Abstract
G309 or S310 mutations on the HER2 extracellular domain II induce receptor activation. Clinically, S310F is most frequent among HER2 extracellular domain mutations and patients with the S310F mutation without HER2 amplification responded to trastuzumab with or without the pertuzumab combination. However, the ability of S310F mutant to form homodimers or heterodimers with wild-type HER2 and other HER receptors, or their reactivity to trastuzumab and pertuzumab treatments, has not been reported. We overexpressed S310F as well as G309A, G309E and S310Y HER2 mutants and tested their reactivity to trastuzumab and pertuzumab. All mutants reacted to trastuzumab, but S310F mutant did not react to pertuzumab along with S310Y or G309E mutants. Thereafter, we tested the effects of trastuzumab and pertuzumab on 5637 cell line expressing both wild-type HER2 and S310F mutant. The ligand-independent HER2 homodimerization blocking antibody, trastuzumab, did not inhibit the activation of the HER2 receptor, suggesting that the S310F HER2 mutant did not form homodimers or heterodimers with wild-type HER2. Because 5637 cells overexpressed the EGFR, the effects of cetuximab and gefitinib were determined, and both inhibited the activation of HER2 and significantly reduced cell growth. Because pertuzumab did not inhibit the phosphorylation of HER2 while it bound to wild-type HER2, EGFR-mediated phosphorylation is expected to occur on the S310F mutant. To confirm whether the S310F mutant HER2 retained its affinity to the EGFR, single molecule interaction analyses using TIRF microscopy were performed, which showed that S310F mutant successfully formed complexes with EGFR. In conclusion, HER2 S310F mutant can form an active heterodimer with the EGFR and it can be inhibited by cetuximab, but not by trastuzumab in combination with pertuzumab.
Keywords: HER2, Mutation, Pertuzumab, TIR microscope, EGFR, Heterodimerization
1. Introduction
HER2/ErbB2/Neu is a member of the human epidermal growth factor receptor (EGFR, HER) family of homologous transmembrane receptor tyrosine kinases. The HER family is composed of four receptors (EGFR and HER2–4/ErbB1–4) and a number of variants generated by alternative slicing. There are 11 types of ligands reactive to HER family receptors, including those generated by alternative splicing [1]. The roles for these receptors and their ligands have been well described in many types of human cancers, including breast, colon, pancreatic, ovarian, brain, and lung cancers [2]. Binding of the ligand to EGFR or HER3/4 induces conformational changes in the proteins, facilitating receptor dimerization, which results in transphosphorylation of tyrosine residues in the carboxy tail [3]. The phosphotyrosines are docking sites for the recruitment of downstream signaling proteins. Human epidermal growth factor receptor 2 (HER2) has no known ligand but forms heterodimers with other EGFR family members and activates downstream signaling through the phosphoinositide 3-kinase/AKT and MAPK pathways [4].
Amplification/overexpression of HER2 is associated with cell transformation and oncogenesis [5], is observed in 20–25% of breast cancers, and is associated with poor survival [6]. In breast cancer cells with HER2 gene amplification, HER2 receptors exist on the cell surface as monomers, homodimers and heterodimers. The therapeutic antibody trastuzumab, binding to extracellular domain IV, cannot block ligand-induced HER2 heterodimers and has preferential activity against breast cancers driven by HER2 homodimers [7,8]. Meanwhile, pertuzumab binds to extracellular domain II, sterically blocks a binding pocket necessary for receptor dimerization, and inhibits HER2 association with its partner receptors [9]. This property may explain why pertuzumab, unlike trastuzumab, is effective against a broad range of cancers which do not express HER2 at high levels [10,11]. Trastuzumab and pertuzumab could colocalize to HER2 while not augmenting the binding of each other [12]. Moreover, the combination of these two antibodies achieved significant improvement in the survival of HER2 overexpressing breast cancer patients. Antibody drug conjugates (trastuzumab emtansine), and small molecule tyrosine kinase inhibitors (lapatinib, erlotinib, and neratinib) have also dramatically improved the outcomes of HER2-positive cancer patients [13,14,15,16]. Besides the amplification/overexpression of HER2, somatic HER2 gene mutations have been detected in a range of human cancer types, including prostate neuroendocrine cancer, metastatic cutaneous squamous cell carcinoma, and bladder cancer [17,18]. Preclinical data suggest that functionally activating HER2 mutations may drive and maintain cancers in a manner analogous to HER2 gene amplification, and those HER2 mutations may similarly confer changes in sensitivity to HER2-directed drugs [19].
There are more than 20 extracellular domain mutations detected in human cancer tissues (Table 1) [8,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38]. Furthermore, 26 additional extracellular domain mutations have been detected in various human cancer cell lines (Table 2). G309 and S310 mutations of HER2 domain II are well-known activating mutations. Previous studies showed that the G309 mutation leaves unpaired cysteine residues (C299–C311), forming reduction-sensitive dimers [20]. Likewise, the S310 mutation promotes noncovalent dimerization between neighboring molecules, promoting hyperphosphorylation of HER2 [20]. The Ba/F3 cells transfected with an expression vector encoding the S310F HER2 mutant showed hyperphosphorylated HER2 while cells overexpressing G309E mutant did not. However, Ba/F3 cells expressing either G309E or S310F HER2 mutant showed enhanced proliferation compared to cells transfected with wild-type HER2 and were sensitive to trastuzumab, lapatinib, afatinib, and neratinib [20]. Clinically, S310F is the most frequently found mutation in the HER2 extracellular domain [39]. Two HER2-nonamplified breast cancer patients with S310F mutation were successfully treated with the trastuzumab alone or in combination with pertuzumab [19,31]. G309 and S310 residues are located near the binding epitope for pertuzumab, obscuring the reactivity of the antibody [9]. However, the reactivity of the combination of pertuzumab and trastuzumab to the S310F mutant has not been previously reported. In this study, we prepared recombinant proteins corresponding to all reported G309 and S310 HER2 mutants and analyzed their interactions in Enzyme-linked immunosorbent assay (ELISA) with both trastuzumab and pertuzumab. We also used 5637 cells, an EGFR-amplified bladder cancer cell line expressing both S310F HER2 mutant and wild-type HER2 to analyze the downstream signaling of HER2 mutant and found that the S310F HER2 mutant efficiently formed heterodimers with EGFR, which could not be inhibited by the combination of trastuzumab and pertuzumab. And using single molecule level total internal reflection (TIR) microscopy, we confirmed that S310 mutant can form a heterodimer with EGFR at the molecular level.
Table 1.
Human epidermal growth factor receptor 2 (HER2) extracellular domain mutations reported in previous studies from patient samples.
| Protein Change | Pfam Domain [8] | Tumor Type | Impact | HER2 | Phosphorylation | Reference |
|---|---|---|---|---|---|---|
| L12R | - | Breast Cancer | ND | Negative | ND * | [35] |
| A20T | Lung cancer | ND | ND | ND | [24] | |
| L49H | Lung cancer | ND | Negative | ND | [21] | |
| L49H | Glioblastoma | ND | ND | ND | [20] | |
| E139D | Receptor L | Breast Cancer | ND | Negative | ND | [35] |
| E139G | Breast Cancer | ND | Negative | ND | [35] | |
| R157W | Bladder cancer (MPUC) | ND | Negative | ND | [32] | |
| T216S | Furin-like | Lung cancer | ND | Negative | ND | [21] |
| T216S | Glioblastoma | ND | ND | High | [20] | |
| I263T | Colorectal cancer | ND | Negative | ND | [36] | |
| G309A | Breast Cancer | Activating | Negative | ND | [27] | |
| G309E | Breast Cancer | Activating | Positive | ND | [24] | |
| S310F | Gastric cancer | Activating | ND | High | [38] | |
| S310F(2) | Breast Cancer | Negative | High | [26] | ||
| S310F | Lung cancer | Negative | High | [20] | ||
| S310F | Breast Cancer | Negative | High | [25] | ||
| S310F(2) | Colorectal cancer | Negative | High | [37] | ||
| S310F | Breast Cancer | Negative | High | [28] | ||
| S310F | Lung cancer | Positive | High | [22] | ||
| S310F | Breast Cancer | Negative | High | [29] | ||
| S310F(4) | Bladder cancer (MPUC) | Negative | High | [32] | ||
| S310F | Adnexal cancer | Negative | High | [33] | ||
| S310F | Breast Cancer | Negative | High | [29] | ||
| S310F(2) | Breast Cancer | Positive | High | [30] | ||
| S310F | Lung cancer | ND | High | [23] | ||
| S310F | Breast Cancer | Negative | High | [31] | ||
| S310F(4) | Breast Cancer | Positive | ND | [35] | ||
| S310Y | Lung cancer | Activating | ND | High | [24] | |
| S310Y | Lung cancer | ND | High | [20] | ||
| S310Y | Colorectal cancer | Positive | High | [21] | ||
| S310Y | Bladder cancer (MPUC) | Negative | High | [20] | ||
| C311R and E321G | Lung cancer | ND | Negative | ND | [21] | |
| C311R | Glioblastoma | ND | ND | Low | [20] | |
| N319D | Lung cancer | ND | Negative | ND | [21] | |
| N319D | Glioblastoma | ND | ND | ND | [20] | |
| E321G | Glioblastoma | ND | ND | Low | [20] | |
| D326G and C334S | Lung cancer | ND | Negative | ND | [21] | |
| D326G | Glioblastoma | ND | ND | Low | [20] | |
| C334S | Glioblastoma | ND | ND | ND | [20] | |
| A466T | Receptor L | Colorectal cancer | ND | Negative | ND | [36] |
| A466V | Breast Cancer | Activating | Negative | High | [35] | |
| C515R | Growth factor receptor | Breast Cancer | Activating | Negative | High | [35] |
| T526A | Breast Cancer | Activating | Negative | High | [35] |
* ND = no data available.
Table 2.
HER2 extracellular domain mutations reported in cell lines.
| Cell Line | Protein Change | Pfam Domain [8] | Lineage | Impact | Phosphorylation |
|---|---|---|---|---|---|
| SW1271 | S22N | - | Lung | ND * | ND |
| HEC59 | R100W | Receptor L | Endometrium | ND | ND |
| HEC108 | T166M | Endometrium | ND | ND | |
| SBC1 | Q178H | Lung | ND | ND | |
| MOLT16 | R217H | Furin-like | Haematopoietic and lymphoid tissue | ND | ND |
| NALM6 | R226H | Haematopoietic and lymphoid tissue | ND | ND | |
| KM12 | P230L | Large intestine | ND | ND | |
| DSH1 | D277H | Urinary | ND | ND | |
| 5637 | S310F | Urinary | Activating | High | |
| DSH1 | Urinary | ||||
| OACM51 | Oesophagus | ||||
| HRT18 | L313I | Intestine | ND | ND | |
| NCIH2110 | N319Y | Lung | ND | ND | |
| HCC1359 | T328S | Lung | ND | ND | |
| NCIH1563 | S335C | Lung | ND | ND | |
| Jurkat | S335I | - | Haematopoietic and lymphoid tissue | ND | ND |
| OC314 | A386T | Receptor L | Ovary | ND | ND |
| OC316 | A386T | Ovary | ND | ND | |
| NCIN87 | F425L | Stomach | ND | ND | |
| NCIN87 | L436V | Stomach | ND | ND | |
| SET2 | T444S | Haematopoietic and lymphoid tissue | ND | ND | |
| M059J | W452S | Central nervous system | ND | ND | |
| ISTSL1 | R499Q | - | Lung | ND | ND |
| SUPB8 | R499W | - | Haematopoietic and lymphoid tissue | ND | ND |
| Jurkat | T479M | Receptor L | Haematopoietic and lymphoid tissue | ND | ND |
| RL952 | G518V | Growth factor receptor | Endometrium | ND | ND |
| NCIH1793 | V541M | Lung | ND | ND | |
| NCIH740 | R558M | Lung | ND | ND | |
| Karpas45 | A586G | Haematopoietic and lymphoid tissue | ND | ND |
* ND = no data available.
2. Materials and Methods
2.1. Expression and Purification of Recombinant Fusion Proteins
Genes encoding the extracellular domain of wild-type HER2, wild-type HER3 and G309A, G309E, S310F, or S310Y HER2 mutants were chemically synthesized with SfiI restriction sites at the 5′ and 3′ ends (GenScript Biotech, Jiangsu, China). After restriction digestion with SfiI (New England Biolabs, Hertfordshire, UK), the genes were ligated into pCEP4 vectors encoding either human Fc or Cκ, as described previously [40,41]. Genes encoding the scFv form of pertuzumab, trastuzumab, and cetuximab were also synthesized with SfiI restriction sites at the 5′ and 3′ ends (GenScript Biotech) and cloned into the human Cκ vector [42,43,44].
The expression vectors were then transfected into HEK293F cells (FreeStyle 293-F cells) using polyethylenimine (Sigma-Aldrich, St. Louis, MO, USA) as described previously [45]. The transfected cells were grown in FreeStyle 293 expression media (Invitrogen, Carlsbad, CA, USA) as described previously [46]. The culture supernatants were purified by affinity chromatography using either Protein A or KappaSelect resin (GE Healthcare, Buckinghamshire, UK) following the manufacturer’s instructions.
The purified recombinant proteins were subjected to 4%–12% NuPage bis-Tris gels (Invitrogen) according to the manufacturer’s instructions. After the electrophoresis, the gel was stained with Coomassie Brilliant Blue R-250 (Ameresco, Framingham, MA, USA).
2.2. ELISA
Recombinant wild-type HER2, wild-type HER3 or HER2 mutant extracellular domain-human Fc fusion proteins were coated overnight at 4 °C on the wells of microtiter plates (Corning, Corning, NY, USA). The wells were blocked with 130 μL of blocking buffer (3% bovine serum albumin (BSA) in phosphate-buffered saline (PBS)) for 1 h at 37 °C. The plates were then incubated with recombinant pertuzumab or trastuzumab scFv- human Cκ fusion proteins serially diluted four-fold in blocking buffer for 2 h at 37 °C. The plates were then washed with 150 μL of 0.05% Tween 20 in PBS (PBST) for three times. Subsequently, horseradish peroxidase (HRP) conjugated anti-human Cκ light chain antibody (1:5000; Millipore, Hayward, CA, USA) was added to each well. After incubation for 1 h at 37 °C, the plates were washed. Finally, 50 μL of 3,3′,5,5′-tetramethyl benzidine substrate solution (TMB) substrate was added and optical density was measured at 650 nm (LabSystems Diagnostics Oy, Vantaa, Finland).
2.3. Cell Culture
The 5637and AU565 cells were obtained from the Korean Cell Line Bank (Seoul, Republic of Korea). The cells were grown in RPMI-1640 media (Welgene, Seoul, Republic of Korea) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Gibco, Waltham, MA, USA) and 1% penicillin-streptomycin (1000 U/mL) (Thermo Fisher Scientific, Gibco).
2.4. Sequencing of Amplified HER2 Gene Fragments of 5637 Cell
Total RNA was isolated from the 5637 cells using TRIzol reagent (Invitrogen) as previously described [47]. After cDNA was synthesized using a SuperScript III First-Strand Synthesis system (Invitrogen), the HER2 gene fragment encoding from N302 residue to R340 residue was amplified using specific primer sets (HER2 forward: 5′-GCCTCCACTTCAACCACAGTGGC-3′ and HER2 reverse: 5′-CTGTGATCTCTTCCAGAGTCTCAAAC-3′). The PCR conditions were as follows: Preliminary denaturation at 95 °C for 7 min, followed by 25 cycles of 30 s at 95 °C, 30 s at 54 °C, and 1 min at 72 °C. The reaction was ended with an extension step for 5 min at 72 °C. After agarose gel electrophoresis, the amplified DNA was extracted using the Qiagen Gel Extraction Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany) and subjected to Sanger sequencing (Macrogen, Seoul, Korea).
2.5. Flow Cytometry Analysis
The 5637 and AU565 cells (3 × 105 cells/well, Corning) were resuspended in 100 µL of flow cytometry buffer (1% BSA and 0.02% sodium azide in PBS), then incubated with cetuximab, pertuzumab, or trastuzumab scFv- human Cκ fusion proteins at a final concentration of 1 μM for 1 h at 37 °C. The cells were then washed twice with 1% (w/v) BSA in PBS and incubated with FITC-conjugated anti-human Cκ light chain antibody (Thermo Fisher Scientific) for 40 min at 37 °C. After washing with flow cytometry buffer, the fluorescence intensity of cells was measured using a FACS Canto II (BD Bioscience, Heidelberg, Germany) and analyzed with FlowJo data analysis software, version 8.8.4 (Threestar, OR, USA). Non-transfected HEK293T cell and cells transfected with EGFR-mCherry and HER2-eGFP bicistronic expression vector were used as control cells and were analyzed in the same manner as above.
2.6. Immunoprecipitation and Immunoblot Analyses
The AU565 and 5637 cells (1 × 107 cells) were lysed in 100 μL of cold RIPA buffer (150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris-HCl, and 2 mM EDTA at pH 7.5) containing a protease inhibitor cocktail (Roche, Basel, Switzerland). Subsequently, 500 μL of the cell lysate was incubated with either recombinant trastuzumab or pertuzumab scFv-human Cκ fusion protein with a final concentration of 100 μg/mL at 4 °C overnight with rotation. Then, 10 μL of KappaSelect resin (GE Healthcare) was added to the lysate, followed by incubation at 4°C for 4 h with gentle rotations. After centrifugation for 3 min at 1200× g, the resin and supernatant were collected. The cell lysate, resin, and supernatant representing the same number of the cells were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as described above and transferred onto nitrocellulose membranes (Whatman, Dassel, Germany) using a transfer system as previously described [48]. Thereafter, the membrane was blocked with 5% skim milk (BD Biosciences, CA, USA) followed by incubation with rabbit anti-HER2 antibody (Cell Signaling Technology, Danvers, MA, USA) at 4 °C overnight and with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:5000, 111-035-008; Jackson Immuno Research Laboratory, West Grove, PA, USA). After several washes with Tris-buffered saline and Tween [10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% (v/v) Tween 20], the protein bands were visualized using Super Signal Pico West chemiluminescent substrate (#34080; Thermo Fisher Scientific) and a gel doc EZ system (Bio-Rad, Hercules, CA, USA).
2.7. Cell Viability Assay and Immunoblotting
The 5637 cells were seeded at a density of 5.5–10 × 103/well in 96-well culture plates. After incubation for 24 h, cells were treated with 1 μM of cetuximab (Merck, Palo Alto, CA, USA), pertuzumab (Roche), trastuzumab (Roche), gefitinib (AstraZeneca, Cambridge, UK), or lapatinib (GlaxoSmithKline, Brentford, UK). After 96 h, the number of viable cells was determined using a Premix WST-1 kit (Takara, Kyoto, Japan) following the manufacturer’s protocol. For immunoblot analysis, 5637 cells were lysed in ice-cold RIPA buffer containing a protease inhibitor cocktail (Roche) and phosphatase inhibitor (Roche). The cell lysates were cleared by centrifugation for 10 min at 13,000× g and the amount of protein in the supernatants was determined by a BCA assay (Pierce Biotechnology, Waltham, MA, USA). The proteins in the supernatants were separated by SDS-PAGE using 4%–12% bis-Tris gels (Invitrogen) as described above and transferred onto a nitrocellulose membrane. The membrane was blocked by pre-incubation in 5% BSA/0.2% TBST at room temperature for 30 min and then incubated with anti-EGFR antibody (Cell Signaling Technology), anti-phospho-EGFR (Tyr 1068) antibody (Cell Signaling Technology), anti-HER2 antibody (Cell Signaling Technology), anti-phospho-HER2 (Tyr 1221/1222) antibody (Cell Signaling Technology), anti-PARP (Cell Signaling Technology), anti-cleaved PARP (Cell Signaling Technology), or anti-β-actin antibody (Cell Signaling Technology) for overnight at 4 °C. After washing three times with 0.2% TBST, the membrane was incubated with either HRP-conjugated goat anti-rabbit IgG Fc antibody (#31463; Thermo Fisher Scientific) or HRP-conjugated goat anti-mouse IgG antibody (sc-2005; Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at room temperature. The membrane was washed three times with TBST, and the bound antibody was visualized by addition of Super Signal Pico West chemiluminescent substrate (#34080; Thermo Fisher Scientific) following the manufacturer’s instructions. The Image Lab program (BioRad, CA, USA) was used to determine the intensity of bands.
2.8. Single-Molecular Interaction Analysis Using Total Internal Reflection Fluorescence (TIRF) Microscopy
Single-molecular interaction analysis was performed as described in a previous study with appropriate modifications [49]. Genes encoding fusion proteins composed of the extracellular domain of the EGFR fused with mCherry protein (EGFR-mCherry) and that of wild-type HER2 fused with eGFP (HER2-eGFP) were ligated into a bicistronic expression vector. Another expression vector was prepared with the S310F mutant (S310F HER2-eGFP) replacing wild-type HER2.Then, these vectors were transfected into HEK293T cells (FreeStyle 293-T cells). After culture at 37 °Cwith 5% CO2 for 1 day, the cells were dissolved in lysis buffer (1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 10% glycerol) with protease (P8340; Sigma-Aldrich) and phosphatase (P5726; Sigma-Aldrich) inhibitor cocktail. After centrifugation at 15,000× g for 10 min at 4 °C, the supernatant was collected. Recombinant mCherry (4999-100; Biovision, Milpitas, CA, USA) and eGFP (4993-100; Biovision) protein standards were used to determine the concentration of fluorescently tagged proteins in the cell lysates. The fluorescence levels for recombinant eGFP and mCherry proteins of known concentrations (5–25 nM, five different concentration points) were measured using a fluorometer (Enspire 2300; Perkin-Elmer, San Jose, CA, USA) and were used to construct a calibration curve.
Detailed procedures of the flow chamber construction are described in previous studies [50,51]. The flow chamber was washed twice with 200 μL of PBS. Afterwards, 50 μL of NeutrAvidin solution (A2666; Invitrogen) was added to the flow chamber and was incubated for 5 min at room temperature. Subsequently, the flow chambers were washed twice followed by addition of diluted biotinylated polyclonal RFP antibody (34771; Abcam, Cambridge, UK) for 5 min at room temperature. This antibody was reported to be reactive to all RFP variants from Discosoma, including mCherry. After the flow chambers were washed again, cell lysates were added and incubated for 10 min at room temperature. Finally, the flow chambers were washed with 0.1% lysis buffer/PBS. EGFR/HER2 heterodimers were then detected with a TIRF microscope equipped with a 488-nm laser for eGFP detection. The images were recorded at 0.1 s per frame, and 20 frames were recorded. To obtain a time-averaged image, three frames were averaged. EGFP spots were characterized from these time-averaged images. To obtain the intensity of eGFP spots, 10 time-averaged images were used.
3. Results
3.1. The Recombinant S310F Mutant Is Not Reactive to Pertuzumab but Binds to Trastuzumab
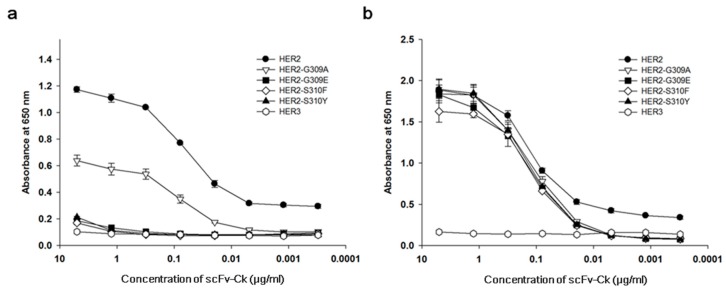
A construct encoding the S310F HER2 extracellular domain fused to a human Fc domain of immunoglobulin heavy chain was prepared and cloned into a mammalian expression vector. For comparison, the expression vectors encoding the extracellular domains of all the other G309 and S310 mutants reported previously (G309A, G309E, S310Fand S310Y HER2 mutants) and those of wild-type HER2 and HER3 were also prepared. After transfection, the recombinant fusion proteins were purified from the culture supernatant using an affinity column reactive to human Fc. The extracellular domain of four mutants and wild-type HER2 was also prepared as a fusion protein of human Cκ through cloning into a mammalian expression vector, transfection and purification using an affinity column reactive to human Cκ from the culture supernatant. To check the purity, the recombinant fusion proteins were subjected to SDS-polyacrylamide gel electrophoresis and the gel was stained. No band except the fusion protein was visualized. In non-reducing conditions, multimers of G309E HER2 mutant fusion proteins were visualized in both fusion protein forms (Figure S1). The purified recombinant Fc fusion proteins were then subjected to an ELISA to test reactivity to pertuzumab and trastuzumab. A microtiter plate was coated with the recombinant Fc fusion proteins, blocked, and incubated with either pertuzumab or trastuzumab expressed as recombinant scFv- human Cκ fusion protein. Then, the amount of bound antibody was determined using anti-human Cκ antibody conjugated to HRP. S310F mutants did not bind to pertuzumab but reacted with trastuzumab in a dose-dependent manner (Figure 1). Pertuzumab did not bind to the G309E, S310F, and S310Y HER2 mutants, but reacted with the S309A HER2 mutant with reduced affinity compared to that of the wild-type HER2. Trastuzumab bound to the wild-type HER2 and all G309 and S310 HER2 mutants in a dose-dependent manner.
Figure 1.
Reactivity of G309 and S310 HER2 mutants to pertuzumab and trastuzumab. Recombinant G309 and S310 mutant, wild-type HER2 or wild-type HER3 human Fc fusion protein was coated onto the wells of microtiter plates. The plate was blocked and subjected to incubation with (a) recombinant pertuzumab scFv-human Cκ fusion protein, or (b) trastuzumab scFv-human Cκ fusion protein at varying concentrations. Wild-type HER3 human Fc fusion protein was used as a negative control, because it does not bind to either antibody. The amount of bound antibody was determined using HRP-conjugated human Cκ light chain antibody and 3,3′,5,5′-tetramethyl benzidine (TMB) substrate solution.
3.2. A Bladder Cancer Cell Line, 5637, Expresses Both Wild-Type HER2 and S310F Mutant
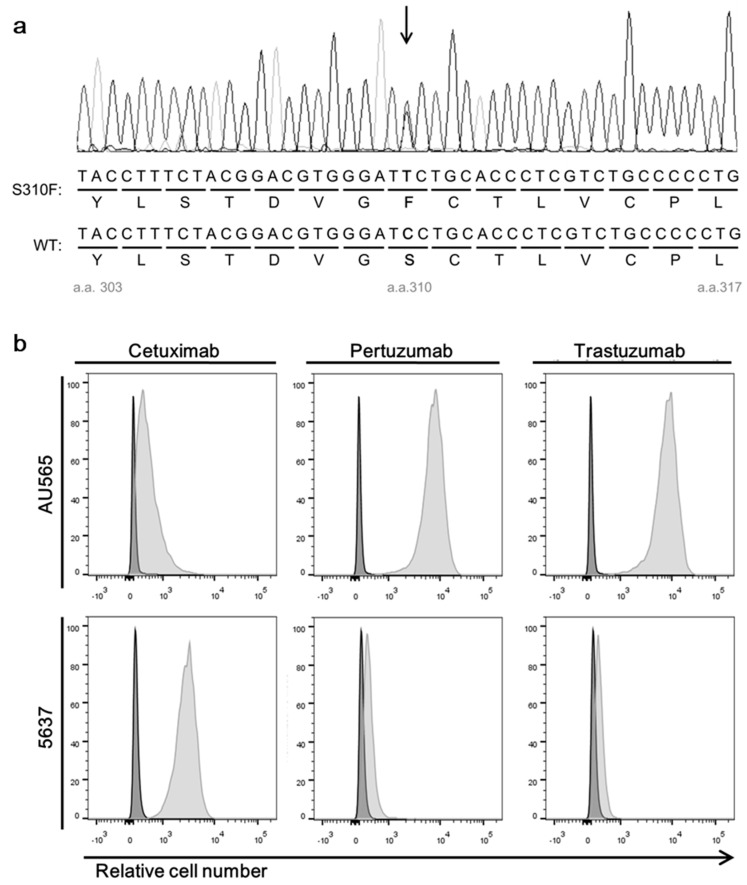
We searched literatures to identify a human cell line expressing the S310F mutant and found that bladder cancer cell line 5637 expressed the mutant. To test the allelic expression of the S310F HER2 mutant in the 5637 cell line, we extracted total RNA from the cells and prepared cDNA. A HER2 gene fragment including the S310 residue was amplified by PCR using specific primers and subjected to Sanger sequencing. A representative sequence chromatogram is shown in Figure 2. We found that both the wild-type (nucleotide C) and the mutant (nucleotide T) peaks co-existed at the corresponding nucleotide position (Figure 2a). To check the expression of the wild-type and mutant HER2 on the cell surface, we used recombinant trastuzumab and pertuzumab scFv-Cκ fusion proteins to avoid the possible non-specific interaction through Fc regions of IgG and Fc receptors on the cell and performed flow cytometric analyses. As expected, the combination of pertuzumab and the trastuzumab scFv-Cκ fusion protein reacted with the 5637 cells (Figure 2b), which proved the presence of the wild-type HER2 on the cell surface of 5637 cells.
Figure 2.
Expression of HER2 and epidermal growth factor receptor (EGFR) in 5637 and AU565 cells. (a) A representative sequence chromatogram showing the presence of two transcripts encoding wild-type HER2 and the S310F mutant in 5637 cells. (b) Flow cytometry analysis of two cancer cell lines assessing their reactivity to cetuximab, pertuzumab, and trastuzumab. The cells were incubated with individual antibody using the recombinant scFv-human Cκ fusion protein. The amount of bound antibody was determined using Allophycocyanin (APC)-labeled anti-human Cκ antibody.
To confirm the expression of the S310F HER2 mutant in 5637 cells, we used immunoprecipitation experiments. We hypothesized that if the cells expressed the S310F HER2 mutant, it would not be immunoprecipitated by pertuzumab and would remain in the cell lysate. To determine the saturable amount of pertuzumab for immunoprecipitation experiments, we screened cell lines expressing higher amounts of HER2 than 5637 cells and identifiedAU565 cells (Figure 2b). The amount of pertuzumab and trastuzumab scFv-Cκ fusion protein guaranteeing the immunoprecipitation of all of HER2 in AU565 cell lysates was then determined (Figure S2). This predetermined amount of scFv-human Cκ fusion protein was then incubated with the 5637 cell lysate, with added affinity resin reactive to Cκ. After centrifugation, pellets and supernatants were collected. Thereafter, the cell lysate, precipitated resin, and supernatant representing the same number of 5637 cells were subjected to SDS-polyacrylamide gel electrophoresis and immunoblot analysis using anti-HER2 antibody reactive to the HER2 domain III. Trastuzumab scFv-Cκ fusion protein immunoprecipitated nearly all HER2 moleucules in the 5637 cell lysate, but pertuzumab scFv-human Cκ fusion protein immunoprecipitated only a fraction of HER2 (Figure S2). With this observation, we concluded that 5637 cells expressed significant amounts theS310F mutant.
3.3. EGFR Activates S310F HER2 Mutant in 5637 Cell
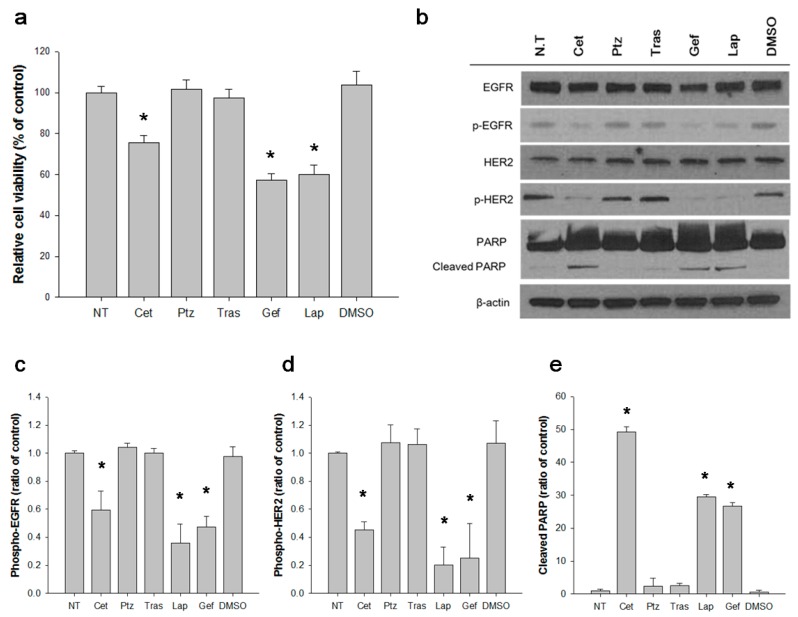
We tested the effects of anti-HER2 agents on 5637 cell proliferation and the level of HER2 phosphorylation at Y1221 and Y1222 residues. The cells were incubated with pertuzumab, trastuzumab and lapatinib for 96 h, lysed and subjected to SDS-PAGE and immunoblot analysis using anti-phospho-HER2 antibody. In parallel, the number of viable cells after the incubation was determined. Inhibiting the ligand-independent dimerization of wild-type and S310F HER2 mutant by trastuzumab did not reduce the cell proliferation or HER2 phosphorylation (Figure 3). Inhibiting the ligand-dependent dimerization of wild-type HER2 by pertuzumab also failed to show any effect (Figure 3). However, lapatinib inhibited the cell proliferation and reduced HER2 phosphorylation (Figure 3a,b,d). Because lapatinib inhibits the catalytic activity of both the EGFR and HER2, we then tested the effects of cetuximab and gefitinib, and both effectively inhibited the cell proliferation and reduced the levels of phosphorylated HER2and EGFR (Figure 3). The apoptosis induced by anti-EGFR and anti-HER2 agents was also determined (Figure 3b,d). In accordance with the cell proliferation data, only cetuximab, gefitinib, and lapatinib induced the cleavage of PARP (Figure 3b,e). Based on these data, we concluded that EGFR induced phosphorylation of S310F mutant in 5637 cells.
Figure 3.
Inhibition of 5637 cell proliferation and receptor activation by anti-HER2 and anti-EGFR agents. Cells were treated with HER2 or EGFR inhibitors for 96 h. (a) Cell viability was measured by WST-1 kit as described in materials and methods. Relative cell viability as a percent was determined as ((absorbance in each treatment set—absorbance in untreated set)/absorbance in non-treated set X 100). Results represent the mean ± SD obtained from three independent experiments. * p < 0.05. (b) The cell lysate was subjected to immunoblot analysis to visualize the relative phosphorylation level EGFR (c) and HER2 (d) and the level of cleaved PARP (e) Each of these levels were quantified by densitometry and plotted, individually. * p < 0.005. NT; not treated, Cet; Cetuximab, Ptz; Pertuzumab, Gef; Gefitinib, Lap; Lapatinib, DMSO; dimethyl sulfoxide, PARP; poly (ADP-ribose) polymerase.
3.4. Single-Molecular Interaction Analysis Demonstrated That the S310F HER2 Mutant Formed Heterodimers with the EGFR
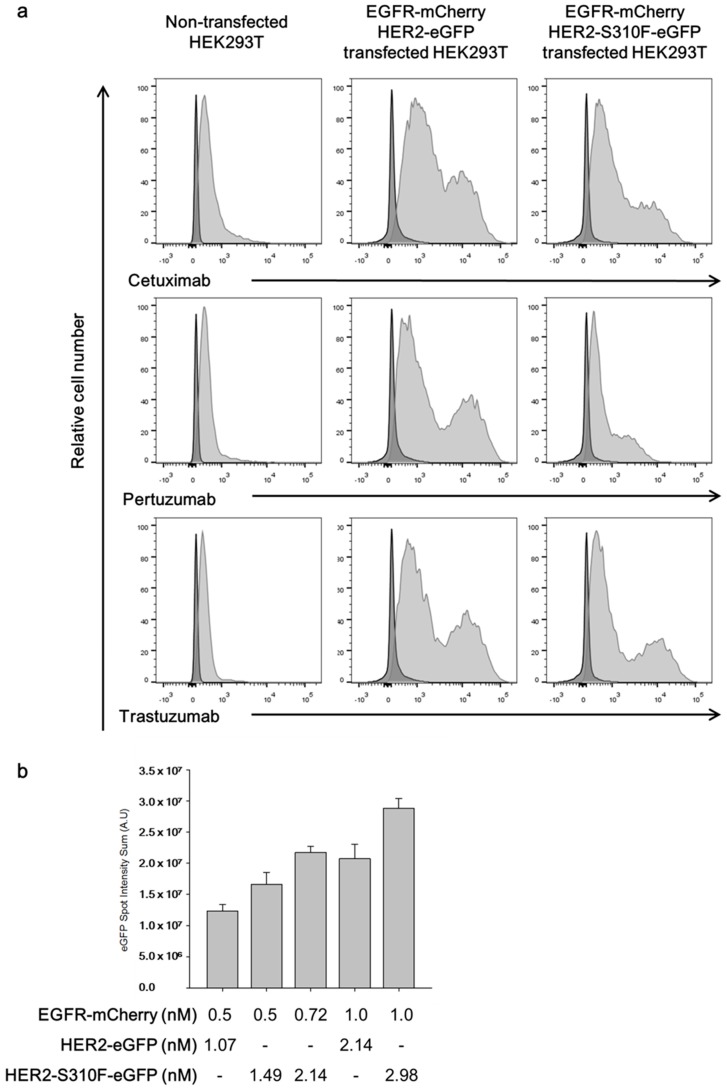
Single molecule interaction analysis was performed to determine whether the S310F HER mutant formed a heterodimer with the EGFR. We constructed a bicistronic mammalian expression vector encoding EGFR-mCherry and S310F HER2-eGFP fusion proteins. For comparison, we also prepared another expression vector encoding EGFR-mCherry and wild-type HER2-eGFP fusion proteins. After transfection of the expression vector into HEK293T cells, the expression of EGFR, S310F HER2 mutant, and wild-type HER2 were confirmed by flow cytometry analysis; using recombinant cetuximab, trastuzumab and pertuzumab scFv-human Cκ fusion proteins, the HEK293T cells showed basal expression of EGFR and HER2 (Figure 4a). The transfected cells showed significantly increased reactivity to cetuximab and trastuzumab. In HEK293T cells transfected with the expression vector encoding EGFR-mCherry and S310F HER2-eGFP, the reactivity with pertuzumab was significantly lower compared to the cells transfected with that encoding EGFR-mCherry and wild-type HER2-eGFP.
Figure 4.
Single-molecular interaction analysis for the S310F mutant and EGFR heterodimerization. HEK293T cells were transfected with expression vectors encoding either EGFR-mCherry and wild-type HER2-eGFP or EGFR-mCherry and S310F HER2-eGFP fusion proteins. (a) After transfection, the cell surface expression of EGFR and HER2 or HER2 was checked by flow cytometry analysis using cetuximab, pertuzumab, and trastuzumab in the scFv-human Cκ format and APC-labeled anti-human Cκ antibody. (b) The lysates were subjected to a flow chamber coated with anti-mCherry antibody. Then, the summed intensity of eGFP spots was measured and plotted. Results represent the mean ± SD obtained from experiments performed in triplicate.
After transfection, the cells were allowed to grow for 24 h and then lysed. The level of EGFR-mCherry and wild-type and S310F HER2-eGFP in the cell lysate was determined by measuring the fluorescence intensity. After a calibration curve was prepared using recombinant mCherry and eGFP protein standard with a florometer, the ratios of EGFR-mCherry vs. wild-type HER2-eGFP and EGFR-mCherry vs. S310F HER2-eGFP in the cell lysates were determined to be 1:2.14 and 1:2.98, respectively. For single molecule interaction analysis, the lysate was subjected to a flow chamber coated with anti-mCherry antibody. Using the TIRF microscope, eGFP spots were identified and their intensity was determined and summed. The eGFP spot intensity summation value was increased in a dose-dependent manner as the concentration of EGFR-mCherry injected into the flow cell was increased in both the cell lysates of wild-typeHER2-eGFP and S310F HER2-eGFP transfectants (Figure 4b). Considering the ratio of wild-typeHER2-eGFP and S310F HER2-eGFP vs. EGFR-mCherry in the cell lysates of the two transfectants, the ratio of eGFP vs. mCherry in the S310F HER2-eGFP transfectants was not significantly different from that of the wild-typeHER2-eGFP transfectants (Figure 4b). Two additional independent experiments yielded similar results. Based on these observations, we concluded that S310F HER2 interacted with the EGFR and its efficiency was comparable to that of the EGFR and wild-type HER2 interactions.
4. Discussion
After the initiation of the Cancer Genome Anatomy Project, new genetic alterations and new driver events were found in human cancers [52]. Somatic mutations of HER2 have been identified in a wide range of solid tumors, including breast, lung, colorectal, bladder, gastric, and cervical cancers [53,54,55,56,57,58]. HER2 activating mutations have been identified mostly in the kinase domain. C515R, T526A, G776R, L755S, L869R, R897G, and P1074S mutations were reported to increase phosphorylation levels of HER2. D769H and V777L mutations increased the phosphorylation of both HER2 and EGFR [27,35]. L755S and V777L were the most studied mutations in clinical cases [59,60]. Cells with HER2 L775S mutation showed increased levels of phospho-EGFR, -HER2, and -HER3 [27]. Patients with the HER2 L755S mutation were resistant to trastuzumab and lapatinib treatment, and a non-small cell lung cancer patient with HER2 N813D mutations was resistant to afatinib [61,62]. HER2 positive metastatic colorectal cancer patients with HER2 T784G mutation did not respond to cetuximab therapy [63]. A recent study showed that cells with the HER2 L755S mutation were resistant to lapatinib [64]. HER2 activating mutations were also found on the extracellular domain. Seven mutations, including G309 and S310 mutations, were found to increase the phosphorylation level of HER2 (Table 1) [32,35]. However, the mechanism of how these extracellular mutations increase the phosphorylation of HER2 has not been clarified.
In our study, ligand-independent HER2 dimerization blocking antibody, trastuzumab, did not inhibit the phosphorylation of HER2 in 5637 cells (Figure 3a), although it bound to the S310F mutant and the wild-type HER2 (Figure 1b). Trastuzumab treatment also did not hinder the proliferation of the cells (Figure 3b). This observation proved that either S310F mutant or wild-type HER2 can be activated without forming heterodimer or homodimers among the mutant and wild-type HER2. Because 5637 cells overexpress the EGFR, we hypothesized that the S310F mutant or wild-type HER2 formed heterodimers with the EGFR. As expected, the ligand binding-blocking antibody, cetuximab, almost completely abolished the phosphorylation of not only EGFR but also HER2 (Figure 3a). As a ligand-dependent HER2 heterodimerization blocking antibody, pertuzumab did not inhibit the phosphorylation of HER2 (Figure 3a), while it bound to wild-type HER2 (Figure 1a), EGFR-mediated phosphorylation should occur on the S310F mutant HER2. In this study, we were unable to determine the level of wild-type and S310F mutant in 5637 cells at the protein level; however, the limited amount of wild-type HER2 compared to the S310F mutant might be the reason why pertuzumab is not effective. To determine whether the S310F mutant retained affinity for the EGFR, we performed single molecule interaction analyses using a TIRF microscope as described previously [49]. The S310F HER2 mutant showed an affinity equivalent to that of wild-type HER2 in the formation of EGFR/HER2 complexes (Figure 4b).
In this study, we confirmed the loss or reduction of pertuzumab reactivity to G309 and S310 mutated HER2 proteins (Figure 1a). Complementary determining region 3 (CDR3) of pertuzumab heavy chain makes hydrophobic and hydrogen bond contacts with residues K333 and H318 of HER2 [9]. Therefore, mutations on G309 and S310 would induce critical structural changes on the epitope of pertuzumab. Interestingly, pertuzumab showed reduced binding activity to G309A mutated proteins but no reactivity to the G309E mutated proteins. The loss of binding activity could be explained by the structural difference between the two mutated amino acids; alanine and glutamic acid. G309 is located close to the C299-C311 disulfide bond [65]. While both glycine and alanine have a small nonpolar side chain, the glutamic acid has a long and charged side chain, influencing the structure of the HER2 domain II by breaking the disulfide bond. This hypothesis was tested by using SDS-polyacrylamide gel electrophoresis of the mutated recombinant HER2 proteins. G309E mutated protein formed multimers in non-reducing conditions, while the wild-type and G309A mutated HER2 proteins formed monomers (Figure S1). This observation was in agreement with that of a previous study reporting that the G309E HER2 mutant formed homodimers under non-reducing conditions in SDS-polyacrylamide gel electrophoresis analysis, which could be resolved by reducing agents [20].
Our study showed that a HER2 S310F mutation induced structural changes in domain II of HER2, abolishing its reactivity to pertuzumab, but its ability to form EGFR/HER2 dimers was not affected. HER2 heterodimerization with ligand-bound HERs was through a domain II-mediated dimerization interface [66]. In contrast, HER2 heterodimerization with ligand-free HERs may be mainly involved in domain IV [67]. After activation of EGFR, trastuzumab failed to reduce the phosphorylation level of the EGFR bound to HER2 [68]. Meanwhile, pertuzumab reduced the phosphorylation level of EGFR bound to HER2 [69]. The retained reactivity of S310F HER2 mutant to EGFR observed in our study suggested the unaffected structure of HER2 domain II on the interface of HER2/EGFR heteromdimer. C213, H215, and E243 residues of HER2 that are located on the top of the dimerization arm, are reported to stabilize the heterodimer by hydrogen bonding and salt bridges, and hence HER2 S310F mutation may not have affected the heterodimerization [70].
Acknowledgments
This study was supported by Seoul National University College of Medicine Foundation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/9/10/629/s1, Figure S1: SDS-polyacrylamide gel electrophoresis analysis of recombinant G309 mutant extracellular domains fused with the human Fc or Cκ domain The expression vectors encoding the fusion proteins were transfected into HEK293 cells. The recombinant proteins were purified from the culture supernatants using the affinity resin reactive to either Fc or human Cκ and subjected to 4%–12% (w/v) SDS-polyacrylamide gel electrophoresis either with or without a reducing agent. After electrophoresis, the protein bands were visualized with Coomassie Blue staining, Figure S2. Immunoprecipitations of HER2 in AU565 and 5637 cell lysates using trastuzumab and pertuzumab. Cell lysates were incubated with trastuzumab or pertuzumab scFv- human Cκ fusion protein followed by affinity resin reactive to human kappa light chain. After centrifugation, the amount of HER2 in the supernatant, resin, and cell lysate representing the same number of cells was determined by immunoblot analysis using anti-HER2 antibody.
Author Contributions
Conceptualization, J.C.; Formal analysis, J.W.S.; Funding acquisition, J.C.; Investigation, J.C.; Methodology, J.W.S., S.H., B.C. and T.-Y.Y.; Resources, S.K. (Soohyun Kim), S.K. (Seongyeong Kim) and S.-A.I.; Supervision, J.C.; Validation, J.W.S., S.K. (Soohyun Kim) and S.H.; Visualization, J.W.S., B.C. and T.-Y.Y.; Writing—Original Draft, J.W.S., S.K. (Soohyun Kim) and J.C.; Writing—Review & Editing, J.W.S., S.H. and J.C.
Funding
This study was supported by Seoul National University College of Medicine Foundation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Shepard H.M., Brdlik C.M., Schreiber H. Signal integration: A framework for understanding the efficacy of therapeutics targeting the human EGFR family. J. Clin. Invest. 2008;118:3574–3581. doi: 10.1172/JCI36049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yarden Y., Sliwkowski M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon M.A., Schlessinger J., Ferguson K.M. The EGFR family: Not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 2014;6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarden Y., Pines G. The ERBB network: At last, cancer therapy meets systems biology. Nat. Rev. Cancer. 2012;12:553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- 5.Moasser M.M. The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26:6469–6487. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh R., Narasanna A., Wang S.E., Liu S., Chakrabarty A., Balko J.M., Gonzalez-Angulo A.M., Mills G.B., Penuel E., Winslow J., et al. Trastuzumab has preferential activity against breast cancers driven by HER2 homodimers. Cancer Res. 2011;71:1871–1882. doi: 10.1158/0008-5472.CAN-10-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho H.S., Mason K., Ramyar K.X., Stanley A.M., Gabelli S.B., Denney D.W., Jr., Leahy D.J. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 9.Franklin M.C., Carey K.D., Vajdos F.F., Leahy D.J., de Vos A.M., Sliwkowski M.X. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–328. doi: 10.1016/S1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 10.Agus D.B., Akita R.W., Fox W.D., Lewis G.D., Higgins B., Pisacane P.I., Lofgren J.A., Tindell C., Evans D.P., Maiese K., et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell. 2002;2:127–137. doi: 10.1016/S1535-6108(02)00097-1. [DOI] [PubMed] [Google Scholar]
- 11.Mann M., Sheng H., Shao J., Williams C.S., Pisacane P.I., Sliwkowski M.X., DuBois R.N. Targeting cyclooxygenase 2 and HER-2/neu pathways inhibits colorectal carcinoma growth. Gastroenterology. 2001;120:1713–1719. doi: 10.1053/gast.2001.24844. [DOI] [PubMed] [Google Scholar]
- 12.Lua W.H., Gan S.K., Lane D.P., Verma C.S. A search for synergy in the binding kinetics of Trastuzumab and Pertuzumab whole and F(ab) to Her2. NPJ Breast Cancer. 2015;1:15012. doi: 10.1038/npjbcancer.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burris H.A., 3rd, Rugo H.S., Vukelja S.J., Vogel C.L., Borson R.A., Limentani S., Tan-Chiu E., Krop I.E., Michaelson R.A., Girish S., et al. Phase II study of the antibody drug conjugate trastuzumab-DM1 for the treatment of human epidermal growth factor receptor 2 (HER2)-positive breast cancer after prior HER2-directed therapy. J. Clin. Oncol. 2011;29:398–405. doi: 10.1200/JCO.2010.29.5865. [DOI] [PubMed] [Google Scholar]
- 14.Goss P.E., Smith I.E., O’Shaughnessy J., Ejlertsen B., Kaufmann M., Boyle F., Buzdar A.U., Fumoleau P., Gradishar W., Martin M., et al. Adjuvant lapatinib for women with early-stage HER2-positive breast cancer: A randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14:88–96. doi: 10.1016/S1470-2045(12)70508-9. [DOI] [PubMed] [Google Scholar]
- 15.Kelly K., Altorki N.K., Eberhardt W.E., O’Brien M.E., Spigel D.R., Crino L., Tsai C.M., Kim J.H., Cho E.K., Hoffman P.C., et al. Adjuvant Erlotinib Versus Placebo in Patients With Stage IB-IIIA Non-Small-Cell Lung Cancer (RADIANT): A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2015;33:4007–4014. doi: 10.1200/JCO.2015.61.8918. [DOI] [PubMed] [Google Scholar]
- 16.Chan A., Delaloge S., Holmes F.A., Moy B., Iwata H., Harvey V.J., Robert N.J., Silovski T., Gokmen E., von Minckwitz G., et al. Neratinib after trastuzumab-based adjuvant therapy in patients with HER2-positive breast cancer (ExteNET): A multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2016;17:367–377. doi: 10.1016/S1470-2045(15)00551-3. [DOI] [PubMed] [Google Scholar]
- 17.Connell C.M., Doherty G.J. Activating HER2 mutations as emerging targets in multiple solid cancers. ESMO Open. 2017;2:e000279. doi: 10.1136/esmoopen-2017-000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiss B., Wyatt A.W., Douglas J., Skuginna V., Mo F., Anderson S., Rotzer D., Fleischmann A. Her2 alterations in muscle-invasive bladder cancer: Patient selection beyond protein expression for targeted therapy. Sci. Rep. 2017;7:42713. doi: 10.1038/srep42713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chumsri S., Weidler J., Ali S., Balasubramanian S., Wallweber G., DeFazio-Eli L., Chenna A., Huang W., DeRidder A., Goicocheal L., et al. Prolonged Response to Trastuzumab in a Patient With HER2-Nonamplified Breast Cancer With Elevated HER2 Dimerization Harboring an ERBB2 S310F Mutation. J. Natl. Compr. Cancer Netw. 2015;13:1066–1070. doi: 10.6004/jnccn.2015.0132. [DOI] [PubMed] [Google Scholar]
- 20.Greulich H., Kaplan B., Mertins P., Chen T.H., Tanaka K.E., Yun C.H., Zhang X., Lee S.H., Cho J., Ambrogio L., et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc. Natl. Acad. Sci. USA. 2012;109:14476–14481. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L., Getz G., Wheeler D.A., Mardis E.R., McLellan M.D., Cibulskis K., Sougnez C., Greulich H., Muzny D.M., Morgan M.B., et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia Y., Ali S.M., Saad S., Chan C.A., Miller V.A., Halmos B. Successful treatment of a patient with Li-Fraumeni syndrome and metastatic lung adenocarcinoma harboring synchronous EGFR L858R and ERBB2 extracellular domain S310F mutations with the pan-HER inhibitor afatinib. Cancer Biol. Ther. 2014;15:970–974. doi: 10.4161/cbt.29173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eng J., Hsu M., Chaft J.E., Kris M.G., Arcila M.E., Li B.T. Outcomes of chemotherapies and HER2 directed therapies in advanced HER2-mutant lung cancers. Lung Cancer. 2016;99:53–56. doi: 10.1016/j.lungcan.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kan Z., Jaiswal B.S., Stinson J., Janakiraman V., Bhatt D., Stern H.M., Yue P., Haverty P.M., Bourgon R., Zheng J., et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 25.Shah S.P., Roth A., Goya R., Oloumi A., Ha G., Zhao Y., Turashvili G., Ding J., Tse K., Haffari G., et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerji S., Cibulskis K., Rangel-Escareno C., Brown K.K., Carter S.L., Frederick A.M., Lawrence M.S., Sivachenko A.Y., Sougnez C., Zou L., et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bose R., Kavuri S.M., Searleman A.C., Shen W., Shen D., Koboldt D.C., Monsey J., Goel N., Aronson A.B., Li S., et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross J.S., Wang K., Sheehan C.E., Boguniewicz A.B., Otto G., Downing S.R., Sun J., He J., Curran J.A., Ali S., et al. Relapsed classic E-cadherin (CDH1)-mutated invasive lobular breast cancer shows a high frequency of HER2 (ERBB2) gene mutations. Clin. Cancer Res. 2013;19:2668–2676. doi: 10.1158/1078-0432.CCR-13-0295. [DOI] [PubMed] [Google Scholar]
- 29.Ali S.M., Alpaugh R.K., Downing S.R., Stephens P.J., Yu J.Q., Wu H., Buell J.K., Miller V.A., Lipson D., Palmer G.A., et al. Response of an ERBB2-mutated inflammatory breast carcinoma to human epidermal growth factor receptor 2-targeted therapy. J. Clin. Oncol. 2014;32:e88-91. doi: 10.1200/JCO.2013.49.0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endo Y., Dong Y., Kondo N., Yoshimoto N., Asano T., Hato Y., Nishimoto M., Kato H., Takahashi S., Nakanishi R., et al. HER2 mutation status in Japanese HER2-positive breast cancer patients. Breast Cancer. 2016;23:902–907. doi: 10.1007/s12282-015-0659-y. [DOI] [PubMed] [Google Scholar]
- 31.Jasra S., Opyrchal M., Norton L., Mehta R. A Rare Case of S310F Somatic ERBB2 Mutation in a HER2-Nonamplified Breast Cancer. Clin. Breast Cancer. 2017;17:e37–e41. doi: 10.1016/j.clbc.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Ross J.S., Wang K., Gay L.M., Al-Rohil R.N., Nazeer T., Sheehan C.E., Jennings T.A., Otto G.A., Donahue A., He J., et al. A high frequency of activating extracellular domain ERBB2 (HER2) mutation in micropapillary urothelial carcinoma. Clin. Cancer Res. 2014;20:68–75. doi: 10.1158/1078-0432.CCR-13-1992. [DOI] [PubMed] [Google Scholar]
- 33.Vornicova O., Hershkovitz D., Yablonski-Peretz T., Ben-Itzhak O., Keidar Z., Bar-Sela G. Treatment of metastatic extramammary Paget’s disease associated with adnexal adenocarcinoma, with anti-HER2 drugs based on genomic alteration ERBB2 S310F. Oncologist. 2014;19:1006–1007. doi: 10.1634/theoncologist.2014-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang T., Xu Y., Sheng S., Yuan H., Ouyang T., Li J., Wang T., Fan Z., Fan T., Lin B., et al. HER2 somatic mutations are associated with poor survival in HER2-negative breast cancers. Cancer Sci. 2017;108:671–677. doi: 10.1111/cas.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muzny D.M., Bainbridge M.N., Chang K., Dinh H.H., Drummond J.A., Fowler G., Kvar C.L., Lewis L.R. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han S.W., Kim H.P., Shin J.Y., Jeong E.G., Lee W.C., Lee K.H., Won J.K., Kim T.Y., Oh D.Y., Im S.A., et al. Targeted sequencing of cancer-related genes in colorectal cancer using next-generation sequencing. PLoS ONE. 2013;8:e64271. doi: 10.1371/journal.pone.0064271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K., Kan J., Yuen S.T., Shi S.T., Chu K.M., Law S., Chan T.L., Kan Z., Chan A.S., Tsui W.Y., et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 39.Petrelli F., Tomasello G., Barni S., Lonati V., Passalacqua R., Ghidini M. Clinical and pathological characterization of HER2 mutations in human breast cancer: A systematic review of the literature. Breast Cancer Res. Treat. 2017;166:339–349. doi: 10.1007/s10549-017-4419-x. [DOI] [PubMed] [Google Scholar]
- 40.Lee Y., Kim H., Chung J. An antibody reactive to the Gly63-Lys68 epitope of NT-proBNP exhibits O-glycosylation-independent binding. Exp. Mol. Med. 2014;46:e114. doi: 10.1038/emm.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park S., Lee D.H., Park J.G., Lee Y.T., Chung J. A sensitive enzyme immunoassay for measuring cotinine in passive smokers. Clin. Chim. Acta. 2010;411:1238–1242. doi: 10.1016/j.cca.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 42.Takeshi K., Hitomi S., Yasuto A., Shun’ichiro T. In situ delivery and production system of trastuzumab scFv with Bifidobacterium. Biochem. Biophy. Res. Commun. 2017;493:306–312. doi: 10.1016/j.bbrc.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 43.Akbari V., Sadeghi H.M.M., Jafrian-Dehkordi A., Abedi D., Chou C.P. Functional expression of a single-chain antibody fragment against human epidermal growth factor receptor 2 (HER2) in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2014;41:947–956. doi: 10.1007/s10295-014-1437-0. [DOI] [PubMed] [Google Scholar]
- 44.Judith N., Christoph S., Hannes B., Grit H.T., Rolf F., Georg M., Rainer F., Stefan B. Novel EGFR-specific immunotoxins based on panitumumab and cetuximab show in vitro and ex vivo activity against different tumor entities. J. Cancer Res. Clin. Oncol. 2015;141:2079–2095. doi: 10.1007/s00432-015-1975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldi L., Muller N., Picasso S., Jacquet R., Girard P., Thanh H.P., Derow E., Wurm F.M. Transient gene expression in suspension HEK-293 cells: Application to large-scale protein production. Biotechnol. Prog. 2005;21:148–153. doi: 10.1021/bp049830x. [DOI] [PubMed] [Google Scholar]
- 46.Yoon S., Kim Y.H., Kang S.H., Kim S.K., Lee H.K., Kim H., Chung J., Kim I.H. Bispecific Her2 x cotinine antibody in combination with cotinine-(histidine)2-iodine for the pre-targeting of Her2-positive breast cancer xenografts. J. Cancer Res. Clin. Oncol. 2014;140:227–233. doi: 10.1007/s00432-013-1548-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H., Hong S.H., Kim J.Y., Kim I.C., Park Y.W., Lee S.J., Song S.W., Kim J.J. Preclinical development of a humanized neutralizing antibody targeting HGF. Exp. Mol. Med. 2017;49:e309. doi: 10.1038/emm.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee S., Yoon I.H., Yoon A., Cook-Mills J.M., Park C.G., Chung J. An antibody to the sixth Ig-like domain of VCAM-1 inhibits leukocyte transendothelial migration without affecting adhesion. J. Immunol. 2012;189:4592–4601. doi: 10.4049/jimmunol.1103803. [DOI] [PubMed] [Google Scholar]
- 49.Lee H.W., Ryu J.Y., Yoo J., Choi B., Kim K., Yoon T.Y. Real-time single-molecule coimmunoprecipitation of weak protein-protein interactions. Nat. Protoc. 2013;8:2045–2060. doi: 10.1038/nprot.2013.116. [DOI] [PubMed] [Google Scholar]
- 50.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selvin P.R., Ha T. Single-Molecule Techniques: A Labatory Manual. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2008. [Google Scholar]
- 52.Strausberg R.L. Cancer Genome Anatomy Project. Wiley Online Library; New York, New York, USA: 2006. [DOI] [Google Scholar]
- 53.Shigematsu H., Takahashi T., Nomura M., Majmudar K., Suzuki M., Lee H., Wistuba I.I., Fong K.M., Toyooka S., Shimizu N., et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 54.The Cancer Genome Atlas Research Network. Weinstein J.N., Akbani R., Broom B.M., Wang W., Verhaak R.G.W., Lerner S., Morgan M., Creighton C.J., Kwiatkowski D.J., et al. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen W., Chen W.S., Xiao N., Bender R., Ghazalpour A., Tan Z., Swensen J., Millis S.Z., Basu G., Gatalica Z., et al. Mutations in the Kinase Domain of the HER2/ERBB2 Gene Identified in a Wide Variety of Human Cancers. J. Mol. Diagn. 2015;17:487–495. doi: 10.1016/j.jmoldx.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuo W.J., Jiang Y.Z., Wang Y.J., Xu X.E., Hu X., Liu G.Y., Wu J., Di G.H., Yu K.D., Shao Z.M. Dual Characteristics of Novel HER2 Kinase Domain Mutations in Response to HER2-Targeted Therapies in Human Breast Cancer. Clin. Cancer Res. 2016;22:4859–4869. doi: 10.1158/1078-0432.CCR-15-3036. [DOI] [PubMed] [Google Scholar]
- 57.Kavuri S.M., Jain N., Galimi F., Cottino F., Leto S.M., Migliardi G., Searleman A.C., Shen W., Monsey J., Trusolino L., et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015;5:832–841. doi: 10.1158/2159-8290.CD-14-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang L., Jiang W., Ye S., He T., Pei X., Li J., Chan D.W., Ngan H.Y.S., Li F., Tao P., et al. ERBB2 mutation: A promising target in non-squamous cervical cancer. Gynecol. Oncol. 2018;148:311–316. doi: 10.1016/j.ygyno.2017.12.023. [DOI] [PubMed] [Google Scholar]
- 59.Endo Y., Dong Y., Yoshimoto N., Asano T., Hato Y., Yamashita H., Sato S., Takahashi S., Fujii Y., Toyama T. HER2 mutation status in Japanese HER2-negative breast cancer patients. Jpn. J. Clin. Oncol. 2014;44:619–623. doi: 10.1093/jjco/hyu053. [DOI] [PubMed] [Google Scholar]
- 60.Weigelt B., Reis-Filho J.S. Activating mutations in HER2: Neu opportunities and neu challenges. Cancer Discov. 2013;3:145–147. doi: 10.1158/2159-8290.CD-12-0585. [DOI] [PubMed] [Google Scholar]
- 61.Kancha R.K., von Bubnoff N., Bartosch N., Peschel C., Engh R.A., Duyster J. Differential sensitivity of ERBB2 kinase domain mutations towards lapatinib. PLoS ONE. 2011;6:e26760. doi: 10.1371/journal.pone.0026760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park C.K., Hur J.Y., Choi C.M., Kim T.O., Cho H.J., Shin H.J., Lim J.H., Choi Y.D., Kim Y.C., Oh I.J. Efficacy of Afatinib in a Previously-Treated Patient with Non-Small Cell Lung Cancer Harboring HER2 Mutation: Case Report. J. Korean Med. Sci. 2018;33:e7. doi: 10.3346/jkms.2018.33.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gharib E., Salmanipour R., Nazemalhosseini M.E., Yaghoob T.M., Sarlak S., Malekzade-Moghani M., Nasrabadi P.N., Meiary M.A., Asadzadeh A.H., AZali M.R. HER2+ mCRC patients with exon 20 R784G substitution mutation do not respond to the cetuximab therapy. J. Cell Physiol. 2018 doi: 10.1002/jcp.27984. [DOI] [PubMed] [Google Scholar]
- 64.Zabransky D.J., Yankaskas C.L., Cochran R.L., Wong H.Y., Croessmann S., Chu D., Kavuri S.M., Red Brewer M., Rosen D.M., Dalton W.B., et al. HER2 missense mutations have distinct effects on oncogenic signaling and migration. Proc. Natl. Acad. Sci. USA. 2015;112:E6205–E6214. doi: 10.1073/pnas.1516853112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garrett T.P., McKern N.M., Lou M., Elleman T.C., Adams T.E., Lovrecz G.O., Kofler M., Jorissen R.N., Nice E.C., Burgess A.W., et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol. Cell. 2003;11:495–505. doi: 10.1016/S1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 66.Kumagai T., Katsumata M., Hasegawa A., Furuuchi K., Funakoshi T., Kawase I., Greene M.I. Role of extracellular subdomains of p185c-neu and the epidermal growth factor receptor in ligand-independent association and transactivation. Proc. Natl. Acad. Sci. USA. 2003;100:9220–9225. doi: 10.1073/pnas.1633546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Junttila T.T., Akita R.W., Parsons K., Fields C., Lewis Phillips G.D., Friedman L.S., Sampath D., Sliwkowski M.X. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 68.Maadi H., Nami B., Tong J., Li G., Wang Z. The effects of trastuzumab on HER2-mediated cell signaling in CHO cells expressing human HER2. BMC Cancer. 2018;18:238. doi: 10.1186/s12885-018-4143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laux I., Jain A., Singh S., Agus D.B. Epidermal growth factor receptor dimerization status determines skin toxicity to HER-kinase targeted therapies. Br. J. Cancer. 2006;94:85–92. doi: 10.1038/sj.bjc.6602875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arkhipov A., Shan Y., Kim E.T., Dror R.O., Shaw D.E. Her2 activation mechanism reflects evolutionary preservation of asymmetric ectodomain dimers in the human EGFR family. Elife. 2013;2:e00708. doi: 10.7554/eLife.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.