial (Waters). Chromatographic separation was performed on a nano-UPLC system (Dionex Ultimate 3000 UPLC system, Thermo Fisher Scientific) with a 2-buffer system (buffer A: 0.1% FA in water, buffer B: 0.1% FA in acetonitrile, both at pH 3). Attached to the UPLC was a reversed-phase column (Acclaim PepMap 100 C18; 100 µm × 2 cm, 100-Å pore size, 5-µm particle size) for desalting a purification followed by a reversed-phase column (Acclaim PepMap 100 C18; 75 µm × 50 cm, 100-Å pore size, 2-µm particle size). Peptides were separated using a 60-min gradient with increasing acetonitrile concentration from 2 to 30%. The eluted peptides were analyzed on a tribrid mass spectrometer (Fusion; Thermo Fisher Scientific) in data-dependent acquisition (DDA) and data-independent acquisition (DIA) mode.
In DDA mode, randomly chosen samples from each age group were used to build a reference spectral library for data extraction of samples acquired in DIA mode. For DDA, the 12 most intense ions per precursor scan (2 × 105 ions, 120,000 resolution, 120-ms fill time) were analyzed by MS/MS (higher energy collision induced dissociation [HCD] at 30 normalized collision energy, 1 × 105 ions, 15,000 resolution, 60-ms fill time) in a range of 400 to 1,300 m/z. A dynamic precursor exclusion of 20 s was used. For DIA, each sample was analyzed using a 32 sequential 25-Da fixed window method covering the mass range from 400 to 1,200 m/z. Two precursor scans (2 × 105 ions, 60,000 resolution, 50-ms fill time, m/z range from 390 to 1,210 m/z) and 32 MS/MS scans (HCD at 28 normalized collision energy, 1 × 105 ions, 30,000 resolution, 50-ms fill time) were performed per cycle. After the first precursor scan, 16 MS/MS scans were performed covering the precursor mass range from 400 to 800 m/z followed by the second precursor scan and another 16 MS/MS scans ranging from 800 to 1,200 m/z.
Acquired DDA LC-MS/MS data were searched against the mouse SwissProt protein database downloaded from Uniprot (released August 2017; 16,909 protein entries) using the Sequest algorithm integrated in the Proteome Discoverer software version 2.0. Mass tolerances for precursors was set to 10 ppm and 0.02 Da for fragments. Carbamidomethylation was set as a fixed modification for cysteine residues, while the oxidation of methionine, pyro-glutamate formation at N-terminal glutamine residues, N-terminal methionine loss, and the acetylation before and after methionine loss at the protein N terminus were allowed as variable modifications. Only peptides with a high confidence FDR of <1% using a decoy database approach were accepted as identified.
Proteome Discoverer search results were imported into Skyline software version 4.2 allowing only high confidence peptides with more than 4 fragment ions. A maximum of 5 fragment ions per peptide were used for information extraction from DIA files for peptides with a dot product of >0.85. Peptide peak areas were summed to generate protein areas, which were then used for relative abundance comparison. Protein areas were normalized to the median of all detected proteins within each sample.
For comparisons of the relative quantities of RPs only, the RPs amounts are represented as a fraction of the total RPs in each dataset. For comparisons, the same peptides for each RP were used, despite detecting more peptides for some RPs in some samples (SI Appendix, Table S1). The normalization was performed using in-house written scripts in Python.
Immunoblot Analysis.
The immunoblot analysis was performed with liver samples using the capillary electrophoresis system (Jess, ProteinSimple). Frozen tissues (n = 4 to 6 biological replicates) were homogenized separately and fractionated in monosomal and polysomal fractions as prepared for the mass spectrometry analysis (see above) and loaded on the Jess separation module (2 to 40 kDa and 12 to 230 kDa). Antibodies against RACK1, uS3, and eL22 were purchased from Santa Cruz Biotechnology. The corresponding RP peaks were normalized to the total protein concentration using a protein normalization kit (ProteinSimple) and quantified using the Jess quantification module.
PCA Analysis.
PCA was applied to the normalized abundances of the RPs to calculate the modes of the largest variance in the datasets. Columns were standardized, following calculation of a covariance matrix. Diagonalization of the covariance matrix provided a set of their eigenvectors (PCs), arranged in descending order of their corresponding eigenvalues, which reflected the magnitude of variance along the PCs. RP levels were projected onto the PC1 and PC2 to reveal the distribution of either RPs or tissues along the 2 orthogonal directions of the highest variance.
We applied the Robust PCA (RPCA) algorithm that consistently removes outliers. RPCA decomposes the initial data matrix into a low-rank matrix and a sparse matrix, in which the latter one contains outlying values. Thus, after decomposition, the sparse matrix is discarded while the low-rank matrix is used as an input for the conventional PCA. The slight separation of the tissues of 4- and 7-mo-old mice from those of 3-wk- and 12-mo-old mice is likely due to the different storage time required in order to perform the LC-MS/MS analysis on the same day (see above).
The Scikit-learn Python module was used for PCA, and the RPCA (53) was used to separate low-rank and sparse matrices.
Statistical Analysis and Data Deposition.
Two-tailed Student’s t test (α = 0.05), Wilcoxon rank-sum test, Pearson correlation, and FDR analysis were used for statistical analysis of the data. The statistical analyses were performed using in-house written scripts in R.
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (54) partner repository with the dataset identifier PXD014138.
Supplementary Material
Acknowledgments
We thank Paul Saffert for the initial protocol for dissection and isolation of tissues and polysomes from brain tissues, Andreas Czech for his help with organizing the mice experiments, and Chrsitian Lohr and his coworkers for helping with maintanance of the mouse colony. This work was supported by Deutsche Forschungsgemeinschaft Grant FOR1805 (to Z.I.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository, https://www.ebi.ac.uk/pride/archive/ (dataset identifier PXD014138).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912060116/-/DCSupplemental.
References
- 1.Petrov A. S., et al. , Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 111, 10251–10256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts E., Sethi A., Montoya J., Woese C. R., Luthey-Schulten Z., Molecular signatures of ribosomal evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 13953–13958 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wohlbach D. J., Thompson D. A., Gasch A. P., Regev A., From elements to modules: Regulatory evolution in Ascomycota fungi. Curr. Opin. Genet. Dev. 19, 571–578 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guimaraes J. C., Zavolan M., Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 17, 236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franks A., Airoldi E., Slavov N., Post-transcriptional regulation across human tissues. PLoS Comput. Biol. 13, e1005535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhavsar R. B., Makley L. N., Tsonis P. A., The other lives of ribosomal proteins. Hum. Genomics 4, 327–344 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dörrbaum A. R., Kochen L., Langer J. D., Schuman E. M., Local and global influences on protein turnover in neurons and glia. eLife 7, e34202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price J. C., Guan S., Burlingame A., Prusiner S. B., Ghaemmaghami S., Analysis of proteome dynamics in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 107, 14508–14513 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adamla F., Ignatova Z., Somatic expression of unc-54 and vha-6 mRNAs declines but not pan-neuronal rgef-1 and unc-119 expression in aging Caenorhabditis elegans. Sci. Rep. 5, 10692 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumgart M., et al. , RNA-seq of the aging brain in the short-lived fish N. furzeri: Conserved pathways and novel genes associated with neurogenesis. Aging Cell 13, 965–974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahn J. M., et al. , AGEMAP: A gene expression database for aging in mice. PLoS Genet. 3, e201 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dice J. F., Schimke R. T., Turnover and exchange of ribosomal proteins from rat liver. J. Biol. Chem. 247, 98–111 (1972). [PubMed] [Google Scholar]
- 13.Pulk A., et al. , Ribosome reactivation by replacement of damaged proteins. Mol. Microbiol. 75, 801–814 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Komili S., Farny N. G., Roth F. P., Silver P. A., Functional specificity among ribosomal proteins regulates gene expression. Cell 131, 557–571 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malinovskaya E. M., et al. , Copy number of human ribosomal genes with aging: Unchanged mean, but narrowed range and decreased variance in elderly group. Front. Genet. 9, 306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parks M. M., et al. , Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 4, eaao0665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta V., Warner J. R., Ribosome-omics of the human ribosome. RNA 20, 1004–1013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaillou T., Zhang X., McCarthy J. J., Expression of muscle-specific ribosomal protein L3-like impairs myotube growth. J. Cell. Physiol. 231, 1894–1902 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugihara Y., et al. , Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J. Proteome Res. 9, 1351–1366 (2010). [DOI] [PubMed] [Google Scholar]
- 20.O’Leary M. N., et al. , The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 9, e1003708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong Q. W., et al. , RPL39L is an example of a recently evolved ribosomal protein paralog that shows highly specific tissue expression patterns and is upregulated in ESCs and HCC tumors. RNA Biol. 11, 33–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondrashov N., et al. , Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue S., et al. , RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werner A., et al. , Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525, 523–527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeidan Q., Wang Z., De Maio A., Hart G. W., O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol. Biol. Cell 21, 1922–1936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H. W., Kim H. D., Kim T. S., Kim J., Senescent cells differentially translate senescence-related mRNAs via ribosome heterogeneity. J. Gerontol. A Biol. Sci. Med. Sci. 74, 1015–1024 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Xue S., Barna M., Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cenik E. S., et al. , Maternal ribosomes are sufficient for tissue diversification during embryonic development in C. elegans. Dev. Cell 48, 811–826.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khajuria R. K., et al. , Ribosome levels selectively regulate translation and lineage commitment in human Hematopoiesis. Cell 173, 90–103.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lodish H. F., Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251, 385–388 (1974). [DOI] [PubMed] [Google Scholar]
- 31.Mills E. W., Green R., Ribosomopathies: There’s strength in numbers. Science 358, eaan2755 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Brekke T. D., Steele K. A., Mulley J. F., Inbred or outbred? Genetic diversity in laboratory rodent colonies. G3 (Bethesda) 8, 679–686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuttle A. H., Philip V. M., Chesler E. J., Mogil J. S., Comparing phenotypic variation between inbred and outbred mice. Nat. Methods 15, 994–996 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slavov N., Semrau S., Airoldi E., Budnik B., van Oudenaarden A., Differential stoichiometry among core ribosomal proteins. Cell Rep. 13, 865–873 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakano K., Sidransky H., Age-related changes in ribosomal profiles and in vitro protein synthesis in skeletal muscle during fasting and subsequent refeeding of rats. J. Nutr. 108, 399–409 (1978). [DOI] [PubMed] [Google Scholar]
- 36.Mori N., Mizuno D., Goto S., Conservation of ribosomal fidelity during ageing. Mech. Ageing Dev. 10, 379–398 (1979). [DOI] [PubMed] [Google Scholar]
- 37.Rattan S. I., Synthesis, modifications, and turnover of proteins during aging. Exp. Gerontol. 31, 33–47 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Gonskikh Y., Polacek N., Alterations of the translation apparatus during aging and stress response. Mech. Ageing Dev. 168, 30–36 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Flurkey K., Currer J. M., Harrison D., “Mouse models in aging research” in The Mouse in Biomedical Research, Fox G. E., Barthold S. W., Davisson M. T., Newcomer C. E., Quimby F. W., Smith A. L., Eds (Elsevier, New York, 2007), vol. 3, pp. 637–672. [Google Scholar]
- 40.Zhao L., Mao Z., Woody S. K., Brinton R. D., Sex differences in metabolic aging of the brain: Insights into female susceptibility to Alzheimer’s disease. Neurobiol. Aging 42, 69–79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finlay B. L., Darlington R. B., Linked regularities in the development and evolution of mammalian brains. Science 268, 1578–1584 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Emmott E., Jovanovic M., Slavov N., Ribosome stoichiometry: From form to function. Trends Biochem. Sci. 44, 95–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reschke M., et al. , Characterization and analysis of the composition and dynamics of the mammalian riboproteome. Cell Rep. 4, 1276–1287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson A., Zakon H. H., Kirkpatrick M., Compensatory drift and the evolutionary dynamics of dosage-sensitive duplicate genes. Genetics 202, 765–774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steele R. E., Thomas P. S., Reeder R. H., Anucleolate frog embryos contain ribosomal DNA sequences and a nucleolar antigen. Dev. Biol. 102, 409–416 (1984). [DOI] [PubMed] [Google Scholar]
- 46.Kurylo C. M., et al. , Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep. 25, 236–248.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song W., et al. , Divergent rRNAs as regulators of gene expression at the ribosome level. Nat. Microbiol. 4, 515–526 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Wang Y. J., et al. , Lso2 is a conserved ribosome-bound protein required for translational recovery in yeast. PLoS Biol. 16, e2005903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee C. H., et al. , A regulatory response to ribosomal protein mutations controls translation, growth, and cell competition. Dev. Cell 46, 807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imami K., et al. , Phosphorylation of the ribosomal protein RPL12/uL11 affects translation during mitosis. Mol. Cell 72, 84–98.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Mazumder B., et al. , Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115, 187–198 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Sinturel F., et al. , Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169, 651–663.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Candès E. J., Li X., Ma Y., Wright J., Robust principal component analysis? J. Assoc. Comp. Mach. 58, 11 (2011). [Google Scholar]
- 54.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.