Significance
We introduce a key role for low doses of DNA methyltransferase inhibitors (DNMTis) in reprogramming the DNA repair transcriptome and creating a homologous recombination defect (HRD), sensitizing to poly (ADP-ribose) polymerase (PARP) inhibitors (PARPis) in breast cancer gene (BRCA)-proficient nonsmall cell lung cancer (NSCLC), which are not clinically responsive to PARPis. The nonhomologous end-joining pathway is also downregulated by DNMTis, generating sensitivity to radiation therapy (RT). The significant increases in antitumor effects with DNMTi and PARPis in combination with RT suggest a potentially potent therapeutic strategy for patients with BRCA-proficient NSCLC, for whom limited therapy is available.
Keywords: DNA repair, lung cancer, homologous recombination defect, nonhomologous end-joining, poly (ADP-ribose) polymerase inhibitors
Abstract
A minority of cancers have breast cancer gene (BRCA) mutations that confer sensitivity to poly (ADP-ribose) polymerase (PARP) inhibitors (PARPis), but the role for PARPis in BRCA-proficient cancers is not well established. This suggests the need for novel combination therapies to expand the use of these drugs. Recent reports that low doses of DNA methyltransferase inhibitors (DNMTis) plus PARPis enhance PARPi efficacy in BRCA-proficient AML subtypes, breast, and ovarian cancer open up the possibility that this strategy may apply to other sporadic cancers. We identify a key mechanistic aspect of this combination therapy in nonsmall cell lung cancer (NSCLC): that the DNMTi component creates a BRCAness phenotype through downregulating expression of key homologous recombination and nonhomologous end-joining (NHEJ) genes. Importantly, from a translational perspective, the above changes in DNA repair processes allow our combinatorial PARPi and DNMTi therapy to robustly sensitize NSCLC cells to ionizing radiation in vitro and in vivo. Our combinatorial approach introduces a biomarker strategy and a potential therapy paradigm for treating BRCA-proficient cancers like NSCLC.
Inhibitors of poly (ADP-ribose) polymerase 1 (PARP1) have shown promise for targeting cancer cells harboring mutations in the double-strand break (DSB) repair breast cancer genes BRCA1 and BRCA2, where these drugs induce synthetic lethality (1). PARP inhibitors (PARPi) function by dual mechanisms: 1) inhibition of the catalytic activity of PARP1 and 2) allosteric enhancement of PARP1 binding into DNA, leading to PARP trapping at single-strand break (SSB) and DSB sites (2). Upon the initiation of DNA replication, the presence of trapped PARP causes replication fork stalling (3) that can collapse the replication fork into a 1-ended DSB (4). Homologous recombination (HR) is the major pathway required for high-fidelity repair of replication-associated DSBs and is also critical in replication fork restart (5). As both prolonged fork stalling and unrepaired DSBs are potent triggers of apoptosis, PARPis rapidly produce synthetic lethality where DSB repair is impaired, such as with the HR defect associated with BRCA1 or BRCA2 mutations (6, 7). While PARPi success in the clinic has, to date, excluded BRCA-proficient cancers, several preclinical and clinical studies are attempting to refine and expand this narrow therapeutic scope. Strategies include identifying cancers with an inherited or acquired defect in other HR genes that produces a BRCA-like phenotype (termed BRCAness) (8) or combining PARPi with traditional DNA damaging agents such as chemotherapy and/or radiotherapy (9).
In BRCA-proficient nonsmall cell lung cancer (NSCLC), studies have shown that the PARPi veliparib acts as a radiosensitizer in xenograft models, effectively decreasing tumor volume using a 50% lower total dose of ionizing radiation (RT) compared to RT alone (10). Furthermore, olaparib was shown to increase levels of DSBs, as measured by γH2AX foci following 2 Gy RT exposure, which correlated with decreased cell survival and slowed tumor growth in xenografts pretreated with olaparib prior to RT treatment (11). Clinical investigations have also shown efficacy of PARPi in NSCLC used in combination with RT, compared with standard chemotherapeutic agents and RT, and RT alone (12). Notably, early stage clinical trials are underway with the new generation PARPi talazoparib (TAL), which exhibits increased potency of PARP trapping activity and enhanced cytotoxic effects (13, 14) that may correlate with increased clinical efficacy (15, 16).
Abnormal DNA methylation patterns and associated transcriptional silencing are observed in many cancers, including in NSCLC (17), and can be targeted by inhibitors of DNA methyltransferase (18). DNA methyltransferase inhibitors (DNMTi) are cytidine analogs that are incorporated as a base in DNA and targeted for methylation, resulting in covalent entrapment of the maintenance methylation enzyme DNMT1 (19, 20), promoting DNMT1 proteasomal degradation (21), and inducing widespread methylation changes. Nanomolar doses of the DNA demethylating agents Vidaza (5-AZA) or Dacogen (DAC) can reprogram cancer cell gene expression, exerting durable antitumor effects on leukemia, breast, and lung cancer cells in vitro and in vivo (22, 23). These effects are accompanied by sustained decreases in genome-wide promoter methylation, alterations in heritable gene expression patterns, and antiproliferative changes in key cellular pathways, including DNA repair pathways (18). Accordingly, in combination with DNA damage induction by RT, these agents increase cellular toxicity and enhance treatment effectiveness, including in NSCLC (24). Furthermore, interaction between PARP1 and DNMT1 has been described, specifically by 1) localization of PARP1 at the DNMT1 promoter to protect from CpG island methylation and hence prevent transcriptional silencing (25) and 2) noncovalent interaction between PARylated PARP1 and DNMT1 that modulates methylation activity (26, 27). We recently reported a novel combinatorial approach to enhance cytotoxicity in BRCA-proficient triple negative breast cancer (TNBC) and acute myeloid leukemia (AML) (28). Exposure to 5-AZA or DAC results in increased PARP1-chromatin binding, which is further enhanced by addition of the PARP-trapping PARPi TAL (28). DNA damage induction by laser microirradiation suggests that this binding may occur specifically at damage sites (29, 30) and is dependent upon the PARP1-DNMT1 interaction given that depletion of either protein abolishes PARP1 localization to damage. In keeping with enhanced and prolonged PARP trapping, the PARPi-DNMTi combination is associated with increased accumulation of cytotoxic DSBs, as measured by γH2AX foci, compared to either drug alone. Of translational importance, the combination demonstrates a well-tolerated and potent in vivo antitumor response both in an immune-deficient, BRCA-proficient MDA-MB-231 TNBC model and in orthotopic models of MV411 and MOLM14 AML cells (28). We also recently reported that this drug combination is efficacious in BRCA-proficient ovarian cancer (OC), suggesting that this therapeutic strategy may be extended to other BRCA-proficient cancers (31).
In this study, we aimed to determine in NSCLC whether nanomolar doses of DNMTis can perturb the DNA damage response as a mechanism underlying a therapy response to potent PARP-trapper TAL. Furthermore, we sought to delineate whether this DNMTi reprograming response would also augment RT treatment effects. Finally, we assayed whether efficacy of the noncytotoxic doses of DNMTis and TAL would be further enhanced by combination with RT.
Results
5-AZA in Combination with TAL Decreases Clonogenicity and Exhibits Synergistic Cytotoxicity.
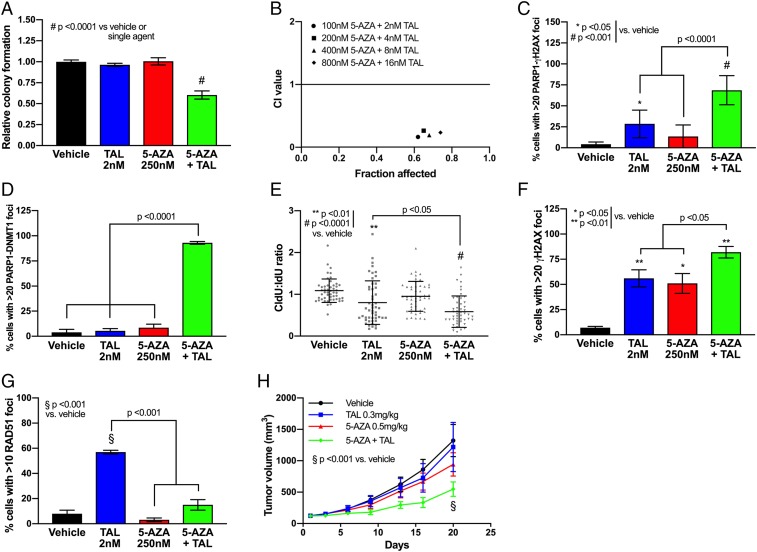
We have previously established the efficacy of DNMTi and PARPi combination therapy in TNBC and AML (28), and thus, we aimed to determine whether this therapeutic strategy could be extended to BRCA-proficient NSCLC. Lung cancer represents the most common cause of cancer death in the United States, exceeding death rates from breast, colon, and prostate cancers combined (32). Accordingly, we first sought to establish the phenotypic implications of DNMTi and PARPi through use of clonogenic assays across 4 NSCLC cell lines (A549, H460, H358, and H838) treated with 5-AZA (250 nM) and TAL (2 nM), alone and in combination. In accordance with BRCA-proficient status, all cell lines tested were insensitive to single-agent TAL, while 1 cell line (H838) demonstrated significant (P < 0.0001) sensitivity to 5-AZA treatment alone (Fig. 1A and SI Appendix, Fig. S1). In contrast, when exposed to combination 5-AZA+TAL, a significant decrease in clonogenicity was observed in all cell lines tested, ranging from ∼75% in A549 (P < 0.01) to >95% in H838 (P < 0.0001) (Fig. 1A and SI Appendix, Fig. S1). In a key association with the above data, 3 NSCLC cell lines (A549, H460, and H358) treated with the combination of 5-AZA+TAL exhibit synergistic cytotoxicity, as assessed by Chou–Talalay method (33, 34) (Fig. 1B and SI Appendix, Fig. S2).
Fig. 1.
Combination treatment with 5-azacytidine and talazoparib produces synergistic cytotoxicity in NSCLC models. (A) Colony forming assay in H460 NSCLC cells in presence of vehicle (DMSO), TAL (2 nM), 5-AZA (250 nM), or combination (day 10, n = 9 from 3 experiments performed in triplicate). Data are represented as mean number of colonies ± SEM. P value is calculated using 1-way ANOVA. (B) Combination index (CI) plot for 5-AZA+TAL in H460 cells (n = 9 from 3 experiments performed in triplicate). (C) Proximity ligation assay for PARP-γH2AX foci formation in H460 following treatment with vehicle (DMSO), TAL (2 nM), 5-AZA (250 nM), or combination (day 4, n = 100 per condition; 25 cells counted per condition from 4 experimental replicates). Data are represented as mean number of cells with >20 PLA foci ± SEM. P value is calculated using 1-way ANOVA. (D) Proximity ligation assay for DNMT1-PARP1 foci formation in H460 following treatment with vehicle (DMSO), TAL (2 nM), 5-AZA (250 nM), or combination (day 4, n = 100 per condition; 25 cells counted per condition from 4 experimental replicates). Data are represented as mean number of cells with >20 PLA foci ± SEM. P value is calculated using 1-way ANOVA. (E) DNA fiber assay in H460 following treatment with vehicle (DMSO), TAL (2 nM), 5-AZA (250 nM), or combination (day 4, n = 50 per condition; 25 fibers measured per condition from 2 experimental replicates). Data are represented as ratio of CldU fiber length to IdU fiber length for individual fiber tracts, overlaid with group mean ± SEM. P value is calculated using 1-way ANOVA. (F) Detection of γH2AX foci in H460 following treatment with vehicle (DMSO), TAL (2 nM), 5-AZA (250 nM), or combination (day 4, n = 100 per condition; 25 cells counted per condition from 4 experimental replicates). Data are represented as mean number of cells with >20 foci ± SEM. P value is calculated using 1-way ANOVA. (G) Detection of RAD51 foci in H460 following treatment with vehicle (DMSO), TAL (2 nM), 5-AZA (250 nM), or combination (day 4, n = 100 per condition; 25 cells counted per condition from 4 experimental replicates). Data are represented as mean number of cells with >20 foci ± SEM. P value is calculated using 1-way ANOVA. (H) Tumor volume in in vivo H460 NSCLC model. H460 xenograft (107 cells per mouse) was delivered via flank injection (n = 8 per group). Treatment with 5-AZA (0.5 mg/kg SC) and/or TAL (0.3 mg/kg PO) was initiated once tumor volume reached 100 mm3 and continued until endpoint. Data are represented as mean external tumor volume ± SEM. P value is calculated using 1-way ANOVA.
5-AZA+TAL Increases γH2AX Foci Formation, Increases PARP1 Binding at DNA Damage Sites, and Decreases RAD51 Recruitment.
The PARPi TAL is recognized to be a potent trapper of PARP1 (35). We previously demonstrated that 5-AZA further increases the retention of PARP1 by TAL at sites of laser-induced DNA damage in TNBC cells (28). In the present study, we assessed for localization of PARP1 at DSB sites using a proximity ligation assay (PLA), which detects the presence of 2 antibody-bound epitopes within a 40-nm range (36). Pretreatment with TAL (P < 0.05) or 5-AZA+TAL (P < 0.001) significantly increased the proportion of cells exhibiting PARP1-γH2AX foci, indicating increased PARP1 localization within the chromatin at sites of γH2AX-associated DNA damage, which includes DSBs and stalled replication forks (37) (Fig. 1C and SI Appendix, Fig. S3). Furthermore, a PLA assay indicated a significant (P < 0.05) increase in nuclear colocalization of PARP1 and DNMT1 following treatment with the 5-AZA+TAL combination compared to vehicle or single-agent treatment (Fig. 1D and SI Appendix, Fig. S3).
Retention of PARP1-PARPi complexes in DNA is an effective block to replication fork progression (3). Based on the above data that 5-AZA+TAL maximizes PARP1 trapping into DNA, we predicted our combination would reduce the DNA replication rate. Replication was assessed by thymidine analog CldU incorporation in the presence of single agent or 5-AZA+TAL and compared as a ratio to incorporation of a second thymidine analog, IdU, in an untreated environment. In agreement with the PLA assay, both single-agent TAL and 5-AZA+TAL combination significantly (P < 0.05) impaired replication fork progression, producing DNA fiber tract patterns indicative of fork stalling (Fig. 1E and SI Appendix, Fig. S4). Both the persistent stalling of replication forks and subsequent fork regression with collapse into DSBs are associated with Ser139 phosphorylation of the histone protein H2AX (γH2AX) (38). We therefore assessed γH2AX foci formation at 4 h following drug exposure. As predicted from the replication fiber analysis, single-agent TAL significantly (P < 0.01) increased γH2AX foci burden, and 5-AZA+TAL further increased DSB levels (Fig. 1F and SI Appendix, Fig. S5). Despite the absence of significant replication fork stalling, an accumulation of γH2AX foci was also observed following single-agent 5-AZA treatment (Fig. 1F). In keeping with the above data, we observed a 2-fold increase (P < 0.05) in apoptosis with 5-AZA+TAL combination treatment (SI Appendix, Fig. S6).
Given that HR is the predominant pathway for repairing replication-associated DSBs and contributes to the restart of stalled replication forks, we assayed whether our treatment paradigm produced an accumulation of RAD51 nuclear foci, a surrogate HR functionality marker. Single-agent TAL treatment increased RAD51 foci in ∼55% of cells (P < 0.01) compared to vehicle controls, indicative of an accumulation of PARP-trapping lesions requiring HR-mediated resolution (Fig. 1G and SI Appendix, Fig. S5). In contrast, neither single-agent 5-AZA nor 5-AZA+TAL produced an increase in RAD51 foci, despite the presence of DNA damage as indicated by γH2AX foci. These data suggest the absence of a robust HR response in 5-AZA–treated cells.
As further evidence for the important translational implications of the present studies, potent antitumor responses were observed in an in vivo model treated with low-dose 5-AZA+TAL. In the H460 flank xenograft model, we assayed the effects of TAL (0.3 mg/kg) and/or 5-AZA (0.5 mg/kg) on both tumor growth kinetics and terminal burden. While single-agent treatment had no discernible effects on tumor growth, the drug combination showed a significant 2.5-fold (P < 0.01) reduction in tumor burden at day 20 after treatment (Fig. 1H), confirmed by 50% reduction in tumor weight at endpoint (SI Appendix, Fig. S7A). Importantly, the treatment was well tolerated as no significant change in body weight was observed during the study (SI Appendix, Fig. S7B).
DNMTi Treatment Decreases Expression of DSB Repair Genes and Reduces HR Capacity.
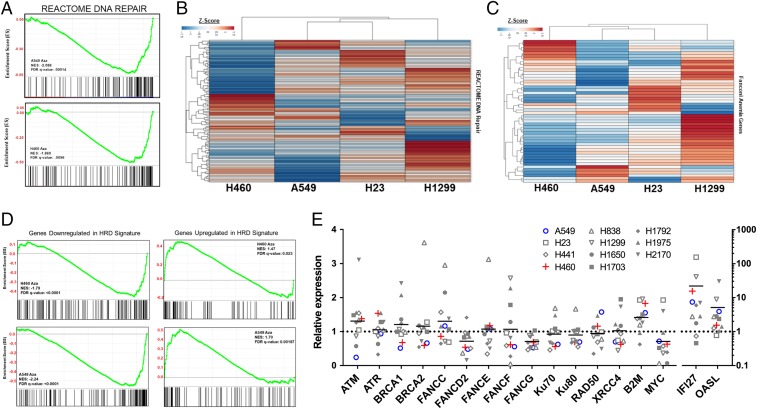
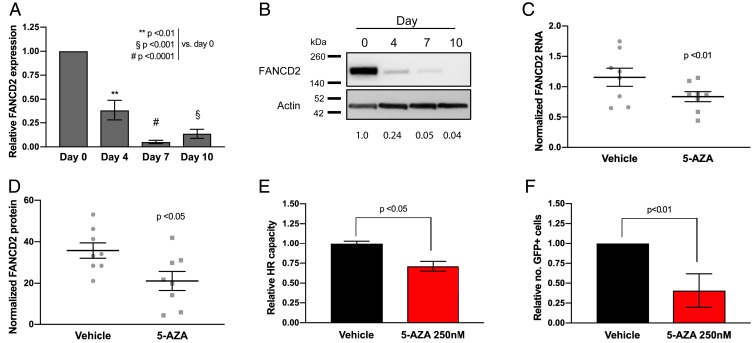
Our finding that RAD51 foci formation is reduced with 5-AZA exposure suggests that DSB repair may be impacted by epigenetic reprogramming associated with low-dose DNMTi treatment (23). To undertake assessment of 5-AZA mediated reprogramming of the DNA repair response, we conducted a reanalysis of published expression array data that included NSCLC cell lines (A549, H460, H23, and H1299) pre– and post–5-AZA treatment (500 nM) (Gene Expression Omnibus [GEO] accession no. GSE104244). Interrogation of these data revealed a DNA repair pathway-related transcriptional defect as the result of 5-AZA treatment, most pronounced in H460 and A549 cell lines (Fig. 2 A and B). DNA repair-related gene sets represent several of the top down-regulated gene sets across 5-AZA treated cell lines (SI Appendix, Table S1 A–D). Further evaluation of DNA repair genes revealed a robust repression of Fanconi anemia (FA)-related genes involved in interstrand crosslink (ICL) repair and DNA replication restart, specifically as the result of 5-AZA treatment (Fig. 2C and SI Appendix, Table S2 A and B). Additionally, 5-AZA treatment induced significant perturbation of genes included a published PARPi sensitization gene signature (39). These data in the aggregate provide evidence of a 5-AZA–induced DNA repair gene defect as a sensitization mechanism to PARPi (Fig. 2D). We next sought to expand our assessment of a 5-AZA–induced DNA repair defect to a panel of 11 NSCLC cell lines (A549, H23, H441, H460, H838, H1299, H1650, H1703, H1792, H1975, and H2170). While there is a broad heterogeneity of transcriptional response, a common feature is down-regulation of FA pathway-related genes (Fig. 2E). We confirm, at both the mRNA (Fig. 3A) and protein level (Fig. 3B), FANCD2 down-regulation beginning at day 4 of 5-AZA treatment and persisting through day 10 in H460 cells. Similar results were observed in A549 cells (SI Appendix, Fig. S8). This noted down-regulation of FANCD2 was also confirmed in H460 xenografts after treatment with 5-AZA at mRNA (Fig. 3C) and protein levels (Fig. 3D), suggesting our paradigm can induce similar repair defects in the in vivo setting.
Fig. 2.
The 5-Azacitidine treatment of NSCLC cell lines significantly down-regulates DNA repair associated genes. (A) Normalized enrichment score plot for REACTOME DNA Repair Pathway derived from preranked GSEA of relative RNA expression in H460 and A549 NSCLC cell lines. (B) Unsupervised hierarchical clustering by Euclidean distance of Z score distribution for relative RNA expression 5-AZA treated over Mock for REACTOME DNA Repair Pathway associated genes in H460, A549, H23, and H1299 NSCLC cell lines. Full gene list with associated Z score is available in SI Appendix, Table S2A. (C) Unsupervised hierarchical clustering by Euclidean distance of Z score distribution for relative RNA expression 5-AZA treated over Mock for Fanconi Anemia Pathway associated genes in H460, A549, H23, and H1299 NSCLC cell lines. Full gene list with associated Z score is available in SI Appendix, Table S2B. (D) Normalized enrichment score plots for HRD gene set derived from preranked GSEA of relative RNA expression in H460 and A549 NSCLC cell lines. (E) Relative RNA expression plot for select DNA repair genes (n = 9 from 3 experimental replicates performed in triplicate). Negative control c-MYC and positive controls B2M, IFI27, and OASL have previously been validated in Topper et al. (53). Above microarray data are derived from reanalysis of publicly deposited data GEO accession no. GSE104244 for NSCLC cell lines (A549, H23, H441, H460, H838, H1299, H1650, H1703, H1792, H1975, and H2170) treated with 500 nM 5-AZA as detailed in Topper et al. (53).
Fig. 3.
The 5-Azacytidine induces an FA-associated DNA repair defect. (A) FANCD2 RNA expression in H460 cells following 5-AZA treatment (250 nM). Data are represented as mean expression ± SEM, normalized against pretreatment expression level (n = 12 from 4 experimental replicates performed in triplicate). P value is calculated using 1-way ANOVA. (B) FANCD2 protein immunoblot in H460 cell lysates following 5-AZA treatment (250 nM). Relative protein level normalized against pretreatment level represented by figures under FANCD2 bands. (C) FANCD2 RNA expression in samples from in vivo H460 NSCLC model (n = 8 animals) treated with 5-AZA (0.5 mg/kg SC). Data are represented as mean expression per sample (from 3 experimental replicates performed in triplicate) and normalized against actin, overlaid with group mean ± SEM. P value is calculated using 2-way ANOVA. (D) FANCD2 protein level in samples from H460 in vivo H460 NSCLC model (n = 8 animals) treated with 5-AZA (0.5 mg/kg SC). Data are represented as mean expression (from 3 experimental replicates performed in triplicate) and normalized against actin, overlaid with group mean ± SEM. P value is calculated using 2-way ANOVA. (E) Homologous recombination repair capacity by extrachromosomal assay in H460 cells following 5-AZA treatment (day 7; 250 nM). Data are represented as mean HR capacity normalized against vehicle-treated cells (n = 6 from 3 experimental replicates performed in duplicate) ± SEM. P value is calculated using unpaired Student’s t test. (F) Homologous recombination repair capacity by in vitro DR-GFP flow cytometry in H460 cells following 5-AZA treatment (day 7; 250 nM). Data are represented as mean HR capacity normalized against vehicle-treated cells (10,000 cells analyzed per condition; 4 experimental replicates) ± SEM. P value is calculated using Student’s t test.
In addition to canonical ICL repair functions, FANCD2 also has key interactions with HR pathway members, including BRCA1 and the MRE11-RAD50-NBS1 (MRN) damage-sensing complex, and has been ascribed as having a role as a BRCAness gene (40). In this regard, 5-AZA–associated down-regulation of FANCD2 is associated with a reduction in HR capacity. Pretreatment with 5-AZA significantly (P < 0.05) reduced recombination efficiency in H460 cells (Fig. 3E) as assessed by extrachromosomal HR assay, which employs plasmids that form a functional LacZ sequence detected by PCR in the presence of a proficient HR system (41). These results were substantiated using a chromosomally integrated DR-GFP reporter plasmid that relies on reconstitution of GFP sequences in the presence of competent HR, as detected by flow cytometry (42). Stable clones containing the DR-GFP integrant treated with 5-AZA (250 nM, 7 d) prior to transient transfection with I-SceI manifested a 75% reduction (P < 0.01) in GFP fluorescence compared to vehicle-treated cells (Fig. 3F). Similar results were also obtained using nuclear protein extracts from 5-AZA–treated tumor tissue (SI Appendix, Fig. S9).
Functional Confirmation for the Role of FANCD2 in 5-AZA–induced HR Defect.
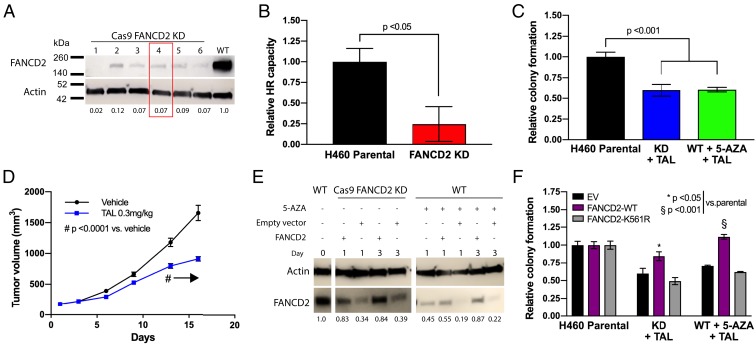
Our gene expression data indicate that 5-AZA induces down-regulation of FANCD2, which is recruited to stalled replication forks to participate in ICL repair and HR and which, when reduced, produces a BRCAness phenotype that induces synthetic lethality with PARPi (43). To validate these data, we generated an H460 FANCD2 knockdown (KD) model by Cas9/CRISPR targeting (Fig. 4A) and utilized a cell population (FANCD2 clone 4) that persistently exhibits a baseline FANCD2 protein expression of 5% as compared to wild-type cells. This reduction is similar to the level observed following 5-AZA exposure in the parental cell line. As predicted by the biological function of FANCD2, HR capacity in this KD cell line was significantly (P < 0.05) reduced (Fig. 4B), approximating the defect observed in 5-AZA–treated parental cells. In addition, TAL sensitivity was increased in the KD cells to a degree equivalent to that observed when the parental cell line was treated with the 5-AZA+TAL, as indicated by reduced colony formation capacity post-drug treatment (Fig. 4C). Interestingly, this degree of TAL sensitivity occurs with KD of FANCD2 alone, despite this gene being one of multiple HR genes that are underexpressed following low-dose DNMTi. Importantly, TAL sensitivity was replicated in a xenograft model using the FANCD2 KD cell line described above (Fig. 4D and SI Appendix, Fig. S10A). In agreement with our previous findings, no toxicity as defined by evaluation of mouse body weight was observed, further validating the tolerability of our paradigm (SI Appendix, Fig. S10B).
Fig. 4.
FANCD2 depletion sensitizes NSCLC to talazoparib treatment. (A) Immunoblotting for FANCD2 knockdown (FANCD2-KD) in H460 Cas9/CRISPR clones. Relative protein level normalized against wild-type level represented by figures under FANCD2 bands. Red box indicates clone selected for further studies and in vivo model. (B) Homologous recombination repair capacity by extrachromosomal assay in FANCD2-KD H460 cells. Data are represented as mean HR capacity normalized against vehicle-treated cells (n = 9 from 3 experimental replicates performed in triplicate) ± SEM. P value is calculated using unpaired Student’s t test. (C) Colony forming assay in H460 FANCD2-KD cells in presence of TAL (2 nM) (day 10, n = 9 from 3 experimental replicates performed in triplicate) compared to H460 wild-type cells treated with TAL+5-AZA (250 nM) combination. Data are represented as mean number of colonies ± SEM. P value is calculated using 1-way ANOVA. (D) On study tumor volume in in vivo H460 FANCD2-KD NSCLC model. H460 FANCD2-KD xenograft (107 cells per mouse) was delivered via flank injection (n = 8 per group). Treatment with TAL (0.3 mg/kg PO) was initiated once tumor volume reached 100 mm3 and continued until endpoint. #P < 0.001 from day 13 onward as indicated by arrow. (E) Immunoblotting for FANCD2 in FANCD2-KD or 5-AZA treated (day 4; 250 nM) H460 cells following transfection of FANCD2-wild-type vector or empty vector. Relative protein level normalized against untreated H460 wild-type level represented by figures under FANCD2 bands. (F) Colony forming assay in H460 FANCD2-KD cells or wild-type H460 cells pretreated with 5-AZA (250 nM) for 4 d, followed by transfection with FANCD2-wild-type or FANCD2-mutant vector in presence of TAL (2 nM) (day 10, n = 9 from 3 experimental replicates performed in triplicate). Data are represented as mean number of colonies ± SEM. P value is calculated using 1-way ANOVA.
To further define the role of FANCD2 down-regulation after 5-AZA treatment, we overexpressed wild-type FANCD2 (FANCD2-WT) protein in FANCD2 KD and 5-AZA–treated parental cells (Fig. 4E). In both systems, FANCD2-WT overexpression was able to rescue TAL sensitivity (Fig. 4F). Monoubiquitination of FANC2 is critical to enable recruitment to stalled replication forks, and importantly, expression of a FANCD2-K561R monoubiquitination-dead mutant did not alter sensitivity compared to expression of the empty vector (Fig. 4F).
DNMTi Treatment Produces a Nonhomologous End-Joining Defect.
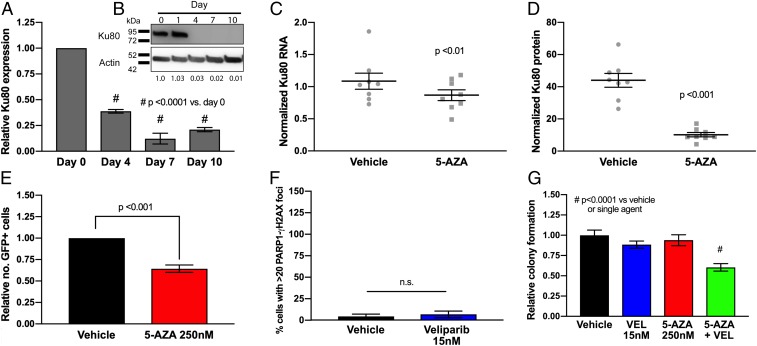
As stated earlier, interrogation of mRNA levels across a panel of NSCLC cell lines (Fig. 2E) revealed that 5-AZA exposure down-regulates a number of genes involved in DNA repair, including several key members involved in nonhomologous end-joining (NHEJ), the pathway required for DSB repair throughout the cell cycle (44). These genes included Ku70 (SI Appendix, Fig. S11A), XRCC4 (SI Appendix, Fig. S11B), and Ku80 (Fig. 5A), the latter of which was also found to be strongly down-regulated at a protein level in H460 cells in vitro (Fig. 5B) and in our xenograft model (Fig. 5 C and D). Finally, 5-AZA reduced activity of NHEJ activity in a functional assay which utilizes an EJ5-GFP reporter plasmid in which tandem I-SceI restriction enzyme cuts produce blunt ends that produce GFP expression when ligated by NHEJ (42). Pretreatment with 5-AZA (250 nM, 7 d) produces a 33% reduction in GFP-positive cells (as determined by flow cytometry) compared to vehicle controls, indicating a significant (P < 0.001) defect in NHEJ activity (Fig. 5E).
Fig. 5.
The 5-Azacytidine induces a nonhomologous end-joining DNA repair defect in NSCLC. (A) Ku80 RNA expression in H460 cells following 5-AZA treatment (250 nM). Data are represented as mean expression ± SEM, normalized against pretreatment expression level (n = 12 from 4 experimental replicates performed in triplicate). P value is calculated using 1-way ANOVA. (B) Ku80 protein immunoblot in H460 cell lysates following 5-AZA treatment (250 nM). Relative protein level normalized against pretreatment level represented by figures under Ku80 bands. (C) Ku80 RNA expression in samples from in vivo H460 NSCLC model (n = 8 animals) treated with 5-AZA (0.5 mg/kg SC). Data are represented as mean expression (from 3 experimental replicates performed in triplicate) and normalized against actin, overlaid with group mean ± SEM. P value is calculated using 2-way ANOVA. (D) Ku80 protein level in samples from H460 in vivo H460 NSCLC model (n = 8 animals) treated with 5-AZA (0.5 mg/kg SC). Data are represented as mean expression (from 3 experimental replicates performed in triplicate) and normalized against actin, overlaid with group mean ± SEM. P value is calculated using 2-way ANOVA. (E) Nonhomologous end-joining repair capacity by EJ5-GFP flow cytometry in H460 cells following 5-AZA treatment (day 7; 250 nM). Data are represented as mean NHEJ capacity normalized against vehicle-treated cells (10,000 cells analyzed per condition; 4 experimental replicates) ± SEM. P value is calculated using Student’s t test. (F) Proximity ligation assay for PARP1-γH2AX colocalization in H460 cells following treatment with veliparib (day 4, n = 50 per condition; 25 cells counted per condition from 3 experimental replicates). Data are represented as mean number of cells with >20 PLA foci ± SEM. P value is calculated using 1-way ANOVA. (G) Colony forming assay in H460 NSCLC cells in presence of vehicle (DMSO), veliparib (VEL; 15 nM), 5-AZA (250 nM), or combination (day 10, n = 9 from 3 experiments performed in triplicate). Data are represented as mean number of colonies ± SEM. P value is calculated using 1-way ANOVA.
The data presented in the above sections present 2 mechanisms by which our combination exerts its cytotoxic effect: 1) enhanced trapping of PARP1 by 5-AZA with subsequent accumulation of cytotoxic DSBs and 2) 5-AZA–induced reduction in HR- and NHEJ-mediated repair. Confirmation of a role for both mechanisms can be illustrated by use of the PARP1 catalytic inhibitor veliparib (15 μM), which possesses minimal PARP-trapping activity (2), as indicated by absence of significant PARP1-γH2AX PLA foci accumulation (Fig. 5F and SI Appendix, Fig. S12). Despite limited PARP trapping, however, veliparib significantly potentiates the cytotoxicity of 5-AZA (Fig. 5G), in keeping with 5-AZA–induced synthetic lethality via a DSB repair (DSBR) defect.
Radiation Potentiates the 5-AZA+TAL Combination In Vitro and In Vivo.
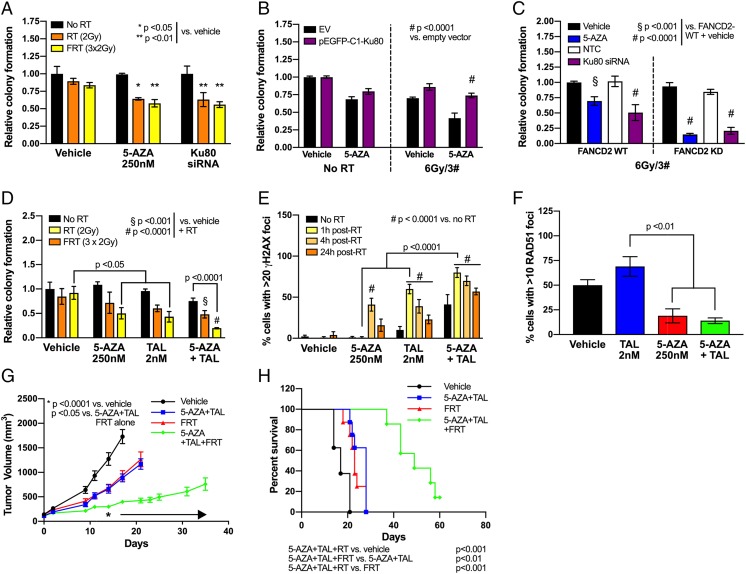
From a translational standpoint, much of the data in previous sections have an important ramification in the context of ionizing radiation, a standard therapeutic modality for NSCLC. For example, Ku80 is a component of the Ku heterodimer that binds and stabilizes DSBs during the initial damage recognition step of NHEJ (45) and has been a target for knockdown or small molecule inhibition to enhance radiosensitivity (46). The application of Ku80 siRNA inhibition in NSCLC cell lines (SI Appendix, Fig. S13) potently sensitized to ionizing radiation (single 2-Gy fraction or 6 Gy in 3 daily fractions) as detected by colony-forming assay (Fig. 6A). An equivalent level of radiosensitization was observed when cells without siRNA were pretreated with 5-AZA (250 nM, 7 d). This radiosensitization could be partially rescued by transfection of a Ku80 expression plasmid (Fig. 6B and SI Appendix, Fig. S14). In the context of the above defects in both HR and NHEJ following DNMTi treatment, Ku80 siRNA in our FANCD2 CRISPR/Cas9 KD cell lines produced a similar radiosensitization effect to 5-AZA treatment on colony-forming capacity as seen with dual FANCD2/Ku80 knockdown (Fig. 6C).
Fig. 6.
The 5-Azacytidine–mediated impairment of DSB repair sensitizes NSCLC to combination treatment with talazoparib and radiation. (A) Colony forming assay in H460 cells transfected with Ku80 siRNA or treated with 5-AZA (250 nM), followed by single 2 Gy (RT) or fractionated 3 × 2 Gy (FRT) irradiation (n = 9 from 3 experimental replicates performed in triplicate). Data are represented as mean number of colonies ± SEM. P value is calculated using 1-way ANOVA. (B) Colony forming assay in H460 cells pretreated with 5-AZA (250 nM; 4 d) and nucleofected with Ku80-wild-type or empty vector, followed by fractionated 3 × 2 Gy irradiation (n = 9 from 3 experimental replicates performed in triplicate). Data are represented as mean number of colonies ± SEM. P value is calculated using 1-way ANOVA. (C) Colony forming assay in H460 FANCD2-wild-type or FANCD2-KD cells, pretreated with 5-AZA (250 nM; 4 d) or transfected with Ku80 siRNA (or nontargeting control [NTC]), followed by fractionated 3 × 2 Gy irradiation (n = 9 from 3 experimental replicates performed in triplicate). Data are represented as mean number of colonies ± SEM. P value is calculated using 1-way ANOVA. (D) Colony forming assay in H460 NSCLC cell lines in presence of vehicle (DMSO), TAL (2 nM), 5-AZA (250 nM), or combination, followed by single 2 Gy (RT) or fractionated 3 × 2 Gy (FRT) irradiation (n = 9 from 3 experimental replicates performed in triplicate). Data are represented as mean number of colonies ± SEM. P value is calculated using 1-way ANOVA. (E) Detection of γH2AX foci in H460 after 2 Gy irradiation, following pretreatment with vehicle (DMSO), TAL (2 nM), 5-AZA (250 nM), or combination (day 4; n = 100 per condition; 25 cells counted per condition from 4 experimental replicates). Data are represented as mean number of cells with >20 foci ± SEM. P value is calculated using 1-way ANOVA. (F) Detection of RAD51 foci in H460 at 4 h after 2 Gy irradiation, following treatment with vehicle (DMSO), TAL (2 nM), 5-AZA (250 nM), or combination (day 4, n = 100 per condition; 25 cells counted per condition from 4 experimental replicates). Data are represented as mean number of cells with >20 foci ± SEM. P value is calculated using 1-way ANOVA. (G) Tumor volume in in vivo H460 NSCLC model. H460 xenograft (107 cells per mouse) was delivered via flank injection (n = 8 per group). Treatment with 5-AZA (0.5 mg/kg SC) and TAL (0.3 mg/kg PO) was initiated once tumor volume reached 100 mm3 and continued until endpoint. Fractionated radiation was delivered at days 7, 8, and 9 after initiation of TAL+5-AZA. *P < 0.05 from day 18 onward as indicated by arrow. (H) Kaplan–Meier survival plot of in vivo H460 NSCLC model treated with 5-AZA+TAL and/or fractionated radiation (n = 8 per group).
Our data suggest that 5-AZA+TAL efficacy relies upon impaired PARP activity at sites of DNA damage and that this might be augmented by RT-induced DNA damage. As compared with untreated controls or RT alone, single-agent TAL or 5-AZA in combination with fractionated RT (FRT; 3 × 2 Gy) significantly reduced H460 colony formation by ∼50% (P < 0.05), although no significant effect was observed with single-dose RT (2 Gy). In contrast, 5-AZA+TAL in combination with either RT or FRT, led to a significant decrease in colony formation, with FRT-treated cells exhibiting a 75% reduction in colonies (RT, P < 0.001; FRT, P < 0.0001) (Fig. 6D). Combination 5-AZA+TAL also sensitized to RT and FRT in A549 cells (SI Appendix, Fig. S15). Combination index analysis indicated a synergistic effect of the 5-AZA+TAL+FRT combination at most drug concentration combinations tested, whereas single-agent plus FRT combinations did not exhibit the same potentiation (SI Appendix, Fig. S16).
Effects of RT on DNA damage accumulation indicated that γH2AX burden in TAL-treated H460 cells peaked at 60% at 1 h post-RT (2 Gy) (Fig. 6E; P < 0.0001 compared to RT+vehicle–treated controls). A significant increase in γH2AX-positive cells following RT was also observed in 5-AZA–treated cells, reaching maximal levels at 4 h post-RT (41%; P < 0.0001 compared to RT+vehicle–treated controls). Peak staining levels (80%) in the 5-AZA+TAL treatment group were observed at 1 h post-RT, representing a significant increase over vehicle or single-agent treatment (P < 0.0001). Notably, at 24 h post-RT, levels of γH2AX-positive cells persist in 5-AZA+TAL group, compared with single-agent treatment or controls (P < 0.0001). Despite this increased γH2AX-associated DNA damage burden, 5-AZA treatment (alone or in combination with TAL) significantly decreased RAD51 foci formation following RT (Fig. 6F and SI Appendix, Fig. S17). In agreement with our previous data, these results suggest that 5-AZA treatment may play a key role in decreasing HR activity in NSCLC cells.
Importantly, the above in vitro results indicating that radiation can potentiate our combination drug treatment can also be observed in vivo. Compared to vehicle controls, treatment of mice bearing H460 cells as NSCLC xenograft flank heterotransplants with either 5-AZA+TAL or FRT showed a similar, nonsignificant reduction in tumor volume (Fig. 6G). In contrast, the group treated with 5-AZA+TAL in combination with FRT showed a significant (P < 0.05) decrease in tumor growth and an increase in survival (Fig. 6 G and H). A similar but less potent survival response was also observed when 5-AZA+TAL was combined with a single 2-Gy RT dose (SI Appendix, Fig. S18).
Discussion
The introduction of PARPis into the clinic has been an exciting development for the treatment of breast and ovarian cancers harboring BRCA mutations. For the majority of BRCA-proficient cancers, including NSCLC, PARPis fail as single agents; thus, the discernment of whether combination strategies can expand the use of these inhibitors is a priority. The data presented in this study suggest that low doses of DNMTi can reprogram the DNA damage repair response in NSCLC. We establish that this repair defect can be leveraged to induce synthetic lethality when deployed in combination with PARPi with or without radiation. Given that these therapeutic modalities are already available for clinical use, this approach represents an accessible treatment strategy for NSCLC patients.
In the present study, we find that PARPi synergizes with DNMTi in the setting of BRCA-proficient NSCLC (Fig. 1B and SI Appendix, Fig. S2). This noted synergy is correlated with the large-scale perturbation of DNA repair-related genes (Fig. 2 A and B). Further delineation of the 5-AZA–induced DNA repair-associated transcriptional changes revealed significant alteration of a PARPi sensitization gene signature and significant down-regulation of FA-related genes (Fig. 2C). Critical to these findings is the known function of the FA pathway and associated genes for repair of ICL (47), resolution of replication blocking lesions, and restarting of the replication machinery (48). These results extend into NSCLC a published report of synergistic cytotoxicity between 5-AZA and TAL in BRCA-proficient breast, ovarian, and AML model systems (28). Together, these data support the validity of therapeutic HR repression, through perturbation of FA-related genes, as a potential strategy for expanding PARPi utility, and point to DNMTi as a potent arbiter of this sensitization phenotype.
An area of heightened translational importance is the collision of our treatment paradigm with RT (Fig. 6D), which represents a widely deployed therapeutic modality for NSCLC (49). RT-induced DNA damage commonly features nonligatable ends (50) and is not associated with replication (51). Therefore, the major pathway for repair of RT-associated DSBs is cell cycle-independent NHEJ (51, 52). NHEJ deficiency induced by DNMTi prevents the resolution of RT-associated DSBs (Fig. 6F) and is driven through the down-regulation of NHEJ factors, most notably Ku80 (Fig. 5 A–D). These data indicate that 5-AZA treatment induces an NHEJ defect mediated in part through Ku80 depletion, which potently sensitizes to RT-induced DNA damage, leading to cytotoxicity.
Future work must be directed toward deciphering the contributions of the 5-AZA component to the cytotoxicity of our combination by elucidating how down-regulation of DNA damage repair is mediated. One important consideration is the effect 5-AZA has on key oncogenic functions, including the oncogene c-MYC. We previously reported that low-dose 5-AZA produces, in all of the NSCLC lines studied herein, a significant down-regulation of the transcription factor c-MYC and MYC-regulated targets (53). Furthermore, we and others have directly linked MYC dysregulation to alterations in DNA repair gene expression and capacity (54), including impairment of HR (55) and NHEJ (56) pathway factors. The specificity of the effect on homologous recombination defect (HRD) for 5-AZA over other DNMTis should also be considered. Both 5-AZA and the structurally related DNMTi 5-aza-2′-deoxycytidine (decitabine/DAC) require intracellular activation by kinases, but while DAC is sequentially phosphorylated to its active metabolite 5-aza-deoxycytidine-triphosphate (5-aza-dCTP), 5-AZA requires an intermediate reduction reaction by ribonucleotide reductase (57). Only ∼10 to 20% of 5-AZA is metabolized in this manner, with the remainder being phosphorylated to 5-azacytidine-triphosphate (5-aza-CTP), which is incorporated instead into RNA (58). Accordingly, 5-AZA has been specifically linked to DNMT2 trapping in RNA and subsequent hypomethylation (59), which may affect tRNA folding and stability (60) and hence protein expression, potentially including DNA repair proteins. Furthermore, 5-AZA may impact stability and/or function of noncoding RNAs or RNA–DNA hybrids, which have been implicated in a wide range of important roles in promoting and coordinating the DNA damage response (61–63). However, work from our laboratory in AML cell lines provides evidence that treatment with DAC produces a similar effect upon DNA repair gene expression to 5-AZA (SI Appendix, Fig. S19), suggesting that a 5-AZA–specific effect upon RNA metabolism is unlikely to mediate the observed HRD.
In summary, the preclinical work presented indicates that low-dose DNMTi treatment induces a DSB repair defect that encompasses both HR/FA, represented by FANCD2 depletion, and NHEJ, observed as a reduction in Ku80 expression. The HR repair deficiency can be targeted in a synthetic lethality approach using the PARPi talazoparib, whose potent PARP-trapping ability is further enhanced by the presence of colocalized DNMT1-DNMTi complexes. Furthermore, DNMTi-induced impairment of NHEJ also enhances the cytotoxicity of DNA damage induced by radiation, a standard treatment for NSCLC. As these agents are already in clinical use, these results suggest an exciting possibility for expanding the therapeutic potential of PARPis in BRCA-proficient NSCLC in combination with DNMTi and RT.
Experimental Procedures
Cell Culture.
Human NSCLC cell lines A549, H460, H838, and H358 were cultured in RPMI1640 + l-Glutamine (Mediatech Inc.) with 10% FBS (Sigma–Aldrich). Transient Ku80 knockdown was achieved by Lipofectamine 3000 (ThermoFisher) transfection of ON-TARGETplus SMARTpool siRNA (Dharmacon). Ku80 overexpression was achieved by nucleofection of pEGFP-C1-FLAG-Ku80, a gift from Steve Jackson, The Wellcome Trust/Cancer Research UK Gurdon Institute and Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom (Addgene no. 46958) (64). FANCD2 CRISPR knockout H460 cells were generated using the lentiviral plasmid lentiCRISPR-FANCD2-KO vector (Addgene no. 111099) as described in SI Appendix, Supplemental Experimental Procedures. FANCD2 overexpression was achieved by Lipofectamine 3000 transfection of pMMP-FANCD2-WT or pMMP-FANCD2-K561R (a gift from Alan D’Andrea, Department of Radiation Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA) (65).
Cytotoxic Treatments.
The 5-azacytidine (Sigma–Aldrich) was prepared at 500 μM in PBS. Talazoparib (BMN-673; AbMole BioScience) was prepared at 5 mM in DMSO. For RT studies, cells were exposed to 2 Gy X-ray radiation using a Pantak HF320 X-ray machine (250 kV peak, 13 mA; half-value layer, 1.65 mm copper) at a dose rate of 2.4 Gy/min.
Antibodies.
See SI Appendix, Supplementary Experimental Procedures Table S1.
Primers for qRT-PCR.
See SI Appendix, Supplementary Experimental Procedures Table S2.
Gene Expression Array Analysis.
Gene expression array text files from GEO accession no. GSE104244 were analyzed as depicted in Fig. 2. The R/Bioconductor package limma was used to process expression data. Within- and between-array normalizations were performed using the loess and aquantile methods, respectively. The normexp option was used for background correction. Raw files read in using the read.maimages function. Log2 fold change in transcription for drug treated conditions over mock treated was obtained for each sample at each time point studied. Ranked lists of log2 fold change were analyzed using Gene Set Enrichment Analysis (GSEA) by the Broad Institute and data packages (Reactome) and a custom HRD associated genes as defined in ref. 39. This gene set was split into up-regulated and down-regulated in HRD as used as input into GSEA (66). Log2 fold change gene expression for 5-AZA over Mock was used for unsupervised hierarchical clustering by Euclidean distance and Z score for gene expression. For heat maps depicted, the following gene lists were used as input: DNA repair, REACTOME DNA REPAIR PATHWAY; Fanconi anemia, MATHEW FANCONI ANEMIA GENES, PID FANCONI PATHWAY, and REACTOME FANCONI ANEMIA PATHWAY.
Quantitative Real-Time PCR.
NSCLC cells were treated for the indicated number of days with vehicle or 5-AZA. RNA was isolated by NucleoSpin RNA (Macherey-Nagel) and converted to cDNA by High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) per the manufacturers’ instructions. Quantitative real-time PCR was performed using Power SYBR Green Master Mix (Applied Biosystems) on a CFX96 Real Time System (BioRad). Analysis was performed using CFX Maestro software (BioRad).
Immunoblotting.
Refer to SI Appendix, Supplemental Experimental Procedures, for details.
Colony-Forming Assay.
NSCLC cells were plated in triplicate in 6-well plates at low density (200 cells per well) and treated for 10 d with 5-AZA and/or TAL (or vehicle). Irradiation was initiated on day 8 as a 2-Gy single dose or for 3 consecutive days (totalling 6 Gy exposure). Colonies were stained with 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyltetrazolium chloride 10% (wt/vol) in 70% ethanol for 2 min and counted using an automated colony counter (Synbiosis).
Determination of Synergism.
Cells were plated on 96-well plates and treated daily with indicated concentrations of drugs alone or in combination at a constant ratio (1:20 TAL:5-AZA). On day 7, assays were terminated using CellTiter96 MTS Reagent (Promega). Absorbance values were used to determine the fraction of cells affected in each treatment and determine combination indices according to the Chou–Talalay method using CompuSyn software.
Immunofluorescence.
Refer to SI Appendix, Supplemental Experimental Procedures, for details.
Proximity Ligation Assay (PLA).
Refer to SI Appendix, Supplemental Experimental Procedures, for details.
DNA Fiber Analysis.
Refer to SI Appendix, Supplemental Experimental Procedures, for details.
Extrachromosomal Homologous Recombination and Nonhomologous End-Joining Assays.
Refer to SI Appendix, Supplemental Experimental Procedures, for details.
DSB Repair Reporter Assays.
Refer to SI Appendix, Supplemental Experimental Procedures, for details.
Determination of Apoptosis by Annexin V Staining.
Refer to SI Appendix, Supplemental Experimental Procedures, for details.
Xenograft Models.
Refer to SI Appendix, Supplemental Experimental Procedures, for details.
Statistical Analysis.
Statistical analysis for biological assays and xenograft studies was determined using Graphpad Prism software to calculate 2-tailed unpaired t test or 1-way or 2-way ANOVA as appropriate. All data are presented as mean ± SEM.
Supplementary Material
Acknowledgments
Our studies have been supported by funding from the National Cancer Institute (NCI) University of Maryland Cancer Canter Support Grant (P30CA134274 to R.A. and F.V.R.), NCI Grant R25CA186872 (to R.G.), Van Andel Research Institute-Stand Up 2 Cancer (R.A., M.J.T., S.B.B., and F.V.R.), the Adelson Medical Research Foundation (R.A., M.J.T., F.V.R., and S.B.B.), and Maryland Cigarette Restitution funds (R.A. and F.V.R.).
Footnotes
The authors declare no competing interest.
See Commentary on page 22429.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903765116/-/DCSupplemental.
References
- 1.Lord C. J., Ashworth A., PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murai J., et al. , Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ying S., Hamdy F. C., Helleday T., Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 72, 2814–2821 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Saleh-Gohari N., et al. , Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 25, 7158–7169 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helleday T., The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol. Oncol. 5, 387–393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farmer H., et al. , Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Bryant H. E., et al. , Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Lord C. J., Ashworth A., BRCAness revisited. Nat. Rev. Cancer 16, 110–120 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Shall S., Gaymes T., Farzaneh F., Curtin N. J., Mufti G. J., “The use of PARP inhibitors in cancer therapy: Use as adjuvant with chemotherapy or radiotherapy, use as a single sgent in susceptible patients, and techniques used to identify susceptible patients” in Poly(ADP-Ribose) Polymerase: Methods and Protocols, Tulin A. V., Ed. (Methods in Molecular Biology, Springer New York, New York, NY, 2017), pp. 343–370. [DOI] [PubMed] [Google Scholar]
- 10.Albert J. M., et al. , Inhibition of poly(ADP-ribose) polymerase enhances cell death and improves tumor growth delay in irradiated lung cancer models. Clin. Cancer Res. 13, 3033–3042 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Senra J. M., et al. , Inhibition of PARP-1 by olaparib (AZD2281) increases the radiosensitivity of a lung tumor xenograft. Mol. Cancer Ther. 10, 1949–1958 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levra M. G., Olaussen K. A., Novello S., Soria J. C., PARP inhibitors: An interesting pathway also for non-small cell lung cancer? Curr. Pharm. Des. 20, 3875–3882 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Shen Y., et al. , BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res. 19, 5003–5015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murai J., et al. , Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 13, 433–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith M. A., et al. , Synergistic activity of PARP inhibition by talazoparib (BMN 673) with temozolomide in pediatric cancer models in the pediatric preclinical testing program. Clin. Cancer Res. 21, 819–832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bono J., et al. , Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov. 7, 620–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiffmann I., Greve G., Jung M., Lübbert M., Epigenetic therapy approaches in non-small cell lung cancer: Update and perspectives. Epigenetics 11, 858–870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo C. B., Jones P. A., Epigenetic therapy of cancer: Past, present and future. Nat. Rev. Drug Discov. 5, 37–50 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Santi D. V., Norment A., Garrett C. E., Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc. Natl. Acad. Sci. U.S.A. 81, 6993–6997 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Champion C., et al. , Mechanistic insights on the inhibition of c5 DNA methyltransferases by zebularine. PLoS One 5, e12388 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel K., et al. , Targeting of 5-aza-2’-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 38, 4313–4324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juergens R. A., et al. , Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 1, 598–607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai H. C., et al. , Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 21, 430–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H. J., et al. , DNMT (DNA methyltransferase) inhibitors radiosensitize human cancer cells by suppressing DNA repair activity. Radiat. Oncol. 7, 39 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zampieri M., et al. , Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One 4, e4717 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reale A., Matteis G. D., Galleazzi G., Zampieri M., Caiafa P., Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene 24, 13–19 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Caiafa P., Guastafierro T., Zampieri M., Epigenetics: Poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. 23, 672–678 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Muvarak N. E., et al. , Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents—A potential therapy for cancer. Cancer Cell 30, 637–650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haince J.-F., et al. , PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197–1208 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Ha K., et al. , Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery. Hum. Mol. Genet. 20, 126–140 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pulliam N., et al. , An effective epigenetic-PARP inhibitor combination therapy for breast and ovarian cancers independent of BRCA mutations. Clin. Cancer Res. 24, 3163–3175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.American Cancer Society, Key statistics for lung cancer, https://www.cancer.org/cancer/non-small-cell-lung-cancer/about/key-statistics.html. Accessed 16 November 2018.
- 33.Chou T. C., “The median-effect principle and the combination index for quantitation of synergism and antagonism” in Synergism and Antagonism in Chemotherapy, Chou T. C., Rideout D. C., Eds. (Academic Press, San Diego, CA, 1991), pp. 61–102. [Google Scholar]
- 34.Chou T.-C., Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70, 440–446 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Shen Y., Aoyagi-Scharber M., Wang B., Trapping poly(ADP-ribose) polymerase. J. Pharmacol. Exp. Ther. 353, 446–457 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Söderberg O., et al. , Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Petermann E., Orta M. L., Issaeva N., Schultz N., Helleday T., Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo L. J., Yang L. X., Gamma-H2AX—A novel biomarker for DNA double-strand breaks. In Vivo 22, 305–309 (2008). [PubMed] [Google Scholar]
- 39.Peng G., et al. , Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat. Commun. 5, 3361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogliolo M., Surrallés J., The Fanconi Anemia/BRCA Pathway: FANCD2 at the Crossroad between Repair and Checkpoint Responses to DNA Damage (Landes Bioscience, 2013). https://www.ncbi.nlm.nih.gov/books/NBK6087/. Accessed 5 November 2018. [Google Scholar]
- 41.Vasileva A., Linden R. M., Jessberger R., Homologous recombination is required for AAV-mediated gene targeting. Nucleic Acids Res. 34, 3345–3360 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierce A. J., Johnson R. D., Thompson L. H., Jasin M., XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan W., et al. , Fanconi anemia repair pathway dysfunction, a potential therapeutic target in lung cancer. Front. Oncol. 4, 368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bee L., Fabris S., Cherubini R., Mognato M., Celotti L., The efficiency of homologous recombination and non-homologous end joining systems in repairing double-strand breaks during cell cycle progression. PLoS One 8, e69061 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mari P.-O., et al. , Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. U.S.A. 103, 18597–18602 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weterings E., et al. , A novel small molecule inhibitor of the DNA repair protein Ku70/80. DNA Repair (Amst.) 43, 98–106 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Kais Z., et al. , FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 15, 2488–2499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michl J., Zimmer J., Tarsounas M., Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 35, 909–923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.NCCN Guidelines for Patients , Early and Locally Advanced Lung Cancer, https://www.nccn.org/patients/guidelines/lung-early-stage/32/index.html. Accessed 24 September 2019.
- 50.Mahaney B. L., Meek K., Lees-Miller S. P., Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417, 639–650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Branzei D., Foiani M., Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9, 297–308 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Rothkamm K., Krüger I., Thompson L. H., Löbrich M., Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23, 5706–5715 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Topper M. J., et al. , Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171, 1284–1300.e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campaner S., Amati B., Two sides of the Myc-induced DNA damage response: From tumor suppression to tumor maintenance. Cell Div. 7, 6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luoto K. R., et al. , Tumor cell kill by c-MYC depletion: Role of MYC-regulated genes that control DNA double-strand break repair. Cancer Res. 70, 8748–8759 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Adachi S., et al. , c-Myc is necessary for DNA damage-induced apoptosis in the G(2) phase of the cell cycle. Mol. Cell. Biol. 21, 4929–4937 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Issa J.-P. J., Kantarjian H. M., Targeting DNA methylation. Clin. Cancer Res. 15, 3938–3946 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L. H., Olin E. J., Buskirk H. H., Reineke L. M., Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer Res. 30, 2760–2769 (1970). [PubMed] [Google Scholar]
- 59.Schaefer M., Hagemann S., Hanna K., Lyko F., Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 69, 8127–8132 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Alexandrov A., et al. , Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21, 87–96 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Francia S., et al. , Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488, 231–235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu W.-T., et al. , Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 9, 532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thapar R., Regulation of DNA double-strand break repair by non-coding RNAs. Molecules 23, E2789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Britton S., Coates J., Jackson S. P., A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 202, 579–595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richardson C. D., et al. , CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 50, 1132–1139 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.