Abstract
Genomic organization impacts accessibility and movement of information processing systems along DNA. DNA-bound proteins dynamically dictate gene expression and provide regulatory potential to tune transcription rates to match ever-changing environmental conditions. Archaeal genomes are typically small, circular, gene dense, and organized either by histone proteins that are homologous to their eukaryotic counterparts, or small basic proteins that function analogously to bacterial nucleoid proteins. We review here how archaeal genomes are organized and how such organization impacts archaeal gene expression, focusing on conserved DNA-binding proteins within the clade and the factors that are known to impact transcription initiation and elongation within protein-bound genomes.
Keywords: RNA polymerase, histone, Alba, archaea, transcription regulation
Introduction
The regulation imposed on gene expression by chromatin or nucleoid structures in Eukarya and Bacteria, respectively, has a long and rich history [1–10]. Organization of the genome can facilitate or impair the ability of the transcription apparatus to recognize promoter elements, to form an open complex and to transition into stable elongation. Once transcription elongation complexes (TECs) are established, they must traverse a protein-bound template [11–14]. The dynamic associations of DNA-bound proteins and the resultant larger structures formed by cooperative interactions of such hinder translocation. Both bacterial nucleoid and eukaryotic chromatin structures involve the formation of loops, connecting spatially distant locations on the genome via protein-DNA interactions [15,16], and the formation and stability of such topologically-constrained regions can be controlled to alter expression of single loci or very large regions of the genome. Regulation of gene expression through alteration of genomic architecture offers the potential to tailor gene expression to maximize fitness gains in changing environments.
The role of genomic architecture in modulating gene expression in archaeal species has only more recently been investigated with the scrutiny applied to bacterial and eukaryotic systems. Archaeal genomes are typically circular, small (< 5 Mbp), gene dense (~ 80–90% coding sequence), and many genes are organized within operons [17–21]. Despite sharing many hallmarks of typical bacterial genomes, archaeal genomes are expressed with a single RNA polymerase (RNAP) that shares more similarity in overall structure, subunit composition, and basal-transcription factor requirements with eukaryotic RNAPs, in particular Pol II [22–34]. The archaeal RNAP lacks the C-terminal repeats found on Pol II and is not known to be post-translationally modified, but the archaeal RNAP is still directed to the transcription start site in a manner comparable to Pol II. The archaeal transcription system is a component simplified version of the Pol II apparatus, requiring only interactions with Transcription Factor B (TFB - TFIIB in Eukarya), Transcription Factor E (TFE - TFIIE in Eukarya) and TATA Binding Protein (TBP) to recognize core archaeal promoter elements – TATA box and BRE – that share sequence conservation with eukaryotic promoter elements [34–38]. Minimal evidence for long-range interactions between transcription factors and promoter elements is known, and substantial evidence has instead emerged that demonstrates that most archaeal promoters are regulated by bacterial-like repressors or activators that bind immediately adjacent to or overlapping core promoter elements [39–49]. Studies suggest that core promoters are generally devoid of organized chromatin structures [50,51], and that when present, the binding affinity of transcription regulators outcompetes the binding of histones or nucleoid-associated proteins to permit regulation within an organized and protein-bound genome [40,43,47,52].
Following transcription initiation, TECs must stably associate with and transcribe the template for long periods (e.g. minutes or hours at ~40 nt/sec), necessarily displacing DNA-bound proteins that impede translocation. Transcription initiation and elongation in eukaryotes is facilitated by the combinatorial activities of transcription factors and chromatin remodeling and modification machinery. Given the absence of obvious chromatin remodeling and modification machinery in archaeal genomes, transcription factors likely play the dominant role in aiding archaeal transcription during initiation and elongation. The rates of elongation and pausing of archaeal transcription are regulated by conserved archaeal-eukaryotic factors Spt4/Spt5 and TFS (TFIIS in eukaryotes) [13]. Spt5, homologous to bacterial NusG, is the only universally conserved transcription factor. Spt5-RNAP interactions facilitate formation of the closed-clamp configuration of RNAP that aids in processive elongation. Pausing is inevitable, and when collisions with DNA-bound proteins stalls forward translocation of RNAP, reverse translocation can inactivate RNAP. The cleavage-stimulatory activity of TFS/TFIIS [53] – analogous to the cleavage stimulatory activities of GreA and GreB in Bacteria [54,55]– helps rescue such backtracked complexes, and the activity of TFS is essential for archaeal species [13].
In this review, we discuss recent advancements in archaeal chromatin and genome organization in the context of transcription regulation. We first examine the architectural mechanisms and regulatory implications of genome compaction dominated by archaeal histone proteins. Most archaeal clades encode histone proteins that generate DNA structures remarkably similar to eukaryotic nucleosomes, albeit with only the core histone-fold and often with only a single histone isoform. We next identify and outline important advancements in the identification of transcription factors and basal transcription mechanisms that facilitate transcription in the context of an archaeal histone-based chromatin landscape. We then focus on the archaeal clades that lack histone proteins and instead encode a suite of small basic proteins that presumably function like bacterial nucleoid-associated proteins to condense and organize the archaeal genome. Finally, we consider the major bottlenecks within the archaeal transcription field in the context of chromatin organized genomic architectures. We conclude with discussion of current debates within the field and highlight the future potential of studies investigating the influence of genomic architecture on archaeal gene expression.
Archaeal histone-based chromatin
Structure of archaeal histone-based chromatin
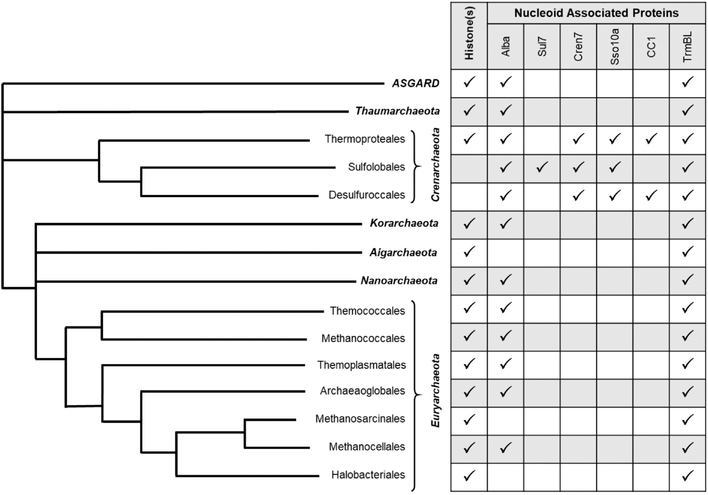
Whole genome sequencing of many cultured and many-more environmentally-isolated, but not yet cultured Archaea suggests that most archaeal lineages encode one or more histone proteins (Figure 1) [56–64] – six histone isoforms can be identified in Methanocaldococcus jannaschii [65] – that are likely to organize the genome into structures that mimic DNA organization by eukaryotic nucleosomes [56,66,67]. Although not universally encoded (typically to the exclusion of Crenarchaeota [56,64]), in archaeal species with histone proteins, a chromatin landscape presents barriers to initiation [42,52,65,68–70], elongation [12,13], and likely influences termination. Archaeal histones are composed of only the core-histone fold and lack the N- and C-terminal tails and extensions common to the canonical eukaryotic histones [66,67,71–75] (e.g. H2A, H2B, H3, and H4). Archaeal genomes do not encode obvious linker histones (e.g. H1), nor chromatin-remodeling complexes that are abundant and essential for gene expression in eukaryotes. Unlike the mandatory eukaryotic histone heterodimer partnerships, archaeal chromatin can be spontaneously assembled with a single histone protein [51,66,67,72,76,77], and there is currently no evidence for post-translational modification of archaeal histones.
Figure 1. Distribution of chromatin associated proteins identified across the Archaea.
Histone proteins and nucleoid-associated proteins (NAPs; right) encoded in each phylum according to the schematic evolutionary tree of Archaea (left).
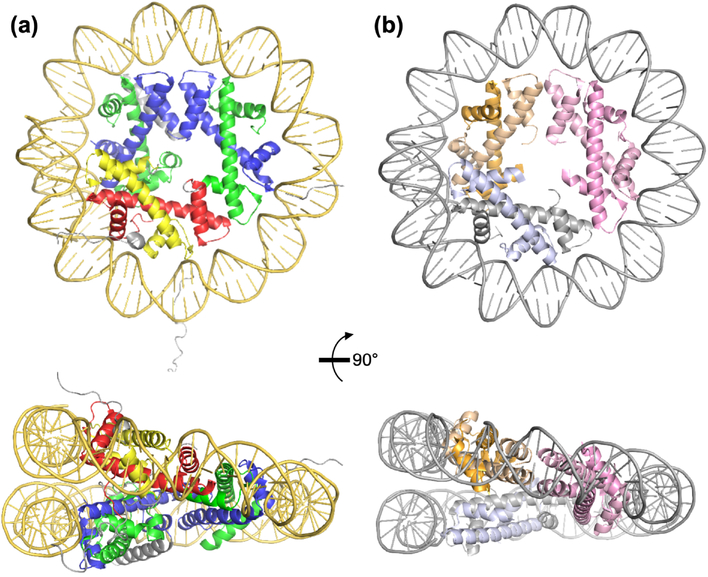
Despite this minimalist approach to histone-based chromatin architecture, archaeal histone-DNA interactions align to the same nucleosome positioning code that was established for Eukarya [10,51,63], and the constrained structure of DNA bound by archaeal histones is nearly identical to the structure of DNA in the eukaryotic nucleosome (Figure 2) [66,67,78,79]. The superhelically-wrapped DNA shares the geometry, diameter, pitch, and writhe of the eukaryotic nucleosomal superhelix, and specific protein-DNA contacts that stabilize archaeal histone-based chromatin are conserved in eukaryotes [56,66,67,79]. The structure of archaeal histone-based chromatin suggests the architectural function of histones (i.e. the ability to bend DNA into the nucleosomal superhelix) was established long (>1 bya) ago, and that the ‘signaling functions’ (i.e. addition of histone extensions and epigenetic modifications) were a secondary addition that came with the expansion to four canonical histones in eukaryotes [66,67].
Figure 2. The structure of histone-based chromatin in Archaea mirrors that of the eukaryotic nucleosome.
(a) The eukaryotic nucleosome hexamer containing two H3-H4 dimers (blue, green respectively) and one H2A-H2B dimer (yellow, red respectively) with wrapped DNA (gold) from a top-down and side view. N and C terminal extensions, specific to eukaryotic histones, are shown in grey. (b) Histone based-chromatin in Archaea can form from varied numbers of histone dimers (three dimers are shown here for comparison to the eukaryotic hexasome), with wrapped DNA (silver) from a top-down and side view. The archaeal histone-based chromatin structure formed with three histone dimers is almost identical to the eukaryotic hexasome without the N- and C-terminal extensions.
Local histone-binding is known to sterically compete with binding of transcription components and offers regulatory potential [12,40,42,43,80], and the extended structure of archaeal histone-based chromatin may also offer regulatory potential. Perhaps the most striking feature of the structure of archaeal histone-based chromatin is the continuous helical ramp of histone dimers and the close association of adjacent layers of the complex that result in a tightly-packed 3D chromatin structure [66,67]. The extensions common to eukaryotic histones normally radiate into solution and facilitate nucleosome-nucleosome interactions. The absence of such extensions on archaeal histones in part permits the close association of adjacent layers of archaeal chromatin. The resultant superstructure places the L1 loops of histone-dimers 1 and 4 along the helical ramp in closest-proximity to each other. Apart from four helix-bundles that link the histone dimers, the only region of close contact between the adjacent layers of archaeal chromatin is where the L1 regions of dimers 1 & 4 meet. L1 sequences almost always retain a central glycine at the point of closest approach and substitution of this glycine with larger side-chains impedes tight packing of archaeal chromatin, impairs gene expression in vivo, and reduces overall fitness [67].
Extension of the structure by one additional histone-dimer extends the length of DNA protection by ~30 bp, resulting in extended polymers that protect DNA from minimally ~60 bp (two histone dimers) to ~480 bp, in 30bp increments [81]. Comparisons of archaeal histone sequences with the atomic-resolution structure of archaeal chromatin reveals that most archaeal histones retain the residues that directly interact with the DNA backbone, use nearly identical residues to stabilize histone-histone and histone- DNA interactions, and that close association of chromatin gyres is likely possible due to minimal side chains in the L1-L1 interface. The eukaryotic nuclear RNA polymerases (RNAPs) and the archaeal RNAP thus regularly encounter – and must overcome – nearly identical histone-DNA contacts that present barriers to transcription elongation [67,82–85].
In contrast to eukaryotic histones, there is no evidence of post-translational modifications to archaeal histones. Although there are many acetyl- and methyl-transferases encoded throughout the Archaea, no activity towards histone proteins has been reported, and the bulk of characterized acetyl- and methyl-transferases are active on DNA or RNA [86–88]. A minority of archaeal organisms encode histones which contain sequences beyond the core histone fold. Excluding single histone isoforms that contain a fused second histone fold (effectively a histone-dimer within a single polypeptide) extended histone sequences are rarely observed [56]. Such extensions are not homologous to those found in eukaryotes but are ‘eukaryote-like’ in being rich in charged residues, especially lysine. Investigation of one such extended archaeal histone variant, MJ1647, a C-terminal extension-containing histone in M. jannaschii, demonstrated that the C-terminal extension was critical for DNA binding and the formation of higher order structures [89]. Modeling the C-terminal extension of MJ1647 into the atomic structure of archaeal histone-based chromatin suggests that the C-terminal extension might impair continued polymerization and impact the global structure of archaeal chromatin. The discovery of new histone variants and histone proteins with extensions in the Heimdallarchaeota and ASGARD archaeal clades hints at the expansion to four canonical histones, the exchange of a histone-polymer for discrete nucleosome particles, and the regulation imposed by post-translational modifications of the histone proteins in all Eukarya [56,57,61,66,67]. Structural modeling of Heimdall LC 3 histones, which contain tails similar in length and sequence composition to extensions on eukaryotic H4, suggests that the extended archaeal histone-based chromatin structure will not be impacted by inclusion of such tails [56]. It will now be important to elucidate the expression, abundance, and function of these archaeal histone variants, included extended-histone variants, in controlling genomic architecture and gene expression.
Global regulation of transcription by archaeal histone-based chromatin
A consensus surrounding the role of archaeal histones in transcription regulation is dubious. This is highlighted by the varying essentiality of histone proteins across archaeal species. Controversy on the role of archaeal histones in controlling gene expression exists at the total transcriptome level when genetically-accessible archaeal species have their histone-encoding loci deleted or modified. In the euryarchaeaon Thermococcus kodakarensis, two histone variants are encoded, and while each individually is not essential, attempts at deleting both histones have been unsuccessful indicating histone-based chromatin is critical for regulation of cellular processes. The importance of regulated genomic architecture was revealed by changes – up to ~10-fold – in the expression of ~5% of genes upon deletion of either histone isoform [90]. The importance of tightly-packed 3D archaeal histone-based chromatin was demonstrated by introduction of histone variants with specific mutations to residues in the L1-L1 interface [67]. Replacing G17 with bulkier amino acid residues does not disrupt local DNA binding but does disrupt extended chromatin structures that in turn impact gene expression. Disruption of extended histone-based chromatin structures also abrogates adaptive gene expression necessary to respond to changing environmental conditions. Histone-proteins are not encoded in all species (Figure 1) and histones are not essential for some extant histone-encoding archaeal species. The sole histone encoded in the methanogen Methanosarcina mazei is dispensable but deletion results in reduced growth, increased sensitivity to DNA damaging agents, reduced overall transcription for many genes, and an altered overall transcriptome [91]. Thus, although non-essential, deletion of the histone-encoding locus, and thus the presumptive loss of histone-based genomic organization – does significantly impact global transcription. Changes to global gene expression and growth were restored upon complementation of M. mazei strains with exogenously produced histone protein, suggesting that histone-based genomic architecture is important, but not essential in some archaeal species. A potentially different view of the role of archaeal histones emerges from studies of halophilic (e.g. salt-loving) archaea. Halobacterium salinarum encodes just one histone protein with several unique attributes. Unlike the typical basic pI of most histone proteins, the high-intracellular salt concentrations of halophiles (~ 4 M) has likely resulted in retention of a histone with an acidic pI. The halophilic histone proteins are also typically a single polypeptide containing two tandemly-repeated histone folds. The single H. salinarum histone, like the single M. mazei histone, is dispensable and deletion results in globally significant, but mild fold-changes in gene expression [91]. Interestingly, these mild changes are growth-phase dependent and, although often small at the transcriptome level, result in significant changes in overall cell morphology. These results were interpreted as indicating a transcription factor-like function of histone proteins in Halobacterium, with global architecture imparted by histone-proteins as largely unimportant to regulating transcriptome-wide expression but select loci with critical histone-binding positions displaying differential expression due to loss of histone production in deletion strains.
Regulation of transcription initiation and elongation with archaeal histone-based chromatin.
Genome-wide impacts of archaeal histone-based chromatin on regulation of gene expression implies that histones are important, often essential, and that changes in histone expression, or histone-induced genomic architecture, impact cellular fitness [9]. To determine how the histone-based landscape directly impacts gene expression, most studies have taken advantage of purified transcription systems and the capacity of archaeal histones to spontaneously bind DNAs in vitro at the same positions utilized in vivo and to form structures that match in vivo 3D chromatin architectures. Early in vitro transcription experiments using components from Methanothermobacter thermautotrophicus demonstrated a repressive effect of histone-addition on transcript production, with complete inhibition of transcription when histone proteins were provided at levels that would theoretically saturate DNA binding (~1 histone dimer per 30 bp of DNA) [12,52,72,92]. These in vitro results were later extended and confirmed using components from Pyrococcus furiosus [69].
Transcription regulation must normally occur within a chromatin landscape. Most archaeal transcription regulators mimic bacterial transcription regulators and bind within or immediately adjacent to core promoter elements to impact formation of initiation complexes. DNA binding positions upstream of the rb2 gene in M. jannaschii were shown to act as histone-nucleating sites, localizing histones whose binding reduces transcription by blocking formation of pre-initiation complexes [65]. Histones are non-specific DNA binding proteins, and unsurprisingly, precision in vitro hydroxyl radical footprinting revealed that the site-specific DNA binding transcription factor Ptr2 effectively competes with localized histone binding – even at saturating histone levels – to activate transcription.
Transcription elongation is also affected by archaeal histone-based chromatin. In vitro transcription assays have been used to establish that the archaeal RNAP is unable to achieve elongation rates that are physiologically relevant through an archaeal chromatin barrier [12,13]. Using DNA templates capable of binding M. thermautotrophicus histone proteins, the M. thermautotrophicus RNA polymerase transcribed template DNA at a rate of ~20 nts/sec in the absence of histone, but just ~2–5 nts/sec when archaeal histones were added to template. The initial collision between the TEC and the histone-barrier results in the greatest obstacle, causing RNA polymerase to pause and likely backtrack. The duration of the initial pause is much greater than subsequent pauses which occur every ~10–15 bp after the transcription elongation complex (TEC) escapes the initial collision. The rate limiting step of transcription through these archaeal histone-based barriers is translocation through the initial DNA-histone contacts.
The first data supporting factors that facilitate elongation through chromatin barriers is supportive of the congruent nature of the simplified archaeal transcription system and the more component complex Pol II apparatus [13]. In vitro transcription experiments, using factors purified from T. kodakarensis, demonstrate that the activities of the conserved transcription factor TFS (TFIIS in Eukarya), and an Spt4/5 complex (also termed Spt4/5 in Eukarya) accelerate the archaeal transcription apparatus through histone-bound templates. The archaeal RNAP often backtracks due to downstream chromatin barriers, and archaeal TFS stimulated endonucleolytic cleavage of transcripts within backtracked complexes results in formation of a new RNA 3’-OH in the active center of RNAP [93–96]. Reactivation of backtracked TECs permits elongation restart and another opportunity for the TEC to transcribe up to and through a downstream chromatin barrier. The Spt4/5 complex, but neither factor individually, also aided in vitro transcription through archaeal histone-based chromatin, presumably due to their stabilizing effects of a closed-clamp configuration of the TEC in aiding proper alignment and retention of the 3’-OH in the RNAP active center [32,95,97–99].
Given the observations of archaeal histone-based chromatin controlling the initiation and elongation aspects of transcription, it is likely that the local chromatin environment also plays a role in termination and proper 3’ end formation of transcripts.
Nucleoid-Associated Proteins (NAPs) in Archaea
The regulation imposed by genomic architecture in archaeal species that do not encode histone proteins has also been investigated in diverse clades. Perhaps the best studied protein is the well-conserved Alba (Sac10b-homologues), but abundant small basic proteins are encoded in both histone- and non-histone encoding archaea that likely impact genomic architectures. We focus first on Alba, then on more recently identified and emerging NAPs in diverse species.
Alba, a conserved chromatin protein, with controversial roles in genomic architecture
Substantial and contentious debate surrounds the Sac10b family of proteins, commonly termed Alba for ‘acetylation lowers binding affinity’, which dominates studies of the non-histone-based organization and regulation of archaeal genomes [100]. Sac10b is a general nucleic-acid binding protein, with affinity for both single-stranded and double-stranded RNA and DNA. Evidence for Sac10b-mediated roles in DNA compaction and organization are recognized, although near equal evidence supports a role for Sac10b in RNA metabolism and binding. A contentious debate surrounds Sac10b, its role in DNA versus RNA binding, and whether acetylation or methylation is the post-translational modification that may impact function of Sac10b proteins in vivo. The focus of many studies was the modification of lysine 16, a well-conserved residue in Sac10b homologues, and identification of proteins that could add or remove a reported acetyl group to impact Sac10b activity. Post-translational modification of K16 within Sac10b proteins was initially described as an acetylation event, hence the common Alba acronym (acetylation lowers binding affinity), but this modification has more recently been identified as a trimethylation [101]. Due to the limited research regarding other nucleoid associated proteins, examination of this paradox is presented here from a historical perspective in the context of newer findings and argues for the further examination of other potential chromatin protein targets.
The Sac10b family of nucleic acid-binding proteins are highly conserved within Archaea, especially species that thrive in (hyper)thermophilic environments. Sac10b family members are encoded in both histone-encoding and non-histone-encoding archaea and are thought to play a major structural role in archaeal chromatin. Most research has focused within the Crenarchaeota, specifically the Sulfolobales. Much of the initial biochemical analyses focused on Alba-DNA interactions. The Sac10b homologue from Sulfolobus shibatae (Ssh10b) is a highly abundant protein (~4% of total protein), was shown to bind dsDNA and influence DNA topology at physiological temperatures [102]. Both electron microscopy (EM) and atomic force microscopy (AFM) experiments revealed an Alba concentration-dependent compaction of archaeal DNA [103–105].
Sac10b proteins are typically encoded in archaeal genomes in the form of Alba1 but some species encode an additional paralog (Alba2) that is typically expressed at lower steady-state protein levels [103]. More detailed investigations detailed that Sac10b bound DNA as a homodimer, and when Alba2 isoforms were present, that Alba heterodimers could also bind and compact DNA [103–105]; Alba2 forms obligate heterodimers with Alba1 and is found exclusively associated with Alba1 in vivo. At lower Alba:DNA ratios, Alba1 homodimers bridge DNA duplexes, slightly compacting DNA by promoting the formation of loop structures [104,105]. At higher concentrations Alba1 homodimers form rigid protein-bound DNA structures [105]. Much like Alba1 homodimers at low concentration, Alba1/Alba2 heterodimers form looped, slightly contracted DNA structures [103]. However, at higher Alba:DNA ratios, the Alba1/Alba2 heterodimers induced highly compacted DNA structures that differed significantly from the rigidified linear chromatin structure of Alba1 homodimers [105]. Crystal structures of Sac10b protein homologues from Aeropyrum pernix K1, Sulfolobus solfataricus, and Pyrococcus horikoshii OT3 all confirm a dimeric mode of nucleic acid interaction [106–111].
In addition to forming distinct protein:DNA complexes that impact DNA topology based on concentration and dimeric partnerships, Sac10b proteins were shown to have high affinity for RNA [112,113]. In Eukarya, Alba-like proteins have diverse RNA metabolism roles [112], suggesting Sac10b proteins may be involved in RNA stability or degradation pathways. Localization of Sac10b to the cytoplasm with no observable association with the nucleoid suggested interaction with RNA rather than DNA in vivo [114]. This suggestion was corroborated by in vivo cross-linking studies with Ssh10b that resulted in the co-purification of primarily ribosomal RNA and mRNA over DNA [113]. Finally, addition of Ssh10b was demonstrated to directly destabilize RNA secondary structure in vitro [115]. The in vitro binding affinity of Sac10b is comparable between RNA, ssDNA, and dsDNA and Sac10b can protect both RNA and DNA from RNase and DNase digestion.
Phyla specific modes of action have also been observed for Sac10b homologues, and particular notice should be taken to studies in mesophilic species versus (hyper)thermophilic archaea. Current evidence suggests that the biological role of Sac10b proteins may have diverged between mesophilic and thermophilic archaea. In contrast to the abundance of Sac10b in (hyper)thermophiles, studies of the Sac10b protein homolog Mmo10b in the mesophilic species Methanococcus maripaludis revealed that Mmo10b is present only in low abundance and bound specific DNA sequences rather than displaying general DNA affinity [116,117]. Deletion of a Sac10b homolog from Methanococcus voltae resulted in changes to protein expression patterns that overlapped with a histone B deletion in the same species [116] and in T. kodakarensis deletion of histone B resulted in altered Sac10b homolog expression [90]. Taken together, these results suggest Sac10b homologues may share an overlapping regulatory role with histones in archaea, and that the presence of histones may reduce the impact of Sac10b regulation of genomic architecture.
Post-translational modification of Alba may impact genomic architecture and gene expression in vivo.
The post-translational modification (PTM) of Sac10b was shown to impact DNA binding affinity and was extrapolated to suggest that PTM of Alba provided regulation akin to PTMs of histone residues common in eukaryotes [100,107]. Recombinant preparations of Alba lacking PTMs displayed greater affinity for DNA than natively purified, PTM-Alba populations. The increased affinity of unmodified Alba also impeded transcription elongation to a greater extent than native, PTM-Alba preparations, consistent with Alba-mediated regulation of genomic structure based on PTM of Alba.
Initial MALDI-TOF mass spectrometry analysis identified lysine 16 (K16) in the Sac10b protein from Sulfolobus solfataricus P2 as the primary site of acetylation. In vitro acetylation by protein acetyltransferase 1 (Pat1) and in vitro deacetylation by the silent information regulator (Sir2) were shown to modify Sac10b imparting a mechanism of Sac10b binding control [100,108,118]. However, K16 is not well-conserved in Sac10b homologues [117], and the initial identification of K16 as the site of modification, and even the PTM itself are now in question. More recent studies have identified Sac10b as a target for both methylation and N-terminal acetylation, but not K16 acetylation [119]. Post-translational modification of the N-terminus of Sac10b by N-acetyl transferase (NAT) has been demonstrated in vitro and is proposed to be the primary site of Sac10b acetylation in vivo. Recent mass-spectrometry (NanoLC-MS-MS) data of a Sac10b homologue from S. islandicus has revealed methylation, acetylation, and deamination of this protein [101]. Strikingly, K16 was trimethylated, not acetylated. The improvements in mass spectrometry and identification of K16 trimethylation challenges the core assertion of Sac10b:DNA interactions being controlled by acetylation at K16. Taken together, the conflicting information on the PTM status of K16, the likely role of Sac10b homologues in binding DNA and RNA, and the differential abundance and importance of Sac10b homologues in diverse species argues that PTM(s) of Sac10b members may also be diverse and likely impact aspects of both RNA and DNA binding.
Variety in archaeal NAPs may shape genomes in diverse environments.
While the biological importance of mechanisms governing Sac10b nucleic acid interactions are heavily debated, it is important to consider the roles of the many other NAPs encoded in archaeal genomes. In addition to Sac10b, most crenarchaea encode small ~7 kDa proteins, with Sul7 and Cren7 dominating the literature. Cren7 is a 7kDa, basic protein that has been found associated with DNA in vivo. The abundant and basic Cren7 protein has high affinity for double-stranded DNA, suggesting a primary role in genomic organization [120]. Although no obvious relationship is present at the primary amino acid level, Sul7 is structurally homologous to Cren7, and both are known to induce DNA compaction in vitro [121]. Crenarchaeal species such as Pyrobaculum aerophilum and Thermoproteus tenax lack obvious Cren7 or Sul7 homologues and instead encode the chromatin protein CC1. Like Sac10b proteins, CC1 is able to bind double stranded and single stranded DNA, suggesting a role in chromatin organization [122].
In the euryarchaeal Thermococcales the TrmBL2 family is an abundant DNA-associated protein [123]. At likely physiological salt concentrations (~300mM KCl) TrmBL2 binds DNA in a site-specific manner, while displaying non-specific DNA binding at lower salt concentrations. Non-specific DNA binding results in a filamentous structure that can compete with histone-binding [124]. In T. kodakarensis the abundance of TrmBL2 changes with the growth phase, and the interplay/competition between histones and TrmBL2 may offer an additional path to regulate genomic architecture and thus gene expression in response to environmental conditions. TrmBL2 occupancy of promoter regions can impact transcription, whereas TrmBL2 minimally impacts transcription elongation [125]. Deletion of TrmBL2 is possible and results in reduced condensation of chromatin and altered expression of approximately the same percentage of genes as deletion of a histone isoform [125].
Conclusions and future perspectives
Archaea are ecologically and metabolically diverse and thus it is perhaps not surprising that substantial differences in genomic architecture and regulation are imposed in different clades. Most species encode proteins with the core histone-fold, and archaeal chromatin thus dominates the landscape of regulation in archaeal species. The overall structural similarities between archaeal histone-based chromatin and eukaryotic chromatin are obvious, but the regulatory potential of the latter far exceeds the potential of the former. Archaeal chromatin is often formed with only one histone isoform, and given the absence of identifiable PTMs, it is likely that archaeal histones are not subject to repositioning or changes in DNA affinity that could increase or decrease transcription levels at specific loci. Significant questions remain for species that encode multiple histone isoforms and whether regulated assembly or binding of unique heterodimers impacts genomic architecture and thus gene regulation. The identification of PTMs or factors that could impact the normally tight association of adjacent gyres of archaeal histone-based chromatin may provide a route to regulate chromatin structure and transcriptional output. Identification of any such factors may help reveal the evolutionary origin of remodeling and modification machineries found ubiquitously in eukaryotes.
The identification of archaeal species that encode extensions on the core histone fold is an exciting new revelation in the context of histone-based regulation of gene expression. The expansion beyond the core histone-fold, and the retention of discrete histone isoforms in many archaeal species provides tantalizing evidence in support of the expansion that must have occurred to provide all extant eukaryotes with the canonical four histones. The length and stability of extended nucleosome-like structures formed with archaeal histones is likely impacted by histone isoforms and the presence of extensions beyond the core histone-fold. Given that disrupting the tight-association of archaeal histone-based chromatin results in massive fitness defects, it is plausible to predict that more fine-tuned and regulated mechanisms may exist to control and adjust chromatin formation or limit the length of the extended histone-based polymers to control gene expression in vivo. The timing of and expansion to defined heterodimeric histone partnerships that lead to the transition from an extended histone-based polymer structure to the discrete particles that define the eukaryotic nucleosome is a major outstanding question.
In addition to chemical modification machinery, eukaryotes encode a wealth of complexes to reposition nucleosomes. Repositioning nucleosomes or altering histone-DNA affinity may help or hinder transcription in eukaryotic cells. It remains possible that archaeal encoded modification or repositioning complexes exist, but current evidence suggests instead that archaeal TECs are reliant on conserved transcription factors to aid in overcoming histone-induced barriers to transcription elongation. To fully illustrate the evolution of the transcription apparatus, the roles of other conserved and potentially novel transcription factors and effectors will need to be characterized. The noted effects of Spt4/5 and TFS suggest that direct modification of the transcription apparatus may suffice for unmodified and relatively uniform histone-based chromatin structures, but that more powerful chromatin remodeling complexes and modification machinery are required for the diverse landscape of extant eukaryotic chromatin landscapes.
Despite many archaea encoding both NAPs and histone proteins, only limited information is available regarding the combinatorial regulation provided by the interplay of architectures produced by binding of both classes of proteins [125]. It is logical to predict that the length and stability of extended histone-based structures may be regulated by NAP binding or NAP-mediated formation of DNA loops that impact overall topology and DNA flexibility. While minimally conserved at an amino acid sequence level, the structural conservation and functionality of archaeal NAPs suggests a conserved strategy for organizing DNA structure [126]. Clarity surrounding the role of the nearly ubiquitous Sac10b family of proteins with respect to RNA versus DNA binding – and clarification of the locations, identity and impacts of potential PTMs – should illuminate the role of this often-abundant protein in organizing and providing dynamic regulation of archaeal genomes.
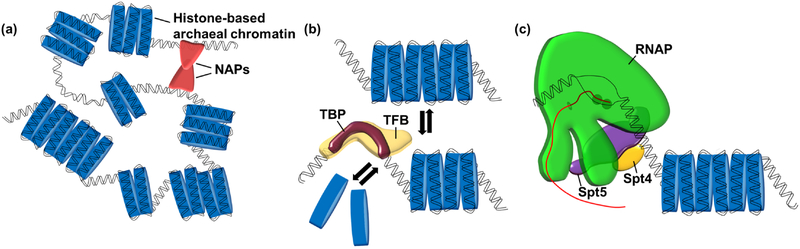
Figure 3. The archaeal chromatin landscape is dynamic.
a) Wrapping of DNA by archaeal histones forms various sizes of extended histone-based chromatin structures. The regulation and depositions of these structures is unknown, but nucleoid associated proteins (NAPs) may play a role in both looping of DNA and size restriction of extended histone polymers. b) Transcription initiation factors TFB and TBP compete with histone proteins for the promoter element in archaea allowing transcription initiation upstream of a chromatinized gene body. c) RNAP must traverse a chromatinized gene body. Spt4-Spt5 permit the transition from initiation to early elongation by displacing TFE and facilitating processive elongation through a chromatin landscape.
Highlights.
Archaea encode histone proteins and nucleoid-associated proteins to organize DNA
Archaeal and eukaryotic histones wrap DNA in a similar geometry
Extended histone-based chromatin structures regulate archaeal gene expression
Conserved transcription factors facilitate transcription through archaeal chromatin
Unique archaeal histone-isoforms may provide regulatory potential
Nucleoid associated proteins (NAPs) are diverse, especially in the Crenarchaeota
Archaeal NAPs likely bind and influence both RNA and DNA structure and dynamics
Post-translational modification control of NAP activity is heavily debated
Acknowledgements
We thank members of the Santangelo laboratory for assistance with manuscript preparation and editing. Studies in the Santangelo lab are supported with funding from the NIH (GM100329) and the Department of Energy (DE-SC0014597).
Abbreviations:
- TEC
Transcription elongation complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- [1].Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution., Nature. 389 (1997) 251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [2].Dame RT, The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin., Mol. Microbiol 56 (2005) 858–70. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- [3].Farnung L, Vos SM, Cramer P, Structure of transcribing RNA polymerase II-nucleosome complex., Nat. Commun 9 (2018) 5432. doi: 10.1038/s41467-018-07870-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Krogh TJ, Møller-Jensen J, Kaleta C, Impact of Chromosomal Architecture on the Function and Evolution of Bacterial Genomes., Front. Microbiol 9 (2018) 2019. doi: 10.3389/fmicb.2018.02019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dillon SC, Dorman CJ, Bacterial nucleoid-associated proteins, nucleoid structure and gene expression., Nat. Rev. Microbiol 8 (2010) 185–95. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- [6].Spurio R, Falconi M, Brandi A, Pon CL, Gualerzi CO, The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending., EMBO J. 16 (1997) 1795–805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Arold ST, Leonard PG, Parkinson GN, Ladbury JE, H-NS forms a superhelical protein scaffold for DNA condensation., Proc. Natl. Acad. Sci. U. S. A 107 (2010) 15728–32. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Becker PB, Hörz W, ATP-dependent nucleosome remodeling., Annu. Rev. Biochem 71 (2002) 247–73. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- [9].Koster MJE, Snel B, Timmers HTM, Genesis of chromatin and transcription dynamics in the origin of species., Cell. 161 (2015) 724–36. doi: 10.1016/j.cell.2015.04.033. [DOI] [PubMed] [Google Scholar]
- [10].Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang J-PZ, Widom J, A genomic code for nucleosome positioning, Nature. 442 (2006) 772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chang CH, Luse DS, The H3/H4 tetramer blocks transcript elongation by RNA polymerasem II in vitro, J. Biol. Chem 272 (1997) 23427–23434. doi: 10.1074/jbc.272.37.23427. [DOI] [PubMed] [Google Scholar]
- [12].Xie Y, Reeve JN, Transcription by an archaeal RNA polymerase is slowed but not blocked by an archaeal nucleosome, J. Bacteriol 186 (2004) 3492–3498. doi: 10.1128/JB.186.11.3492-3498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sanders TJ, Lammers M, Marshall CJ, Walker JE, Lynch ER, Santangelo TJ, TFS and Spt4/5 accelerate transcription through archaeal histone-based chromatin, Mol. Microbiol (2018). doi: 10.1111/mmi.14191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].V Kotlajich M, Hron DR, Boudreau BA, Sun Z, Lyubchenko YL, Landick R, Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria., Elife. 4 (2015). doi: 10.7554/eLife.04970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].van der Valk RA, Vreede J, Crémazy F, Dame RT, Genomic looping: a key principle of chromatin organization., J. Mol. Microbiol. Biotechnol 24 (2014) 344–59. doi: 10.1159/000368851. [DOI] [PubMed] [Google Scholar]
- [16].Bickmore WA, van Steensel B, Genome architecture: domain organization of interphase chromosomes., Cell. 152 (2013) 1270–84. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- [17].Sela I, Wolf YI, Koonin EV, Theory of prokaryotic genome evolution., Proc. Natl. Acad. Sci. U. S. A 113 (2016) 11399–11407. doi: 10.1073/pnas.1614083113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Woese CR, Kandler O, Wheelis ML, Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya., Proc. Natl. Acad. Sci. U. S. A 87 (1990) 4576–9. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=54159&tool=pmcentrez&rendertype=abstract (accessed October 23, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chan PP, Holmes AD, Smith AM, Tran D, Lowe TM, The UCSC Archaeal Genome Browser: 2012 update, Nucleic Acids Res. 40 (2012). doi: 10.1093/nar/gkr990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schneider KL, The UCSC Archaeal Genome Browser, Nucleic Acids Res. 34 (2006) D407–D410. doi: 10.1093/nar/gkj134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brochier-Armanet C, Forterre P, Gribaldo S, Phylogeny and evolution of the Archaea: one hundred genomes later., Curr. Opin. Microbiol 14 (2011) 274–81. doi: 10.1016/j.mib.2011.04.015. [DOI] [PubMed] [Google Scholar]
- [22].Wojtas MN, Abrescia NGA, Archaeal transcription: making up for lost time, Biochem. Soc. Trans 41 (2013) 356–361. doi: 10.1042/BST20120305. [DOI] [PubMed] [Google Scholar]
- [23].Wojtas MN, Mogni M, Millet O, Bell SD, Abrescia NGA, Structural and functional analyses of the interaction of archaeal RNA polymerase with DNA, Nucleic Acids Res. 40 (2012) 9941–9952. doi: 10.1093/nar/gks692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Werner F, Structural evolution of multisubunit RNA polymerases., Trends Microbiol. 16 (2008) 247–50. doi: 10.1016/j.tim.2008.03.008. [DOI] [PubMed] [Google Scholar]
- [25].Werner F, Grohmann D, Evolution of multisubunit RNA polymerases in the three domains of life, Nat. Rev. Microbiol 9 (2011) 85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- [26].Gehring AM, Walker JE, Santangelo TJ, Transcription Regulation in Archaea., J. Bacteriol 198 (2016) 1906–1917. doi: 10.1128/JB.00255-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jun S-H, Hirata A, Kanai T, Santangelo TJ, Imanaka T, Murakami KS, The X-ray crystal structure of the euryarchaeal RNA polymerase in an open-clamp configuration, Nat. Commun 5 (2014) 5132. doi: 10.1038/ncomms6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hirata A, Klein BJ, Murakami KS, The X-ray crystal structure of RNA polymerase from Archaea, Nature. 451 (2008) 851–854. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hirata A, Kanai T, Santangelo TJ, Tajiri M, Manabe K, Reeve JN, Imanaka T, Murakami KS, Archaeal RNA polymerase subunits E and F are not required for transcription in vitro, but a Thermococcus kodakarensis mutant lacking subunit F is temperature-sensitive., Mol. Microbiol 70 (2008) 623–33. doi: 10.1111/j.1365-2958.2008.06430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Santangelo TJ, Cubonová L, James CL, Reeve JN, TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro., J. Mol. Biol 367 (2007) 344–57. doi: 10.1016/j.jmb.2006.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Blombach F, Smollett KL, Grohmann D, Werner F, Molecular Mechanisms of Transcription Initiation-Structure, Function, and Evolution of TFE/TFIIE-Like Factors and Open Complex Formation., J. Mol. Biol 428 (2016) 2592–2606. doi: 10.1016/j.jmb.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Blombach F, Daviter T, Fielden D, Grohmann D, Smollett K, Werner F, Archaeology of RNA polymerase: factor swapping during the transcription cycle., Biochem. Soc. Trans 41 (2013) 362–7. doi: 10.1042/BST20120274. [DOI] [PubMed] [Google Scholar]
- [33].Fouqueau T, Blombach F, Werner F, Evolutionary Origins of Two-Barrel RNA Polymerases and Site-Specific Transcription Initiation., Annu. Rev. Microbiol 71 (2017) 331–348. doi: 10.1146/annurev-micro-091014-104145. [DOI] [PubMed] [Google Scholar]
- [34].Reeve JN, Archaeal chromatin and transcription., Mol. Microbiol 48 (2003) 587–98. http://www.ncbi.nlm.nih.gov/pubmed/12694606 (accessed September 27, 2017). [DOI] [PubMed] [Google Scholar]
- [35].Gehring AM, Santangelo TJ, Manipulating Archaeal Systems to Permit Analyses of Transcription Elongation-Termination Decisions In Vitro, in: Methods Mol. Biol, 2015: pp. 263–279. doi: 10.1007/978-1-4939-2392-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Grohmann D, Werner F, Recent advances in the understanding of archaeal transcription., Curr. Opin. Microbiol 14 (2011) 328–34. doi: 10.1016/j.mib.2011.04.012. [DOI] [PubMed] [Google Scholar]
- [37].Gietl A, Holzmeister P, Blombach F, Schulz S, von Voithenberg LV, Lamb DC, Werner F, Tinnefeld P, Grohmann D, Eukaryotic and archaeal TBP and TFB/TF(II)B follow different promoter DNA bending pathways., Nucleic Acids Res. 42 (2014) 6219–31. doi: 10.1093/nar/gku273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Aravind L, Koonin EV, DNA-binding proteins and evolution of transcription regulation in the archaea., Nucleic Acids Res. 27 (1999) 4658–70. http://www.ncbi.nlm.nih.gov/pubmed/10556324 (accessed February 25, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Peeters E, Charlier D, The Lrp Family of Transcription Regulators in Archaea, Archaea. 2010 (2010) 1–10. doi: 10.1155/2010/750457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Peeters E, Driessen RPC, Werner F, Dame RT, The interplay between nucleoid organization and transcription in archaeal genomes., Nat. Rev. Microbiol 13 (2015) 333–41. doi: 10.1038/nrmicro3467. [DOI] [PubMed] [Google Scholar]
- [41].Bell SD, Cairns SS, Robson RL, Jackson SP, Transcriptional regulation of an archaeal operon in vivo and in vitro., Mol. Cell 4 (1999) 971–82. http://www.ncbi.nlm.nih.gov/pubmed/10635322 (accessed February 25, 2019). [DOI] [PubMed] [Google Scholar]
- [42].Pritchett MA, Wilkinson SP, Geiduschek EP, Ouhammouch M, Hybrid Ptr2- like activators of archaeal transcription., Mol. Microbiol 74 (2009) 582–93. doi: 10.1111/j.1365-2958.2009.06884.x. [DOI] [PubMed] [Google Scholar]
- [43].Wilkinson SP, Ouhammouch M, Geiduschek EP, Transcriptional activation in the context of repression mediated by archaeal histones, Proc. Natl. Acad. Sci 107 (2010) 6777–6781. doi: 10.1073/pnas.1002360107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Krug M, Lee S-J, Boos W, Diederichs K, Welte W, The three-dimensional structure of TrmB, a transcriptional regulator of dual function in the hyperthermophilic archaeon Pyrococcus furiosus in complex with sucrose, Protein Sci. 22 (2013) 800–808. doi: 10.1002/pro.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lie TJ, Hendrickson EL, Niess UM, Moore BC, Haydock AK, Leigh JA, Overlapping repressor binding sites regulate expression of the Methanococcus maripaludis glnK(1) operon., Mol. Microbiol 75 (2010) 755–62. doi: 10.1111/j.1365-2958.2009.07016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lipscomb GL, Keese AM, Cowart DM, Schut GJ, Thomm M, Adams MWW, Scott RA, SurR: a transcriptional activator and repressor controlling hydrogen and elemental sulphur metabolism in Pyrococcus furiosus., Mol. Microbiol 71 (2009) 332–49. doi: 10.1111/j.1365-2958.2008.06525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Karr EA, Transcription Regulation in the Third Domain, in: Adv. Appl. Microbiol, 2014: pp. 101–133. doi: 10.1016/B978-0-12-800259-9.00003-2. [DOI] [PubMed] [Google Scholar]
- [48].Lee S-J, Surma M, Hausner W, Thomm M, Boos W, The role of TrmB and TrmB-like transcriptional regulators for sugar transport and metabolism in the hyperthermophilic archaeon Pyrococcus furiosus., Arch. Microbiol 190 (2008) 247–56. doi: 10.1007/s00203-008-0378-2. [DOI] [PubMed] [Google Scholar]
- [49].Kanai T, Akerboom J, Takedomi S, van de Werken HJG, Blombach F, van der Oost J, Murakami T, Atomi H, Imanaka T, A global transcriptional regulator in Thermococcus kodakaraensis controls the expression levels of both glycolytic and gluconeogenic enzyme-encoding genes., J. Biol. Chem 282 (2007) 33659–70. doi: 10.1074/jbc.M703424200. [DOI] [PubMed] [Google Scholar]
- [50].Maruyama H, Harwood JC, Moore KM, Paszkiewicz K, Durley SC, Fukushima H, Atomi H, Takeyasu K, Kent NA, An alternative beads-on-a-string chromatin architecture in Thermococcus kodakarensis, EMBO Rep. 14 (2013) 711–717. doi: 10.1038/embor.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nalabothula N, Xi L, Bhattacharyya S, Widom J, Wang J-P, Reeve JN, Santangelo TJ, Fondufe-Mittendorf YN, Archaeal nucleosome positioning in vivo and in vitro is directed by primary sequence motifs., BMC Genomics. 14 (2013) 391. doi: 10.1186/1471-2164-14-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Reeve JN, Sandman K, Daniels CJ, Archaeal histones, nucleosomes, and transcription initiation., Cell. 89 (1997) 999–1002. http://www.ncbi.nlm.nih.gov/pubmed/9215621 (accessed February 25, 2019). [DOI] [PubMed] [Google Scholar]
- [53].Fish RN, Kane CM, Promoting elongation with transcript cleavage stimulatory factors., Biochim. Biophys. Acta 1577 (2002) 287–307. http://www.ncbi.nlm.nih.gov/pubmed/12213659 (accessed December 17, 2018). [DOI] [PubMed] [Google Scholar]
- [54].Laptenko O, Lee J, Lomakin I, Borukhov S, Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase., EMBO J. 22 (2003) 6322–34. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S, Intrinsic transcript cleavage activity of RNA polymerase., Proc. Natl. Acad. Sci. U. S. A 92 (1995) 4596–600. http://www.ncbi.nlm.nih.gov/pubmed/7538676 (accessed February 25, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Henneman B, van Emmerik C, van Ingen H, Dame RT, Structure and function of archaeal histones, PLOS Genet. 14 (2018) e1007582. doi: 10.1371/journal.pgen.1007582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJG, Asgard archaea illuminate the origin of eukaryotic cellular complexity., Nature. 541 (2017) 353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- [58].Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG, Archaea and the origin of eukaryotes, Nat. Rev. Microbiol 15 (2017) 711–723. doi: 10.1038/nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- [59].Reeve JN, Bailey KA, Li W-T, Marc F, Sandman K, Soares DJ, Archaeal histones: structures, stability and DNA binding., Biochem. Soc. Trans 32 (2004) 227–30. doi:10.1042/. [DOI] [PubMed] [Google Scholar]
- [60].Sandman K, Reeve JN, Chromosome packaging by archaeal histones., Adv. Appl. Microbiol 50 (2001) 75–99. http://www.ncbi.nlm.nih.gov/pubmed/11677690 (accessed December 17, 2018). [DOI] [PubMed] [Google Scholar]
- [61].Mariño-Ramírez L, Levine KM, Morales M, Zhang S, Moreland RT, Baxevanis AD, Landsman D, The Histone Database: an integrated resource for histones and histone fold-containing proteins., Database (Oxford). 2011 (2011) bar048. doi: 10.1093/database/bar048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Visone V, Vettone A, Serpe M, Valenti A, Perugino G, Rossi M, Ciaramella M, Chromatin structure and dynamics in hot environments: architectural proteins and DNA topoisomerases of thermophilic archaea., Int. J. Mol. Sci 15 (2014) 17162–87. doi: 10.3390/ijms150917162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ammar R, Torti D, Tsui K, Gebbia M, Durbic T, Bader GD, Giaever G, Nislow C, Chromatin is an ancient innovation conserved between Archaea and Eukarya., Elife. 1 (2012) e00078. doi: 10.7554/eLife.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cubonova L, Sandman K, Hallam SJ, DeLong EF, Reeve JN, Histones in Crenarchaea, J. Bacteriol 187 (2005) 5482–5485. doi: 10.1128/JB.187.15.5482-5485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wilkinson SP, Ouhammouch M, Geiduschek EP, Transcriptional activation in the context of repression mediated by archaeal histones., Proc. Natl. Acad. Sci. U. S. A 107 (2010) 6777–81. doi: 10.1073/pnas.1002360107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bhattacharyya S, Mattiroli F, Luger K, Archaeal DNA on the histone merry-go-round, FEBS J. 285 (2018) 3168–3174. doi: 10.1111/febs.14495. [DOI] [PubMed] [Google Scholar]
- [67].Mattiroli F, Bhattacharyya S, Dyer PN, White AE, Sandman K, Burkhart BW, Byrne KR, Lee T, Ahn NG, Santangelo TJ, Reeve JN, Luger K, Structure of histone-based chromatin in Archaea, Science (80-.). 357 (2017) 609–612. doi: 10.1126/science.aaj1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ouhammouch M, Dewhurst RE, Hausner W, Thomm M, Geiduschek EP, Activation of archaeal transcription by recruitment of the TATA-binding protein., Proc. Natl. Acad. Sci. U. S. A 100 (2003) 5097–102. doi: 10.1073/pnas.0837150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Soares D, Dahlke I, Li WT, Sandman K, Hethke C, Thomm M, Reeve JN, Archaeal histone stability, DNA binding, and transcription inhibition above 90 degrees C., Extremophiles. 2 (1998) 75–81. http://www.ncbi.nlm.nih.gov/pubmed/9672681 (accessed September 27, 2017). [DOI] [PubMed] [Google Scholar]
- [70].Bailey KA, Marc F, Sandman K, Reeve JN, Both DNA and histone fold sequences contribute to archaeal nucleosome stability., J. Biol. Chem 277 (2002) 9293–301. doi: 10.1074/jbc.M110029200. [DOI] [PubMed] [Google Scholar]
- [71].Bintu L, Ishibashi T, Dangkulwanich M, Wu Y-Y, Lubkowska L, Kashlev M, Bustamante C, Nucleosomal elements that control the topography of the barrier to transcription., Cell. 151 (2012) 738–49. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Grayling RA, Sandman K, Reeve JN, Histones and chromatin structure in hyperthermophilic Archaea., FEMS Microbiol. Rev 18 (1996) 203–13. doi: 10.1111/j.1574-6976.1996.tb00237.x. [DOI] [PubMed] [Google Scholar]
- [73].Starich MR, Sandman K, Reeve JN, Summers MF, NMR structure of HMfB from the hyperthermophile, Methanothermus fervidus, confirms that this archaeal protein is a histone., J. Mol. Biol 255 (1996) 187–203. doi: 10.1006/jmbi.1996.0016. [DOI] [PubMed] [Google Scholar]
- [74].Decanniere K, Sandman K, Reeve JN, Heinemann U, Crystallization and preliminary X-ray characterization of the Methanothermus fervidus histones HMfA and HMfB., Proteins. 24 (1996) 269–71. doi:. [DOI] [PubMed] [Google Scholar]
- [75].Decanniere K, Babu AM, Sandman K, Reeve JN, Heinemann U, Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus, J. Mol. Biol 303 (2000) 35–47. doi: 10.1006/jmbi.2000.4104. [DOI] [PubMed] [Google Scholar]
- [76].Sandman K, Soares D, Reeve JN, Molecular components of the archaeal nucleosome., Biochimie. 83 (2001) 277–81. http://www.ncbi.nlm.nih.gov/pubmed/11278079 (accessed February 20, 2017). [DOI] [PubMed] [Google Scholar]
- [77].Sandman K, Reeve JN, Archaeal histones and the origin of the histone fold., Curr. Opin. Microbiol 9 (2006) 520–5. doi: 10.1016/j.mib.2006.08.003. [DOI] [PubMed] [Google Scholar]
- [78].Soares DJ, Sandman K, Reeve JN, Mutational analysis of archaeal histone- DNA interactions, J. Mol. Biol 297 (2000) 39–47. doi: 10.1006/jmbi.2000.3546. [DOI] [PubMed] [Google Scholar]
- [79].Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution., Nature. 389 (1997) 251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [80].Ouhammouch M, Werner F, Weinzierl ROJ, Geiduschek EP, A fully recombinant system for activator-dependent archaeal transcription., J. Biol. Chem 279 (2004) 51719–21. doi: 10.1074/jbc.C400446200. [DOI] [PubMed] [Google Scholar]
- [81].Marc F, Sandman K, Lurz R, Reeve JN, Archaeal histone tetramerization determines DNA affinity and the direction of DNA supercoiling., J. Biol. Chem 277 (2002) 30879–86. doi: 10.1074/jbc.M203674200. [DOI] [PubMed] [Google Scholar]
- [82].Kujirai T, Ehara H, Fujino Y, Shirouzu M, Sekine S, Kurumizaka H, Structural basis of the nucleosome transition during RNA polymerase II passage, Science (80-.). (2018) eaau9904. doi: 10.1126/science.aau9904. [DOI] [PubMed] [Google Scholar]
- [83].Ehara H, Yokoyama T, Shigematsu H, Yokoyama S, Shirouzu M, Sekine S, Structure of the complete elongation complex of RNA polymerase II with basal factors, Science (80-.). 357 (2017) 921–924. doi: 10.1126/science.aan8552. [DOI] [PubMed] [Google Scholar]
- [84].Teves SS, Weber CM, Henikoff S, Transcribing through the nucleosome, Trends Biochem. Sci 39 (2014) 577–586. doi: 10.1016/j.tibs.2014.10.004. [DOI] [PubMed] [Google Scholar]
- [85].Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M, Nature of the nucleosomal barrier to RNA polymerase II., Mol. Cell 18 (2005) 97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- [86].Höfer K, Jäschke A, Epitranscriptomics: RNA Modifications in Bacteria and Archaea, Microbiol. Spectr 6 (2018). doi: 10.1128/microbiolspec.RWR-0015-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ouellette M, Gogarten J, Lajoie J, Makkay A, Papke R, Characterizing the DNA Methyltransferases of Haloferax volcanii via Bioinformatics, Gene Deletion, and SMRT Sequencing, Genes (Basel). 9 (2018) 129. doi: 10.3390/genes9030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Couturier M, Lindås A-C, The DNA Methylome of the Hyperthermoacidophilic Crenarchaeon Sulfolobus acidocaldarius., Front. Microbiol 9 (2018) 137. doi: 10.3389/fmicb.2018.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li WT, Sandman K, Pereira SL, Reeve JN, MJ1647, an open reading frame in the genome of the hyperthermophile Methanococcus jannaschii, encodes a very thermostable archaeal histone with a C-terminal extension., Extremophiles. 4 (2000) 43–51. http://www.ncbi.nlm.nih.gov/pubmed/10741836 (accessed February 25, 2019). [DOI] [PubMed] [Google Scholar]
- [90].Čuboňováa L, Katano M, Kanai T, Atomi H, Reeve JN, Santangelo TJ, An archaeal histone is required for transformation of Thermococcus kodakarensis., J. Bacteriol 194 (2012) 6864–74. doi: 10.1128/JB.01523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Weidenbach K, Glöer J, Ehlers C, Sandman K, Reeve JN, Schmitz RA, Deletion of the archaeal histone in Methanosarcina mazei Gö1 results in reduced growth and genomic transcription, Mol. Microbiol 67 (2008) 662–671. doi: 10.1111/j.1365-2958.2007.06076.x. [DOI] [PubMed] [Google Scholar]
- [92].Sandman K, Reeve JN, Archaeal chromatin proteins: different structures but common function?, Curr. Opin. Microbiol 8 (2005) 656–61. doi: 10.1016/j.mib.2005.10.007. [DOI] [PubMed] [Google Scholar]
- [93].Hausner W, Lange U, Musfeldt M, Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase, J. Biol. Chem 275 (2000) 12393–12399. doi: 10.1074/jbc.275.17.12393. [DOI] [PubMed] [Google Scholar]
- [94].Lange U, Hausner W, Transcriptional fidelity and proofreading in Archaea and implications for the mechanism of TFS-induced RNA cleavage, Mol. Microbiol 52 (2004) 1133–1143. doi: 10.1111/j.1365-2958.2004.04039.x. [DOI] [PubMed] [Google Scholar]
- [95].Sanders TJ, Lammers M, Marshall CJ, Walker JE, Lynch ER, Santangelo TJ, TFS and Spt4/5 accelerate transcription through archaeal histone-based chromatin, Mol. Microbiol (2019). doi: 10.1111/mmi.14191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Fouqueau T, Blombach F, Hartman R, Cheung ACM, Young MJ, Werner F, The transcript cleavage factor paralogue TFS4 is a potent RNA polymerase inhibitor., Nat. Commun 8 (2017) 1914. doi: 10.1038/s41467-017-02081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hirtreiter A, Damsma GE, Cheung ACM, Klose D, Grohmann D, Vojnic E, Martin ACR, Cramer P, Werner F, Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif, Nucleic Acids Res. 38 (2010) 4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P, Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity, EMBO J. 30 (2011) 1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Guo G, Gao Y, Zhu Z, Zhao D, Liu Z, Zhou H, Niu L, Teng M, Structural and biochemical insights into the DNA-binding mode of MjSpt4p:Spt5 complex at the exit tunnel of RNAPII., J. Struct. Biol 192 (2015) 418–425. doi: 10.1016/j.jsb.2015.09.023. [DOI] [PubMed] [Google Scholar]
- [100].Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF, The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation., Science. 296 (2002) 148–51. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- [101].Cao J, Wang Q, Liu T, Peng N, Huang L, Insights into the post-translational modifications of archaeal Sis10b (Alba): lysine-16 is methylated, not acetylated, and this does not regulate transcription or growth., Mol. Microbiol 109 (2018) 192–208. doi: 10.1111/mmi.13973. [DOI] [PubMed] [Google Scholar]
- [102].Xue H, Guo R, Wen Y, Liu D, Huang L, An abundant DNA binding protein from the hyperthermophilic archaeon Sulfolobus shibatae affects DNA supercoiling in a temperature-dependent fashion., J. Bacteriol 182 (2000) 3929–33. http://www.ncbi.nlm.nih.gov/pubmed/10869069 (accessed February 12, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Jelinska C, Conroy MJ, Craven CJ, Hounslow AM, Bullough PA, Waltho JP, Taylor GL, White MF, Obligate heterodimerization of the archaeal Alba2 protein with Alba1 provides a mechanism for control of DNA packaging., Structure. 13 (2005) 963–71. doi: 10.1016/j.str.2005.04.016. [DOI] [PubMed] [Google Scholar]
- [104].Lurz R, Grote M, Dijk J, Reinhardt R, Dobrinski B, Electron microscopic study of DNA complexes with proteins from the Archaebacterium Sulfolobus acidocaldarius., EMBO J. 5 (1986) 3715–21. http://www.ncbi.nlm.nih.gov/pubmed/16453745 (accessed February 12, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Laurens N, Driessen RPC, Heller I, Vorselen D, Noom MC, Hol FJH, White MF, Dame RT, Wuite GJL, Alba shapes the archaeal genome using a delicate balance of bridging and stiffening the DNA., Nat. Commun 3 (2012) 1328. doi: 10.1038/ncomms2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Tanaka T, Padavattan S, Kumarevel T, Crystal structure of archaeal chromatin protein Alba2-double-stranded DNA complex from Aeropyrum pernix K1., J. Biol. Chem 287 (2012) 10394–402. doi: 10.1074/jbc.M112.343210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wardleworth BN, Russell RJM, Bell SD, Taylor GL, White MF, Structure of Alba: an archaeal chromatin protein modulated by acetylation., EMBO J. 21 (2002) 4654–62. http://www.ncbi.nlm.nih.gov/pubmed/12198167 (accessed February 23, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Zhao K, Chai X, Marmorstein R, Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA binding., J. Biol. Chem 278 (2003) 26071–7. doi: 10.1074/jbc.M303666200. [DOI] [PubMed] [Google Scholar]
- [109].Kumarevel T, Sakamoto K, Gopinath SCB, Shinkai A, Kumar PKR, Yokoyama S, Crystal structure of an archaeal specific DNA-binding protein (Ape10b2) from Aeropyrum pernix K1., Proteins. 71 (2008) 1156–62. doi: 10.1002/prot.21807. [DOI] [PubMed] [Google Scholar]
- [110].Cui Q, Tong Y, Xue H, Huang L, Feng Y, Wang J, Two conformations of archaeal Ssh10b. The origin of its temperature-dependent interaction with DNA., J. Biol. Chem 278 (2003) 51015–22. doi: 10.1074/jbc.M308510200. [DOI] [PubMed] [Google Scholar]
- [111].Hada K, Nakashima T, Osawa T, Shimada H, Kakuta Y, Kimura M, Crystal structure and functional analysis of an archaeal chromatin protein Alba from the hyperthermophilic archaeon Pyrococcus horikoshii OT3., Biosci. Biotechnol. Biochem 72 (2008) 749–58. http://www.ncbi.nlm.nih.gov/pubmed/18323660 (accessed February 12, 2019). [DOI] [PubMed] [Google Scholar]
- [112].Aravind L, Iyer LM, Anantharaman V, The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism., Genome Biol. 4 (2003) R64. doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Guo R, Xue H, Huang L, Ssh10b, a conserved thermophilic archaeal protein, binds RNA in vivo., Mol. Microbiol 50 (2003) 1605–15. http://www.ncbi.nlm.nih.gov/pubmed/14651642 (accessed February 12, 2019). [DOI] [PubMed] [Google Scholar]
- [114].Bohrmann B, … E.K.-J. of S., undefined 1994, Localization of histone-like proteins in thermophilic Archaea by immunogold electron microscopy, Elsevier; (n.d.). https://www.sciencedirect.com/science/article/pii/S1047847784710082 (accessed February 25, 2019). [Google Scholar]
- [115].Guo L, Ding J, Guo R, Hou Y, Wang D-C, Huang L, Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea., J. Biol. Chem 289 (2014) 1478–90. doi: 10.1074/jbc.M113.521351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Heinicke I, Müller J, Pittelkow M, Klein A, Mutational analysis of genes encoding chromatin proteins in the archaeon Methanococcus voltae indicates their involvement in the regulation of gene expression., Mol. Genet. Genomics 272 (2004) 76–87. doi: 10.1007/s00438-004-1033-5. [DOI] [PubMed] [Google Scholar]
- [117].Liu Y, Guo L, Guo R, Wong RL, Hernandez H, Hu J, Chu Y, Amster IJ, Whitman WB, Huang L, The Sac10b homolog in Methanococcus maripaludis binds DNA at specific sites., J. Bacteriol 191 (2009) 2315–29. doi: 10.1128/JB.01534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Marsh VL, Peak-Chew SY, Bell SD, Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba., J. Biol. Chem 280 (2005) 21122–8. doi: 10.1074/jbc.M501280200. [DOI] [PubMed] [Google Scholar]
- [119].Vorontsov EA, Rensen E, Prangishvili D, Krupovic M, Chamot-Rooke J, Abundant Lysine Methylation and N-Terminal Acetylation in Sulfolobus islandicus Revealed by Bottom-Up and Top-Down Proteomics., Mol. Cell. Proteomics 15 (2016) 3388–3404. doi: 10.1074/mcp.M116.058073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Guo L, Feng Y, Zhang Z, Yao H, Luo Y, Wang J, Huang L, Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea., Nucleic Acids Res. 36 (2008) 1129–37. doi: 10.1093/nar/gkm1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Driessen RPC, Meng H, Suresh G, Shahapure R, Lanzani G, Priyakumar UD, White MF, Schiessel H, van Noort J, Dame RT, Crenarchaeal chromatin proteins Cren7 and Sul7 compact DNA by inducing rigid bends., Nucleic Acids Res. 41 (2013) 196–205. doi: 10.1093/nar/gks1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Luo X, Schwarz-Linek U, Botting CH, Hensel R, Siebers B, White MF, CC1, a novel crenarchaeal DNA binding protein., J. Bacteriol 189 (2007) 403–9. doi: 10.1128/JB.01246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Wierer S, Daldrop P, Ud Din Ahmad M, Boos W, Drescher M, Welte W, Seidel R, TrmBL2 from Pyrococcus furiosus Interacts Both with Double-Stranded and Single-Stranded DNA., PLoS One. 11 (2016) e0156098. doi: 10.1371/journal.pone.0156098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Efremov AK, Qu Y, Maruyama H, Lim CJ, Takeyasu K, Yan J, Transcriptional Repressor TrmBL2 from Thermococcus kodakarensis Forms Filamentous Nucleoprotein Structures and Competes with Histones for DNA Binding in a Salt- and DNA Supercoiling-dependent Manner., J. Biol. Chem 290 (2015) 15770–84. doi: 10.1074/jbc.M114.626705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Maruyama H, Shin M, Oda T, Matsumi R, Ohniwa RL, Itoh T, Shirahige K, Imanaka T, Atomi H, Yoshimura SH, Takeyasu K, Histone and TK0471/TrmBL2 form a novel heterogeneous genome architecture in the hyperthermophilic archaeon Thermococcus kodakarensis., Mol. Biol. Cell 22 (2011) 386–98. doi: 10.1091/mbc.E10-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Driessen RPC, Dame RT, Nucleoid-associated proteins in Crenarchaea., Biochem. Soc. Trans 39 (2011) 116–21. doi: 10.1042/BST0390116. [DOI] [PubMed] [Google Scholar]