Abstract
Purpose
Circular RNAs (circRNAs) play important roles in the development and progression of various human cancers. hsa-circ-0046600 is a circRNA of unknown function. The purpose of this study was to investigate the biological function of hsa-circ-0046600 in hepatocellular carcinoma (HCC) and elucidate the possible molecular mechanisms of this circRNA.
Materials and methods
GSE97332, quantitative reverse transcription polymerase chain reaction (qRT-PCR) and fluorescence in situ hybridization (FISH) were used to detect the expression of hsa-circ-0046600 in HCC tissues and cells. A dual-luciferase reporter assay was used to confirm the interaction between hsa-circ-0046600 and miR-640, and a meta-analysis confirmed the expression of miR-640 in HCC. Bioinformatics was used for the functional analysis of miR-640 target genes. N-cadherin and HIF-1α expression was measured by Western blot analysis.
Results
The expression level of hsa-circ-0046600 in HCC tissue was significantly higher than that in adjacent normal tissue (P < 0.05) and was associated with tumour size, TNM stage and pathological vascular invasion. Moreover, the downregulated expression of hsa-circ-0046600 significantly inhibited the migration of HepG2 and SK-HEP-1 cells. hsa-circ-0046600 is present mainly in the cytoplasm and promotes the expression of proteins such as HIF-1α by competitively binding to miR-640 in HCC, thereby affecting the malignant biological behaviour of liver cancer cells.
Conclusion
hsa-circ-0046600 can be used as a new biomarker for HCC diagnosis and disease progression and provides a potential target for targeted therapy.
Keywords: hsa-circ-0046600, miR-640, HIF-1α, migration, HCC
Introduction
Hepatocellular carcinoma (HCC) is a malignant tumour with a high incidence and risk of death worldwide and is related to obesity, alcohol abuse, dyslipidaemia, metabolic syndrome and hepatitis.1,2 Currently, although several studies have shown that many genes are involved in HCC and many treatment strategies have been developed for this disease, the 5-year survival rate of patients is still very low.3,4 Therefore, it is still necessary to explore the mechanism and therapeutic targets in further research on HCC.
Circular RNAs (circRNAs) are non-coding RNAs with a length of approximately 100 bp to 4 kb and are found in almost all major eukaryotes (such as animals, plants and fungi), including in the blood, different subcellular compartments (including nuclei and cytoplasms), and extracellular exosomes.5 At present, most studies focus on the mode of action of circRNAs (which is similar to that of long non-coding RNAs (lncRNAs)), as microRNA (miRNA) sponges affect the negative regulatory effect of miRNAs on mRNAs, thereby regulating gene expression.6
In this study, we discovered a functionally unknown circRNA, hsa-circ-0046600, which is associated with HCC, by using the bioinformatics software circBase (http://www.circbase.org/).7 hsa-circ-00446600 is a circRNA consisting of the anti-shear of exons 4, 5, and 6 of its maternal gene B3GNTL1 and has a length of 227 bp. We found that hsa-circ-0046600 is highly expressed in HCC and is involved in regulating the migration of HCC cells. As hsa-circ-0046600 is mainly expressed in the cytoplasm, we mainly studied the competitive endogenous RNA (ceRNA) mechanism of hsa-circ-0046600. Studies have shown that hsa-circ-004660 regulates the functions of its target genes, such as HIF, by acting as a sponge for miR-640. These results provide an important basis for the use of hsa-circ-0046600 as a potential target for the diagnosis and treatment of HCC.
Materials And Methods
Human Tissue Samples And Cell Culture
A total of 20 paired HCC tissue and adjacent normal tissue samples were obtained from patients who underwent surgical procedures at HENAN PROVINCIAL PEOPLE’S HOSPITAL of Zhengzhou University. All of the patients provided written consent, and approval was obtained from the Ethics Committee of HENAN PROVINCIAL PEOPLE’S HOSPITAL of Zhengzhou University. The patient consent was written informed consent, and that this was conducted in accordance with the Declaration of Helsinki. The HL-7702 (L-02), SK-HEP-1, SMMC-7721, HepG2, HuH7, and 293T cell lines were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). HepG2, SMMC-7721, SK-HEP-1, and HuH7 cells were cultured in Dulbecco’s modified Eagle medium (Gibco, USA), and L-02 and SMMC-7721 cells were cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% foetal bovine serum (Gibco, USA), 50 U/mL penicillin, and 50 μg/mL streptomycin (Gibco, USA). The cells were maintained at 37°C in a humidified incubator at 5% CO2.
Small Interfering RNA (siRNA) And miRNA Transfection
hsa-circ-0046600 siRNA and miR-640 mimics were all purchased from RiboBio (Guangzhou, China). SK-HEP-1 and HepG2 cells were seeded 24 h before transfection. The siRNA and miRNA mimics were transfected using Lipofectamine 3000 (Invitrogen, USA) with serum-free medium. After 48 h, the cells were collected for testing.
Quantitative Reverse Transcription PCR (qRT-PCR)
Total RNA was extracted from cell lines or the 20 paired HCC tissue and adjacent normal tissue samples using TRIzol Reagent (Invitrogen, USA). A total of 1 µg of total RNA was reverse transcribed into cDNA using a Thermo Fisher Scientific RevertAid First Star and cDNA Synthesis Kit (Thermo Fisher Scientific, USA) (the circRNA primer was a random hexamer primer; the miRNA primer was synthesized by RIBO). Then, qRT-PCR amplification of the cDNA was performed using PowerUpTM SYBRTM Green Master Mix (Applied Biosystems, USA). The sequences of the miRNA primers synthesized by RiboBio and of the circRNA and mRNA primers were as follows:
hsa-circ-0046600, 5`-GCTCCTAACCCAGTCGGATAC-3` (forward), 5`-CCGAATCCAAAAAGCAAAGGTA-3` (reverse);
B3GNTL1 mRNA, 5`-TCGGATACGCTAAAAATCAAG-3` (forward), 5`-CTGGGTTAGGAGCTGCTCCG-3` (reverse); and
β-actin, 5`-CCTGTACGCCAACACAGTGC-3` (forward), 5`-ATACTCCTGCTTGCTGATCC-3` (reverse).
Fluorescence In Situ Hybridization (FISH)
Hybridization was performed overnight with hsa-circ-0046600 probes, sections were washed, and DAPI stain was added and incubated for 8 min in the dark. The sections were observed under a Nikon upright fluorescence microscope, and images were acquired. The nuclei stained with DAPI appeared blue under UV excitation and contained hsa-circ-0046600, which was positively labelled with Cy3 (red emission). The sequence of the hsa-circ-0046600 probe used for FISH was 5ʹ-TTTAG CGTAT CCGAC TGGGT TAGGA GCTGC-3ʹ.
Wound Scratch Assay
SK-HEP-1 and HepG2 cells were seeded into 6-well plates and transfected on the second day. Cells were then incubated at 5% CO2/37°C until they fused into a monolayer, and the cell monolayer was scraped along a straight line with a 200 µl pipette tip to create a gap. The migration distance was calculated using an inverted microscope at 0 h, 24 h, and 48 h.
Western Blot Analysis
When cells reached 80% confluence, the medium was discarded, and the cells were washed with pre-chilled 1× phosphate-buffered saline (PBS). Then, 320 μL of cell lysis buffer (RIPA buffer supplemented with 3.2 µL PMSF) was added to the cells to extract cellular protein. After a 30-min incubation on ice, the cells were scraped into a 1.5-mL centrifuge tube and subjected to centrifugation at 4°C for 15 min at 12,000 rpm. Protein quantification was performed using a NanoDrop ND-1000. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and electrotransferred to a polyvinylidene fluoride membrane. The membrane was incubated with a primary antibody at 4°C overnight, washed extensively with 1% TBST, and incubated with a secondary antibody conjugated to horseradish peroxidase (1:1000) at room temperature for 3 h. Immunolabelling was visualized using an electrochemiluminescence system. The primary antibodies included anti-N-cadherin (dilution 1:1000, AF4039) and anti-HIF-1α (dilution 1:200, AF1009) antibodies, which were purchased from Affinity (USA), and an anti-GAPDH antibody (dilution 1:2000, AF7021).
Acquisition Of hsa-circ-0046600, miR-640 And Target mRNAs
The GSE97332 dataset in the Gene Expression Omnibus (GEO) database was used to screen for differentially expressed circRNAs in HCC.8 R software was used to screen circRNAs with |logFC |> 2 and adjusted P-value <0.05. CircInteractome (https://circinteractome.nia.nih.gov/), TargetScan 7.2 (http://www.targetscan.org/vert_72/), miRDB (http://www.mirdb.org/) and mirDIP (http://ophid.utoronto.ca/mirDIP/index_confirm.jsp) were used to predict the assembly sites of circRNA-miRNA or miRNA-mRNA.9–11 For the meta-analysis, GEO datasets were screened to verify the expression of miR-640 in HCC (dataset screening keywords: HCC and (microRNAs OR miRNA) and Homo sapiens). R software was used to screen mRNAs with |logFC |> 1 and adjusted P-value <0.05 in The Cancer Genome Atlas (TCGA)-HCC data.12
Functional Enrichment Analysis And Protein-Protein Interaction (PPI) Network Construction
To understand the underlying biological processes and pathways of the aberrantly expressed mRNAs, Gene Ontology (GO) biological process term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted using the Cytoscape plug-in ClueGO and Metascape (Database for Annotation, Visualization, and Integrated Discovery; http://metascape.org).13 The GO enrichment analysis was based on a P-value < 0.05 and a kappa score of 0.3, which were applied as the thresholds to divide the tree into term clusters. Significant enrichment results were visualized using Cytoscape software 3.3.0. The STRING database (http://string-db.org/)14 was used as background data to construct a PPI network using proteins with an interaction score greater than 0.4.
Statistical Analysis
Statistical analysis was performed using SPSS (version 25.0), and data are recorded as the mean ± standard deviation (SD). Differences between the two groups were analysed using Student’s t-test. Differences between the three groups were analysed using one-way analysis of variance (ANOVA). Pearson’s correlation analysis was used to study the correlation between circRNAs and miRNAs. P < 0.05 was considered statistically significant.
Results
hsa-circ-0046600 Expression Is Upregulated In HCC And Associated With Clinicopathological Variables
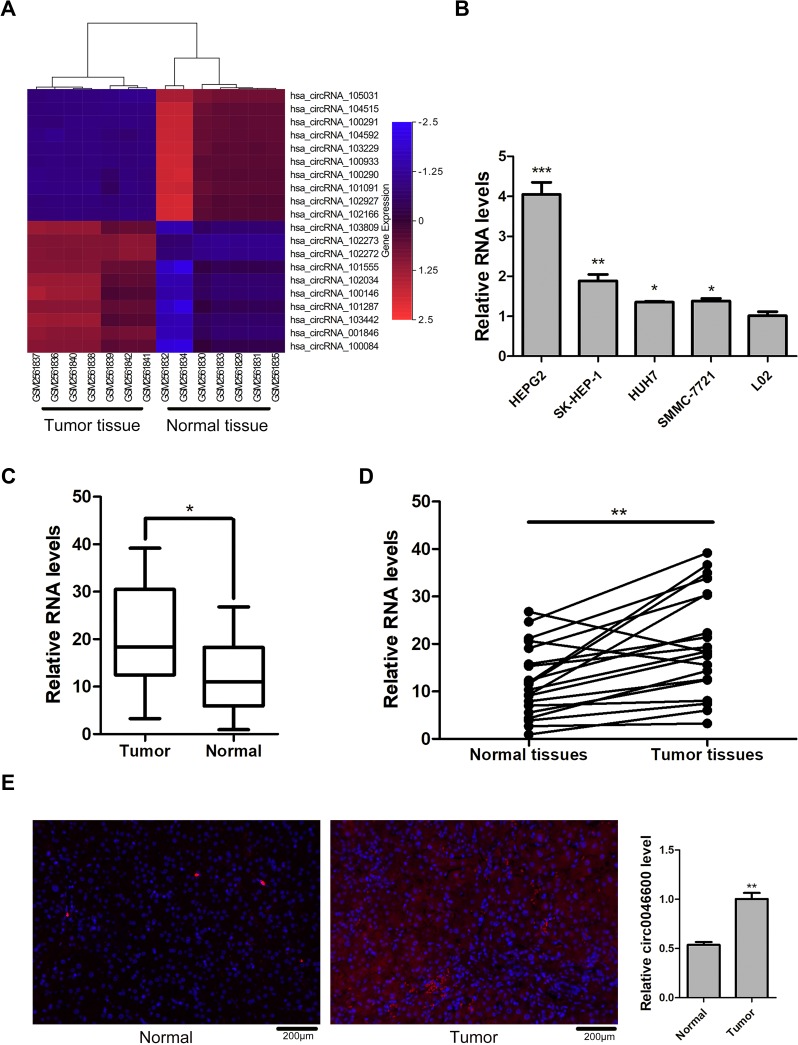
The GSE97332 dataset showed that hsa-circ-0046600 was highly expressed in HCC tissues (Figure 1A). The qRT-PCR results showed that the expression level of hsa-circ-0046600 was upregulated by approximately 1.35–4.04-fold in the tumour cell line compared to that in the normal liver cell line (L-02) (Figure 1B). hsa-circ-0046600 was highly expressed in 90% of the HCC tissue samples relative to the corresponding adjacent tissue samples (P = 0.003; Figure 1C and D). In addition, the expression of hsa-circ-0046600 was significantly upregulated in patients with tumours ≥5 cm (P = 0.003) and stage III/IV (P = 0.028). High hsa-circ-0046600 expression was significantly associated with pathological vascular invasion (P = 0.019). However, tumour differentiation, patient age and sex were irrelevant (all P > 0.05, Table 1). We used FISH to locate the expression of hsa-circ-0046600 in HCC and adjacent tissue samples and randomly selected 5 fields of view to count. The results showed that the expression in the tumour group was approximately 1.83 times higher than that in the adjacent normal tissue group.
Figure 1.
hsa-circ-0046600 expression in HCC. (A) The top 20 differentially expressed circRNAs in the GSE97332 dataset. In circBase, hsa-circ-102272 is named hsa-circ-0046600. (B) The expression levels of hsa-circ-0046600 in HepG2, SK-HEP-1, HuH7, and SMMC-7721 cells were compared with the expression level of hsa-circ-0046600 in L-02 cells: the fold change was 4.04, 1.88, 1.35, and 1.38, respectively. (C) The expression level of hsa-circ-0046600 was upregulated in 20 HCC tissue samples compared with matched noncancerous tissue samples (fold change≈1.7). (D) hsa-circ-0046600 expression was upregulated in 90% (18/20) of liver tissue samples. (E) The localization of hsa-circ-0046600 was assessed in HCC and adjacent tissues. The results showed that hsa-circ-0046600 was highly expressed in tumour tissue (fold change ≈1.9). Data are reported as the mean ± SD. *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001.
Table 1.
Correlation Between hsa-circ-0046600 Expression And The Clinicopathological Features Of HCC Patients
| Clinicopathological Feature | Number (n=20) | P-value |
|---|---|---|
| Age (years) | 0.951 | |
| <60 | 16 | |
| ≥60 | 4 | |
| Sex | 0.574 | |
| Male | 16 | |
| Female | 4 | |
| Tumour size (cm) | 0.004** | |
| <5 | 9 | |
| ≥5 | 11 | |
| TNM stage | 0.032* | |
| I | 7 | |
| II | 2 | |
| III | 10 | |
| IV | 1 | |
| Tumour differentiation | 0.706 | |
| Low | 3 | |
| Moderate | 16 | |
| High | 1 | |
| Pathological vascular invasion | 0.019* | |
| Negative | 13 | |
| Positive | 7 |
Notes: * P-value < 0.05, ** P-value < 0.01.
hsa-circ-0046600 Promotes The Migration Of HCC Cells
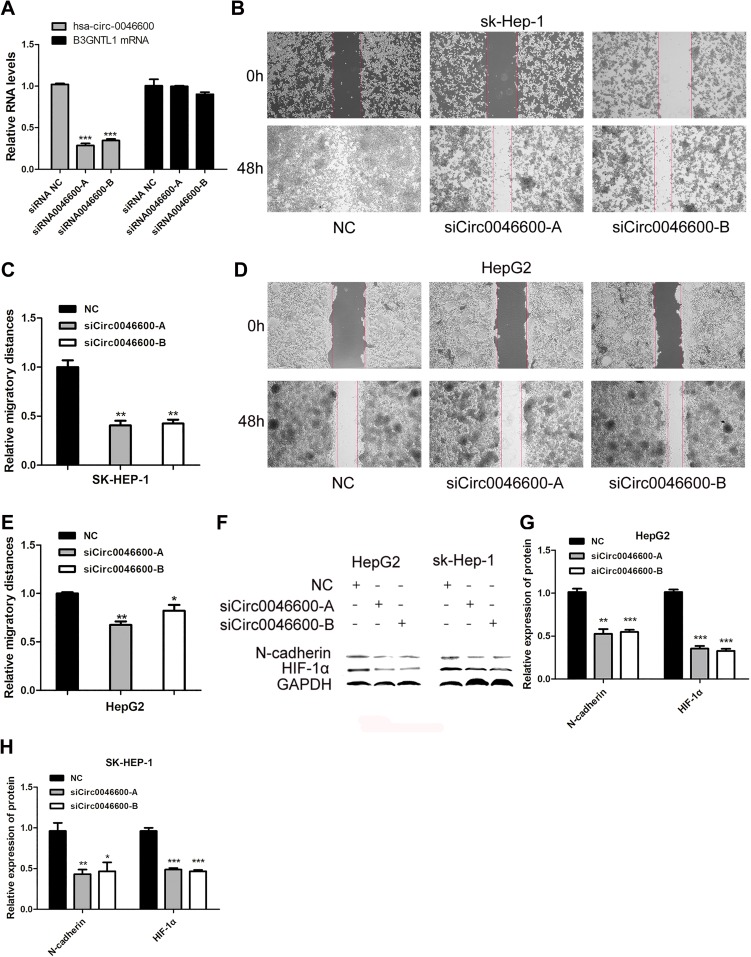
To reveal the effect of hsa-circ-0046600 in HCC, SK-HEP-1 and HepG2 cells were transfected with an hsa-circ-0046600 siRNA inhibitor or the corresponding negative control (NC). The qRT-PCR results indicated that the siRNA affected only hsa-circ-0046600 and not B3GNTL1 mRNA (Figure 2A). A wound scratch test showed that hsa-circ-0046600 knockdown inhibited the migration of SK-HEP-1 cells (siCirc0046600-A group, the migration distance was 40.33% relative to the NC group, P=0.002; siCirc0046600-B group, the migration distance was 42.33% relative to the NC group, P=0.002) and HepG2 cells (siCirc0046600-A group, the migration distance was 67.33% relative to the NC group, P=0.001; siCirc0046600-B group, the migration distance was 88.25% relative to the NC group, P=0.045) (Figure 2B–E). In SK-HEP-1 and HepG2 cells, Western blot analysis showed that the relative expression level of N-cadherin was significantly lower in the siCirc0046600-A and siCirc0046600-B groups than in the NC group (P<0.05), indicating that hsa-circ-0046600 expression is positively correlated with N-cadherin expression (Figure 2F–H). GAPDH was used as an internal control. Taken together, these results indicate that hsa-circ-0046600 is overexpressed in HCC cells and promotes the migration of these cells.
Figure 2.
The role of hsa-circ-0046600 in HCC migration. (A) SiCirc0046600 had a specific effect on hsa-circ-0046600, and the corresponding mRNA was not affected. (B) The migration rates of SK-HEP-1 cells were determined by a wound scratch assay. The migratory ability of the experimental group was lower than that of the control group. (C) Quantification of relative migration in SK-HEP-1 cells transfected with NC, siCirc0046600-A, siCirc0046600-B. (D) The migration rates of HepG2 cells were determined by a wound scratch assay. The migratory ability of the experimental group was lower than that of the control group. (E) Quantification of relative migration in HepG2 cells transfected with NC, siCirc0046600-A, siCirc0046600-B. (F) Western blot analysis showed that N-cadherin and HIF-1α expression was significantly reduced in the experimental group. (G, H) Quantification of relative protein in HepG2 and SK-HEP-1 cells transfected with NC, siCirc0046600-A, siCirc0046600-B. Data are reported as the mean ± standard deviation. *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001.
Features Of hsa-circ-0046600
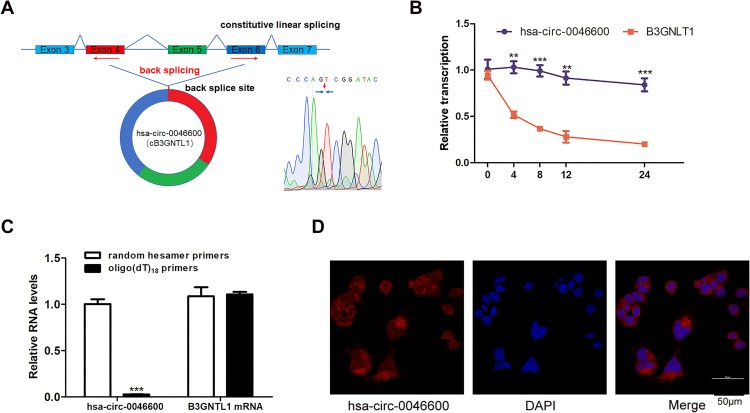
To confirm the circular characteristics of hsa-circ-0046600, we conducted experiments with 3 different approaches. Sanger sequencing confirmed that hsa-circ-0046600 is a head-to-tail splice product of exon 4 and exon 6 of the PCR product (Figure 3A). Next, we used actinomycin D to inhibit the transcription of hsa-circ-0046600 and B3GNTL1 mRNA in HepG2 cells and then measured the half-lives of these molecules. The results showed that hsa-circ-0046600 was more stable than B3GNTL1 mRNA (Figure 3B). Unlike mRNAs, circRNAs do not have a poly-A tail; therefore, we used random hexamer or oligo(dT) 18 primers for reverse transcription RNA experiments in HepG2 cells. When the oligo(dT) 18 primers were used, the relative expression of hsa-circ-0046600 was significantly lower than that measured with the random hexamer primers, while B3GNTL1 mRNA expression was not, confirming that hsa-0046600 has no poly-A tail (Figure 3C). Furthermore, FISH for hsa-circ-0046600 in HepG2 cells showed that hsa-circ-0046600 has a primarily cytoplasmic distribution (Figure 3D).
Figure 3.
Features of hsa-circ-0046600. (A) The genomic locus of the B3GNTL1 gene and hsa-circ-0046600 is shown. The reverse splicing product of exon 4, exon 5, and exon 6 of B3GNTL1 was confirmed by Sanger sequencing. (B) Total RNA was extracted from HepG2 cells treated with actinomycin D for 0, 4, 8, 12, and 24 h, and the levels of hsa-circ-0046600 and B3GNTL1 mRNA were evaluated by qRT-PCR. (C) qRT-PCR analysis of hsa-circ-0046600 and B3GNTL1 mRNA in HepG2 cells was performed with different reverse transcription primers. (D) A FISH assay was performed to determine the subcellular localization of hsa-circ-0046600. Data are reported as the mean ± SD. **P-value < 0.01, ***P-value < 0.001.
hsa-circ-0046600 Acts As A Molecular Sponge Of miR-640
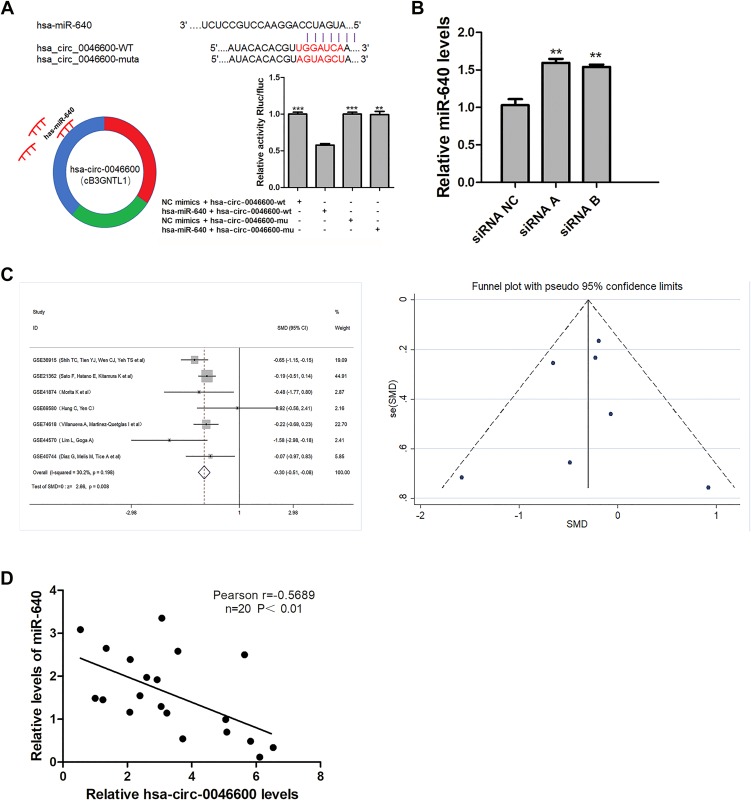
To explore the ceRNA mechanism of hsa-circ-0046600, we used CircInteractome and miRDB to predict miRNAs targeted by circRNAs. miR-640 scored high in both prediction methods (the context+ score percentile was 97, the site type was 8 mer-1a). Subsequently, reduced luciferase activity was observed in cells co-transfected with the hsa-circ-0046600 WT/miR-640 construct; however, the luciferase activity in cells transfected with the hsa-circ-0046600-Mut construct was not affected (Figure 4A). miR-640 expression was upregulated in HepG2 and SK-HEP-1 cells after transfection with si-circRNA-0046600 (Figure 4B). Next, we conducted a meta-analysis of seven sets of microarrays containing miR-640 in the GEO database. A total of 403 HCC samples and 134 control samples were included. The heterogeneity between the datasets was small (P = 0.198, I 2 = 30.2%), and the combined standard mean difference (SMD) calculated by the fixed effects model was −0.30 (95% confidence interval (CI): −0.61- −0.08, P = 0.008). There were no significant publication biases between the datasets (Figure 4C). These results indicate that miR-640 is significantly downregulated in HCC. In addition, qRT-PCR and a correlation analysis showed a negative correlation between the expression of hsa-circ-0046600 and miR-640 in 20 HCC tissue samples (r = −0.5689, P = 0.0089) (Figure 4D). The above results indicate that hsa-circ-0046600 acts by targeting miR-640 in HCC.
Figure 4.
Correlation of miR-640 with hsa-circ-0046600 and the expression of miR-640 in HCC. (A) Relative luciferase activity was reduced after the co-transfection of hsa-circ-0046600-WT and miR-640 mimics. The predicted binding sites, sequences and models of hsa-circ-0046600-Mut are shown. (B) siCircRNA0046600 transfection caused a corresponding change in miR-640 in HepG2 cells. (C) The meta-analysis of miR-640 and HCC gene microarrays in the GEO database is shown. (D) miR-640 is negatively correlated with hsa-circ-0046600 in HCC. Data are reported as the mean ± SD. **P-value < 0.01, ***P-value < 0.001.
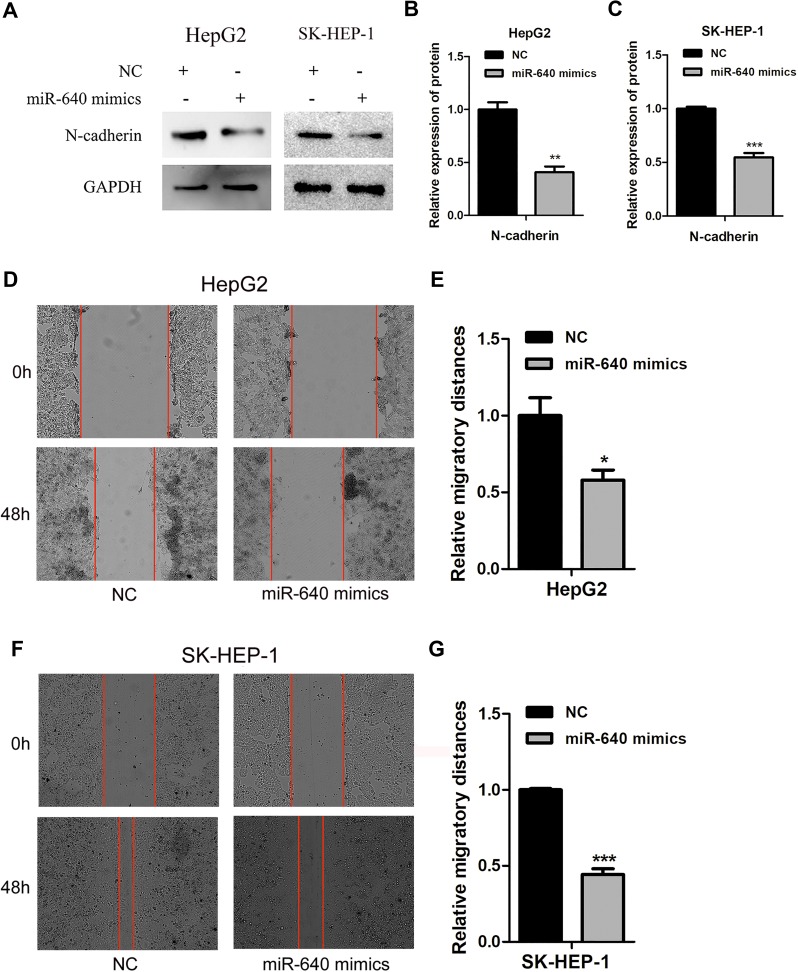
miR-640 Affects The Migration Of HCC Cells
HepG2 and SK-HEP-1 cells were seeded into 6-well plates, transfected with miR-640 mimics and harvested or scraped after transfection for the indicated times. Nontransfected cells served as a control. Western blotting showed that when the expression level of miR-640 was increased, the expression of N-cadherin was downregulated compared with that in the control group (P < 0.05, Figure 5A). Wound scratch tests showed that after 48 h, the increased expression of miR-640 reduced the migration of SK-HEP-1 cells (the migration distance was 75.10% relative to the NC group, P=0.0003) and HepG2 cells (the migration distance was 58.00% relative to the NC group, P = 0.034, Figure 5B–E).
Figure 5.
miR-640 inhibits the migration of HepG2 and SK-HEP-1 cells. (A) After transfection of miR-640 mimics, the expression level of N-cadherin in HepG2 and SK-HEP-1 cells was lower than that in NC group. (B, C) Quantification of relative protein in HepG2 and SK-HEP-1 cells transfected with NC, miR-640 mimics. (D) The migration rates of HepG2 cells were determined by a wound scratch assay. The migratory ability of miR-640 mimics group was lower than that of the control group. (E) Quantification of relative migration in HepG2 cells transfected with NC, miR-640 mimics. (F) The migration rates of SK-HEP-1 cells were determined by a wound scratch assay. The migratory ability of miR-640 mimics group was lower than that of the control group. (G) Quantification of relative migration in SK-HEP-1 cells transfected with NC, miR-640 mimics. *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001.
Prediction Of miR-640 Target Genes
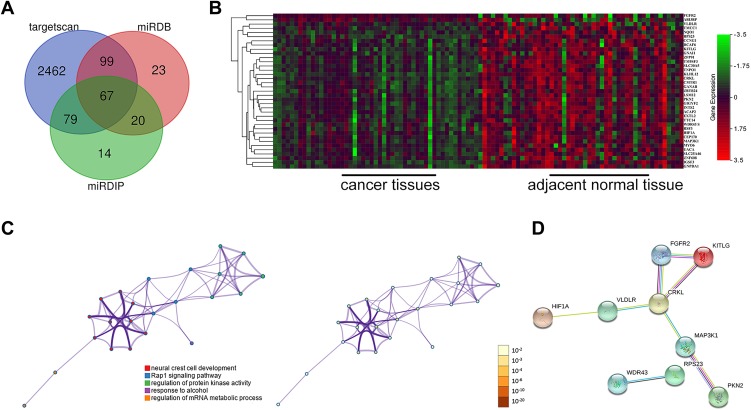
An analysis of the TCGA database using R software revealed 879 differentially expressed mRNAs in HCC. Target gene prediction of miR-640 was performed using three prediction platforms, and a total of 64 mRNAs were predicted to be repeated (Figure 6A). By intersecting these 64 mRNAs with the 879 differentially expressed mRNAs, we obtained a total of 37 miR-640-targeted mRNAs differentially expressed between HCC and control samples, of which 35 mRNAs were significantly upregulated in HCC (Figure 6B).
Figure 6.
Functional enrichment analysis and PPI network of miR-640 target genes. (A) The intersection of miR-640 target genes was predicted with bioinformatics software. (B) The intersection of the differentially expressed genes and the miR-640 target genes in the TCGA liver hepatocellular carcinoma (LIHC) dataset is shown, and 35 of the 37 genes were highly expressed in HCC. (C) GO and KEGG analyses of the 37 genes are shown. (D) The PPI network of the 37 genes is shown.
Functional Enrichment Analysis And PPI Network Of The Target Genes
Table 2 shows the results of GO enrichment and KEGG pathway analyses by Metascape for the 37 target genes. Representative pathways revealed by the GO analysis included four items: neural crest cell development, protein kinase activity regulation, response to alcohol, and mRNA metabolic process regulation. The KEGG analysis showed that the Rap1 signalling pathway was the main representative pathway (Table 2, Figure 6C). Among the pathways included, the Rap1 signalling pathway, protein kinase regulation and mRNA metabolic process regulation are the most relevant to tumours. The PPI network indicated that of the 37 mRNAs, the downstream proteins of 9 mRNAs formed 8 PPI pairs (Figure 6D). Upon combining this result with those from existing studies on miR-640, we verified HIF-1α as the downstream protein of hsa-circ-0046600. Consistent with the predicted results, the experimental results showed that in HepG2 and SK-HEP-1 cells, HIF-1α expression was significantly downregulated in the si-circ-004600 group compared to the NC group (P<0.05, Figure 2D).
Table 2.
GO Enrichment Terms And KEGG Pathway Enrichment Analysis Of The Target Genes
| GO/KEGG ID | Term | Ontology | Count | Log P | |
|---|---|---|---|---|---|
| GO:0014032 | Neural crest cell development | BP | 5 | −3.6 | |
| GO:0045859 | Regulation of protein kinase activity | BP | 6 | −3.0 | |
| GO:0097305 | Response to alcohol | BP | 3 | −2.3 | |
| GO:1903311 | Regulation of mRNA metabolic process | BP | 3 | −2.1 | |
| KEGG:04015 | Rap1 signalling pathway | — | 4 | −3.5 | |
Abbreviations: GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process.
Discussion
As a large portion of the non-coding RNA family, circRNAs have generated strong interest in the past few years.15 Second-generation sequencing techniques have demonstrated that circRNAs are abundant non-coding RNAs that are ubiquitous throughout eukaryotes.16 Similar to lncRNAs and miRNAs, circRNAs function as transcriptional competitors or enhancers of parental genes and translatable transcripts, interacting with RNA-binding proteins (RBPs) and even translating directly into proteins.17 Because of their ubiquitous existence and diversity, they are particularly important in cancer research. At present, an increasing number of circRNAs related to HCC, such as circMTO1, cSMARCA5 and Cul2, have been described.18–20 However, there are still many unknown HCC-related circRNAs that have not been identified. We performed a bioinformatic analysis and explored the mechanism of the newly identified circRNA, hsa-circ-0046600, in HCC. First, we compared the microarray data of Cao et al, who reported that hsa-circ-004660 (hsa-circ-102272) was among the top 10 differentially expressed genes. According to our data, hsa-circRNA-0046600 is upregulated in most hepatoma cell lines, particularly in the HepG2 and SK-HEP-1 cell lines. Therefore, to obtain clearer results, we chose to verify the biological function and ceRNA mechanism of hsa-circ-0046600 in HepG2 and SK-HEP-1 cells. In addition, hsa-circRNA-0046600 expression was significantly upregulated in 90% of our tested HCC tissue samples compared with that in the paired adjacent normal tissue samples, as measured by qRT-PCR, and was positively correlated with tumour size, stage and pathological vascular invasion. Next, to verify whether there was a functional difference associated with hsa-circ-0046600, an hsa-circ-0046600 siRNA was constructed, and its specificity was verified. In vitro experiments showed that the migration of HCC cells after siRNA transfection and the expression of the migration-related protein N-cadherin were reduced. The effect of hsa-circ-0046600 on migration was not as obvious in HepG2 cells as it was in SK-HEP-1 cells. After repeated verification, we believe that the difference may be due to the poor migration of HepG2 cells themselves, resulting in a diminishing migration distance after 48 h. In addition, we demonstrated the proliferation and apoptosis of these two cell types after siRNA regulation. There was no significant difference between the experimental and control groups. However, hsa-circ-0046600 is closely related to tumour size, which contradicts the experimental results. We believe that this discrepancy may exist because tumour proliferation is a complex process in which hsa-circ-0046600 has little effect despite its involvement.
Unlike linear RNAs, circRNAs are produced by a covalent linkage between the 5ʹ and 3ʹ ends of an RNA molecule.21 circRNAs can consist of a single exon, multiple exons or multiple exons and introns, and they lack 5ʹ and 3ʹ ends and the circular structure of the poly-A tail.19 Therefore, circRNAs can avoid degradation by exonucleases and RNase R, making these molecules conservative and stable. According to the sequence of hsa-circ-0046600, we found that hsa-circ-0046600 consists of three exons from its parent gene, B3GNTL1. Due to the nature of circRNAs, hsa-circ-0046600 degraded much more slowly than did B3GNTL1 mRNA following treatment with actinomycin D, and hsa-circ-0046600 could not undergo reverse transcription with oligo(dT) 18 primers. We conclusively demonstrated that the molecules detected in the experiment were circRNAs instead of mRNAs. In addition, we also determined by FISH that hsa-circ-0046600 is present mainly in the cytoplasm. Therefore, we speculate that hsa-circ-0046600 can act as a sponge.
The ceRNA hypothesis posits that mRNAs, lncRNAs and circRNAs can release miRNA-targeted mRNA transcripts by competing with base-complementary miRNA response elements (using a sponge mechanism).22 Using bioinformatic software, we predicted that miR-640 is the miRNA most likely to bind to hsa-circ-0046600. Furthermore, the combination of hsa-circ-0046600-miR-640 was confirmed with the transfection of siCirc0046600 and a dual-luciferase reporter assay. However, the difference in the expression of miR-640 was not confirmed in HCC; therefore, we performed a meta-analysis of relevant microarrays to confirm that miRNA-640 is expressed at a higher level in normal tissue than in HCC tissue. In summary, we determined that the hsa-circ-0046600-miR-640 pathway plays an important role in the migration of HCC cells.
Next, we predicted the possible downstream mRNA binding partners of miR-640 and compared our results with those from the TCGA HCC dataset. This approach confirmed that the expression of 35 of the 37 closely related HCC mRNAs was upregulated in HCC tissue compared with normal tissue, and this result was in accordance with the expected results. Subsequently, we performed a functional analysis of the 37 mRNAs and constructed a PPI network and confirmed that there are 9 core mRNAs in the hsa-circ-0046600-miR-640 pathway. Yi-Chun Zhu et al verified that miR-640 bound to the 3ʹ-untranslated region (UTR) of HIF-1α mRNA and then inhibited the expression of HIF-1α with dual-luciferase reporter and Western blot experiments. Based on the functional enrichment analysis and the PPI network, as well as the results described by Yi-Chun Zhu et al, we selected HIF-1α as the downstream protein of hsa-circ-0046600 for verification by Western blot.23 We used Western blotting to confirm that HIF-1α expression was downregulated after transfection with siCirc-0046600. The above results demonstrate that the hsa-circ-0046600-miR-640 combination can affect the migration of HCC cells through a variety of mechanisms.
Conclusion
Our study indicates that hsa-circ-0046600 is highly expressed in HCC tissues and cells and is closely related to tumour size, TNM stage, and pathological vascular invasion in HCC patients. hsa-circ-0046600 promotes the expression of HIF-1α by acting as a sponge of miR-640, further affecting the migration of HCC cells. hsa-circ-0046600 may be a potential new target for the diagnosis and treatment of HCC.
Acknowledgment
We would like to thank the patients who participated in this study and the Institute of Genetics of Henan Provincial People’s Hospital, which cooperated to provide the experimental environment.
Abbreviations
circRNA, circular RNA; HCC, hepatocellular carcinoma; qRT-PCR, quantitative reverse transcription polymerase chain reaction; FISH, fluorescence in situ hybridization; ceRNA, competitive endogenous RNA; TCGA, The Cancer Genome Atlas; PPI, protein-protein interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GEO, Gene Expression Omnibus; LIHC, liver hepatocellular carcinoma.
Disclosure
The authors report no conflicts of interest with this work.
References
- 1.Seehawer M, Heinzmann F, D’Artista L, et al. Necroptosis microenvironment directs lineage commitment in liver cancer. Nature. 2018;562(7725):69–75. doi: 10.1038/s41586-018-0519-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150(4):835–853. doi: 10.1053/j.gastro.2015.12.041 [DOI] [PubMed] [Google Scholar]
- 3.Ringelhan M, Pfister D, O’Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19(3):222–232. doi: 10.1038/s41590-018-0044-z [DOI] [PubMed] [Google Scholar]
- 4.Febbraio MA, Reibe S, Shalapour S, Ooi GJ, Watt MJ, Karin M. Preclinical models for studying NASH-driven HCC: how useful are they? Cell Metab. 2019;29(1):18–26. doi: 10.1016/j.cmet.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: functions and implications. Cancer Med. 2018;7:3101–3109. doi: 10.1002/cam4.2018.7.issue-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu LP, Wu YH, Yu XF, Tang Q, Chen L, Chen KP. The emerging role of circular RNAs in hepatocellular carcinoma. J Cancer. 2018;9(9):1548–1559. doi: 10.7150/jca.24566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. doi: 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Research. 2015;43(D1):D146–D152. doi: 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokar T, Pastrello C, Rossos AEM, et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360–D370. doi: 10.1093/nar/gkx1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi S, Pohl Marie O, Zhou Y, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–735. doi: 10.1016/j.chom.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 16.Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi: 10.1038/s41419-018-0454-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Li X, Yang L, Chen -L-L. The biogenesis, functions, and challenges of circular RNAs. Molecular Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034 [DOI] [PubMed] [Google Scholar]
- 18.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi: 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 19.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi: 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 20.Meng J, Chen S, Han JX, et al. Twist1 regulates vimentin through cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78(15):4150–4162. doi: 10.1158/0008-5472.CAN-17-3009 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res 2017;36(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, Nie C, Liu B, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin Cancer Res. 2018;37(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Li XH, Zhang CC, et al. Hydrogen sulfide promotes angiogenesis by downregulating miR-640 via the VEGFR2/mTOR pathway. Am J Physiol Cell Physiol. 2016;310(4):C305–C317. doi: 10.1152/ajpcell.00230.2015 [DOI] [PubMed] [Google Scholar]