Abstract
Background
The association between interferon lambda-3 (IFNL3,also known as interleukin 28B, IL28B) rs12979860 polymorphism and the development of hepatocellular carcinoma (HCC) has been investigated in recent studies with inconclusive and inconsistent results. IFNL3 rs12979860 polymorphism has been shown a marked differential distribution with regional and ethnic variation. Whether this single nucleotide polymorphism influences susceptibility to hepatitis C virus (HCV)-related HCC remains elusive.
Methods
In this case–control study, a total of 157 Chinese Han patients with chronic HCV infection were enrolled, including 62 HCV-related HCC patients and 95 chronic hepatitis C (CHC) patients without HCC, and the genetic polymorphism of IFNL3 rs12979860 was genotyped via a DNA microarray-based assay. The logistic regression analysis was employed to determine the correlation between the genetic polymorphism and risk of HCV-related HCC.
Results
A higher proportion of CT/TT genotype and T allele was observed in HCC patients compared to the CHC group. Under the genetic model of allele frequency, the T allele was associated with elevated risk of HCV-related HCC in the Chinese population compared to C allele after an adjustment for age, gender, body mass index, HCV infection duration, and HCV genotypes (P=0.046). In the subgroup analysis stratified by HCV genotype, subjects with CHC genotype 1b infection carrying rs12979860 T allele and CT+TT genotype had higher susceptibility to HCC than those with C allele and CC genotype (P=0.020, P=0.037, respectively).
Conclusion
IFNL3 rs12979860 polymorphism with T allele could be a factor that increases the risk of HCV-related HCC in the Chinese population, especially those subjects with CHC genotype 1b infection.
Keywords: IFNL3, rs12979860, polymorphism, HCV, HCC
Introduction
Hepatitis C virus (HCV) infection is a major public health problem throughout the world. Chronic HCV infection may eventually progress to liver cirrhosis (LC) or even develop to hepatocellular carcinoma (HCC).1,2 The risk of development from chronic hepatitis C (CHC) to HCC involves a complex interplay between the viral and host genetic factors.3 Our previous study4 together with others’ have revealed that host gene polymorphisms may play important roles in the development and progression of HCV-related HCC.
IFNL3 encodes IFN lambda-3 (IFN-λ3), also known as interleukin 28B, which belongs to the type III IFN-λ family consisting of IL29/IFNL1, IL28A/IFNL2, IL28B/IFNL3, and newly discovered IFNL4.5–7 It has been investigated that rs12979860 (C/T), located near the IFNL3 gene, involves in a number of aspects of HCV infection and disease progress,8–19 including response to therapy, natural elimination of the virus, viral clearance rate, changes in gene expression and lipid metabolism, hepatocyte death rate, inflammatory activity, fibrosis risk, cirrhosis and hepatocarcinogenesis.
Several studies have been recently carried out to explore the association between IFNL3 rs12979860 polymorphism and HCC risk in different geographic regions and ethnic populations, but with inconclusive and inconsistent results (Table 1).20–36 Some reports demonstrated that the carriage of T allele was a risk factor for HCC development.20–29 In contrast, one study showed opposite results,30 while other studies did not find any significant association between IFNL3 rs12979860 polymorphism and HCC risk.31–36 Therefore, we sought to conduct a case–control study to investigate the role of genetic polymorphism of IFNL3 rs12979860 in the susceptibility to HCV-related HCC in a Chinese population.
Table 1.
Characteristics of previous studies on the association of IFNL3 rs12979860 polymorphism and HCC risk
| Study | Year | HCV genotype | Risk factor associated with HCC | Reference |
|---|---|---|---|---|
| Buivydiene et al. | 2018 | GT1 GT2 GT3 | rs12979860 CC genotype | 30 |
| Suo et al. | 2013 | ND | rs12979860 TT genotype | 20 |
| Attallah et al. | 2018 | GT4 | rs12979860 TT genotype | 21 |
| Chang et al. | 2015 | GT1 non-GT1 | rs12979860 CT+TT genotype | 22 |
| Lee et al. | 2015 | GT1 non-GT1 | rs12979860 CT+TT genotype | 23 |
| Ibrahim et al. | 2016 | ND | rs12979860 CT+TT genotype | 24 |
| Chang et al. | 2018 | GT1 non-GT1 | rs12979860 CT+TT genotype | 25 |
| Fabris et al. | 2011 | ND | rs12979860 T allele | 26 |
| El-Awady et al. | 2012 | GT4a | rs12979860 T allele | 27 |
| Eurich et al. | 2012 | GT1b non-GT1b | rs12979860 T allele | 28 |
| Zhang et al. | 2016 | ND | rs12979860 T allele | 29 |
| Agúndez et al. | 2012 | GT1 non-GT1 | No association | 31 |
| Bochud et al. | 2012 | GT1 GT2 GT3 GT4 | No association | 32 |
| Joshita et al. | 2012 | GT1 GT2 | No association | 33 |
| Miura et al. | 2012 | GT1b | No association | 34 |
| Akkiz et al. | 2014 | ND | No association | 35 |
| Zekri et al. | 2014 | ND | No association | 36 |
Abbreviations: IFNL3, interferon lambda-3; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; GT, genotype; ND, not determined.
Materials and methods
Patients
A total of 157 patients with chronic HCV infection, including 62 HCV-related HCC patients, and 95 HCV-infected patients without HCC as the control subjects, were recruited from Tianjin Second People’s Hospital and Tianjin institute of Hepatology from July 2015 to February 2017. The diagnostic criteria for CHC with or without HCC were based on the combination of clinical history, physical examination, imaging, and laboratory data, and/or histology. Seropositive patients for other hepatitis viruses such as hepatitis B virus or human immunodeficiency virus were excluded from this study. The written informed consent and the ethical approval for this study were obtained from all subjects and the Faculty of Health Science Ethics Committee of Tianjin Second People’s Hospital. The following data were collected from all HCC patients and control subjects including gender, age, body mass index (BMI), the known duration of HCV infection, and infected HCV genotype.
Genotyping
Genotyping for rs12979860 polymorphism was performed via a DNA microarray-based assay, as described in our previous study.37
Statistical analysis
All analyses were performed using the SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Chi-square test and Student t-test were used where appropriate. The logistic regression analysis was employed to analyze the correlation between the genetic polymorphism and risk of HCC. A P-value of less than 0.05 was considered to indicate a significant difference.
Results
Demographic and clinical characteristics of the study population
The demographic and clinical characteristics of HCV-related HCC patients and control subjects are reported in Table 2. No significant differences were found between the two groups in terms of gender, age, BMI, duration of HCV infection, and HCV genotype distribution (P>0.05, all).
Table 2.
Demographic and clinical characteristics of the study population
| Characteristics | HCC patients N=62 (%) | CHC patients N=95 (%) | P-value |
|---|---|---|---|
| Gender | 0.467 | ||
| Male | 35(56.45%) | 48(50.53%) | |
| Female | 27(43.55%) | 47(49.47%) | |
| Age (years) | 0.118 | ||
| ≤60 | 30(48.39%) | 58(61.05%) | |
| >60 | 32(52.61%) | 37(38.95%) | |
| BMI | 0.329 | ||
| <24 | 33(53.23%) | 43(45.26%) | |
| ≥24 | 29(46.77%) | 52(54.74%) | |
| Duration of HCV infection (years) | 0.825 | ||
| ≤20 | 25(40.32%) | 40(42.11%) | |
| >20 | 37(59.68%) | 55(57.89%) | |
| HCV genotype | 0.416 | ||
| 1b | 51(82.26%) | 73(76.84%) | |
| Non-1b | 11(17.74%) | 22(23.16%) |
Abbreviations: HCC, hepatocellular carcinoma; CHC, chronic hepatitis C; BMI, body mass index; HCV, hepatitis C virus.
Genotype and allele frequencies of IFNL3 rs12979860
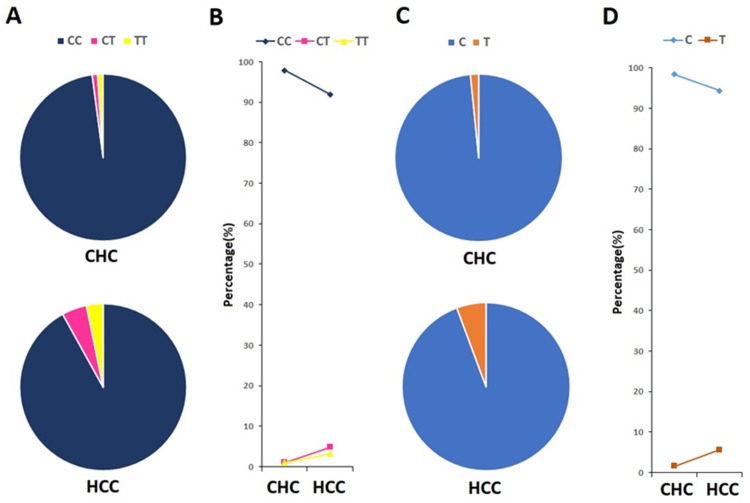
The genotype and allele frequencies of IFNL3 rs12979860 polymorphism are shown in Figure 1. Three genotypes were detected in rs12979860, CC, CT, and TT. The frequencies of these three genotypes were 91.94%, 4.84%, and 3.22% in HCC group (Figure1A), 97.90%, 1.05%, and 1.05% in control group (Figure 1A). The C and T allele frequencies of the two groups were 94.35% and 5.65%, 98.42% and 1.58%, respectively (Figure 1C). Though the distribution of IFNL3 rs12979860 genotypes and alleles was not statistically different between HCC patients and control subjects (P>0.05), a higher proportion of CT/TT genotype (Figure 1B) and T allele (Figure 1D) was observed in HCC patients.
Figure 1.
Genotype and allele frequencies of IFNL3 rs12979860 in CHC and HCC groups.
Notes: (A) Genotype frequencies of IFNL3 rs12979860 in CHC and HCC groups; (B) comparison of genotype proportions between CHC group and HCC group; (C) allele frequencies of IFNL3 rs12979860 in CHC and HCC groups; (D) comparison of allele proportions between CHC group and HCC group.
Abbreviations: CHC, chronic hepatitis C; HCC, hepatocellular carcinoma; IFNL3, interferon lambda-3.
Association analysis of HCV-related HCC risk
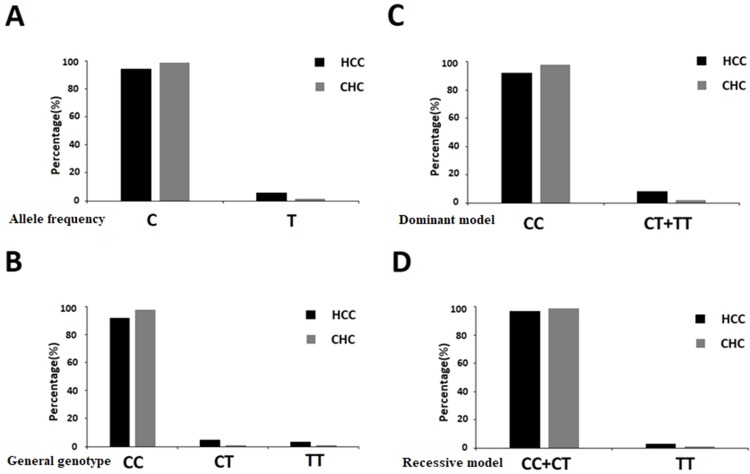
We further analyzed the correlation between IFNL3 rs12979860 polymorphism and susceptibility to HCV-related HCC using logistic regression analysis according to four models (i.e., allele frequency, general genotype, dominant, and recessive models). The OR and P-values of all the genetic models were adjusted on age, gender, BMI, HCV infection duration, and HCV genotypes. Under the genetic model of allele frequency (Figure 2A), the T allele of rs12979860 was associated with elevated risk of HCC compared to the C allele (OR=4.166, 95% CI: 1.024–16.959; P=0.046). Under the dominant model (Figure 2C), the CT+TT genotype increased the risk of developing HCC with a tendency toward statistical significance (OR=4.643, 95% CI: 0.826–26.098; P=0.081). No statistical significances were found under genetic model of general genotype (Figure 2B) as well as recessive model (Figure 2D).
Figure 2.
Association between IFNL3 rs12979860 polymorphism and HCC risk under different genetic models.
Notes: (A) Genetic model of allele frequency; (B) genetic model of general genotype; (C) dominant model; (D) recessive model.
Abbreviations: HCC, hepatocellular carcinoma; CHC, chronic hepatitis C; IFNL3, interferon lambda-3.
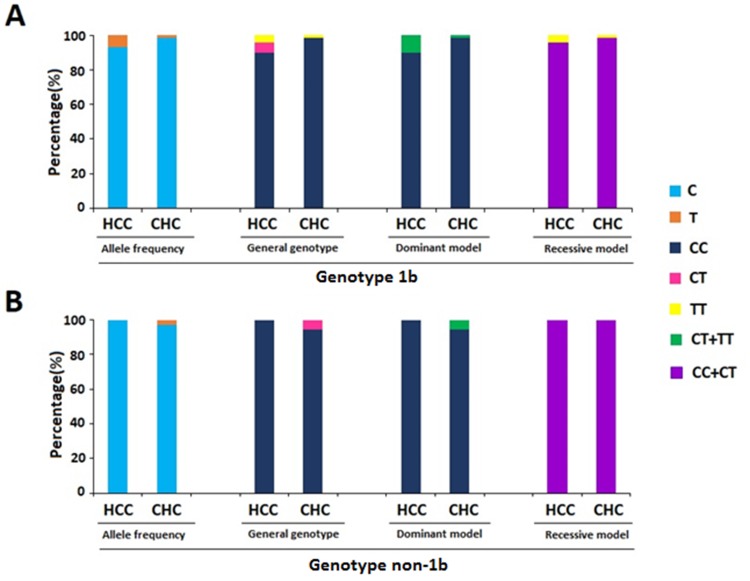
Furthermore, to explore whether HCV genotype would play a role in the association, we separated all the subjects of this study into HCV genotype 1b and non-1b groups (Figure 3). We found that rs12979860 T allele and CT+TT genotype were associated with elevated risk of HCC compared to C allele and CC genotype in HCV 1b genotype group (P=0.020; P=0.037, respectively).
Figure 3.
Association between IFNL3 rs12979860 polymorphism and HCC risk stratified by genotype.
Notes: (A) HCV genotype 1b group; (B) HCV genotype non-1b group.
Abbreviations: HCC, hepatocellular carcinoma; CHC, chronic hepatitis C; IFNL3, interferon lambda-3; HCV, hepatitis C virus.
Discussion
In this case–control study, we attempted to investigate the association between IFNL3 rs12979860 polymorphism and the HCV-related HCC in a Chinese population. As a result, we found a higher proportion of CT/TT genotype and T allele in HCC patients compared to the CHC group. The T allele was associated with elevated risk of HCV-related HCC compared to C allele under the genetic model of allele frequency. In the subgroup analysis stratified by HCV genotype, subjects with CHC genotype 1b infection carrying rs12979860 T allele and CT+TT genotype had higher susceptibility to HCC than those with C allele and CC genotype.
Our data add new insights into the association between genetic polymorphism of IFNL3 rs12979860 and the susceptibility to HCV-related HCC, and provide further evidence in a Chinese population. Our findings are consistent with the previous observations,20–29 whereas different from some other reports.30–36 The following factors might account for the inconsistent results: 1) geographic and racial differences, 2) diversity of viral genotypes.
IFNL3 encodes IFN-λ3, one of the type III IFN-λ family member, inducing signaling through binding to the specific IFN-λ receptor chain 1 and the shared IL-10 receptor chain 2. Accumulating evidence strongly suggests that IFN-λ plays a major role in the control of viral infection and antitumor activity,38–40 including activation immune cells and suppression of HCC cell proliferation and growth in HCC models.41,42 The allelic variants of the IFNL3 polymorphism may affect the efficiency of the immunomodulatory process, which could lead to HCC. The exact molecular mechanisms by which IFNL3 rs12979860 polymorphism influence susceptibility to HCV-related HCC warrant further elucidation.
There are also some potential limitations in the study, principally limited to a relatively small sample size in a Chinese population at a single center. Further studies involving large-scale samples from different centers should be performed.
Conclusion
In summary, our results indicate that IFNL3 rs12979860 T allele could be a factor that increases the risk of HCV-related HCC, especially those subjects with CHC genotype 1b infection in the Chinese population. Screening genetic polymorphism of IFNL3 rs12979860 might be helpful in designing effective and efficient HCC surveillance programs for chronic HCV-infected patients.
Acknowledgments
We would like to thank all participants for their participation in this study. This work was presented in the 27th Annual Conference of APASL, March 14–18, 2018, New Delhi, India and was published in abstract form in Hepatol Int (2018) 12 (Suppl 2): S388, HCC-24. This work was supported by the National Natural Science Foundation of China (grant nos. 30800974 and 81271845) and Tianjin Municipal Health Bureau of Science and Technology Fund (grant nos. 2012KR02 and 12KG118). Resources were provided by the Ralph H Johnson VAMC, Charleston, South Carolina.
Abbreviations
IFNL3, interferon lambda-3; IL28B, interleukin 28B; HCC, hepatocellular carcinoma; SNP, single nucleotide polymorphism; CHC, chronic hepatitis C; HCV, hepatitis C virus; LC, liver cirrhosis; IFN-λ3, IFN lambda-3; SNP, single nucleotide polymorphism; HBV, hepatitis B virus; HIV, human immunodeficiency virus; BMI, body mass index.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Ethics approval and consent to participate
Written informed consent was obtained from all participants included in the present study. Ethical approval to carry out the study was obtained from the Ethics Committee of Tianjin Second People’s Hospital. This study was conducted in accordance with the Declaration of Helsinki.
Author contributions
WH and WKS conceived and designed the study. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Pradat P, Virlogeux V, Trépo E. Epidemiology and elimination of HCV-related liver disease. Viruses. 2018;10(10):pii: E545. doi: 10.3390/v10100545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axley P, Ahmed Z, Ravi S, Singal AK. Hepatitis C virus and hepatocellular carcinoma: a narrative review. J Clin Transl Hepatol. 2018;6(1):79–84. doi: 10.14218/JCTH.2017.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuura K, Tanaka Y. Host genetic variations associated with disease progression in chronic hepatitis C virus infection. Hepatol Res. 2018;48(2):127–133. doi: 10.1111/hepr.13042 [DOI] [PubMed] [Google Scholar]
- 4.Zhang S, Qiao K, Trieu C, et al. Genetic polymorphism of epidermal growth factor rs4444903 influences susceptibility to HCV-related liver cirrhosis and hepatocellular carcinoma in a Chinese Han population. Clin Lab. 2017;63(4):845–850. doi: 10.7754/Clin.Lab.2016.161203 [DOI] [PubMed] [Google Scholar]
- 5.Prokunina-Olsson L, Muchmore B, Tang W, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45(2):164–171. doi: 10.1038/ng.2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibert S, Roger T, Calandra T, et al.; Swiss Hepatitis C Cohort Study. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210(6):1109–1116. doi: 10.1084/jem.20130012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terczyńska-Dyla E, Bibert S, Duong FH, et al.; Swiss Hepatitis C Cohort Study Group. Reduced IFNλ4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat Commun. 2014;5:5699. doi: 10.1038/ncomms5972 [DOI] [PubMed] [Google Scholar]
- 8.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309 [DOI] [PubMed] [Google Scholar]
- 9.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41(10):1105–1109. doi: 10.1038/ng.449 [DOI] [PubMed] [Google Scholar]
- 11.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41(10):1100–1104. doi: 10.1038/ng.447 [DOI] [PubMed] [Google Scholar]
- 12.Rauch A, Kutalik Z, Descombes P, et al.; Swiss Hepatitis C Cohort Study; Swiss HIV Cohort Study. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138(4):1338–45, 1345.e1-7. doi: 10.1053/j.gastro.2009.12.056 [DOI] [PubMed] [Google Scholar]
- 13.Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139(6):1865–1876. doi: 10.1053/j.gastro.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann WP, Zeuzem S. A new standard of care for the treatment of chronic HCV infection. Nat Rev Gastroenterol Hepatol. 2011;8(5):257–264. doi: 10.1038/nrgastro.2011.49 [DOI] [PubMed] [Google Scholar]
- 15.Hayes CN, Imamura M, Aikata H, Chayama K. Genetics of IL28B and HCV–response to infection and treatment. Nat Rev Gastroenterol Hepatol. 2012;9(7):406–417. doi: 10.1038/nrgastro.2012.101 [DOI] [PubMed] [Google Scholar]
- 16.Eslam M, George J. Genome-wide association studies and hepatitis C: harvesting the benefits of the genomic revolution. Semin Liver Dis. 2015;35(4):402–420. doi: 10.1055/s-0035-1567830 [DOI] [PubMed] [Google Scholar]
- 17.Eslam M, McLeod D, Kelaeng KS, et al.; International Liver Disease Genetics Consortium (ILDGC). IFN-λ3, not IFN-λ4, likely mediates IFNL3-IFNL4 haplotype-dependent hepatic inflammation and fibrosis. Nat Genet. 2017;49(5):795–800. doi: 10.1038/ng.3836 [DOI] [PubMed] [Google Scholar]
- 18.Chinnaswamy S. Gene-disease association with human IFNL locus polymorphisms extends beyond hepatitis C virus infections. Genes Immun. 2016;17(5):265–275. doi: 10.1038/gene.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heim MH, Bochud PY, George J. Host - hepatitis C viral interactions: the role of genetics. J Hepatol. 2016;65(1 Suppl):S22–S32. doi: 10.1016/j.jhep.2016.07.037 [DOI] [PubMed] [Google Scholar]
- 20.Suo GJ, Zhao ZX. Association of the interleukin-28B gene polymorphism with development of hepatitis virus-related hepatocellular carcinoma and liver cirrhosis: a meta-analysis. Genet Mol Res. 2013;12(3):3708–3717. doi: 10.4238/2013.September.19.1 [DOI] [PubMed] [Google Scholar]
- 21.Attallah AM, Omran D, Marie MS, et al. IL-28B rs12979860 polymorphism affect the course of chronic hepatitis and the development of HCC in Egyptian patients with hepatitis C type 4. Br J Biomed Sci. 2018;75(4):157–162. doi: 10.1080/09674845.2018.1489599 [DOI] [PubMed] [Google Scholar]
- 22.Chang KC, Tseng PL, Wu YY, et al. A polymorphism in interferon L3 is an independent risk factor for development of hepatocellular carcinoma after treatment of hepatitis C virus infection. Clin Gastroenterol Hepatol. 2015;13(5):1017–1024. doi: 10.1016/j.cgh.2014.10.035 [DOI] [PubMed] [Google Scholar]
- 23.Lee MH, Yang HI, Lu SN, et al. Polymorphisms near the IFNL3 gene associated with HCV RNA spontaneous clearance and hepatocellular carcinoma risk. Sci Rep. 2015;5:17030. doi: 10.1038/srep17030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim MK, Salama H, Abd El Rahman M, et al. Three gene signature for predicting the development of hepatocellular carcinoma in chronically infected hepatitis C virus patients. J Interferon Cytokine Res. 2016;36(12):698–705. doi: 10.1089/jir.2016.0042 [DOI] [PubMed] [Google Scholar]
- 25.Chang KC, Ye YH, Wu CK, et al. Risk factors for development of hepatocellular carcinoma in patients with chronic hepatitis C without sustained response to combination therapy. J Formos Med Assoc. 2018;117(11):1011–1018. doi: 10.1016/j.jfma.2017.11.008 [DOI] [PubMed] [Google Scholar]
- 26.Fabris C, Falleti E, Cussigh A, et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J Hepatol. 2011;54(4):716–722. doi: 10.1016/j.jhep.2010.07.019 [DOI] [PubMed] [Google Scholar]
- 27.El-Awady MK, Mostafa L, Tabll AA, et al. Association of IL28B SNP with progression of Egyptian HCV genotype 4 patients to end stage liver disease. Hepat Mon. 2012;12(4):271–277. doi: 10.5812/hepatmon.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eurich D, Boas-Knoop S, Bahra M, et al. Role of IL28B polymorphism in the development of hepatitis C virus-induced hepatocellular carcinoma, graft fibrosis, and posttransplant antiviral therapy. Transplantation. 2012;93(6):644–649. doi: 10.1097/TP.0b013e318244f774 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Zhu SL, Chen J, Li LQ. Meta-analysis of associations of interleukin-28B polymorphisms rs8099917 and rs12979860 with development of hepatitis virus-related hepatocellular carcinoma. Onco Targets Ther. 2016;9:3249–3257. doi: 10.2147/OTT.S104904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buivydiene A, Liakina V, Kashuba E, et al. Impact of the Uridine-Cytidine kinase like-1 protein and IL28B rs12979860 and rs8099917 SNPs on the development of hepatocellular carcinoma in cirrhotic chronic hepatitis C patients – a pilot study. Medicina (Kaunas). 2018;54(5):pii: E67. doi: 10.3390/medicina54050067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agúndez JA, García-Martin E, Maestro ML, et al. Relation of IL28B gene polymorphism with biochemical and histological features in hepatitis C virus-induced liver disease. PLoS One. 2012;7(5):e37998. doi: 10.1371/journal.pone.0037998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochud PY, Bibert S, Kutalik Z, et al.; Swiss Hepatitis C Cohort Study Group; ANRS HC EP 26 Genoscan Study Group. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology. 2012;55(2):384–394. doi: 10.1002/hep.24678 [DOI] [PubMed] [Google Scholar]
- 33.Joshita S, Umemura T, Katsuyama Y, et al. Association of IL28B gene polymorphism with development of hepatocellular carcinoma in Japanese patients with chronic hepatitis C virus infection. Hum Immunol. 2012;73(3):298–300. doi: 10.1016/j.humimm.2011.12.021 [DOI] [PubMed] [Google Scholar]
- 34.Miura M, Maekawa S, Kadokura M, et al. Analysis of viral amino acids sequences and the IL28B SNP influencing the development of hepatocellular carcinoma in chronic hepatitis C. Hepatol Int. 2012;6(1):386–396. doi: 10.1007/s12072-011-9307-6 [DOI] [PubMed] [Google Scholar]
- 35.Akkiz H, Kuran S, Akgöllü E, Usküdar O, Bekar A, Bayram S. The role of interleukin 28B gene polymorphism in Turkish patients with hepatocellular carcinoma. Ann Hepatol. 2014;13(6):788–795. [PubMed] [Google Scholar]
- 36.Zekri AR, Salama H, Medhat E, et al. IL28B rs12979860 gene polymorphism in Egyptian patients with chronic liver disease infected with HCV. Asian Pac J Cancer Prev. 2014;15(17):7213–7218. doi: 10.7314/apjcp.2014.15.17.7213 [DOI] [PubMed] [Google Scholar]
- 37.Qiao K, Trieu C, Huo Z, Du Y, Hou W. Distribution of IL28B rs12979860 and rs8099917 genotypes in patients with chronic hepatitis C virus infection in Tianjin, China. Clin Lab. 2018;64(4):543–550. doi: 10.7754/Clin.Lab.2017.171036 [DOI] [PubMed] [Google Scholar]
- 38.Wack A, Terczyńska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol. 2015;16(8):802–809. doi: 10.1038/ni.3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lasfar A, Zloza A, de la Torre A, Cohen-Solal KA. IFN-λ: a new inducer of local immunity against cancer and infections. Front Immunol. 2016;7:598 eCollection 2016. doi: 10.3389/fimmu.2016.00598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lasfar A, Zloza A, Silk AW, Lee LY, Cohen-Solal KA. Interferon lambda: toward a dual role in cancer. J Interferon Cytokine Res. 2019;39(1):22–29. doi: 10.1089/jir.2018.0046 [DOI] [PubMed] [Google Scholar]
- 41.Abushahba W, Balan M, Castaneda I, et al. Antitumor activity of type I and type III interferons in BNL hepatoma model. Cancer Immunol Immunother. 2010;59(7):1059–1071. doi: 10.1007/s00262-010-0831-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Y, Wang L, He J, et al. Synergy with interferon-lambda 3 and sorafenib suppresses hepatocellular carcinoma proliferation. Biomed Pharmacother. 2017;88:395–402. doi: 10.1016/j.biopha.2017.01.077 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.