Abstract
It is postulated that the complexity and heterogeneity in cancer may hinder most efforts that target a single pathway. Thus, discovery of novel therapeutic agents targeting multiple pathways, such as microRNAs (miRs), holds promise for future cancer therapy. One such miR, miR-489, is downregulated in majority of breast cancer cells and several drug-resistant breast cancer cell lines, but its role and underlying mechanism for tumor suppression and drug resistance needs further investigation. The current study identifies autophagy as a novel pathway targeted by miR-489 and reports Unc-51 like autophagy activating kinase 1 (ULK1) and lysosomal protein transmembrane 4 beta (LAPTM4B) to be direct targets of miR-489. Furthermore, the data demonstrate autophagy inhibition and LAPTM4B down-regulation as a major mechanism responsible for miR-489-mediated doxorubicin sensitization. Finally, miR-489 and LAPTM4B levels were inversely correlated in human tumor clinical specimens, and more importantly, miR-489 expression levels predict overall survival in patients with 8q22 amplification (the region in which LAPTM4B resides).
Keywords: miR-489, autophagy, breast cancer, starvation, doxorubicin, chemoresistance, ULK1, LAPTM4B, 8q22, autophagosome accumulation
Introduction
Despite recent advances in chemotherapy and novel treatment strategies for breast cancer, the development of broad-range treatments has been restricted due to the discrepancy in transcriptional and genomic heterogeneity of the disease, and the complexity of resistance mechanisms. Therefore, identification of a single therapeutic agent targeting multiple oncogenic and pro-survival pathways would provide a promising future cancer therapy (1,2). Since microRNAs (miRNAs) target multiple pathways and control gene expression of their targets, they could offer considerable therapeutic options. miRNAs are small non-coding RNAs that post transcriptionally regulate expression of their target genes (3). Because miRNAs regulate many genes, their dysregulation have been shown to cause various pathological conditions including cancer. Identifying such dysregulated miRNAs and their targets might provide insights for the development of novel therapy.
Previously, we have established miR-489 as a tumor suppressor miRNA in breast cancer by targeting HER2 signaling pathway (4). Since then, several groups have demonstrated its tumor suppressive role in many different cancers including gastric cancer, lung cancer, ovarian cancer, hepatic cancer, osteosarcoma and bladder cancer (4–12). Remarkably, miR-489 has been reported to induce cell cycle arrest and apoptosis, inhibit metastasis and epithelial to mesenchymal transition in context dependent manner. A thorough understanding of miR-489 mediated tumor suppression in a specific cancer will be valuable in evaluating the possibility of miRNA-489 based therapy. Hence, the current study was aimed to identify novel pathways and molecular targets affected by miR-489 which will lead to better understanding of miR-489 mediated tumor suppression.
Macroautophagy (referred to as autophagy now onwards) has recently received great attention in the field of cancer and chemo-resistance due to its pro-survival role under stressful conditions. Autophagy is a highly conserved process by which cells capture intracellular proteins, lipids and organelles, and deliver them to the lysosomal compartment for degradation (13). The role of autophagy in cancer remains controversial, as it exhibits tumor suppressive and tumor promoting activity in context and molecular subtype dependent manner. For example, autophagy has been shown to inhibit tumor initiation and progression by protecting cells from ROS induced DNA damage and mutagenesis. Conversely, other studies reported that autophagy protects cancer cells from metabolic stress such as starvation or hypoxia (14). Due to its cytoprotective function it enables cancer cells to cope with cytotoxic or other stress induced by treatment. Indeed, autophagy inhibition has been shown to reverse resistance to several chemotherapeutic agents. Currently, chloroquine, an anti-malarial drug, is under clinical trials for adjuvant therapy to reduce resistance via autophagy inhibition in various cancers. Many miRNAs have also been shown to sensitize cancer cells to chemotherapy by inhibiting autophagy. For example, miR-200b has been shown to reverse autophagy mediated resistance to docetaxel by targeting ATG12 (15), while miR-140–5p disrupts cancer stem cell growth in colorectal cancer via autophagy inhibition (16,17). Understanding the role of miRNA in autophagy mediated cancer cell survival and resistance may provide detailed insight on potential miRNA based therapies.
In the present study, for the first time we reported that miR-489 inhibits autophagy by affecting multiple genes involved in the process. We further found that miR-489 reduces tumor cell survival and sensitizes tumor cells under metabolic stress induced by starvation via inhibiting autophagosome degradation. Moreover, we found that miR-489 can also sensitize breast cancer cells to doxorubicin via autophagy inhibition. From the combination of our in-vitro and in-vivo studies, we report that miR-489 inhibits autophagy by targeting ULK1 and LAPTM4B and sensitizes tumor cells to doxorubicin by inhibiting doxorubicin induced cytoprotective autophagy and LAPTM4B expression.
Materials and Methods
The detailed procedures about cell lines and reagents, plasmid construction, cell culture, western blot, immunohistochemistry, qRT-PCR, Luciferase assay and MTT assay are described in detail in Supplemental Experimental Procedures. All the primer sequences are listed in Supplementary Table 1 (Table S1).
Microarray analysis
T47D cells were seeded in 6-well culture dish, treated with 28nM scramble miRNA or miR-489 mimic for 72hrs. RNA was extracted with Trizol reagent, followed by clean-up and DNase I treatment with QIAGEN RNeasy mini kit in accordance with the prescribed protocol provided with the kit. Quality control was performed with Agilent Bioanalyser before performing microarray. The data were normalized using the default quantile normalization with R/bioconductor package lumi version 3.2.2. The microarray data in this manuscript is available on the GEO database (GSE99728). A subset of identified genes was validated by q-PCR.
Autophagy and cell viability assays
Breast cancer cells were transfected with 28nM scr, mimic or inh, for 68hrs, and then treated with Bafilomycin A1 (400nM) or DMSO for 4hrs. The levels of LC3B-I/II and SQSTM1 protein expression were assayed by western blot. MDA-MB-231 cells were transfected with scr, mimic or inhibitor for 24hrs and then treated with 3-MA for 48hrs. Autophagic flux was then assessed by examining LC3B-II and p62 expression using western blot. MDA-MB-231 cells stably expressing mCherry-EGFP-LC3B fusion protein were transfected with scr or mimic for 48hrs and assayed for co-localization of red and green punta using confocal microscopy.
To study effect of miR-489 on cell survival, indicated cell lines were first seeded in 96 well plate in triplicate and transfected with 28nM scr or mimic. Cell viability assay was performed using MTT reagent at 72hrs. To assess effect of miR-489 under metabolic stress, MDA-MB-231, HCC1954 or T47D cells were seeded in 96-well plate. Cells were transfected with scr, mimic or inh in complete media or in low serum. Cell viability assay was performed using MTT reagent at indicated time points. Expression of cleaved caspase 3, LC3B and p62 was analyzed using western blot. To study role of autophagy in miR-489 induced sensitization under starvation, MDA-MB-231 cells were transfected with 9.3nM scr or mimic in complete media or low serum in presence or absence of 3-MA, siATG5 or Bafilomycin A1 followed by cell viability assay using MTT reagent and western blot analysis to examine effect on autophagy and apoptosis. To examine effect of miR-489 on doxorubicin induced cytoprotective autophagy, MDA-MB-231 and HCC1954 cells were transfected with 28nM scr or mimic with or without doxorubicin. Protein was isolated 72hrs post treatment and western blot was performed.
Chemo-sensitization Assays
To study doxorubicin localization, cells were treated with 28nM scr or mimic for 24hrs then treated with 0.5uM doxorubicin for 48hrs followed by confocal microscopy. MDA-MB-231 cells were transfected with 9.3nM scr or mimic for 24 hours and treated with indicated concentration of doxorubicin in presence or absence of 3-MA (5mM), siATG5 (50nM) or Bafilomycin A1 (50nM) for 48 hours and cell proliferation was measured by MTT assay to examine role of autophagy inhibition in miR-489 mediated doxorubicin sensitization. To assess lysosomal integrity, MDA-MB-231 cells were transfected with 28nM scr or mimic and stained with Acridine orange (1μg/ml) for 20min followed by flow cytometry and confocal microscopy.
miRNA-nanoparticles preparation
Liposomes were prepared as described elsewhere with slight modifications (53,54). Briefly, cationic liposomes consisting of DOTAP and cholesterol (2:1 molar ratio) were prepared using the thin-film hydration method. The film was hydrated with nuclease free water and sonicated using probe sonicator for 15min. The lipid concentration adjusted at 10mg/ml. For miRNA-nanoparticles preparation, 231.4μls of cationic liposomes and 90μls of miRNA were mixed in final volume of 1 ml nuclease free water. The miRNA-liposomes were allowed to stand at room temperature for 10 min. miRNA-nanoparticles were further decorated with sodium hyaluronate by adding 128.56μls of 1mg/ml sodium hyaluronate solution and kept at room temperature for another 10min. The samples were condensed using Millipore centrifugal filter units.
Xenograft experiment
Six weeks old Athymic Female nude mice were purchased from Envigo. All mice were handled and maintained under supervision of veterinarian in accordance with institutional guidelines and under a University of South Carolina Institutional Animal Care and Use Committee (IACUC) approved protocol. All mice were subcutaneously injected with 1 × 106 MDA-MB-231 cells left flank of each mice (n=5/group). Mouse were randomly distributed in four groups when tumor size reached 50–100mm3. The mice were administered with control nanoparticles or miR-489 loaded nanoparticles every third day. Doxorubicin (4mg/kg) was administered day after injection of miRNA. Tumor volumes were calculated by measuring length and width every third day. After 3 weeks, all mice were sacrificed and tumors were extracted after euthanizing all animals. Tumor volumes were calculated by modified ellipsoidal formula (1/2(lxw2)).
Clinical samples:
Human breast cancer tissue samples were obtained through the South Carolina Tissue Bank with approval from the Institutional Review Board at the University of South Carolina. Tissue samples were randomly collected from patients who were diagnosed with invasive breast ductal carcinoma between 2003 and 2007. RNA was isolated from tumors samples and expression of miR-489, ULK1 and LAPTM4B was analyzed by qRT-PCR. Demographic and histopathology data of the patient samples are listed in Supplementary table 2.
Statistical analyses
The statistical analyses were conducted with R and GraphPad software packages. A Student t test or ANOVA test was used for comparison of quantitative data. The clinical effect of the gene expression profiles of miR-489 in the patients with 8q22 gain/amplified tumor was evaluated using a published data set containing 1302 breast cancer patients (38,39) The median expression value was used as the cutoff to classify miR-489 expression as high or low. Recurrence-free survival was estimated using the Kaplan-Meier method and compared with log-rank test. The linear correlations between miR-489 and potential target genes expression in primary breast cancer tissues were evaluated with Pearson correlation coefficient analysis. Values of P < 0.05 were considered significant.
RESULTS
Identification of novel miR-489 target genes involved in autophagy regulation in breast cancer
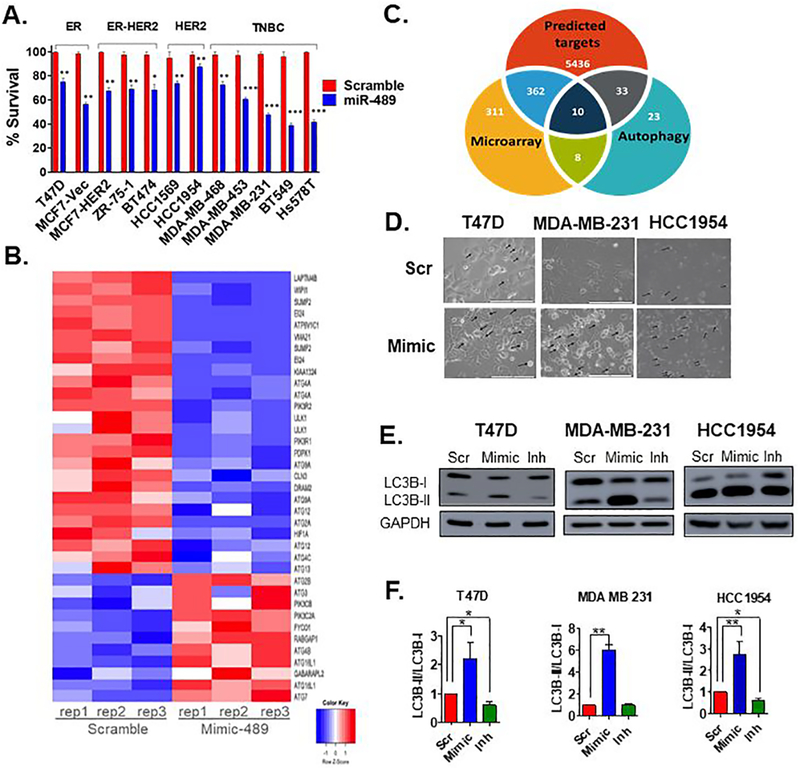
Recent studies revealed that miR-489 is significantly down-regulated in various types of cancer. Previously, through screening of miRNAs dysregulated by HER2, using two isogenic cell lines MCF7-Vec and MCF7-HER2, we identified miR-489 as a novel tumor suppressive miRNA regulated by HER2 and reported double negative feedback loop between miR-489 and HER2 (4,18). However, upon further investigation we found significant cytotoxic and cytostatic effect of miR-489 on other types of breast cancer cell lines as well. (Fig. 1A). This result suggests miR-489 targets other pathways involved in proliferation and survival besides the HER2 signaling pathway. Our gene expression analysis also revealed multiple pathways affected by miR-489. We observed that several genes involved in autophagy were affected by miR-489 reconstitution. (Fig. 1B). We utilized target prediction tools such as miRwalk and TargetScan to identify potential miR-489 targets involved in autophagy. Consistent with our microarray data, target prediction tools also suggested multiple genes involved in regulation of autophagy such as ULK1, ATG4A, LAPTM4B, ATG4C, ATG2B, ATG3, ATG5, ATG7, ATG12 and ATG14 as potential target of miR-489. (Fig. 1C). Interestingly, miR-489 mediated cytotoxicity was associated with distinct morphological features, including the appearance of dense intracellular vacuolar structures (Fig. 1D and S1A). This further indicated a possible perturbation of autophagy. To investigate effect of miR-489 on autophagy, we transfected breast cancer cell lines with non-targeting scramble miRNA (scr) miR-489 mimic (mimic) and miR-489 inhibitor (inh) and monitored the effect on autophagy marker LC3B using western blot. Conversion of non-lipidated soluble LC3B (LC3B-I) to phosphatidylethanolamine-conjugated LC3B (LC3B-II) serves as a sensitive indicator of autophagy (19). We found significant accumulation of LC3B-II upon miR-489 overexpression, while inhibition of miR-489 completely reversed this deposition (Fig. 1E, 1F and S1B). We observed this effect in HER2 positive, estrogen positive and triple negative breast cancer cells suggesting autophagy modulation by miR-489 is independent of breast cancer subtype. In summary, we identified autophagy as a novel pathway modulated by miR-489 expression.
Fig. 1. miR-489 inhibits breast cancer cell growth and modulates multiple genes involved in autophagy.
A. MTT based cell proliferation/viability assay measuring viability of indicated breast cancer cell lines at 72hrs post transfection with 28nM scramble control miRNA (Scr) and miR-489 mimic (Mimic). **, p < 0.01; ***, p < 0.001. Data are representative of three independent experiments. B. Heatmap analysis of microarray data for the genes associated with autophagy. Genes were selected based on their differential expression between the scr transfected cells and mimic transfected cells. Red and blue indicate high and low gene expression, respectively. C. Hypergeometric analysis was performed using microarray data and putative targets identified using microRNA target-predicting software. D. Bright-field microscopy images of Scr or Mimic transfected cells. Arrow indicates intracellular endosomal, possibly vacuolar structures. Data are representative of three independent experiments. E. Indicated breast cancer cell lines were transfected with scr, mimic or miR-489 inhibitor (Inh). Protein was collected 72hrs post transfection and subject to western blot analysis of autophagy markers. GAPDH was used as a loading control. F. Quantification of LC3B-I and LC3B-II ratio.
miR-489 inhibits autophagy mainly by blocking maturation step
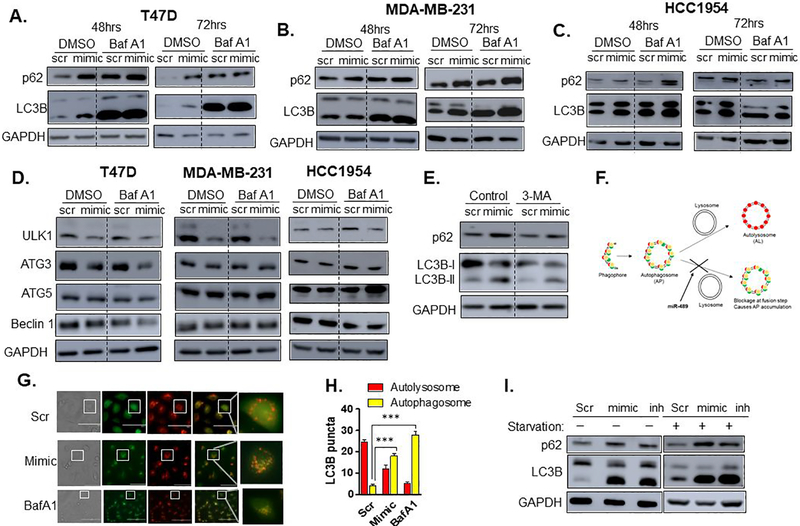
Autophagy is complex multistep process. Numerous assays have been developed to precisely decipher the effect on basal autophagy or autophagic flux. Although change in LC3B-I and II is an indicator of change in autophagy flux, it fails to provide conclusive information about autophagy activation or inhibition (19). Deposition of LC3B-II can be caused by induction of autophagosome (AP) formation or blockage of autophagosome degradation via fusion of autophagosome with lysosome (Autolysosome). To distinguish these two possibilities, we first used Bafilomycin A1 immunoblot. Bafilomycin A1 is a lysomotropic agent which inhibits the V-ATPase responsible for acidification of the lysosome, and prevents fusion of autophagosomes and lysosomes (19). Comparison of LC3-II levels in the absence and presence of Bafilomycin A1 allows the effects of formation and degradation to be uncoupled. As shown in Figure 2A, 2B and 2C, no significant difference was detected between double treatment and lysomotropic agent alone at 48hrs and 72hrs time points, suggesting that miR-489 might be blocking degradation of autophagosome. Moreover, our finding on the clearance of cargo protein p62, another distinctive feature of autophagy, also indicated blockage of autophagosome degradation (Fig. 2A to 2C, and S2A to S2D) (19,20). These results suggest that miR-489 inhibits autophagy by blocking formation of autophagosome and lysosome fusion (Autolysosome). We then examined protein levels of some of the core autophagy genes using western blot and found significant downregulation of ULK1 upon miR-489 overexpression (Fig.2D).
Fig. 2. miR-489 inhibits autophagy mainly by blocking maturation step.
A-C. Bafilomycin A1 blots after reconstituting cells with miR-489. T47D (A.), MDA-MB-231 (B.) and HCC1954 (C.) cell lines were transfected with 28nM scr or mimic, for 48 or 72hrs and p62 and LC3B-II protein expression was assayed by western blot in presence or absence of Bafilomycin A1 (BafA1) (400nM). GAPDH was used as a loading control. D. Western blot analysis of core autophagy genes after 72hrs post transfection. Cells were transfected with 28nM scr or mimic, for 72hrs, and then treated with BafA1 for 4hrs. GAPDH was used as a loading control. E. Western blot analysis of autophagic flux after reconstitution of scr or mimic with or without autophagy inhibitor 3-MA. Cells were transfected with 28nM scr or mimic, for 24hrs, and then treated with 3-MA for 48hrs. Autophagic flux was then analyzed using western blot. GAPDH was used as a loading control. F. Schematic diagram of mCherry-EGFP-LC3B reporter. G. Confocal microscopy of autophagy maturation. MDA-MB-231 cells stably expressing mCherry-EGFP-LC3B fusion protein were transfected with 28nM scr or mimic for 48hrs or Bafilomycin for 4hrs and assayed for co-localization of red and green punta using confocal microscopy. H. Quantitative analysis of red and yellow punta in MDA-MB-231 cells at 48hrs post-transfection of scr or mimic I. miR-489 blocks starvation induced autophagy. MDA-MB-231 cells were transfected with 28nM scr or mimic for 68hrs and treated with EBSS for last 4hrs to induce autophagy. Bafilomycin A1 was used as a control. GAPDH was used as a loading control. **, p < 0.01; ***, p < 0.001. Data are representative of three independent experiments.
We then asked whether autophagy inhibitors that block autophagosome formation such as 3-methyladenine (3-MA) can prevent miR-489 induced autophagosome deposition. Consistent with previous observations, 3-MA indeed attenuated miR-489 induced p62 and LC3B-II deposition (Fig. 2E). To further confirm that miR-489 inhibits formation of autolysosome (AL) by blocking fusion of autophagosome and lysosome, we used common dual fluorescent LC3B reporter, which expresses an N-terminal fusion of mCherry and GFP to human LC3B (19). This reporter system enables real time monitoring of autophagic flux. Since, low pH quenches GFP fluorescence, AL is seen as distinct red aggregates, while AP appears as yellow aggregates due to overlap of red and green fluorescence. (Fig. 2F). If the autophagy pathway is in flux, then these yellow aggregates will be transient as APs fuse with acidic lysosomes to form ALs, in which the low pH quenches GFP fluorescence, resulting in rapid accumulation of dense red aggregates. However, blockage of autophagy at the fusion step results in accumulation of yellow puncta. Consistent with our earlier results, miR-489 mimic transfected cells show presence of more yellow puncta compared to control, further suggesting miR-489 causes accumulation of AP by blocking fusion step (Fig. 2G, 2H). We also examined the effect of miR-489 restoration on starvation-induced autophagy and found substantial accumulation of p62 and LC3B-II. Interestingly, this accumulation was comparable to that of Bafilomycin A1, which also blocks the fusion step (Fig. 2I). Together, these results indicate that miR-489 inhibits autophagy mainly by blocking the fusion of APs to lysosome.
miR-489 directly targets ULK1 and LAMTM4B genes
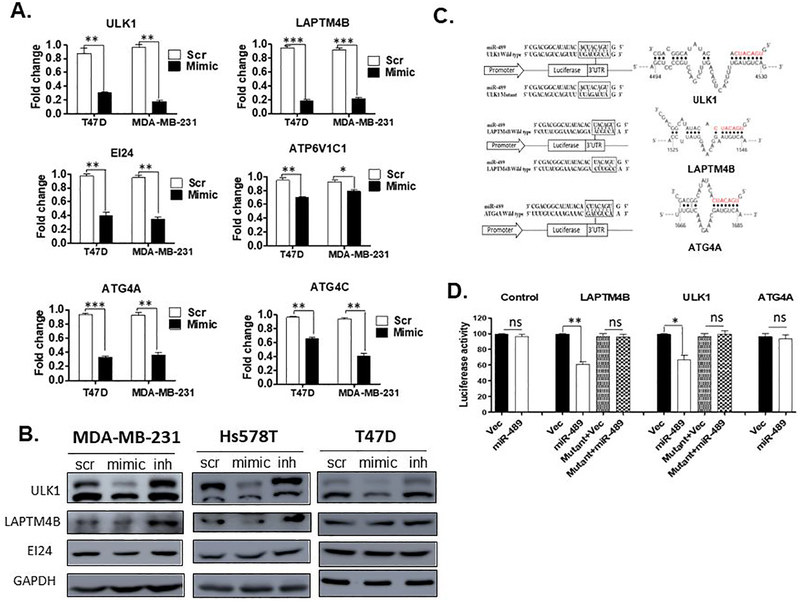
After establishing role of miR-489 in autophagy, we sought to identify direct targets of miR-489 involved in the process. Our microarray and target predication algorithm analyses revealed multiple genes directly involved in autophagy pathway such as ATG4A, ULK1, ATG4C, LAPTM4B, EIG121, VMA21, WIPI1, ATP6V1C1 and EI24 (Fig. 1B, 1C). To validate our microarray results, we performed qRT-PCR after transient transfection of mimic or scr in indicated breast cancer cell lines (Fig. 3A, Fig S3A and S3B). As shown in Figure 3A, the expression of four out of six genes were greatly diminished upon miR-489 reconstitution in multiple breast cancer cell lines. The expression of the other two genes was reduced in a cell line-dependent manner (Fig. S1A). To further confirm, we performed western blot analysis and found downregulation of LAPTM4B and ULK1 but not EI24 in all three cell lines (Fig. 3B). We were also interested in investigating whether knockdown of endogenous miR-489 would affect target genes expression. Indeed, miR-489 inhibitor efficiently increased the amounts of ULK1 and LAPTM4B (Fig. 3B). We then performed 3’UTR luciferase assay to identify direct targets. Our assay revealed that ULK1 (35% reduction, p-value = 0.0184) and LAPTM4B (40% reduction, p-value = 0.0038) are direct target of miR-489 while ATG4A might be an indirect target (Fig. 3C and 3D). Because ULK1 and LAPTM4B are positive regulators of autophagy, these results further suggest that miR-489 negatively regulates the autophagy.
Fig. 3. miR-489 directly targets ULK1 and LAMTM4B genes.
A. MDA-MB-231 and T47D cell lines were transfected with 28nM scr or mimic. RNA was isolated 72hrs post transfection and qRT-PCR was performed to examine expression level of indicated genes. Data are means of three replicates ± SEM. B. Western blot showing expression of potential targets upon transfection of 28nM scr, mimic or inh in indicated cell lines. GAPDH was used as a loading control. C. A schematic representation of the target mRNA with putative miR-489 binding site in the 3’ UTR by S fold database, where the seed region is highlighted in red. D. HEK293T cells were co-transfected with miR-489 expressing vector or empty vector and renilla expressing vector for 72hrs. Firefly luciferase was measured for each condition and normalized with renilla luciferase. Normalized luciferase activity was compared with WT-3’UTR and Mutant 3’ UTR of target mRNA. Data are means of three replicates ± SEM.
miR-489 reduces tumor cell survival and sensitizes tumor cells under metabolic stress induced by starvation via inhibiting autophagosome degradation
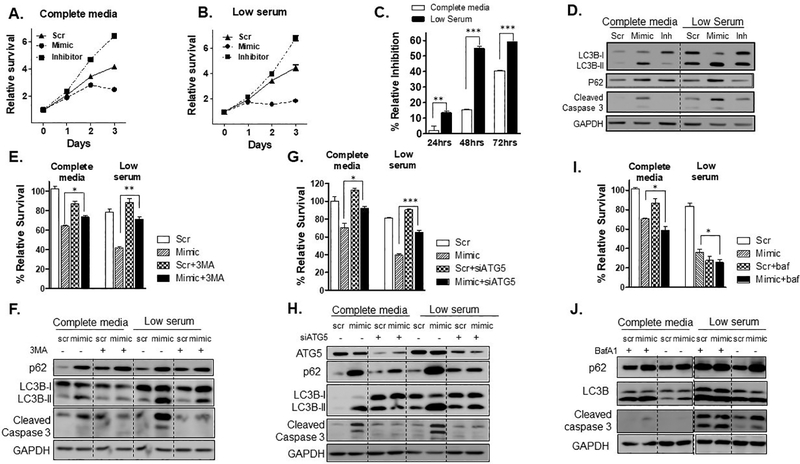
Previous experiments suggest that overexpression of miR-489 may result in increased sensitivity of tumor cells to insults that induce autophagy as survival mechanism. Since starvation is a fairly common insult that requires autophagy for survival, we examined if miR-489 over expression can reduce tumor cell survival under starvation. We selected MDA-MB-231 cells for this study as previous study has showed that MDA-MB-231 cells are more resistant to starvation compared to other breast cancer cell lines. Study also showed that this aggressive cell line is sensitive to autophagic induction and additionally possesses the ability to proliferate following nutrient deprivation (21). Consistent with this study we also observed that MDA-MB-231 cells were much more resistant to starvation compared to T47D and HCC1954 cells. (Fig. 4A, 4B, S4B and S4E). We transfected MDA-MB-231 cells with scr, mimic or inh under complete media or starvation and performed cell survival assay at indicated time points. We found that miR-489 over expression significantly reduced survival under starvation (p-value = 0.0001) (Fig. 4A and 4B). miR-489 over expression caused more than 20% growth inhibition after 72hrs of transfection under starvation compared to nutrient rich condition (Fig. 4C). Interestingly, this cytotoxic and cytostatic effect began as early as 24hrs under starvation. While this was not observed until 48hrs in nutrient rich medium. Consistent with previous observations, starvation indeed induce autophagy and miR-489 restoration showed stronger autophagy inhibition under starvation as indicated by increased accumulation of LC3B-II and p62 in western blot analysis (Fig. 4D). Western blot results also showed significant increase in cleaved caspase 3 upon miR-489 over expression under starvation. We also performed similar experiment on T47D and HCC1954 cells (Fig. S4A to S4F). Both cell lines were significantly affected under starvation. We tested whether miR-489 inhibition can protect these cells under stress induced by starvation. miR-489 inhibition marginally increased survival under starvation in HCC1954. Interestingly, this protective effect was more prominent in T47D cells (Fig. S5C, p-value=0.0017) that possesses higher endogenous miR-489 (Fig. S5).
Fig. 4. miR-489 reduces tumor cell survival and sensitizes tumor cells under starvation by inhibition autophagosome maturation.
MDA-MB-231 cells were transfected with 28nM scr, mimic or inh in complete media (A) or low serum (B). Cell viability assay was performed at indicated time points (0, 24, 28 and 72hrs) using MTT. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data are means of three replicates ± SEM. C. Relative inhibition by mimic under both conditions at indicated time points. D. MDA-MB-231 cells were transfected with 28nM scr, mimic or inh in complete media or low serum for 72hrs and western blot was performed to investigate expression of cleaved caspase 3 and autophagy markers. GAPDH was used as a loading control. E-J. MDA-MB-231 cells were transfected with 9.3nM scr or mimic in complete media or low serum with or without 3-MA (5mM), siATG5 (50nM) or BafA1 (10nM) treatment and cell viability assay (E, G, I) and western blot analysis (F, H, J) was performed to examine autophagic flux and apoptosis *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data are means of three replicates ± SEM.
To examine whether cytotoxic effect of miR-489 is mediated through autophagy inhibition, we blocked autophagy at different stages using pharmacological and RNA interference approach under nutrient rich condition and under starvation and assessed effect of miR-489 on autophagic flux and cell viability. We observed that blocking early stages of autophagy by 3-MA or siATG5 resulted in significant attenuation of cytotoxic effect of miR-489 (Fig. 4E, 4F). This rescue effect was more pronounced under starvation (Fig. 4G). In fact, 3-MA and siATG5 treatment almost completely prevented miR-489 induced apoptosis as evidenced by western blot analysis of cleaved caspase 3 (Fig. 4H). However, autophagy blockage at later stage by Bafilomycin A1 did not rescue cells from miR-489 induced death. Rather, it mildly synergized the growth inhibition. These results suggest that increased autophagosome accumulation by miR-489 may account for its cytotoxic effect on these cells. Previous reports have shown that autophagosome accumulation confers cytotoxicity in cancer cells and blocking autophagosome synthesis by chemical inhibitor or by genetic ablation alleviates this cytotoxicity (22,23). Starvation is known to induce autophagosome formation. Hence, restoring miR-489 under starvation resulted in pronounced increase in autophagosome accumulation. This might explain why miR-489 further sensitized cells under starvation. We also observed greater rescue from miR-489 induced death under starvation upon blocking autophagosome synthesis by 3-MA or siATG5. Further confirming the idea that miR-489 induced autophagosome accumulation is crucial for its cytotoxic effect. Previous studies have also reported similar observations where perturbation of earlier stages of autophagy attenuated effect of autophagy inhibitors that block later stages (24).
In summary, these data suggest that miR-489 mediated autophagosome accumulation is at least partially responsible for its cytotoxic effect and it further sensitizes cancer cell death under metabolic stress induced by starvation due to further increase in autophagosome accumulation.
miR-489 acts as a therapeutic sensitizer in breast cancer cells by inhibiting doxorubicin induced cytoprotective autophagy and directly targeting LAPTM4B
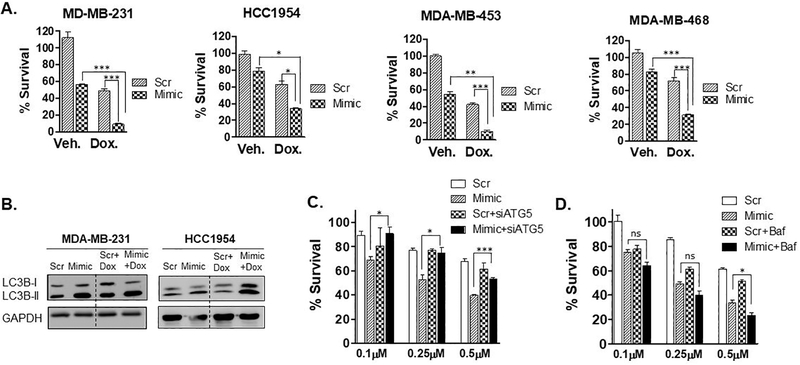
We next tested whether miR-489 could sensitize breast cancer cells to certain front-line chemotherapeutic agents. Few studies have demonstrated that miR-489 is severely down regulated in cisplatin and doxorubicin resistant breast cancer cell lines and tumors (12,25). We hypothesized that restoring miR-489 can sensitize breast cancer cells to these chemotherapeutic agents. Using MTT based cell viability assay, we found miR-489 restoration sensitized MDA-MB-231, MDA-MB-468, HCC1954 and MDA-MB-453 to cisplatin and doxorubicin (Fig. 5A and S6A). We observed substantial sensitization of doxorubicin upon miR-489 restoration, while only a mild sensitization effect was observed with cisplatin. Remarkably, double treatment of miR-489 and doxorubicin resulted in almost complete death of MDA-MB-231 and MDA-MB-453 cells (Fig. 5A).
Fig. 5. miR-489 sensitizes breast cancer cells to doxorubicin induced cell death by inhibiting doxorubicin induced cytoprotective autophagy.
A. Indicated breast cancer cell lines were transfected with 28nM scr or mimic for 72hrs in presence of doxorubicin (0.75μM) for 48hrs and cell proliferation was measured by MTT assay. Data are representative of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. B. MDA-MB-231 and HCC1954 cells were transfected with scr or mimic for 72hrs in absence or presence of doxorubicin and autophagy marker LC3B-I and LC3B-II were monitored to examine autophagic flux. C. MDA-MB-231 cells were transfected with 9.3nM scr or mimic with or without siATG5 for 24hrs and treated with indicated concentration of doxorubicin for 48hrs and cell proliferation was measured by MTT assay. D. MDA-MB-231 cells were transfected with 9.3nM scr or mimic with for 24hrs and treated with indicated concentration of doxorubicin in presence or absence of Bafilomycin A1 (50nM) for 48hrs and cell proliferation was measured by MTT assay. Data are representative of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Doxorubicin has been previously shown to induce cytoprotective autophagy (26,27). We examined autophagic flux upon miR-489 restoration in doxorubicin treated MDA-MB-231 and HCC1954 cells (Fig. 5B). Consistent with previous results, we found that doxorubicin indeed increased autophagic flux. This cytoprotective autophagy was blocked by miR-489 as indicated by increased LC3B-II levels (Fig. 5B). Previous studies have demonstrated that autophagy inhibition sensitizes breast cancer cells to doxorubicin (26–28). Consistent with these results, we observed sensitization with autophagy inhibitor Bafilomycin A1 and siATG5 (Fig. 5C, 5D). Several studies have demonstrated that autophagy inhibition at later stage is more effective in sensitizing tumor cells against chemotherapy (29–31). Consistent with these results, we observed only mild sensitization with siATG5 while miR-489 and Bafilomycin A1 resulted in higher sensitization compared to siATG5. Interestingly, we observed greater sensitization with miR-489 compared to Bafilomycin A1. We then seek to examine role of autophagy in miR-489 mediated doxorubicin sensitization. Blocking autophagy at early step by siATG5 attenuated miR-489 induced drug sensitization while blocking late stage autophagy by Bafilomycin A1 further showed mild sensitization at higher doxorubicin concentration (Fig. 5C). Furthermore, although ATG5 knockdown attenuated sensitization by miR-489, it failed to completely prevent this sensitization (see Fig. 5C, 0.5μM Dox. treatment). These data suggest that miR-489 may also inhibit additional pathway besides autophagy that render cells resistant to doxorubicin. Apart from inducing autophagic flux, LAPTM4B has been shown to promote chemoresistance (32–35). Therefore, we seek to examine role of LAPTM4B in miR-489 mediated doxorubicin sensitization.
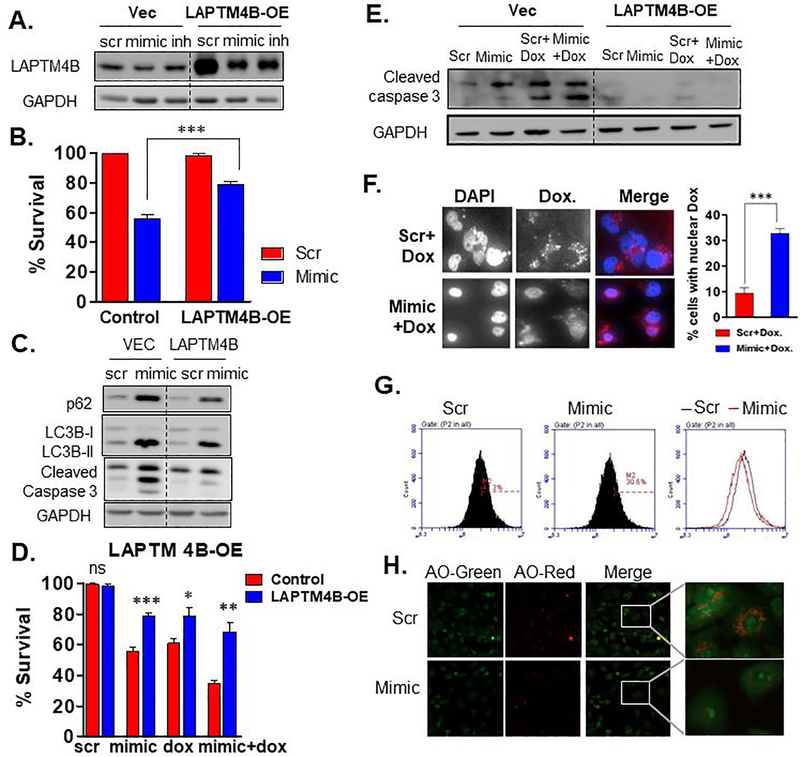
To directly access role of LAPTM4B, we stably overexpressed LAPTM4B in MDA-MB-231 cell lines (Fig. 6A). As suggested by previous studies, LAPTM4B restoration increased autophagic flux and attenuated miR-489 induced autophagy inhibition (Fig. 6C). Surprisingly, LAPTM4B restoration increased survival by 23% (p-value = 0.0005) upon miR-489 overexpression (Fig 6B). LAPTM4B overexpression not only rescued cells from miR-489 induced apoptosis but also increased survival by 17% (p-value = 0.0175) upon doxorubicin treatment and 33% (p-value = 0.001) upon combination treatment (Fig. 6D). To validate this result, we performed western blot analysis to monitor apoptotic marker cleaved caspase 3 and detected significant reduction in cleaved caspase 3 in LAPTM4B overexpressing cell line (Fig. 6E). LAPTM4B has been shown to sequester doxorubicin in lysosomes preventing their entry into nucleus thereby rendering it less effective (32,33,36,37). We tested whether miR-489 can induce redistribution of doxorubicin in MDA-MB-231 cells and found that miR-489 indeed increased doxorubicin localization in nucleus (Fig. 6F, Fig. S6B). We then assessed effect of miR-489 on lysosomal integrity. We utilized Acridine orange dye which specifically stains lysosomal-endosomal organelles red. Our flow cytometry and confocal microscopy results showed reduction in red fluorescence upon miR-489 restoration (Fig. 6G, 6H). These results indicate that miR-489 affects lysosomal integrity and that this may be one of the mechanism through which miR-489 sensitizes cell to doxorubicin. We also examined role of ULK1 in miR-489 induced doxorubicin sensitization. ULK1 overexpression provided mild but significant rescue from the cytotoxic and cytostatic effect of miR-489 (Fig. S7A and S7B), but it could not rescue cells from doxorubicin (Fig. S7C). A modest rescue was observed in double treatment which may be due to rescue from the effect of miR-489. Collectively, these results indicate that miR-489 sensitizes MDA-MB-231 cells to doxorubicin via inhibiting doxorubicin induced cytoprotective autophagy and through doxorubicin redistribution induced by LAPTM4B downregulation.
Fig. 6. miR-489 sensitizes breast cancer cells to doxorubicin partly by targeting LAPTM4B.
A. Western blot indicating stable over expressing LAPTM4B. GAPDH was used as a loading control for western blot analysis. B-C. MDA-MB-231 cells stably expressing LAPTM4B were transfected with 28nM scr or mimic. Cell viability assay (B.) and western blot (C.) was performed 72hrs post transfection to examine autophagic flux and apoptosis. D-E. MDA-MB-231 cells stably expressing LAPTM4B were transfected with 28nM scr or mimic with or without doxorubicin followed by cell viability assay (D.) and western blot analysis (E.) F. Microscopy analysis of subcellular localization of doxorubicin. Confocal microscopy was performed 72hrs after MDA-MB-231 cells were treated with 28nM scr or mimic with doxorubicin. Data are means of three replicates ± SEM. Data are representative of three independent experiments. G. MDA-MB-231 cells were transfected with 28nM scr or mimic and stained with Acridine orange (1mg/ml) for 20min and flow cytometry was performed to examine lysosomal integrity. H. Confocal microscopy of MDA-MB-231 cells after transfection with scr or mimic and staining with Acridine orange (1mug/ml) for 20min. Data are representative of three independent experiments.
miR-489 sensitizes breast cancer cells to chemotherapy in vivo
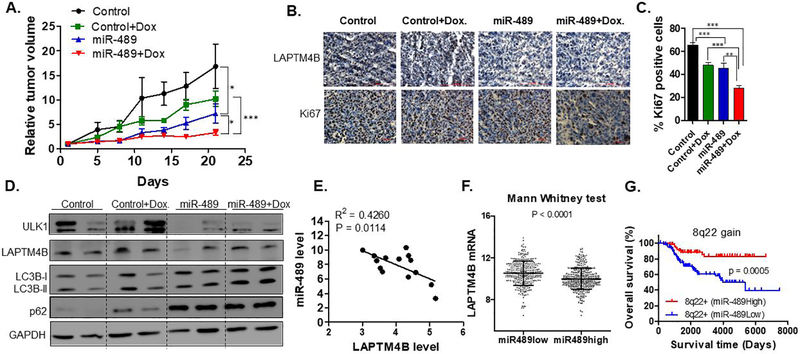
Given the in-vitro findings and clinical significance, we tested whether miR-489 could sensitize tumors to doxorubicin in-vivo. To explore the possibility of using miR-489 for therapy, we developed a nanoparticle delivery system to deliver miR-489 into tumor cells (4). In-vitro treatment with miR-489 packaged nanoparticle inhibited target genes (Fig. S8A), indicating that nanoparticles can effectively deliver miR-489 into tumor cells. Our qRT-PCR analysis on tumor samples confirmed successful delivery of miR-489 and down regulation of ULK1 and LAPTM4B mRNA (Fig. S8B, S8C and S8D). Consistent with our in-vitro data, miR-489 indeed sensitized cells to doxorubicin in-vivo (Fig. 7A). Tumor growth in mice treated with doxorubicin alone was moderately reduced compared to control, while mice treated with miR-489 exhibited a significant reduction in tumor growth compared to control (p-value = 0.0441) and doxorubicin (p-value = 0.0284). Tumor growth in mice treated with the combination of doxorubicin and miR-489 was significantly reduced compared to control (by more than 75%; p-value = 0.0066), monotherapy with doxorubicin (by more than 40%; p-value<0.0001), or miR-489 alone (by more than 20%; p-value = 0.0326) (Fig. 7A). Our immunohistochemistry data revealed reduced cytoplasmic staining of LAPTM4B and significant reduction in number of Ki-67-positive cells in combination treatment when compared to the control tumors (Fig. 7B, 7C, p-value<0.0001). Western blot of tumor samples indicated autophagy inhibition and down-regulation of ULK1 and LAPTM4B (Fig. 7D). Together, these data demonstrated that miR-489 delivered through nanoparticles inhibits tumor growth and sensitizes tumor cells to doxorubicin in xenografts.
Fig. 7. Nanoparticle-delivered miR-489 inhibits tumor growth and sensitized cells against doxorubicin in vivo.
A. miR-489 inhibits tumor growth and sensitized cells against doxorubicin in xenograft animals. After the tumors were palpable, the animals were randomly assigned into four groups (n=5 per group). All animals were injected with miR-489 or control encapsulated in nanoparticle every third day. The treatment starting day was referred to as ‘Day zero’ in the figure. B. IHC analysis revealed reduced expression of LAPTM4B and Ki67 in tumors treated with miR-489 encapsulated nanoparticles. C. Quantification of Ki-67 positive cells in tumors of all four groups. D. Western blot analysis of tumors revealed down regulation of ULK1, LAPTM4B and autophagy inhibition by miR-489. GAPDH was used as a loading control. E. miR-489 and LAPTM4B expression was measured in breast tissues form breast cancer patients (n=14) using qPCR. F. Correlation of miR-489 and its potential target gene expression in primary breast cancers. The linear dependence between miR489 and its potential target genes expression was evaluated by Pearson analysis of a published breast cancer data set (38). G. miR-489 expression predict overall survival of breast cancer patients with 8q22 gain/amplified tumors. Patient survival was estimated using the Kaplan-Meier method and compared with log-rank tests. The Y axis represents the probability of overall survival. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
miR-489 and LAPTM4B expression are inversely correlated in breast cancer tissues
To further define the clinical relevance of our findings, we examined miR-489 and LAPTM4B expression in breast tissues from breast cancer patients (n=14). We found significant inverse correlation between expression of miR-489 and LAPTM4B (Fig.7E). Similarly, in-silico analysis on primary tumors revealed a strong inverse correlation between miR-489 and LAPTM4B expression with a p-value of 0.0000413 (Fig. 7F). In the clinical dataset, although statistically not significant, we found patients with higher miR-489 possess lower ULK1 expression (Fig.S9A).
However, we do not observe inverse correlation between miR-489 and ULK1 in breast tissues from breast cancer patients (n=19) (Fig.S9B).
Considerable clinical evidences suggest that LAPTM4B over-expression promotes autophagy and chemoresistance (32,33). One study show that this over-expression is through amplification of the gene. The gene resides on 8q22 region, which is amplified in 20% of breast cancer patients (32). Therefore, we evaluated the clinical effect of the gene expression profiles of miR-489 in the patients with 8q22 gain/amplified tumor using a published data set containing 1302 breast cancer patients (38,39). Intriguingly, we found that 8q22 amplified patients with high miR-489 have a significantly better survival as compared to low miR-489 (p-value = 0.0005) (Fig. 7H) These results indicate potential application of miR-489 as prognostic biomarker in 8q22 amplified breast cancers.
Discussion
In this study, we show that miR-489 inhibits autophagy in multiple subtypes of breast cancer cell line. Restoration of miR-489 reduces viability of MDA-MB-231 under stress induced by starvation. Lastly, we show miR-489 can sensitize breast cancer cell lines to doxorubicin in-vitro and in-vivo. miR-489 imparts these phenotypic effects partly by directly targeting LAPTM4B and ULK1.
Our gene expression analysis reveals downregulation of many autophagy related genes upon miR-489 reconstitution. Although, we observe that miR-489 affects multiple genes involved at different stages of autophagy, we conclude that miR-489 restoration is mainly associated with defective maturation that leads to AP accumulation. The defect at this fusion step caused by miR-489 is supported by three independent experiment. First, miR-489 restoration leads to concomitant accumulation of LC3B-II and cargo protein p62. Upon completion of the autophagic process, p62 and LC3B-II are degraded via fusion with lysosome. However, blockage at maturation step prevents this degradation and results in simultaneous accumulation of both proteins. Second, no further significant accumulation of LC3B-II was observed in combination treatment of miR-489 and Bafilomycin A1. Last, miR-489 increases AP as shown by more yellow punta demonstrated by microscopic analysis of fluorescent mCherry-EGFP-LC3B reporter. Reduced ULK1 expression levels are commonly associated with decreased autophagic flux due to reduced autophagosome formation. In contrast to this notion, we observed accumulation of AP. However, it is worth to note that while ULK1 is involved in the initiation stage of the autophagy, it is not essential for activation of the LC3 conjugation machinery. Studies show that ULK1 and ULK2 (homolog of ULK1) possess redundant roles in autophagy (40,41). and indeed, it takes a double knockout of Ulk1 and Ulk2 to completely block amino acid starvation-induced autophagy in mouse embryonic fibroblasts (42). This could be one of the reason why, despite ULK1 downregulation by miR-489, collective effect is observed at maturation stage. LAPTM4B is a lysosomal transmembrane protein that has been shown to affect maturation of autophagosome. Studies show that knockdown of LAPTM4B results lysosomal membrane permeabilization (43) and increases lysosomal pH which inhibits lysosome and autophagosome fusion (36). This results in accumulation of autophagosome with simultaneous deposition of LC3B-II and p62. Together, these results indicate that miR-489 inhibits autophagy mainly through inhibiting autophagosome maturation.
Autophagy has a contentious role in tumorigenesis. Loss of autophagy promotes oxidative stress, activation of the DNA damage response and genome instability which are known cause of cancer initiation and progression. (44). However, autophagy has also been seen to promote tumor cell survival. Being a pro-survival mechanism, it allows cell survival under stressful conditions such as starvation and hypoxia. Due to excessive proliferation rate, these conditions are fairly common among tumors (45). Autophagy, in such established tumors, promotes survival of cancer cells and inhibition of autophagy leads to tumor growth inhibition. Furthermore, genotoxic stress induced by various chemotherapeutic agents also induces cytoprotective autophagy. Several studies have reported that inhibiting autophagy in such cases leads to tumor growth inhibition and chemo-sensitization. However, impact of autophagy inhibition on tumor cell survival and chemo-sensitization depends on at which stage autophagy in blocked. Studies show that blocking autophagy at later stage have pronounced cytotoxic effect and sensitization. In fact, few studies suggested that blocking early stage autophagy might promote survival and attenuate effect of chemo-therapeutic agent. One such report explained that this contrasting response of autophagy inhibition at different stages is because this toxic effect is attributed to autophagosome accumulation (22). Since, inhibition of early stage of autophagy inhibits autophagosome synthesis, it alleviates cytotoxicity induced by autophagosome accumulation. Consistent with this result, blocking early stage of autophagy by 3-MA and siATG5 blunted miR-489 induced cell death. These results suggest application of miR-489 in tumors with increased autophagic flux or autophagy dependency. Few studies demonstrate higher basal autophagic flux and increased dependency of TNBC cells on autophagy for their survival (46). Intriguingly, we observed significant cytotoxic effect of miR-489 in TNBC cells compared to cell lines of other breast cancer subtypes. TNBC cell line MDA-MB-231 is sensitive to autophagic induction and additionally possesses the ability to proliferate following nutrient deprivation (21). In the current study, we also observed MDA-MB-231 cells are more resistant to starvation compared to estrogen positive T47D cell line and HER2 positive HCC1954 cell line. miR-489 under these conditions can provide useful therapeutic approach to target such resistant and aggressive tumor cells. Interestingly, several groups including us, have shown that miR-489 is significantly down regulated in TNBC patients and cell lines, which might contribute to increased autophagic flux observed in this subtype (4,25). All these evidences suggest a notion that loss of miR-489 in this subtype might play an important role for their survival. It will be interesting to explore if miR-489 mediated severe cytotoxic effect on these cells is specifically due to their increased sensitivity to autophagy inhibition or if miR-489 shuts down other pro-tumorigenic pathways aberrantly overexpressed in this subtype.
miR-489 has long been reported in drug resistance. (12,25,47,48). Various drug resistant cell lines, such doxorubicin resistant cell line, tamoxifen resistant cell line and cisplatin resistant cell line, display significantly reduced miR-489 expression. A study showed that MCF7/ADR cells have significantly higher basal autophagy level and inhibition of autophagy leads to sensitization of these resistant cells to doxorubicin(49). Interestingly, restoration of miR-489 leads to significant sensitization towards doxorubicin. A study reported that miR-489 sensitizes MCF7/ADR cells by inhibiting epithelial to mesenchymal transition (50) through SMAD3 downregulation (48). Several studies showed cells that underwent EMT require autophagy activation to survive during the metastatic spreading(51). Furthermore TGF-β, a master regulator of SMAD3 and EMT, has also been shown to induce autophagy and knockdown of SMAD3 attenuates TGF-β induced autophagy (52). However, on the other side autophagy has also been shown to contrast the activation of the EMT mainly by selectively destabilizing crucial mediators of this process (51). It will be very interesting to explore cross talk between miR-489, autophagy and EMT to gain better understanding of miR-489’s role as therapeutic sensitizer.
Restoration of miR-489 in some of these cell lines showed sensitization to specific chemotherapeutic agent (12,25,47,48). Many mechanisms exist for drug resistance. The fact that miR-489 sensitizes cells to multiple chemotherapeutic agents indicates that miR-489 regulates several pathways involved in drug resistance. One study showed miR-489 can sensitize doxorubicin resistant cell line via targeting SPIN1-PI3K-Akt pathway (25). In the current study, we establish miR-489-LAPTM4B-Autophagy pathway as an additional mechanism involved in doxorubicin resistance. Our findings are consistent with previous studies that show role of LAPTM4B in increased autophagy and anthracycline resistance. Our clinical analysis found patient with 8q22 amplification, who have higher miR-489 expression, have better survival (p-value =0.0005) than patient with lower miR-489 expression. These data indicate potential clinical significance of miR-489 in 8q22 amplified patients, where it can mitigate effects of amplified LAPTM4B and sensitizes patients to doxorubicin. Together, these data suggest a possible application of using miR-489 as a potential biomarker or therapeutic sensitizer in a defined subgroup of resistant breast cancer patients.
Supplementary Material
Implications.
These findings expand the understanding of miR-489-mediated tumor suppression and chemo-sensitization in and suggest a strategy for using miR-489 as a therapeutic sensitizer in a defined subgroup of resistant breast cancer patients.
Acknowledgement:
The authors thank Dr. Thomas Blom and Dr. Elina Ikonen for providing LAPTM4B-mCherry and pcDNA-3xFLAG-LAPTM4B vectors, Dr. Jayanta Debnath for mCherry-EGFP-LC3B (Addgene # 22418) vector and Dr. Do-Hyung Kim for HA-ULK1 vector (Addgene # 31963).
Funding
This work was partially supported by the NIH grant (5R01 CA178386-04) and the USC ASPIRE-1 grant to HC, the NIH grants (1R15CA188847-01A1 and 1R01AG054839-01A1) to PX, the USC ASPIRE post-doctoral fellowship to SL and the USC SPARC graduate fellowship to MS.
Footnotes
Disclosure of potential conflicts of interest
The authors have no potential conflict of interest to disclosure.
References
- 1.Adams BD, Wali VB, Cheng CJ, Inukai S, Booth CJ, Agarwal S, et al. miR-34a Silences c-SRC to Attenuate Tumor Growth in Triple-Negative Breast Cancer. Cancer research 2016;76(4):927–39 doi 10.1158/0008-5472.CAN-15-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esparza-Lopez J, Escobar-Arriaga E, Soto-Germes S, Ibarra-Sanchez MJ. Breast Cancer Intra-Tumor Heterogeneity: One Tumor, Different Entities. Revista de investigacion clinica; organo del Hospital de Enfermedades de la Nutricion 2017;69(2):66–76. [DOI] [PubMed] [Google Scholar]
- 3.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer research 2005;65(16):7065–70 doi 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 4.Patel Y, Shah N, Lee JS, Markoutsa E, Jie C, Liu S, et al. A novel double-negative feedback loop between miR-489 and the HER2-SHP2-MAPK signaling axis regulates breast cancer cell proliferation and tumor growth. Oncotarget 2016;7(14):18295–308 doi 10.18632/oncotarget.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y, Liu J, Huang Y, Liu D, Zhang G, Kan H. microRNA-489 Plays an Anti-Metastatic Role in Human Hepatocellular Carcinoma by Targeting Matrix Metalloproteinase-7. Translational oncology 2017;10(2):211–20 doi 10.1016/j.tranon.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Q, Yang G, Qian Y. Loss of MicroRNA-489–3p promotes osteosarcoma metastasis by activating PAX3-MET pathway. Molecular carcinogenesis 2017;56(4):1312–21 doi 10.1002/mc.22593. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Qu W, Jiang Y, Sun Y, Cheng Y, Zou T, et al. miR-489 Suppresses Proliferation and Invasion of Human Bladder Cancer Cells. Oncology research 2016;24(6):391–8 doi 10.3727/096504016X14666990347518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao Y, Han T, Zhang T, Ma C, Sun C. LncRNA CHRF-induced miR-489 loss promotes metastasis of colorectal cancer via TWIST1/EMT signaling pathway. Oncotarget 2017. doi 10.18632/oncotarget.16850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, et al. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC). British journal of cancer 2010;103(6):877–84 doi 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Z, Cai L, Li R, Zheng J, Wu H, Yang X, et al. Down-regulation of miR-489 contributes into NSCLC cell invasion through targeting SUZ12. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 2015;36(8):6497–505 doi 10.1007/s13277-015-3340-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Ji S, Ma F, Ma Q, Lu X, Chen X. miR-489 acts as a tumor suppressor in human gastric cancer by targeting PROX1. American journal of cancer research 2016;6(9):2021–30. [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Xiao Z, Zhang H, Wang K, Liu W, Hao Q. MiR-489 modulates cisplatin resistance in human ovarian cancer cells by targeting Akt3. Anti-cancer drugs 2014;25(7):799–809 doi 10.1097/CAD.0000000000000107. [DOI] [PubMed] [Google Scholar]
- 13.Fulda S, Kogel D. Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene 2015;34(40):5105–13 doi 10.1038/onc.2014.458. [DOI] [PubMed] [Google Scholar]
- 14.Eskelinen EL. The dual role of autophagy in cancer. Current opinion in pharmacology 2011;11(4):294–300 doi 10.1016/j.coph.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Pan B, Feng B, Chen Y, Huang G, Wang R, Chen L, et al. MiR-200b regulates autophagy associated with chemoresistance in human lung adenocarcinoma. Oncotarget 2015;6(32):32805–20 doi 10.18632/oncotarget.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140–5p mediated suppression of Smad2 and autophagy. Oncotarget 2015;6(23):19735–46 doi 10.18632/oncotarget.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei R, Cao G, Deng Z, Su J, Cai L. miR-140–5p attenuates chemotherapeutic drug-induced cell death by regulating autophagy through inositol 1,4,5-trisphosphate kinase 2 (IP3k2) in human osteosarcoma cells. Bioscience reports 2016;36(5) doi 10.1042/BSR20160238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel Y, Lee JS, Chen H. Clinicopathological Analysis of miRNA Expression in Breast Cancer Tissues by Using miRNA In Situ Hybridization. J Vis Exp 2016(112) doi 10.3791/53928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12(1):1–222 doi 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010;140(3):313–26 doi 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YI, Libby EF, Lewis MJ, Liu J, Shacka JJ, Hurst DR. Increased autophagic response in a population of metastatic breast cancer cells. Oncology letters 2016;12(1):523–9 doi 10.3892/ol.2016.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Button RW, Roberts SL, Willis TL, Hanemann CO, Luo S. Accumulation of autophagosomes confers cytotoxicity. The Journal of biological chemistry 2017;292(33):13599–614 doi 10.1074/jbc.M117.782276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi J, Jo M, Lee E, Choi D. Induction of apoptotic cell death via accumulation of autophagosomes in rat granulosa cells. Fertility and sterility 2011;95(4):1482–6 doi 10.1016/j.fertnstert.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Geng Y, Kohli L, Klocke BJ, Roth KA. Chloroquine-induced autophagic vacuole accumulation and cell death in glioma cells is p53 independent. Neuro-oncology 2010;12(5):473–81 doi 10.1093/neuonc/nop048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Wang YW, Xing AY, Xiang S, Shi DB, Liu L, et al. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. The Journal of pathology 2016;239(4):459–72 doi 10.1002/path.4743. [DOI] [PubMed] [Google Scholar]
- 26.Chittaranjan S, Bortnik S, Dragowska WH, Xu J, Abeysundara N, Leung A, et al. Autophagy inhibition augments the anticancer effects of epirubicin treatment in anthracycline-sensitive and -resistant triple-negative breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2014;20(12):3159–73 doi 10.1158/1078-0432.CCR-13-2060. [DOI] [PubMed] [Google Scholar]
- 27.Kim DG, Jung KH, Lee DG, Yoon JH, Choi KS, Kwon SW, et al. 20(S)-Ginsenoside Rg3 is a novel inhibitor of autophagy and sensitizes hepatocellular carcinoma to doxorubicin. Oncotarget 2014;5(12):4438–51 doi 10.18632/oncotarget.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Li G, Zheng Y, Shen HM, Hu X, Ming QL, et al. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy 2015;11(8):1259–79 doi 10.1080/15548627.2015.1056970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shingu T, Fujiwara K, Bogler O, Akiyama Y, Moritake K, Shinojima N, et al. Inhibition of autophagy at a late stage enhances imatinib-induced cytotoxicity in human malignant glioma cells. International journal of cancer 2009;124(5):1060–71 doi 10.1002/ijc.24030. [DOI] [PubMed] [Google Scholar]
- 30.Kanematsu S, Uehara N, Miki H, Yoshizawa K, Kawanaka A, Yuri T, et al. Autophagy inhibition enhances sulforaphane-induced apoptosis in human breast cancer cells. Anticancer research 2010;30(9):3381–90. [PubMed] [Google Scholar]
- 31.Li C, Liu Y, Liu H, Zhang W, Shen C, Cho K, et al. Impact of autophagy inhibition at different stages on cytotoxic effect of autophagy inducer in glioblastoma cells. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 2015;35(4):1303–16 doi 10.1159/000373952. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Li Y, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nature medicine 2010;16(2):214–8 doi 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Zhang Q, Tian R, Wang Q, Zhao JJ, Iglehart JD, et al. Lysosomal transmembrane protein LAPTM4B promotes autophagy and tolerance to metabolic stress in cancer cells. Cancer research 2011;71(24):7481–9 doi 10.1158/0008-5472.CAN-11-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Ronde JJ, Lips EH, Mulder L, Vincent AD, Wesseling J, Nieuwland M, et al. SERPINA6, BEX1, AGTR1, SLC26A3, and LAPTM4B are markers of resistance to neoadjuvant chemotherapy in HER2-negative breast cancer. Breast cancer research and treatment 2013;137(1):213–23 doi 10.1007/s10549-012-2340-x. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Wei XH, Pan YP, Li HC, Yang H, He QH, et al. LAPTM4B: a novel cancer-associated gene motivates multidrug resistance through efflux and activating PI3K/AKT signaling. Oncogene 2010;29(43):5785–95 doi 10.1038/onc.2010.303. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Iglehart JD, Richardson AL, Wang ZC. The amplified cancer gene LAPTM4B promotes tumor growth and tolerance to stress through the induction of autophagy. Autophagy 2012;8(2):273–4 doi 10.4161/auto.8.2.18941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng Y, Wang L, Chen D, Chang Y, Zhang M, Xu JJ, et al. LAPTM4B: an oncogene in various solid tumors and its functions. Oncogene 2016;35(50):6359–65 doi 10.1038/onc.2016.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013;497(7449):378–82 doi 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 39.Gray J, Druker B. Genomics: the breast cancer landscape. Nature 2012;486(7403):328–9 doi 10.1038/486328a. [DOI] [PubMed] [Google Scholar]
- 40.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. The Journal of cell biology 2008;181(3):497–510 doi 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Molecular and cellular biology 2009;29(1):157–71 doi 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee EJ, Tournier C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy 2011;7(7):689–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blom T, Li S, Dichlberger A, Back N, Kim YA, Loizides-Mangold U, et al. LAPTM4B facilitates late endosomal ceramide export to control cell death pathways. Nature chemical biology 2015;11(10):799–806 doi 10.1038/nchembio.1889. [DOI] [PubMed] [Google Scholar]
- 44.White E, Mehnert JM, Chan CS. Autophagy, Metabolism, and Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21(22):5037–46 doi 10.1158/1078-0432.CCR-15-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu YL, Jahangiri A, Delay M, Aghi MK. Tumor cell autophagy as an adaptive response mediating resistance to treatments such as antiangiogenic therapy. Cancer research 2012;72(17):4294–9 doi 10.1158/0008-5472.CAN-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maycotte P, Gearheart CM, Barnard R, Aryal S, Mulcahy Levy JM, Fosmire SP, et al. STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer research 2014;74(9):2579–90 doi 10.1158/0008-5472.CAN-13-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. The Journal of biological chemistry 2008;283(44):29897–903 doi 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang L, He D, Yang D, Chen Z, Pan Q, Mao A, et al. MiR-489 regulates chemoresistance in breast cancer via epithelial mesenchymal transition pathway. FEBS letters 2014;588(11):2009–15 doi 10.1016/j.febslet.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 49.Guo B, Tam A, Santi SA, Parissenti AM. Role of autophagy and lysosomal drug sequestration in acquired resistance to doxorubicin in MCF-7 cells. BMC cancer 2016;16(1):762 doi 10.1186/s12885-016-2790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borst P, Schinkel AH, Smit JJ, Wagenaar E, Van Deemter L, Smith AJ, et al. Classical and novel forms of multidrug resistance and the physiological functions of P-glycoproteins in mammals. Pharmacology & therapeutics 1993;60(2):289–99. [DOI] [PubMed] [Google Scholar]
- 51.Gugnoni M, Sancisi V, Manzotti G, Gandolfi G, Ciarrocchi A. Autophagy and epithelial-mesenchymal transition: an intricate interplay in cancer. Cell death & disease 2016;7(12):e2520 doi 10.1038/cddis.2016.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR, et al. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer research 2009;69(23):8844–52 doi 10.1158/0008-5472.CAN-08-4401. [DOI] [PubMed] [Google Scholar]
- 53.Li SD, Chono S, Huang L. Efficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. Journal of controlled release : official journal of the Controlled Release Society 2008;126(1):77–84 doi 10.1016/j.jconrel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Molecular pharmaceutics 2006;3(5):579–88 doi 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.