Abstract
The development of antimicrobial resistance has become a global concern. One approach to overcome the problem of drug resistance is the application of bacteriophages. This study aimed at characterizing three phages isolated from sewage, which show lytic activity against clinical isolates of multidrug-resistant Staphylococcus aureus. Morphology, genetics and biological properties, including host range, adsorption rate, latent time, phage burst size and lysis profiles, were studied in all three phages. As analyzed by transmission electron microscopy (TEM), phages vB_SauM-A, vB_SauM-C, vB_SauM-D have a myovirion morphology. One of the tested phages, vB_SauM-A, has relatively rapid adsorption (86% in 17.5 min), short latent period (25 min) and extremely large burst size (~500 plaque-forming units (PFU) per infected cell). The genomic analysis revealed that vB_SauM-A, vB_SauM-C, vB_SauM-D possess large genomes (vB_SauM-A 139,031 bp, vB_SauM-C 140,086 bp, vB_SauM-D 139,088 bp) with low G+C content (~30.4%) and are very closely related to the phage K (95–97% similarity). The isolated bacteriophages demonstrate broad host range against MDR S. aureus strains, high lytic activity corresponding to strictly virulent life cycle, suggesting their potential to treat S. aureus infections.
Keywords: bacteriophage, Staphylococcus aureus, Kayvirus
1. Introduction
Staphylococcus aureus is the prime cause of hospital-acquired infections. It is associated with a number of diseases in humans, ranging from superficial to invasive infections such as pneumonia and sepsis [1]. S. aureus infections can be difficult to treat because of the prevalence of S. aureus strains resistant to β-lactam antibiotics (methicillin-resistant Staphylococcus aureus—MRSA). In 2017, the percentage of invasive MRSA isolates in Europe ranged from 1% in Norway to 44.4% in Romania, with a population-weighted mean of 16.9% [2]. Treatment of S. aureus infections is a challenge not only because of MRSA resistance towards β-lactams antibiotics but because many isolates exhibit resistance to other antimicrobial agents, for example, macrolides, tetracycline, aminoglycosides, chloramphenicol and fluoroquinolones, leading to the emergence of multidrug resistant S. aureus (MDRSA). The rate at which S. aureus can develop or acquire resistance to new antibiotics, is actually higher than that, at which new antibiotics are discovered and developed [3]. Problems connected with antibiotic treatment necessitate the investigation and development of novel treatment strategies.
A potential solution is the use of bacteriophages–viruses that specifically lyse bacterial cell. Their host specificity ranges from a few strains of a bacterial species to, more rarely, more than one relatively closely related bacterial genus. Thus, they do not destroy beneficial bacterial flora and the risk of developing secondary infections is minimalized [4,5]. Owing to relatively narrow host range of most phages, the number of bacterial types with which the selection for specific phage-resistance mechanisms can occur, is limited [4]. Moreover, spontaneous mutations may confer phage–bacteria coevolution, therefore phages can overcome bacterial resistance [6]. Although phage specificity is an advantage in the case of the minimal impact of natural flora, it can be also seen as a problem in therapy. Because phages exhibit such a narrow host range, there is a need to identify the etiologic agent and select a phage lytic for the targeted bacterial strain before using phages therapeutically. This involves creating an extensive collection of well-characterized phage for a broad range of pathogens [7]. Kayviruses are polyvalent bacteriophages infecting a broad spectrum of S. aureus and other staphylococci. These bacteriophages are ones of the best agents that can be used to treat staphylococcal infection [8,9,10,11]. Therefore, some of them are used in commercial bacteriophage preparations [8,10,12].
The aim of this work was to isolate and characterize S. aureus bacteriophages in a view of their potential application in phage therapy.
2. Materials and Methods
2.1. Bacterial Strains
A total of 88 bacterial strains, including 21 representatives of different species (Table 2) and 67 S. aureus clinical isolates (Table 3) from different clinical materials (wound, pus, furuncle, bronchial tree), of both inpatients (n = 56) and outpatients (n = 11) were used in the study. The isolates originated from the collection of the Department of Medical Microbiology, Medical University of Gdansk and were obtained from 7 clinical centers of Gdansk region between 2013 and 2015. All isolates were identified by conventional bacteriological tests and antimicrobial susceptibility testing was performed by means of the disk diffusion method according to the guidelines of Clinical and Laboratory Standards Institute. All the strains tested were confirmed to be methicillin-resistant. Moreover, the strains were also resistant to erythromycin (96%), clindamycin (91%), norfloxacin (76%) and ciprofloxacin (76%), sulfamethoxazole/trimethoprim (7%), tetracycline (33%), chloramphenicol (27%), gentamicin (10%). All strains assayed were susceptible to fusidic acid, mupirocin and linezolid. Bacteria were stored at −80°C in tryptic soy broth (TSB) supplemented with 10% glycerol. All strains were cultured in Luria-Bertani broth (LB; Biomaxima, Lublin, Poland) or on LB agar plates (LA) solidified with 1.5% (w/v) agar (Becton Dickinson, Franklin Lakes, NJ, USA).
2.2. Isolation and Purification of Phages
Samples (n = 86) collected from wastewater treatment plants were used for isolation of bacteriophages according to Guzmán et al. [13]. Approximately 10 mL of sample was centrifuged at 2500 × g for 20 min and filtered through 0.2 μm Minisart® filters (Sartorius Stedim Biotech, Goettingen, Germany). Sample was mixed with the culture of different strains of Staphylococcus aureus in its mid-exponential phase of growth (an optical density at 600 nm [OD600 nm] of 0.6), growing in 2 × TSB supplemented with 10 mM of MgSO4 and CaCl2. Samples were incubated with rotary shacking (180 rpm, at 37 °C). Chloroform was added to the culture following incubation and samples were placed on a rotor shaker for the next 15 min. Afterwards, tubes were centrifuged 2500 × g for 20 min and filtered through the 0.2 μm filter. Phage activity was tested by a double overlay agar plaque assay.
The clear plaque was picked and phages were suspended with SMG buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl [pH 7.5], 0.01% gelatin). Isolated phages were purified by a triple transfer of a single plaque and propagation until homologous plaques were obtained. Decimal dilutions of lysates in SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl [pH 7.5]) were prepared to define the titer of bacteriophage and subjected to plaque assay using the double agar layer method. Plaques were counted on the dilution plate in spots containing 30–300 plaques. Results were expressed in plaque forming units per mL (PFU/mL).
2.3. Phage Propagation
To amplify the phages, 5 mL of bacterial host strain grown overnight in liquid LB medium were added to 500 mL of LB and incubated at 37 °C with agitation. When the OD of the culture reached 0.1 at a 600 nm wavelength, the bacteria were infected with phages at a multiplicity of infection (MOI) of 0.1 and incubated at 37 °C until lysis occurred [14]. For phage purification, polyethylene glycol (PEG) 8000 (BioShop, Burlington, Ontario, Canada) was added (final concentration 10% w/v) and stirred overnight at 4 °C [15]. The precipitate was collected by centrifugation at 11,000× g for 20 min at 4 °C and suspended in TM buffer (10 mM Tris-HCl, 10 mM MgSO4 [pH 7.2]). PEG8000 was removed by adding the same volume of chloroform, vortexing and centrifugation at 3000× g for 15 min [16]. Phages for Transmission Electron Microscopy (TEM) were purified by centrifugation in CsCl gradients at 95,000× g for 2.5 h [17].
2.4. Electron Microscopy Imaging of Phages
Phage particles in buffer were deposited on Formvar/Carbon-coated copper grids and negatively stained with 1% uranyl acetate in water (pH 4) [18]. Visualization was performed using a transmission electron microscope (TEM) (JEM 1200EX II; Jeol Ltd., Akishima, Tokyo, Japan) operated at 100 kV. The size of the head and length of the tail were calculated from 5 independent measurements of separate virions and reported as a mean value ± standard deviation [19].
2.5. Efficiency of Phage Adsorption
An adsorption assay was performed according to the protocol described before, with some modifications [20]. Overnight liquid cultures of the bacterial strains were diluted 1:100 in LB medium and incubated at 37 °C until the OD600 reached 0.1 value. Samples of 2 mL were centrifuged at 2000× g for 5 min and pellets were suspended in 1.5 mL LB. After 15 min of incubation at 37 °C, the phages were added to each culture at a MOI of 0.01. Samples were withdrawn immediately after the phage addition (time zero) and at indicated time points and centrifuged at 8000× g for 30 s. Subsequently, the supernatants were titrated to determine the number of unabsorbed phages. The whole experiment was replicated thrice. A sample withdrawn immediately after addition of bacteriophages to the cell suspension was considered as 100% non-adsorbed phages.
2.6. One-Step Growth Analysis
Lytic development of phages was studied in one-step growth experiment, following the protocol described previously [20,21], with minor modifications. Host bacteria were grown in LB medium at 37 °C to OD600 = 0.1. Bacterial culture of 10 mL was centrifuged (2000× g for 5 min at 4 °C) and the pellet was suspended in 5 mL of LB medium with 3 mM sodium azide (Sigma Aldrich, Saint Louis, MO, USA). The sample was centrifuged again after 10 min of incubation at 37 °C and the pellet was resuspended in 5 mL of fresh LB medium. Following 10-min incubation at 37 °C, the phage was added to MOI of 0.05 and allowed to adsorb for 5 min. Then, 100 µL of the suspension was added to 20 mL of LB medium and cultivated in an incubator shaker. The number of infection centers was determined at times: 5, 10, 15 min after infection by mixing l0 µL of the sample with 200 µL of an overnight culture of appropriate indicator bacteria and 4 mL of top agar. To determine the number of intracellular progeny phages, samples were shaken vigorously for 1 min with equal volume of chloroform and cleared by centrifugation. The number of PFU (number of phages able to form plaques) per mL was estimated by titration using the double-layer agar plate method. Plates were incubated at 37 °C overnight. Burst size was calculated as a ratio of phage titer to the titer of infection centers. Each experiment was repeated three times.
2.7. Determination of the Host Range
The phage host range was determined using spot test method [22]. The bacterial isolates listed in Tables 2 and 3 were incubated overnight at 37 °C in LB liquid medium. Two hundred microliters of each test bacterial culture were added to 4 mL of molten 0.7% LB agar and the mixture was overlaid on 1.5% LB agar plates. The plates were left to solidify. Then 2.5 µL of ten-fold dilutions of each phage was spotted onto the surface of the double-layer agar plates with a tested host. Initial concentration of phages vB_SauM-A, vB_SauM-C and vB_SauM-D was 8 × 1010 PFU/mL, 2.4 × 1011 PFU/mL, 6.4 × 1011 PFU/mL, respectively. Following incubation at 37 °C for 24 h, the plates were examined for the presence of plaques. All the experiments were performed in triplicate.
2.8. Sensitivity of Phage Particles to Temperature, pH and Chloroform and Virion Stability
The effect of temperature and various pH levels was performed according to the procedure described previously [22]. For thermo-stability testing, equal volumes of bacteriophages were incubated at 40 °C (40 min) and 62 °C (40 min). After incubation, the phage titer was determined using the spot test method as described above. For pH stability testing, phage lysate was mixed (at the volume proportion 1:9) with LB medium of different pH values (2–12, adjusted using NaOH or HCl). Phage titer was assessed after 2 h incubation at 37 °C. Virion stability was assessed by titration of phage lysates over time. Purified phage lysate was stored at 10 °C and titrated over the period of 10 months in order to analyze any changes in phage titer. All the experiments were repeated three times.
2.9. Spectrophotometric Assay of Phage-Treated Liquid
The effects of phages on representative strains of S. aureus in LB medium were observed by measuring the OD600 with a SmartSpec 3000 spectrophotometer (BIO-RAD, Hercules, CA, USA) following the procedure described previously [23], with some modifications. Overnight cultures of the bacterial strains were diluted 1:100 in LB medium and incubated at 37 °C with shaking at 150 rpm until early logarithmic phase (OD600, 0.1) was reached. Then, the phage lysate was added to achieve MOI of 0.1, 0.25, 0.5, 1. The OD600 of each culture was measured every 15 min for a minimum of 4 h following phage addition.
2.10. Lysogeny Test for Verification of Lytic Cycle
Test was performed according to D’Andrea et al. [24] with minor modification. Briefly, bacterial host strain was propagated in TSB broth and infected by bacteriophages at a MOI equal to approx. 1. Cultures were incubated at 37 °C with shaking at 160 rpm for 24 h. The infected culture was then diluted and bacteriophage stock was added at an MOI > 1000 and incubated for 1 h. Subsequently, the culture was plated on TSB and incubated overnight. Single colonies were streaked and the resistance to isolated bacteriophages was tested by spotting 10 µL of phage lysate. The cells which survived this exposure were confirmed to be phage-resistant by PCR screening with primers specific to the Kayvirus DNA polymerase I gene.
2.11. Extraction of Phage DNA
The purified phage lysate (109 PFU/mL) was treated with DNase I (1 U/μL; A&A Biotechnology, Gdynia, Poland) and RNase A (5 µg/μL; A&A Biotechnology, Gdynia, Poland). The mixture was incubated for 30 min at 37 °C in order to digest any exogenous DNA and RNA. Then, DNase I and RNase A were inactivated according to manufacturer protocol. The MasterPure™ Complete DNA and RNA Purification Kit (Epicentre, Madison, WI, USA) was used to isolate the genomic DNA of phages. The DNA concentration and purity was determined spectrophotometrically at 260 nm [25].
2.12. Phage Genome Sequencing and Bioinformatics Analysis
DNA samples were sequenced in MiSeq (Illumina, Inc., San Diego, CA, USA) genome sequencer and assembled with CLCGenomicWorkbench by the Genomed company. Open reading frames (ORFs) prediction and genome annotations were prepared with the use of Geneious Prime 2019.2.1 (https://www.geneious.com). Genome annotations were checked by sequence comparison of protein sequence using the blastn software (https://blast.ncbi.nlm.nih.gov). Whole genome-based phylogeny was prepared with the use of VICTOR software [26]. The pairwise comparisons of the bacteriophage genomes were prepared with applying the Genome-BLAST Distance Phylogeny (GBDP) method with settings optimized for bacteriophages [26,27]. Phylogeny between isolated phages was prepared with the use of CSI phylogeny 1.4 [28]. Prediction of bacteriophage life cycle was performed using the PHACTS software [29]. Sequence similarity of isolated bacteriophages was prepared with the use of Circoletto software [30]. The bacteriophages G1, K, JD007, MCE-2014 and phiIPLA-RODI (NCBI accession numbers: AY954969.1, KF766114.1, JX878671.1, KJ888149.1, KP027446.1 respectively) were used for the generation of sequence similarity graphs.
2.13. Statistical Analysis
The obtained data were analyzed by two-way ANOVA with Tuckey post-hoc tests. p values lower than 0.05 were considered statistically significant. All statistical analyses were carried out using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA). All data are presented as mean from minimum five replicates with standard deviation (SD).
3. Results
3.1. Virion Morphology
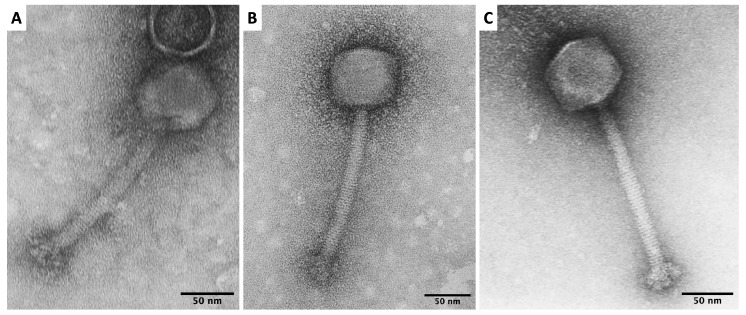
Bacteriophages were isolated from different wastewater treatment plants with initially 9 phages isolated (10.47% positive samples), 6 phages were excluded from this study according to their low lytic activity or genome characteristic showed lysogenic life cycle. The isolated phages were characterized with regard to their morphology. TEM images (Figure 1) revealed that virions of vB_SauM-A, vB_SauM-C and vB_SauM-D have icosahedral heads and long contractile tails of 149–158 nm in the extended state. Phage dimensions are summarized in Table 1. Also, the knob-like appendages, previously described by O’Flaherty et al. [31], extending from the baseplate were evident. Based on their morphology, we assumed that phages vB_SauM-A, vB_SauM-C and vB_SauM-D have myovirids morphotypes.
Figure 1.
Transmission electron micrographs (TEM) of vB_SauM-A (A), vB_SauM-C (B), vB_SauM-D (C).
Table 1.
Dimensions of phages vB_SauM-A, vB_SauM-C and vB_SauM-D obtained from transmission electron micrographs.
| Parameter | vB_SauM-A | vB_SauM-C | vB_SauM-D |
|---|---|---|---|
| Head morphology | Icosahedral | Icosahedral | Icosahedral |
| Head length (nm) | 62.99 ± 9.99 | 76.75 ± 5.04 | 72.80 ± 3.66 |
| Head diameter (nm) | 63.33 ± 5.81 | 68.37 ± 4.14 | 65.98 ± 3.11 |
| Tail length (nm) | 149.70 ± 14.65 | 153.51 ± 15.16 | 158.05 ± 6.56 |
3.2. Analysis of the Bacteriophage Adsorption Rate and One-Step Growth Curves
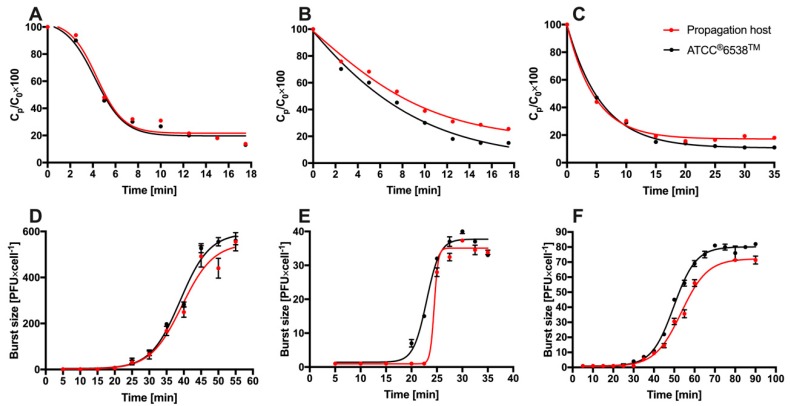
The life cycle and adsorption rate of phages vB_SauM-A, vB_SauM-C and vB_SauM-D were determined on S. aureus 203, 343 and 342 hosts, respectively and on reference S. aureus ATCC®6538™. The adsorption curves of these bacteriophages on reference strain and propagation hosts were very similar (Figure 2) and showed that after 10 min, depending on the strain, 69–73.3%, 61–64.6%, 70–70.9% of bacteriophages vB_SauM-A, vB_SauM-C, vB_SauM-D were adsorbed. Statistical analysis revealed statistically significant difference between adsorption of vB_SauM-C and vB_SauM-A, vB_SauM-D (p > 0.05). There was no significant difference between vB_SauM-A and vB_SauM-D. Bacteriophage vB_SauM-A showed a short latent period of only 25 min and an extremely large burst size of approximately 500 PFU/infected cell. Differences between burst size of these phages were significant (p > 0.05) when compared.
Figure 2.
Characterizations of phages vB_SauM-A, vB_SauM-C, vB_SauM-D. Kinetics of vB_SauM-A (A), vB_SauM-C (B) and vB_SauM-D (C) phages adsorption on propagation and reference host cells. Curve for one-step growth of phages vB_SauM-A (D), vB_SauM-C (E) and vB_SauM-D (F) on propagation and reference host cells. Results are presented as mean values ± SD from three independent experiments.
3.3. Determination of the Host Range
The host ranges of phages vB_SauM-A, vB_SauM-C, vB_SauM-D were determined using 21 different bacterial strains that are listed in Table 2 and 67 MDRSA clinical isolates listed in Table 3. We found out that tested bacteriophages are able to efficiently infect different S. aureus strains and one of reference S. epidermidis strains but no other bacterial genera. Phages vB_SauM-A and vB_SauM-D were found to infect the same MDRSA clinical isolates.
Table 2.
Host range of phages vB_SauM-A, vB_SauM-C, vB_SauM-D on different bacterial species.
| Species | Reference Number | vB_SauM-A | vB_SauM-C | vB_SauM-D |
|---|---|---|---|---|
| Staphylococcus epidermidis | ATCC 12228 | − | − | − |
| Staphylococcus epidermidis | ATCC 14990 | ++ | + | ++ |
| Staphylococcus intermedius | PCM 2405 * | − | − | − |
| Staphylococcus aureus | ATCC 25923 | ++ | +++ | ++ |
| Staphylococcus aureus | ATCC 29213 | ++ | ++ | ++ |
| Escherichia coli | ATCC 11775 | − | − | − |
| Salmonella Typhimurium | ATCC 14028 | − | − | − |
| Salmonella Enteritidis | ATCC 13076 | − | − | − |
| Shigella flexneri | ATCC 12022 | − | − | − |
| Shigella sonnei | ATCC 25931 | − | − | − |
| Proteus vulgaris | ATCC 6380 | − | − | − |
| Proteus mirabilis | ATCC 7002 | − | − | − |
| Yersinia enterocolitica | ATCC 27729 | − | − | − |
| Pseudomonas aeruginosa | ATCC 27853 | − | − | − |
| Enterococcus faecalis | ATCC 29212 | − | − | − |
| Lactococcus lactis | ATCC 19435 | − | − | − |
| Lactobacillus gasseri | ATCC 19992 | − | − | − |
| Lactobacillus acidophilus | ATCC 4356 | − | − | − |
| Bacillus cereus | ATCC 14579 | − | − | − |
| Streptococcus agalactiae | ATCC 12386 | − | − | − |
| Listeria monocytogenes | ATCC 19118 | − | − | − |
* PCM, Polish Collection of Microorganisms. +++ plaques at lysate concentration ≤ 104 PFU/mL, ++ plaques at lysate concentration 105–106 PFU/mL, + plaques at lysate concentration 107 PFU/mL, − no plaques or plaques at lysate concentration ≥108 PFU/mL.
Table 3.
Host range of phages vB_SauM-A, vB_SauM-C, vB_SauM-D on 67 multidrug resistant S. aureus clinical isolates.
| SA 1 | Origin 2 | Antibiotic Resistance 3 | A 4 | C 5 | D 6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | IP | Pus | Fox | P | E | CC | SXT | TE | − | − | − | ||||
| 17 | IP | Wound | Fox | P | E | CC | SXT | TE | − | − | − | ||||
| 19 | IP | Blood | Fox | P | E | CC | NOR | CIP | SXT | TE | − | − | − | ||
| 20 | IP | Wound | Fox | P | E | CC | SXT | TE | − | − | − | ||||
| 21 | IP | drain content | Fox | P | CC | TE | − | − | − | ||||||
| 23 | IP | Wound | Fox | P | E | CC | NOR | CIP | SXT | TE | − | − | − | ||
| 44 | IP | Throat | Fox | P | E | CC | NOR | CIP | TE | +++ | − | +++ | |||
| 70 | IP | Throat | Fox | P | E | CC | NOR | CIP | TE | ++ | +++ | +++ | |||
| 108 | IP | Urine | Fox | P | E | CC | NOR | CIP | +++ | + | +++ | ||||
| 109 | IP | Wound | Fox | P | E | CC | NOR | CIP | ++ | + | +++ | ||||
| 110 | IP | Fistula | Fox | P | E | CC | NOR | CIP | ++ | + | +++ | ||||
| 111 | IP | Wound | Fox | P | E | CC | NOR | CIP | ++ | +++ | +++ | ||||
| 112 | IP | Bedsore | Fox | P | E | CC | NOR | CIP | TE | GM | − | − | − | ||
| 113 | IP | Wound | Fox | P | E | CC | NOR | CIP | C | ++ | +++ | +++ | |||
| 115 | IP | Wound | Fox | P | E | CC | NOR | CIP | ++ | + | +++ | ||||
| 116 | IP | wound | Fox | P | E | CC | NOR | CIP | ++ | + | +++ | ||||
| 118 | IP | blood | Fox | P | E | CC | NOR | CIP | TE | GM | − | + | − | ||
| 120 | IP | bedsore | Fox | P | E | CC | NOR | CIP | TE | GM | − | + | − | ||
| 121 | IP | bronchial tree discharge | Fox | P | E | CC | NOR | CIP | TE | GM | − | + | − | ||
| 122 | IP | bronchial tree discharge | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 124 | IP | bronchial tree discharge | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 126 | OP | sputum | Fox | P | E | CC | NOR | CIP | TE | GM | − | + | − | ||
| 140 | OP | furuncle | Fox | P | E | CC | NOR | CIP | TE | C | − | − | − | ||
| 149 | IP | nose | Fox | P | E | CC | NOR | CIP | ++ | + | +++ | ||||
| 151 | IP | furuncle | Fox | P | E | CC | C | − | + | − | |||||
| 173 | OP | furuncle | Fox | P | E | CC | TE | C | + | + | − | ||||
| 182 | OP | furuncle | Fox | P | E | CC | C | − | + | − | |||||
| 183 | OP | furuncle | Fox | P | E | CC | TE | C | − | + | − | ||||
| 184 | IP | wound | Fox | P | E | CC | NOR | CIP | TE | GM | − | + | + | ||
| 193 | IP | bedsore | Fox | P | E | CC | NOR | CIP | ++ | +++ | +++ | ||||
| 196 | OP | wound | Fox | P | E | CC | TE | C | − | − | − | ||||
| 198 | OP | wound | Fox | P | E | CC | NOR | CIP | TE | C | GM | − | + | + | |
| 199 | OP | wound | Fox | P | E | CC | TE | C | +++ | +++ | +++ | ||||
| 200 | OP | sputum | Fox | P | E | CC | NOR | CIP | +++ | + | +++ | ||||
| 201 | IP | sputum | Fox | P | E | CC | NOR | CIP | − | + | + | ||||
| 202 | IP | bronchial tree discharge | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 203 | IP | vascular catheter | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 204 | OP | wound | Fox | P | E | CC | − | − | − | ||||||
| 205 | OP | bedsore | Fox | P | E | CC | NOR | CIP | − | + | + | ||||
| 258 | IP | nose | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 271 | IP | wound | Fox | P | E | CC | NOR | CIP | C | +++ | +++ | +++ | |||
| 297 | IP | sore | Fox | P | E | CC | NOR | CIP | C | +++ | +++ | +++ | |||
| 298 | IP | tissue | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 301 | IP | ear | Fox | P | E | CC | NOR | CIP | +++ | ++ | +++ | ||||
| 305 | IP | wound | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 311 | IP | wound | Fox | P | E | CC | NOR | CIP | +++ | ++ | +++ | ||||
| 315 | IP | pus from abscess | Fox | P | E | CC | TE | C | − | + | − | ||||
| 316 | IP | wound | Fox | P | E | NOR | CIP | +++ | + | +++ | |||||
| 317 | IP | skin | Fox | P | E | NOR | CIP | +++ | + | +++ | |||||
| 324 | IP | nose | Fox | P | E | CC | NOR | CIP | +++ | ++ | +++ | ||||
| 340 | IP | wound | Fox | P | E | CC | NOR | CIP | +++ | ++ | +++ | ||||
| 341 | IP | urine | Fox | P | E | NOR | CIP | ++ | + | +++ | |||||
| 342 | IP | endotracheal tube content | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 343 | IP | wound | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 344 | IP | wound | Fox | P | E | CC | C | − | + | − | |||||
| 345 | IP | nose | Fox | P | E | CC | C | − | + | − | |||||
| 351 | IP | pus from abscess | Fox | P | E | CC | C | − | + | − | |||||
| 352 | IP | nose | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 353 | IP | nose | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 355 | IP | endotracheal tube content | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 357 | IP | ear | Fox | P | NOR | CIP | +++ | +++ | +++ | ||||||
| 358 | IP | bronchial aspirate | Fox | P | E | CC | NOR | CIP | C | +++ | +++ | +++ | |||
| 366 | IP | wound | Fox | P | E | CC | NOR | CIP | ++ | +++ | +++ | ||||
| 367 | IP | wound | Fox | P | E | CC | NOR | CIP | +++ | +++ | +++ | ||||
| 369 | IP | nose | Fox | P | E | NOR | CIP | − | + | − | |||||
| 370 | IP | endotracheal tube content | Fox | P | E | CC | NOR | CIP | ++ | +++ | +++ | ||||
| 371 | IP | bronchoalveolar lavage fluid | Fox | P | TE | C | +++ | +++ | ++ | ||||||
1 SA, Staphylococcus aureus strain; 2 IP, inpatient, OP, outpatient; 3 FOX, cefoxitin (30 μg); P, penicillin (10 U); E, erythromycin (15 μg); CC, clindamycin (2 μg); NOR, norfloxacin (10 μg); CIP, ciprofloxacin (5 μg); SXT, sulfamethoxazole/trimethoprim (23.75 μg/1.25 μg); TE, tetracycline (30 μg); C, chloramphenicol (30 μg); GM, gentamicin (10 μg); 4 vB_SauM-A; 5 vB_SauM-C; 6 vB_SauM-D; +++ plaques at lysate concentration ≤ 104 PFU/mL, ++ plaques at lysate concentration 105–106 PFU/mL, + plaques at lysate concentration 107 PFU/mL, − no plaques or plaques at lysate concentration ≥108 PFU/mL.
3.4. Sensitivity of Phage Particles to Temperature, pH and Chloroform and Virion Stability Analysis
Phages vB_SauM-A, vB_SauM-C, vB_SauM-D were tested for their sensitivity to physical and chemical factors and the results are listed in Table 4. All of the phages showed strong resistance to CHCl3 and they were stable in pH range between 4 and 8. Unfortunately, exposition to pH lower than 3 and pH equal or higher than 10 resulted in loss of phage activity. Exposition of bacteriophages to higher temperature (40 °C) resulted in approx. 50% reduction of relative phage titer. Furthermore, exposition of bacteriophages to 62 °C resulted in an inactivation of all tested bacteriophages.
Table 4.
Bacteriophages sensitivity to physical and chemical factors.
| Phage Name | Phage Survivability in Studied Conditions (Relative Phage Titer in %) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 40 °C (40 min) | 62 °C (40 min) | pH 2 (2 h) | pH 3 (2 h) | pH 4 (2 h) | pH 6 (2 h) | pH 8 (2 h) | pH 10 (2 h) | pH 12 (2 h) | CHCl3 (4 °C; 1 h) | |
| vB_SauM-A | 58 | 0 | 0 | 0 | 8 | 100 | 83 | 0 | 0 | 97 |
| vB_SauM-C | 59 | 0 | 0 | 0 | 20 | 100 | 100 | 0 | 0 | 80 |
| vB_SauM-D | 43 | 0 | 0 | 0 | 87 | 100 | 95 | 0 | 0 | 100 |
Virion stability tests showed no change in titer of phages vB_SauM-C, vB_SauM-D over 10-month period. In the case of phage vB_SauM-A, a drop from original titer of 1.2 × 1011 PFU/mL to 1.56 × 1010 PFU/mL was observed. However, a decrease in titer did not affect the phage performance. Therefore, all three phages are able to withstand prolonged storage periods without the loss in ability to infect their host.
3.5. Lysis Profiles
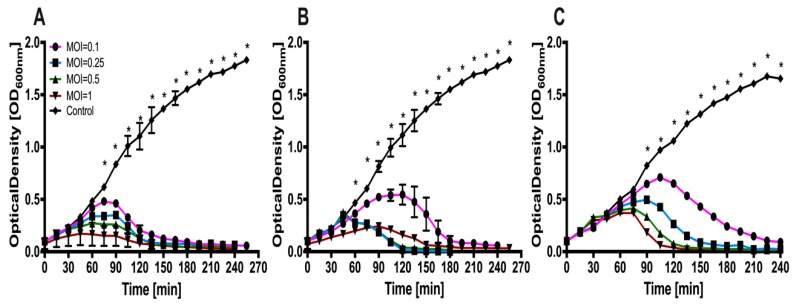
To examine the efficiency of phages vB_SauM-A, vB_SauM-C, vB_SauM-D infection, liquid culture of corresponding propagation host was infected with different MOI values and the bacterial growth was assessed by measuring the optical density of the culture. All of the isolated bacteriophages exhibit strong lytic activity, addition of phage lysate resulted in high reduction of the bacterial culture optical density. Figure 3 illustrates results of bacteriophage mediated reduction of S. aureus liquid culture optical density.
Figure 3.
Bacteriolytic activity of phages vB_SauM-A, vB_SauM-C, vB_SauM-D at different multiplicities of infection (MOIs) (that are 0, 0.1, 0.25, 0.5 and 1) against representative MDR S. aureus strains in liquid culture. Each curve represents the average results for three replicates, error bars represent standard deviation (SD) (A) Growth curves of S. aureus 203 infected with vB_SauM-A (B) Growth curves of S. aureus 343 infected with vB_SauM-C (C) Growth curves of S. aureus 343 infected with vB_SauM-D. Statistically significant difference was highlighted by asterisk (*). Only those time points at which all measured optical density (OD) values were significantly different when compared to control, were indicated.
The lysogenization test showed limited production of bacteriophage insensitive mutants. Lack of amplicons after PCR reaction with primers specific to the Kayvirus DNA polymerase I gene indicate that bacteriophage do not integrate into bacterial chromosome. Bacteriophages genome analysis showed also lack of integrase genes which makes genome integration impossible.
3.6. Phage Genome Analysis
The complete nucleotide sequences of phages vB_SauM-A, vB_SauM-C, vB_SauM-D were determined and deposited in GeneBank under the accession numbers MN539738, MN539737, MN539736, respectively. The detailed bacteriophage genome properties are presented in Table 5. Phages have genome length between 139,031 bp and 140,086 bp with low G+C content: approximately 30%.
Table 5.
Bacteriophages genomes properties.
| Phage Name | Genome Properties | ||
|---|---|---|---|
| Genome Length | GC content (%) | Phage K Similarity (%) | |
| vB_SauM-A | 139,031 | 30.45 | 97 |
| vB_SauM-C | 140,086 | 30.43 | 95 |
| vB_SauM-D | 139,088 | 30.46 | 97 |
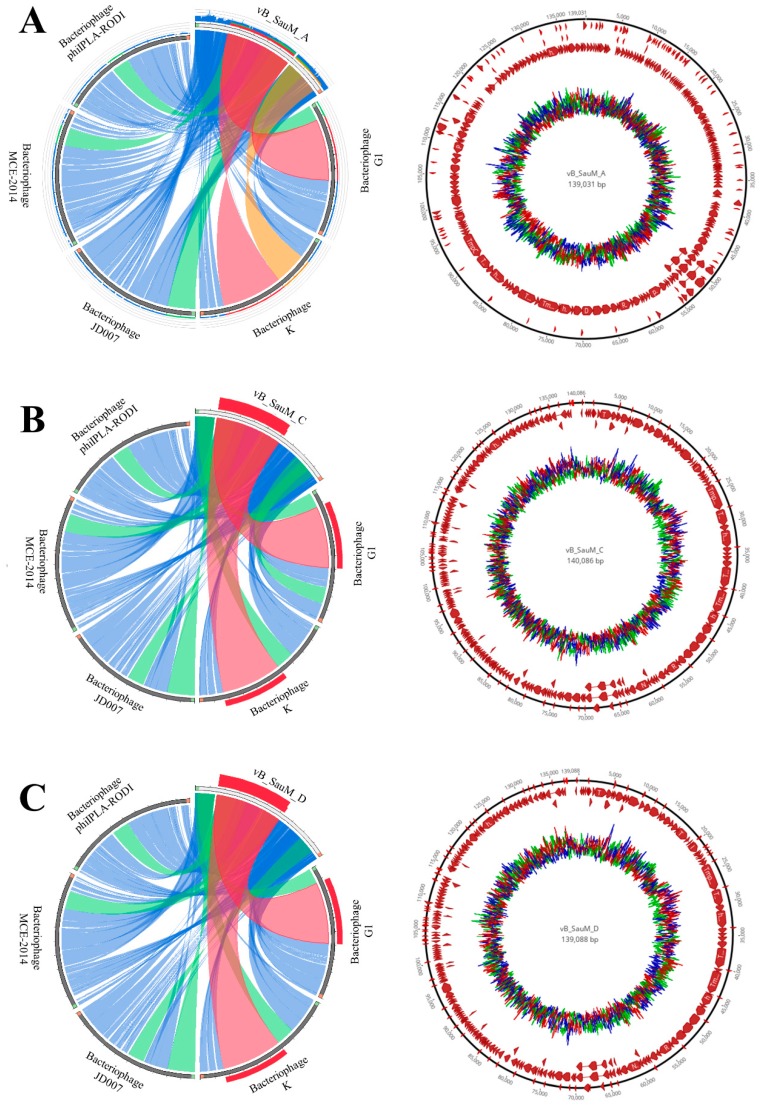
Bacteriophages genome organization is presented in Figure 4 and supplementary Figures S1–S3. Detailed genome annotation is available in supplementary Tables S1–S3. Detailed key information that corresponds to cell lysis as well as to other aspects of phage life cycle is presented in Table 6. The table represents putative lytic genes and their products, as well as other genetic peculiarities for example, those that determine the host specificity. The middle genome regions identified in all isolated bacteriophages carry genes responsible for DNA metabolism, replication and repair. These genes are presented in DNA metabolisms and DNA replication rows of Table 6. The late genome regions are responsible for encoding the structural, cell lysis and DNA packaging proteins. The identified genes are presented in head, neck, tail, baseplate, tail fiber and lysis rows of Table 6. Additionally, four genes that encode Met, Asp, Phe and Trp tRNA were annotated. Integrases were not found in the bacteriophage genomes. Analysis showed similar genome organization of the bacteriophages vB_SauM-C and vB_SauM-D. The genome similarity of isolated bacteriophages showed 95–97% coverage with phage K-type strain of the kayvirus. Figure 4 also shows high similarity of isolated bacteriophages to selected kayviruses like bacteriophage G1, K, JD007, MCE-2014 and phiIPLA-RODI (NCBI accession numbers: AY954969.1, KF766114.1, JX878671.1, KJ888149.1, KP027446.1, respectively). Additionally, the prediction of bacteriophage lifecycle with PHACT software indicates that all of isolated bacteriophages are characterized by lytic life cycle.
Figure 4.
Genome organization of isolated bacteriophages. Comparison of genomes of isolated bacteriophages with selected kayviruses, where: blue represents more than 50% similarity, green more than 75% similarity, orange more than 90% similarity and the red represents more than 99% similarity. (A) genome similarity of bacteriophage vB_SauM-A with other kayviruses and genome organization and G+C screw of vB_SauM-A genome. (B) Genome similarity of bacteriophage vB_SauM-C with other kayviruses and genome organization and GC screw of vB_SauM-C genome. (C) genome similarity of bacteriophage vB_SauM-D with other kayviruses and genome organization and GC screw of vB_SauM-D genome.
Table 6.
Detailed information on bacteriophages.
| Gene Group | vB_SauM-A | vB_SauM-C | vB_SauM-D |
|---|---|---|---|
| DNA metabolisms | nucleotide kinase (orf613), NTP pyrophosphohydrolase (orf612), nucleoside 2-deoxyribosyltransferase (orf602), RNA ligase (orf601), ribonuclease H (orf559), HNH endonuclease (orf587, 580), intron-encoded nuclease (orf1593), endonuclease (orf201, 264), homing endonuclease (orf206), ribose-phosphate pyrophosphokinase (orf280) | HNH endonuclease (orf312,319, 392, 688), ribonuclease H (orf2411), NTP pyrophosphohydrolase (orf343) nucleotide kinase (orf344), ribose-phosphate pyrophosphokinase (orf187), intron-encoded nuclease (orf216), | ribonuclease H (orf594), HNH endonuclease (orf580, orf587), RNA ligase (orf601), NTP pyrophosphohydrolase (orf612), ribonuclease H (orf453), intron-encoded nuclease (orf592), |
| DNA replication | DNA helicase (orf177, 180), recombination-related endonuclease (orf181, 184), anti-sigma factor (orf186), DNA primase (orf187), resolvase (orf190), ribonucleotide reductase stimulatory protein (orf191), ribonucleotide reductase large subunit (orf193), ribonucleotide reductase small subunit (orf195), DNA polymerase I (orf208), recombinase (orf218), sigma factor (orf221), DNA repair exonuclease (orf230), sliding clamp inhibitor (orf252), nucleotidyl transferase (orf642), terminase large subunit (orf101) | nucleotidyl transferase (orf373), recombinase (orf125), endonuclease (orf669), DNA polymerase I (orf107), ribonucleotide reductase large subunit (orf100), ribonucleotide reductase small subunit (orf638), resolvase (orf97), DNA primase (orf94), anti-sigma factor (orf93), recombination-related endonuclease (orf91, 88), helicase (orf87, 84), terminase large subunit (orf8) | DNA polymerase I (orf200), anti-sigma factor (orf186), recombination-related endonuclease (orf181), sigma factor (orf221), DNA helicase (orf177, 180), DNA primase (orf187), DNA repair exonuclease (orf230), ribonucleotide reductase small subunit (orf227), terminase large subunit (orf101), ribonucleotide reductase large subunit (orf193), recombinase (orf218) |
| Head | portal protein (orf110), prohead protease (orf112), major capsid protein (orf115) | major capsid protein (orf22), prohead protease (orf19), portal protein (orf17), putative virion protein (orf49) | portal protein (orf110), major capsid protein (orf115), prohead protease (orf112) |
| neck protein | neck protein (orf120, 122) | putative neck protein (orf29, 27) | neck protein (orf120, 122) |
| Tail | major tail protein (orf38), tail sheath protein (orf127), tail tube protein (orf129), tail assembly chaperone(orf137), tape-measure protein (orf140), tail murein hydrolase (orf145), cysteine protease (orf147), tail protein (orf226) | major tail protein (orf366), tail protein (orf 133), tail central spike (orf55), tail murein hydrolase (orf52), tape measure protein (orf47), tail morphogenetic protein (orf241), tail assembly chaperone (orf44), tail tube protein (orf36), tail sheath protein (orf34), baseplate hub assembly protein (orf31) | major tail protein (orf38), tape-measure protein (orf140), tail murein hydrolase (orf145), nicotinamide phosphoribosyl transferase (orf381), tail morphogenetic protein (orf55), adsorption-associated tail protein (orf162), tail assembly chaperone (orf137), tail sheath (orf127), tail tube protein (orf129), receptor binding protein (orf174, 170), tail central spike (orf148) |
| baseplate | baseplate component (orf151, 155, 173), baseplate wedge subunit (orf154), baseplate protein (orf160), receptor binding protein (orf170), oxidoreductase (orf197) | putative baseplate component (orf80, 60, 58), receptor binding protein (orf81), receptor binding protein (orf77) baseplate protein (orf67) baseplate component (orf62), baseplate wedge subunit (orf61) | peptidoglycan binding protein (orf604), baseplate component (orf153, 155,160, 173), baseplate wedge subunit (orf154), baseplate hub assembly protein (orf124) |
| Tail fiber | tail fiber protein (orf169) | tail fiber protein (orf69), Tail fiber protein complex (orf65) | tail fiber protein (orf169) |
| Lysis | lytic transglycosylase (orf588), N-acetylmuramoyl-L-alanine amidase (orf578), holin (orf575) | holin (orf307), N-acetylmuramoyl-L-alanine amidase (orf313), lytic transglycosylase (orf320) | holin (orf575), N-acetylmuramoyl-L-alanine amidase (orf581, 578) lytic transglycosylase (orf588) |
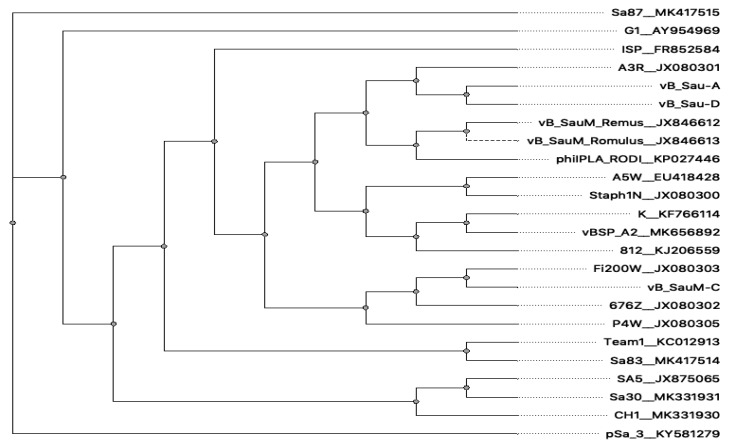
The phylogenetic analysis based on the GBDP of whole phage genomes revealed that isolated bacteriophages are related to members of the genus Kayvirus in subfamily Twortvirinae of the family Herelleviridae (Figure 5). Worth mentioning is that bacteriophages can be grouped in one clade with other bacteriophages isolated in Poland. Phylogenetic relative differences between the three newly isolated phages were shown in the Figure 6.
Figure 5.
Whole genome phylogeny of selected kayviruses and newly isolated bacteriophages described in this work.
Figure 6.
Phylogeny of newly isolated bacteriophages described in this work.
4. Discussion
Phage therapy is seen as a more cost-effective alternative to the isolation and development of new antibiotics against multidrug-resistant nosocomial pathogens such as MRSA [32]. Phages can be found in all habitats and be isolated relatively easy and at low cost. Furthermore the use of phages lacks serious side effects on eukaryotic cells, that some antibiotics may cause [4,32]. However, an in-depth characterization of isolated phages is required in order to assess their therapeutic potential against clinical isolates of pathogenic bacteria. In the last decade, several papers describing the isolation, biological and genomic characterization of S. aureus bacteriophages have been published [9,11,33]. As bacteriophages tend to be more specific than antibiotics [4] and since properties of pathogenic strains of MRSA can differ based on a variety of factors [34,35,36], there is a tendency to focus on strains and phages obtained locally [8,10,37]. So far, there is a lack of information on the bacteriophages active against clinical isolates from Gdansk region, Poland. Thus, this study aimed at characterizing three newly isolated phages that show lytic activity against clinical isolates of multidrug-resistant Staphylococcus aureus in a view of their potential application in therapy. Bacteriophages were characterized according to protocols implemented by other research groups [8,10,11,21] taking the phage morphology, genetics, survivability, stability, host range and growth parameters into consideration.
TEM observation revealed that all three phages have similar morphology with the same head shape, similar tail lengths and diameters. According to previously published criteria [38] phages vB_SauM-A, vB_SauM-C and vB_SauM-D display morphology corresponding to phages of the myoviridae family. However, taking into account genome length, low G+C content, gene organization, it can be stated that isolated viruses are similar to well-studied S. aureus phages such as phage K, G1 or ISP, which have been applied successfully in the treatment of bacterial infections [31,39]. Comparative genomic analysis of isolated bacteriophages showed that these bacteriophages are closely related to bacteriophage K and other kayviruses. Phylogenetically isolated bacteriophages are closely related to other bacteriophages isolated in Poland and widely described by Łobocka et al. [40]. Considering the latest changes in phage taxonomy [41] we conclude that phages vB_SauM-A, vB_SauM-C and vB_SauM-D belong to Kayvirus genus of Herelleviridae family. Additionally, comparison of genomes structure of isolated bacteriophages to well-known viruses representing Kayvirus genus showed high similarity of genome structure of isolated bacteriophages to kayviruses. It is worth mentioning that bacteriophage lifestyle prediction showed strictly virulent cycle also characteristic to other kayviruses and there were no integrases detected in these bacteriophages’ genomes. Lysins gene (lysK) of isolated bacteriophages showed high similarity to lysK gene and lysins of bacteriophage LM12, according to which there is high probability that bacteriophages will exhibit biofilm reducing properties [9,42].
Bacteriophages can be resistant to various unfavorable environmental conditions [18,43]. Based on phage tolerance to various environmental factors as well as other properties their therapeutic potential can be assessed [22]. Performed experiments revealed that isolated phages were resistant to pH between 4 and 8, which is a typical range of pH tolerance for most of environmental phages [22,44,45]. Unfortunately, all of isolated bacteriophages showed limited resistance to the growth temperature. None of them is able to persist 40 min at 62 °C. These results seem to be similar to the results of Zhang et al. (2015) which showed that other Kayvirus, bacteriophage JS25, rapidly decreased its viability when it to 60 °C [43]. However, since isolated phages are considered to be used as a potential treatment against MDRSA, primary focus should be on their performance and survivability at 37 °C, which has been proven to be satisfactory. Isolated phages are also able to survive long storage periods without decrease in titer or activity, which is also a desirable feature of phages used for therapy [45,46,47].
The spot test results indicate that phages vB_SauM-A, vB_SauM-C, vB_SauM-D are specific to Staphylococcus sp. and do not possess the ability to infect other bacterial genera both Gram-negative (such as Salmonella, Shigella or Pseudomonas) and Gram-positive (Lactobacillus, Enterococcus). Furthermore, it was found that isolated phages have a wide host range among MDRSA isolates collected from different clinical sites, both from inpatients and outpatients which is in accordance with other staphylococcal K-like phages [9,16,18]. Though it was proven that selected spot test method, when spotting high titer lysates (109 PFU/mL), can generate false positive results correlated with abortive infection, lysis from without, and enzyme activity [48] it is still used to screen the bactericidal ability of the isolated phages [25,49,50]. Moreover, applying phages at lower concentrations prevents from the overestimation of phage sensitivity [51]. In this study we used 10-fold dilutions of phage lysates (from 1011 PFU/mL to 102 PFU/mL) to ensure that a productive phage infection occurred. Broad lytic spectrum on pathogenic strains of S. aureus and high specificity of isolated phages show that they can be considered a promising tool for therapeutic purposes [9].
The one-step growth experiments displayed that burst size of vB_SauM-C and vB_SauM-D can be compared to other phages but the burst size of vB_SauM-A is considerably higher than that obtained for other anti-staphylococcal bacteriophages like JS25, SAP-26, phage K and DRA88 which produced approx. 21, 107, 125 or 76 PFU per infected cell, respectively [18,43,52]. Broad host range in combination with short latent period and a relatively large burst size confirm the lytic nature of the isolated bacteriophage.
The lytic potential of isolated viruses showed that all isolated bacteriophages possess strong lytic activity. The bacteriolytic assay showed that all of isolated bacteriophages were able to completely lyse S. aureus in in vitro culture at a MOI approx. 0.1. In contrast, bacteriophage JS25 completely lyse the bacterial culture at a MOI = 1 but the other MOI values were not shown [53]. The specific and efficient lysis of the host cell showed that these bacteriophages have a feature that make them potentially good candidates for therapeutic purposes. Additionally, low phage insensitive mutant production, bacteriophage impossibility to integrate into the host genome, lack of Staphylococcus specific DNA and toxins in genome, minimizes the transduction probability.
Taken together, the isolated bacteriophages demonstrate properties desirable for therapeutic phages. Phage vB_SauM-A with its high lytic potential and unusually large burst size shows the most promise in that aspect and its potential should be further investigated in future research.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/7/10/471/s1, Figure S1: Genome organization of bacteriophage vB_SauM_A, Table S1: Genome annotation of bacteriophage vB_SauM_A, Figure S2: Genome organization of bacteriophage vB_SauM_C, Table S2: Genome annotation of bacteriophage vB_SauM_C, Figure S3: Genome organization of bacteriophage vB_SauM_D, Table S3: Genome annotation of bacteriophage vB_SauM_D.
Author Contributions
Conceptualization, N.Ł.; methodology, N.Ł., B.G., A.Z.-P. and K.K.-K.; software, B.G. and N.Ł.; formal analysis, N.Ł.; investigation, N.Ł., B.G., A.Z.-P. and K.K.-K.; resources, N.Ł.; writing—original draft preparation, N.Ł., B.G. and K.K.-K.; writing—review and editing, A.W., L.P., B.D., supervision, N.Ł., L.P.; project administration, N.Ł.; funding acquisition, N.Ł. and L.P.
Funding
This work was supported by MN-01-0320/08/289 grant of Medical University of Gdansk.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Tong S.Y.C., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Centre for Disease Prevention and Control . Surveillance of Antimicrobial Resistance in Europe—Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017. ECDC; Stockholm, Sweden: 2018. [(accessed on 14 May 2019)]. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017. [Google Scholar]
- 3.Foster T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017;41:430–449. doi: 10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- 4.Loc-Carrillo C., Abedon S.T. Pros and cons of phage therapy. Bacteriophage. 2011;1:111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sulakvelidze A., Alavidze Z., Morris J.G.J. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres-Barcelo C. Phage Therapy Faces Evolutionary Challenges. Viruses. 2018;10:323. doi: 10.3390/v10060323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abhilash M., Vidya A.G., Jagadevi T. Bacteriophage Therapy: A War Against Antibiotic Resistant Bacteria. Internet J. Altern. Med. 2008;7:e17744. [Google Scholar]
- 8.Pantůček R., Rosypalová A., Doškař J., Kailerová J., Růžičková V., Borecká P., Snopková Š., Horváth R., Götz F., Rosypal S. The Polyvalent Staphylococcal Phage φ812:Its Host-Range Mutants and Related Phages. Virology. 1998;246:241–252. doi: 10.1006/viro.1998.9203. [DOI] [PubMed] [Google Scholar]
- 9.Melo L.D.R., Brandão A., Akturk E., Santos S.B., Azeredo J. Characterization of a new Staphylococcus aureus Kayvirus harboring a lysin active against biofilms. Viruses. 2018;10:182. doi: 10.3390/v10040182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leskinen K., Tuomala H., Wicklund A., Horsma-Heikkinen J., Kuusela P., Skurnik M., Kiljunen S. Characterization of vB_SauM-fRuSau02, a Twort-Like Bacteriophage Isolated from a Therapeutic Phage Cocktail. Viruses. 2017;9:258. doi: 10.3390/v9090258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abatángelo V., Peressutti Bacci N., Boncompain C.A., Amadio A.A.F., Carrasco S., Suárez C.A., Morbidoni H.R. Broad-range lytic bacteriophages that kill Staphylococcus aureus local field strains. PLoS ONE. 2017;12:e0181671. doi: 10.1371/journal.pone.0181671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandersteegen K., Mattheus W., Ceyssens P.-J., Bilocq F., De Vos D., Pirnay J.-P., Noben J.-P., Merabishvili M., Lipinska U., Hermans K., et al. Microbiological and Molecular Assessment of Bacteriophage ISP for the Control of Staphylococcus aureus. PLoS ONE. 2011;6:e24418. doi: 10.1371/journal.pone.0024418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzmán C., Jofre J., Blanch A.R., Lucena F. Development of a feasible method to extract somatic coliphages from sludge, soil and treated biowaste. J. Virol. Methods. 2007;144:41–48. doi: 10.1016/j.jviromet.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Park M., Lee J.H., Shin H., Kim M., Choi J., Kang D.H., Heu S., Ryu S. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl. Environ. Microbiol. 2012;78:58–69. doi: 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danis-Wlodarczyk K., Olszak T., Arabski M., Wasik S., Majkowska-Skrobek G., Augustyniak D., Gula G., Briers Y., Jang H.B., Vandenheuvel D., et al. Characterization of the Newly Isolated Lytic Bacteriophages KTN6 and KT28 and Their Efficacy against Pseudomonas aeruginosa Biofilm. PLoS ONE. 2015;10:e0127603. doi: 10.1371/journal.pone.0127603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Z., Feng T., Gu F., Li Q., Dong K., Zhang Y., Zhu Y., Han L., Qin J., Guo X. Characterization and complete genome of the virulent Myoviridae phage JD007 active against a variety of Staphylococcus aureus isolates from different hospitals in Shanghai, China. Virol. J. 2017;14:26. doi: 10.1186/s12985-017-0701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J.F., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd ed. Volume 1. Cold Spring Harbor; New York, NY, USA: 2001. [Google Scholar]
- 18.Alves D.R., Gaudion A., Bean J.E., Perez Esteban P., Arnot T.C., Harper D.R., Kot W., Hansen L.H., Enright M.C., Jenkins A.T.A. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl. Environ. Microbiol. 2014;80:6694–6703. doi: 10.1128/AEM.01789-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barylski J., Nowicki G. The Discovery of phiAGATE, A Novel Phage Infecting Bacillus pumilus, Leads to New Insights into the Phylogeny of the Subfamily Spounavirinae. PLoS ONE. 2018;9:e86632. doi: 10.1371/journal.pone.0086632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nejman-Faleńczyk B., Bloch S., Licznerska K., Dydecka A., Felczykowska A., Topka G., Węgrzyn A., Węgrzyn G. A small, microRNA-size, ribonucleic acid regulating gene expression and development of Shiga toxin-converting bacteriophage ϕ24B. Sci. Rep. 2015;5:10080. doi: 10.1038/srep10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dydecka A., Bloch S., Rizvi A., Perez S., Nejman-Faleńczyk B., Topka G., Gasior T., Necel A., Wegrzyn G., Donaldson L.W., et al. Bad Phages in Good Bacteria: Role of the Mysterious orf63 of λ and Shiga Toxin-Converting ϕ24B Bacteriophages. Front. Microbiol. 2017;8:1618. doi: 10.3389/fmicb.2017.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurczak-Kurek A., Gasior T., Nejman-Faleńczyk B., Bloch S., Dydecka A., Topka G., Necel A., Jakubowska-Deredas M., Narajczyk M., Richert M., et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016;6:34338. doi: 10.1038/srep34338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamal M., Andleeb S., Jalil F., Imran M., Nawaz M.A., Hussain T., Ali M., Rahman S., Das C.R. Isolation, characterization and efficacy of phage MJ2 against biofilm forming multi-drug resistant Enterobacter cloacae. Folia Microbiol. 2019;64:101–111. doi: 10.1007/s12223-018-0636-x. [DOI] [PubMed] [Google Scholar]
- 24.D’Andrea M.M., Marmo P., Henrici De Angelis L., Palmieri M., Ciacci N., Di Lallo G., Demattè E., Vannuccini E., Lupetti P., Rossolini G.M., et al. φBO1E, a newly discovered lytic bacteriophage targeting carbapenemase-producing Klebsiella pneumoniae of the pandemic Clonal Group 258 clade II lineage. Sci. Rep. 2017;7:2614. doi: 10.1038/s41598-017-02788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topka G., Bloch S., Nejman-Falenczyk B., Gasior T., Jurczak-Kurek A., Necel A., Dydecka A., Richert M., Wegrzyn G., Wegrzyn A. Characterization of Bacteriophage vB-EcoS-95, Isolated From Urban Sewage and Revealing Extremely Rapid Lytic Development. Front. Microbiol. 2019;9:3326. doi: 10.3389/fmicb.2018.03326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier-Kolthoff J.P., Göker M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier-Kolthoff J.P., Auch A.F., Klenk H.-P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNair K., Bailey B.A., Edwards R.A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics. 2012;28:614–618. doi: 10.1093/bioinformatics/bts014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darzentas N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics. 2010;26:2620–2621. doi: 10.1093/bioinformatics/btq484. [DOI] [PubMed] [Google Scholar]
- 31.O’Flaherty S., Ross R.P., Meaney W., Fitzgerald G.F., Elbreki M.F., Coffey A. Potential of the Polyvalent Anti-Staphylococcus Bacteriophage K for Control of Antibiotic-Resistant Staphylococci from Hospitals. Appl. Environ. Microbiol. 2005;71:1836–1842. doi: 10.1128/AEM.71.4.1836-1842.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golkar Z., Bagasra O., Gene Pace D. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014;8:129–136. doi: 10.3855/jidc.3573. [DOI] [PubMed] [Google Scholar]
- 33.Jensen K.C., Hair B.B., Wienclaw T.M., Murdock M.H., Hatch J.B., Trent A.T., White T.D., Haskell K.J., Berges B.K. Isolation and Host Range of Bacteriophage with Lytic Activity against Methicillin-Resistant Staphylococcus aureus and Potential Use as a Fomite Decontaminant. PLoS ONE. 2015;10:e0131714. doi: 10.1371/journal.pone.0131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson L.O., Murphy C.R., Spratt B.G., Enright M.C., Elkins K., Nguyen C., Terpstra L., Gombosev A., Kim D., Hannah P., et al. Diversity of Methicillin-Resistant Staphylococcus aureus (MRSA) Strains Isolated from Inpatients of 30 Hospitals in Orange County, California. PLoS ONE. 2013;8:e62117. doi: 10.1371/journal.pone.0062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guimarães M.A., Ramundo M.S., Américo M.A., de Mattos M.C., Souza R.R., Ramos-Júnior E.S., Coelho L.R., Morrot A., Melo P.A., Fracalanzza S.E.L., et al. A comparison of virulence patterns and in vivo fitness between hospital- and community-acquired methicillin-resistant Staphylococcus aureus related to the USA400 clone. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:497–509. doi: 10.1007/s10096-014-2253-1. [DOI] [PubMed] [Google Scholar]
- 36.Figueiredo A.M.S. What is behind the epidemiological difference between community-acquired and health-care associated methicillin-resistant Staphylococcus aureus? Virulence. 2017;8:640–642. doi: 10.1080/21505594.2017.1335847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasser A., Azizian R., Tabasi M., Khezerloo J.K., Heravi F.S., Kalani M.T., Sadeghifard N., Amini R., Pakzad I., Radmanesh A., et al. Specification of Bacteriophage Isolated Against Clinical Methicillin-Resistant Staphylococcus Aureus. Osong Public Heal. Res. Perspect. 2019;10:20–24. doi: 10.24171/j.phrp.2019.10.1.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ackermann H.-W. Tailed Bacteriophages: The Order Caudovirales. Adv. Virus Res. 1998;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merabishvili M., Pirnay J.P., Verbeken G., Chanishvili N., Tediashvili M., Lashkhi N., Glonti T., Krylov V., Mast J., Van Parys L., et al. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE. 2009;4:e4944. doi: 10.1371/journal.pone.0004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Łobocka M., Hejnowicz M.S., Dabrowski K., Gozdek A., Kosakowski J., Witkowska M., Ulatowska M.I., Weber-Dabrowska B., Kwiatek M., Parasion S., et al. Advances in Virus Research. Volume 83. Elsevier; Amsterdam, The Netherlands: 2012. Genomics of Staphylococcal Twort-like Phages-Potential Therapeutics of the Post-Antibiotic Era; pp. 143–216. [DOI] [PubMed] [Google Scholar]
- 41.Barylski J., Enault F., Dutilh B.E., Schuller M.B., Edwards R.A., Gillis A., Klumpp J., Knezevic P., Krupovic M., Kuhn J.H., et al. Analysis of spounaviruses as a case study for the overdue reclassification of tailed bacteriophages. Syst. Biol. 2019:220434. doi: 10.1093/sysbio/syz036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao J., Schmelcher M., Harty W.J., Foster-Frey J., Donovan D.M. Chimeric Ply187 endolysin kills Staphylococcus aureus more effectively than the parental enzyme. FEMS Microbiol. Lett. 2013;342:30–36. doi: 10.1111/1574-6968.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q., Xing S., Sun Q., Pei G., Cheng S., Liu Y., An X., Zhang X., Qu Y., Tong Y. Characterization and complete genome sequence analysis of a novel virulent Siphoviridae phage against Staphylococcus aureus isolated from bovine mastitis in Xinjiang, China. Virus Genes. 2017;53:464–476. doi: 10.1007/s11262-017-1445-z. [DOI] [PubMed] [Google Scholar]
- 44.Rodela M.L., Sabet S., Peterson A., Dillon J.G. Broad Environmental Tolerance for a Salicola Host-Phage Pair Isolated from the Cargill Solar Saltworks, Newark, CA, USA. Microorganisms. 2019;7:106. doi: 10.3390/microorganisms7040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jończyk E., Kłak M., Międzybrodzki R., Górski A. The influence of external factors on bacteriophages-review. Folia Microbiol. 2011;56:191–200. doi: 10.1007/s12223-011-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casey E., van Sinderen D., Mahony J. In vitro characteristics of phages to guide ‘real life’ phage therapy suitability. Viruses. 2018;10:163. doi: 10.3390/v10040163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dąbrowska K. Phage therapy: What factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Med. Res. Rev. 2019:2000–2025. doi: 10.1002/med.21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirzaei M.K., Nilsson A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE. 2015;10:e0118557. doi: 10.1371/journal.pone.0127606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cha Y., Chun J., Son B., Ryu S. Characterization and Genome Analysis of Staphylococcus aureus Podovirus CSA13 and Its Anti-Biofilm Capacity. Viruses. 2019;11:54. doi: 10.3390/v11010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliveira H., Pinto G., Oliveira A., Noben J.P., Hendrix H., Lavigne R., Łobocka M., Kropinski A.M., Azeredo J. Characterization and genomic analyses of two newly isolated Morganella phages define distant members among Tevenvirinae and Autographivirinae subfamilies. Sci. Rep. 2017;7:1–14. doi: 10.1038/srep46157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Y., Wahab L., Gill J.J. Development and validation of a microtiter plate-based assay for determination of bacteriophage host range and virulence. Viruses. 2018;10:189. doi: 10.3390/v10040189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rahman M., Kim S., Kim S.M., Seol S.Y., Kim J. Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling. 2011;27:1087–1093. doi: 10.1080/08927014.2011.631169. [DOI] [PubMed] [Google Scholar]
- 53.Zhang L., Bao H., Wei C., Zhang H., Zhou Y., Wang R. Characterization and partial genomic analysis of a lytic Myoviridae bacteriophage against Staphylococcus aureus isolated from dairy cows with mastitis in Mid-east of China. Virus Genes. 2015;50:111–117. doi: 10.1007/s11262-014-1130-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.