Abstract
The Asian house shrew, Suncus murinus, is an insectivore (Eulipotyphla, Mammalia) and an important laboratory animal for life-science studies. The gastrointestinal tract of Suncus is simple: the length of the entire intestine is very short relative to body size, the large intestine is quite short, and there are no fermentative chambers such as the forestomach or cecum. These features imply that Suncus has a different nutritional physiology from those of humans and mice, but little is known about whether Suncus utilizes microbial fermentation in the large (LI) or small (SI) intestine. In addition, domestication may affect the gastrointestinal microbial diversity of Suncus. Therefore, we compared the gastrointestinal microbial diversity of Suncus between laboratory and wild Suncus and between the SI and LI (i.e., four groups: Lab-LI, Lab-SI, Wild-LI, and Wild-SI) using bacterial 16S rRNA gene library sequencing analyses with a sub-cloning method. We obtained 759 cloned sequences (176, 174, 195, and 214 from the Lab-LI, Lab-SI, Wild-LI, and Wild-SI samples, respectively), which revealed that the gastrointestinal microbiota of Suncus is rich in Firmicutes (mostly lactic acid bacteria), with few Bacteroidetes. We observed different bacterial communities according to intestinal region in laboratory Suncus, but not in wild Suncus. Furthermore, the gastrointestinal microbial diversity estimates were lower in laboratory Suncus than in wild Suncus. These results imply that Suncus uses lactic acid fermentation in the gut, and that the domestication process altered the gastrointestinal bacterial diversity.
Keywords: domestication, Eulipotyphla, insectivore, microflora, symbiosis
Introduction
The Asian house shrew, Suncus murinus (Soricidae, Eulipotyphla, Mammalia), has a wide distribution from Southeast to West Asia [17]. In Japan, presumptively native populations are distributed in the Nansei Islands (the Amami, Okinawa, Miyako, and Yaeyama Islands), whereas introduced populations had distributed in Kyushu [29]. This species has been domesticated as a laboratory bioresource since the 1970s [32, 35, 36], and several strains captured from different areas have been developed [18, 34]. As a laboratory animal, the domesticated Asian house shrew is called “Suncus”. It has been used in various fields of science due to its unique characteristics compared with laboratory rodents, including as models of emesis [7, 54], for in vivo motilin studies [42, 51, 52], and for dental studies [20, 57].
These unique characters of Suncus owe a great deal to its evolutionary background. The Asian house shrew belongs to the phylum Eulipotyphla (in superorder Laurasiatheria), one of the oldest mammalian groups. Eulipotyphla appeared before the Cretaceous–Paleogene boundary (ca. 66 million years ago) and diverged from the superorder Euarchontoglires (humans, mice, rats, and rabbits) well before the Cretaceous–Paleogene boundary [1, 10]. Animals in this group possess many primitive mammalian morphological characters. The members of Eulipotyphla are insectivorous and lack fermentative chambers such as the cecum and forestomach in their gastrointestinal tracts, meaning they have a very simple gut.
Interestingly, the ratio of the length of the gastrointestinal tract to body size in Suncus is very small compared with that in other mammalian taxa [25, 49]. Although the small and large intestines are not perfectly separated because Suncus lacks a cecum, the ratio of the length of the large intestine to that of the small intestine is quite small compared with that in other mammals [14, 25]. Histologically, the large intestine extends only 1 cm from the anus [14, 58]. Therefore, Suncus has unique gastrointestinal tract characteristics: a very simple structure (with no fermentative chambers), very short total intestine length, and very short large intestine length. Because these features imply that Suncus has a different nutritional physiology, we wondered what microbes inhabit their intestines.
The symbiotic microbiota of the mammalian gastrointestinal tract provide the host mammals with valuable substances (e.g., enzymes) for nutritional symbiosis [review in 15, 43]. Recently, it was found that the symbiotic microbiota inhabiting the mammalian gastrointestinal tract play substantial roles in maintaining host homeostasis [3, 19, 37, 50]. The diversity of the gut microbiota has been shaped not only by the host evolutionary background but also by its food habits [5, 26, 27, 30]. Ley et al. revealed that the diversity of the fecal microbiota in mammals is higher in herbivores than in omnivores and is lowest in carnivores, based on a network-based analysis of cloned bacterial 16S rRNA gene sequences from 59 mammalian species [26]. This implies that food habits are an important factor in gut microbiota. However, little is known about the gut microbiota of insectivores such as Suncus.
In addition, domestication, a prolonged process of captivity over generations, may alter the mammalian gastrointestinal tract microbiota. There are reports that captivity altered the gut microbiota of certain mammalian species [5, 22, 24, 28, 53]. In mammals, the intestinal microbiota is generally transferred vertically from dam to offspring via the birth canal [2, 38]. Furthermore, laboratory animals are usually housed under very stable and clean conditions to avoid infectious diseases and experimental errors that arise from environmental factors, which may prevent the horizontal acquisition of certain bacterial species from the environment. Therefore, we hypothesized that the gut microbiota of laboratory Suncus would differ from that of wild Suncus.
In this study, we collected the intestinal contents of the small and large intestines of wild and laboratory Suncus and compared the microbiota using libraries of cloned bacterial 16S rRNA gene sequences. The main purposes of this study were to characterize the gut microbiota of Suncus and compare the microbiota between the small and large intestines and between laboratory and wild Suncus to determine how domestication has affected gut microbial diversity. This new knowledge may increase the utility of laboratory Suncus and shed light on the evolution of mammalian gut microbial diversity.
Materials and Methods
Animals
We used six (three female and three male) laboratory Suncus (the standard KAT strain; originally, 1 male and 2 females were collected from Kathmandu, Nepal in 1991 and domesticated at Nagoya University [34]) animals maintained at the Faculty of Agriculture, University of Miyazaki. The animals were housed in accordance with the standard protocol for laboratory Suncus [32, 33] and given fish feed for trout (Masu-Deluxe HI, Nosan, Yokohama, Japan) and tap water ad libitum. In addition, 5 (2 females and 3 males) wild Suncus individuals were captured in Nishihara in Nakagami District in Okinawa Prefecture using Sherman traps in February 2017. The contents of their small and large intestines were removed using autoclaved scissors and tweezers into gamma-ray-sterilized 2.0-ml tubes and stored at −80°C until DNA extraction. The small or large intestines of some of the laboratory Suncus were empty; we ultimately analyzed samples from three small intestines and four large intestines.
All of the protocols used in this study were approved by the Animal Experiment Committee of the University of Miyazaki (Permission No. 2010-512, 2016-520). The capture of wild animals in Okinawa Prefecture was approved by the Okinawa Prefectural Government (Permit No. Dai 28 Honcho 114, Dai 28 Honcho 115).
Laboratory experiments
Total DNA was extracted from samples using ISOFECAL for Beads Beating (Nippon Gene, Tokyo, Japan), according to the manufacturer’s protocol. To construct the 16S rRNA gene libraries, we followed the protocols described in Ley et al. [26]. In brief, bacterial 16S rRNA genes (ca. 1300 bp) were amplified separately for individuals and intestinal sections using the following universal primer pair: 8F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1391R (5’-GACGGGCGGTGWGTRCA-3’). The PCR conditions were 94°C for 2 min, followed by 20 cycles of 94°C for 1 min, 55°C for 45 s, and 72°C for 2 min, with a final 20-min extension at 72°C. The amplified PCR products were pooled for each experimental group: i.e., the large intestines of laboratory Suncus (Lab-LI), the small intestines of laboratory Suncus (Lab-SI), the large intestines of wild Suncus (Wild-LI), and the small intestines of wild Suncus (Wild-SI). The PCR products were then gel-purified using a GENECLEAN II kit (Funakoshi, Tokyo, Japan) and cloned using the pGEM-T Easy vector system (Promega, Tokyo, Japan). From each library, more than 196 colonies containing cloned amplicons were processed for sequencing. The plasmid inserts were sequenced using vector-specific primers, a Big Dye Terminator cycle sequencing kit v3.1 (Applied Biosystems, Foster City, CA, USA), and a genetic analyzer (Model 3730, 3130, Applied Biosystems).
Data analyses
The data analyses were performed using the 16S rRNA gene analysis software package Mothur v. 1.40.3 [44]. First, the obtained cloned sequences (ca. 1300 bp; 179 from Lab-LI, 184 from Lab-SI, 215 from Wild-LI, and 226 from Wild-SI samples) were aligned as SILVA alignments using align.seqs [40] with SILVA reference file release 132 (released in June, 2018), and then putative chimeras were checked using chimera.slayer [13] with the SILVA-based alignment of the template file. The SILVA reference and template files were both obtained from the Mothur web site (https://mothur.org/wiki/Silva_reference_files). As a result, 3, 10, 20, and 12 sequences were identified as putative chimeras from the Lab-LI, Lab-SI, Wild-LI, and Wild-SI samples, respectively, and removed from the dataset. The final dataset included 759 sequences (176 Lab-LI, 174 Lab-SI, 195 Wild-LI, and 214 Wild-SI sequences), which were deposited in the DNA Database of Japan under accession numbers LC463303 to LC464061. The classifications of the 759 cloned sequences were estimated using classify.seqs with SILVA reference file release 132.
Next, pairwise genetic distances were calculated using dist.seqs (set countends=F and output=lt options) following filter.seqs (set vertical=T option). Then, the data were clustered into operational taxonomic units (OTUs) with 98.7% similarity thresholds (according to Stackebrandt and Ebers [48]) using the cluster command for OTU-based analyses. From these clustered data, we estimated Good’s coverage [12], the Shannon (H’) [45] and Simpson (1-D) [47] biodiversity indexes, and the Chao1 [4] species richness estimator using summary.single to compare the alpha diversity estimates among the 4 samples. We also ran rarefaction.single to execute rarefaction analyses of the number of OTUs observed and the coverage index. Finally, the similarity indexes for community membership (Jaccard similarity coefficient) and community structure (θ-YC [59]) among the four samples were estimated using dist.shared following the make.shared command. We also estimated quantitative (weighted UniFrac) and qualitative (unweighted UniFrac) beta diversity measurements based on the phylogenetic tree constructed using clearcut [8].
The correlation between the numbers of sequences and OTUs was analyzed using the Rcmdr package v2.5-1 [11] with R v3.5.1 [41].
Results
We obtained 176 (Lab-LI), 174 (Lab-SI), 195 (Wild-LI), and 214 (Wild-SI) chimera-free 16S rRNA sequences from the small and large intestine contents of laboratory and wild Suncus. Using a 98.7% similarity threshold, the cloned sequences clustered into 12 (Lab-LI), 11 (Lab-SI), 46 (Wild-LI), and 39 (Wild-SI) OTUs (Table 1). We found no significant correlation between the numbers of sequences and OTUs (adjusted R2=0.541, P=0.167). The Good’s coverage for all samples was very high (89.2–99.4%; Table 1), and the rarefaction curve reached a plateau (Fig. 1A), indicating that our sequencing effort was sufficient to obtain perspectives on each microbial community. However, the rarefaction curve analyses of the number of OTUs implied that unsequenced bacteria still existed (Fig. 1B). The results indicate that we successfully revealed the major members of the gastrointestinal microbial communities of laboratory and wild Suncus, but we still cannot reject the existence of minor species. All of the alpha diversity estimates were much lower for laboratory Suncus than for wild Suncus (Table 1).
Table 1. The microbial diversity and species richness of the large (LI) and small (SI) intestinal contents from laboratory Suncus murinus and wild Suncus murinus based on 16S rRNA gene libraries.
| Laboratory | Wild | |||
|---|---|---|---|---|
| LI | SI | LI | SI | |
| Number of clones | 176 | 174 | 195 | 214 |
| Number of OTUs (98.7%) | 12 | 11 | 46 | 39 |
| Good’s Coverage (%) | 97.7 | 99.4 | 89.2 | 93.9 |
| Shannon index (H’) | 1.75 | 1.22 | 3.22 | 3.05 |
| Simpson index (1-D) | 0.77 | 0.49 | 0.94 | 0.93 |
| Chao1 | 15 | 11 | 69.3 | 54.6 |
Number of clones=number of cloned sequences after removing putative chimeras; Number of OTUs (98.7%)=number of operational taxonomic units (OTUs) detected with 98.7% similarity thresholds [48]; Good’s coverage=read depth of each library [12]; Shannon and Simpson indexes=diversity indexes [45 and 47, respectively]; Chao1=species richness estimator [4].
Fig. 1.
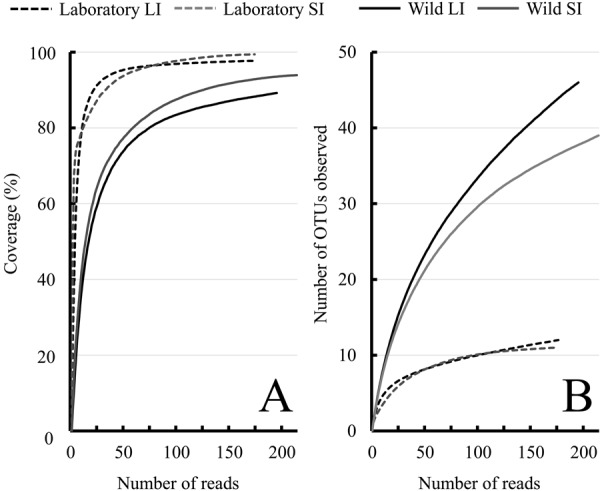
Rarefaction curves for Good’s coverage (A) and the number of OTUs observed (B) for bacterial 16S rRNA gene sequences in the large (LI) and small (SI) intestinal contents from laboratory Suncus murinus and wild Suncus murinus with a cut-off threshold of 98.7% similarity. Good’s coverage indicates read depth of each library [12], and the number of OTUs observed were detected with 98.7% similarity thresholds [48].
In all samples, Firmicutes was the predominant bacterial phylum detected (59.5–87.4%; Fig. 2). Within Firmicutes, Streptococcaceae was the major family detected in laboratory Suncus (53.1% in Lab-LI and 89.5% in Lab-SI), whereas it was less prevalent in wild Suncus (19% in Wild-LI and 38.1% in Wild-SI) (Fig. 3). Of note, all of the Streptococcaceae sequences from laboratory Suncus were identified as Streptococcus spp., whereas most from wild Suncus were identified as Lactococcus spp. (Supplementary Table 1). Peptostreptococcaceae was a second major family group detected within Firmicutes: 34.6% in Lab-LI, 44.8% in Wild-LI, and 39.6% in Wild-SI. Most of these sequences were identified as Paeniclostridium spp. (which were formerly classified in the genus Clostridium) and Romboutsia spp. (Supplementary Table 1). Other groups detected were Clostridiaceae (all sequences were identified as Clostridium [sensu stricto]) and Enterococcaceae (including Enterococcus spp. and Vagococcus spp.) (Supplementary Table 1). These bacterial groups are common in animal gastrointestinal tracts and feces.
Fig. 2.
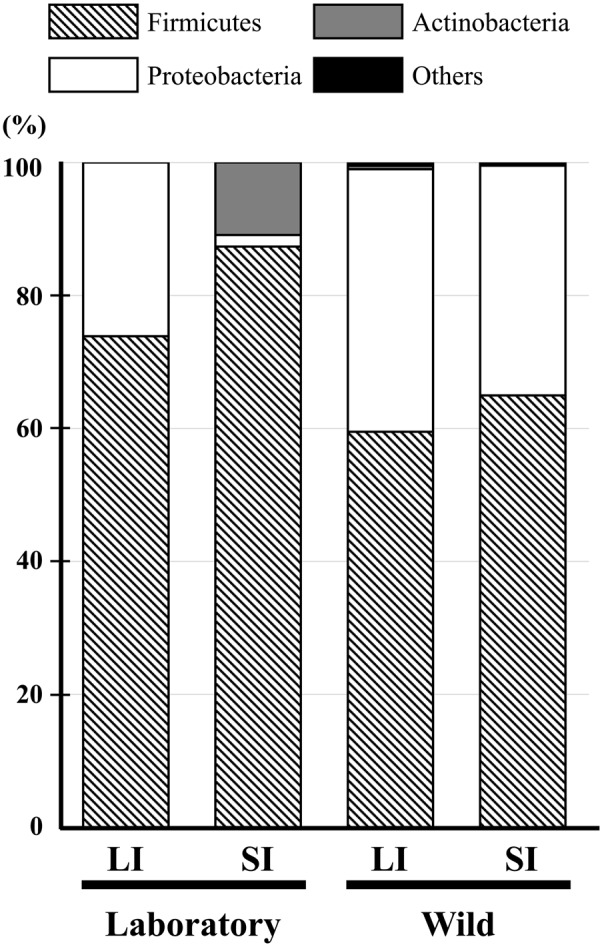
Relative abundance of bacterial 16S rRNA gene sequences in the large (LI) and small (SI) intestinal contents from the laboratory Suncus murinus and wild Suncus murinus classified at the phylum level.
Fig. 3.
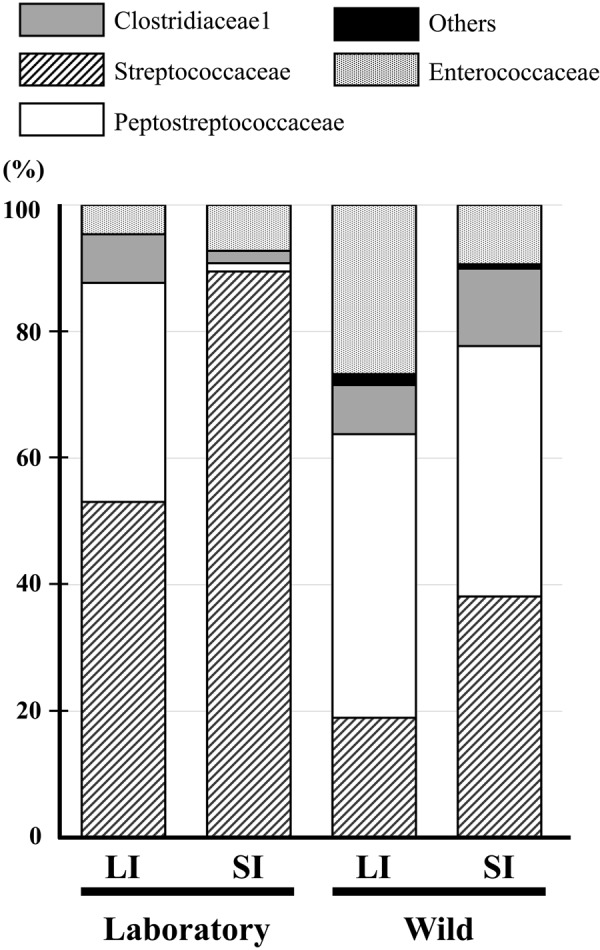
Relative abundances of bacterial 16S rRNA gene sequences in the large (LI) and small (SI) intestinal contents from the laboratory Suncus murinus and wild Suncus murinus classified at the family level within Firmicutes.
Proteobacteria was another major group detected in Lab-LI, Wild-LI, and Wild-SI (26.1–39.5%; Fig. 2), most of which were facultative anaerobes in the family Enterobacteriaceae, another major group in animal gastrointestinal tracts. Of these sequences, the Escherichia/Shigella group was found in all samples, whereas Plesiomonas spp., which is a pathogenic group, was found only in wild Suncus (Supplementary Table 1).
The Lab-SI samples, but not the other samples, contained a relatively high percentage of Actinobacteria (10.9%; Fig. 2), including Actinomycetaceae, Corynebacteriaceae, and Propionibacteriaceae (Supplementary Table 1), which are also found in the gastrointestinal tracts of animals. A single sequence belonging to the phylum Tenericutes was found in Wild-LI and was identified as Mycoplasma spp. (Supplementary Table 1). We did not detect any sequences identified as Bacteroidetes, which is a major bacterial group in the gastrointestinal tracts of humans and mice.
The UniFrac test (Table 2) indicated differences in both community membership (unweighted) and community structure (weighted) among most of the samples (P<0.001), except between Wild-LI and Wild-SI (Table 2), which had smaller Jaccard index (for community membership) and θ-YC (for community structure) values. The difference between Lab-LI and Lab-SI was apparently observed in Actinobacteria (Fig. 2), and numbers of sequences detected were different in some Firmicutes (Fig. 3, see also Supplementary Table 1).
Table 2. Similarity indices of microbial membership (Jaccard index and UniFrac unweighted) and community structure (θ-YC index and UniFrac weighted) of the large (LI) and small (SI) intestinal contents from laboratory Suncus murinus and wild Suncus murinus.
| Jaccard index | θ-YC index | Unifrac test | |||
|---|---|---|---|---|---|
| unweighted | weighted | ||||
| Between intestinal part | |||||
| Laboratory LI vs. Laboratory SI | 0.647 | 0.407 | 0.634 (P<0.001) | 0.672 (P<0.001) | |
| Wild LI vs. Wild SI | 0.583 | 0.220 | 0.625 (P=0.022) | 0.102 (P=0.034) | |
| Between laboratory and wild | |||||
| Laboratory LI vs. Wild LI | 0.885 | 0.974 | 0.767 (P<0.001) | 0.279 (P<0.001) | |
| Laboratory SI vs. Wild SI | 0.913 | 0.965 | 0.797 (P<0.001) | 0.741 (P<0.001) | |
Jaccard index=traditional similarity coefficient based on the number of shared operational taxonomic units (OTUs); θ-YC index=similarity index employing relative abundance of OTUs [59]; Unifrac test=β-diversity measure which describes whether communities have same generic structure based on the phylogenetic tree. Weighted Unifrac test incorporates proportional abundance of the taxa (qualitative), and unweighted Unifrac does not take (quantative).
Discussion
Suncus is the only experimental animal belonging to Eulipotyphla. In this study, we first revealed the major members of the gastrointestinal microbiota of laboratory Suncus and wild Suncus. The microbiotas were rich in Firmicutes, with no Bacteroidetes present (Fig. 2). These two phyla are the most common phyla in the vertebrate gut microbiota [27]; we did not detect any Bacteroidetes sequences, even though we analyzed 759 clones in total. This was not due to experimental error in our laboratory because we previously detected many Bacteroidetes from rodents using the same strategy [46]. Although our sequencing effort was not perfect for detecting rare species, Bacteroidetes may be scarce in the Suncus gut. Characterization of few Bacteroidetes has also been reported in some mammalian species, including bears, cheetahs, giant pandas, red pandas, hedgehogs, and echidnas (see Supplementary Data in Ley et al. [26]). These species are carnivorous, herbivorous, and insectivorous. However, the giant pandas and red pandas are “apparent herbivores” because both species evolved from carnivorous to herbivorous (eating bamboo) with loss of the umami taste receptor [16]; as a result they are poorly adapted for digesting bamboo [6, 23, 55, 56]. Therefore, we suggest that the presence of few Bacteroidetes is not common but may be observed in some insectivorous and carnivorous mammals.
Ley et al. [26] reported that fecal microbiota diversity in mammals was high in herbivores, low in carnivores, and intermediate in omnivores. In this study, we detected only 11–46 OTUs from 174–214 clones, implying that the gut microbiota diversity of Suncus is very low. Although we do not have definitive evidence, we speculate that the low diversity is due to the morphological features of the gastrointestinal tract in Suncus, i.e., its simplicity, short total intestine relative to body size, very short large intestine, and lack of fermentative chambers such as the forestomach and caecum [14, 25, 58]. These characteristics may limit the physiological space and physical area where microbes occur. The carnivorous species containing few Bacteroidetes listed above also had very low alpha diversity estimates compared with other mammals (see the Supplementary Data file in Ley et al. [26]), and each has at least one of the four abovementioned anatomical features [49]. Unfortunately, we could not identify all members of the gut microbiota. A future study with deep-sequencing technology using a next-generation sequencer will provide more detailed analyses.
We did identify 4–10 genera within Firmicutes. The lactic acid bacteria Enterococcus, Lactococcus, Streptococcus, and Vagococcus were abundant (27.2–84.5%) in the Suncus gut, implying that lactic acid fermentation is important in Suncus, as in many mammals. Lactic acid bacteria contribute to the complexity of the gut microbiota and have many beneficial effects on metabolism and immunomodulation, as revealed in human studies [39]. Although Suncus has a short and simple gastrointestinal tract, lactic acid fermentation is probably essential, and the lactic acid bacteria found in this study are likely autochthonous, as reported in carnivores [9]. However, these lactic acid bacteria and other members of Firmicutes are not the core members of the human or mouse gut microbiotas [31], indicating that the gut microbiota in Suncus is quite different from that in humans and mice.
Comparing the gut microbial diversity between wild Suncus and laboratory Suncus, the diversity was lower in the laboratory animals. Although our experimental design was not a strict comparison between captive and wild animals because the original collection locality of the KAT strain (Kathmandu, Nepal [34]) is far from Okinawa, where we captured the wild samples, studies have found that captivity (including domestication) lowers gut microbial diversity in mammals [21,22,23, 28]. In addition, the members of Firmicutes differed between laboratory Suncus and wild Suncus, with Streptococcus being the major genus in the former but not the latter. A recent study that compared fecal microbiotas between wild and captive animals from 41 mammalian species also found a higher abundance of Streptococcus spp. in captive animals [28]. Considering this evidence, our results imply that the gut microbiota of Suncus has changed with domestication. Furthermore, there was differentiation between the large and small intestines in laboratory Suncus, unlike in wild Suncus (Table 2). Moreover, the reduction in gut microbial diversity in the phylum Proteobacteria was remarkable (see Supplementary Table 1); more than 10 genera were found in wild Suncus and only 2 genera in laboratory Suncus. Although we do not have a good explanation for this, it is notable that putative pathogenic bacteria (e.g., Salmonella, Plesiomonas, Kluyvera, and Morganella) were found only in wild Suncus. It is possible that these putative pathogenic bacteria are exogenous, and not autochthonous. An alternative hypothesis is elimination. Because these putative pathogens might be an obstacle to domestication as experimental animals, it is possible that they were eliminated without being noticed by keeping them in stable and clean environment (see for trial history of breeding environment [32, 33]). Further study of captive Suncus from Okinawa will provide more insight into these hypotheses.
Supplementary Material
Acknowledgments
We thank Dr. Tetsuo Morita and Dr. Shinsuke H. Sakamoto for providing the laboratory Suncus animals and for their valuable advice pertaining to this study. We also thank Dr. Atsushi Nakamoto for providing information regarding the field collection site of wild Suncus. We thank Dr. Toshihiro Takahashi and Dr. Seiji Ieiri for their valuable comments on our project. This study was supported by a JSPS Grant-in-Aid for Scientific Research (C) (KAKENHI 15K07191).
References
- 1.Bininda-Emonds O.R.P., Cardillo M., Jones K.E., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A.2007. The delayed rise of present-day mammals. Nature 446: 507–512. doi: 10.1038/nature05634 [DOI] [PubMed] [Google Scholar]
- 2.Blaser M.J., Dominguez-Bello M.G.2016. The human microbiome before birth. Cell Host Microbe 20: 558–560. doi: 10.1016/j.chom.2016.10.014 [DOI] [PubMed] [Google Scholar]
- 3.Burokas A., Moloney R.D., Dinan T.G., Cryan J.F.2015. Microbiota regulation of the Mammalian gut-brain axis. Adv. Appl. Microbiol. 91: 1–62. doi: 10.1016/bs.aambs.2015.02.001 [DOI] [PubMed] [Google Scholar]
- 4.Chao A., Lee S.M.1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87: 210–217. doi: 10.1080/01621459.1992.10475194 [DOI] [Google Scholar]
- 5.Delsuc F., Metcalf J.L., Wegener Parfrey L., Song S.J., González A., Knight R.2014. Convergence of gut microbiomes in myrmecophagous mammals. Mol. Ecol. 23: 1301–1317. doi: 10.1111/mec.12501 [DOI] [PubMed] [Google Scholar]
- 6.Dierenfeld E.S., Hintz H.F., Robertson J.B., Van Soest P.J., Oftedal O.T.1982. Utilization of bamboo by the giant panda. J. Nutr. 112: 636–641. doi: 10.1093/jn/112.4.636 [DOI] [PubMed] [Google Scholar]
- 7.Ebukuro S., Wakana S., Hioki K., Nomura T.2000. Selective breeding of house musk shrew (Suncus murinus) lines in relation to emesis induced by veratrine sulfate. Comp. Med. 50: 281–283. [PubMed] [Google Scholar]
- 8.Evans J., Sheneman L., Foster J.2006. Relaxed neighbor joining: a fast distance-based phylogenetic tree construction method. J. Mol. Evol. 62: 785–792. doi: 10.1007/s00239-005-0176-2 [DOI] [PubMed] [Google Scholar]
- 9.Endo A., Futagawa-Endo Y., Dicks L.M.T.2010. Diversity of Lactobacillus and Bifidobacterium in feces of herbivores, omnivores and carnivores. Anaerobe 16: 590–596. doi: 10.1016/j.anaerobe.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 10.Foley N.M., Springer M.S., Teeling E.C.2016. Mammal madness: is the mammal tree of life not yet resolved? Philos. Trans. R. Soc. Lond. B Biol. Sci. 371: 20150140. doi: 10.1098/rstb.2015.0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox J.2005. The R Commander: A basic statistics graphical user interface to R. J. Stat. Softw. 14: 1–42. doi: 10.18637/jss.v014.i09 [DOI] [Google Scholar]
- 12.Good I.J.1953. The population frequencies of species and the estimation of population parameters. Biometrika 40: 237–264. doi: 10.1093/biomet/40.3-4.237 [DOI] [Google Scholar]
- 13.Haas B.J., Gevers D., Earl A.M., Feldgarden M., Ward D.V., Giannoukos G., Ciulla D., Tabbaa D., Highlander S.K., Sodergren E., Methé B., DeSantis T.Z., Petrosino J.F., Knight R., Birren B.W., Human Microbiome Consortium2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21: 494–504. doi: 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanamura H., Mineda T., Kurohmaru M., Kitoh J., Miyaki T., Isomura G., Yamashita K.1985. Digestive Organs. pp. 210–228. In: Suncus murinus: Biology of the Laboratory Shrew (Oda, S., Kitoh, J., Ota, K., and Isomura, G. eds.), Japan Scientific Societies Press, Tokyo (in Japanese). [Google Scholar]
- 15.Hooper L.V., Midtvedt T., Gordon J.I.2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22: 283–307. doi: 10.1146/annurev.nutr.22.011602.092259 [DOI] [PubMed] [Google Scholar]
- 16.Hu Y., Wu Q., Ma S., Ma T., Shan L., Wang X., Nie Y., Ning Z., Yan L., Xiu Y., Wei F.2017. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad. Sci. USA 114: 1081–1086. doi: 10.1073/pnas.1613870114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutterer R.2005. Order Soricomorpha. pp. 220–311. In: Mammal Species of the World: a taxonomic and geographic reference (Wilson, D.E. and Reeder, D.M. eds.). 3rd ed. Johns Hopkins University Press, Baltimore, Maryland. [Google Scholar]
- 18.Iseki R.1985. Strains and marker genes. pp. 117–120. In: Suncus murinus: Biology of the Laboratory Shrew (Oda, S., Kitoh, J., Ota, K. and Isomura, G. eds.), Japan Scientific Societies Press, Tokyo (in Japanese). [Google Scholar]
- 19.Jandhyala S.M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., Nageshwar Reddy D.2015. Role of the normal gut microbiota. World J. Gastroenterol. 21: 8787–8803. doi: 10.3748/wjg.v21.i29.8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jogahara T., Koyasu K., Oda S., Kawai T., Hanamura H.2007. Quest for the cause of oligodontia in Suncus murinus (Soricomorpha, Soricidae): Morphological re-examination. Arch. Oral Biol. 52: 836–843. doi: 10.1016/j.archoralbio.2007.01.011 [DOI] [PubMed] [Google Scholar]
- 21.Kohl K.D., Dearing M.D.2014. Wild-caught rodents retain a majority of their natural gut microbiota upon entrance into captivity. Environ. Microbiol. Rep. 6: 191–195. doi: 10.1111/1758-2229.12118 [DOI] [PubMed] [Google Scholar]
- 22.Kohl K.D., Skopec M.M., Dearing M.D.2014. Captivity results in disparate loss of gut microbial diversity in closely related hosts. Conserv. Physiol. 2: cou009. doi: 10.1093/conphys/cou009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong F., Zhao J., Han S., Zeng B., Yang J., Si X., Yang B., Yang M., Xu H., Li Y.2014. Characterization of the gut microbiota in the red panda (Ailurus fulgens). PLoS One 9: e87885. doi: 10.1371/journal.pone.0087885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreisinger J., Cížková D., Vohánka J., Piálek J.2014. Gastrointestinal microbiota of wild and inbred individuals of two house mouse subspecies assessed using high-throughput parallel pyrosequencing. Mol. Ecol. 23: 5048–5060. doi: 10.1111/mec.12909 [DOI] [PubMed] [Google Scholar]
- 25.Kurohmaru M.1985. Characteristics of the gastrointestinal tract of the house musk shrew, Suncus murinus pp. 389–396. In: Suncus murinus: Biology of the Laboratory Shrew (Oda, S., Kitoh, J., Ota, K. and Isomura, G. eds.), Japan Scientific Societies Press, Tokyo (in Japanese with English summary). [Google Scholar]
- 26.Ley R.E., Hamady M., Lozupone C., Turnbaugh P.J., Ramey R.R., Bircher J.S., Schlegel M.L., Tucker T.A., Schrenzel M.D., Knight R., Gordon J.I.2008. Evolution of mammals and their gut microbes. Science 320: 1647–1651. doi: 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ley R.E., Lozupone C.A., Hamady M., Knight R., Gordon J.I.2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6: 776–788. doi: 10.1038/nrmicro1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenzie V.J., Song S.J., Delsuc F., Prest T.L., Oliverio A.M., Korpita T.M., Alexiev A., Amato K.R., Metcalf J.L., Kowalewski M., Avenant N.L., Link A., Di Fiore A., Seguin-Orlando A., Feh C., Orlando L., Mendelson J.R., Sanders J., Knight R.2017. The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 57: 690–704. doi: 10.1093/icb/icx090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motokawa M.2015. Suncus murinus (Linnaeus, 1766). pp. 26–27. In: the Wild Mammals of Japan, Second edition (Ohdachi, S.D., Ishibashi, Y., Iwasa, M., Fukui, D., and Saitoh, T. eds.), Shouladoh, Kyoto, Japan. [Google Scholar]
- 30.Muegge B.D., Kuczynski J., Knights D., Clemente J.C., González A., Fontana L., Henrissat B., Knight R., Gordon J.I.2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974. doi: 10.1126/science.1198719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen T.L., Vieira-Silva S., Liston A., Raes J.2015. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 8: 1–16. doi: 10.1242/dmm.017400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oda S.1985. Management, keeping, and breeding. pp.103–111. In: Suncus murinus: Biology of the Laboratory Shrew (Oda, S., Kitoh, J., Ota, K., and Isomura, G. eds.), Japan Scientific Societies Press, Tokyo (in Japanese with English summary). [Google Scholar]
- 33.Oda S.2011. Getting, keeping, and breeding. pp. 69–72. In: Biology of Suncus (Oda, S., Tohya, K., and Miyaki, T. eds.), Japan Scientific Societies Press, Tokyo (in Japanese). [Google Scholar]
- 34.Oda S., Jogahara T.2011. Strains/lines derived from local populations and mutations. pp. 78–83. In: Biology of Suncus (Oda, S., Tohya, K., and Miyaki, T. eds.), Japan Scientific Societies Press, Tokyo (in Japanese). [Google Scholar]
- 35.Oda S., Kondo K.1976. Progress in domestication of Suncus murinus riukiuanus for experiments. Honyurui Kagaku 16: 13–30(Mammalian Science). [Google Scholar]
- 36.Oda S., Kondo K.1977. [Usefulness of wild insectivores as laboratory animals (author’s transl)]. Jikken Dobutsu 26: 273–280(in Japanese). [DOI] [PubMed] [Google Scholar]
- 37.O’Hara A.M., Shanahan F.2006. The gut flora as a forgotten organ. EMBO Rep. 7: 688–693. doi: 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pantoja-Feliciano I.G., Clemente J.C., Costello E.K., Perez M.E., Blaser M.J., Knight R., Dominguez-Bello M.G.2013. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 7: 1112–1115. doi: 10.1038/ismej.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pessione E.2012. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front. Cell. Infect. Microbiol. 2: 86. doi: 10.3389/fcimb.2012.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruesse E., Peplies J., Glöckner F.O.2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. doi: 10.1093/bioinformatics/bts252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R core team. 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL; http://www.R-project.org.
- 42.Sakahara S., Xie Z., Koike K., Hoshino S., Sakata I., Oda S., Takahashi T., Sakai T.2010. Physiological characteristics of gastric contractions and circadian gastric motility in the free-moving conscious house musk shrew (Suncus murinus). Am. J. Physiol. Regul. Integr. Comp. Physiol. 299: R1106–R1113. doi: 10.1152/ajpregu.00278.2010 [DOI] [PubMed] [Google Scholar]
- 43.Savage D.C.1986. Gastrointestinal microflora in mammalian nutrition. Annu. Rev. Nutr. 6: 155–178. doi: 10.1146/annurev.nu.06.070186.001103 [DOI] [PubMed] [Google Scholar]
- 44.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F.2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75: 7537–7541. doi: 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon C.E., 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27: 379–423, 623–656. doi: 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- 46.Shinohara A., Uchida E., Shichijo H., Sakamoto H.S., Morita T., Koshimoto C.2016. Microbial diversity in forestomach and caecum contents of the greater long-tailed hamster Tscherskia triton (Rodentia: Cricetidae). Mamm. Biol. 81: 46–52. doi: 10.1016/j.mambio.2014.10.007 [DOI] [Google Scholar]
- 47.Simpson E.H.1949. Measurement of diversity. Nature 163: 688. doi: 10.1038/163688a0 [DOI] [Google Scholar]
- 48.Stackebrandt E., Ebers J.2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33: 152–155. [Google Scholar]
- 49.Stevens C.E., Hume I.D.1995. Comparative Physiology of the Vertebrate Digestive System, 2nd ed. Cambridge University Press, Cambridge. [Google Scholar]
- 50.Tropini C., Earle K.A., Huang K.C., Sonnenburg J.L.2017. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 21: 433–442. doi: 10.1016/j.chom.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsutsui C., Kajihara K., Yanaka T., Sakata I., Itoh Z., Oda S., Sakai T.2009. House musk shrew (Suncus murinus, order: Insectivora) as a new model animal for motilin study. Peptides 30: 318–329. doi: 10.1016/j.peptides.2008.10.006 [DOI] [PubMed] [Google Scholar]
- 52.Tsutsui C., Ishida Y., Sakahara S., Sakai T.2011. Identification of motilin and ghrelin in the house musk shrew (Suncus murinus) cDNA: cloning and tissue distribution. pp. 335–341. In: Biology of Suncus (Oda, S., Tohya, K., and Miyaki, T. eds.), Japan Scientific Societies Press, Tokyo (in Japanese with English summary). [Google Scholar]
- 53.Uenishi G., Fujita S., Ohashi G., Kato A., Yamauchi S., Matsuzawa T., Ushida K.2007. Molecular analyses of the intestinal microbiota of chimpanzees in the wild and in captivity. Am. J. Primatol. 69: 367–376. doi: 10.1002/ajp.20351 [DOI] [PubMed] [Google Scholar]
- 54.Ueno S., Matsuki N., Saito H.1987. Suncus murinus: a new experimental model in emesis research. Life Sci. 41: 513–518. doi: 10.1016/0024-3205(87)90229-3 [DOI] [PubMed] [Google Scholar]
- 55.Wei F., Feng Z., Wang Z., Zhou A., Hu J.1999. Use of the nutrients in bamboo by the red panda (Ailurus fulgens). J. Zool. (Lond.) 248: 535–541. doi: 10.1111/j.1469-7998.1999.tb01053.x [DOI] [Google Scholar]
- 56.Xue Z., Zhang W., Wang L., Hou R., Zhang M., Fei L., Zhang X., Huang H., Bridgewater L.C., Jiang Y., Jiang C., Zhao L., Pang X., Zhang Z.2015. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. MBio 6: e00022–e15. doi: 10.1128/mBio.00022-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamanaka A., Yasui K., Sonomura T., Uemura M.2007. Development of heterodont dentition in house shrew (Suncus murinus). Eur. J. Oral Sci. 115: 433–440. doi: 10.1111/j.1600-0722.2007.00499.x [DOI] [PubMed] [Google Scholar]
- 58.Yamanaka A., Hanamura H., Moriguchi K., Shuang-Qin Y.I., Takeuchi K., Somazawa F., Isomura G., Kiso Y., Miyaki T.2011. Digestive organs. pp. 107–136. In: Biology of Suncus (Oda, S., Tohya, K. and Miyaki, T. eds.), Japan Scientific Societies Press, Tokyo (in Japanese). [Google Scholar]
- 59.Yue J.C., Clayton M.K.2005. A similarity measure based on species proportions. Commun. Stat. Theory Methods 34: 2123–2131. doi: 10.1080/STA-200066418 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


