Significance
After RNA synthesis is initiated by RNA polymerase II (pol II) and general transcription factors (GTFs), pol II dissociates from GTFs and promoter DNA and undergoes the transition to productive elongation. This transition is generally referred to as promoter escape and is a major rate-limiting step at many mRNA genes, but key details have remained unclear. We have developed an in vitro reconstituted transcription system that has high initiation efficiency, which allowed us to dissect the timing of this transition. We found that the formation of the initially transcribing complex, containing pol II, general transcription factors, and a nascent transcript, is long persisting and that 2 elongation factors conserved across eukaryotes have robust and positive biochemical activities on this transition.
Keywords: transcription, RNA polymerase II, preinitiation complex, promoter escape, capping enzyme
Abstract
After synthesis of a short nascent RNA, RNA polymerase II (pol II) dissociates general transcription factors (GTFs; TFIIA, TFIIB, TBP, TFIIE, TFIIF, and TFIIH) and escapes the promoter, but many of the mechanistic details of this process remain unclear. Here we developed an in vitro transcription system from the yeast Saccharomyces cerevisiae that allows conversion of the preinitiation complex (PIC) to bona fide initially transcribing complex (ITC), elongation complex (EC), and reinitiation complex (EC+ITC). By biochemically isolating postinitiation complexes stalled at different template positions, we have determined the timing of promoter escape and the composition of protein complexes associated with different lengths of RNA. Almost all of the postinitiation complexes retained the GTFs when pol II was stalled at position +27 relative to the transcription start site, whereas most complexes had completed promoter escape when stalled at +49. This indicates that GTFs remain associated with pol II much longer than previously expected. Nevertheless, the long-persisting transcription complex containing RNA and all of the GTFs is unstable and is susceptible to extensive backtracking of pol II. Addition of the capping enzyme and/or Spt4/5 significantly increased the frequency of promoter escape as well as assembly of a follow-on PIC at the promoter for reinitiation. These data indicate that elongation factors play an important role in promoter escape and that ejection of TFIIB from the RNA exit tunnel of pol II by the growing nascent RNA is not sufficient to complete promoter escape.
In eukaryotic transcription, RNA polymerase II (pol II) and a set of general transcription factors (GTFs), including TFIIA, TBP, TFIIB, TFIIE, TFIIF, and TFIIH, assemble in a preinitiation complex (PIC) that is responsible for promoter opening and scanning of transcription start sites (TSSs) (1, 2). Once a TSS is recognized, pol II begins to synthesize a nascent RNA, thereby converting the PIC into the initially transcribing complex (ITC), which is composed of all of the GTFs, pol II, and RNA. The transcription bubble is propagated downstream until the nascent RNA reaches a certain length. The upstream segment of the bubble then abruptly reanneals, resulting in dissociation of all of the GTFs except TFIIF from the ITC (3). This causes the ITC to be converted to an elongation complex (EC), which only contains pol II, TFIIF, and RNA, and this conversion step is known as promoter escape. Additional pol II and TFIIF can subsequently be recruited to the promoter to enable reinitiation of transcription using the other GTFs that remained committed to the template (4).
Once the nascent transcript reaches a length of ∼20 to 30 nt in vivo (5, 6) and ∼20 nt in vitro, 5′ capping of the nascent RNA occurs (7–9). This is shortly after the 5′ end of the transcript has emerged from the pol II RNA exit tunnel. In yeast (Saccharomyces cerevisiae), RNA 5′ capping involves 3 steps: 1) removal of a gamma phosphate from the 5′ end of the RNA by Cet1, 2) transfer of guanosine monophosphate (GMP) by Ceg1, and 3) methylation of the guanosine by Abd1 (10, 11). Cet1 and Ceg1 form a heterodimer and are recruited to the transcription complex upon binding phosphorylated Ser5 of the pol II C-terminal domain (CTD) (12–14). Shortly after recruitment of Cet1-Ceg1, Spt4/5 (the yeast homolog of DSIF) also binds pol II (15) and facilitates productive elongation in vivo. It has been long assumed that promoter escape occurs after synthesis of 9 to 15 nt RNA, thus preceding the recruitment of Cet1-Ceg1 and Spt4/5, but this assumption has not been experimentally proven.
To better characterize the early steps in transcription, we previously developed an in vitro reconstituted system from yeast (S. cerevisiae) purified factors in which ∼10 to 30% of the assembled PICs were capable of RNA synthesis (0.1 to 0.3 transcripts per template) (16, 17). Single-molecule analysis using this system indicated that pol II can remain associated with promoter DNA (via interactions with GTFs) even when transcribing RNAs of ∼50 nt in length (18). This result is inconsistent with promoter escape occurring when the RNA reaches 9 to 15 nt in length. Nevertheless, as only pol II and GTFs were present in these reactions, we hypothesized that additional factors, e.g., capping enzymes and/or Spt4/5, may play a role in promoter escape.
In this study, we first improved our in vitro transcription system and have now achieved at least 90% efficiency (by determining the extent of template usage). We then used this system to reexamine the mechanism of transcription initiation, specifically the characteristics of the ITC in the absence and presence of capping enzymes (Cet1-Ceg1 and Abd1) and Spt4/5. Transcription complexes were stalled at various positions on a G-less SNR20 promoter and isolated by glycerol gradient sedimentation. This revealed the composition of transcription complexes associated with different lengths of RNA. Our data indicate that the ITC, which contains all of the GTFs, pol II, and RNA, persists at least until Cet1-Ceg1 and Spt4/5 are recruited. Cet1-Ceg1 and Spt4/5 then facilitate the transition from initiation to elongation by promoting promoter escape.
Results
In Vitro Reconstituted Transcription System with ∼90% Efficiency.
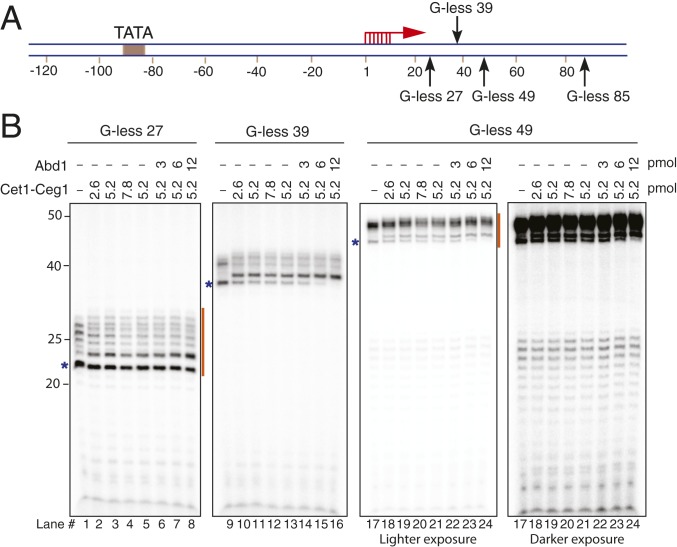
We previously developed a transcription initiation reconstituted system using yeast proteins that had been purified from Escherichia coli or yeast (SI Appendix, Fig. S1) and showed that 10 to 30% of the assembled PICs were active (16, 17). To reveal additional insights into the transition from initiation to elongation, we set out to further optimize this system and achieve higher transcription efficiency. We generated a set of U2 snRNA promoter (SNR20) variants that have a G-free region between the TSS (+1) and a G-stop at +27, +39, +49, or +85 (named G-less 27, G-less 39, G-less 49, and G-less 85, respectively) (Fig. 1A). Inclusion of chain-terminating 3′-O-methyl GTP instead of GTP in the reactions enabled pol II to be efficiently stalled at the end of the G-free region. We thus reasoned that this approach should allow accurate quantification of the efficiency of initiation, including independent measurements of the efficiencies of the first round of transcription vs. reinitiation. It should nevertheless be noted that the SNR20 promoters yielded various lengths of transcripts due to multiple TSSs at positions +1 to +7 (Fig. 1B).
Fig. 1.
Transcription initiation assay with a series of G-less SNR20 promoter variants. (A) Schematic diagram of the SNR20 promoter variants. The TSSs (red arrows) and G-stops (black arrows) are indicated. (B) PIC was formed on the indicated SNR20 promoter DNA variants with TFIIA, TFIIB, TBP, TFIIE, TFIIF, TFIIH, Sub1, and pol II. Transcription was initiated by addition of ATP, CTP, 3′-O-methyl GTP, UTP, and [α-32P]UTP and then incubated for 20 min at 30 °C. Varying concentrations of Cet1-Ceg1 and Abd1 were added at the time of addition of NTPs. Orange lines indicate the sets of transcripts that were used to calculate transcription efficiency in Table 1. The bands indicated by blue asterisks are used for quantification of the shift in RNA mobility in SI Appendix, Fig. S3A.
By adjusting the concentrations of factors added to the reactions (SI Appendix, Fig. S2 A–C), we identified conditions that allowed ∼90% efficiency (by determining the extent of template usage, defined as the percentage of DNA templates that were transcribed) from the G-less SNR20 templates as measured by incorporation of [α-32P] UTP into nascent transcripts (Table 1). As in our previous studies (16, 17), we use almost equimolar amounts of the GTFs (TFIIA, TFIIB, TBP, TFIIE, TFIIH, and TFIIK) and promoter DNA. Here, however, we found that the efficiency increased by ∼2- to 3-fold upon addition of 4-fold molar excess pol II and TFIIF relative to DNA (SI Appendix, Fig. S2A) and by ∼1.4-fold in the presence of Sub1 (SI Appendix, Fig. S2B), the yeast homolog of PC4 (19, 20). No increase in efficiency was observed upon titrating other GTFs (TFIIA, TFIIB, TFIIE, TFIIH, or TFIIK) (SI Appendix, Fig. S2C), and thus, all subsequent analyses were done using reactions that contain Sub1 along with excess TFIIF and pol II. As expected, the level of nascent transcripts produced increased over time but peaked once the reactions had been incubated for 20 min (SI Appendix, Fig. S2 D and E). At this time point, the efficiency of the first round of transcription was very high, ranging from 0.9 to 0.98 depending on the promoter variant examined (Table 1).
Table 1.
Transcription efficiency (efficiency of template usage) with SNR20 promoter
| DNA template | Efficiency from first round of initiation (%) ± SEM | Efficiency from second round of initiation (%) ± SEM |
| G-less 27 | 98.9 ± 6.6 | NA |
| G-less 49 | 90.8 ± 10.0 | 28.4 ± 1.4 |
Transcription was initiated in the presence of Sub1 (3 pmol) and 4-fold molar excess pol II and TFIIF (5.2 pmol) relative to DNA (1.3 pmol). The efficiency of template usage, defined as the percentage of templates that were transcribed, was calculated based on the incorporation of radiolabeled UTP (Methods). G-less 27, n = 3; G-less 49, n = 5.
Reconstituted Transcription Initiation System Supports 5′ Capping and Reinitiation.
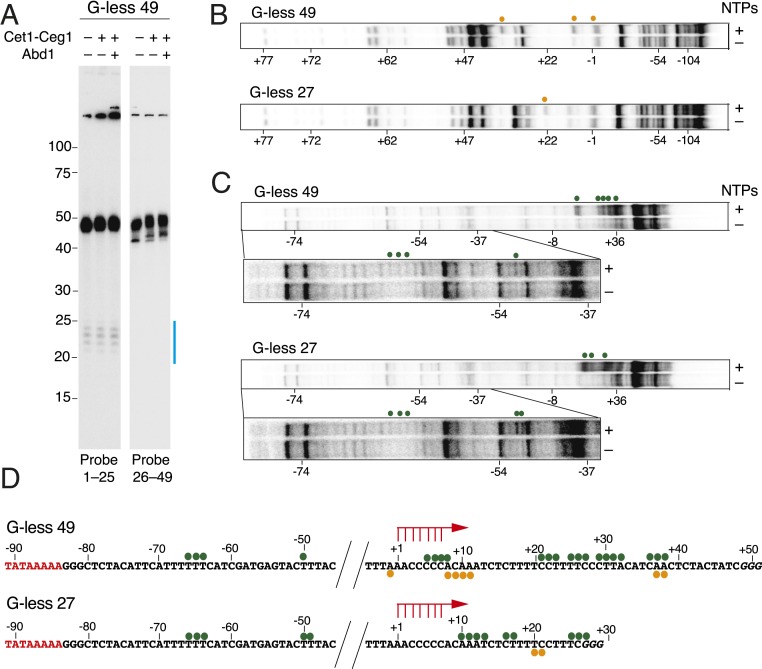
To further define the capabilities of the optimized transcription initiation system, we tested whether transcripts generated from the G-less templates become capped by 3′-O-methyl GTP and recombinant Cet1-Ceg1 and Abd1, which were added to the initiation reactions at the time of NTP addition. Given that Abd1 might have roles in transcription initiation independent of its methylation activities (11, 21), S-adenosyl methionine (SAM), which is required for methylation of the cap, was not included unless otherwise noted. A shift in RNA mobility by ∼1 nt was observed upon addition of Cet1-Ceg1, consistent with addition of a 5′ cap, and this effect was enhanced by addition of Abd1, Spt4/5, and/or SAM (Fig. 1B and SI Appendix, Fig. S3 A and B). To quantitate the extent of 5′ capping, RNAs were isolated from the transcription reactions and then treated with CIP/PNK followed by digestion with a 5′-3′ exonuclease (SI Appendix, Fig. S4A). In the absence of Cet1-Ceg1, RNAs had triphosphorylated 5′ ends and were susceptible to digestion by the exonuclease, as expected (SI Appendix, Fig. S4 B and C). In contrast, when a 4-fold molar excess of Cet1-Ceg1 relative to DNA was added to the reactions, ∼41% of the G-less 27 (SI Appendix, Fig. S4B) and ∼80% of the G-less 49 (SI Appendix, Fig. S4C) transcripts from the first round of initiation became capped, consistent with the efficiencies estimated from the ∼1-nt shifts in RNA mobility (SI Appendix, Fig. S3A). The difference in capping efficiency between templates is likely due to the 5′ ends of the shorter transcripts being less accessible for capping (discussed further below). It should be noted that a similar ∼1-nt shift in RNA mobility was observed when capping enzymes were added after transcription had been completed, confirming that the mobility shift was indeed due to capping and not a shift in the TSS (SI Appendix, Fig. S4D). Based on these results, we conclude that our transcription system supports 5′ capping.
Upon further examining the transcripts generated from the different promoter templates, we noted that the G-less 49 (Fig. 1B, lanes 17 to 24) and G-less 85 templates (SI Appendix, Fig. S5) produced transcripts with ∼25 nt stepwise decrease in length. These results are analogous to a previous study that showed successive pol II stacking when pol II was stalled in an in vitro human transcription system (22). This ∼25-nt decrease in length is also in good agreement with previous in vitro footprinting analysis of 2 colliding pol II ECs (23). We thus reasoned that the shorter transcripts may represent products of transcription reinitiation. To address this hypothesis, we focused on the G-less 49 template that yielded ∼49- and ∼25-nt transcripts (Fig. 1B, lanes 17 to 24). Consistent with reinitiation from the same TSS as the 49-nt transcripts, Northern blot analysis revealed that the ∼25-nt transcripts could be detected with a probe antisense to nt 1 to 25 but not with a probe to nt 26 to 49 (Fig. 2A). These data strongly suggest that transcription from the G-less 49 template results in a pol II EC that has escaped the promoter and is stalled at +49 and that another pol II has initiated transcription by reutilizing the promoter to generate the ∼25-nt RNAs.
Fig. 2.
Reconstituted transcription initiation system supports 5′ capping and reinitiation. (A) To confirm reinitiation from the G-less 49 promoter, Northern blot analysis was performed on G-less 49 transcripts that were generated in the presence or absence of Cet1-Ceg1 and Abd1. Probes antisense to nt 1 to 25 or nt 26 to 49 were used. Transcripts generated by the second round of transcription are indicated by a blue line. (B and C) Potassium permanganate (KMnO4) footprinting assays with the G-less 49 (Upper) and the G-less 27 (Lower) templates (−122/+97) to detect single-stranded regions. The 5′ ends of the template (B) or nontemplate (C) strands were labeled with 32P. After incubation of the initiation reactions for 20 min, 18 mM KMnO4 was added. In C, an ∼50-bp region downstream of the TATA box is enlarged and shown with darker exposure (Lower). KMnO4 reactive positions on template and nontemplate strands are indicated by orange and green dots, respectively. (D) G-less 49 (Top) and G-less 27 (Bottom) DNA sequences showing KMnO4 reactive residues. The TATA box is shown in red, and TSSs are indicated by red arrows.
To further support reinitiation, we mapped the location of pol II molecules on the G-less templates by performing potassium permanganate (KMnO4) footprinting. G-less 27 and G-less 49 DNA with the 5′ ends of template (Fig. 2B) or nontemplate (Fig. 2C) strands were radiolabeled, and the DNA bound by stalled transcription complexes were reacted with KMnO4. The DNA templates were then cleaved at reactive residues by treatment with piperidine and analyzed by denaturing PAGE gel electrophoresis (Fig. 2 B and C). The increase in KMnO4 reactivity in the presence of NTPs (i.e., posttranscription complexes) compared to in the absence of NTPs (i.e., PIC) was observed at residues downstream of TSS where pol II is stalled (Fig. 2D). On the G-less 27 template, an ∼17-bp KMnO4 hyperreactive region from residues +10 to +27 was observed (Fig. 2 B and C, Bottom), which is consistent with the pol II active center being localized at the stall position. A more extended KMnO4 hyperreactive region (residues −1 to +38) was observed on the G-less 49 template (Fig. 2 B and C, Top). Although the signals are faint, residues ∼20 to 40 bp downstream of the TATA box (residues −66 to −49) were also slightly reactive to KMnO4 on both the G-less 27 and G-less 49 templates (Fig. 2 B–D). This reactive region is consistent with the location where initial melting occurs through the translocase activity of TFIIH (17, 18, 24, 25) and could be indicative of the presence of ITCs (24, 26, 27). Taken together, we conclude that the improved transcription initiation system can support 5′ capping as well as reinitiation (and validation by gradient sedimentation is described below that further supports that reinitiation is indeed occurring). Of note, the efficiency of the first round of initiation from the G-less 49 template is ∼90%, but reinitiation is only ∼28% efficient (Table 1).
Promoter Escape Is Nearly Completed When Pol II Is Stalled at +49.
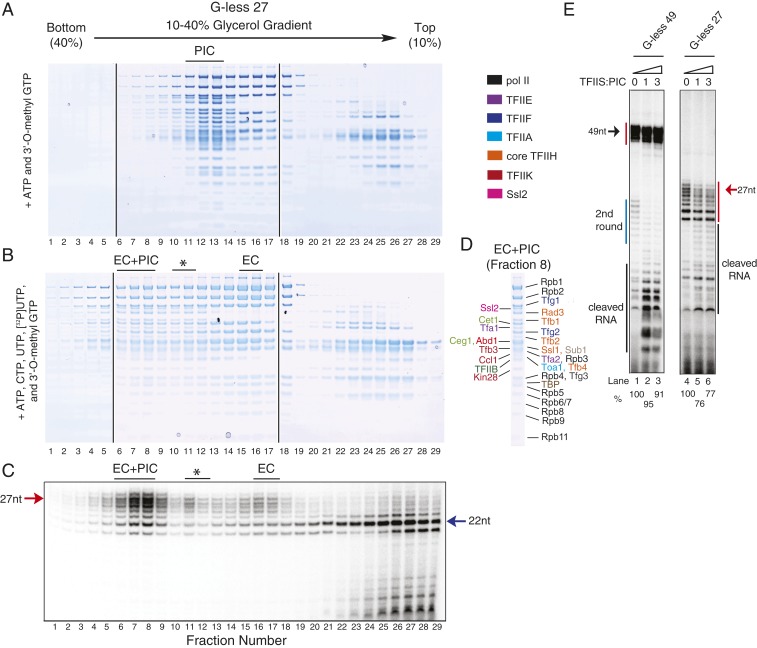
To define the timing of promoter escape as well as how quickly the subsequent pol II can associate with the template, we sought to use glycerol gradient sedimentation to isolate complexes stalled at different template positions. After completion of the transcription reactions but before gradient sedimentation, nonhydrolyzable ATP (AMP-PNP) was added to potentially inhibit the translocase activity of TFIIH (26). This was done to minimize structural changes during glycerol gradient sedimentation while retaining the structural assembly (28). Sedimentation gradients were then fractionated and protein stoichiometry in each fraction was analyzed by SDS/PAGE. As a control, we first analyzed the sedimentation of the PIC and capping enzymes on the G-less 49 template under conditions where only ATP and 3′-O-methyl GTP (but not CTP or UTP) were added, and thus there should be only abortive RNA (∼2 nt) synthesis (Fig. 3A). Note that ATP was added because the PIC may behave as a slightly larger complex in the presence of ATP due to the binding of the capping enzymes through the phosphorylated pol II CTD (12, 13, 29). In a glycerol gradient, we found that the capping enzymes and pol II interacted at nearly a 1:1 molar ratio irrespective of presence of transcripts that extend outside the RNA exit tunnel of pol II (Fig. 3 A–C). In contrast, when TFIIK activity was inhibited, no CTD phosphorylation was observed (SI Appendix, Fig. S6 A–C), and the capping enzymes no longer cosedimented with the PIC (SI Appendix, Fig. S6 D and E). It should be noted that excess pol II and TFIIF present in the reactions had no effect on sedimentation of the PIC (compare the sedimentation profiles in Fig. 3A to Fig. 4A, fractions 10 to 13) and instead accumulated in fractions 14 to 17 (Fig. 3 A and C).
Fig. 3.
Separation of the postinitiation complexes stalled at +49. (A) PIC was assembled with 4-fold excess pol II and TFIIF relative to G-less 49 template DNA and then combined with ATP, 3′-O-methyl GTP, 4-fold excess Cet1-Ceg1, and 8-fold excess Abd1 (relative to the DNA). Reactions were incubated for 20 min at 30 °C, combined with 2 mM AMP-PNP, and sedimented on a 10 to 40% glycerol gradient; ∼130 µL per fraction were isolated and then analyzed by SDS/PAGE. (B and C) Protein identification of the PIC in fraction 11 (B) and free pol II-TFIIF in fraction 15 (C). (D) PIC was assembled in the same manner as in A. Transcription was initiated by addition of ATP, CTP, 3′-O-methyl GTP, UTP, [α-32P]UTP, Cet1-Ceg1, and Abd1. The postinitiation complexes were combined with 2 mM AMP-PNP, sedimented on a gradient, and analyzed as in A. (E) RNA analysis of fractions isolated in D by denaturing Urea-PAGE. The transcripts (∼25 nt) from the second round of initiation are indicated by a blue line. (F–H) Protein identification of the EC+PIC and EC+ITC in fraction 8 (F), EC in fraction 15 (G), and ITC in fraction 11 (H). Asterisk in D, E, and H indicates the presence of the ITC.
Fig. 4.
Separation of the postinitiation complexes stalled at +27 that are prone to extensive backtracking. (A and B) Separation and SDS/PAGE analysis of PICs (A) and postinitiation complexes (B) with G-less 27 DNA template were performed in the same manner as in Fig. 3 except that the control PIC in A was assembled with only 1.4-fold excess pol II and TFIIF relative to G-less 27 DNA template DNA and received 1.5-fold excess Cet1-Ceg1 and Abd1 (relative to the DNA). (C) RNA analysis of fractions from B by denaturing Urea-PAGE. The 27- and 22-nt transcripts are indicated by red and blue arrows, respectively. Asterisk in B and C indicates the presence of the ITC. (D) Protein identification of EC+PIC in fraction 8. (E) TFIIS cleavage assay of complexes stalled at promoter proximal positions. Transcription initiation assays with G-less 49 (Left) or G-less 27 (Right) DNA templates were performed as described in Fig. 1. After 20 min, reactions were combined with the indicated concentrations of TFIIS and incubated for another 6 min, and cleaved transcripts were analyzed by denaturing PAGE. Note that the cleaved RNAs indicated by black lines were not used for determining how far pol II backtracks as many of these RNAs are 3′ fragments that were generated by partial backtracking followed by TFIIS-induced cleavage and subsequent resumption of transcription. Thus full-length transcripts indicated by red lines were quantified, and the amount compared to the control (0 pmol TFIIS) is shown below the gel.
We next compared this sedimentation profile to that obtained from G-less 49 transcription reactions that had postinitiation complexes stalled at +49 (Fig. 3D). Analysis of the RNAs present in each fraction revealed 2 major (fractions 5 to 8 and 14 to 17) and 1 minor (fractions 11 and 12) postinitiation complex (Fig. 3E). Notably, the complex that migrated fastest (fractions 5 to 8) contained all of the GTFs, pol II, and capping enzymes (Fig. 3F) but with pol II and the TFIIF subunits present at ∼1.6- and 1.5-fold molar excess, respectively, compared to their levels in the PIC from Fig. 3A (quantification in SI Appendix, Fig. S7). This suggests that this postinitiation complex underwent the transition from initiation to promoter escape, thereby allowing binding of a second pol II. Indeed, both ∼49- and ∼25-nt transcripts, representing the first and second rounds of transcription, respectively, were observed in fractions 5 to 8 (Fig. 3E and SI Appendix, Fig. S8). The molar ratio of the first and second rounds of transcripts was estimated as ∼5:1.8 based on the band intensities, indicating that the complex is a mixture of EC+PIC (∼64%) and EC+ITC (∼36%) (SI Appendix, Fig. S8). The second major postinitiation complex (fractions 14 to 17 in Fig. 3 D and E) is EC containing pol II, TFIIF, Cet1-Ceg1, Abd1, and ∼49 nt transcripts (but not TFIIE, TFIIH, TFIIA, and TBP) (Fig. 3G). Note that TFIIB, which is not compatible with EC, appeared in these fractions due to comigration of free pol II-TFIIF-TFIIB that did not engage in transcription (compare Fig. 3 C and G). The third postinitiation complex (fractions 11 and 12 in Fig. 3 D and E) migrated as fast as the control PIC (Fig. 3 A and B) and contained the entire complement of the PIC polypeptides, capping enzymes, and ∼49-nt RNAs (Fig. 3H). This complex is thus likely to be the ITC, which has not undergone promoter escape, as evidenced by the presence of all PIC components, in contrast to the other 2 postinitiation complexes. However, the third complex is much less populated than the other 2 postinitiation complexes, and thus, we conclude that promoter escape usually occurs before +49.
Promoter Escape Is Often Completed When the RNA Length Is Longer than 22 nt.
To then further clarify the timing of promoter escape, we sedimented G-less 27 PIC (Fig. 4A) and postinitiation complexes (Fig. 4 B and C) and analyzed whether promoter escape had been completed when pol II was stalled at +27. Similar to the results obtained with the G-less 49 template, a fast migrating postinitiation complex was observed (fractions 6 to 9 in Fig. 4B) that contained mainly ∼24- to 27-nt transcripts (Fig. 4C) as well as all of the GTFs, pol II, and capping enzymes (Fig. 4D). Subunits of pol II and TFIIF were present at 1.9- and 1.6-fold molar excess compared to those in PIC from Fig. 4A (quantification in SI Appendix, Fig. S9), suggesting that the first pol II underwent promoter escape to allow a second pol II to bind. Given the ∼25-nt spacing between pol II molecules that was observed on the G-less 49 template (Figs. 1 and 2), any reinitiated products from the G-less 27 template should be ∼3 nt or shorter. This length is too short to be retained in a transcription complex (3), and thus, we assigned this complex exclusively to EC+PIC (hereafter referred to as the reinitiation complex). Besides this major complex present in fractions 6 to 9, we observed a smaller number of G-less 27 transcription complexes that converted to ECs (Fig. 4 B and C, fractions 15 to 17) and ITC (Fig. 4 B and C, fraction 11).
The 24- to 27-nt transcripts were predominately retained in the reinitiation complex (Fig. 4C, fractions 6 to 9), but 21- and 22-nt transcripts (derived from initiation from TSSs downstream of +1) largely remained on the top of the gradient after sedimentation (Fig. 4C, fractions 25 to 29). These RNAs may have been released from either ITCs or ECs, but we thought that ITCs seemed more likely as it has been previously suggested that the EC is very stable (30). To confirm these prior observations about the EC, we formed artificial ECs on the G-less 27 and G-less 49 templates by preannealing a 9-mer RNA at the TSS and then adding TFIIF, TFIIB, ATP, CTP, UTP, and 3′-O-methyl GTP to allow the 9-nt RNA to be elongated in the same conditions as the transcription initiation assay (SI Appendix, Fig. S10 A–C). Glycerol gradient sedimentation revealed that the 22-, 27-, and 49-nt RNAs were all predominantly present in the center of the gradient (SI Appendix, Fig. S10C), confirming that elongated RNAs are stably retained in ECs. Based on these data, it is highly likely that the 21- and 22-nt RNAs that did not migrate into the gradient (Fig. 4C, fractions 25 to 29) were associated with ITCs but were then released from the complex during their isolation. These data suggest that promoter escape usually happens after the nascent RNA is longer than 22 nt in length.
Pol II in the ITC Is Susceptible to Extensive Backtracking.
A reconstituted human system previously demonstrated that 1) short (<9-nt) RNAs can be released from the ITC when it reconverts to PIC upon addition of nonhydrolyzable ATP (26) and 2) complexes stalled at promoter proximal positions (up to ∼32 nt) are susceptible to extensive backtracking (31, 32). We, therefore, reasoned that RNA release from the yeast ITC may similarly be due to extensive pol II backtracking that is caused by the addition of nonhydrolyzable ATP at the end of the transcription reaction and/or removal of ATP during glycerol gradient sedimentation. To explore this idea, we first used TFIIS cleavage assays to directly examine whether pol II backtracking occurs in the stalled transcription complexes (Fig. 4E). TFIIS stimulates the intrinsic activity of pol II to cleave the 3′ end of the transcript when pol II backtracks, allowing for replacement of the new 3′ end at the poI II active site and transcription restart (33, 34). Transcription complexes that had been stalled on the G-less 27 or G-less 49 template were combined with TFIIS and incubated for an additional 6 min, and the resulting transcripts were analyzed on a denaturing RNA gel (Fig. 4E). The level of ∼49-nt transcripts derived from the G-less 49 template was largely insensitive to addition of TFIIS, whereas about 24% of the transcripts derived from the G-less 27 template were degraded upon addition of TFIIS (Fig. 4E, lanes 4 to 6). The short (∼25-nt) transcripts from the second round of transcription on the G-less 49 template were also highly sensitive to TFIIS (Fig. 4E, lanes 1 to 3). These results indicate that transcription complexes stalled at promoter proximal positions (up to approximately +27) are prone to extensive backtracking, whereas little backtracking is observed with the EC transcribing ∼49-nt RNA.
Next, to address whether the observed extensive backtracking is an inherent feature of the ITC but not the EC, the artificial EC on the G-less 27 template was subjected to TFIIS cleavage assays (SI Appendix, Fig. S10 D and E). The ∼27-nt transcripts in the artificial EC were, as expected, cleaved by ∼1 nt at their 3′ termini but were otherwise insensitive to addition of TFIIS (SI Appendix, Fig. S10E, lanes 1 to 3). Note that substantial amounts of cleaved RNA were observed in the artificial EC (SI Appendix, Fig. S10E) as well as from G-less 49 (Fig. 4 E, Left), which are probably due to repetitive partial backtracking followed by TFIIS reactivation of transcription and not complete backtracking that would result in transcript release. Last, we tested whether more extensive pol II backtracking occurs in the ITC when the translocase activity of TFIIH, which exerts a forward force on pol II (17, 18, 24, 25), is impeded by addition of a nonhydrolyzable ATP analog. Unlike in the human system (26), addition of 2 mM AMP-PNP (in the presence of 800 μM ATP) had no effect on pol II backtracking (lanes 7 to 9 vs. 10 to 12 in SI Appendix, Fig. S10E). This suggests that the continuous translocase activity of TFIIH may be maintained after addition of 2 mM AMP-PNP and that the large amounts of RNA that we observed to be released from the ITC with the G-less 27 template (Fig. 4C) were likely caused by removal of ATP during gradient sedimentation.
Cet1-Ceg1 and Spt4/5 Facilitate the Transition from Initiation to Elongation.
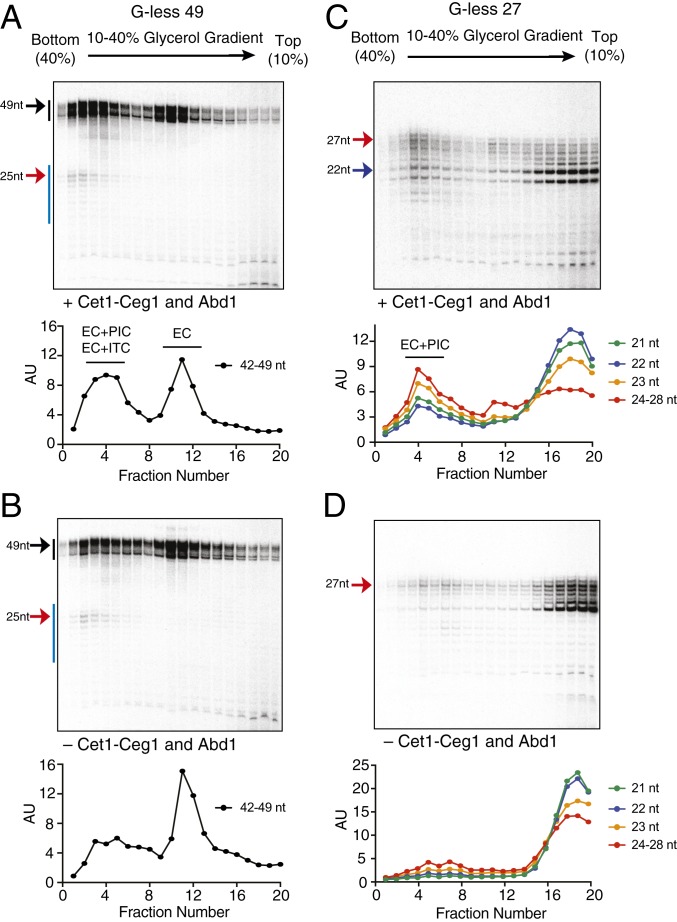
Considering that the ITC persists until a nascent transcript reaches a length of ∼22 and 23 nt, which is roughly when the capping enzymes bind (9), we hypothesized that the capping enzyme may play a role in promoter escape and recruitment of a new incoming pol II for reinitiation. To test this model, we used gradient sedimentation to compare transcription complexes stalled at +49 or +27 in the presence (Fig. 5 A and C) or absence of the capping enzymes (Fig. 5 B and D). Omission of Cet1-Ceg1 and Abd1 from the reactions reduced the population of G-less 49 reinitiation complex (EC+PIC) (fractions 2 to 6 in Fig. 5 A and B), while increasing the population of the ECs that failed to assemble a follow-on PIC (fractions 10 to 12 in Fig. 5 A and B). Analogous experiments revealed that addition of Cet1-Ceg1 alone without Abd1 (SI Appendix, Fig. S11A) or SAM (SI Appendix, Fig. S11B) was sufficient to promote formation of the reinitiation complex. Omission of Cet1-Ceg1 and Abd1 likewise reduced the population of the G-less 27 reinitiation complex (EC+PIC) (fractions 3 to 5 in Fig. 5 C and D), while increasing the population of ITCs as indicated by increased amounts of released RNAs (fractions 17 to 20 in Fig. 5 C and D). Notably, omission of the capping enzymes resulted in release of almost all lengths of the nascent transcripts from ITCs (Fig. 5D), whereas only shorter transcripts (21 to 23 nt in length) were released in the presence of the capping enzymes (Fig. 5C). This suggests that RNA length may play a critical role in promoter escape, likely by contributing to recruitment of Cet1-Ceg1 to the transcription complex. Taken together, our results suggest that Cet1-Ceg1 facilitates promoter escape and the assembly of a follow-on PIC for reinitiation.
Fig. 5.
Cet1-Ceg1 facilitates promoter escape and assembly of a follow-on PIC for reinitiation. Transcription was initiated with the (A and B) G-less 49 or (C and D) G-less 27 template DNA in the presence (A and C) or absence (B and D) of Cet1-Ceg1 and Abd1. All reactions received 2 mM AMP-PNP prior to 10 to 40% glycerol gradient sedimentation. The gradients were fractionated, and ∼180 µL per fraction was isolated prior to analyzing RNA by denaturing PAGE. The levels of RNA in each fraction were quantified by ImageJ and normalized to fraction 1, and the relative RNA levels were plotted.
Spt4/5, the yeast homolog of DSIF, is recruited soon after recruitment of Cet1-Ceg1 (15). We thus asked whether Spt4/5 also promotes promoter escape and the association of a new incoming pol II for reinitiation. Indeed, glycerol gradient sedimentation of G-less 27 transcription complexes showed that formation of the reinitiation complex (EC+PIC) was enhanced upon addition of Spt4/5 in a similar manner to addition of Cet1-Ceg1 (SI Appendix, Fig. S12). We thus conclude that both Cet1-Ceg1 and Spt4/5 act to promote the transition from initiation to elongation.
Arresting Pol II in the ITC Facilitates the Conversion to the EC but Not the Assembly of a Follow-On PIC for Reinitiation.
Pol II in the ITC stalled at +27 is prone to extensive backtracking, but not arrested, as evidenced by RNA release upon gradient sedimentation (Figs. 4C and 5 C and D). We thus sought a way to isolate the ITC with the G-less 27 template by inducing pol II arrest and thereby preventing RNA release. Previous studies of bacterial RNA polymerase (RNAP) demonstrated that nucleotide analogs incorporated at the 3′ terminus of RNA generally induce RNAP backtracking followed by stable arrest via destabilizing the 3′-proximal RNA–DNA hybrid (35). By taking advantage of successive U residues clustered at +17, +18, +19, +20, +23, +24, +25, and +26 of the G-less 27 template (Fig. 2D), we screened UTP analogs and found that 4′-thio UTP can be incorporated by pol II without reducing initiation activity and that the resulting 4′-thio RNAs in the postinitiation complexes are less sensitive to addition of TFIIS than RNAs without 4′-thio UTP (SI Appendix, Fig. S13 A and B). Using G-less 27 template DNA, we then stalled postinitiation complexes at +27 in the absence of the capping enzymes, sedimented them on a glycerol gradient, and analyzed 4′-thio RNAs by denaturing PAGE (SI Appendix, Fig. S13C, Top). Whereas ∼21- to 27-nt RNAs containing standard uridine were largely released from the ITC and were present on the top of the gradient (Fig. 5D), 4′-thio RNAs were near completely present in fractions corresponding to ECs (SI Appendix, Fig. S13C, Top). No released transcripts were observed, as indicated by the absence of RNA on the top of the gradient (SI Appendix, Fig. S13C, Top). Incorporation of 4′-thio UTP thus strongly prevented release of nascent transcripts during gradient sedimentation presumably by inducing pol II arrest before +17. Notably, almost no reinitiation complexes were observed with 4′-thio UTP (SI Appendix, Fig. S13C, Top), although promoter escape occurred as shown by the presence of ECs. These data suggest that promoter escape is not necessarily followed by the assembly of a follow-on PIC even in the presence of excess pol II and TFIIF and further highlight the critical roles of the capping enzyme and Spt4/5 in both promoter escape and the assembly of a follow-on PIC.
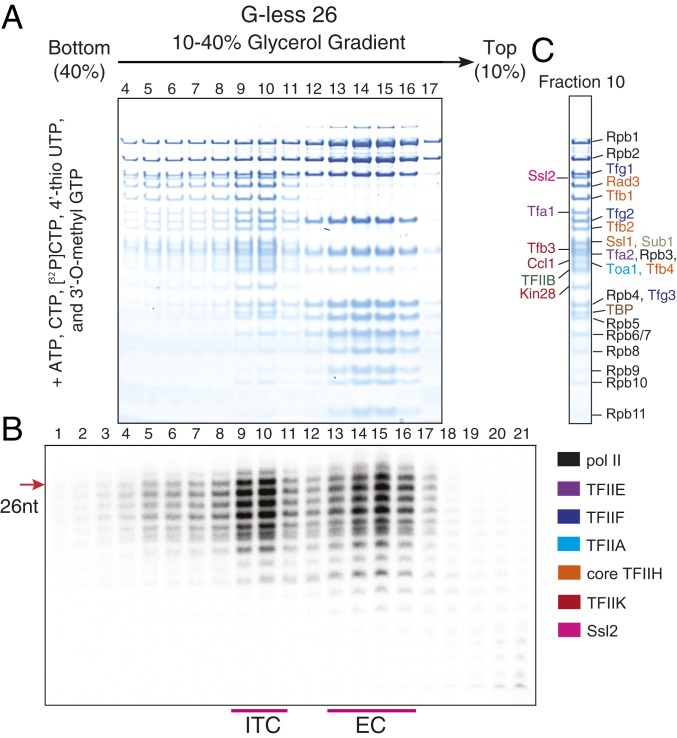
Given that 4′-thio UTP can prevent RNA release from the ITC, we attempted to isolate the ITC by stalling pol II upstream of +27. When pol II was stalled at +26 in the presence of 4′-thio UTP, we for the first time observed a major peak of RNA at the position where the PIC migrates (Fig. 6 A and B, fractions 9 and 10; SI Appendix, Fig. S13C, Bottom) along with a peak for the EC (Fig. 6 A and B, fractions 14 to 16). All of the GTFs, pol II, and RNA are present in fraction 10 (Fig. 6 B and C) indicating the presence of the ITC with 26 nt RNA. Taken together, these results definitively confirm that ITC can persist longer than previously expected (26).
Fig. 6.
The 4′-Thio UTP suppresses RNA release and allows transcripts to be trapped in the ITC. PIC assembly and initiation reactions were performed in the same manner as in Fig. 3 D and E except that pol II was stalled at +26 using G-less 26 template DNA and that the capping enzymes were omitted, transcription was initiated in the presence of [32P]CTP and 630 µM 4′-thio UTP instead of [32P]UTP and 500 µM UTP, and the concentration of CTP in the reaction was lowered to 630 µM from 800 µM. Centrifugation was performed in the same manner as before. The gradient was fractionated, and ∼130 μL per fraction was isolated. (A) Proteins in fractions 4 to 17 were analyzed by SDS/PAGE. (B) RNA in fractions 1 to 21 was analyzed by denaturing PAGE. (C) Protein identification of fraction 10.
Discussion
The transition from transcription initiation to elongation is a major rate-limiting step at many mRNA genes (36), but key details of how the PIC transitions through the ITC to the EC have remained unclear. Here we gained important insights into this transition by using an improved in vitro transcription system and yeast SNR20 promoter DNA. Compared to our prior in vitro system (16, 17), we were able to increase the transcription efficiency by ∼3- to 4-fold. By then isolating and characterizing naturally generated postinitiation complexes, we found that the ITC, which contains pol II, GTFs, and a nascent transcript, often persists much longer than previously expected. In particular, we find that promoter escape and assembly of a follow-on PIC are facilitated by the capping enzyme and Spt4/5.
Previous crystal structures have shown that the RNA 5′ end begins to clash with the N-terminal region of TFIIB (TFIIBN) when the nascent RNA reaches a length of 9 to 15 nt (37–40). It has thus been thought that TFIIB and then other GTFs (TFIIA, TBP, TFIIE, and TFIIH) dissociate from the ITC while the upstream end of the initial bubble collapses (3, 24, 41). In contrast to this model, our results indicate that all of the GTFs, including TFIIB, can be associated with complexes transcribing 26-nt- (Fig. 6 A–C) or even 49-nt-long RNAs (Fig. 3 D and E). Nevertheless, the ITC is generally unstable and susceptible to long-range backtracking (Fig. 4E), and it undergoes the transition to the EC in an RNA length-dependent manner (Figs. 3 and 4). Human pol II complexes associated with short RNAs up to ∼50 nt in length are also known to be prone to backtracking (31, 32), suggesting an evolutionarily conserved feature of the early stages of eukaryotic transcription. There are important differences between human and yeast ITC, however. Backtracked human transcription complexes can be arrested and restart transcription with assistance from TFIIS (31). In contrast, we found that backtracking of yeast ITCs led to RNA release from the complex and termination of transcription (Fig. 5 C and D), unless pol II arrest is induced, e.g., by the use of 4′-thio UTP (Fig. 6). It remains unclear why the RNA is released from the ITC only in the yeast system, but it may be due to fundamental differences in promoter architecture between human and yeast genes, for example, the spacing between the TATA box and TSS (42).
In this study, the ITC (Fig. 5 C and D), but not the EC (SI Appendix, Fig. S10), dissociated the RNA from the complex during glycerol gradient sedimentation. This difference allowed us to determine the timing of promoter escape as a function of position downstream of the TSS (Figs. 3 and 4). When transcription complexes were stalled at +49, glycerol gradient sedimentation revealed that a majority of the pol II escaped the promoter, as indicated by the presence of EC, EC+PIC, and EC+ITC (Fig. 3 D–G). Interestingly, when pol II was stalled at +27, a larger proportion of transcription complexes transcribing 24 to 27 nt RNA escaped the promoter than those transcribing shorter (21 to 23 nt) RNAs (Fig. 4 B–D). This indicates that, at least with SNR20 promoter DNA we tested in this study, a major structural change may occur when the RNA length reaches ∼23 nt. The timing of promoter escape we observed differs from the prevailing assumption, which was based on in vitro KMnO4 footprinting (24, 26). We found that KMnO4 footprinting was not sensitive enough to assess the timing of promoter escape (at least by bulk measurements; Fig. 2 B–D) as KMnO4 reactivity was observed mainly within the ∼15-bp transcription bubble surrounding the pol II active center (Fig. 2D). Addition of KMnO4, a strong oxidizing agent, may collapse the extended bubble in the ITC (18), whereas the extremely stable ∼15-bp transcription bubble (43) remains unwound. We instead found that glycerol gradient sedimentation gave clear indications of what factors are present in transcription complexes transcribing various lengths of RNA and thus a clearer view of when promoter escape occurs.
Upon addition of capping enzymes to the in vitro system, we noticed that the capping efficiency of ∼27-nt transcripts was ∼2-fold lower than that of ∼49-nt transcripts (SI Appendix, Fig. S4 B and C). The differences in capping efficiency may be due to the fact that ∼79% of G-less 49 (Fig. 3E) transcription complexes escaped the promoter, compared to only ∼43% of G-less 27 (Fig. 4C) complexes. This is in contrast to a previous study that used a reconstituted mammalian system and showed that the capping efficiency of short (23 nt) and long (223 nt) transcripts was indistinguishable (44). In our study, the capping enzyme was added at the same time as addition of NTPs (i.e., transcription initiation), whereas the capping enzyme was added in the previous study to stalled transcription complexes that had been washed with high salt. High salt should result in dissociation of some GTFs from the ITC, essentially forming ECs (8, 44) Thus, the differences in capping efficiency between the G-less 27 and G-less 49 transcription complexes may suggest that recruitment of the capping enzymes to the pol II surface is restricted due to the presence of GTFs. Nilson et al. (8) indeed observed that the capping efficiency is much lower on human transcription complexes containing 21-nt RNAs after a low-salt wash (which should not remove GTFs) compared to after a high-salt wash. Alternatively, backtracking of the ITC (Fig. 4E) may cause the 5′ ends of transcripts to be pulled back inside the RNA exit tunnel of pol II, limiting the access of the capping enzyme on RNAs and resulting in less efficiently capped ∼27-nt RNAs than ∼49-nt RNAs.
Our results further indicate that the conversion of the ITC to EC is assisted by Cet1-Ceg1 and Spt4/5 (Fig. 5 and SI Appendix, Fig. S12). In light of recent studies that mapped binding sites for Cet1-Ceg1 immediately adjacent to Rpb4/7 of pol II, which overlap with those for TFIIE in the PIC (perhaps also in ITC) (9, 44), we propose that the recruitment of Cet1-Ceg1 to pol II with an emerging nascent RNA may lead to ejection of TFIIE from the ITC and promoter escape. Similarly, interactions between Spt4/5 and an emerging nascent RNA (45, 46) in the ITC may likewise aid promoter escape. In addition to Cet1-Ceg1 and Spt4/5, some stress-responsive transcription factors that stimulate transcription restart from pol II backtracking in early transcription may also play a role in the transition from initiation to elongation (47). Moreover, the transition could be regulated by the +1 nucleosome (48). It will be of considerable interest to explore in the future how a variety of transcription factors and chromatin factors positively or negatively regulate the transition from transcription initiation to elongation.
Finally, our improved system represents a significant technical advance that will be highly useful for biochemical and structural studies of transcription initiation as well as the transition to elongation. By taking advantage of this highly efficient transcription initiation system, structural determination of the naturally formed ITC can be now pursued to provide further molecular insights into the mechanism of promoter escape.
Methods
Detailed method descriptions are in SI Appendix.
Expression and Purification of Proteins.
TFIIA, TFIIB, TBP, and Sub1 were purified from bacteria, and TFIIE, TFIIF, and TFIIH were purified from yeast as previously described (17, 49). pSBET-His7-ABD1 and pSBET-His7-CET1-CEG1 plasmids were provided by Dr. Stephen Buratowski, Harvard Medical School, Boston, MA. Abd1 and Cet1-Ceg1 were separately expressed from bacteria and purified as previously described with minor modifications (50, 51). pST69-His6-Spt5-Strep-Spt4 plasmid was a gift from Dr. Joseph Reese, The Pennsylvania State University, University Park, PA. Recombinant Spt4/5 was prepared as described (52) with some modifications. Detailed purification methods for Cet1-Ceg1, Abd1, and Spt4/5 are in SI Appendix.
In Vitro Transcription Initiation Assays.
SNR20 promoter DNA was obtained as described (17). A series of point mutations were performed using QuikChange Site-Directed Mutagenesis (New England Biolabs). SNR20 (−122/+97) was amplified by PCR and purified as previously published (17). All transcription assays were performed as previously described (16, 17) with modifications. PIC was formed on 1.3 pmol of DNA fragment with 2 pmol of TFIIA, 3 pmol of TFIIB, 1.5 pmol of TBP, 3 pmol of TFIIE, 5.2 pmol of TFIIF, 1.5 pmol of TFIIH, 1.8 pmol of TFIIK, 3 pmol of Sub1, and 5.2 pmol of pol II in 5 µL of buffer 300 (50 mM Hepes [pH 7.6], 300 mM potassium acetate, 5 mM DTT, and 5% glycerol). The mixture was diluted with 5 µL of buffer 10 (20 mM Hepes [pH 7.6], 10 mM potassium acetate, 5 mM magnesium sulfate, 5 mM DTT) and incubated on ice for 24 h. After 20 min of preincubation at 30 °C, the reaction was initiated by addition of 10 µL of 2× NTPs containing 1.6 mM ATP, 1.6 mM CTP, 1 mM UTP, 0.5 mM 3′-O-methyl GTP, 1 unit of RNaseOUT, 66 to 132 nM [α-32P] UTP (2 to 4 µCi), 5.2 pmol Ceg1-Ceg1, and 10.4 pmol of Abd1 in buffer 10. Transcription initiation samples without capping enzymes received an equal volume of the capping enzyme buffer. The reaction was carried out for 20 min and quenched by addition of 190 µL of stop buffer (300 mM sodium acetate [pH 5.5], 5 mM EDTA, 0.7% SDS, 0.1 mg/mL glycogen, 0.013 mg/mL of proteinase K [Sigma]) followed by 15 min incubation at 41 °C. RNAs were recovered by ethanol precipitation, dried, and dissolved in formamide before running a urea acrylamide denaturing gel. In TFIIS-induced cleavage assays, transcription reactions were performed as described above and were followed by addition of 1 µL of TFIIS (1.5 or 4.5 µM) and 6-min incubation at 30 °C. Methods for calculation of the transcription efficiency (template usage) are in SI Appendix.
Northern Blotting.
RNAs were separated by 15% denaturing polyacrylamide gel electrophoresis (National Diagnostics) and electroblotted/UV crosslinked to Hybond N+ membrane (GE Healthcare RPN303B). ULTRAhyb-oligo Hybridization Buffer (Thermo Fisher Scientific AM8663) was used as per the manufacturer’s instructions. Oligonucleotide probes were designed to anneal to the 5′ end (nt 1 to 25; 5′-AAAGGAAAAGAGATTTGTGGGGGTT-3′) or the 3′ end (nt 26 to 49; 5′- CGATAGTAGAGTTGATGTAAGGGA-3′) of the G-less 49 transcripts. Blots were viewed with the Typhoon 9500 scanner (GE Healthcare).
Separation of Postinitiation Complexes.
As described above, 45.5 pmol of PICs were assembled. The reaction was initiated by adding 2× NTPs containing 4-fold excess Cet1-Ceg1 and 8-fold excess Abd1 relative to DNA. The reaction was stopped by addition of 2 mM AMP-PNP and loaded on a 10 to 40% glycerol gradient (vol/vol) containing 80 mM potassium acetate, 20 mM Hepes (pH7.6), 5 mM DTT, and 2 mM MgOAc. All of the gradients in this study were prepared at 82.6° tilt except the gradient in Fig. 6, which was prepared at 74° tilt. All centrifugation was performed for 14 h at 30,000 rpm in a Beckman SW60 Ti rotor. After the gradient was fractionated (∼130 µL per fraction) using a PGF Piston Gradient Fractionator (BioComp Instruments, Inc.), 90 µL was subjected to TCA precipitation for protein analysis by SDS/PAGE, and 20 µL was subjected to ethanol precipitation for RNA analysis as described above. When only RNA analysis was performed without protein analysis, 5.2 pmol of PICs were assembled, initiated, and sedimented as described above. After the gradient was fractionated (∼180 µL per fraction), 150 µL was subjected to ethanol precipitation for RNA analysis. Note that the gradients in Fig. 5 and SI Appendix, Figs. S6 and S11A, are fractionated using a peristaltic pump, and the rest of the gradients in this study are fractionated using a fractionator.
KMnO4 Footprinting Assays.
The 5′ end of the template or nontemplate strand of SNR20 (−122/+97) DNA was 32P-labeled by T4 PNK. Transcription initiation assay was performed in the presence of Abd1 and Cet1-Ceg1 as described above except that 20 mM Hepes (pH 7.6) in buffer 10 and buffer 300 was substituted with 20 mM potassium/sodium phosphate (pH 7.5) and that DTT was removed. After 20 min reaction at 30 °C, the reaction mixture received 18 mM KMnO4 (as indicated) and was incubated for 2 min 30 s at 30 °C. KMnO4 was quenched by addition of 1.2 M 2-mercaptoethanol. RNA was extracted by ethanol precipitation, and the dried pellet was incubated with 150 µL of 0.1 M piperidine for 30 min at 90 °C. The sample was mixed with 1 µL of 12.5 mg/mL yeast tRNA, 15 µL of 3 M sodium acetate pH 5.5, and 600 µL of 100% ethanol and incubated at −80 °C for 1.5 h before centrifugation at 15,000 rpm for 30 min. The pellet was washed with 200 µL of 80% ethanol and dried. RNA was analyzed on a denaturing 8% polyacrylamide gel.
Reconstitution of artificial EC and exonuclease experiments are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Roger Kornberg for initial support for this project; Deirdre Tatomer for advice on experiments; and Peter Geiduschek, Furqan Fazal, and Hillary Nelson for critical reading of the manuscript. We thank Dr. Steve Buratowski and Dr. Joseph Reese for providing us with Cet1-Ceg1, Abd1, and Spt4/5 expression plasmids; and members of the K.M. and J.E.W. laboratories for discussion. This research was supported by National Institutes of Health grants GM123233 to K.M. and R35-GM119735 to J.E.W.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.L. is a guest editor invited by the Editorial Board.
See Commentary on page 22426.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905449116/-/DCSupplemental.
References
- 1.Conaway R. C., Conaway J. W., General initiation factors for RNA polymerase II. Annu. Rev. Biochem. 62, 161–190 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Kornberg R. D., The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. U.S.A. 104, 12955–12961 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luse D. S., Promoter clearance by RNA polymerase II. Biochim. Biophys. Acta 1829, 63–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yudkovsky N., Ranish J. A., Hahn S., A transcription reinitiation intermediate that is stabilized by activator. Nature 408, 225–229 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen E. B., Lis J. T., In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. U.S.A. 90, 7923–7927 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tome J. M., Tippens N. D., Lis J. T., Single-molecule nascent RNA sequencing identifies regulatory domain architecture at promoters and enhancers. Nat. Genet. 50, 1533–1541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandal S. S., et al. , Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 101, 7572–7577 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilson K. A., et al. , THZ1 reveals roles for Cdk7 in co-transcriptional capping and pausing. Mol. Cell 59, 576–587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Rucobo F. W., et al. , Molecular basis of transcription-coupled pre-mRNA capping. Mol. Cell 58, 1079–1089 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Mao X., Schwer B., Shuman S., Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 15, 4167–4174 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroeder S. C., Zorio D. A., Schwer B., Shuman S., Bentley D., A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell 13, 377–387 (2004). [DOI] [PubMed] [Google Scholar]
- 12.McCracken S., et al. , 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11, 3306–3318 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho E. J., Takagi T., Moore C. R., Buratowski S., mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11, 3319–3326 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez C. R., et al. , Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20, 104–112 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lidschreiber M., Leike K., Cramer P., Cap completion and C-terminal repeat domain kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol. Cell. Biol. 33, 3805–3816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami K., et al. , Formation and fate of a complete 31-protein RNA polymerase II transcription preinitiation complex. J. Biol. Chem. 288, 6325–6332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami K., et al. , Uncoupling promoter opening from start-site scanning. Mol. Cell 59, 133–138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fazal F. M., Meng C. A., Murakami K., Kornberg R. D., Block S. M., Real-time observation of the initiation of RNA polymerase II transcription. Nature 525, 274–277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge H., Roeder R. G., Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78, 513–523 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Henry N. L., Bushnell D. A., Kornberg R. D., A yeast transcriptional stimulatory protein similar to human PC4. J. Biol. Chem. 271, 21842–21847 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Schroeder S. C., Schwer B., Shuman S., Bentley D., Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14, 2435–2440 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szentirmay M. N., Sawadogo M., Sarkosyl block of transcription reinitiation by RNA polymerase II as visualized by the colliding polymerases reinitiation assay. Nucleic Acids Res. 22, 5341–5346 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobson D. J., Wei W., Steinmetz L. M., Svejstrup J. Q., RNA polymerase II collision interrupts convergent transcription. Mol. Cell 48, 365–374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pal M., Ponticelli A. S., Luse D. S., The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell 19, 101–110 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Fishburn J., Tomko E., Galburt E., Hahn S., Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc. Natl. Acad. Sci. U.S.A. 112, 3961–3966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holstege F. C., Fiedler U., Timmers H. T., Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16, 7468–7480 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi W. S., Lin Y. C., Gralla J. D., The Schizosaccharomyces pombe open promoter bubble: Mammalian-like arrangement and properties. J. Mol. Biol. 340, 981–989 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Dvir A., Conaway R. C., Conaway J. W., A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc. Natl. Acad. Sci. U.S.A. 94, 9006–9010 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suh M. H., et al. , A dual interface determines the recognition of RNA polymerase II by RNA capping enzyme. J. Biol. Chem. 285, 34027–34038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komissarova N., Kireeva M. L., Becker J., Sidorenkov I., Kashlev M., Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 371, 233–251 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Pal M., McKean D., Luse D. S., Promoter clearance by RNA polymerase II is an extended, multistep process strongly affected by sequence. Mol. Cell. Biol. 21, 5815–5825 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ujvári A., Pal M., Luse D. S., RNA polymerase II transcription complexes may become arrested if the nascent RNA is shortened to less than 50 nucleotides. J. Biol. Chem. 277, 32527–32537 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Sigurdsson S., Dirac-Svejstrup A. B., Svejstrup J. Q., Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol. Cell 38, 202–210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung A. C., Cramer P., Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471, 249–253 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Shaevitz J. W., Abbondanzieri E. A., Landick R., Block S. M., Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature 426, 684–687 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade J. T., Struhl K., The transition from transcriptional initiation to elongation. Curr. Opin. Genet. Dev. 18, 130–136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bushnell D. A., Westover K. D., Davis R. E., Kornberg R. D., Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science 303, 983–988 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Kostrewa D., et al. , RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Liu X., Bushnell D. A., Wang D., Calero G., Kornberg R. D., Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 327, 206–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sainsbury S., Niesser J., Cramer P., Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 493, 437–440 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Čabart P., Újvári A., Pal M., Luse D. S., Transcription factor TFIIF is not required for initiation by RNA polymerase II, but it is essential to stabilize transcription factor TFIIB in early elongation complexes. Proc. Natl. Acad. Sci. U.S.A. 108, 15786–15791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang C., Bolotin E., Jiang T., Sladek F. M., Martinez E., Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 389, 52–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gnatt A. L., Cramer P., Fu J., Bushnell D. A., Kornberg R. D., Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science 292, 1876–1882 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Noe Gonzalez M., Sato S., Tomomori-Sato C., Conaway J. W., Conaway R. C., CTD-dependent and -independent mechanisms govern co-transcriptional capping of Pol II transcripts. Nat. Commun. 9, 3392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehara H., et al. , Structure of the complete elongation complex of RNA polymerase II with basal factors. Science 357, 921–924 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Bernecky C., Plitzko J. M., Cramer P., Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat. Struct. Mol. Biol. 24, 809–815 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Damodaren N., et al. , Def1 interacts with TFIIH and modulates RNA polymerase II transcription. Proc. Natl. Acad. Sci. U.S.A. 114, 13230–13235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagai S., Davis R. E., Mattei P. J., Eagen K. P., Kornberg R. D., Chromatin potentiates transcription. Proc. Natl. Acad. Sci. U.S.A. 114, 1536–1541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibbons B. J., et al. , Subunit architecture of general transcription factor TFIIH. Proc. Natl. Acad. Sci. U.S.A. 109, 1949–1954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho E. J., Rodriguez C. R., Takagi T., Buratowski S., Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev. 12, 3482–3487 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takase Y., Takagi T., Komarnitsky P. B., Buratowski S., The essential interaction between yeast mRNA capping enzyme subunits is not required for triphosphatase function in vivo. Mol. Cell. Biol. 20, 9307–9316 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crickard J. B., Fu J., Reese J. C., Biochemical analysis of yeast suppressor of Ty 4/5 (Spt4/5) reveals the importance of nucleic acid interactions in the prevention of RNA polymerase II arrest. J. Biol. Chem. 291, 9853–9870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.