Abstract
Invasive rodents impact biodiversity, human health and food security worldwide. The biodiversity impacts are particularly significant on islands, which are the primary sites of vertebrate extinctions and where we are reaching the limits of current control technologies. Gene drives may represent an effective approach to this challenge, but knowledge gaps remain in a number of areas. This paper is focused on what is currently known about natural and developing synthetic gene drive systems in mice, some key areas where key knowledge gaps exist, findings in a variety of disciplines relevant to those gaps and a brief consideration of how engagement at the regulatory, stakeholder and community levels can accompany and contribute to this effort. Our primary species focus is the house mouse, Mus musculus, as a genetic model system that is also an important invasive pest. Our primary application focus is the development of gene drive systems intended to reduce reproduction and potentially eliminate invasive rodents from islands. Gene drive technologies in rodents have the potential to produce significant benefits for biodiversity conservation, human health and food security. A broad-based, multidisciplinary approach is necessary to assess this potential in a transparent, effective and responsible manner.
Keywords: rodent, mice, rat, biodiversity, gene drive, island
1. Introduction
Rodents are common and usually unwelcome ‘fellow travellers’ of humans across the world, often making them important pests. The key invasive rodent pests include house mice (Mus musculus) and three species of rats (Rattus norvegicus, R. rattus and R. exulans [1]). Their impacts are varied, and include pre- and post-harvest agricultural losses, zoonotic disease threats and biodiversity impacts. Rodents cause 5–15% losses each year to agricultural production [2–6] including enough rice in South Asia to feed an estimated 180 million people per year [3] and tens of millions of dollars in losses during ‘mouse plagues’ in Australia [7,8]. Rodents are significant vectors for a variety of diseases including leptospirosis and Lyme disease [9,10]. Lastly and as the focus of this paper, rodents are key threats to biodiversity, especially on islands [11–14]. This review focuses on knowledge gaps for gene drives that could potentially address the threat invasive rodents pose to biodiversity, particularly in island ecosystems where they are introduced and non-native, and where the success of existing technologies (primarily rodenticides) is limited [15]. Functional gene drives are not yet available for rodents, but efforts to develop them are active, and the larger genome editing field is moving rapidly, so assessing knowledge needs before a rodent gene drive could be tested in an island situation is useful.
Islands are biodiversity hotspots with approximately 5% of Earth's land area, but 20% of species including 37% of critically endangered and 61% of extinct species [16]. Invasive mammalian predators are key agents in these extinctions [17,18]. Records are sparser for invertebrate and plant losses on islands, but these are thought to be significant as well [19,20]. Indirect effects can also be important, as documented by high seabird densities increasing coral reef productivity on rat-free islands and rat removal also eliminating the disease vector mosquito Aedes albopictus [21,22]. Several features contribute to the vulnerability of island faunas to invasive mammalian predators [23]. These include their limited geographic distributions on most islands and often consequently small population sizes, as well as lack of or evolutionary loss of anti-predator defences (e.g. flightlessness in some birds). Because the effects of invasive mammalian predators on island ecosystems are often significant and pervasive, removal of these predators has proven a very effective conservation measure. Predator removal on 181 islands worldwide produced conservation benefits for approximately 236 terrestrial insular species [24].
Eradication of invasive rodents on islands larger than approximately five hectares has primarily relied on the aerial broadcast of anticoagulant rodenticide compounds formulated into pelleted baits [1]. We emphasize that this approach has been quite successful and generated impressive conservation benefits (see [13,15,24–26]), but it also has significant drawbacks and limitations. These include high fixed costs and a relatively inflexible financial model, lack of species-specificity leading to potential non-target species impacts, toxicant persistence, potential failure because toxicants must be consumed by essentially every female and negative public perceptions due to the mode of action. Additionally, inhabited islands are very challenging because of potential impacts to humans and domestic animals and approximately 50% of endangered terrestrial vertebrates are on inhabited islands [27].
These challenges for rodenticide use have prompted a search for alternatives. This has coincided with increased attention to potential genetic pest control approaches generally following a seminal paper by Burt in 2003 [28] exploring this possibility. Subsequently, other authors have examined the potential application of engineered drives for rodents [29–33]. This paper reviews some of these contributions with a focus on knowledge gaps for gene drive approaches in the island context. The authors are part of an interdisciplinary consortium effort called ‘Genetic Biocontrol of Invasive Rodents' or GBIRd (www.geneticbiocontrol.org). This review takes this broad perspective for knowledge gaps and some promising areas genome editing capabilities are rapidly developing. The sections below roughly correspond with levels of biological organization, extending briefly beyond this framework at the end to discuss the potential application of this technology in a responsible and ethical way.
2. Naturally occurring and synthetic selfish genetic mechanisms for gene drives
Various potential gene drive mechanisms have been suggested for rodents ranging from naturally occurring selfish genetic elements to synthetic CRISPR-based systems [31,32,34,35]. We approach this at the molecular/genetic level, the stage of technological development of these systems, potential variations that could be useful and efforts aimed at providing spatial restriction of gene drive function. Efforts to date have focused on house mice as a tractable genetic model, but some systems could potentially also be used in other invasive mammalian predators (e.g. [36]).
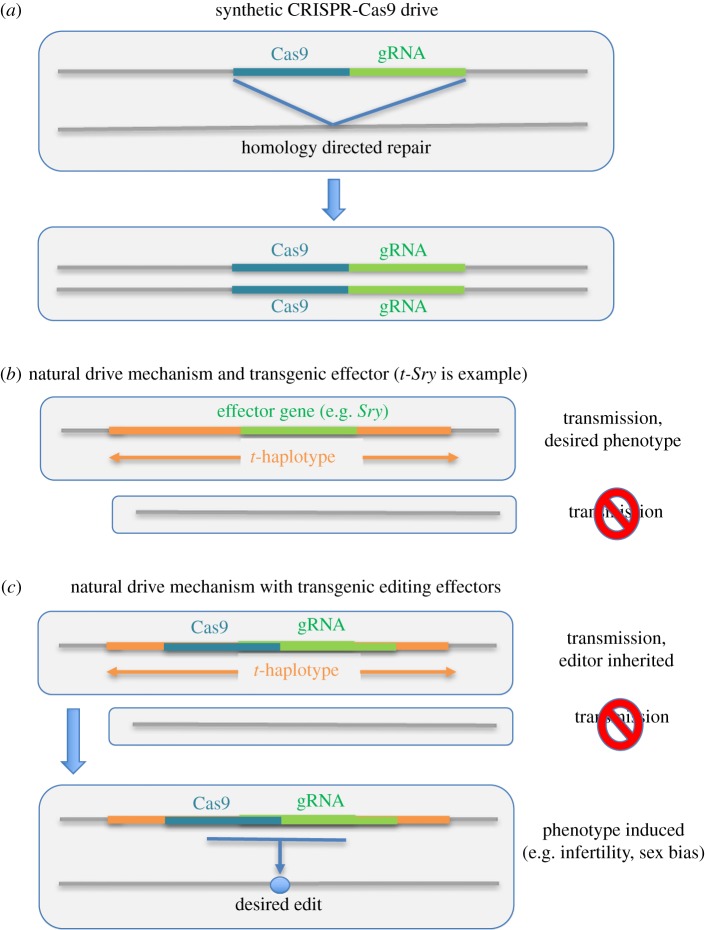
The first drive system considered in rodents was the t-haplotype (also termed the t-allele or t-complex), a well-characterized system in mice first described in 1927 and intensively investigated since [37–39]. Males carrying two copies of the t-haplotype are either non-viable (embryonic lethal) or sterile, but heterozygous males (+/t) are viable and the t-haplotype is inherited in crosses at rates ranging up to 95% or even higher (Mendelian expectation is 50%) [38,40–42]. Initial efforts have focused on inserting the male sex determination gene Sry into the t-haplotype (figure 1b; explained in detail in [35]; see also [15,26,47] and particularly [48]) as Sry is necessary and sufficient to induce male sex determination in mammals [49,50]. Homozygotes for the tw2 variant are sterile, but 94–96% of offspring in crosses between heterozygous males and both laboratory and wild-derived females inherit the t-haplotype [48,51]. Insertion of Sry should result in the production of both fertile (XY-t-Sry) and sterile (XX-t-Sry) males. t-Sry mice have not yet been produced, but this should be feasible [48] and the system may also be useful for carrying other effector genes (see below).
Figure 1.
Gene drive designs incorporating either synthetic or naturally occurring drive mechanisms. (a) A ‘standard’ CRISPR-based gene drive that relies on homing and HDR [43–46]. (b) The t-Sry approach in which spread depends on the naturally occurring t-haplotype system and a transgenic insertion of the masculinizing Sry gene [35,47]. Sperm that do not carry a t-haplotype are compromised in function and fertilization occurs with sperm carrying the t-haplotype (termed transmission ratio distortion). (c) A system that would spread through a natural drive mechanism (e.g. the t-haplotype) but incorporate CRISPR system effectors to produce genome edits and desired phenotypes. (Online version in colour.)
Synthetic drive systems are under development in mice but are not robustly functional in any mammal to date with some results suggesting this may be more challenging in mammals than insects [52]. Some synthetic gene drives rely on high rates of homology-directed repair (HDR) to effectively convert the heterozygous parental germline to homozygous form for the gene drive construct (figure 1a). HDR uses the introduced transgenic cassette as a template to repair the double-stranded break (DSB) created on the homologous chromosome. The gene drive process can fail if the endonuclease fails to cut at the intended recognition site or if the DSB is repaired by end joining. Grunwald et al. [52] assessed these functions using a system similar to a functional gene drive and found cutting by Cas9 (generation of DSBs) was efficient, but repair occurred by end joining rather than HDR. Restricting Cas9 activity to the germline produced some evidence of effective HDR, but only in females for reasons that are not clear. Appropriately timing Cas9 expression for effective HDR may be more difficult than in insect systems [43–46], which is perhaps unsurprising given the evolutionary divergence between these groups.
The major knowledge gap for synthetic drives in mammals is clearly the lack of a functional and efficient design. Assuming this hurdle can be overcome, it will be critical to characterize the system in terms of editing rates, relative rates of HDR versus end joining and fitness of end joining mutants at the target site as well as off-target activity of editors. The proportions of HDR versus end joining are critical because end joining both represents a failure of drive and generates mutant haplotypes likely to be resistant to further editing since the recognition sequence is changed. Use of multiple gRNAs, as first suggested by Burt [28], has been modelled in mammals [36] and is discussed further below. Other potentially useful endonucleases include Cas12a (Cpf1) and both naturally occurring and engineered variants of the Cas endonucleases (e.g. [53,54]).
Because a key failure mechanism of gene drives is the lack of effective HDR, mechanisms that do not require homing may be more attractive. One alternative would use a natural drive system such as the t-haplotype. For example, other effector genes could be introduced to the t-haplotype to induce a desired phenotype (e.g. infertility). The t-haplotype system could also be the drive mechanism with the insertion of an endonuclease and gRNAs targeting genes necessary for female development or fertility (figure 1c). Such a system would rely on an evolutionarily persistent meiotic drive mechanism (e.g. the t-haplotype in mice for an estimated 2.9 Myr [55]) and therefore probably be less prone to resistance development than the synthetic mechanisms developed to date. Second, modelling suggests that at least the t-Sry system would probably require multiple introductions in order to effectively suppress an invasive mouse population [47,56]. While this could reduce effectiveness and increase cost, concerns have been expressed about the uncontrolled spread of drive carriers [57,58]. So, lower drive effectiveness and invasiveness could be desirable. Of note, t-haplotype frequencies are typically lower in house mouse populations than predicted from transmission ratio distortion rates alone [59–61]. Incorporating an endonuclease and gRNAs should also make it possible to target this sort of hybrid natural/engineered drive system to only a population of interest as explained below. Importantly, the t-haplotype is known only from mice, but other selfish genetic elements occur in mice [62] and are widespread in nature (multiple papers in this issue).
Other non-homing drive mechanisms have been proposed, but not yet tested in mammals. These include Y-linked editors, X-shredder systems and the ‘cleave and rescue’ mechanism [63–65]. The X-shredder in mosquitoes would rely on editors inserted on the Y-chromosome that target repetitive sequences on the X chromosome, thereby ‘shredding’ it. Y-shredder mechanisms have been demonstrated in vitro and in vivo for mice [66,67] and modelled for natural populations [68], but an X-shredder would instead prevent female production and therefore be potentially more effective for reducing invasive mouse populations.
3. Population genetics and spatial limitation of drive function
The sequence specificity of CRISPR-based editing renders synthetic drive designs vulnerable to the evolution of resistance, but this sequence specificity could also be used to achieve spatial limitation of drive function by targeting locally fixed alleles (LFA) [69]. Island populations are often derived from few initial colonizers and genetic diversity is therefore typically reduced relative to mainland populations (e.g. [70]). This reduced diversity means there may be drive recognition sequences that are fixed in a target population, but absent in nearby non-target populations. Preliminary investigations do show these suitable LFAs are abundant in invasive mouse populations on islands [71]. An appealing feature of the LFA approach is that it relies on basic principles of population genetics (i.e. founder effects) to target an effector to a specific and localized population. This approach could be useful for both synthetic drive mechanisms that require HDR and those that do not.
4. Behavioural and ecological considerations for gene drives in rodents
It will perhaps be surprising to readers that some key gaps regarding the implementation of any gene drive for rodents are related to basic aspects of ecology, behaviour and genetics in island ecosystems. The spread of house mice and rats into islands worldwide is due largely to their impressive adaptability. This adaptability and the resultant systematic differences between island and related mainland populations in a range of characteristics including morphology, demography, behaviour and reproductive biology were termed the ‘island syndrome’ [72]. House mice introduced to the Faroe Islands first received attention for this type of local morphological adaptation from Darwin and later Huxley [73]. Extreme adaptation to non-commensal habitats is seen on islands like Gough Island, where the invasive mice reach very large sizes [74,75]. There are also behavioural adaptations on islands, examples of which include novel predatory behaviour on sea birds termed ‘scalping’, reduced aggression and reduced dispersal [72,76].
A particularly important knowledge gap for gene drive implementation for rodents concerns what Moro et al. termed ‘translocation biology’ [33]. As part of a knowledge gap analysis for Australia, these authors defined translocation biology as the factors influencing the survival and success of individuals newly transplanted into an established invasive population. This is likely to be especially important with gene drive carriers and models highlight the importance of drive carrier fitness relative to wild-type individuals (e.g. [47]). The available knowledge of translocation biology was assessed as ‘minimal’ for house mice and as ‘lacking’ for rats.
Evidence is mixed regarding the invasibility of established invasive house mouse populations. Some findings suggest established invasive mouse populations are relatively resistant to introgression by later-arriving animals with a study by Hardouin et al. in the Kerguelen Archipelago [77]. An experimental finding of sorts from an invasive rat population is consistent with an established invasive rodent population limiting the introgression of later-arriving individuals. Russell et al. [78] found that ship rats and Norway rats on Pearl Island were genetically distinct from nearby New Zealand mainland populations, suggesting a lack of connectivity. However, rapid reinvasion following an eradication suggested rats swam to the island more frequently than the genetic patterns had suggested [79].
Contrasting results from other studies suggest invasive mouse populations on islands are permeable to newly invading individuals, although likely more so to males than females. The population genetic patterns for mice on Madeira indicate a northern European origin for maternally inherited mtDNA, but a Portuguese origin for nuclear DNA, suggesting an early colonization (perhaps by Vikings) and later secondary invasion as connections with Portugal developed [80,81]. Consistent with this finding, a Y-chromosome linked genetic marker in mice experimentally introduced to the Isle of May spread rapidly through the established population while a matrilineally spread marker did not [82,83]. This result suggested that males may have a greater ability to invade a new population than females. A follow-up behavioural study found that Isle of May mice were strikingly less aggressive than mice from a mainland, commensal population [84]. A sex difference in invasion ability is supported by other genetic data for at least some populations. Jones & Searle [85] found evidence of a greater number of Y-chromosome introductions than mitochondrial DNA introductions in island contexts and inferred that introgression into established populations was more difficult for females than for males, suggesting this was due to behavioural differences. This could be an important consideration for gene drive approaches as the introduced carriers would be males for a male-biasing drive (e.g. t-Sry), but females for a female-biasing drive (e.g. [68]).
The relative paucity of studies and conflicting evidence regarding the permeability of established populations suggest this should be an important focus if an effective gene drive is developed. As a drive would likely be developed in laboratory mice, determining the degree of backcrossing necessary to ensure the competitiveness of introduced individuals would also be needed. Serr et al. found that F1 hybrid laboratory/wild-derived strain males were surprisingly successful in competitive mating trials with purely wild-derived island males [86]. However, trials in larger and complex naturalistic (but highly biosecure) environments will be needed. Simulating island conditions while also maintaining high biosecurity represents a challenge and another important area of development (A. B. Shiels et al., unpublished). We described the impressive degree of behavioural adaptation rodents show in island ecosystems at the beginning of this section. Such behavioural adaptations could represent mechanisms of resistance to gene drive spread as well in the form of inbreeding, mate choice and patterns of multiple mating (see [68] for polyandry and [56] for polygyny).
5. Mathematical modelling
Modelling of gene drives in rodents is more limited than for other systems, but there has been important progress. Modelling a t-Sry drive in mice defined a predicted range of conditions where this approach could effectively suppress an island-invasive mouse population [47]. The relative fitness of drive carriers strongly influenced whether a single introduction or instead multiple, repeated introductions would be necessary to achieve eradication. Because the introduction of drive carriers could transiently increase the overall mouse numbers, there is likely to be a tradeoff between speed of eradication and potential negative ecological consequences. Prowse et al. modelled synthetic drives for mice, rats and rabbits [36] and found that embryonic non-viability and female sterility mechanisms would be more effective than a sex-biasing strategy. Consistent with other studies, incorporating multiple gRNAs was critical to prevent the evolution of resistance in these simulations. This group also modelled mouse eradication using a Y-shredder, female-biasing approach [68]. Drive performance variables including homing rates and efficiency of Y-shredding were critical, but the likelihood of eradication was also crucially dependent on the number of mates per male. These simulations indicated that this could be an effective population suppression mechanism even if males mated with up to five females.
A separate effort modelling spatial limitation of drive function through targeting LFA [69] indicates that even in what might be considered a ‘worst-case scenario’ where the target allele is present at 99% frequency in non-target populations, the presence of resistance alleles leads to an ‘escaped drive’ producing only limited and transient population suppression in these non-target populations (figure 2). The drive-resistant alleles have a significant fitness advantage in this situation and this development of resistance is the key feature usually predicted and/or observed to render a drive ineffective [36,87].
Figure 2.
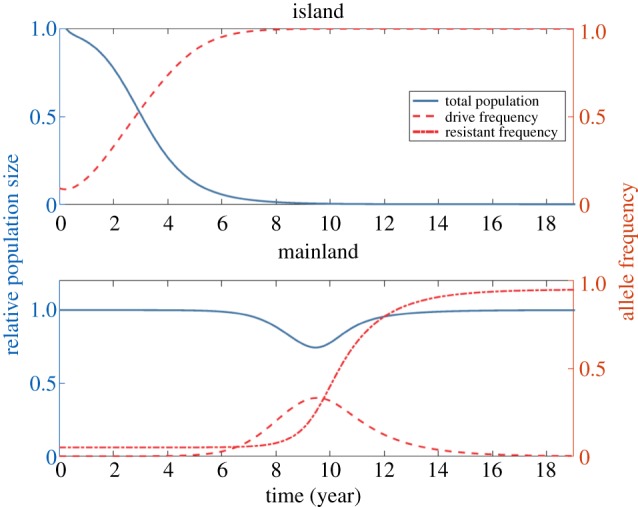
Island and mainland population dynamics and genetics for a drive targeting a locally fixed allele on an island (figure redrawn from Sudweeks et al. [69]). This scenario models a drive with no invasion threshold, meaning there is no minimum frequency that a drive must reach in order to spread. Blue curves and axes denote population sizes, measured relative to pre-release equilibria, while red curves and axes denote allele frequencies. A small release (five homozygous drive individuals) occurs at time t = 0. Resistance is assumed to be very low on the mainland (allele frequency of just 5%)—a quite pessimistic ‘worst-case’ scenario in terms of the susceptibility of the mainland population to the drive. The drive spreads to fixation and suppresses the island population. Migration to the mainland (on average, one island individual travels to the mainland a month) means that the drive is introduced to the mainland, where it can spread through the susceptible population but not the resistant population. The total population undergoes a temporary suppression as the drive spreads through the susceptible population. The frequency of resistant alleles increases as a result of drive, and density-dependent population regulation returns the mainland population to the pre-release equilibrium level. (Online version in colour.)
The knowledge gaps for gene drive modelling in rodents remain numerous and only major ones will be addressed below. Immigration to or escape from an island is a stochastic event and stochastic modelling approaches are needed to address the LFA strategies above. Additionally, incorporating the ecological and social system variation known from invasive mouse populations will be useful and these data will likely be important to obtain for a given target population before any field trial. For example, we know many invasive mouse populations are highly cyclical (e.g. [20,88,89]), but the impact of such population variation on gene drive spread has not yet been modelled. Also, mice typically show social structures and space use characterized as reproductive demes and territories that can be defended aggressively [90]. This subdivision of populations and complex reproductive interactions could impact gene drive spread significantly but has also not yet been addressed in models.
Finally, two other factors that are particularly relevant to gene drive use for mice and likely other mammals need to be explored. The first of these is the common and reasonable assumption that gene drive carriers will have lower relative fitness than non-drive carriers in the target population. As noted above, Serr et al. [86] found that hybrid males of mixed lab/wild origin may instead have fitness advantages relative to wild males, and males from commensal origins could have advantages over island males generally based on comparisons to mice from the Isle of May [83,84]. Before any field test of a gene drive mouse, it will be necessary to decide what the genetic background of the gene drive carriers should be—that of the invasive population of the island or potentially a background that would confer a competitive advantage on gene drive carriers relative to the target population. The very limited results available so far suggest mice from commensal habitat backgrounds may be more competitive than mice from invasive island populations [83,84], but this needs more investigation and seems likely to vary depending on the specific island population considered. Additionally, any competitive advantage based on a different genetic background may need to be balanced against the potential negative consequences of introducing new alleles to island populations that show reduced genetic diversity [70] and potentially enhanced invasiveness elsewhere should gene drive carriers escape the target island. Runge & Lindholm [91] found that t-haplotype carriers showed increased migratory propensity within and from their naturalistic barn colony.
A second factor that could be important are the consequences of a changing sex ratio on space use and behavioural interactions. As this is a new and particularly applied question specific to the gene drive context, it is unsurprising it has not apparently been addressed for mice or other mammals to our knowledge. However, as animal movement and dispersal generally are strongly affected by mating opportunities, it is reasonable to suggest that these behaviours would change with shifts in both population density and sex ratios, and in ways that could affect gene drive spread.
The behavioural and ecological characteristics of invasive mouse populations would also likely be impacted by conventional control measures (e.g. rodenticides). From a strongly applied perspective, it would be valuable to better understand how conventional and genetic approaches could potentially be combined to increase the chances for eradication success.
6. Regulatory, stakeholder and community engagement
Although the focus of this review is primarily on biological questions, we close with a brief summary of key issues and needs in the engagement at the regulatory, stakeholder and community levels. We do so for three primary reasons. First, regulatory frameworks that govern biotechnology have not kept pace with the rapid pace of technological development, and debates about how to effectively govern these technologies persist [92–94]. Second, in part because of regulatory uncertainty and in part because these technologies are designed to be deployed in shared environments with no ‘opt out’ potential, informed and deliberative engagement will need to guide testing and potential implementation of these technologies well before their environmental release [28,30,31,95]. Lastly, when deliberative engagement parallels rather than follows the research, feedback from engagement efforts can inform and guide research directions to address specific questions and concerns that can give rise to approaches that are more likely to be both technically feasible and acceptable from regulatory and social perspectives.
As mentioned above, the regulatory systems that govern biotechnology are ill-equipped to effectively manage gene drive organisms. While gene drive organisms will indeed be released at the local scale and should thus be governed accordingly, global governing bodies are also an important scale to set broader norms around their governance [93]. Decisions about how to regulate gene drives at the global scale [94], how to consider field trials [96], and how to meaningfully operate with free, prior and informed consent [97] are all in progress, and are all important for the broader governance of gene drive technologies.
How to balance a normative commitment to democratic decision-making and what may be considered a pressing environmental concern creates the need for deliberative engagement that effectively integrates complexity, scientific uncertainty, as well as explicit attention to human values [95]. In part, deliberative engagement can also help to fill important knowledge gaps: how different communities, stakeholders and public audiences perceive the use of these emerging technologies for wildlife conservation. As we approach gene editing and genetic engineering for species conservation, deep abiding ethical questions have been raised about the fundamental nature of the human relationship to non-human nature. Deliberative engagement can offer insight into how different groups are grappling with these important considerations and potentially offer empirical evidence for broader questions about the social acceptability of using these biotechnologies. Another perennial question in public engagement scholarship also remains: particularly in an uncertain regulatory environment, how might outputs from deliberative engagement meaningfully influence decision-making?
While channelling deliberative engagement outputs into shaping discourse in the policy arena may be more challenging, one suite of solutions emerging out of scholarship that attends to the governance of emerging technologies is identifying ways in which feedback from engagement efforts can shape the innovation process itself. Examples such as Mice Against Ticks [98] or the stakeholder workshop that several authors organized and participated in [99] provide illustrations of how this work is being conducted. Both of the projects mentioned above have developed specific design strategies to engage during the development of the technology. Yet outstanding questions remain, including how to identify the right time in that innovation process to engage, and how to effectively harness that early opportunity to meaningfully shape project outcomes. The Mice Against Ticks project has been exploring the use of open science in collaboration with potentially impacted communities, whereas organizers of the GBIRd engagement efforts have conducted a landscape analysis based on stakeholder interviews [100], participated in multiple workshops with scientists developing and testing the technology, organized a stakeholder workshop to create direct communication between diverse stakeholders and the innovation team [99] and have been experimenting with the use of fictional scenarios to explore underlying values and tradeoffs regarding future island selection for potential field trials. Together, these efforts demonstrate new strategies to incorporate diverse viewpoints and public values into innovation processes that have historically been visible only to narrow groups of scientists and funders.
Supplementary Material
Acknowledgements
The authors wish to thank the organizers of the ‘Evolution of Resistance to Gene Drive’ workshop from which this special issue developed, A. K. Lindholm and T. A. R. Price, and the invitation to participate. Several of the authors received support from the DARPA Safe Genes program under project SAFE-FP-005. The ideas discussed were developed and refined with participation from the broader Genetic Biocontrol of Invasive Rodents consortium and input from interested stakeholders. This is a contribution of the Genetic Engineering and Society Center and W. M. Keck Center for Behavioral Biology at North Carolina State University.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Capizzi D, Bertolino S, Mortelliti A. 2014. Rating the rat: global patterns and research priorities in impacts and management of rodent pests. Mammal Rev. 44, 148–162. ( 10.1111/mam.12019) [DOI] [Google Scholar]
- 2.John A. 2014. Rodent outbreaks and rice pre-harvest losses in Southeast Asia. Food Security 6, 249–260. ( 10.1007/s12571-014-0338-4) [DOI] [Google Scholar]
- 3.Singleton GR.2003. Impacts of rodents on rice production in Asia. Discussion Paper Series No. 45. Los Baños, Philippines: IRRI.
- 4.Singleton GR, Belmain S, Brown PR, Aplin K, Htwe NM. 2010. Impacts of rodent outbreaks on food security in Asia. Wildl. Res. 37, 355–359. ( 10.1071/WR10084) [DOI] [Google Scholar]
- 5.Stenseth N, et al. 2003. Mice, rats, and people: the bio-economics of agricultural rodent pests. Front. Ecol. Environ. 1, 367–375. ( 10.1890/1540-9295(2003)001[0367:MRAPTB]2.0.CO;2) [DOI] [Google Scholar]
- 6.Swanepoel LH, et al. 2017. A systematic review of rodent pest research in Afro-Malagasy small-holder farming systems: are we asking the right questions? PLoS ONE 12, e0174554 ( 10.1371/journal.pone.0174554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown PR, Singleton G. 2000. Impacts of house mice on crops in Australia—costs and damage. See https://digitalcommons.unl.edu/nwrchumanconflicts/6.
- 8.Brown PR, Huth NI, Banks PB, Singleton GR. 2007. Relationship between abundance of rodents and damage to agricultural crops. Agric. Ecosyst. Environ. 120, 405–415. ( 10.1016/j.agee.2006.10.016) [DOI] [Google Scholar]
- 9.Meerburg BG, Singleton GR, Kijlsta A. 2009. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 35, 221–270. ( 10.1080/10408410902989837) [DOI] [PubMed] [Google Scholar]
- 10.Vanasco NB, Sequeira MD, Sequeira G, Tarabla HD. 2003. Associations between leptospiral infection and seropositivity in rodents and environmental characteristics in Argentina. Prev. Vet. Med. 60, 227–235. ( 10.1016/S0167-5877(03)00144-2) [DOI] [PubMed] [Google Scholar]
- 11.Doherty T, Glen A, Nimmo D, Ritchie E, Dickman C. 2016. Invasive predators and global biodiversity loss. Proc. Natl Acad. Sci. USA 113, 11 261–11 265. ( 10.1073/pnas.1602480113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris DB. 2009. Review of negative effects of introduced rodents on small mammals on islands. Biol. Invasions 11, 1611–1630. ( 10.1007/s10530-008-9393-0) [DOI] [Google Scholar]
- 13.Howald G, et al. 2007. Invasive rodent eradication on islands. Conserv. Biol. 21, 1258–1268. ( 10.1111/j.1523-1739.2007.00755.x) [DOI] [PubMed] [Google Scholar]
- 14.Towns D, Atkinson I, Daugherty C. 2006. Have the harmful effects of introduced rats on islands been exaggerated? Biol. Invasions 8, 863–891. ( 10.1007/s10530-005-0421-z) [DOI] [Google Scholar]
- 15.Campbell KJ, et al. 2015. The next generation of rodent eradications: innovative technologies and tools to improve species specificity and increase their feasibility on islands. Biol. Conserv. 185, 47–58. ( 10.1016/j.biocon.2014.10.016) [DOI] [Google Scholar]
- 16.Tershy BR, Shen KW, Newton KM, Holmes ND, Croll DA. 2015. The importance of islands for the protection of biological and linguistic diversity. Bioscience 65, 592–597. ( 10.1093/biosci/biv031) [DOI] [Google Scholar]
- 17.Bellard C, Cassey P, Blackburn TM. 2016. Alien species as a driver of recent extinctions. Biol. Lett. 12, 20150623 ( 10.1098/rsbl.2015.0623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spatz DR, Zilliacus KM, Holmes ND, Butchart SHM, Genovesi P, Ceballos G, Tershy BR, Croll DA. 2017. Globally threatened vertebrates on islands with invasive species. Sci. Adv. 3, e1603080 ( 10.1126/sciadv.1603080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Priddel D, Carlile N, Humphrey M, Fellenberg S, Hiscox D. 2003. Rediscovery of the ‘extinct’ Lord Howe Island stick-insect (Dryococelus australis (Montrouzier)) (Phasmatodea) and recommendations for its conservation. Biodivers. Conserv. 12, 1391–1403. ( 10.1023/A:1023625710011) [DOI] [Google Scholar]
- 20.U.S. Fish and Wildlife Service. 2013. South Farallon islands invasive house mouse eradication project: revised draft environmental impact statement. Fremont, CA: United States Fish and Wildlife Service, San Francisco Bay National Wildlife Refuge Complex; See https://www.fws.gov/uploadedFiles/South_Farallon_Island_%20Invasive_House_Mouse_Eradication_Project_Final%20EIS.pdf. [Google Scholar]
- 21.Graham NAJ, Wilson SK, Carr P, Hoey AS, Jennings S, Macneil MA. 2018. Seabirds enhance coral reef productivity and functioning in the absence of invasive rats. Nature 559, 250–253. ( 10.1038/s41586-018-0202-3) [DOI] [PubMed] [Google Scholar]
- 22.Lafferty KD, et al. 2018. Local extinction of the Asian tiger mosquito (Aedes albopictus) following rat eradication on Palmyra Atoll. Biol. Lett. 14, 20170743 ( 10.1098/rsbl.2017.0743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oppel S, Beaven B, Bolton M, Vickery J, Bodey TW. 2011. Eradication of invasive mammals on islands inhabited by humans and domestic animals. Conserv. Biol. 25, 232–240. ( 10.1111/j.1523-1739.2010.01601.x) [DOI] [PubMed] [Google Scholar]
- 24.Jones HP, et al. 2016. Invasive mammal eradication on islands results in substantial conservation gains. Proc. Natl Acad. Sci. USA 113, 4033–4038. ( 10.1073/pnas.1521179113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell KJ, et al. 2019. A potential new tool for the toolbox: assessing gene drives for eradicating invasive rodent populations. In Island invasives: scaling up to meet the challenge (eds Veitch CR, Clout MN, Martin AR, Russell JC, West CJ), pp. 6–14. Gland, Switzerland: IUCN. [Google Scholar]
- 26.Leitschuh CM, Kanavy D, Backus GA, Valdez RX, Serr M, Pitts EA, Threadgill D, Godwin J. 2018. Developing gene drive technologies to eradicate invasive rodents from islands . J. Responsible Innov. 5, S121–S138. ( 10.1080/23299460.2017.1365232) [DOI] [Google Scholar]
- 27.TIB Partners. 2012. Threatened Island Biodiversity database. Version 2012.1. See http://tib.islandconservation.org/.
- 28.Burt A. 2003. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. R. Soc. Lond. B 270, 921–928. ( 10.1098/rspb.2002.2319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dearden PK, et al. 2017. The potential for the use of gene drives for pestcontrol in New Zealand: a perspective. J. Royal Soc. N. Z. 48, 225–244. ( 10.1080/03036758.2017.1385030) [DOI] [Google Scholar]
- 30.Esvelt KM, Smidler AL, Catteruccia F, Church GM. 2014. Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3, e03401 ( 10.7554/eLife.03401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gould F. 2008. Broadening the application of evolutionarily based genetic pest management. Evolution 62, 500–510. ( 10.1111/j.1558-5646.2007.00298.x) [DOI] [PubMed] [Google Scholar]
- 32.Harvey-Samuel T, Ant T, Alphey L. 2017. Towards the genetic control of invasive species. Biol. Invasions 19, 1683–1703. ( 10.1007/s10530-017-1384-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moro D, Byrne M, Kennedy M, Campbell S, Tizard M. 2017. Identifying knowledge gaps for gene drive research to control invasive animal species: the next CRISPR step. Glob. Ecol. Conserv. 13, e00363 ( 10.1016/j.gecco.2017.e00363) [DOI] [Google Scholar]
- 34.Lindholm AK, et al. 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31, 315–326. ( 10.1016/j.tree.2016.02.001) [DOI] [PubMed] [Google Scholar]
- 35.Piaggio AJ, et al. 2017. Is it time for synthetic biodiversity conservation? Trends Ecol. Evol. 32, 97–107. ( 10.1016/j.tree.2016.10.016) [DOI] [PubMed] [Google Scholar]
- 36.Prowse TA, Cassey P, Ross JV, Pfitzner C, Wittmann TA, Thomas P. 2017. Dodging silver bullets: good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Proc. R. Soc. B 284, 20170799 ( 10.1098/rspb.2017.0799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ardlie KG, Silver LM. 1998. Low frequency of t haplotypes in natural populations of house mice (Mus musculus domesticus). Evolution 52, 1185–1196. ( 10.1111/j.1558-5646.1998.tb01844.x) [DOI] [PubMed] [Google Scholar]
- 38.Lyon MF. 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37, 393–408. ( 10.1146/annurev.genet.37.110801.143030) [DOI] [PubMed] [Google Scholar]
- 39.Silver LM. 1985. Mouse t haplotypes. Annu. Rev. Genet. 19, 179–208. ( 10.1146/annurev.ge.19.120185.001143) [DOI] [PubMed] [Google Scholar]
- 40.Baker AEM. 2008. Mendelian inheritance of t haplotypes in house mouse (Mus musculus domesticus) field populations. Genet. Res. 90, 331–339. ( 10.1017/S0016672308009439) [DOI] [PubMed] [Google Scholar]
- 41.Bauer H, Willert J, Koschorz B, Herrmann BG. 2005. The t complex–encoded GTPase-activating protein Tagap1 acts as a transmission ratio distorter in mice. Nat. Genet. 37, 969–973. ( 10.1038/ng1617) [DOI] [PubMed] [Google Scholar]
- 42.Herrmann BG, Bauer H. 2012. The mouse t-haplotype: a selfish chromosome: genetics, molecular mechanism, and evolution. In Evolution of the house mouse (eds Macholan M, Baird SJE, Munclinger P, Pialek J), pp. 297–314. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 43.Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl Acad. Sci. USA 112, E6736–E6743. ( 10.1073/pnas.1521077112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gantz VM, Bier E. 2015. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444. ( 10.1126/science.aaa5945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammond A, et al. 2015. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83. ( 10.1038/nbt.3439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, Beaghton AK, Nolan T, Crisanti A. 2018. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066. ( 10.1038/nbt.4245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Backus GA, Gross K. 2016. Genetic engineering to eradicate invasive mice on islands: modeling the efficiency and ecological impacts. Ecosphere 7, e01589 ( 10.1002/ecs2.1589) [DOI] [Google Scholar]
- 48.Kanavy DM. 2018. Genetic pest management technologies to control invasive rodents. Doctoral thesis, North Carolina State University, Raleigh, NC. [Google Scholar]
- 49.Kashimada K, Koopman P. 2010. Sry: the master switch in mammalian sex determination. Development 137, 3921–3930. ( 10.1242/dev.048983) [DOI] [PubMed] [Google Scholar]
- 50.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351, 117–121. ( 10.1038/351117a0) [DOI] [PubMed] [Google Scholar]
- 51.Serr M, Copio J, Dyke M, Gopal M, Heard N, Pandya N, Sears R, Godwin J. In review. Male mate competition studies to inform a gene drive approach in house mice (Mus musculus).
- 52.Grunwald HA, Gantz VM, Poplawski G, Xu X-RS, Bier E, Cooper KL. 2019. Super-Mendelian inheritance mediated by CRISPR–Cas9 in the female mouse germline. Nature 566, 105–109. ( 10.1038/s41586-019-0875-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu JH, et al. 2018. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. ( 10.1038/nature26155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murugan K, Babu K, Sundaresan R, Rajan R, Sashital DG. 2017. The revolution continues: newly discovered systems expand the CRISPR-Cas toolkit. Mol. Cell. 68, 15–25. ( 10.1016/j.molcel.2017.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morita T, et al. 1992. Evolution of the mouse t haplotype: recent and worldwide introgression to Mus musculus. Proc. Natl Acad. Sci. USA 89, 6851–6855. ( 10.1073/pnas.89.15.6851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manser A, Cornell SJ, Sutter A, Blondel DV, Serr M, Godwin J, Price TAR. 2019. Controlling invasive rodents via synthetic gene drive and the role of polyandry. Proc. R. Soc. B 286, 20190852 ( 10.1098/rspb.2019.0852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Esvelt KM, Gemmell NJ. 2017. Conservation demands safe gene drive. PLoS Biol. 15, e2003850 ( 10.1371/journal.pbio.2003850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noble C, Olejarz J, Esvelt KM, Church GM, Nowak MA. 2017. Evolutionary dynamics of CRISPR gene drives. Sci. Adv. 3, e1601964 ( 10.1126/sciadv.1601964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunn LC, Levene H. 1961. Population dynamics of a variant t-allele in a confined population of wild house mice. Evolution 15, 385–393. ( 10.1111/j.1558-5646.1961.tb03168.x) [DOI] [Google Scholar]
- 60.Sutter A, Lindholm AK. 2015. Detrimental effects of an autosomal selfish genetic element on sperm competitiveness in house mice. Proc. R. Soc. B 282, 20150974 ( 10.1098/rspb.2015.0974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sutter A, Lindholm AK. 2016. Meiotic drive changes sperm precedence patterns in house mice: potential for male alternative mating tactics? BMC Evol. Biol. 16, 133 ( 10.1186/s12862-016-0710-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Didion JP, et al. 2016. R2d2 drives selfish sweeps in the house mouse. Mol. Biol. Evol. 33, 1381–1395. ( 10.1093/molbev/msw036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burt A, Deredec A. 2018. Self-limiting population genetic control with sex-linked genome editors. Proc. R. Soc. B 285, 20180776 ( 10.1098/rspb.2018.0776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galizi R, et al. 2016. A CRISPR-Cas9 sex-ratio distortion system for genetic control. Sci. Rep. 6, 31139 ( 10.1038/srep31139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oberhofer G, Ivy T, Hay BA. 2019. Cleave and rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl Acad. Sci. USA 116, 6250–6259. ( 10.1073/pnas.1816928116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adikusuma F, Williams N, Grutzner F, Hughes J, Thomas P. 2017. Targeted deletion of an entire chromosome using CRISPR/Cas9. Mol. Ther. 25, 1736–1738. ( 10.1016/j.ymthe.2017.05.021.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuo E, et al. 2017. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 18, 224 ( 10.1186/s13059-017-1354-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prowse TA, Adikusuma F, Cassey P, Thomas P, Ross JV. 2019. A Y-chromosome shredding gene drive for controlling pest vertebrate populations. Elife 2019, e41873 ( 10.7554/eLife.41873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sudweeks J, et al. 2019. Locally fixed alleles: a method to localize gene drive to island populations. See https://www.biorxiv.org/content/10.1101/509364v1 .
- 70.Morgan AP, Didion JP, Hughes JJ, Searle JB, Jolley WJ, Campbell KJ, Threadgill DW, de Villena FP.2018. Genetic characterization of invasive house mouse populations on small islands. See https://www.biorxiv.org/content/10.1101/332064v2 . [DOI] [PMC free article] [PubMed]
- 71.Oh K, et al. In preparation. Population genomics of invasive rodents on islands: genetic consequences of colonization and prospects for localized synthetic gene drive. [DOI] [PMC free article] [PubMed]
- 72.Adler GH, Levins R. 1994. The island syndrome in rodent populations. Q. Rev. Biol. 69, 473–490. ( 10.1086/418744) [DOI] [PubMed] [Google Scholar]
- 73.Berry RJ, Jakobson ME. 1975. Adaptation and adaptability in wild-living house mice (Mus musculus). J. Zool. 176, 391–402. ( 10.1111/j.1469-7998.1975.tb03210.x) [DOI] [Google Scholar]
- 74.Gray MM, Parmenter MD, Hogan CA, Ford I, Cuthbert RJ, Ryan PG, Broman KW, Payseur BA. 2015. Genetics of rapid and extreme size evolution in island mice. Genetics 201, 213–228. ( 10.1534/genetics.115.177790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rowe-Rowe DT, Crafford JE. 1992. Density, body size, and reproduction of feral house mice on Gough Island. S. Afr. J. Zool. 27, 1–5. ( 10.1080/02541858.1992.11448252) [DOI] [Google Scholar]
- 76.Dilley BJ, Schoombie S, Schoombie J, Ryan PG. 2016. ‘Scalping'of albatross fledglings by introduced mice spreads rapidly at Marion Island. Antarct. Sci. 28, 73–80. ( 10.1017/S0954102015000486) [DOI] [Google Scholar]
- 77.Hardouin EA, Chapuis J-L, Stevens MI, Van Vuuren J, Quillfeldt P, Scavetta RJ, Teschke M, Tautz D. 2010. House mouse colonization patterns on the sub-Antarctic Kerguelen Archipelago suggest singular primary invasions and resilience against re-invasion. BMC Evol. Biol. 10, 325 ( 10.1186/1471-2148-10-325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Russell JC, Miller SD, Harper GA, Macinnes HE, Wylie MJ, Fewster RM. 2010. Survivors or reinvaders? Using genetic assignment to identify invasive pests following eradication. Biol. Invasions 12, 1747–1757. ( 10.1007/s10530-009-9586-1) [DOI] [Google Scholar]
- 79.Fraser CI, Banks SC, Waters JM. 2015. Priority effects can lead to underestimation of dispersal and invasion potential. Biol. Invasions 17, 1–8. ( 10.1007/s10530-014-0714-1) [DOI] [Google Scholar]
- 80.Britton-Davidian J, Miller SD, Harper GA, Macinnes HE, Wylie MJ, Fewster RM. 2007. Patterns of genic diversity and structure in a species undergoing rapid chromosomal radiation: an allozyme analysis of house mice from the Madeira archipelago. Heredity 99, 432–442. ( 10.1038/sj.hdy.6801021) [DOI] [PubMed] [Google Scholar]
- 81.Forster DW, Gündüz I, Nunes AC, Gabriel S, Ramalhinho MG, Mathias ML, Britton-Davidian J, Searle JB. 2009. Molecular insights into the colonization and chromosomal diversification of Madeiran house mice. Mol. Ecol. 18, 4477–4494. ( 10.1111/j.1365-294X.2009.04344.x) [DOI] [PubMed] [Google Scholar]
- 82.Berry RJ, Triggs GS, King P, Nash HR, Noble LR. 1991. Hybridization and gene flow in house mice introduced into an existing population on an island. J. Zool. 225, 615–632. ( 10.1111/j.1469-7998.1991.tb04329.x) [DOI] [Google Scholar]
- 83.Jones CS, Noble LR, Jones JS, Tegelström H, Triggs GS, Berry RJ. 1995. Differential male genetic success determines gene flow in an experimentally manipulated mouse population. Proc. R. Soc. Lond. B 260, 251–256. ( 10.1098/rspb.1995.0088) [DOI] [PubMed] [Google Scholar]
- 84.Gray S, Hurst J. 1998. Competitive behaviour in an island population of house mice, Mus domesticus. Anim. Behav. 56, 1291–1299. ( 10.1006/anbe.1998.0890) [DOI] [PubMed] [Google Scholar]
- 85.Jones EP, Searle JB. 2015. Differing Y chromosome versus mitochondrial DNA ancestry, phylogeography, and introgression in the house mouse. Biol. J. Linnean Soc. 115, 348–361. ( 10.1111/bij.12522) [DOI] [Google Scholar]
- 86.Serr M, Heard N, Godwin J. 2019. Towards a genetic approach to invasive rodent eradications: assessing reproductive competitiveness between wild and laboratory mice. In Island invasives: scaling up to meet the challenge (eds Veitch CR, Clout MN, Martin AR, Russell JC, West CJ), pp. 64–70. Gland, Switzerland: IUCN; ( 10.2305/IUCN.CH.2019.SSC-OP.62.en) [DOI] [Google Scholar]
- 87.Champer J, Reeves R, Oh SY, Liu C, Liu J, Clark AG, Messer PW. 2017. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 13, e1006796 ( 10.1371/journal.pgen.1006796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choquenot D, Ruscoe WA. 2000. Mouse population eruptions in New Zealand forests: the role of population density and seedfall. J. Anim. Ecol. 69, 1058–1070. ( 10.1046/j.1365-2656.2000.00462.x) [DOI] [Google Scholar]
- 89.Moro D, Morris K. 2000. Population structure and dynamics of sympatric house mice, Mus domesticus, and Lakeland Downs short-tailed mice, Leggadina lakedownensis, on Thevenard Island, Western Australia. Wildl. Res. 27, 257–268. ( 10.1071/WR99019) [DOI] [Google Scholar]
- 90.Latham N, Mason G. 2004. From house mouse to mouse house: the behavioural biology of free-living Mus musculus and its implications in the laboratory. Appl. Anim. Behav. Sci. 86, 261–289. ( 10.1016/j.applanim.2004.02.006) [DOI] [Google Scholar]
- 91.Runge JN, Lindholm AK. 2018. Carrying a selfish genetic element predicts increased migration propensity in free-living wild house mice. Proc. R. Soc. B. 285, 20181333 ( 10.1098/rspb.2018.1333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barnhill-Dilling SK, Serr M, Blondel DV, Godwin J. 2019. Sustainability as a framework for considering gene drive mice for invasive rodent eradication. Sustainability 11, 1334 ( 10.3390/su11051334) [DOI] [Google Scholar]
- 93.Kofler N, et al. 2018. Editing nature: local roots of global governance. Science 362, 527–529. ( 10.1126/science.aat4612) [DOI] [PubMed] [Google Scholar]
- 94.Kuzma J, et al. 2018. A roadmap for gene drives: using institutional analysis and development to frame research needs and governance in a systems context. J. Responsible Innov. 5, S13–S39. ( 10.1080/23299460.2017.1410344) [DOI] [Google Scholar]
- 95.National Academies of Sciences, Engineering, and Medicine. 2016. Gene drives on the horizon: advancing science, navigating uncertainty, and aligning research with public values. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- 96.Kolopack PA, Lavery JV. 2017. Informed consent in field trials of gene-drive mosquitoes. Gates Open Res. 1, 1–12. ( 10.12688/gatesopenres.12771.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.George D, Kuiken T, Delborne J. In review. Free, prior, and informed consent for engineered gene drives: sharing power and respecting communities. [DOI] [PMC free article] [PubMed]
- 98.Buchthal J, Evans SW, Lunshof J, Telford SR III, Esvelt KM. 2019. Mice Against Ticks: an experimental community guided effort to prevent tick-borne disease by altering the shared environment. Phil. Trans. R. Soc. B 374, 20180105 10.1098/rstb.2018.0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Farooque M, Barnhill-Dilling SK, Shapiro J, Delborne J. 2019. Exploring stakeholder perspectives on the development of a gene drive mouse for biodiversity protection on islands: workshop report. See http://go.ncsu.edu/ges-gene-drive-workshop.
- 100.Delborne J, et al. 2019. Exploring stakeholder perspectives on the development of a gene drive mouse for biodiversity protection on islands: summary report of stakeholder interviews. See https://go.ncsu.edu/ges-gene-drive-landscape.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.