Abstract
The nuclear receptor corepressor Hairless (HR) interacts with nuclear receptors and controls expression of specific target genes involved in hair morphogenesis and hair follicle cycling. Patients with HR gene mutations exhibit atrichia, and in rare cases, immunodeficiency. Pigs with HR gene mutations may provide a useful model for developing therapeutic strategies because pigs are highly similar to humans in terms of anatomy, genetics, and physiology. The present study aimed to knockout the HR gene in pigs using the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated-9 (Cas9) system and to investigate the molecular and structural alterations in the skin and thymus. We introduced a biallelic mutation into the HR gene in porcine fetal fibroblasts and generated nine piglets via somatic cell nuclear transfer. These piglets exhibited a lack of hair on the eyelids, abnormalities in the thymus and peripheral blood, and altered expression of several signaling factors regulated by HR. Our results indicate that introduction of the biallelic mutation successfully knocked out the HR gene, resulting in several molecular and structural changes in the skin and thymus. These pigs will provide a useful model for studying human hair disorders associated with HR gene mutations and the underlying molecular mechanisms.
Keywords: clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated-9 (Cas9), hairless, pig model, somatic cell nuclear transfer (SCNT), thymus
Introduction
There are several forms of hereditary human hair loss, collectively known as alopecia, which may be caused by dysregulation of the hair growth cycle and tissue remodeling [1, 10]. Mutations in 28 codons of hairless (HR) have been reported in numerous families [26], and these people uniquely exhibit atrichia, follicular cysts, and, in rare cases, immunodeficiency [15]. However, the molecular basis of alopecia remains largely unexplored [5].
HR, also known as lysine demethylase and nuclear receptor corepressor, interacts with nuclear receptors, including the thyroid hormone receptor, the retinoic acid orphan receptor α, and the vitamin D receptor, to control the expression of specific target genes involved in hair morphogenesis and hair follicle cycling [11, 38]. Mice with homozygous knockout (KO) of the Hr gene not only exhibit complete and persistent hair loss, but also immunological abnormalities, similar to the hair follicle and thymus defects associated with HR mutations in humans [12, 28].
Hr mutant mice provide a useful model for studying various aspects of skin physiology, including skin aging, pharmacokinetic evaluation of drug activity and cutaneous absorption, skin carcinogenesis, and skin toxicology [8, 21, 34]. However, mice and humans differ in many respects, including physiological traits and gene expression. Thus, mouse models cannot adequately mimic human diseases in some cases, and various animal models are urgently needed [23, 40, 41].
Pigs have recently been used as models of human diseases because they are more similar to humans than mice in terms of anatomy, neurobiology, cardiac vasculature, gastrointestinal tract, and genome [4]. Pigs are especially useful for skin studies [24, 35]. In contrast with rodent skin, the gross, microscopic, and ultrastructural features of pig skin are analogous to those of human skin. Moreover, the epidermal thickness and dermal: epidermal thickness ratio are comparable in pigs and humans. These similarities suggest that the skin of pigs and humans will respond similarly to stimuli [24].
In this study, we successfully generated nine HR-KO (HR−/−) piglets using the clustered regularly interspaced short palindromic repeat/CRISPR-associated-9 (CRISPR/Cas9) system and somatic cell nuclear transfer (SCNT). To investigate changes in the skin and thymus of these piglets, gene expression, thymus structure, and peripheral lymphocyte subsets were analyzed. These pigs are good models for studying the functions of HR and may provide insights into the pathophysiology of human hair disorders associated with disruption of this gene.
Materials and Methods
Ethics approval and consent to participate
Nine HR−/− piglets (0, 2, and 4 days old) and age-matched WT pigs were used in this research. All protocols of the animal studies were approved by the Committee on the Ethics of Animal Experiments at Yanbian University (Yanji, China), and all procedures were performed instrict accordance with the Guide for the Care and Use of Laboratory Animals.
Design and construction of CRISPR/Cas9 plasmids
The Cas9-encoding vector used in this study was described previously [7]. This vector contains a constitutive CMV promoter driving Cas9 expression and the C-terminus of Cas9 protein is tagged with nuclear localization signal and HA epitope. We used crRNA and tracrRNA pair (dual guide RNA, dgRNA) rather than single guide RNA (sgRNA) with Cas9 protein for targeted gene modification [2]. dgRNA-encoding vector transcribes crRNA and tracrRNA under the control of the U6 promoter. The crRNA sequence was designed for targeting exon 1 of the HR gene. The target site was selected based on microhomology patterns to introduce frame shifting mutations at a high frequency [2]. For enrichment of cells containing Cas9-induced mutations, we employed surrogate reporter system [18]. We used magnetic reporters, which express both eGFP and H-2Kk when frame-shifting mutations are introduced in the target sequence of the reporter [17]. Targeted enrichment of cells containing Cas9-induced mutations can be achieved by magnetic separation using a magnetic bead-conjugated with anti- H-2Kk antibody.
Cell culture and transfection
Porcine primary fetal fibroblasts were cultured and transfected as previously described [14]. For inducing targeted mutation at the hairless gene in the transgenic fibroblasts, we employed a modified CRISPR/Cas9 system using dgRNA. The plasmids for Cas9 and guide RNA (total 10 µg, 1:3 ratio, respectively) were transfected into porcine primary fetal fibroblast cells. The transfected cells were cultured for 2 days at 37°C and subjected to magnetic separation. Briefly, trypsinized cell suspensions were incubated with magnetic bead-conjugated antibody against H-2Kk (MACSelect Kk microbeads; Miltenyi Biotech, Cologne, Germany) for 15 min at 4°C. Labeled cells were separated using a MACS LS column (Miltenyi Biotech), according to the manufacturer’s protocol.
SCNT and embryo transfer
SCNT was performed as previously described [44]. Briefly, mature eggs with the first polar body were cultured for 1 h in medium supplemented with 0.4 mg/ml demecolcine and 0.05 M sucrose. Treated eggs with a protruding membrane were transferred to medium containing 5 mg/ml cytochalasin-B and 0.4 mg/ml demecolcine, and the protrusions were removed with a beveled pipette. A single donor cell was injected into the perivitelline space of each oocyte and electrically fused and then oocytes were activated. Activated oocytes were cultured in medium for 2 days. Cloned embryos were surgically transferred to oviducts of sows within 1 day of estrus onset. In the first round of SCNT, recipient pigs were euthanized on day 25 of gestation. The fetuses were collected and HR mutations were confirmed. Fibroblasts were obtained from HR−/− fetuses for the second round of SCNT. Pregnancy was assessed ultrasonographically on day 25. Cloned piglets were delivered naturally or by inducing labor via intramuscular injections of prostaglandin F2 alpha (Ningbo, China) on day 113 of gestation.
T7E1 assay and sequencing
The T7E1 assay was performed as described previously [19]. Briefly, genomic DNA was isolated using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The region of DNA containing the nuclease-targeting site was PCR-amplified. The amplicons were denatured by heating and annealed to form heteroduplex DNA, which was treated with 5 units of T7E1 (New England Biolabs, Ipswich, MA, USA) for 20 min at 37°C and electrophoresed on agarose gels. To confirm that the mutation had been introduced into the target allele, PCR amplicons spanning the target sites were purified using aGel Extraction Kit (Qiagen, Hilden, Germany) and cloned into the T-Blunt vector using a T-Blunt PCR Cloning Kit (SolGent, Daejeon, Korea). The cloned inserts were amplified using the same primers and sequenced. The cloned inserts were sequence using M13F primers.
Morphological analysis of skin
Skin samples for scanning electron microscopy analysis were acquired rom the dorsal and eyelid regions of 3 HR−/− piglets and 3 WT piglets at postnatal (PN) day 0 or 2. Samples were fixed in 2% glutaraldehyde at 4°C for 4h, dehydrated via the critical point drying methods inganalcohol gradient (50%, 70%, 95%, and 100%, 15min each), and finally soaked in isoamyl acetate for 30 min. Treated samples were fitted into a vacuum chamber and visualized using a transmission electron microscope (Hitachi, Tokyo, Japan).
Quantitative RT-PCR
Eyelid skin was carefully disecsted from two HR−/− piglets and two WT piglets at PN day 4. Total RNA was extracted from 100 mg of each sample using an Eastep® Super Total RNA Extraction Kit (Promega, Madison, WI, USA) according to the manufacturer’s recommended method. cDNA was synthesized from 1 µg RNA using a PrimeScriptTM RT Reagent Kit (Takara Bio, Kusatsu, Japan) according to the manufacturer’s protocol. Primers were synthesized from the oligo synthesis group (Invitrogen, Carlsbad, CA, USA) and are listed in Supplementary Table 1. Quantitative RT-PCR was performed using a real-time thermocycler (Agilent Technology, San Francisco, CA, USA) and each 20 µl reaction comprised 10 µl of SYBR® Premix Ex TaqTM mix (Takara Bio), 1 µl cDNA, and 0.5 µl of the appropriate primers. The cycling conditions were initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. Melting curve analysis was performed to confirm the amplification specificity. Data were normalized against expression of an endogenous reference gene (GAPDH) using the comparative threshold cycle method. All experiments were performed in triplicate.
Histology and immunofluorescence staining
Samples were obtained from the thymus of HR−/− and WT piglets at PN day 4. For hematoxylin and eosin staining, samples were fixed in 10% neutral buffered formalin, and embedded in paraffin. Thereafter, samples were cut at a thickness of 5 µm, deparaffinized, rehydrated, and stained with hematoxylin and eosin. Stained sections were dehydrated, mounted, and examined on a light microscope (Olympus, Tokyo, Japan). Cryostat sections (6 µm thick) were stained with In Situ Cell Death detection kit (TUNEL Apoptosis Detection Kit; Roche, Mannheim, Germany) to detect the apoptosis of thymus tissue. And DNA was counterstained with 25 µg/ml Hoechst 33342. The samples were treated according to the manufacturer’s directions and images were acquired using a fluorescent microscope (Olympus, Tokyo, Japan).
Western blot analysis
Eyelid skin of HR−/− and WT piglets at PN day 4 was frozen, crushed, and homogenized in RIPA buffer (Millipore, Boston, MA, USA). Lysed samples were centrifuged at 13,000 rpm for 15 min at 4°C and the supernatant was carefully collected. Protein concentrations were determined using a Pierce® BCA protein assay kit (Thermo Scientific). Thereafter, 50 µg protein was separated on a 10% SDS-PAGE at 100V for 1 h and then transferred to a polyvinylidene fluoride membrane at 70 V for 180 min in transfer buffer (25mM Tris, 190 mM glycine, and 20% methanol, pH 8.1). Membranes were blocked with 5% skimmed milk and incubated with the following primary antibodies overnight at 4°C: rabbit polyclonal anti-HR (1:1,000; Eterlife, Birmingham, UK) and goat polyclonal anti-actin (1:1,000; Santa CruzBiotechnology, Redwood, CA, USA). After washing with Tris-buffered saline (10 mM Tris-HCl and 150 mM NaCl, pH 7.5), membranes were incubated with an anti-rabbit IgG HRP-conjugated secondary antibody (1:1,000; Cell Signaling Technology, Danvers, MA, USA). Signals were detected using chemiluminescence (Millipore). Images were processed using Image J software.
Flow cytometry
Blood was drawn into vacuum tubes treated with the anticoagulant EDTA. For flow cytometry, whole blood was lysed in ACK lysing buffer for 5 min and then samples were washed with phosphate-buffered saline (PBS) via centrifugation, suspended in PBS, filtered through a 70 µm cell strainer (Biologix, Jinan, China), and stained with anti-CD3, anti-CD4, and anti-CD8 monoclonal antibodies conjugated with FITC, PE, and Alexa Fluor (wave length 488 nm), respectively, (all from BD, Franklin Lakes, NJ, USA) for 20 min at 4°C in the dark. After washing with 1 ml of PBS, 300 µl of each sample was analyzed using a BD FACS Verse flow cytometer (BD).
Statistical analysis
Results are expressed as mean ± SEM. Groups were compared using the Student’s t-test. P<0.05 was considered significant.
Results
Generation of HR−/− piglets via SCNT
HR−/− cells were used as a source of donor nuclei for SCNT. In the first round of SCNT, recipient pigs were euthanized on day 25 of gestation, and two morphologically normal fetuses were dissected to analyze HR gene expression and to obtain a pure population of Hr−/− cells. A cell line harboring a 20 bp deletion in one allele and a 1 bp insertion in the other allele was generated (Figs. 1A and B) and used as a source of donor nuclei. Additionally, the collection efficiency of HR mutation cell in magnetic separation was 32%. After the second round of SCNT, four recipients gave birth to 9 live HR−/− piglets and two recipients gave birth to 12 live WT piglets (Table 1). The HR−/− piglets were weak, suffered from diarrhea, and had difficulties suckling, and were therefore sacrificed at PN day 4 in subsequent experiments. As expected, western blotting with a rabbit anti-porcine HR antibody detected a lower level of HR protein in the eyelid skin of HR−/− piglets than in that of WT piglets (Fig. 2).
Fig. 1.
Generation and analysis of an hairless (HR)−/− fetus. (A) T7E1 assay using genomic DNA obtained from an HR−/− fetus. The arrows indicate the DNA fragments produced following T7E1 digestion. (B) DNA sequences of the HR locus in the HR−/− fetus. “−” and “+” denote nucleotide deletions and insertions, respectively. (C) Primers used in the T7E1 assay.
Table 1. Production of piglets by somatic cell nuclear transfer (SCNT).
| Donor cell | Recipient no. | No. of embryos transferred |
No. of fetuses obtained |
No. of piglets born alive |
|---|---|---|---|---|
| HR−/− | H-1 | 232 | 2 (killed on day 25) | |
| H-2 | 189 | 2 | ||
| H-3 | 176 | 1 | ||
| H-4 | 160 | 4 | ||
| H-5 | 140 | 2 | ||
| WT | W-1 | 158 | 7 | |
| W-2 | 165 | 5 | ||
HR, hairless.
Fig. 2.
Western blot analysis of hairless (HR) in eyelid skin samples of HR−/− and WT piglets. HR was detected using a polyclonal anti-Hr antibody. A polyclonal anti-actin antibody was used as a loading control.
Morphological changes in the skin of HR−/− piglets
At birth, HR−/− piglets exhibited significant hair loss specifically on their eyelids. Transmission electron microscopy was performed to determine the density of hairs on the eyelids of HR−/− and WT piglets at PN day 2 (Figs. 3A and B). The hair density was significantly lower on the eyelids of HR−/− piglets than on those of WT piglets (57.13 ± 10.99 vs. 133.30 ± 10.99 hairs/cm2, P<0.05) (Table 2). In addition, this analysis demonstrated that hairs were lost from follicles on the eyelids of HR−/− piglets.
Fig. 3.
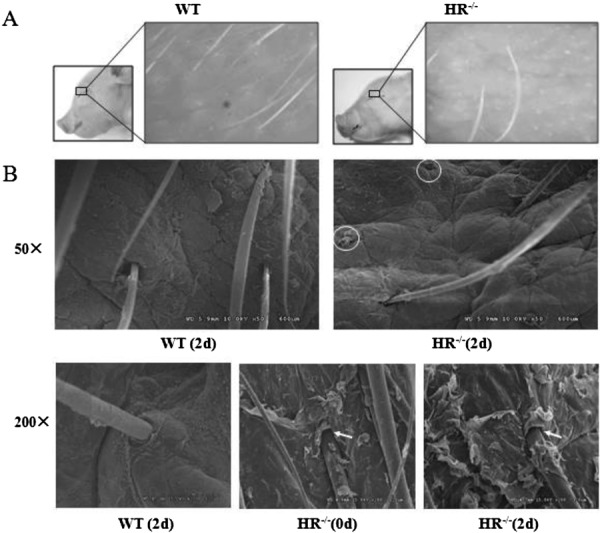
Hair loss and accelerated keratinization in hairless (HR)−/− piglets. (A) Eyelid skin (boxed area) of HR−/− and WT piglets at postnatal (PN) day 2. (B) Morphological changes in the back skin of HR−/− piglets. Transmission electron microscopy analysis of the back skin of HR−/− and WT piglets at the indicated PN day. The circles denote loss of hairs from the follicles. The arrows indicate keratinized stratified squamous epithelium and detachment of the inner root sheath from the hair shaft.
Table 2. Eyelid hair density, thymus weight, thymus weight index, and percentage of peripheral CD4+ CD8+ lymphocytes in WT and hairless (HR)−/− piglets.
| HR−/− piglets (mean ± SEM) |
WT piglets (mean ± SEM) |
P | |
|---|---|---|---|
| Eyelid hair density (hair number/cm2) | 57.13 ± 10.99 | 133.30 ± 10.99 | <0.05 |
| Thymus weight (g) | 2.07 ± 0.17 | 3.94 ± 1.07 | >0.05 |
| Thymus weight index (mg thymus weight/g body weight) | 1.95 ± 0.24 | 3.22 ± 0.37 | <0.05 |
| CD4+ CD8+ lymphocytes (%) | 5.07 ± 0.22 | 2.43 ± 0.14 | <0.05 |
To determine whether hair follicles of HR−/− piglets had an abnormal phenotype, back skin of HR−/−piglets at PN day 0 and 2 and of WT piglets at PN day 2 was imaged by scanning electron microscopy. HR−/− piglets exhibited a large amount of keratinized stratified squamous epithelium around the entrance of hair follicles, and the inner root sheath was detached from the hair shaft at the infundibulum (Fig. 3B).
Transcriptional changes in the skin of HR−/− piglets
Real-time PCR was performed to examine expression of the following genes in the eyelid skin of HR−/− piglets: the Wnt inhibitor Wise, the Wnt-responsive gene AXIN2, the HR-responsive genes apoptosis-related cysteine peptidase 14 (Caspase14) and homolog of the Drosophila muscle segment homeo box gene 2 (MSX2), and the Msx2-responsive gene Forkhead box protein N1 (FOXN1). As expected, expression of Wise, MSX2, and FOXN1 was upregulated in the eyelid skin of HR−/− piglets. Specifically, expression of Wise, a Wnt inhibitor, and of MSX2 and FOXN1, which play a putative role in hair follicle morphogenesis, was 1.2 ± 0.09-fold, 1.55 ± 0.08-fold, and 1.88 ± 0.28-fold higher, respectively, in eyelid skin of HR−/− piglets than in that of WT piglets at PN day 4 (P<0.05). Expression of Caspase14 and HR was not significantly changed. However, AXIN2 expression was 2-fold lower in eyelid skin of HR−/− piglets than in that of WT piglets at PN day 4 (P<0.05) (Fig. 4).
Fig. 4.
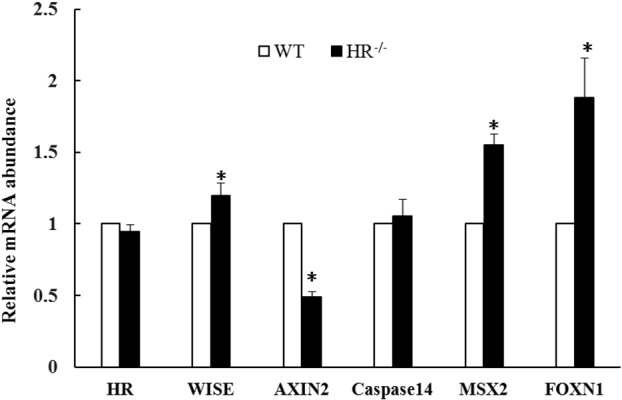
mRNA expression of hairless (HR), Wise, AXIN2, Caspase14, MSX2, and FOXN1 in eyelid skin of HR−/− and WT piglets at postnatal (PN) day 4. Expression of each gene was normalized against that of GAPDH. *P<0.05. Error bars indicate SEM.
Histological analysis of the thymus of HR−/− piglets
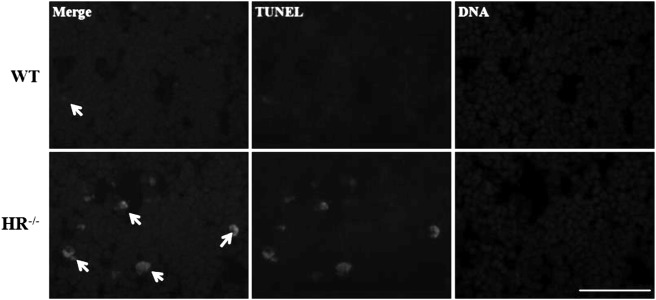
The thymus weight index of HR−/− piglets was significantly lower than that of age-matched WT piglets (1.95 ± 0.25 vs. 3.22 ± 0.37 mg thymus weight/g body weight, P<0.05) (Table 2). This showed evidence of that HR plays a role in thymus organogenesis. HR−/− piglets had many more pores in the thymic cortex than WT piglets at PN day 4. The level of apoptotic cells, characterized by cell shrinkage and blebbing, in these pores was significantly higher in HR−/− piglets than in WT piglets (Fig. 5). And TUNEL staining analysis detected a markedly increased apoptotic cells in HR−/− piglets compared with WT piglets (Fig. 6).
Fig. 5.

Histological analysis of apoptotic cells in the thymus of hairless (HR)−/− and WT piglets at postnatal (PN) day 4. The arrows indicate apoptotic cells.
Fig. 6.
TUNEL and Hoechst staining of apoptotic cells in the thymus of HR−/− and WT piglets at PN day 4. TUNEL of apoptotic cells (green) marked using arrow heads and DNA was stained by Hoechst 33342 (blue). Scale bar, 50 µm.
Changes in peripheral lymphocyte subpopulations in HR−/− piglets
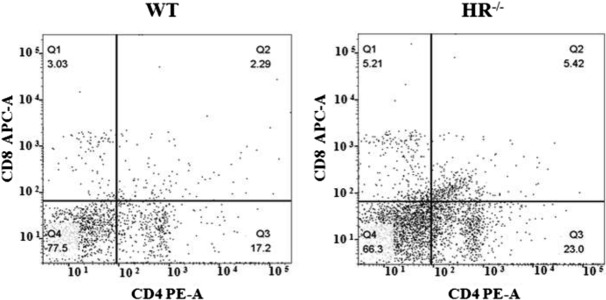
The percentages of lymphocyte subpopulations in the blood were analyzed by flow cytometry. Flow cytometric profiles of CD4 and CD8 expression on CD3+ lymphocytes in HR−/− and WT piglets at PN day 4. Q2 indicates the percentage of CD4+ CD8+ lymphocytes in peripheral blood. The percentages of CD4+ CD8− and CD4− CD8+ lymphocytes did not significantly differ between HR−/− and WT piglets at PN day 4 (data not shown). However, the percentage of CD4+ CD8+ lymphocytes was ~2-fold higher in HR−/− piglets than in WT piglets (P<0.05) (Fig. 7 and Table 2).
Fig. 7.
Flow cytometric analysis of CD4+ CD8+ lymphocytes in peripheral blood. These percentages are presented in Table 2.
Gene expression differences in the thymus of HR−/− piglets. All thymuses used in this experiment were taken from the end of the thymus. mRNA expression levels of MSX2 and FOXN1 in the thymus were measured by real-time PCR (Fig. 8). Expression of MSX2 was 2.73 ± 0.60-fold higher in the thymus of HR−/− piglets than in that of WT piglets, whereas expression of FOXN1 was 0.28 ± 0.15-fold lower. This suggests that MSX2 and FOXN1 are abnormally expressed in the thymus of HR−/− piglets, which may contribute to the observed changes in the weight and structure of the thymus.
Fig. 8.
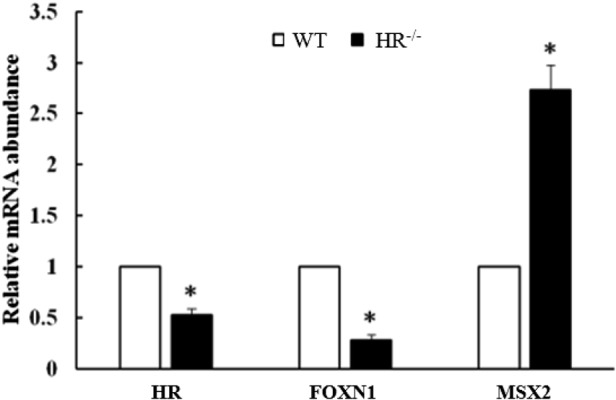
mRNA expression of FOXN1 and MSX2 in the thymus of HR−/− and WT piglets at postnatal (PN) day 4. *P<0.05. Error bars indicate SEM.
Discussion
In this study, we generated HR−/− piglets using the CRISPR/Cas9 system and SCNT. The T7 endonuclease I (T7E1) assay and sequence analysis revealed that the HR gene was successfully mutated (Fig. 1). We analyzed the potential off-target sites in genomes from cell pools treated with CRISPR/Cas9 nucleases,and no obvious off-target effects were observed (data not shown). Furthermore, western blotting (Fig. 2) demonstrated that the introduced mutations (a 20 bp deletion in one allele and a 1 bp insertion in the other allele) decreased protein expression of HR. These data confirmed that HR−/− piglets were successfully generated. This study is the first to mutate HR in pigs to the best of our knowledge, and this animal model could be useful for studying human hair disorders associated with HR gene mutations and the underlying molecular mechanisms.
Regulation of the hair cycle and hair shaft differentiation involves changes in expression of signaling molecules including Wnts, Bmps, Fgfs, and Shh [22]. Among them, the Wnt signaling pathway is important for skin organogenesis and morphogenesis [42]. Gain- and loss-of-function studies in vivo and in vitro demonstrated that HR plays a critical role in the Wnt signaling pathway in mice [3, 20]. Moreover, Hr-overexpressing and HR-KO mice exhibit hair follicle abnormalities and alopecia [16, 38]. Various transcription factors, including Msx2 and FoxN1, were recently reported to be downregulated upon overexpression of Hr in “hair poor” mice [16]. The present study reports that loss of HR function results in hair loss, accelerated keratinization, and changes in the expression of specific genes in pigs.
In YYHL mice with homozygous mutations in the Hr gene, progressive hair loss begins around the eyes and all hair is lost within 2 weeks [46]. Similarly, HR−/− piglets exhibited accelerated keratinization and progressive hair loss starting from the eyelids. Furthermore, gene expression changes in the skin of Hr−/− mice precede morphological alterations such as development of a keratinized stratified epithelium [25], an abnormal utricle, or cysts [45]. Therefore, we speculated that gene expression changes were responsible for the progressive hair loss observed in HR−/− piglets. However, the hair follicle changes in piglets were only detected in the eye skin, and we could not observe the changes in other parts of piglets’ hair follicles at this stage. Therefore, the HR−/− piglets still exhibit different phenotype from that of human patients.
Changes in gene expression have been extensively studied in mouse models lacking hair, but not in similar pig models. Here, we characterized gene expression changes in HR−/− piglets. Eyelid skin of piglets at PN day 4 was analyzed because it exhibited hair follicle disintegration. The Wnt-responsive gene AXIN2 was downregulated in the skin of HR−/− piglets, while MSX2 and FOXN1, which encode transcription factors involved in regulating differentiation of the cuticle, inner root sheath, and hair shaft [6], were upregulated. Axin2 gene expression is not detected in Hr−/− mouse hair follicles at the growing stage, during which the Wnt signaling pathway is switched off [3]. Based on these results, we propose that Wnt signaling is downregulated in HR−/− piglets. Previous research reported that expression of Msx2 and its target FoxN1 is downregulated in Hr-overexpressing mice [16], which is consistent with our findings. Similar to Hr−/− mice, HR−/− piglets exhibited several morphological changes and the misregulation of several genes at PN day 4, which will likely lead to severe morphological defects. Our findings indicate that regulation of the Wnt and Msx2 signaling pathways is perturbed in the skin of HR−/− piglets, in addition to that of mice with Hr mutations.
Hr−/− mice exhibit a progressive decrease in thymus weight, thymic cortex atrophy [31, 47], and immunological abnormalities [36]. Although immunohistochemical analyses have provided insight into the morphological changes that occur in the thymus of mice with Hr mutations [31], the underlying molecular mechanisms remain largely unknown.
HR−/− piglets exhibited structural and gene expression changes in the thymus and an altered level of CD4+ CD8+ lymphocytes in peripheral blood. The thymus weight index was decreased and the level of apoptosis in the thymic cortex was increased in HR−/− piglets, indicating that similar morphological changes occur in the thymus of HR−/− pigs and mice. The increased level of apoptosis in the thymic cortex of HR−/− piglets may cause thymic atrophy. In the thymus, double positive (DP) thymocytes are derived from immature double negative cells originating from bone marrow [43]. These DP thymocytes interact with thymus complex cell lines [37] and mature into functional single positive (SP) thymocytes [30], which are released into the bloodstream [13]. During normal thymocyte differentiation, CD4+ CD8+ lymphocytes are massively depleted and are therefore almost completely absent from peripheral blood [29, 32, 33]. In addition, a subset of DP T cells has been identified in autoimmune and chronic inflammatory disorders [29]. The thymus of BALB/c mice with acute or chronic Trypanosoma cruzi infections exhibits severe atrophy, mainly due to depletion of CD4+ CD8+ thymocytes via apoptosis, and the number of CD4+ CD8+ cells in peripheral T-lymphocyte subsets is significantly increased. Moreover, these DP T cells may contribute to autoimmunity [27]. Based on our results, we hypothesize that the increased number of CD4+ CD8+ lymphocytes in HR−/− piglets is due to a change in the microenvironment of the thymus, as observed in Hr−/− mice [39], and may contribute to autoimmunity in HR−/− piglets.
In our experiment the mRNA expression of HR was decreased in the thymus (Fig. 8). It has been reported that mutations in HR gene not only cause damage to the hair follicles, but also cause immune deficiency [15]. In 4 days, our HR−/− piglets showed the weakness as expressed above. We suspect this could be from the immune deficiency because of HR gene mutation, so the mRNA expression in thymus was decreased. FOXN1 and MSX2 were downregulated and upregulated, respectively, in the thymus of HR−/− piglets (Fig. 8). Because thymus weight was decreased in HR−/− piglets, as observed in mice lacking hair, we speculate that genes related to thymus organogenesis are aberrantly expressed. FOXN1 plays a major role in thymus organogenesis [39], and the thymus fails to develop when it is knocked out [9, 48]. FoxN1 is regulated via a mechanism involving Msx2 in skin [16]. However, our finding that mRNA expression of FOXN1 and MSX2 were differently affected in the thymus of HR−/− piglets indicates that a similar mechanism does not operate in the thymus. A further study is required to elucidate the molecular mechanism underlying the decreased expression of FOXN1.
Conclusion
The HR−/− piglets can be successfully produced using the CRISPR/Cas9 system and SCNT. All the morphological changes observed in HR−/− piglets are similar to those previously described in Hr−/− mice and humans. FOXN1 mRNA expression is decreased in the thymus of HR−/− piglets, and the underlying mechanism does not involve MSX2.
Supplementary Material
Supplementary Table 1-List of primers used for quantitative RT-PCR.
Acknowledgments
This work was supported by the Science and Technology Development Projects of Jilin Province of China (20170204035N Y). The authors would like to thank Hanji and Longxing pig farms for providing surrogate pig recipients.
References
- 1.Ahmad W., Faiyaz ul Haque M., Brancolini V., Tsou H.C., ul Haque S., Lam H., Aita V.M., Owen J., deBlaquiere M., Frank J., Cserhalmi-Friedman P.B., Leask A., McGrath J.A., Peacocke M., Ahmad M., Ott J., Christiano A.M.1998. Alopecia universalis associated with a mutation in the human hairless gene. Science 279: 720–724. doi: 10.1126/science.279.5351.720 [DOI] [PubMed] [Google Scholar]
- 2.Bae S., Kweon J., Kim H.S., Kim J.S.2014. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11: 705–706. doi: 10.1038/nmeth.3015 [DOI] [PubMed] [Google Scholar]
- 3.Beaudoin G.M., 3rd, Sisk J.M., Coulombe P.A., Thompson C.C.2005. Hairless triggers reactivation of hair growth by promoting Wnt signaling. Proc. Natl. Acad. Sci. USA 102: 14653–14658. doi: 10.1073/pnas.0507609102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendixen E., Danielsen M., Larsen K., Bendixen C.2010. Advances in porcine genomics and proteomics--a toolbox for developing the pig as a model organism for molecular biomedical research. Brief. Funct. Genomics 9: 208–219. doi: 10.1093/bfgp/elq004 [DOI] [PubMed] [Google Scholar]
- 5.Bergfeld W.F.1995. Androgenetic alopecia: an autosomal dominant disorder. Am. J. Med. 98:(1A): 95S–98S. doi: 10.1016/S0002-9343(99)80065-5 [DOI] [PubMed] [Google Scholar]
- 6.Cai J., Lee J., Kopan R., Ma L.2009. Genetic interplays between Msx2 and Foxn1 are required for Notch1 expression and hair shaft differentiation. Dev. Biol. 326: 420–430. doi: 10.1016/j.ydbio.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S.W., Kim S., Kim J.M., Kim J.S.2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 230–232. doi: 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- 8.Davies R.E., Austin W.A., Logani M.K.1971. The rhino mutant mouse as an experimental tool. Trans. N. Y. Acad. Sci. 33: 680–693. doi: 10.1111/j.2164-0947.1971.tb02632.x [DOI] [PubMed] [Google Scholar]
- 9.Frank J., Pignata C., Panteleyev A.A., Prowse D.M., Baden H., Weiner L., Gaetaniello L., Ahmad W., Pozzi N., Cserhalmi-Friedman P.B., Aita V.M., Uyttendaele H., Gordon D., Ott J., Brissette J.L., Christiano A.M.1999. Exposing the human nude phenotype. Nature 398: 473–474. doi: 10.1038/18997 [DOI] [PubMed] [Google Scholar]
- 10.Hardy M.H.1992. The secret life of the hair follicle. Trends Genet. 8: 55–61. doi: 10.1016/0168-9525(92)90044-5 [DOI] [PubMed] [Google Scholar]
- 11.Hsieh J.C., Sisk J.M., Jurutka P.W., Haussler C.A., Slater S.A., Haussler M.R., Thompson C.C.2003. Physical and functional interaction between the vitamin D receptor and hairless corepressor, two proteins required for hair cycling. J. Biol. Chem. 278: 38665–38674. doi: 10.1074/jbc.M304886200 [DOI] [PubMed] [Google Scholar]
- 12.Ignat’eva E.L., Malashenko A.M., Blandova Z.K.1988. [Morphofunctional characteristics of the reproductive system in female mice of the mutant strain B10-hr rhY]. Biull. Eksp. Biol. Med. 106: 602–603. (in Russian) doi: 10.1007/BF00840860 [DOI] [PubMed] [Google Scholar]
- 13.Jho E.H., Zhang T., Domon C., Joo C.K., Freund J.N., Costantini F.2002. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22: 1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang J.D., Kim S., Zhu H.Y., Jin L., Guo Q., Li X.C., Zhang Y.C., Xing X.X., Xuan M.F., Zhang G.L., Luo Q.R., Kim Y.S., Cui C.D., Li W.X., Cui Z.Y., Kim J.S., Yin X.J.2017. Generation of cloned adult muscular pigs with myostatin gene mutation by genetic engineering. RSC Advances 7: 12541–12549. doi: 10.1039/C6RA28579A [DOI] [Google Scholar]
- 15.Kanzler M.H., Rasmussen J.E.1986. Atrichia with papular lesions. Arch. Dermatol. 122: 565–567. doi: 10.1001/archderm.1986.01660170095027 [DOI] [PubMed] [Google Scholar]
- 16.Kim B.K., Yoon S.K.2013. Hairless down-regulates expression of Msx2 and its related target genes in hair follicles. J. Dermatol. Sci. 71: 203–209. doi: 10.1016/j.jdermsci.2013.04.019 [DOI] [PubMed] [Google Scholar]
- 17.Kim H., Kim M.S., Wee G., Lee C.I., Kim H., Kim J.S.2013. Magnetic separation and antibiotics selection enable enrichment of cells with ZFN/TALEN-induced mutations. PLoS One 8: e56476. doi: 10.1371/journal.pone.0056476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H., Um E., Cho S.R., Jung C., Kim H., Kim J.S.2011. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat. Methods 8: 941–943. doi: 10.1038/nmeth.1733 [DOI] [PubMed] [Google Scholar]
- 19.Kim H.J., Lee H.J., Kim H., Cho S.W., Kim J.S.2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 19: 1279–1288. doi: 10.1101/gr.089417.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J.K., Kim E., Baek I.C., Kim B.K., Cho A.R., Kim T.Y., Song C.W., Seong J.K., Yoon J.B., Stenn K.S., Parimoo S., Yoon S.K.2010. Overexpression of Hr links excessive induction of Wnt signaling to Marie Unna hereditary hypotrichosis. Hum. Mol. Genet. 19: 445–453. doi: 10.1093/hmg/ddp509 [DOI] [PubMed] [Google Scholar]
- 21.Kligman L.H.1996. The hairless mouse model for photoaging. Clin. Dermatol. 14: 183–195. doi: 10.1016/0738-081X(95)00154-8 [DOI] [PubMed] [Google Scholar]
- 22.Krause K., Foitzik K.2006. Biology of the hair follicle: the basics. Semin. Cutan. Med. Surg. 25: 2–10. doi: 10.1016/j.sder.2006.01.002 [DOI] [PubMed] [Google Scholar]
- 23.Li X.J., Li W.2012. Beyond mice: genetically modifying larger animals to model human diseases. J. Genet. Genomics 39: 237–238. doi: 10.1016/j.jgg.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Chen J.Y., Shang H.T., Liu C.E., Wang Y., Niu R., Wu J., Wei H.2010. Light microscopic, electron microscopic, and immunohistochemical comparison of Bama minipig (Sus scrofa domestica) and human skin. Comp. Med. 60: 142–148. [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., Sundberg J.P., Das S., Carpenter D., Cain K.T., Michaud E.J., Voy B.H.2010. Molecular basis for hair loss in mice carrying a novel nonsense mutation (Hrrh-R ) in the hairless gene (Hr). Vet. Pathol. 47: 167–176. doi: 10.1177/0300985809352970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maatough A., Whitfield G.K., Brook L., Hsieh D., Palade P., Hsieh J.C.2018. Human Hairless Protein Roles in Skin/Hair and Emerging Connections to Brain and Other Cancers. J. Cell. Biochem. 119: 69–80. [DOI] [PubMed] [Google Scholar]
- 27.Mendes-da-Cruz D.A., de Meis J., Cotta-de-Almeida V., Savino W.2003. Experimental Trypanosoma cruzi infection alters the shaping of the central and peripheral T-cell repertoire. Microbes Infect. 5: 825–832. doi: 10.1016/S1286-4579(03)00156-4 [DOI] [PubMed] [Google Scholar]
- 28.Morrissey P.J., Parkinson D.R., Schwartz R.S., Waksal S.D.1980. Immunologic abnormalities in HRS/J mice. I. Specific deficit in T lymphocyte helper function in a mutant mouse. J. Immunol. 125: 1558–1562. [PubMed] [Google Scholar]
- 29.Parel Y., Chizzolini C.2004. CD4+ CD8+ double positive (DP) T cells in health and disease. Autoimmun. Rev. 3: 215–220. doi: 10.1016/j.autrev.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 30.Penit C., Vasseur F.1989. Cell proliferation and differentiation in the fetal and early postnatal mouse thymus. J. Immunol. 142: 3369–3377. [PubMed] [Google Scholar]
- 31.San Jose I., García-Suárez O., Hannestad J., Cabo R., Gauna L., Represa J., Vega J.A.2001. The thymus of the hairless rhino-j (hr/rh-j) mice. J. Anat. 198: 399–406. doi: 10.1046/j.1469-7580.2001.19840399.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprent J., Kishimoto H.2002. The thymus and negative selection. Immunol. Rev. 185: 126–135. doi: 10.1034/j.1600-065X.2002.18512.x [DOI] [PubMed] [Google Scholar]
- 33.Starr T.K., Jameson S.C., Hogquist K.A.2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21: 139–176. doi: 10.1146/annurev.immunol.21.120601.141107 [DOI] [PubMed] [Google Scholar]
- 34.Sundberg J.P., Beamer W.G., Shultz L.D., Dunstan R.W.1990. Inherited mouse mutations as models of human adnexal, cornification, and papulosquamous dermatoses. J. Invest. Dermatol. 95:(Suppl): 62S–63S. doi: 10.1111/1523-1747.ep12505816 [DOI] [PubMed] [Google Scholar]
- 35.Swindle M.M., Makin A., Herron A.J., Jr, Clubb F.J., Jr, Frazier K.S.2012. Swine as models in biomedical research and toxicology testing. Vet. Pathol. 49: 344–356. doi: 10.1177/0300985811402846 [DOI] [PubMed] [Google Scholar]
- 36.Takaoki M., Kawaji H.1980. Impaired antibody response against T-dependent antigens in rhino mice. Immunology 40: 27–32. [PMC free article] [PubMed] [Google Scholar]
- 37.Takayama E., Kina T., Katsura Y., Tadakuma T.1998. Enhancement of activation-induced cell death by fibronectin in murine CD4+ CD8+ thymocytes. Immunology 95: 553–558. doi: 10.1046/j.1365-2567.1998.00636.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson C.C., Sisk J.M., Beaudoin G.M.J., 3rd2006. Hairless and Wnt signaling: allies in epithelial stem cell differentiation. Cell Cycle 5: 1913–1917. doi: 10.4161/cc.5.17.3189 [DOI] [PubMed] [Google Scholar]
- 39.Vaidya H.J., Briones Leon A., Blackburn C.C.2016. FOXN1 in thymus organogenesis and development. Eur. J. Immunol. 46: 1826–1837. doi: 10.1002/eji.201545814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verma N., Rettenmeier A.W., Schmitz-Spanke S.2011. Recent advances in the use of Sus scrofa (pig) as a model system for proteomic studies. Proteomics 11: 776–793. doi: 10.1002/pmic.201000320 [DOI] [PubMed] [Google Scholar]
- 41.Walters E.M., Agca Y., Ganjam V., Evans T.2011. Animal models got you puzzled?: think pig. Ann. N. Y. Acad. Sci. 1245: 63–64. doi: 10.1111/j.1749-6632.2011.06345.x [DOI] [PubMed] [Google Scholar]
- 42.Widelitz R.B.2008. Wnt signaling in skin organogenesis. Organogenesis 4: 123–133. doi: 10.4161/org.4.2.5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Würch A., Biro J., Falk I., Mossmann H., Eichmann K.1999. Reduced generation but efficient TCR beta-chain selection of CD4+8+ double-positive thymocytes in mice with compromised CD3 complex signaling. J. Immunol. 162: 2741–2747. [PubMed] [Google Scholar]
- 44.Yin X.J., Tani T., Yonemura I., Kawakami M., Miyamoto K., Hasegawa R., Kato Y., Tsunoda Y.2002. Production of cloned pigs from adult somatic cells by chemically assisted removal of maternal chromosomes. Biol. Reprod. 67: 442–446. doi: 10.1095/biolreprod67.2.442 [DOI] [PubMed] [Google Scholar]
- 45.Zarach J.M., Beaudoin G.M.J., 3rd, Coulombe P.A., Thompson C.C.2004. The co-repressor hairless has a role in epithelial cell differentiation in the skin. Development 131: 4189–4200. doi: 10.1242/dev.01303 [DOI] [PubMed] [Google Scholar]
- 46.Zhang J.T., Fang S.G., Wang C.Y.2005. A novel nonsense mutation and polymorphisms in the mouse hairless gene. J. Invest. Dermatol. 124: 1200–1205. doi: 10.1111/j.0022-202X.2005.23744.x [DOI] [PubMed] [Google Scholar]
- 47.Zhu K.C., Zhang J.T., Wang C.Y.2010. Skin abnormalities, female reproductive disorders and shorter life span with a mutation in the hairless gene. Life Sci. 7: 105–111. [Google Scholar]
- 48.Žuklys S., Handel A., Zhanybekova S., Govani F., Keller M., Maio S., Mayer C.E., Teh H.Y., Hafen K., Gallone G., Barthlott T., Ponting C.P., Holländer G.A.2016. Foxn1 regulates key target genes essential for T cell development in postnatal thymic epithelial cells. Nat. Immunol. 17: 1206–1215. doi: 10.1038/ni.3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1-List of primers used for quantitative RT-PCR.