Significance
Following infection and inflammation, activation of the transcription factor NF-κB and stimulation of mRNA translation initiation remodel cellular gene expression. In contrast, how translation elongation factor 2, which performs the translocation step in protein synthesis, responds is not understood. We show that eEF2 Thr56 phosphorylation, which slows elongation, and eEF2 kinase (eEF2K) abundance are reduced by multiple NF-κB–activating stimuli. Significantly, NF-κB activation repressed eEF2K transcription to stimulate eEF2 by preventing eEF2 phosphorylation. In addition, eEF2K abundance regulates protein synthesis upon eEF2 inactivation by a bacterial toxin. This demonstrates how eEF2 is integrated into NF-κB innate immune response networks and reveals an unexpected mechanism whereby regulated nuclear transcription impacts translation elongation.
Keywords: translation elongation, eEF2 and eEF2K, NF-κB activation, HCMV infection, double-strand DNA-sensing
Abstract
Gene expression is rapidly remodeled by infection and inflammation in part via transcription factor NF-κB activation and regulated protein synthesis. While protein synthesis is largely controlled by mRNA translation initiation, whether cellular translation elongation factors are responsive to inflammation and infection remains poorly understood. Here, we reveal a surprising mechanism whereby NF-κB restricts phosphorylation of the critical translation elongation factor eEF2, which catalyzes the protein synthesis translocation step. Upon exposure to NF-κB–activating stimuli, including TNFα, human cytomegalovirus infection, or double-stranded DNA, eEF2 phosphorylation on Thr56, which slows elongation to limit protein synthesis, and the overall abundance of eEF2 kinase (eEF2K) are reduced. Significantly, this reflected a p65 NF-κB subunit-dependent reduction in eEF2K pre-mRNA, indicating that NF-κB activation represses eEF2K transcription to decrease eEF2K protein levels. Finally, we demonstrate that reducing eEF2K abundance regulates protein synthesis in response to a bacterial toxin that inactivates eEF2. This establishes that NF-κB activation by diverse physiological effectors controls eEF2 activity via a transcriptional repression mechanism that reduces eEF2K polypeptide abundance to preclude eEF2 phosphorylation, thereby stimulating translation elongation and protein synthesis. Moreover, it illustrates how nuclear transcription regulation shapes translation elongation factor activity and exposes how eEF2 is integrated into innate immune response networks orchestrated by NF-κB.
Infection and inflammation profoundly impact ongoing protein synthesis and often trigger activation of the nuclear factor kappa light-chain enhancer of activated B cells (NF-κB) transcription factor family (1). Diverse effectors, including inflammatory cytokines like TNFα and nucleic acid pathogen-associated molecular patterns (PAMPs) indicative of infection, stimulate NF-κB, which normally is contained in an inhibitory complex within the cytoplasm. Upon subsequent translocation into the nucleus, activated NF-κB transcription factors orchestrate a bevy of cellular responses by regulating transcription of hundreds of genes (1). Indeed, NF-κB activation figures prominently in the development and pathogenesis of autoimmunity and inflammation (2–7), aging and cancer (1, 8–12), and the deployment of innate host defenses, including type I IFN production and induction of IFN-stimulated genes (ISGs) (1, 13–15). Significantly, many ISGs encode functions vital for cell-intrinsic innate defenses that curb intracellular pathogen reproduction by restricting infected cell protein synthesis, foiling efforts by infectious agents to co-opt control of the cellular protein synthesis machinery and subvert host defenses (16, 17). While NF-κB activation indirectly impacts protein synthesis via stimulating IFN production and the subsequent activities of induced ISG-encoded products, whether NF-κB acts through ancillary mechanisms to regulate protein synthesis remains largely unknown.
Posttranscriptional control of gene expression at the level of mRNA translation enables swift changes to the proteome in response to physiological and environmental challenges and plays a key role in infection and immune responses (18). Integrating cellular translation factors into a variety of cell signaling pathways, especially those responsive to the mechanistic target of rapamycin complex 1 (mTORC1), allows protein synthesis to be tuned to different fundamental parameters like energy, nutrient, oxygen, and growth factor sufficiency (19). Translation initiation is a highly regulated, rate-limiting step whereby a multisubunit m7GTP cap-binding complex assembles on the mRNA 5′-end to recruit the 40S ribosome subunit (20). This process is regulated in part by the 4E-BP family of translational repressor proteins, which are substrates of mTORC1 (21). Following AUG start codon recognition and 60S subunit joining, translation elongation commences and is dependent upon eukaryotic elongation factor 2 (eEF2), which mediates ribosome translocation along the mRNA and is regulated by eEF2 kinase (eEF2K) (22, 23). In response to physiological stress including inhibition of mTORC1, eEF2K is activated and phosphorylates eEF2 T56, thereby reducing the affinity of eEF2 for the ribosome and slowing translation elongation (24, 25). Viral and bacterial pathogens also target eEF2 to regulate protein synthesis. Human cytomegalovirus (HCMV) remains a significant cause of morbidity and mortality in transplant recipients and a leading cause of congenital infections (26–30). In response to HCMV-induced mTORC1 activation, eEF2 mRNA translation is stimulated to raise intracellular levels of the critical elongation factor eEF2 (31). In contrast, toxins produced by Corynebacterium diphtheriae and Pseudomonas aeruginosa ADP ribosylate eEF2 on diphthamide, a modified histidine residue unique to eEF2, to block elongation and host protein synthesis (32).
To investigate how protein synthesis responds to different NF-κB–activating stimuli, including TNFα exposure, HCMV infection, and dsDNA-sensing, we utilize a primary human fibroblast model. Tissue-resident fibroblasts are critical long-lived sentinel cells that play a fundamental role coordinating acute resolving vs. chronic inflammation, adaptive immunity, and tissue remodeling and repair. This is accomplished in part by conditioning tissue microenvironments through changes in gene expression, some of which are regulated by cytokines or PAMPs that stimulate NF-κB (33–35). Here, we show that TNFα treatment stimulates global protein synthesis in fibroblasts. Besides promoting initiation via inhibiting the 4E-BP1 translation repressor, we establish that TNFα unexpectedly regulates elongation by preventing phosphorylated eEF2 accumulation and reducing eEF2K abundance. Infection with HCMV also effectively reduced eEF2K protein levels and eEF2 Thr56 phosphorylation, as did exposure of uninfected primary fibroblasts to immunostimulatory dsDNA. Significantly, the reduction in eEF2K protein was accompanied by a corresponding decrease in eEF2K mRNA transcription that was dependent upon the NF-κB subunit p65. Furthermore, eEF2K abundance regulates protein synthesis upon exposure to a bacterial toxin that inactivates eEF2. Overall, this work reveals a surprising mechanism whereby transcriptional repression by NF-κB might modulate translation elongation in response to pathogens or inflammatory cytokines.
Results
TNFα Stimulates Fibroblast Protein Synthesis and Translation Factors.
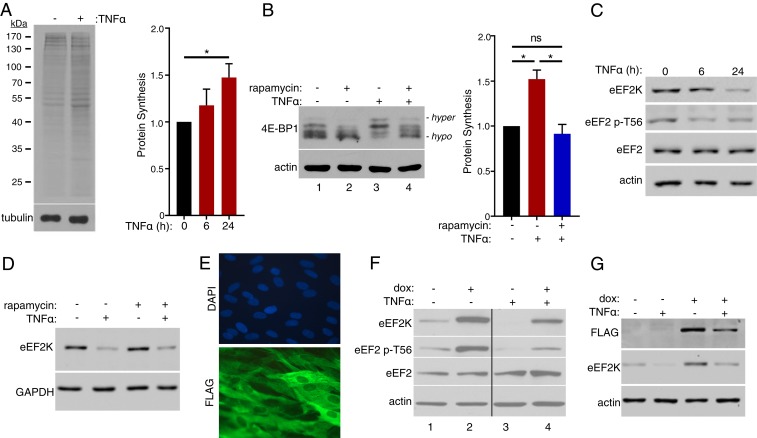
To investigate whether exposure to TNFα impacts global protein synthesis, normal human dermal fibroblasts (NHDFs) treated with TNFα for 24 h were metabolically labeled with 35S-containing amino acids (35S-aa). Fractionation of total protein by SDS/PAGE followed by autoradiography and quantification of acid-insoluble radioactivity by counting in liquid scintillant revealed that TNFα treatment increased overall protein synthesis by ∼50% after 24 h compared to untreated cells (Fig. 1A). Next, we examined how key regulators of mRNA translation responded to TNFα treatment. Phosphorylation of 4E-BP1, a repressor of cap-dependent translation initiation, was evaluated by SDS/PAGE followed by immunoblotting, which distinguishes hypophosphorylated 4E-BP1 active repressor isoforms (faster-migrating species) from hyperphosphorylated, inactivated isoforms (slower-migrating species). Fig. 1B shows that TNFα treatment reduced hypophosphorylated and increased hyperphosphorylated 4E-BP1 abundance (Fig. 1B, compare lanes 1 to 3). The mTORC1-selective inhibitor rapamycin antagonized 4E-BP1 hyperphosphorylation induced by TNFα (Fig. 1B, compare lanes 3 and 4), consistent with reported TNFα-dependent activation of mTORC1 (36–38), and inhibited the TNFα-induced stimulation of protein synthesis (Fig. 1B). Rapamycin also reduced 4E-BP1 phosphorylation in control untreated NHDFs (Fig. 1B, compare lanes 1 and 2). Despite being reduced by rapamycin, 4E-BP1 phosphorylation in TNFα-treated compared to untreated cells was potentially less sensitive to rapamycin. The exact reasons remain unclear, but could reflect TNFα-induced modifications to mTORC1 that might influence the efficacy with which rapamycin inhibits mTORC1 action on select substrates like 4E-BP1 (39, 40).
Fig. 1.
Control of fibroblast protein synthesis by TNFα. (A) NHDFs were untreated or TNFα-treated prior to metabolic labeling with 35S-radioactive amino acids for 30 min. (Left) Cultures were metabolically labeled after 24 h. Following fractionation of total protein by SDS/PAGE, the gel was fixed, dried, and exposed to X-ray film (Top) or analyzed by immunoblotting using an antibody specific for tubulin (Bottom). (Right) Cultures were metabolically labeled after 6 and 24 h. Amino acid incorporation was quantified by counting acid-insoluble radioactivity in liquid scintillant (*P < 0.05 by Student’s t test). (B) NHDFs were treated with rapamycin and TNFα as indicated. (Left) At 24 h posttreatment, total protein was isolated, separated by SDS/PAGE, and analyzed by immunoblotting using antibodies specific for 4E-BP1 and actin. (Right) Cultures were metabolically labeled after 24 h and amino acid incorporation quantified as in A (*P < 0.05; ns, not significant by Student’s t test). (C) As in A except immunoblotting was performed using antibodies specific for eEF2K, eEF2 phospho-T56, eEF2, and actin. (D) As in B except immunoblotting was performed using antibodies specific for eEF2K and GAPDH. (E) NHDFs transduced with a doxycycline (dox)-inducible FLAG-eEF2K lentivirus were stained with DAPI to visualize nuclei, and indirect immunofluorescence staining was performed using a FLAG-specific antibody. (F) NHDFs transduced as in E were treated with dox and TNFα as indicated. At 24 h post-TNFα treatment, total protein was collected and immunoblotting was performed as in C. (G) As in F except immunoblotting was performed using antibodies specific for FLAG, eEF2K, and actin.
Protein synthesis is also regulated by phosphorylation of the critical translation elongation factor eEF2 on T56 by eEF2K, which slows translation and allows it to be tuned in response to physiological and environmental changes (24, 25). To establish whether TNFα regulates eEF2 phosphorylation, total protein isolated from untreated or TNFα-treated NHDFs was fractionated by SDS/PAGE and analyzed by immunoblotting. Compared to untreated cultures, overall levels of T56-phosphorylated eEF2 were reduced by TNFα while the abundance of total eEF2 was not detectably lowered (Fig. 1C). While eEF2K is inhibited upon phosphorylation by p70S6K, which is activated by mTORC1 or p90 RSK1, the reduction in eEF2 phosphorylation was unexpectedly accompanied by a substantial decline in eEF2K protein levels (Fig. 1C) that was not detectably influenced by inhibiting mTORC1 using rapamycin (Fig. 1D). This raised the possibility that eEF2 phosphorylation status in response to TNFα was controlled in part by a different mechanism involving eEF2K abundance instead of solely via the canonical posttranslational modification mechanism involving p70S6K.
Regulation of eEF2K mRNA Abundance by TNFα Is Dependent upon Canonical NF-κB Signaling and the p65 NF-κB Subunit.
NHDFs expressing doxycycline (dox)-inducible, FLAG-tagged eEF2K were used to investigate whether eEF2K abundance regulated TNFα-induced eEF2 phosphorylation. Following dox addition, 100% of the cells exhibited uniform cytoplasmic FLAG-antigen staining with nuclear sparing by indirect immunofluorescence (Fig. 1E). Overall eEF2K and eEF2 p-T56 levels increased in dox-treated cultures while total eEF2 levels remained similar (Fig. 1F, compare lanes 1 and 2). Significantly, whereas the abundance of eEF2K and eEF2 p-T56 was reduced in response to TNFα (Fig. 1F, compare lanes 1 to 3), dox-induced eEF2K expression reversed this effect (Fig. 1F, compare lanes 3 and 4) and largely restored eEF2 p-T56 levels to those observed in cells not exposed to TNFα (Fig. 1F, compare lanes 4 and 1). Taken together, these findings suggest that eEF2 phosphorylation in response to TNFα is regulated by eEF2K abundance. As FLAG-eEF2K levels in dox-induced cells were also reduced by TNFα (Fig. 1G), posttranscriptional or combinatorial mechanisms involving transcriptional regulation could be involved.
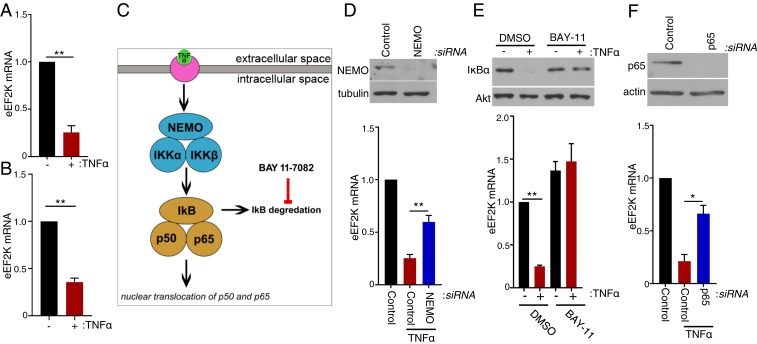
Although the ubiquitin proteasome system reportedly regulates eEF2K abundance in response to DNA damage checkpoint activation (41), TNFα treatment is known to substantially sculpt the cellular transcriptome by impacting transcriptional activation, repression, and mRNA decay (42, 43). Following exposure to TNFα, analysis of total RNA isolated from NHDFs by RT-qPCR revealed a ∼75% reduction in eEF2K mRNA after 6 h and a 65% reduction after 24 h compared to untreated cells (Fig. 2 A and B). Overall eEF2K protein abundance also declined over this same time period, as did levels of T-56 phosphorylated eEF2 (Fig. 1C). Next, the possibility that the TNFα-induced decrease in eEF2K mRNA abundance was dependent upon stimulation of the canonical NF-κB pathway by TNFα was investigated (Fig. 2C). RNA interference was used to deplete IKKγ/NEMO, an IκB kinase (IKK) complex subunit required for degradation of NF-κB inhibitor alpha (IκB) (44). Preventing IκB degradation interferes with nuclear translocation of the NF-κB transcription factors p65 and p50 and restricts NF-κB activation. Indeed, NEMO abundance was reduced in NHDFs transfected with NEMO siRNA compared to control, nonsilencing (ns) siRNA (Fig. 2D). Furthermore, while eEF2K mRNA abundance was reduced in ns siRNA-treated cells exposed to TNFα, eEF2K mRNA levels were significantly greater in NEMO-depleted NHDFs following TNFα exposure (Fig. 2D). Similarly, treatment of cells with BAY-11–7082, a chemical inhibitor that blocks IκB degradation, effectively prevented the TNFα-induced reduction in IκB protein abundance and eEF2K mRNA levels (Fig. 2E). To test directly whether the decrease in eEF2K mRNA abundance in response to TNFα was dependent upon NF-κB, the p65 transcription factor NF-κB subunit was depleted using RNAi. Fig. 2F shows that p65 depletion antagonized the TNFα-induced reduction in eEF2K mRNA compared to cultures treated with nonsilencing siRNA. Taken together, this indicates that the reduction of eEF2K mRNA abundance following TNFα exposure is dependent upon IκB degradation, which regulates NF-κB p50/65 subunit nuclear translocation, and the NF-κB transcription factor subunit p65. It further suggests that p65 controls translation elongation by regulating eEF2K expression.
Fig. 2.
Regulation of eEF2K mRNA abundance by the canonical NF-κB–activating pathway and p65. (A) NHDFs were untreated or TNFα-treated. At 6 h posttreatment, total RNA was isolated and RT-qPCR analysis was performed for eEF2K mRNA. (B) As in A except cells were treated with TNFα for 24 h. (C) Diagram representing the canonical pathway by which TNFα activates NF-κB transcription factors. (D) NHDFs were transfected with nonsilencing (control) or NEMO-targeting siRNAs. (Upper) At 3 d posttransfection, total protein was collected, separated by SDS/PAGE, and analyzed by immunoblotting using antibodies specific for NEMO and tubulin. (Lower) Cultures were untreated or TNFα-treated. At 6 h posttreatment, total RNA was isolated and RT-qPCR was performed for eEF2K mRNA. (E) NHDF cells were treated with DMSO or BAY 11–7082 (10 μM) with or without TNFα. (Upper) At 1 h posttreatment, total protein was fractionated by SDS/PAGE and analyzed by immunoblotting using antibodies specific for IκBα and Akt. (Lower) Total RNA was isolated from cells 6 h posttreatment, and RT-qPCR analysis was performed for eEF2K mRNA. (F) NHDFs were transfected with ns siRNA or p65/RELA siRNA. (Upper) At 3 d posttransfection, total protein was collected, separated by SDS/PAGE, and analyzed by immunoblotting using antibodies specific for p65 and actin. (Lower) NHDFs were untreated or TNFα-treated. At 6 h posttreatment, total RNA was isolated, and RT-qPCR was performed for eEF2K mRNA. Error bars (A, B, and D–F) indicate SEM (*P ≤ 0.05 and **P ≤ 0.01, Student’s t test).
DNA-Sensing Inhibits eEF2 Phosphorylation by Regulating eEF2K mRNA Abundance.
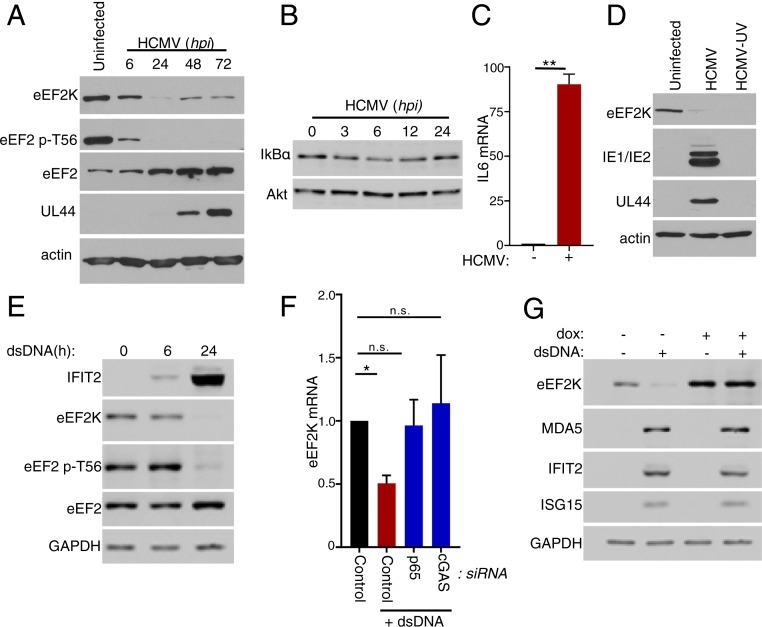
As NF-κB activation integrates responses to numerous inflammatory and infectious agents, the capacity of NF-κB to regulate eEF2 phosphorylation following exposure to stimuli other than TNFα, such as virus infection, was evaluated. Although NF-κB is rapidly activated in HCMV-infected cells (45) and eEF2 levels increase via mTORC1-dependent translational control (31), the impact of infection on eEF2 phosphorylation has not been investigated. Fig. 3A shows that levels of T56 phosphorylated eEF2 decline precipitously as early as 6 hpi, well in advance of the substantial virus-induced rise in eEF2 abundance that is readily detectable by 24 hpi. By 6 hpi, a modest decline in the abundance of the NF-κB inhibitor, IκB, was observed along with induction of a canonical NF-κB–responsive gene (IL6), consistent with NF-κB being activated under these experimental conditions (Fig. 3 B and C). The decline in phosphorylated eEF2 is accompanied by a reduction in eEF2K levels that is apparent by 6 hpi and proceeds to its nadir by 24 hpi (Fig. 3A). Phosphorylated eEF2 remains below detection limits through 72 hpi despite a limited recovery in eEF2K levels (Fig. 3A). Infection of NHDFs with UV-inactivated HCMV, which disables viral gene expression, was at least as effective as active virus in reducing overall eEF2K levels (Fig. 3D). This suggested that an HCMV virion protein component or the viral dsDNA genome itself, a potent PAMP capable of inducing a panoply of inflammatory responses, might represent the underlying trigger for reducing eEF2K abundance.
Fig. 3.
Regulation of eEF2 phosphorylation by dsDNA-sensing. (A) NHDFs were infected with HCMV (multiplicity of infection = 3 pfu per cell). At the indicated times, total protein was collected, fractionated by SDS/PAGE, and analyzed by immunoblotting using the indicated antibodies. (B) As in A. (C) NHDFs were mock-infected (uninfected) or infected with HCMV as in A. After 6 h, total RNA was isolated and RT-qPCR was performed using primers specific for IL6 mRNA (**P < 0.01 by Student’s t test). (D) As in A except cells were infected with HCMV or UV-inactivated HCMV and total protein harvested at 24 hpi. (E) NHDFs were transfected with no DNA or synthetic immunostimulatory dsDNA. At the specified times posttransfection, total protein was collected and immunoblotting was performed using the indicated antibodies. (F) NHDFs were transfected with nonsilencing siRNA (control), p65/RELA siRNA, or cGAS siRNA and transfected with no DNA or dsDNA. At 24 h posttreatment, total RNA was isolated and RT-qPCR analysis was performed for eEF2K mRNA. Error bars indicate SEM (*P ≤ 0.05; **P ≤ 0.01; and n.s., nonsignificant by Student’s t test). (G) NHDFs transduced with a doxycycline (dox)-inducible FLAG-eEF2K lentivirus were transfected with (+) or without (−) synthetic dsDNA and treated with dox as indicated. After 24 h, total protein was collected and analyzed by immunoblotting with the indicated antibodies.
To determine if dsDNA could regulate eEF2 phosphorylation, uninfected primary fibroblasts were transfected with and without a synthetic, immunostimulatory dsDNA, and levels of eEF2K, eEF2, and T56 phosphorylated eEF2 were measured by immunoblotting. Although eEF2 pT56 and eEF2K were readily detected in uninfected, transfected NHDFs that were not exposed to dsDNA, their overall abundance was markedly reduced over 24 h by dsDNA exposure, while levels of total eEF2 did not detectably decline (Fig. 3E). The dsDNA treatment effectively stimulated type I IFN production, a well-characterized response to dsDNA-sensing as evidenced by the accumulation of the IFN-stimulated gene product IFIT2 (Fig. 3E). The reduction in eEF2K protein levels was accompanied by a decline in eEF2K mRNA abundance and was abrogated by depleting either the NF-κB transcription factor subunit p65 or the dsDNA sensor cGAS (Fig. 3F). Even though eEF2K levels decreased in response to dsDNA, artificially counteracting the decline in eEF2K abundance using dox-inducible eEF2K-expressing cells to overexpress eEF2K did not detectably change the overall abundance of 3 representative proteins encoded by IFN-stimulated genes (Fig. 3G). This does not preclude the possibility, however, that their rates of synthesis might be impacted. Nevertheless, these findings together demonstrate that dsDNA exposure and sensing regulates phosphorylation of the translation elongation factor eEF2 and reduces the abundance of its primary regulator eEF2K. Moreover, it establishes that multiple inflammatory triggers including TNFα and dsDNA modulate eEF2 phosphorylation via an NF-κB–dependent mechanism that regulates eEF2K mRNA abundance.
Repression of eEF2K mRNA Transcription by NF-κB p65.
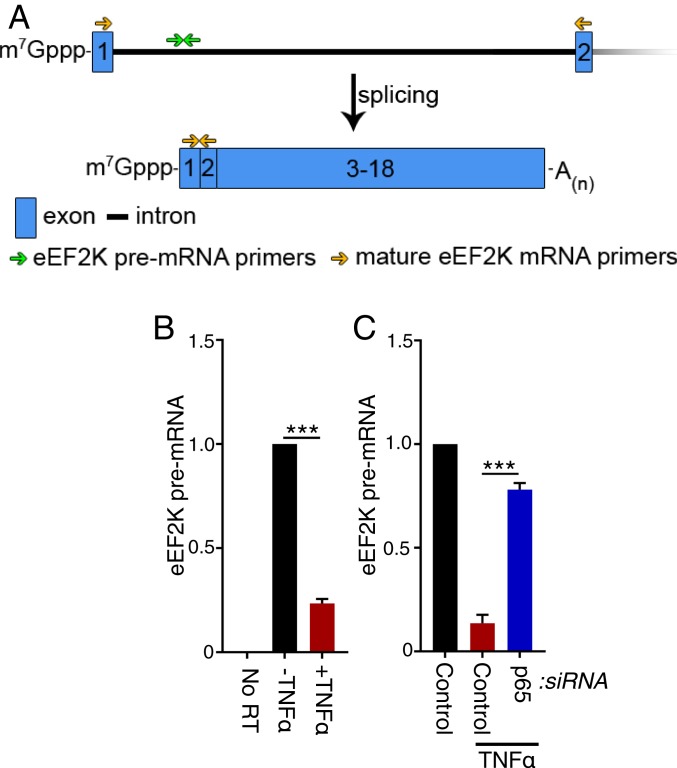
Although p65 largely functions as a transcriptional activator, NF-κB also reportedly can function as a transcriptional repressor of select target genes (46, 47). This raised the possibility that the reduction in steady-state eEF2K mRNA levels in response to NF-κB activation resulted from reduced transcription as opposed to posttranscriptional control by mRNA decay. To test whether eEF2K mRNA transcription might be repressed by p65, the abundance of mRNA containing the first intron of eEF2K pre-mRNA were measured by RT-qPCR (Fig. 4A). As intron removal by mRNA splicing happens cotranscriptionally, this method provides a suitable proxy for transcription rates (48). In response to TNFα exposure, eEF2K pre-mRNA levels declined substantially in a manner that was largely counteracted by p65 depletion (Fig. 4 B and C). This demonstrates that eEF2K pre-mRNA levels are reduced in a manner dependent upon the p65 NF-κB transcription factor subunit. Furthermore, it is consistent with eEF2K transcription being repressed by p65 and with eEF2K being a negatively regulated NF-κB–responsive gene. Indeed, data from chromatin immunoprecipitation studies available on the ENCODE database map p65 (RelA) occupancy to the eEF2K promoter, supporting the notion that eEF2K is an NF-κB–responsive gene (49, 50).
Fig. 4.
Repression of eEF2K mRNA transcription by TNFα is p65-dependent. (A, Top) eEF2K pre-mRNA exons (1, 2; blue boxes) and intron 1 (solid black line). The location of primers specific for the pre-mRNA intron (green arrows) and those specific for exons 1 and 2 (gold arrows) are shown. (A, Bottom) Mature eEF2K mRNA, which is comprised of 18 exons. The 5-terminal mRNA cap is represented by m7Gppp, and the polyadenylated 3′-end is shown as A(n). (B) NHDFs were untreated or TNFα-treated. At 6 h posttreatment, total RNA was isolated and RT-qPCR analysis was performed for eEF2K pre-mRNA. (C) NHDFs were transfected with nonsilencing siRNA (control) or p65/RELA siRNA and treated with TNFα. At 6 h posttreatment, total RNA was isolated and RT-qPCR analysis was performed for eEF2K pre-mRNA. (A and B) A control siRNA-treated culture was not exposed to TNFα as a reference for eEF2K mRNA levels in untreated NHDFs. Error bars indicate SEM (***P ≤ 0.001, Student’s t test).
eEF2K Regulates the Responsiveness of Protein Synthesis to Exotoxin A.
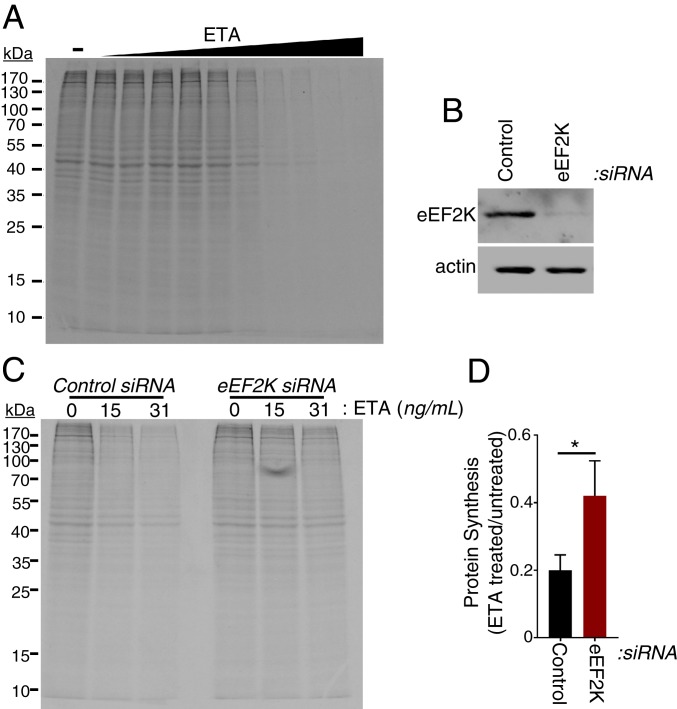
To test whether NF-κB–dependent eEF2K repression could impact host responses, we sought a model to evaluate whether fibroblast protein synthesis would respond to levels of active versus inactive, T-56 phosphorylated eEF2. Determining whether the stimulation of TNFα-induced protein synthesis was dependent upon eEF2 elongation activity was difficult because (i) eEF2 acts catalytically and is among the most abundant of cellular proteins (51) and (ii) TNFα also stimulates protein synthesis initiation by promoting 4E-BP1 hyperphosphorylation (Fig. 1B and refs. 36–38). To circumvent these confounding parameters, protein synthesis following exposure to a bacterial toxin selective for eEF2 was investigated. P. aeruginosa is a Gram-negative bacteria known to activate NF-κB (52–54) that produces an ADP ribosylating enzyme, exotoxin A (ETA), which irreversibly inactivates eEF2 (55). Incubation of NHDFs with ETA concentrations greater than 8 ng/mL profoundly inhibited global protein synthesis as measured by metabolic labeling with 35S amino acids (Fig. 5A). Having defined concentrations at which ETA efficiently reduced protein synthesis, we next asked whether ETA toxicity was impacted by eEF2K abundance by comparing overall protein synthesis in NHDFs where eEF2K was depleted using siRNA to NHDFs treated with nonsilencing siRNA (Fig. 5B). Remarkably, eEF2K depletion significantly interfered with ETA-induced inhibition of protein synthesis at 2 toxin concentrations (Fig. 5 C and D). This demonstrates that the sensitivity of fibroblast protein synthesis to ETA is dependent upon eEF2K abundance. Thus, regulating eEF2K abundance can in fact control protein synthesis and change the sensitivity of fibroblasts to an ADP ribosylating toxin that selectively targets eEF2.
Fig. 5.
eEF2K depletion renders cells refractory to protein synthesis inhibition by exotoxin A. (A) NHDFs were treated with increasing concentrations (0 to 2,000 ng/mL) of exotoxin A (ETA). At 24 h posttreatment, cells were metabolically radiolabeled with 35S-containing amino acids for 30 min. Total protein was isolated and separated by SDS/PAGE, and the fixed, dried gel was exposed to X-ray film. (B) NHDFs were transfected with nonsilencing (control) or eEF2K targeting siRNAs. At 3 d posttransfection, total protein was collected, separated by SDS/PAGE, and analyzed by immunoblotting using antibodies specific for eEF2K and actin. (C) As in B, except that, 3 d after siRNA transfection, cells were treated with the indicated concentrations of ETA. (D) As in B, except that, 3 d after siRNA transfection, cells were treated with 0 ng/μL or 31 ng/μL ETA. At 24 h posttreatment, cells were metabolically radiolabeled with 35S-containing amino acids for 30 minutes, and the amount of acid-insoluble radioactivity was quantified by counting in liquid scintillant. *P < 0.05 by Student’s t test.
Discussion
By virtue of the rapid responses engendered, regulating mRNA translation is ideally suited to tune gene expression to inflammatory and infectious insults. While translation initiation is rate-limiting and has been intensively investigated (56), much less is known regarding how infection and inflammation impact cellular translation elongation factors. Here, we reveal an unexpected mechanism whereby the critical translation elongation factor eEF2 is stimulated upon activation of the transcription factor NF-κB, a universal coordinator of cell-intrinsic responses to inflammation and infection (1). Phosphorylation of eEF2 on T56 by eEF2K regulates translation elongation by reducing the affinity of eEF2 for the 80S ribosome and thereby slowing the translocation step in protein synthesis (24, 25). In response to different NF-κB–activating stimuli including TNFα, HCMV infection, and synthetic immunostimulatory dsDNA, we show that the overall abundance of T56 phosphorylated eEF2 is reduced together with a corresponding decrease in eEF2K overall levels. The decrease in eEF2K protein abundance was dependent upon the classical NF-κB activation pathway and the p65 NF-κB transcription factor. Surprisingly, the reduction in eEF2K pre-mRNA levels was dependent upon p65, suggesting that eEF2K transcription is repressed by NF-κB activation. Finally, we demonstrate that manipulating eEF2K abundance regulates protein synthesis in response to a bacterial toxin that inactivates eEF2. This establishes that NF-κB activation by physiological effectors controls eEF2 activity via a transcriptional repression mechanism that limits eEF2K abundance. Moreover, it illustrates how eEF2, which catalyzes the translocation step in translation elongation, can be integrated into the network of innate immune responses orchestrated by NF-κB and provides a powerful example of how nuclear transcription regulation shapes translation elongation in eukaryotes.
Previous reports on how TNFα and NF-κB impact protein synthesis have been conflicting, with some demonstrating an enhancement in protein synthesis and others inhibition (57–60), perhaps due to differing cell types and culture conditions. In addition, prior genome-wide studies did not identify eEF2K among genes transcriptionally repressed by TNF (43). Here, we focus exclusively on human fibroblasts and show that NF-κB activation effectively stimulates translation by preventing eEF2K from slowing elongation. Preserving eEF2 activity by limiting eEF2K abundance could better enable fibroblasts to coordinate responses that resolve inflammation and repair tissue damage. Besides translation elongation, initiation is also stimulated by TNFα as mTORC1 activation inhibits the cellular repressor of cap-dependent translation 4E-BP1, which is consistent with published reports (36–38). Furthermore, mTORC1 also stimulates p70S6K1, which phosphorylates eEF2K S366 (61) and inhibits eEF2K activity. Due to the quick kinetics of TNFα-induced mTORC1 activation (62), this could represent a rapid initial response to stimulate protein synthesis by inactivating 4E-BP1 and/or eEF2K through posttranscriptional modification. Coupling this initial response to an NF-κB–dependent, transcriptionally mediated decrease in eEF2K protein abundance could maintain high eEF2 activity as time progresses and allow unrestricted elongation even if initiation on mRNAs relying upon a cap-dependent translation mechanism was tempered by inhibiting mTORC1. Likewise, limiting eEF2K abundance could allow maximal eEF2 activity on transcripts that rely upon noncanonical initiation mechanisms when functional eIF4F levels are limited by inhibiting mTORC1. This could explain earlier studies showing that that insulin stimulates muscle protein synthesis in neonates during acute endotoxemia despite suppression of translation initiation, suggesting that an mTOR-independent process regulates muscle protein synthesis (63). Additionally, as eEF2K inhibits the translation rate of TNFα mRNA (64), TNFα-induced reduction in eEF2K protein abundance could positively regulate TNFα synthesis to amplify inflammatory signaling.
Using NF-κB to repress eEF2K likely represents a mechanism to support translation of the hundreds of transcripts induced by NF-κB in response to infection or nucleic acid sensing (1, 65, 66). Limiting eEF2K levels prevents eEF2 T56 phosphorylation, which would slow elongation and cellular protein synthesis and potentially impact infection outcome. Our finding that eEF2K depletion influences the inhibition of protein synthesis in response to ETA is consistent with this hypothesis and supports a role for eEF2K in influencing host responses to pathogens. Translation rates have also been shown to inversely correlate with cotranslational folding efficiency as well as translational accuracy (67). While accelerating elongation by repressing eEF2K is expected to enable rapid protein production, this potentially compromises translational accuracy and fidelity (68) upon NF-κB activation by inflammatory or infection stress. It also provides a means to reduce accuracy of mRNA decoding through elongation while mTORC1 is inactivated. Significantly, regulating eEF2K levels by NF-κB allows infection and/or inflammation to influence elongation rates independent of mTORC1/p70S6K. This distinguishes regulation of elongation in response to infection stress from energy insufficiency or nutrient insufficiency (69). Reducing eEF2K levels by NF-κB activation could reduce fidelity of viral mRNA translation as part of a broader defense strategy or impact synthesis of cellular and or viral proteins from alternate reading frames and/or non-AUG initiator codons. Indeed, analysis of cis-acting sequences controlling translation of innate immune effectors including MAVS revealed they did not fit classic Kozak consensus sites (70). Instead, there were hundreds of transcripts that are predicted to encode more than one protein product. Alternatively, speeding up elongation would resolve ribosome queuing, which can form in response to protein synthesis inhibitors or occur naturally to regulate polyamine synthesis via translation of an inhibitory upstream ORF (71). In either case, reducing eEF2K levels in direct response to NF-κB–activating stimuli provides a powerful mechanism to tune translation elongation to a wide range of physiological and pathological processes impacted by NF-κB, including cancer, infection biology, development, inflammation, and immunity.
Materials and Methods
Cell Culture, Viruses, Transfections, Metabolic Labeling with Radioactive Amino Acids, RT-qPCR, and Immunoblotting.
Normal human dermal fibroblasts (Lonza; CC-2509) were maintained and serum-deprived as previously described (72). Human cytomegalovirus (AD169-GFP) (73) propagation and infections, transfections of dsDNA and siRNA, immunoblotting, metabolic labeling with radioactive amino acids, and RT-qPCR were previously described (74). GAPDH was used for normalization of all RT-qPCR experiments. UV-inactivated HCMV was prepared as described (72) using 6 pulses of 125 mJ/cm2. The pINDUCER21-plasmid encoding dox-inducible eEF2K (75) was generously provided by Fabio Martinon (University of Lausanne, Lausanne, Switzerland) and used to generate lentivirus as described previously (76).
Nucleotide Sequences.
Primer sequences used in this study were eEF2K forward (5′-GCACAGCCAGAGTTACTGGAC-3′), eEF2K reverse (5′-CTCTGCCAACTCCTCTGGAC-3′), eEF2K pre-mRNA forward (5′-CGCTTCTCCTCTGGTTGTT-3′), eEF2K pre-mRNA reverse (5′-TTCCATATCCACATCGCCTTTA-3′), GAPDH forward (5′-TCTTTTGCGTCGCCAGCCGA-3′), and GAPDH reverse (5′-ACCAGGCGCCCAATACGACC-3′).
Sequences of siRNAs used in this study were NEMO/IKBKG (5′-GAGAGACUCGGCCUGGAGA[dT][dT]-3′), p65/RELA (5′-GGAAUCCAGUGUGUGAAGA[dT][dT]-3′), cGAS (5′-GCUACUAUGAGCACGUGAA[dT][dT]-3′), and eEF2K (5′-CUCAUCACAUCCUAGCCGA [dT][dT]-3′).
Synthetic double-strand DNA (dsDNA) was produced by annealing sense and anti-sense 60-mer oligonucleotides derived from HSV-1 sequences as previously described (77). Briefly, sense (5′-TAAGACACGATGCGATAAAATCTGTTTGTAAAATTTATTAAGGGTACAAATTGCCCTAGC-3′) and anti-sense (5′- GCTAGGGCAATTTGTACCCTTAATAAATTTTACAAACAGATTTTATCGCATCGTGTCTTA-3′) strands were annealed by heating at 95 °C for 3 min and cooling to room temperature in annealing buffer (10 mM Tris⋅HCl, pH 7.5, 100 mM NaCl, and 1 mM EDTA).
Antibodies, Chemicals, and Cytokines.
Antibodies used in this study: anti-4E-BP1 (Bethyl; A300-501A), anti-tubulin (Sigma; T5168), anti-eEF2K (Cell Signaling; 3692), anti-eEF2 (Cell Signaling; 2332), anti-eEF2 phospho Thr-56 (Cell Signaling; 2331), anti-actin (Cell Signaling; 3700), anti-NEMO/IKKγ (Cell Signaling; 2685), anti-IκBα (Proteintech; 10268–1-AP), anti-p65/RELA (Proteintech; 10745–1-AP), anti-Akt (Cell Signaling; 9272), and anti-Flag (Cell Signaling; 2368S). Doxycycline (Sigma; D3072) was used at 0.125 µg/mL tumor necrosis factor α (PeproTech; 300–02A) was dissolved in water and used at 50 ng/mL P. aeruginosa exotoxin A was obtained from List Biological Labs (no. 160) and dissolved in water. Rapamycin was dissolved in DMSO and used at 100 ng/mL.
Acknowledgments
We thank members of the Mohr laboratory, Michael Garabedian, and Angus Wilson for helpful discussions and Dr. Fabio Martinon for the eEF2K expression vector. This work was supported by National Institutes of Health Grants GM056927 and AI073898 to I.M. Public Health Service Institutional Research Training Awards (AI007180, AI07647) in part supported both C.B. and L.T.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
References
- 1.Zhang Q., Lenardo M. J., Baltimore D., 30 Years of NF-κB: A blossoming of relevance to human pathobiology. Cell 168, 37–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan F. K., Luz N. F., Moriwaki K., Programmed necrosis in the cross talk of cell death and inflammation. Annu. Rev. Immunol. 33, 79–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalliolias G. D., Ivashkiv L. B., TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 12, 49–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun S. C., The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manthiram K., Zhou Q., Aksentijevich I., Kastner D. L., The monogenic autoinflammatory diseases define new pathways in human innate immunity and inflammation. Nat. Immunol. 18, 832–842 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Afonina I. S., Zhong Z., Karin M., Beyaert R., Limiting inflammation-the negative regulation of NF-κB and the NLRP3 inflammasome. Nat. Immunol. 18, 861–869 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Frakes A. E., Dillin A., The UPRER: Sensor and coordinator of organismal homeostasis. Mol. Cell 66, 761–771 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Karin M., Lin A., NF-kappaB at the crossroads of life and death. Nat. Immunol. 3, 221–227 (2002). [DOI] [PubMed] [Google Scholar]
- 9.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudkov A. V., Komarova E. A., p53 and the carcinogenicity of chronic inflammation. Cold Spring Harb. Perspect. Med. 6, a026161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munn L. L., Cancer and inflammation. Wiley Interdiscip. Rev. Syst. Biol. Med. 9, e1370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hrdinka M., Gyrd-Hansen M., The met1-linked ubiquitin machinery: Emerging themes of (De)regulation. Mol. Cell 68, 265–280 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Pfeffer L. M., The role of nuclear factor κB in the interferon response. J. Interferon Cytokine Res. 31, 553–559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan X., Sun L., Chen J., Chen Z. J., Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 72, 447–478 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Ablasser A., Chen Z. J., cGAS in action: Expanding roles in immunity and inflammation. Science 363, eaat8657 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Schoggins J. W., Rice C. M., Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1, 519–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohr I., Sonenberg N., Host translation at the nexus of infection and immunity. Cell Host Microbe 12, 470–483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stern-Ginossar N., Thompson S. R., Mathews M. B., Mohr I., Translational control in virus-infected cells. Cold Spring Harb. Perspect. Biol. 11, a033001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Merrick W. C., Pavitt G. D., Protein synthesis initiation in Eukaryotic cells. Cold Spring Harb. Perspect. Biol. 10, a033092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gingras A. C., et al. , Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15, 2852–2864 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dever T. E., Dinman J. D., Green R., Translation elongation and recoding in eukaryotes. Cold Spring Harb. Perspect. Biol. 10, a032649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter J. D., Coller J., Pausing on polyribosomes: Make way for elongation in translational control. Cell 163, 292–300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proud C. G., Regulation and roles of elongation factor 2 kinase. Biochem. Soc. Trans. 43, 328–332 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Proud C. G., Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb. Perspect. Biol. 11, a033050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cannon M. J., Schmid D. S., Hyde T. B., Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 20, 202–213 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Ljungman P., Hakki M., Boeckh M., Cytomegalovirus in hematopoietic stem cell transplant recipients. Infect. Dis. Clin. North Am. 24, 319–337 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Boeckh M., Geballe A. P., Cytomegalovirus: Pathogen, paradigm, and puzzle. J. Clin. Invest. 121, 1673–1680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramanan P., Razonable R. R., Cytomegalovirus infections in solid organ transplantation: A review. Infect. Chemother. 45, 260–271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawlinson W. D., et al. , Congenital cytomegalovirus infection in pregnancy and the neonate: Consensus recommendations for prevention, diagnosis, and therapy. Lancet Infect. Dis. 17, e177–e188 (2017). [DOI] [PubMed] [Google Scholar]
- 31.McKinney C., Perez C., Mohr I., Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 109, 5627–5632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jørgensen R., Merrill A. R., Andersen G. R., The life and death of translation elongation factor 2. Biochem. Soc. Trans. 34, 1–6 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Buckley C. D., et al. , Fibroblasts regulate the switch from acute resolving to chronic persistent inflammation. Trends Immunol. 22, 199–204 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Van Linthout S., Miteva K., Tschöpe C., Crosstalk between fibroblasts and inflammatory cells. Cardiovasc. Res. 102, 258–269 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Pilling D., Vakil V., Cox N., Gomer R. H., TNF-α-stimulated fibroblasts secrete lumican to promote fibrocyte differentiation. Proc. Natl. Acad. Sci. U.S.A. 112, 11929–11934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozes O. N., et al. , A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc. Natl. Acad. Sci. U.S.A. 98, 4640–4645 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glantschnig H., Fisher J. E., Wesolowski G., Rodan G. A., Reszka A. A., M-CSF, TNFalpha and RANK ligand promote osteoclast survival by signaling through mTOR/S6 kinase. Cell Death Differ. 10, 1165–1177 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Lee D. F., et al. , IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell 130, 440–455 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Feldman M. E., et al. , Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thoreen C. C., et al. , An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruiswijk F., et al. , Coupled activation and degradation of eEF2K regulates protein synthesis in response to genotoxic stress. Sci. Signal. 5, ra40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loupasakis K., et al. , Tumor necrosis factor dynamically regulates the mRNA stabilome in rheumatoid arthritis fibroblast-like synoviocytes. PLoS One 12, e0179762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulsen M. T., et al. , Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proc. Natl. Acad. Sci. U.S.A. 110, 2240–2245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothwarf D. M., Zandi E., Natoli G., Karin M., IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature 395, 297–300 (1998). [DOI] [PubMed] [Google Scholar]
- 45.DeMeritt I. B., Milford L. E., Yurochko A. D., Activation of the NF-kappaB pathway in human cytomegalovirus-infected cells is necessary for efficient transactivation of the major immediate-early promoter. J. Virol. 78, 4498–4507 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell K. J., Rocha S., Perkins N. D., Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol. Cell 13, 853–865 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Beauchef G., et al. , The p65 subunit of NF-κB inhibits COL1A1 gene transcription in human dermal and scleroderma fibroblasts through its recruitment on promoter by protein interaction with transcriptional activators (c-Krox, Sp1, and Sp3). J. Biol. Chem. 287, 3462–3478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ponzio T. A., Yue C., Gainer H., An intron-based real-time PCR method for measuring vasopressin gene transcription. J. Neurosci. Methods 164, 149–154 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kent W. J., et al. , The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenbloom K. R., et al. , ENCODE data in the UCSC genome browser: Year 5 update. Nucleic Acids Res. 41, D56–D63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beck M., et al. , The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DiMango E., Ratner A. J., Bryan R., Tabibi S., Prince A., Activation of NF-kappaB by adherent Pseudomonas aeruginosa in normal and cystic fibrosis respiratory epithelial cells. J. Clin. Invest. 101, 2598–2605 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schroeder T. H., et al. , CFTR is a pattern recognition molecule that extracts Pseudomonas aeruginosa LPS from the outer membrane into epithelial cells and activates NF-kappa B translocation. Proc. Natl. Acad. Sci. U.S.A. 99, 6907–6912 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tran C. S., et al. , Host cell polarity proteins participate in innate immunity to Pseudomonas aeruginosa infection. Cell Host Microbe 15, 636–643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jørgensen R., et al. , Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature 436, 979–984 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Herdy B., et al. , Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat. Immunol. 13, 543–550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lang C. H., Frost R. A., Nairn A. C., MacLean D. A., Vary T. C., TNF-alpha impairs heart and skeletal muscle protein synthesis by altering translation initiation. Am. J. Physiol. Endocrinol. Metab. 282, E336–E347 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Williamson D. L., Kimball S. R., Jefferson L. S., Acute treatment with TNF-alpha attenuates insulin-stimulated protein synthesis in cultures of C2C12 myotubes through a MEK1-sensitive mechanism. Am. J. Physiol. Endocrinol. Metab. 289, E95–E104 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Hiraoka E., et al. , TNF-alpha induces protein synthesis through PI3-kinase-Akt/PKB pathway in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 280, H1861–H1868 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Plaisance I., Morandi C., Murigande C., Brink M., TNF-alpha increases protein content in C2C12 and primary myotubes by enhancing protein translation via the TNF-R1, PI3K, and MEK. Am. J. Physiol. Endocrinol. Metab. 294, E241–E250 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Wang X., et al. , Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 20, 4370–4379 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karonitsch T., et al. , mTOR senses environmental cues to shape the fibroblast-like synoviocyte response to inflammation. Cell Rep. 23, 2157–2167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orellana R. A., et al. , Amino acids augment muscle protein synthesis in neonatal pigs during acute endotoxemia by stimulating mTOR-dependent translation initiation. Am. J. Physiol. Endocrinol. Metab. 293, E1416–E1425 (2007). [DOI] [PubMed] [Google Scholar]
- 64.González-Terán B., et al. , Eukaryotic elongation factor 2 controls TNF-α translation in LPS-induced hepatitis. J. Clin. Invest. 123, 164–178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jan E., Mohr I., Walsh D., A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol. 3, 283–307 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Schneider W. M., Chevillotte M. D., Rice C. M., Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conn C. S., Qian S. B., Nutrient signaling in protein homeostasis: An increase in quantity at the expense of quality. Sci. Signal. 6, ra24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie J., et al. , Regulation of the elongation phase of protein synthesis enhances translation accuracy and modulates lifespan. Curr. Biol. 29, 737–749.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Leprivier G., et al. , The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153, 1064–1079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brubaker S. W., Gauthier A. E., Mills E. W., Ingolia N. T., Kagan J. C., A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell 156, 800–811 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ivanov I. P., et al. , Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol. Cell 70, 254–264.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walsh D., Mohr I., Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18, 660–672 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Terhune S., et al. , Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81, 3109–3123 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McKinney C., et al. , Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell Rep. 6, 1175 (2014). [DOI] [PubMed] [Google Scholar]
- 75.De Gassart A., et al. , Pharmacological eEF2K activation promotes cell death and inhibits cancer progression. EMBO Rep. 17, 1471–1484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bianco C., Mohr I., Restriction of human cytomegalovirus replication by ISG15, a host effector regulated by cGAS-STING double-stranded-DNA sensing. J. Virol. 91, e02483-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Unterholzner L., et al. , IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]