Significance
Currently, there are no successful HIV-1 vaccines. The membrane proximal external region (MPER) of the HIV-1 gp41 subunit is an attractive target for immunogen design given its highly conserved linear epitope sequence. While precise understanding of native trimeric envelope glycoprotein structure will benefit vaccine design to elicit broadly neutralizing antibodies (bNAbs), the hydrophobic nature of the MPER at the viral membrane interface makes obtention of its structural configuration in the prefusion state of the native trimeric envelope [(gp120/41)3] challenging. Here, integrative approaches combining biophysical, structural, and immunological analyses show that bNAbs bind to intact trimeric viral envelope spikes expressed on cell surfaces in a manner consistent with targeting of a partially membrane-embedded monomeric MPER, thus defining relevant protective vaccine immunogens.
Keywords: MPER, HIV-1, Env, transmembrane domains, nanodiscs
Abstract
The membrane proximal external region (MPER) of HIV-1 envelope glycoprotein (gp) 41 is an attractive vaccine target for elicitation of broadly neutralizing antibodies (bNAbs) by vaccination. However, current details regarding the quaternary structural organization of the MPER within the native prefusion trimer [(gp120/41)3] are elusive and even contradictory, hindering rational MPER immunogen design. To better understand the structural topology of the MPER on the lipid bilayer, the adjacent transmembrane domain (TMD) was appended (MPER-TMD) and studied. Membrane insertion of the MPER-TMD was sensitive both to the TMD sequence and cytoplasmic residues. Antigen binding of MPER-specific bNAbs, in particular 10E8 and DH511.2_K3, was significantly impacted by the presence of the TMD. Furthermore, MPER-TMD assembly into 10-nm diameter nanodiscs revealed a heterogeneous membrane array comprised largely of monomers and dimers, as enumerated by bNAb Fab binding using single-particle electron microscopy analysis, arguing against preferential trimeric association of native MPER and TMD protein segments. Moreover, introduction of isoleucine mutations in the C-terminal heptad repeat to induce an extended MPER α-helical bundle structure yielded an antigenicity profile of cell surface-arrayed Env variants inconsistent with that found in the native prefusion state. In line with these observations, electron paramagnetic resonance analysis suggested that 10E8 inhibits viral membrane fusion by lifting the MPER N-terminal region out of the viral membrane, mandating the exposure of residues that would be occluded by MPER trimerization. Collectively, our data suggest that the MPER is not a stable trimer, but rather a dynamic segment adapted for structural changes accompanying fusion.
HIV-1 envelope glycoprotein (Env) is the primary target for antibody-mediated viral neutralization. The trimeric Env spike, the only viral protein expressed on the HIV-1 membrane surface, consists of 3 pairs of noncovalently associated gp120 and gp41 subunits (gp120/41)3. The gp120 mediates attachment and entry into human CD4+ T lymphocytes upon binding to its primary cellular receptor, CD4, and the CCR5 or CXCR4 coreceptor, leading to a cascade of conformational rearrangements in gp41 (1–3). The N-terminal fusion peptide of gp41 inserts into the target cell membrane, forming an extended, prehairpin trimer intermediate. Subsequent folding of the N- and C-terminal heptad repeat (NHR and CHR) regions of gp41 into a hairpin conformation creates the postfusion, antiparallel 6-helix bundle, bringing together viral and cellular membranes to initiate fusion and virus entry (4–6).
Given its critical role in the virus fusion, the gp41 subunit has been a target for prophylactic and therapeutic intervention (7–9). The gp41 membrane proximal external region (MPER) contains immunologically vulnerable epitopes targeted by several of the most broadly neutralizing antibodies (bNAbs) developed during the course of natural HIV-1 infection (10–13). Insertion, deletion, and mutations of residues in the MPER defined the functional importance of the MPER in Env incorporation, viral fusion, and infectivity (14–16).
Early NMR and electron paramagnetic resonance (EPR) structural data revealed that the MPER segment, consisting of 2 α-helices separated by a flexible hinge region, was semiembedded in lipid membrane (17, 18). In line with these data, the tryptophan-rich, hydrophobic residues in the MPER interact with the membrane, contributing to membrane destabilization (19, 20). During gp41 6-helix bundle formation, structural and functional analyses indicated that interaction between the hydrophobic residues of MPER and the fusion peptide proximal region stabilizes the membrane associative component of the 6-helix bundle conformation of gp41, synergistically contributing to the initiation of membrane fusion (21, 22). Along with its functional importance in virus fusion and infectivity, sequence variations in the MPER appear to affect Env gp160 quaternary structural configuration of antigenic determinant at both distal and proximal sites, modulating antibody binding and neutralization sensitivity (23–26).
Further extensive molecular and functional analyses of MPER-specific bNAbs have suggested that the paratopes of MPER-specific bNAbs include lipid binding sites, which are essential for neutralization breadth as well as potency (18, 27–37). However, the detailed atomic structure of the MPER in the prefusion state of the gp120/gp41 trimer on the membrane surface still remains elusive. In a recent 4.2-Å resolution cryoelectron microscopy (cryo-EM) structure of native HIV-1 JR-FL Env trimer (ΔCT) in complex with antibody PGT151, the micelle-embedded transmembrane domain (TMD) and MPER were poorly defined (38). Moreover, while the secondary structures of the MPER segment alone or in conjunction with the TMD in a membrane environment have been elucidated in pursuit of MPER immunogen design, the inferred structural configuration of the MPER relative to the membrane (as well as its oligomeric state preceding the TMD) is inconsistent among various studies (18, 39–41).
To elucidate the immunologically relevant quaternary structural configuration of the MPER in the prefusion state of the gp120/gp41 trimer, we examined both the topology of the isolated MPER-TMD reconstituted in liposomes and nanodiscs (NDs), as well as in the context of native and variant envelope protein trimers expressed on the cell surface. EPR and NMR studies, in addition to single-particle negative-stain EM stoichiometric analysis of MPER-specific bNAbs binding to the MPER-TMD incorporated into NDs, suggest a tripod-like topology of the MPER. In line with these observations, antigenicity profiling of Env trimer variants disfavors an extended trimeric MPER conformation, supporting the view that a partially membrane-embedded monomeric MPER is the immunologically relevant target rather than an extended trimeric MPER immunogen.
Results
The Lipid Bilayer Orientation of the gp41 MPER-TMD Is Modulated by the TM R696 Residue as Well as the Charged Residues at the N Terminus of the Cytoplasmic Domain.
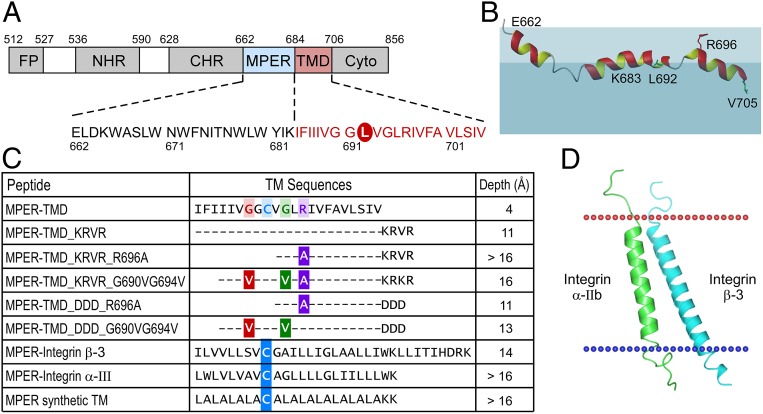
Given the importance of the MPER as a vaccine target against which to elicit bNAbs, we investigated its configuration in the MPER-TMD context. NMR study of HxB2 MPER-TMD peptide (sequence shown in Fig. 1A) revealed that the peptide adopts a mostly helical conformation in lyso-myristoylphosphatidylglycerol (LMPG) micelles, but with 2 breaks or kinks in the middle (Fig. 1B and SI Appendix, Fig. S1). The N-terminal helix region (K665 to N671) of the MPER-TMD is similar to that of the MPER peptide alone (18). The hinge region previously observed in the MPER (amino acids 671 to 674) is unwound further in MPER-TMD, while the C-terminal helix from MPER is extended by an additional 7 residues from the TMD region, becoming a conjoint cMPER-nTM central helix (W678 to I688). This is followed by another helical kink region conferred by the G690xxxG694 motif, and finally a well-defined C-terminal TMD helix (L695 to I704). The MPER hinge region residues are highly dynamic as shown by some of the weak resonance peaks in the 15N-HSQC spectrum (heteronuclear single quantum coherence) (SI Appendix, Fig. S1B). The second kink region is consistent with another NMR study using a shorter MPER-TMD fragment (42).
Fig. 1.
Effect of cytoplasmic and TM sequences on the immersion depth of MPER-TMD in a membrane environment. (A) Domain architecture of gp41. Cyto, cytoplasmic domain; FP, fusion peptide. The peptide sequence used in this study comprised a L692C mutation for R1 spin labeling circled in red. (B) NMR structure of the HxB2 MPER-TMD segment simulated in the liposome (43). The colored region representing the lipid membrane is for illustration only. Lipid headgroup is represented in light blue and aliphatic region is shaded in darker blue. R1 spin labels are placed at the position of green L692 and V705. R696 side-chain position is shown in red relative to the lipid headgroup. (C) Immersion depths of L692 residue in various MPER-TMD segments from EPR measurements. Mutations are color-coded on the wild-type sequence. Cysteine mutations for R1 spin labeling in the MPER-TMD chimeras are highlighted in blue. (D) A model structure of TMD from integrin α-IIb (green) and from integrin β-3 (cyan) used for chimeric MPER-TMD peptide. The models were adapted from 2K9J of the Orientations of Proteins in Membranes (OPM) database.
Next we assessed the manner in which this segment is arrayed on a lipid bilayer. In this regard, EPR spectroscopy was used to investigate the transmembrane orientation of MPER-TMD in POPC/POPG [1-palmitoyl-2-oleoyl-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)] liposome bilayer membranes. The immersion depths of the TMD residues were measured by power saturation techniques using site-directed mutant peptides with spin-labeled side chains (R1), as described previously (43, 44). The results showed that the membrane immersion depths of residues L692(R1) and V705(R1) in the TMD were only 4 Å and 5 Å inside the acyl-chain region (43), implicating a shallower membrane partition of the TMD, as shown in Fig. 1B. Given that the arginine (R696) is located in the center of TMD sequence, the “snorkeling effect” of this positively charged residue may lift up the TMD peptide from a greater depth in the acyl-chain region toward the membrane headgroup region, assuming a shallow insertion (45–47). Moreover, the kink in the middle of the TMD through a G690xxxG694 motif, as observed in an NMR study of MPER-TMD in micelles, may contribute to the shallow orientation of the TMD relative to the membrane (Fig. 1B). Crucially, the absence of membrane flanking charged residues at the C terminus of TMD may result in a less favorable membrane insertion of TMD helices. In this regard, other studies suggest that the orientation of TMD helices depends on the hydrophobicity and length of the sequences and the thickness of lipid bilayers to minimize the energy of consequential hydrophobic mismatch by avoiding unfavorable exposure of a hydrophobic TMD surface to the hydrophilic environment (48–50).
To test those effects on the tilt angle of the TMD, a series of MPER-TMD variants peptides were synthesized, each incorporating a cysteine mutation at L692 for a spin label adduct (R1) as the reference residue. The presence of a native cytoplasmic sequence KRVR was shown to position the L692(R1) residue in the TMD deeper into the lipid acyl-chain region of the bilayer with a depth value of 11 Å (Fig. 1C). The addition of an R696A mutation further mediated efficient membrane insertion of L692(R1) with its immersion depth value of >16 Å in the POPC/POPG liposome, abrogating the postulated “snorkeling effect” from R696. However, while the flanking-charged residues at the C terminus of TMD segment facilitated more efficient insertion of TMD segment, the degree of insertion was affected by sequence differences between KRVR from the native cytoplamic sequence of gp41 and a negatively charged artificial DDD sequence: The immersion depths of L692(R1) are >16 Å for the MPER-TMD_KRVR_R696A mutant and 11 Å for the MPER-TMD_DDD_R696A mutant, respectively. The electrostatic charges of KRVR and DDD were expected to favor an aqueous environment to facilitate the TMD membrane partition. Additionally, the interaction between the positively charged native KRVR and the negatively charged phospholipid headgroup might further stabilize the membrane orientation of the TMD, compared to the negatively charged DDD sequence. Furthermore, the orientations of chimeric MPER-integrin α-III and MPER-integrin β-3 segments (sequences in Fig. 1C) were compared with MPER-TMD variants based on the membrane emersion depth of the reference residue. Residue G9(R1) in the MPER-integrin α-III and residue M9(R1) in the MPER-integrin β-3, equivalent to L692(R1) in MPER-TMD variants, was deeply buried in the membrane of each with the immersion depth values of >16 Å and 14 Å, respectively. The tilt of MPER-integrin β-3 positioned the labeled residues in the TMD to be less membrane-immersed than that of MPER-integrin α-III, similar to a modeled TMD structure of integrin α-IIb and integrin β-3, as depicted in Fig. 1D (51–54), indicating that the membrane tilt angle of TMD was not influenced by the MPER. The integrin β-3 showed a longer membrane-spanning helix than α-IIb, corresponding to a more tilted orientation of TMD. When the TMD segment was substituted by a 22-residue-long artificial leucine-alanine repeat (LA) with a well-defined straight TM orientation from modeling (55), the immersion depth of the equivalent reference residue L9(R1) was similar to that of MPER-integrin α-III chimera and MPER-TMD_KRVR_R696A. Overall, the results suggest that the hydrophobicity of TMD sequence and the presence of charged residues at the C terminus of TMD can be used to modulate the membrane tilt angle of MPER-TMD segment in lipid bilayers. Such variations in TMD tilt angle may influence the membrane orientation of MPER residues, and consequently may affect the quality and the quantity of MPER-specific immune responses elicited by MPER-TMD/liposome immunogens.
The TMD Linked to the MPER Modulates Optimal Binding of the Anti-MPER bNAbs 10E8 and DH511.2_K3.
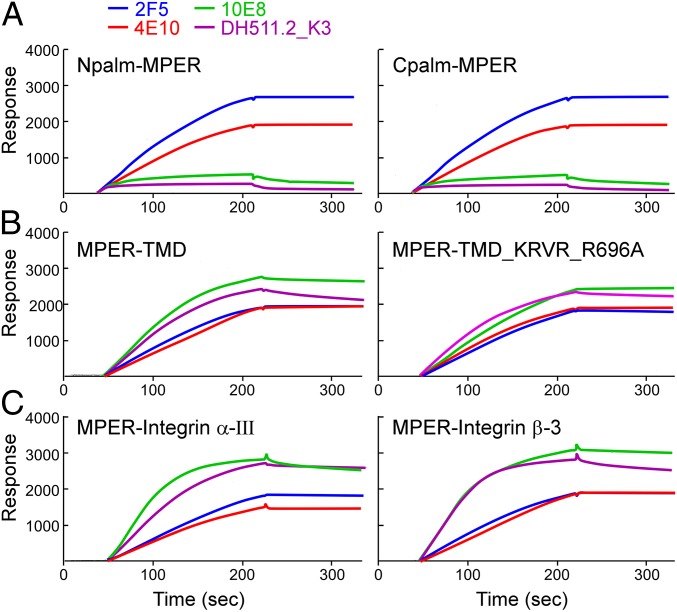
To investigate the effect of the various TMD helical tilt angles on the binding of MPER-specific bNAbs (i.e., antigenicity), the relative binding affinity of bNAbs was measured against those MPER-TMD segments in the membrane environment of DOPC/DOPG [1,2-dioleoyl-sn-glycero-3-phosphocholine/1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)] liposomes using L1 chips and surface plasmon resonance (BIAcore) analysis. For comparison, the bNAbs binding to N- and C-terminally palmitoylated MPER displayed on the surface of liposome was also tested. While a relatively high-affinity binding for N- and C-terminally palmitoylated MPER was achieved by 2F5 and 4E10, 10E8 and DH511.2_K3 binding was remarkably low (Fig. 2A). On the other hand, when MPER was anchored onto the membrane via the HxB2 gp41 TMD domain (MPER-TMD), 10E8 and DH511.2_K3 binding was significantly enhanced (Fig. 2B). In fact, their binding response units were higher than that of 2F5 and 4E10. Note that bNAbs’ specificity to the MPER was confirmed relative to bare liposome and by an irrelevant 1A3 Ab (gp120 V3-specific) binding to various MPER-TMD/liposomes (SI Appendix, Fig. S2). Moreover, although the immersion depth of L692(R1) between MPER-TMD and MPER-TMD_KRVR_R696A peptides differed by 7 Å (4 Å versus 11 Å), the bNAb binding was not significantly affected by the membrane tilt angle of the TMDs. Similar results were obtained when the gp41 TMD was replaced with that of integrin α-III or β-3 (Fig. 2C). However, a relatively faster binding association rate of 10E8 and DH511.2_K3 was observed in sensograms of MPER-integrin α-III TMD and MPER-integrin β-3 TMD, compared to those of MPER-TMD and MPER-TMD_KRVR_R696A (Fig. 2C versus Fig. 2B). This may imply subtle differences in membrane orientation of MPER C-terminal residues constrained by various TMD sequences, leading to different bNAb binding kinetics. Overall, the results suggested that the binding of 10E8 and DH511.2_K3 was highly dependent on the presence of the TMD segment, but less sensitive to the depth and orientation of the TMD relative to the membrane.
Fig. 2.
(A–C) Modulation of MPER-specific bNAb recognition linked to the TMD and its sequence. Antigenicity analysis of indicated antibodies to the denoted MPER segments arrayed on liposomes as measured by BIAcore. DOPC:DOPG liposomes were prepared at a peptide-to-lipid ratio of 1:50. Then, 30 μL of 20 μg/mL antibodies was injected to monitor the binding. See Materials and Methods for details. As shown in SI Appendix, Fig. S2, bNAbs binding to bare liposome and an irrelevant mAb 1A3 (anti-ADA gp120 V3 loop) binding to various MPER-TMD/liposomes were used as a negative control.
MPER-TMD Incorporation into NDs Reveals No Preferential Trimerization.
The structural topology of MPER-TMD in the native Env trimer is of fundamental interest as well as important for immunogen design. The arrangement may be as monomeric segments separated in a tripod configuration, consistent with a prior solution NMR structure and paramagnetic relaxation enhancement measurements (Fig. 3 A, Left) (39) or, alternatively, as separated MPER segments preceding a trimeric TMD suggested by solid state NMR studies (Fig. 3 A, Right) (41). If the MPER formed a trimer in the context of the MPER-TMD, that would result in unfavorable bNAb binding due to a steric blockade of epitope accessibility, as shown in another solution NMR study (40). In fact, any observed bNAb binding to a trimerized MPER would either be the consequence of partial trimer disassembly or the result of different MPER-TMD oligomeric structures incorporating into a spherical 100-nm-sized liposome. To define the topology, we employed an orthogonal approach exploring MPER-TMD assembly in planar 10-nm NDs, nanoparticles with a lipid bilayer stabilized by 2 copies of the membrane scaffold protein (MSP) (56). Specifically, MPER-TMD incorporated into NDs was used to examine the oligomeric state of the MPER-TMD, to ascertain whether a trimer association is preferred over monomer or dimer forms and to assess its impact on bNAb binding.
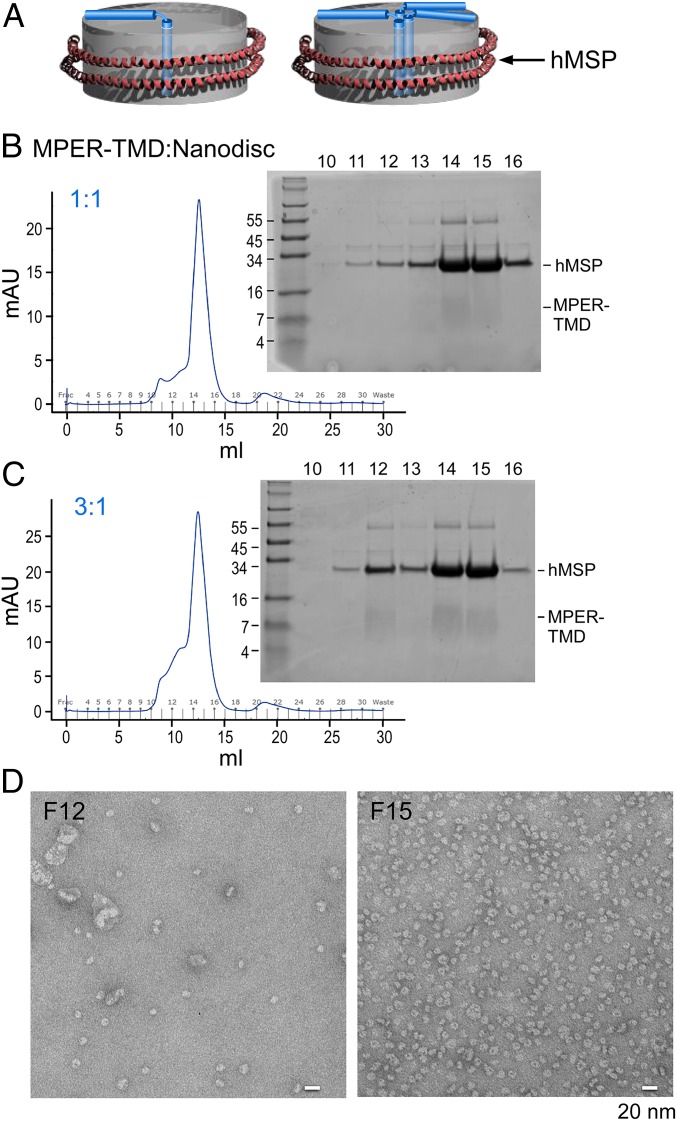
Fig. 3.
MPER-TMD ND assembly. (A) Models of MPER-TMD in NDs in monomeric (Left) versus trimeric (Right) configurations. The MPER-TMD segment is simplified for illustration purposes and does not imply any precise structural and membrane-immersion information. (B and C) MPER-TMD was assembled into NDs as 1:1 (B) and 3:1 (C) MPER-TMD segment to ND ratios. In both, peptides were first codried with POPC:POPG (3:2), and then the mixture was solubilized and incubated with MSP. After dialysis, the assembled mixture was purified by size-exclusion column and peak corresponding fractions were analyzed on tricine SDS/PAGE gels with positions of hMSP and MPER-TMD relative to molecular weight markers given. (D) Minor peak at fraction 12 and major peak at fraction 15, both from C, were visualized by negative-stain EM. (Scale bar, 20 nm.)
The MPER-TMD derived from the clade D HIV-1 isolate 92UG024.2, the same protein construct used for earlier MPER-TMD trimer NMR studies (57), as well as HxB2 MPER-TMD containing 40 residues of cytoplasmic domain (MPER-TMD-Cyto40) instead of 5 residues were compared following expression and purification (SI Appendix, Fig. S3A). The purified clade D MPER-TMD and MPER-TMD-Cyto40, dissolved by methanol, migrated on SDS/PAGE with an apparent size incompatible with trimer and more similar to that of the dimer (SI Appendix, Fig. S3B). Any discordance in observed molecular weight on the SDS/PAGE between this result and the previous study may be due to the different experimental conditions for MPER-TMD purification (40). Subsequently, clade D MPER-TMD was incorporated into NDs at a peptide-to-ND ratio of 1:1 or 3:1. To limit the concentration-dependent oligomeric state of the peptides, MPER-TMD was kept at a fixed concentration of 60 μM during MPER-TMD/ND assembly. Following the removal of detergent by dialysis, MPER-TMD/NDs were formed via self-assembly and then the MPER-TMD/ND particles of proper size were purified by size-exclusion column (Fig. 3 B and C). More MPER-TMD peptides were incorporated into NDs at 3:1 versus 1:1 ratio, judged by the relative band intensity of MPER-TMD and MSP in major peaks (fractions 14 and 15) on SDS/PAGE. Based on the qualitative difference in band intensity of various MPER-TMD peptides-to-MSP ratios (SI Appendix, Fig. S3C), it appears that the MPER-TMD peptides and the MSP comigrated roughly in a molar ratio between 1:1 and 2:1 on the SDS/PAGE. Furthermore, the MPER-TMD/ND particles collected from the major peak in the 3:1 ratio were relatively homogeneous, with the expected size of 10 nm, judged by negative-stain EM analysis, whereas many assembled ND particles collected from the leading minor peak (fraction 12) were significantly enlarged (Fig. 3D), consistent with the different elution volume on the Superdex 200 column.
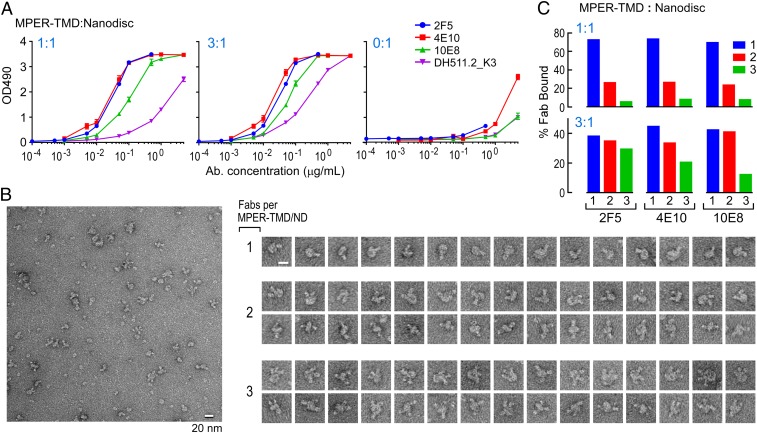
Antigenicity of the clade D MPER-TMD/NDs was tested against a panel of MPER-specific bNAbs by ELISA with empty NDs as a negative control (0:1) (Fig. 4 A, Right). Although DH511.2_K3 and 10E8 reactivity to MPER-TMD/NDs was lower than that of 2F5 and 4E10, with DH511.2_K3 being the lowest, all MPER-specific bNAbs could access their epitopes (Fig. 4 A, Left and Center), confirming MPER-TMD assembly into NDs and including the MPER function-relevant structural configuration. Note that the differences in binding of bNAbs for MPER-TMD observed between BIAcore and ELISA analysis are in part due to different lipid compositions (DOPC/DOPG in a 4:1 ratio for liposomes in BIAcore and POPC/POPG in a 3:2 ratio for NDs in ELISA).
Fig. 4.
Characterization of MPER-TMD/NDs. (A) The binding of MPER-specific bNAbs to MPER-TMD/NDs was measured by ELISA. NDs with MPER-TMD at 1:1 (Left) and 3:1 (Center) ratio was coated onto plates at a ND concentration of 50 nM. The lipid polyreactivity of bNAbs was analyzed by examining bNAb binding to empty ND (peptide-to-MSP ratio at 0:1) (Right) and served as a negative control. (B) 2F5 Fab binding to the ND was visualized by negative-stain EM. Then, 2 μM of MPER-TMD/ND at 1:1 ratio was incubated with 10 μM Fab at 4 °C for 1 h and then diluted accordingly for grid preparation. The field images (Left) and the selected gallery (Right) of ND images represent NDs with different numbers of associated Fab molecules. (Scale bars, 20 nm.) (C) Heterogeneity of MPER-TMD incorporation into ND was visualized by binding of bNAb Fabs, quantitated and graphically represented. Fab fragments of each bNAbs were complexed with MPER-TMD/ND samples at both peptide assembly ratios and the complexes were imaged by negative-stain EM, as above. The particles were categorized based on the number of bound Fabs (1 to 3). Over 3,000 particles were characterized in each Fab-MPER-TMD/ND combination.
To examine the stoichiometric binding of MPER-specific bNAbs at the single-molecule level by negative-stain EM, both ND samples were incubated with a 5-fold molar excess of 2F5, 4E10, and 10E8 Fabs (MPER-TMD/NDs at 2 μM and Fabs at 10 μM), respectively. We imaged the Fab-bound MPER-TMD/NDs at 1-h postincubation to generate a visual binding profile and to semiquantitatively assess the occupancy of Fab per ND from raw images. Representative views of a raw image and galleries of individual NDs with 1, 2, and 3 2F5 Fab bound are shown (Fig. 4B) rather than 2D class averages, as the latter were difficult to achieve given the heterogeneity of subpopulations of Fab/MPER-TMD/ND complexes. Fabs appeared stochastically either on 1 side or both sides of the bilayer for MPER-TMD/NDs with multiple Fabs associated.
Of over 3,000 ND particles counted, about 70 to 72% of MPER-TMD/NDs were occupied by single 2F5, 4E10, or 10E8 Fab, followed by 22 to 25% of particles with 2 Fabs and ∼5% of particles occupied with 3 Fabs (Fig. 4 C, Upper) at a 1:1 ratio after subtraction of corresponding bNAb Fab binding to empty NDs (SI Appendix, Fig. S3D), respectively. For clade D MPER-TMD/NDs formed at a 3:1 ratio, an increased peptide amounts resulted in enriched population of particles with 2 and 3 Fabs bound, compared to those MPER-TMD/NDs with a 1:1 ratio (Fig. 4 C, Lower) (36 to 47% for 1; 33 to 45% for 2; 10 to 30% for 3 Fabs, respectively). The increased stoichiometry of Fab binding per particle is consistent with the higher number of MPER-TMD incorporated into the ND. Pointedly, the heterogeneous stoichiometry of Fab binding to the MPER is not due to the effects of Fab concentration (in large excess and kept constant) or incubation time (58), since similar results were observed after a 24-h coincubation (SI Appendix, Fig. S3E). In addition, the same trend was observed for MPER-TMD-Cyto40/NDs as well (SI Appendix, Fig. S3F). The percentage of 3 Fab complexes is always lower than those 1 Fab and 2 Fabs, especially evident at the 1:1 MPER-TMD/ND ratio. These findings are in contrast to what one would expect if robust trimeric MPER-TMD were formed. Overall, the results suggest that trimers of clade D MPER-TMD are not preferentially incorporated into NDs as revealed by Fab-binding stoichiometry.
Antigenicity Profiling of Wild-Type and Variant Env Trimers Are Inconsistent with Strong MPER Protomer Associations in the Prefusion State.
Given current difficulties in the structural elucidation of the quaternary configuration of the MPER in the context of native Env trimer, we next extended our analysis to investigate a putative MPER oligomeric state in the native Env protein. To determine relevant MPER topology by measuring bNAb binding to the Env expressed on the surface of 293T cells, Env variants of ADA gp145 (cleavable ADA_wt, truncated at cytoplasmic domain A723) were engineered to create a surface-exposed, extended trimeric MPER helical structure. We reasoned that if the antigenicity of Env variants with a panel of bNAbs was diminished relative to that of wild-type Env trimer, it would suggest that the trimeric topology of the MPER in the native Env trimer is disfavored.
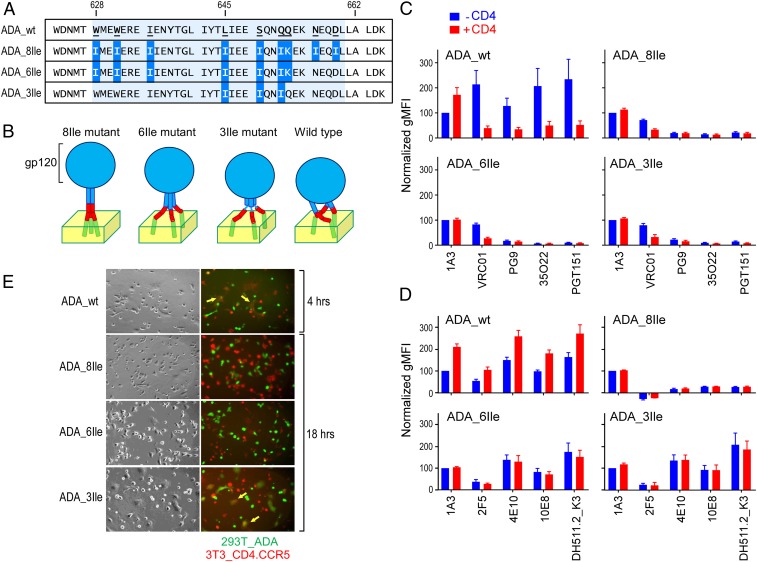
Previous studies demonstrated that yeast transcription factor GCN4 could form a stable trimeric coiled-coil structure by substituting the residues at a and d positions of the heptad repeat with isoleucine (59, 60). Thus, we introduced Ile mutations into the CHR of gp41 at a and d positions to enforce an uninterrupted helical structure extending to the MPER and to facilitate the trimeric association of the MPER N helix in the construct termed ADA_8Ile (Fig. 5 A and B). Q653K was mutated to form an interchain salt-bridge with the upstream E648 from a neighboring helix to maintain a trimeric coiled-coil bundle register. The corresponding CHR peptide with 8Ile mutations was demonstrated to form a stable trimer determined by SDS/PAGE analysis after ethylene glycol bis(succinimidyl succinate) (EGS) chemical cross-linking (SI Appendix, Fig. S4 A and B). The trimeric configuration was also confirmed by circular dichroism (CD) and size-exclusion chromatography analysis in solution under native condition (SI Appendix, Fig. S4 C and D). For comparison, ADA_6Ile was also generated, preserving 6Ile mutations included in ADA_8Ile but maintaining the wild-type residues N656 and D659 immediately N terminal to the MPER (amino acids 662 to 684) (Fig. 5 A and B). These native residues in contradistinction to the Ile mutations therein in ADA_8Ile may break the uninterrupted extended helical structure between CHR and MPER, introducing a hinge that may permit some flexibility in MPER helical movement. Finally, a minimally trimerizing motif at the C-terminal region of CHR was constructed by mutating only L645, S649, and Q652 to Ile (referred as ADA_3Ile), in comparison with the 6Ile mutant (Fig. 5 A and B). Note that the C-terminal MPER segments splayed out from the N-terminal segments of the MPER protomers in the context of a GCN–MPER fusion protein structure determined previously by NMR (61). Therefore, along with our results above, we speculate that the MPER protomers may have a weaker association in the ADA_6Ile and 3Ile mutants (Fig. 5B).
Fig. 5.
Impact of Ile mutations in the gp41 CHR region on Env trimer antigenicity and fusogenicity. (A) CHR-MPER junction sequence comparison of ADA Env (ΔCT) cell expression constructs for the Ile mutants. Introduced mutations are highlighted in blue and the CHR region is shaded. (B) Schematic diagram of the hypothesized conformation for mutant Env trimers on the membrane with gp140 shown in blue, MPER in red, and TMD in green. (C) Antigenicity of 293T cell expressing ADA Env (ΔCT) variants by flow cytometry based on gp140 reactive bNAbs. Cells were incubated without (blue) or with (red) sCD4 and subsequently incubated with the indicated primary antibody followed by staining with fluorochrome-labeled anti-IgG secondary antibody. Geometric mean fluorescence intensity (gMFI) of the single live cell population of each mutant was measured. Normalized gMFI was the percentage of net gMFI for each antibody relative to that of pre-CD4 1A3, where net gMFI was calculated by subtracting MFI of untransfected cells from that of Env-expressing cells. Four replicates were included, and SEMs are shown as error bars. (D) Comparable antigenicity analysis as in C but probed with the designated anti-MPER bNAbs. (E) Fluorescence microscopy analysis of cell–cell fusion. Env-expressing 293T effector cells (green, Calcein-AM) and 3T3.CD4.CCR5 target cells (red, CMTMR) were stained and coincubated. Bright field (Left) and fluorescence images (Right) were collected 4 h (for wild-type) or 18 h postcoincubation for Ile mutants. Fused effector targets are indicated by yellow arrows. (Magnification, 40×.)
The ADA_wt, ADA_8Ile, ADA_6Ile, and ADA_3Ile mutants were all expressed on the surface of 293T cells individually and their antigenicity was assessed by flow cytometry using various bNAbs. By transient transfection, all variants expressed comparable levels of Env with similar extents of cleavage between gp120 and gp41 as judged by V3-specific 1A3 mAb binding by flow cytometry and Western blot analysis (SI Appendix, Fig. S5 A and B). To determine whether the mutations in CHR influenced the quaternary structural configuration of the gp120/gp41 trimer ectodomain, binding of the V1/V2-specific bNAb PG9 and the gp120/gp41 interface targeting 35O22 and PGT151 mAbs was examined against ADA Env variants (Fig. 5C). PG9, 35O22, and PGT151 binding was significantly diminished in ADA_8Ile, ADA_6Ile, and ADA_3Ile, in comparison to the prefusion state of the wild-type Env trimer. As predicted, binding of those trimer-specific antibodies was also diminished after CD4 engagement in the ADA_wt, the result of a CD4-induced open configuration of Env trimer. In contrast, no significant CD4-induced mAb binding changes were observed with Ile mutants. These results suggest that all 3 Ile mutant Env trimers adopt open quaternary structural conformation, likely due to mutations in CHR that allosterically alter the interaction of gp41with gp120 (38, 62).
Next, we assessed the accessibility of MPER-specific bNAbs to their epitopes in these mutants (Fig. 5D and SI Appendix, Fig. S6). The increased MPER-specific bNAbs binding to the wild-type Env was observed after CD4 attachment, as previously described (63). The 293T cell surface-expressed Env captured by MPER-specific bNAbs, such as 10E8, were also recognized by PG9, 35O22, and PGT151 antibodies (SI Appendix, Fig. S5C). While 2F5 binding was abolished by 8Ile mutations in the CHR, it was partially rescued by reversion of Ile mutation in ADA_6Ile and 3Ile mutants. The lack of 2F5 binding in the 8Ile mutant was not due to epitope residue D659I mutation, since the 2F5 binding was not affected by the single mutation (SI Appendix, Fig. S7). In comparison with wild-type, 4E10, 10E8, and DH511.2_K3 binding was also reduced in the 8Ile mutant but recovered in 6Ile and 3Ile mutants. Notwithstanding, no further enhancement in antibody binding mediated by the CD4 receptor was observed with these 3 bNAbs in ADA_6Ile and ADA_3Ile mutants. Overall, the results suggest that the uninterrupted and extended CHR-MPER helical stalk structure introduced by 8Ile mutations may spatially constrain bNAb access to MPER protomers, distinguishing its quaternary structural topology from that in the unliganded wild-type trimer. A trimerized C terminus of CHR might also limit the accessibility of 2F5 to the N helix of MPER.
We next examined the effect of these Ile mutations on gp41-mediated fusion by a fluorescence cell–cell fusion assay. Env-expressing 293T effector cells stained with the cytoplasmic dye calcein-AM (Fig. 5E, shown in green) were cocultured with 3T3.CD4.CCR5 target cells stained with CellTracker orange CMTMR (Fig. 5E, shown in red) (17). Fusion was indicated by the exchange of cytoplasmic dyes between effector and target cells through fluorescence microscopy. After 4 h of coincubation, ADA_wt was able to efficiently mediate the cell–cell fusion, resulting in the formation of large multinuclear fused cell syncytia (Fig. 5E). The conserved hydrophobic pocket (W628, W631, and I635) in the gp41 CHR interacts with the NHR helix and is essential for the stabilization of the gp41 6-helix bundle (4). Two of those residues were mutated in the 8Ile and 6Ile mutants. Accordingly, no fusion activity was observed in cells expressing 8Ile and 6Ile mutants even after an extended 18-h coincubation. The 3Ile mutant was shown to be fusogenic but with delayed kinetics, with cell–cell fusion only observed after 18 h. In summary, it seems unlikely that the extended trimeric MPER helical structure would afford immunogenicity and functional profiles required of this membrane proximal segment in the prefusion state.
10E8 Binding Raises the N-Terminal Region of the MPER up from the Membrane in Support of a Tripod-Like Topology.
While our experimental results above disfavor trimeric association of the MPER segment in the trimeric Env spike, a prior crystallographic study of the 10E8 Fab-bound MPER modeled the epitope to be tilted 75 to 80° from the plane of the membrane (64). Another 10E8-bound MPER structure with appended TMD residues also suggested an oblique tilt of the MPER C helix relative to the micelle membrane (65). Whether the observed orientations represented the native MPER tilt angle relative to the membrane in the prefusion Env state or, alternatively, were a consequence of 10E8 Fab binding is unknown. Based on the results above, we speculate that the MPER may be configured on the membrane interface, as previously shown by our NMR study (18), and is only transiently solvent-exposed extending out of the membrane.
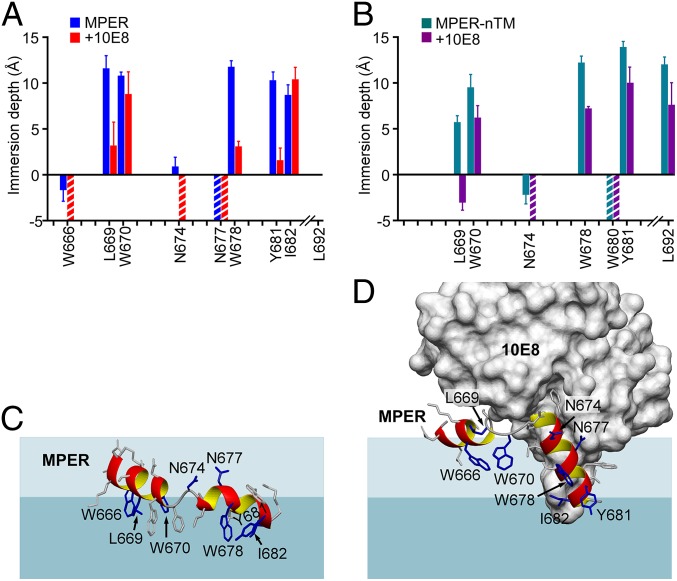
To better define the membrane orientation of the MPER prior to and following bNAb ligation, we determined the membrane immersion depths of spin-labeled MPER residues in the absence and presence of 10E8 Fab using EPR. Since the MPER-TMD was poorly soluble due to its hydrophobicity yielding low spin-labeling efficiency, the TMD was partially truncated (referred as MPER-nTM) (amino acids 662 to 693) and used in this study as a comparison to the MPER alone. The representative spin-labeled residues were selected based on the BIAcore epitope mapping using serial single alanine mutants for retention of greater than 50% of 10E8 binding affinity relative to control MPER, despite cysteine substitutions required for R1 spin-labeled adducts (66).
In the absence of 10E8 Fab, residues L669(R1), W670(R1), W678(R1), and Y681(R1) were deeply buried in the acyl-chain region both in the MPER and the MPER-nTM (Fig. 6 A and B, respectively). 2F5 and 4E10 were previously found to extract the MPER N helix and hinge with significant depth changes of the residues around these regions (18, 44). Here, 10E8 was also able to extract buried residues from liposome-arrayed MPER and MPER-nTM (Fig. 6 A and B). Significant depth changes upon 10E8 binding were found for residues at the MPER N helix and around the hinge region. W666R1 and N674R1 were exposed on the membrane surface upon binding to 10E8. Deeply buried L669R1 was significantly lifted up with reduced membrane depth upon 10E8 binding. A similar trend was also observed for W670R1. 10E8 also caused reduced immersion depths of residues probed on the MPER C helix, W678 and Y681. The binding kinetics of 10E8 and DH511.2_K3 (Fig. 2) are not the determining factor of the antibody-induced MPER conformational changes revealed by the EPR depth measurement, which were conducted at steady state with a relatively high Fab concentration. A docking model was produced based on the EPR depth values and published NMR and crystal structures, which illustrated that the MPER N helix and hinge are extracted from the membrane in the 10E8-bound state (Fig. 6 C and D). Along with the stoichiometry of Fab in complex with MPER-TMD/NDs (Fig. 4C) and the antigenicity of variant Env trimers (Fig. 5D), the depth comparison further supports a model of MPER conformation, where reorientation relative to the membrane was induced or captured by antibody binding.
Fig. 6.
10E8-induced conformational change of the MPER. (A) Membrane immersion depth analysis of the MPER in the absence and presence of 10E8 Fab by EPR. A cysteine mutation was introduced at the site of measurement where an R1 spin label was coupled. (B) Membrane immersion depth measurement of MPER-nTM (amino acids 662 to 693) in the absence and presence of 10E8 Fab. The residues of interest were 2,2,6,6-tetramethyl-N-oxyl-4-amino-4-carboxylic acid (TOAC)-labeled for EPR analysis. Residues measured in both MPER and MPER-nTM analysis are highlighted. Depth values between −5 Å and 0 Å and larger than 0 Å correspond to lipid headgroup region and acyl-chain region, respectively. The precise depths of residues exposed to aqueous phase (depth < −5 Å) cannot be determined experimentally and are thus indicated by the striped bars. (C) MPER peptide docked to lipid bilayer surface based on EPR membrane immersion depth data. (D) 10E8-bound MPER peptide docked to lipid bilayer based on EPR data. The dark blue-shaded area represents lipid aliphatic region and the light blue-shaded area represents lipid head-group region.
Discussion
Early cryoelectron tomography of native HIV-1 and simian immunodeficiency virus (SIV) Env spikes suggested 2 structurally distinct morphological models. Zhu et al. (67, 68) previously reported that SIV and HIV-1 virion-associated Env spikes showed a unique tripod-like leg configuration at the gp41 stalk region. Subsequently, another group using similar technology and virions reported a spike model displaying a compact gp41 stalk, suggesting an extended structural architecture for the MPER and TM region (69, 70). The extended topology of MPER-TMD (amino acids 662 to 710) was further elucidated in a recent solution NMR structure in bicelles (40). The latter study showed that the MPER is solvent-exposed outside the membrane, forming a trimer via a hydrophobic cluster at the N-terminal region of the MPER, while the TMD spans the lipid bilayer also as a trimer stabilized by N-terminal coiled-coil interactions and a C-terminal hydrophilic core (40, 57). In this trimeric organization, binding of MPER-specific bNAbs is not possible given the occlusion of the antibody-binding epitopes by the neighboring MPER protomers. The antigenicity profile of the MPER-TMD reconstituted in the bicelle membrane revealed that hours were required for detectable binding to be achieved, in stark contrast to the rapid 2F5, 4E10, 10E8, and DH511.2_K3 binding observed here with the same segments assembled into liposomes and NDs, both native-like membrane mimics. Furthermore, the stoichiometry of bNAb Fab-bound clade D MPER-TMD/NDs visualized by negative-stain EM analysis showed that the majority of NDs contained only 1 or 2 MPER-TMD peptides, suggesting no clear evidence of strong MPER- or TMD-mediated trimerization in POPC/POPG lipid bilayers. In a small fraction of the nanoparticles, we did observe 3 bNAb Fabs bound to the MPER with a tripod-like shape on the same plane of the membrane. It is unlikely that the TMD in this topology adopts the trimeric conformation predicted from the solid NMR structure of MPER-TMD, given that the steric hindrance of the Fabs in that configuration would not allow 3 Fabs binding to the each MPER protomers (41).
To gain further insights into the quaternary structural organization of the MPER subdomain, we exploited deductive approaches by assessing the antigenic properties of the perturbed MPER configuration in the context of the Env 8Ile mutant expressed on the cell surface. The lack of observed 2F5 binding may result from the rigidity of the 2F5 epitope configured by abutment of an uninterrupted extended helical structure. As a consequence, this mutant segment may not be able to accommodate a structural transition necessary for 2F5 binding (31, 33, 71). In addition, the steric clash of 2F5 with MPER protomers may also result in loss of binding. In line with these notions, 2F5 binding is largely restored in ADA_6Ile and 3Ile mutants. In those, the 2 wild-type residues N656 and D659 residing in the CHR of gp41that may function as a hinge in the native Env were reverted, facilitating independent movement of CHR and MPER (38, 72). The binding of antibodies specific to the MPER C-terminal region (such as 4E10, 10E8, and DH511.2_K3) was also reduced in the 8Ile mutant, supporting the view that the quaternary structural arrangement of an extended MPER trimer additionally disfavors optimal binding by the C-terminal–specific bNAbs.
Given the equivalent binding of all bNAbs, excepting 2F5, to 6Ile and 3Ile mutants compared to the wild-type, we speculate that the N-terminal region of 3 MPER helices may partially be constrained in their conformational flexibility, and limited the 2F5 access spatially or because of weak protomer association in the ADA_6Ile and 3Ile mutants. On the other hand, perhaps because the hinge and the C-terminal region of MPER helices may gradually splay out and associate with the membrane, less impact on the 4E10, 10E8, and DH511.2_K3 bNAbs binding is observed for ADA_6Ile and 3Ile. Taken together, our results disfavored a fixed trimeric organization for the MPER segment in the relevant prefusion state of gp120/41 trimers, as previously suggested by other solution NMR and X-ray crystal structures (40, 73). While a structurally compliant MPER tripod-like base region, as compared to a rigid trimer stalk, would be more responsive in accommodating intricate coordination of gp160/CD4/CCR5 receptor–ligand interactions necessary for binding and fusion and required for HIV-1 viral entry into host cells (74), the role of a rigid stalk conformation at some stage of Env biology is not excluded.
The reduced 4E10 binding to ADA_8Ile suggests that the MPER hinge region in this mutant forms part of an extended α-helices impeding bipartite movement required for epitope exposure. In addition, the extended helices in the 8Ile mutant may hinder 4E10 paratope interaction with lipid membrane important for the core epitope extraction by this bNAb when ligating the wild-type Env (18). In line with this notion, our EPR analysis supports a membrane-embedded configuration of the MPER preceding the TMD. Therefore, 10E8 likely inhibits gp41 membrane fusion by lifting the N helix residues of the MPER out of the membrane as observed in the cryo-EM structure of the 10E8 Fab-bound Env trimer (38) and the NMR structure of Fab/peptide complex in micelles (75). However, our EPR data fit a model of 10E8 binding to membrane-associated MPER in an orientation ∼50° relative to the membrane plane, close to that proposed by Irimia et al. (64), but in contrast to the near horizontal orientation proposed by Rujas et al. (65). The orientation of 10E8 in the cryo-EM structure is hard to decipher given the curvature of the detergent micelle to which Env with MPER-TMD was inserted (38). One possibility is that in the context of the MPER-TMD, the C helix of MPER will be propped up more vertically as a conjoint cMPER-nTM central helix to allow for a more horizontal approach of 10E8 Fab. Alternatively, our EPR data may be more pertinent to the action of 10E8 during an intermediate stage of viral fusion, where the bNAb holds on to the MPER hinge region, preventing it from slipping away into the lipid bilayer to induce membrane fusion.
The MPERs in the glycoprotein of enveloped viruses, such as Ebola and α-viruses, are also recognized as a potential target domain to elicit neutralizing antibodies to inhibit virus entry or virus budding (76, 77). Immunization with HR2-MPER peptide vaccine was reported to induce weakly neutralizing antibody responses against Ebola viruses in a rabbit model (77). In addition, recent cryo-EM analysis of a full-length influenza A virus hemagglutinin spike glycoprotein reveals a strain-specific neutralizing antibody directed to a flexible linker region near the junction between ectodomain and the TMD (78).
While epitope-based immunogen design can be an attractive approach to elicit bNAb responses directed to the conserved linear HIV-1 MPER epitope sequences, it has proven challenging in significant part due to an incomplete understanding of the quaternary structural configuration of the MPER in the prefusion state of Env trimer. The linkage between CHR and MPER was described as a flexible loop in the cryo-EM structure of the 10E8-bound Env trimer in detergent micelles (38). In line with the observation, collectively the structures of the MPER in various contexts (13, 29, 33, 64) suggest that the MPER possesses considerable conformational plasticity. The gp41-mediated fusion likely requires MPER flexibility at the membrane to accommodate conformational transitions that are structurally independent from but working in tandem with the CHR and TMD segments during the biological process of viral entry into host cells. Predicated on these collective results, a membrane-adapted and dynamic monomeric MPER segment appears to be a functionally more relevant immunogen for vaccine design than an extended trimeric conformation.
Ab accessibility to MPER is sterically hindered on the virion surface. The viral membrane and the gp120/gp41 ectodomain on the virion surface limit antibody access to the MPER, resulting in the weakly immunogenic nature of the MPER. Therefore, ideally the dominant immune response to gp120/gp41 must be averted on the one hand, while steric hindrance to MPER epitope access must be integrated into the vaccine design to elicit bNAbs on the other. In that regard, epitope-focused immunogens can be advantageous in redirecting dominant antibody responses to the target epitope. However, while bNAbs can benefit from membrane interaction developing into functionally more potent broadly neutralizing antibodies, the proper MPER orientation relative to the membrane is required. An extended configuration of MPER immunogens is unlikely to capture the native conformation at the membrane surface. Rigid and stabilized MPER peptide immunogens per se may not be sufficient, and may limit the expansion of antibodies with the correct approach angles to slot into a space constrained by the trimer ectodomains above and membrane below. Our findings using integrative structural and functional analyses have implications for immunogen designs focusing on those neutralizing target epitopes located at the membrane interface on the surface of HIV-1 and other viruses.
Materials and Methods
Constructs, Cell Lines, Antibodies, and Peptides.
The stable NIH 3T3 cell line expressing CD4 and CCR5 (3T3.CD4.CCR5) was obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH; NIH-3T3CD4+CCR5+ cells were from Dan R. Littman, NYU School of Medicine, New York, NY (79). Plasmids for VRC01, PG9, 35O22, and PGT151 expression and 4E10 antibody were requested through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH. Antibody 2F5 and 10E8 were gifts from Peter D. Kwong, National Institutes of Allergy & Infectious Diseases, Bethesda, MD. Antibody DH511.2_K3 was a gift from Barton F. Haynes, Duke University School of Medicine, Durham, NC. Peptides used in EPR and BIAcore studies were synthesized, purified, and palmitic acid or spin label-conjugated by the Koch Institute Biopolymers and Proteomics Facility at the Massachusetts Institute of Technology, as described previously (43).
Liposome Preparation and EPR.
Liposomes were prepared as described in Hope et al. (80) and Szoka et al. (81) (SI Appendix). The EPR measurements were performed as previously described (44) (SI Appendix). EPR spectra were collected using a Bruker E680 spectrometer with a High Sensitivity cavity (ER 4119HS) at the National High Magnetic Field Laboratory. The 10E8 Fabs were added to 100-µM peptides on liposomes with a molar ratio of 1.2 to 1. The immersion depth values were calculated by the ratio of the accessibility values of O2 to 50-mM nickel (II) ethylenediaminediacetic acid (NiEDDA). To obtain an EPR signal, various MPER-TMD and MPER-nTM were labeled as described in SI Appendix, Materials and Methods.
NMR.
The NMR backbone assignment experiments (HNCA, HNCOCA, HNCO, HNCACB, HNCOCACB) were performed using a 0.9 mM 15N-13C-labeled MPER-TMD sample with 10% D2O pH 6.6 and 110 mM d27-LMPG at 35 °C, on a Bruker Avance 750 MHz spectrometer equipped with a cryoprobe. Data were processed using NMRPipe (82) and analyzed with the software CARA (Keller Computer aided resonance assignment tutorial, 2004) (83) and TALOS+ (84).
Surface Plasmon Resonance Analysis.
BIAcore analysis was done as previously described (43) (SI Appendix). Next, 30 μL of the MPER-TMD/liposome complex at 0.2-mg/mL concentration of lipids was applied to a Pioneer L1 sensor chip in a BIAcore 3000 and binding of antibodies was then tested by passage of 30 μL antibody over the peptide–liposome complex at 10 μL/min.
Expression, Purification, and Assembly of MPER-TMD into the NDs.
Various constructs of thioredoxin (Trx)-tagged MPER-TMD was produced as described in SI Appendix, Materials and Methods. For ND incorporation, peptide was codried with POPC: POPG (3:2) at different ratios. Then the lipid film was dissolved in 20 mM Tris pH7.5, 100 mM NaCl, 0.5 mM EDTA, 100 mM sodium cholate, 1% dodecylphosphocholine, 8 M Urea, and 0.5 M l-arginine. After mixing with hMSP, NDs were assembled by gradually removing detergent through dialysis in PBS at 4 °C.
Negative-Stain EM Analysis for Fab-Bound NDs.
ND samples were incubated with 5 times excess MPER-specific Fab at 4 °C for 1 h. Then the mixture was diluted and applied to glow-charged 400 mesh, carbon-coated copper grids (Electron Microscopy Science), and stained with 1.5% uranyl formate. The images were acquired with JEM-1400Falsh (JEOL). Numbers of Fab per ND were counted manually from 2 independent preparations. More than 3,000 particles were categorized for each sample.
Antigenicity and Fusogenicity Characterization of Cell-Expressing Env Mutant.
The 293T cells were transiently transfected with ADA gp145 Env variants plasmid. Cells were harvested 48 h posttransfection, and FACS analysis was set up and analyzed as described in SI Appendix, Materials and Methods. The cell–cell fusion assay was conducted as described previously (17) (SI Appendix).
Structural Modeling.
MPER alone (PDB ID code 2PV6) and 10E8-bound MPER (PDB ID code 4G6F) structures were used as templates to create artificial polylysine model peptides. EPR membrane emersion data of individual cysteine-MTSL (S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) mutant peptides were compiled and applied to the model polylysine peptide with the NZeta atom approximate the nitroxide spin-label position. The model peptides were then docked to lipid bilayer simulated by parallel planes based on these emersion depth data using the software X-PLOR-NIH (85). This procedure does not take into account perturbation caused by cysteine mutations and also the flexibility and hydrophobicity of the MTSL side-chain, and therefore serves mainly for the purpose of illustration.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant PO1AI26901 (to E.L.R. and colleagues). L.S. was supported in part by NIH Grant AI122860, National High Magnetic Field Laboratory User Collaboration Grants Program Grant 5080, and NSF DMR-1157490 and 1644779 grants (to the National High Magnetic Field Laboratory). G.W. and M.L.N. were supported in part by NIH Grant AI0237581.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: NMR chemical shift assignments of MPER-TMD have been deposited in the Biological Magnetic Resonance Data Bank, www.bmrb.wisc.edu (accession no. 50028).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912427116/-/DCSupplemental.
References
- 1.Deng H., et al. , Identification of a major co-receptor for primary isolates of HIV-1. Nature 381, 661–666 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Feng Y., Broder C. C., Kennedy P. E., Berger E. A., HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272, 872–877 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Maddon P. J., et al. , The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47, 333–348 (1986). [DOI] [PubMed] [Google Scholar]
- 4.Chan D. C., Fass D., Berger J. M., Kim P. S., Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Tan K., Liu J., Wang J., Shen S., Lu M., Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. U.S.A. 94, 12303–12308 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C., Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Liu H., Su X., Si L., Lu L., Jiang S., The development of HIV vaccines targeting gp41 membrane-proximal external region (MPER): Challenges and prospects. Protein Cell 9, 596–615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molinos-Albert L. M., Clotet B., Blanco J., Carrillo J., Immunologic insights on the membrane proximal external region: A major human immunodeficiency virus type-1 vaccine target. Front. Immunol. 8, 1154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan C., Liu S., Jiang S., HIV-1 gp41 fusion intermediate: A target for HIV therapeutics. J. Formos. Med. Assoc. 109, 94–105 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Buchacher A., et al. , Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10, 359–369 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Huang J., et al. , Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muster T., et al. , A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67, 6642–6647 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams L. D., et al. , Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci. Immunol. 2, eaal2200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muñoz-Barroso I., Salzwedel K., Hunter E., Blumenthal R., Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73, 6089–6092 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzwedel K., West J. T., Hunter E., A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73, 2469–2480 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vishwanathan S. A., Hunter E., Importance of the membrane-perturbing properties of the membrane-proximal external region of human immunodeficiency virus type 1 gp41 to viral fusion. J. Virol. 82, 5118–5126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Z. Y., et al. , Disruption of helix-capping residues 671 and 674 reveals a role in HIV-1 entry for a specialized hinge segment of the membrane proximal external region of gp41. J. Mol. Biol. 426, 1095–1108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Z. Y., et al. , HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28, 52–63 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Suárez T., Gallaher W. R., Agirre A., Goñi F. M., Nieva J. L., Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: Putative role during viral fusion. J. Virol. 74, 8038–8047 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suárez T., Nir S., Goñi F. M., Saéz-Cirión A., Nieva J. L., The pre-transmembrane region of the human immunodeficiency virus type-1 glycoprotein: A novel fusogenic sequence. FEBS Lett. 477, 145–149 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Buzon V., et al. , Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 6, e1000880 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lay C. S., et al. , Role for the terminal clasp of HIV-1 gp41 glycoprotein in the initiation of membrane fusion. J. Biol. Chem. 286, 41331–41343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradley T., et al. , Amino acid changes in the HIV-1 gp41 membrane proximal region control virus neutralization sensitivity. EBioMedicine 12, 196–207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim A. S., Leaman D. P., Zwick M. B., Antibody to gp41 MPER alters functional properties of HIV-1 Env without complete neutralization. PLoS Pathog. 10, e1004271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyo T., et al. , Molecular basis of unusually high neutralization resistance in tier 3 HIV-1 strain 253-11. J. Virol. 92, e02261-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narasimhulu V. G. S., et al. , Distinct functions for the membrane-proximal ectodomain region (MPER) of HIV-1 gp41 in cell-free and cell-cell viral transmission and cell-cell fusion. J. Biol. Chem. 293, 6099–6120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alam S. M., et al. , Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U.S.A. 106, 20234–20239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardoso R. M., et al. , Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22, 163–173 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Irimia A., Sarkar A., Stanfield R. L., Wilson I. A., Crystallographic identification of lipid as an integral component of the epitope of HIV broadly neutralizing antibody 4E10. Immunity 44, 21–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Julien J. P., et al. , Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J. Virol. 84, 4136–4147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim M., et al. , Antibody mechanics on a membrane-bound HIV segment essential for GP41-targeted viral neutralization. Nat. Struct. Mol. Biol. 18, 1235–1243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ofek G., et al. , Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J. Virol. 84, 2955–2962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ofek G., et al. , Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78, 10724–10737 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sánchez-Martínez S., et al. , Specific phospholipid recognition by human immunodeficiency virus type-1 neutralizing anti-gp41 2F5 antibody. FEBS Lett. 580, 2395–2399 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Scherer E. M., Leaman D. P., Zwick M. B., McMichael A. J., Burton D. R., Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc. Natl. Acad. Sci. U.S.A. 107, 1529–1534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H., et al. , Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity. J. Virol. 84, 1076–1088 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwick M. B., et al. , The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J. Virol. 78, 3155–3161 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J. H., Ozorowski G., Ward A. B., Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiliveri S. C., Louis J. M., Ghirlando R., Baber J. L., Bax A., Tilted, uninterrupted, monomeric HIV-1 gp41 transmembrane helix from residual dipolar couplings. J. Am. Chem. Soc. 140, 34–37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Q., et al. , Structure of the membrane proximal external region of HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 115, E8892–E8899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon B., Lee M., Waring A. J., Hong M., Oligomeric structure and three-dimensional fold of the HIV gp41 membrane-proximal external region and transmembrane domain in phospholipid bilayers. J. Am. Chem. Soc. 140, 8246–8259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apellániz B., et al. , The atomic structure of the HIV-1 gp41 transmembrane domain and its connection to the immunogenic membrane-proximal external region. J. Biol. Chem. 290, 12999–13015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim M., et al. , Immunogenicity of membrane-bound HIV-1 gp41 membrane-proximal external region (MPER) segments is dominated by residue accessibility and modulated by stereochemistry. J. Biol. Chem. 288, 31888–31901 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song L., et al. , Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc. Natl. Acad. Sci. U.S.A. 106, 9057–9062 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chamberlain A. K., Lee Y., Kim S., Bowie J. U., Snorkeling preferences foster an amino acid composition bias in transmembrane helices. J. Mol. Biol. 339, 471–479 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Mishra V. K., Palgunachari M. N., Segrest J. P., Anantharamaiah G. M., Interactions of synthetic peptide analogs of the class A amphipathic helix with lipids. Evidence for the snorkel hypothesis. J. Biol. Chem. 269, 7185–7191 (1994). [PubMed] [Google Scholar]
- 47.Ulmschneider M. B., et al. , Transmembrane helices containing a charged arginine are thermodynamically stable. Eur. Biophys. J. 46, 627–637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Planque M. R., et al. , Sensitivity of single membrane-spanning alpha-helical peptides to hydrophobic mismatch with a lipid bilayer: Effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry 40, 5000–5010 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Krishnakumar S. S., London E., Effect of sequence hydrophobicity and bilayer width upon the minimum length required for the formation of transmembrane helices in membranes. J. Mol. Biol. 374, 671–687 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren J., Lew S., Wang Z., London E., Transmembrane orientation of hydrophobic alpha-helices is regulated both by the relationship of helix length to bilayer thickness and by the cholesterol concentration. Biochemistry 36, 10213–10220 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Lau T. L., Dua V., Ulmer T. S., Structure of the integrin alphaIIb transmembrane segment. J. Biol. Chem. 283, 16162–16168 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lau T. L., Partridge A. W., Ginsberg M. H., Ulmer T. S., Structure of the integrin beta3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry 47, 4008–4016 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J., et al. , Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc. Natl. Acad. Sci. U.S.A. 106, 17729–17734 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y. P., Lewis R. N., Hodges R. S., McElhaney R. N., Peptide models of the helical hydrophobic transmembrane segments of membrane proteins: Interactions of acetyl-K2-(LA)12-K2-amide with phosphatidylethanolamine bilayer membranes. Biochemistry 40, 474–482 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Bayburt T. H., Sligar S. G., Membrane protein assembly into nanodiscs. FEBS Lett. 584, 1721–1727 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dev J., et al. , Structural basis for membrane anchoring of HIV-1 envelope spike. Science 353, 172–175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nogal B., Bowman C. A., Ward A. B., Time-course, negative-stain electron microscopy-based analysis for investigating protein-protein interactions at the single-molecule level. J. Biol. Chem. 292, 19400–19410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harbury P. B., Kim P. S., Alber T., Crystal structure of an isoleucine-zipper trimer. Nature 371, 80–83 (1994). [DOI] [PubMed] [Google Scholar]
- 60.Harbury P. B., Zhang T., Kim P. S., Alber T., A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262, 1401–1407 (1993). [DOI] [PubMed] [Google Scholar]
- 61.Reardon P. N., et al. , Structure of an HIV-1-neutralizing antibody target, the lipid-bound gp41 envelope membrane proximal region trimer. Proc. Natl. Acad. Sci. U.S.A. 111, 1391–1396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pancera M., et al. , Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rathinakumar R., Dutta M., Zhu P., Johnson W. E., Roux K. H., Binding of anti-membrane-proximal gp41 monoclonal antibodies to CD4-liganded and -unliganded human immunodeficiency virus type 1 and simian immunodeficiency virus virions. J. Virol. 86, 1820–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Irimia A., et al. , Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design. PLoS Pathog. 13, e1006212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rujas E., et al. , Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface. Sci. Rep. 6, 38177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donius L. R., et al. , Generation of long-lived bone marrow plasma cells secreting antibodies specific for the HIV-1 gp41 membrane-proximal external region in the absence of polyreactivity. J. Virol. 90, 8875–8890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu P., et al. , Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Zhu P., Winkler H., Chertova E., Taylor K. A., Roux K. H., Cryoelectron tomography of HIV-1 envelope spikes: Further evidence for tripod-like legs. PLoS Pathog. 4, e1000203 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.White T. A., et al. , Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: Strain-dependent variation in quaternary structure. PLoS Pathog. 6, e1001249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zanetti G., Briggs J. A., Grünewald K., Sattentau Q. J., Fuller S. D., Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2, e83 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bryson S., Julien J. P., Hynes R. C., Pai E. F., Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: Vaccine design implications. J. Virol. 83, 11862–11875 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guttman M., et al. , CD4-induced activation in a soluble HIV-1 Env trimer. Structure 22, 974–984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J., Deng Y., Dey A. K., Moore J. P., Lu M., Structure of the HIV-1 gp41 membrane-proximal ectodomain region in a putative prefusion conformation. Biochemistry 48, 2915–2923 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shaik M. M., et al. , Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature 565, 318–323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oakes V., et al. , Exposure of the HIV-1 broadly neutralizing antibody 10E8 MPER epitope on the membrane surface by gp41 transmembrane domain scaffolds. Biochim. Biophys. Acta Biomembr. 1860, 1259–1271 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Byrd E. A., Kielian M., An alphavirus E2 membrane-proximal domain promotes envelope protein lateral interactions and virus budding. MBio 8, e01564-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flyak A. I., et al. , Broadly neutralizing antibodies from human survivors target a conserved site in the Ebola virus glycoprotein HR2-MPER region. Nat. Microbiol. 3, 670–677 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benton D. J., et al. , Influenza hemagglutinin membrane anchor. Proc. Natl. Acad. Sci. U.S.A. 115, 10112–10117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deng H. K., Unutmaz D., KewalRamani V. N., Littman D. R., Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388, 296–300 (1997). [DOI] [PubMed] [Google Scholar]
- 80.Hope M. J., Bally M. B., Webb G., Cullis P. R., Production of large unilamellar vesicles by a rapid extrusion procedure: Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim. Biophys. Acta 812, 55–65 (1985). [DOI] [PubMed] [Google Scholar]
- 81.Szoka F., et al. , Preparation of unilamellar liposomes of intermediate size (0.1-0.2 mumol) by a combination of reverse phase evaporation and extrusion through polycarbonate membranes. Biochim. Biophys. Acta 601, 559–571 (1980). [DOI] [PubMed] [Google Scholar]
- 82.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 83.Keller R. L. J., Computer Aided Resonance Assignment Tutorial (Cantina, Goldau, Switzerland, 2004). [Google Scholar]
- 84.Shen Y., Delaglio F., Cornilescu G., Bax A., TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M., The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.