Abstract
Development of new synthetic catalysts for CO2 reduction has been a central focus of chemical research efforts towards mitigating rising global carbon dioxide levels. In parallel with generating new molecular systems, characterization and benchmarking of these compounds across well-defined catalytic conditions are essential. Nickel(II) cyclam is known to be an active catalyst for CO2 reduction to CO. The degree of selectivity and activity has been found to differ widely across electrodes used and upon modification of the ligand environment, though without a molecular-level understanding of this variation. Moreover, while proton transfer is key for catalytic activity, the effects of varying the nature of the proton donor remain unclear. In this work, a systematic investigation of the electrochemical and light-driven catalytic behaviour of nickel(II) cyclam under different aqueous reaction conditions has been performed. The activity and selectivity are seen to vary widely depending on the nature of the buffering agent, even at a constant pH, highlighting the importance of proton transfer for catalysis. Buffer binding to the nickel center is negatively correlated with selectivity, and cationic buffers show high levels of selectivity and activity. These results are discussed in the context of molecular design principles for developing increasingly efficient and selective catalysts. Moreover, identifying these key contributors towards activity has implications for understanding the role of the conserved secondary coordination environments in naturally occurring CO2-reducing enzymes, including carbon monoxide dehydrogenase and formate dehydrogenase.
Introduction
Since the dawn of the industrial age, atmospheric carbon dioxide has increased monotonically, resulting in detrimental global changes.1–4 As these levels have now surpassed 400 ppm, there is renewed interest in developing systems that are capable of converting this greenhouse gas into value-added products such as carbon monoxide, formate, methanol, methane, and more complex carbon-based compounds such as acetate and oxalate.5–8 Of these, conversion to carbon monoxide (CO) is of particular interest due to downstream use of CO in industrial processes, including the Fischer-Tropsch process and the water-gas shift reaction.9 Two highly efficient natural systems capable of accomplishing the two-electron reduction of carbon dioxide selectively to CO and formate are carbon monoxide dehydrogenase (CODH) and formate dehydrogenase (FDH), respectively. However, CODH and FDH are quite large, which limits current densities, suffer from oxygen intolerance, require complex cofactors, and are difficult to express heterologously.2,10,11 Thus, the large-scale application of these naturally occurring enzymes for biotechnological or industrial devices remains impractical. To circumvent some of these issues, the development of synthetic catalysts that perform analogous chemistry has been a key area of research.8,12–14
A primary challenge for the conversion of carbon dioxide to carbon monoxide is selectivity over proton reduction or generation of other reduced products.2 The relative thermodynamic potentials for the competing proton reduction or formate generation reactions are more favorable than that for CO2 reduction to CO (Equations 1–3 vs. NHE, pH 7.0).
| (1) |
| (2) |
| (3) |
Developing synthetic compounds that can overcome this thermodynamic bias through kinetic control, as native enzymes are said to do, is a primary goal in this field of research. One synthetic catalyst of particular interest is the nickel-bound macrocycle [Ni(cyclam)]2+, where cyclam is 1,4,8,11-tetraazacyclotetradecane, due to its reported selectivity in water.15 However, much of this initial work was conducted using a hanging mercury drop electrode.16–18 When a glassy carbon working electrode is used instead, the activity and selectivity drop significantly.19 Substantial efforts have since been extended to improve the selectivity of this catalyst in water using a non-mercury–based electrode, including attachment to a poly(allylamine) backbone,20 attachment to ZnSe quantum dots21 and incorporation into a protein scaffold.22 Alternative approaches for enhancing catalysis have centered around varying the reaction conditions, with recent examples indicating changes in reduction potentials or product selectivity depending on the choice of reaction medium.23–28 Each parameter can be carefully adjusted to optimize catalyst behavior, though comparing activity and selectivity across compounds is made substantially more challenging when different experimental conditions are employed as the thermodynamics and kinetics can vary in different solvents.29,30,31
In characterizing a new catalytic system, it is important to select an appropriate medium for analysis. Ideally, the medium would not coordinate to the molecule, would maintain the intended local and bulk pH,32 and would not interfere with the catalytic reactions that are occurring. Because of this, Good proposed a series of buffers that seemingly met all of these criteria by introducing different functional groups to mitigate negative interactions,33,34 though a number of exceptions have now been observed even for the Good’s buffers.35–37 On the other hand, catalytic reduction of CO2 is often performed in aqueous solution using high concentrations of carbonate or bicarbonate ions, in which the medium itself is also the substrate.38,39 When switching from organic solvents to aqueous reaction conditions, unbuffered water containing just electrolyte is often used,15,17,19,40–42 which, for highly active systems, can result in dramatic local pH changes.32 An alternative strategy is to simply use one buffer for all reported experiments.39,42,43 Under such varied reaction conditions, apparent catalytic properties can change significantly, even for the same compound. Building from prior observations in our lab in which phosphate buffer was shown to bind directly to a catalytic intermediate,23 inhibiting electrocatalysis, we hypothesized that the chemical composition of the buffering agent could affect activity. In this work, we systematically investigate the effects of changing only buffer identity on the catalytic behavior of [Ni(cyclam)]2+. Electrochemical and photochemical characterization of activity and selectivity across eight small-molecule buffering systems has been performed in fully aqueous solutions. Buffers that bind with high affinity to the NiIII state are generally less active for CO2 reduction, consistent with previous studies that analyze [Ni(cyclam)]2+ activity in phosphate buffer,22,23 while cationic buffers increase selectivity and activity, implicating an important role for outer-sphere proton transfer in the catalytic mechanism. Moreover, conditions have been identified under which [Ni(cyclam)]2+ acts as a completely selective catalyst for CO2 reduction, indicating a role for the buffer in establishing a rudimentary secondary coordination environment. These results are discussed in the context of molecular design principles for developing and characterizing increasingly efficient, active, and selective catalysts and shed light into the role of carefully placed residues in the outer coordination spheres of naturally occurring enzymes that perform CO2 reduction.
Materials and Methods
All reagents were used as purchased, unless specified. Buffers used in this study were prepared using deionized water (18.2 MΩ, Elga Technologies), and the pH was adjusted accordingly using KOH or HCl to achieve a final pH of 7.0 for all experiments. The following buffers were used: sodium bicarbonate (Sigma-Aldrich), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; GoldBio), imidazole (Im; Alfa Aesar‡), 3-(N-morpholino)propanesulfonic acid (MOPS; GoldBio), potassium phosphate dibasic (Sigma-Aldrich), potassium phosphate monobasic (VWR), piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; BDH), triethanolamine (TEOA; J.T Baker), and tris(hydroxymethyl)aminomethane (Tris; VWR).
Synthesis of [Ni(cyclam)]2+
[Ni(cyclam)]2+ (cyclam = 1,4,8,11-tetraazacyclotetradecane) was synthesized following a published protocol.44 The cyclam ligand (Acros Organics) was dissolved in ethanol, and NiIICl2*6H2O (Alfa Aesar) was added in a 1:1 molar ratio. Upon addition of NiIICl2•6H2O (Alfa Aesar), the solution immediately turned mauve. This solution was then stirred for 15 minutes. [Ni(cyclam)]2+•Cl2 was precipitated by adding diethyl ether, and the solid purple material was collected by vacuum filtration.
Electrochemistry experiments
All cyclic voltammetry electrochemistry experiments were conducted using a CHI 760E potentiostat (CH Instruments) or a WaveNow potentiostat (Pine Instruments) under an inert or carbon dioxide atmosphere, as indicated. A typical three-electrode set up was employed using a 3 mm glassy carbon working electrode (CH Instruments), a platinum wire counter electrode, and a mini Ag/AgCl (sat. KCl) reference electrode (Pine Instruments). The glassy carbon working electrode was polished for 60 seconds with 1.0 micron alumina powder, extensively rinsed with deionized water, then polished for 60 seconds with 0.05 micron alumina powder (CH Instruments). The electrode was again rinsed and then sonicated for three minutes in deionized water prior to all electrochemistry experiments.
For electrochemical experiments, 100 μM [Ni(cyclam)]2+ was used with 100 mM KCl as a supporting electrolyte. For experiments conducted under an inert atmosphere, a final buffer concentration of 10, 50, or 100 mM was used as indicated. For experiments in the presence of carbon dioxide, 100 mM buffer was used and was pH-adjusted to a final pH of 7.0 following saturation with CO2.
Determination of buffer binding constants
The binding constant, KN, for each buffer was determined using cyclic voltammetry and the following equation (Equation 4):
| (4) |
Where N is the oxidation state to which preferential binding occurs, [X] is the concentration of the buffer under analysis, ΔE is the difference in the NiIII/II reduction potential in water compared to the reduction potential in the buffer of interest (EH2O-EX), F is Faraday’s constant, n is the number of electrons, R is the gas constant and T is the temperature.27,45,46 All reduction potentials were compared to that of [Ni(cyclam)]2+ in pH-adjusted, unbuffered water, and the reported binding constant for each buffer reflects analysed data averaged for each concentration over at least three trials, given with the standard deviation (Figure S1).
Electrochemistry analysis
All potentials were reported against NHE by the addition of +198 mV to the experimentally measured potentials. Baseline corrections were performed using the QSOAS program.47 This program was also used to identify the anodic and cathodic peak potentials, which were then averaged for the midpoint potentials (E1/2). Reduction potentials and errors reported reflect the mean and standard deviation of at least three independent trials. Due to the absence of a clear catalytic current plateau in the cyclic voltammograms of [Ni(cyclam)]2+ across all buffered solutions, a different metric has been established for comparison of onset potentials and turnover frequencies. For experiments performed under a carbon dioxide atmosphere, the onset potential of catalysis (Eonset) is defined as the inflection point of the voltammogram, or the point at which the first derivative is maximized (Figure S2). This procedure provides an objective point to analyze the non-ideal CVs often observed for catalysis in aqueous conditions.
The rate constants and corresponding turnover frequencies (TOFs) were determined using Equation 5:19
| (5) |
where k is the rate constant of interest, ic is the catalytic current, n is the number of electrons for the reaction, F is Faraday’s constant, A is the area of the electrode, [Ni(cyclam)] is the concentration of catalyst, D is the diffusion coefficient, and [CO2] is the concentration of CO2. After subtracting the capacitative current, the catalytic current was obtained by measuring the current at the determined onset potential (Eonset) for each buffer. The diffusion coefficient for [Ni(cyclam)]2+ has been previously reported to be 5.6 × 10−6 cm2 s−1.48 Analysis was done under a saturating carbon dioxide atmosphere ([CO2] = 36 mM).49 The TOF was obtained by multiplying the rate constant by the substrate concentration.
Determination of CO2 binding constants (KCO2)
Analysis was performed in a solution of 1 M buffer containing 100 mM KCl at a final adjusted pH of 7.2. Reactions were conducted in a total volume of 10 mL and contained 150 μM [Ni(cyclam)]Cl2. A three electrode setup was used, which consisted of a glassy carbon working electrode, platinum wire counter electrode, and a Ag/AgCl (sat. KCl) reference electrode. In order to achieve the desired concentrations of CO2, solutions of saturated CO2 were prepared by sparging buffer in a septum-capped GC vial. Aliquots of the CO2-saturated solution were combined with the appropriate amount of Ar-saturated buffer solution to reach a total volume of 10 mL at the desired CO2 concentration (0, 0.25, 0.5, 1, 2.5, 5, 10, 15, 20, 25, 30, 36 mM). Cyclic voltammograms were recorded at a scan rate of 50 mV/s for all trials. Catalytic onset potentials were determined by identifying the local maximum of the first derivative (the inflection point of the voltammogram) as described above. Potentials were then plotted as a function of the concentration of CO2 for each sample and fit to the modified Nernst equation (Equation 6):
| (6) |
where EN2 was allowed to vary (the non-catalytic peak is masked in aqueous solutions due to background proton reduction), ECO2 is the measured potential at a given concentration of CO2, K1,CO2 is the equilibrium binding constant for CO2 to [Ni(cyclam)]+, K2,CO2 is the equilibrium binding constant for CO2 to [Ni(cyclam)]2+, and all other terms are as defined previously.
Light-initiated photochemical assays
The experimental conditions for the light-driven photo-assays were adapted from previously published protocols.22,50 Samples were prepared in a septum-capped gas chromatography (GC) vial containing a stir bar, and all assays were kept at 4 °C using an ice water bath. Assays were carried out under a CO2 atmosphere in a solution of 1 M of the indicated buffer adjusted to a final pH of 7.0. Samples were extensively sparged before use with a high-purity carbon dioxide gas cylinder (Praxair) and maintained under a CO2 atmosphere. Each assay mixture contained 10 μM [Ni(cyclam)]2+, 1 mM [RuII(bpy)3]2+ (Aldrich) as a photosensitizer, and 100 mM ascorbate (Sigma-Aldrich) as a sacrificial electron donor. For photoexcitation, four LUXEON Rebel ES LEDs (447.5 nm) were spaced evenly from the GC vial, which resulted in a power of 4.5 mW. To quantify product formation, headspace samples were removed as a function of time and injected into the gas chromatograph. All values were corrected for background activity by subtracting the CO and H2 produced by the corresponding control of 1 mM [RuII(bpy)3]2+ with 100 mM ascorbate under a CO2-saturated atmosphere in solutions of 1 M buffer. Assays were performed in triplicate, and results reported are the mean values with standard deviations. The “% CO selective” value was obtained for each sample by comparing the amount of product that was produced using Equation 7:
| (7) |
Gas chromatography analysis
GC analysis was performed using a Shimadzu GC-2014 fuel cell analyzer system equipped with a thermal conductivity detector and a flame ionization detector coupled to a methanizer. Argon was used as the carrier gas for all experiments. Separation was achieved using a temperature gradient with the use of the following columns: HayeSep-N (3 m, 80/100 mesh), HayeSep-T (2 m, 80/100 mesh), Shimalite Q (0.2 m, 100/180), Shimalite Q (0.25 m, 100/180), Shimalite Q (0.15 m, 100/180), and a 5-Ångstrom molecular sieve (2.5 m, 60/80). Standard curves were generated using injections of Scotty standard gas calibration mixture (Figure S3; Product #A0908910).
Results
Electrochemical characterization of buffer interactions with [Ni(cyclam)]2+
Prior studies have demonstrated that interactions between [Ni(cyclam)]2+ and buffer components can impact catalysis.23 To quantitatively address the nature and effects of these interactions on activity, cyclic voltammograms (CVs) and photodriven assays have been measured to characterize [Ni(cyclam)]2+ in multiple buffering systems. These buffers ranged from anionic to neutral to cationic under the reaction conditions and included inorganic buffering agents as well as the purportedly non-interacting Good’s buffers (Figure 1). In the electrochemical experiments, a baseline for comparison was established by studying the electrochemical behavior of [Ni(cyclam)]2+ in a pH-adjusted, unbuffered aqueous solution containing only 100 mM KCl as electrolyte. Under an inert atmosphere, a reversible redox transition reflecting the NiIII/II couple is observed at +0.825 V vs. NHE. A reversible NiII/I redox couple under an inert atmosphere is masked by the reduction of protons by both [Ni(cyclam)]2+ and the electrode (Figure S4), resulting in relatively featureless CVs that resemble those previously reported.19 As buffer is introduced into the electrochemical cell, systematic variation in the signal is observed in both the positive and negative potential regimes. The first buffering agent investigated was bicarbonate, which is frequently used to buffer both the pH and the substrate concentration in CO2 reduction cells. As shown in Figure 2A, the NiIII/II couple shifts to lower potentials with increasing concentration of bicarbonate buffer added, indicative of preferential binding to the NiIII state. Following the square scheme established in Figure S5 and using Equation 4, an estimated differential affinity of 210 ± 50 M−1 is obtained for bicarbonate binding to the NiIII state. Additionally, as increasing amounts of bicarbonate are added, background proton reduction currents are reduced, though signals corresponding to the reduction of CO2 do not appear. The addition of a CO2 atmosphere was necessary to observe the standard catalytic signals for CO2 reduction at −1.21 V vs. NHE (Figure S6), which is consistent with slow interconversion between bicarbonate and dissolved CO2.
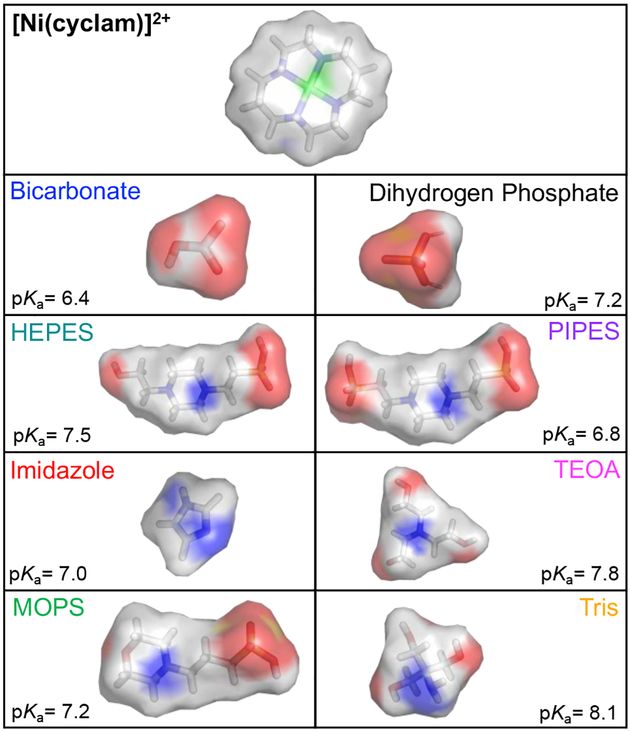
Figure 1.
Space-filling representations of [Ni(cyclam)]2+ in the trans-III configuration and the different buffering components used in this study with reported pKa values. Models were generated using Pymol.
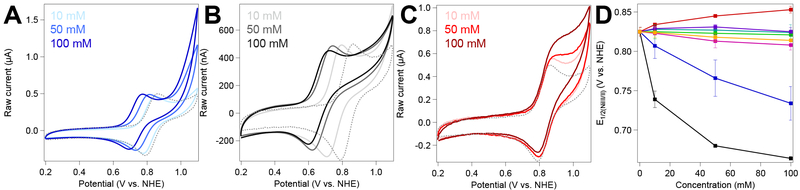
Figure 2.
Cyclic voltammograms (ν = 10 mV/s) of [Ni(cyclam)]2+ under an inert atmosphere in increasing concentrations of (A) sodium bicarbonate, (B) phosphate, and (C) imidazole buffers. Dashed gray line represents [Ni(cyclam)]2+ in pH-adjusted, unbuffered water. (D) E1/2 values of the NiIII/II couple of [Ni(cyclam)]2+ as a function of increasing concentrations of the following buffers: bicarbonate (blue); HEPES (teal); imidazole (red); MOPS (green); phosphate (black); PIPES (purple); TEOA (pink); and Tris (orange). All samples contained 100 μM [Ni(cyclam)]2+ and 100 mM KCl in buffer, pH 7.0, at the indicated concentrations.
To resolve how each of the buffering components individually interact with [Ni(cyclam)]2+, the NiIII/II reduction potentials were initially investigated for all buffers under an inert atmosphere. A trend similar to that seen for bicarbonate is observed when a phosphate buffer is used, with greater peak-to-peak separation and significant shifts of the NiIII/II couple to lower potentials as the phosphate concentration increases (Figure 2B and Table 1). The primary component in this buffer under the experimental conditions is the dihydrogen phosphate species, which for simplicity will be referred to as “phosphate” throughout the manuscript. The presence of the Good’s buffers, which feature sulfonate buffering moieties, show only weak interaction with the NiIII state, with little change in reduction potential or peak separation observed relative to those seen in unbuffered, pH-adjusted water (Figure S7–S9). Addition of imidazole as a buffering agent to the aqueous solution resulted in a positive shift of the NiIII/II midpoint potential, indicating preferential binding of this molecule to the NiII state relative to the NiIII state. However, this overall shift is only due to a change in the anodic peak potential (Figure 2C); the cathodic peak potential remains unchanged across varying amounts of added imidazole. As described above, the shifts in midpoint potential (Figure 2D) and binding constants, KN, for each buffer were extracted using Equation 4 (Table 1). The greatest potential changes and corresponding binding affinities are observed when the buffer added is phosphate and bicarbonate, the two polyanionic buffers analyzed. For the three cationic buffering solutions, the binding affinities are a similar order of magnitude to each other, with imidazole displaying the largest of the three. HEPES, MOPS, and PIPES all displayed insignificant binding, as indicated by NiIII/II reduction potentials that are nearly identical to that in water (Figure S10).
Table 1.
Summary of cyclic voltammetry data on the NiIII/II redox couple for [Ni(cyclam)]2+ in 100 mM buffer (ν = 10 mV/s). Average midpoint reduction potentials (E1/2) and peak separation (ΔEP) values given with standard deviation (n ≥ 3).
| Buffer | Chargea | E1/2 (V vs. NHE) | ΔEp (V vs. NHE) | KN (M−1) |
|---|---|---|---|---|
| H2O | NA | 0.825 ± 0.006 | 0.08 ± 0.02 | - |
| Bicarbonate | −1 | 0.73 ± 0.02 | 0.09 ± 0.03 | KIII = 210 ± 50 |
| HEPES | 0 | 0.82 ± 0.01 | 0.07 ± 0.02 | KIII = 1b |
| Imidazole | +1 | 0.853 ± 0.004 | 0.12 ± 0.01 | KII = 22 ± 3 |
| MOPS | 0 | 0.821 ± 0.005 | 0.06 ± 0.01 | KIII = 1.7 ± 0.1 |
| Phosphate | −1 | 0.664 ± 0.002 | 0.13 ± 0.02 | KIII = 4440 ± 800 |
| PIPES | −1 | 0.825 ± 0.005 | 0.08 ± 0.01 | KIII = 1b |
| TEOA | +1 | 0.810 ± 0.006 | 0.06 ± 0.02 | KIII = 10 ± 1 |
| Tris | +1 | 0.814 ± 0.002 | 0.07 ± 0.01 | KIII = 5.6 ± 0.3 |
The overall charge of the dominant buffering species at pH 7.0.
No preferential binding to either oxidation state.
The reversibility of the transition was also investigated using the peak separation as a reporter. For a completely reversible redox system operating at the diffusion limit, a peak-to-peak separation (ΔEp) of 57 mV is expected at slow scan rates.51 Most of the buffer systems investigated show ΔEp values within this limit at a scan rate of 10 mV/s. However, imidazole and phosphate buffers induce ΔEp values of 0.12 V and 0.13 V, respectively, significantly greater than those expected for a reversible system (Table 1, and Figures S11–S14). With imidazole buffer introduced, the peak separation and anodic peak position also show an unusual dependence on scan rate. Both decrease notably as scan rate is increased, opposite behavior as that observed with the other buffering agents (Figures S13–S14). This dependence on scan rate suggests that a chemical process, such as ligand binding or loss, occurs on the same timescale as electron transfer at the slower scan rates.51
Electrochemical characterization of catalysis
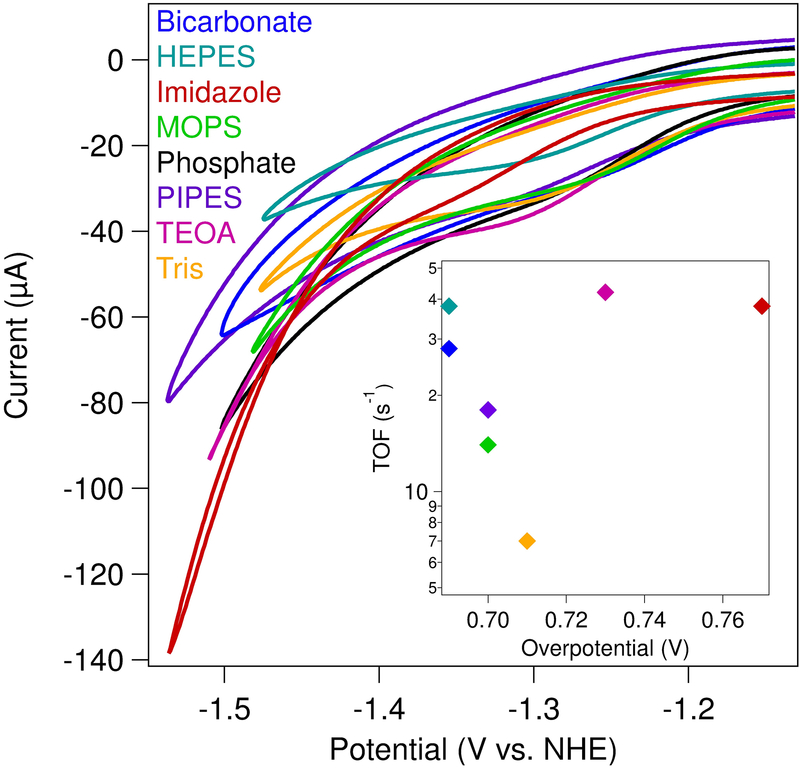
Following characterization of the NiIII/II couple under non-catalytic conditions, [Ni(cyclam)]2+ was analyzed in each buffering system at low potentials under saturating carbon dioxide concentrations. The catalytic electrochemistry was performed with 100 mM of the indicated buffer to maintain an appropriate buffering capacity, and the pH was adjusted to establish a final pH of 7.0. As previously mentioned, cyclic voltammograms of [Ni(cyclam)]2+ in aqueous solutions at negative potentials under an inert atmosphere shows signals that are dominated by proton reduction (Figure S4). However, once carbon dioxide is introduced into the system, the background reductive catalytic current decreases, and a sigmoidal feature appears using most buffers (Figure 3 and Figure S4). The only buffer in which the catalytic current does not decrease as buffer is added is bicarbonate. The onset potential for catalysis, defined as the potential where the slope of the cyclic voltammogram is maximized (Figure S2), spans a range of 80 mV depending on the buffer identity (Table 2). The least negative onset potential occurs in bicarbonate buffer, with catalysis beginning at −1.21 V vs. NHE; this corresponds to an overpotential of 0.69 V relative to the thermodynamic value of −0.52 V for the CO2/CO couple at pH 7.0.2 At the other end of the range, the electrocatalytic overpotential for CO2 reduction by [Ni(cyclam)]2+ in imidazole buffer is 0.77 V, the largest measured in the suite studied here.
Figure 3.
Cyclic voltammograms (ν = 1 V/s) of [Ni(cyclam)]2+ under a CO2-saturated atmosphere. All reactions contained 100 μM [Ni(cyclam)]2+, 100 mM KCl, and 100 mM buffer (as indicated) at a final pH of 7.0. (Inset) TOF as a function of overpotential for each buffer.
Table 2.
Summary of cyclic voltammetry data for the catalytic wave for [Ni(cyclam)]2+ under a CO2-saturated atmosphere (ν = 1 V/s). Samples contained 100 μM [Ni(cyclam)]2+, 100 mM KCl and 100 mM of the indicated buffer, pH 7.0.
| Buffer | Eonset (V vs. NHE) | Overpotential (V) | TOF (s−1) |
|---|---|---|---|
| Bicarbonate | −1.21 ± 0.01 | 0.69 | 28 |
| HEPES | −1.26 ± 0.04 | 0.74 | 38 |
| Imidazole | −1.29 ± 0.04 | 0.77 | 38 |
| MOPS | −1.22 ± 0.03 | 0.70 | 14 |
| Phosphate | −1.22 ± 0.02 | 0.70 | 18 |
| PIPES | −1.22 ± 0.01 | 0.70 | 51 |
| TEOA | −1.25 ± 0.01 | 0.73 | 42 |
| Tris | −1.23 ± 0.05 | 0.71 | 12 |
The turnover frequency (TOF) for CO2 reduction by [Ni(cyclam)]2+ in each buffer was determined using Equation 5 (Table 2), previously used by Kubiak to calculate the TOF for [Ni(cyclam)]2+ in water.19,52 The currents for the TOF calculation were obtained at a scan rate at which the catalytic currents are independent of scan rate (Figures S15–S18, Tables S2–S4), with catalysis occurring in the kinetic catalytic regime.29,51,53,54 From this analysis, the highest TOF of ~50 s−1 is observed in PIPES buffer, while the slowest TOF of ~12 s−1 occurs in Tris buffer. The rates of CO2 reduction in the other buffers fall between these values. Both the range of rates and the catalytic overpotentials observed across the different buffers are relatively small. Importantly, little correlation is seen between overpotential and turnover frequency (Figure 3, inset).
Carbon dioxide binding to [Ni(cyclam)]+
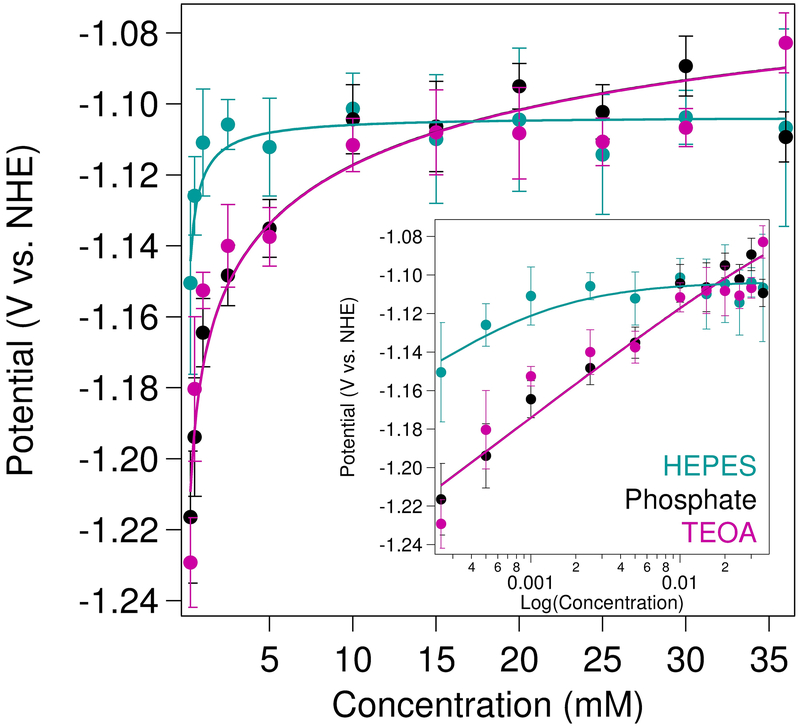
In order to better understand the differences in electrocatalytic overpotential and activity, the binding affinity (KCO2) of CO2 to [Ni(cyclam)]+ at negative potentials was determined in three different buffers by monitoring the shift in onset potential with increasing CO2 concentration (Figure 4). A cationic (TEOA), a neutral (HEPES), and an anionic (phosphate) buffering agent were chosen for analysis at a constant pH of 7.2 to probe the effects of buffer charge state. Due to the background proton reduction that masks the NiII/I couple under an inert atmosphere in aqueous solutions, the measured potentials at 250 μM CO2 were used as reference points (“EN2”) for the analysis. Similar absolute potentials were observed introducing either TEOA and phosphate buffers, with shifts of +140 mV and +130 mV, respectively, in catalytic onset potentials across the accessible CO2 concentration range. These changes in potential correspond to binding affinities of CO2 to [Ni(cyclam)]+ (K1,CO2) of 3 × 105 M−1 in TEOA and phosphate buffers, with a low value of 10 for K2,CO2, though decreasing the value of K2,CO2 to 1 had only minor effects on the quality of the fit (Figure S20 and Table S1). The changes seen in the electrocatalytic waves for increasing concentrations of CO2 using HEPES as the buffer, on the other hand, were much smaller; the onset potential shifted only +40 mV upon saturation with CO2. The shape of the titration curve in HEPES required significantly greater affinity for CO2 binding to the [Ni(cyclam)]2+ state, with best-fit parameters of K1,CO2 = 2 × 105 M−1 and K2,CO2 = 1 × 103 M−1, respectively.
Figure 4.
Binding affinity of CO2 to [Ni(cyclam)]+ measured from voltammetric (ν = 50 mV/s) shifts in catalytic onset potential as [CO2] was varied. Data were fit (solid lines) to Eqn 6. Samples contained 150 μM [Ni(cyclam)]2+ in 1 M buffer, maintained at a constant pH of 7.2, with 100 mM KCl. (Inset) Data shown on log scale to highlight changes at low CO2 concentrations.
Buffer identity affects photochemical turnover of [Ni(cyclam)]2+
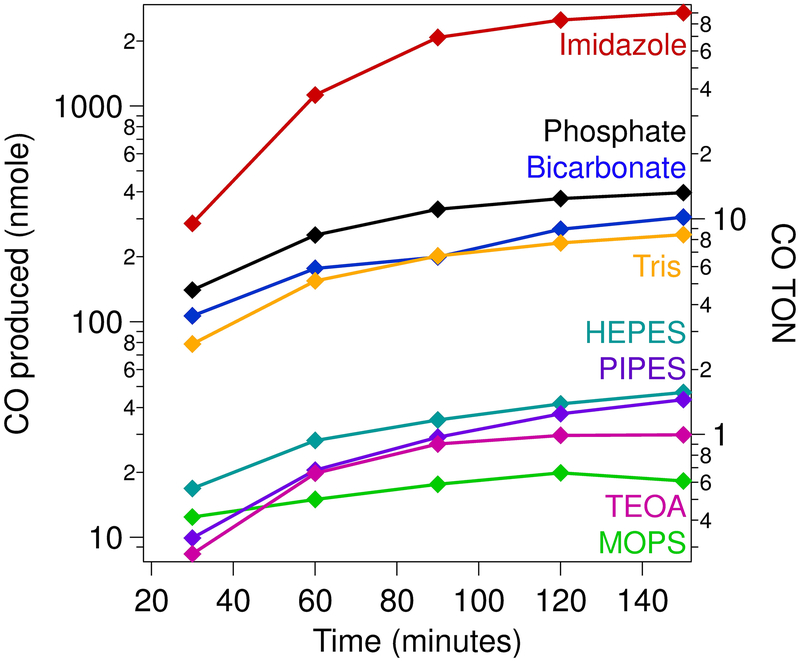
It has recently been emphasized that electrocatalytic analyses must be complemented with solution-phase studies to fully characterize the activity of a new molecular system.55 Towards this end, a light-driven assay coupled with gas chromatography was employed to analyze the product distribution of [Ni(cyclam)]2+-mediated catalysis across the different buffering systems. All experiments were conducted in 1 M buffer under a saturating carbon dioxide atmosphere using 1 mM [Ru(bpy)3]2+ as the photosensitizer and 100 mM ascorbate as the sacrificial electron donor. As with the electrochemical results, the product formation and distribution varied widely across the different buffers (Figure 5, Figures S21–S28). Assays conducted using imidazole as the buffer resulted in the highest amount of carbon monoxide produced, with [Ni(cyclam)]2+ achieving a turnover number (TON) of 90 following 150 minutes of irradiation, which corresponds to a turnover frequency (TOF) of ~36 hr−1. While this rate is substantially lower than the TOF obtained from the electrochemical data, it is common to see a substantial discrepancy across the two sets of experiments.56–59 The product distribution across all buffers also spans a wide range (Table 3 and Table S5). The lowest degree of selectivity is observed when the assays are conducted with phosphate as the buffer, with CO formation only representing ~20% of the two-electron-reduced product; the other 80% of the electrons are shuttled towards H2 evolution. This low selectivity is consistent with previous reports of [Ni(cyclam)]2+ in photochemical assays60,61 as well as recent electrolysis experiments.23 On the other hand, photoassays conducted in the presence of any of the three cationic buffers resulted in ~100% selectivity for CO production, with no detectable H2 produced. The other buffering agents resulted in intermediate levels of selectivity, independent of functional buffering group or pKa. Collectively, these results suggest there are multiple contributing factors that modulate both activity and selectivity, as discussed further below.
Figure 5.
Photochemical CO production by [Ni(cyclam)]2+ (λex = 447.5 nm; 4 °C). Samples contained 10 μM [Ni(cyclam)]2+, 1 mM [RuII(bpy)3]2+, and 100 mM ascorbate in 1 M buffer (as indicated) at pH 7.0, under a saturating carbon dioxide atmosphere.
Table 3.
Product formation following photocatalytic assays of [Ni(cyclam)]2+ following 2.5 hours of irradiation.
| Buffer | CO (nmol) | CO TON | % CO selective |
|---|---|---|---|
| Bicarbonate | 310 ± 20 | 10.2 ± 0.7 | 98 ± 5 |
| HEPES | 47 ± 17 | 1.6 ± 0.6 | 67 ± 9 |
| Imidazole | 2700 ± 600 | 90 ± 20 | 100a |
| MOPS | 18 ± 4 | 0.6 ± 0.1 | 100a |
| Phosphate | 390 ± 60 | 13 ± 2 | 23 ± 4 |
| PIPES | 44 ± 9 | 1.5 ± 0.3 | 30 ± 10 |
| TEOA | 30 ± 9 | 1.0 ± 0.3 | 100a |
| Tris | 250 ± 98 | 8 ± 3 | 100a |
No hydrogen was detected above the background levels.
Discussion
The dominant molecular factors that impact activity are buffer charge and size
The highly variable behavior observed for [Ni(cyclam)]2+-catalyzed CO2 reduction across the range of buffers studied offers insight into the molecular factors dictating activity and selectivity of the catalyst. Since analysis is performed at a constant pH using all buffering systems, the only difference is the identity of the proton donor, leading us to investigate which of the salient distinctions between the molecules (e.g., pKa, size, protonatable site, charge) have the greatest impact. The activity of [Ni(cyclam)]2+ with buffers with the exact same pKa, such as MOPS and phosphate, shows little similarity across the two experiments. Comparing electrocatalytic current at a given overpotential as a function of pKa also shows no correlation, indicating that the ratio of protonated to unprotonated buffer molecules is not a primary contributor to catalysis (Figure S29–S30).
A closer examination of the set of buffering compounds reveals a key fundamental difference—size. The buffers vary greatly in their molecular size, which governs the ability of the buffer to interact with nickel cyclam across different oxidation and substrate-bound states. The effects of size are initially reflected in the binding strengths of these buffers to the NiIII state, with the small inorganic buffers exhibiting the greatest affinity (Table 1). It was previously noted that phosphate binds strongly to the metal center of [Ni(cyclam)]3+, with differential binding to the NiIII intermediate state that is generated during H+ reduction.23 This results in preferential H2 evolution over CO2 reduction, though an inhibited state with a bound phosphate moiety can also accumulate. While the neutral sulfonic acid buffering groups of the Good’s buffers are similar in size to phosphate, they also feature large organic functional groups. Thus, while the Good’s buffers may directly protonate the reduced NiI center, as evidenced by production of hydrogen and decreased selectivity in HEPES and PIPES, the absolute photochemical activity levels are low, potentially due to steric interference between buffer and catalyst.
The most significant contributor to selectivity appears to be overall charge of the buffering component. The cyclam ligand is a neutral species, even when bound to the metal center. As such, cationic buffers such as Tris, imidazole, and TEOA are not likely to interact with the positively charged NiI center due to electrostatic repulsion. The NiIII-H species that is implicated in H2 evolution by [Ni(cyclam)]2+ is thus less likely to be generated in the presence of these buffering moieties when compared to the other functional groups analyzed. However, CO2 binding to the NiI center forms a negatively charged carboxylate species. This species can then be protonated by one of these cationic buffers due to increased electrostatic attraction, leading to release of H2O followed by CO. In contrast, phosphate and bicarbonate remain negatively charged overall at pH 7, even in the protonated form. On this premise, these anionic compounds should be able to protonate a cationic NiI state, and H2 is indeed observed during photocatalysis. Size also appears to play a role in modulating absolute activity; while all of the cationic buffering species are fully selective for CO over H2, the smallest buffer, imidazole, is substantially more active than the largest buffer, TEOA, with Tris falling between those limits. Thus, the ability to access the catalyst is also important.
Another consideration for the difference in selectivity may be that interaction with the different buffers alters the conformation of the cyclam ligand. It is well-established that there are 5 different nickel cyclam conformations that are interchanging on the NMR timescale,62 though only two are dominant in aqueous solution.63 A prior hypothesis for the extreme selectivity seen towards CO2 reduction using a hanging mercury drop electrode was that the electrode interaction with the catalyst biased the conformational preferences.64–66 Looking at the thermodynamics of binding, it was found that [Ni(cyclam)]2+ selectively adsorbs to the mercury electrode in the trans-III conformer, which is the most active species.66 This interaction results in the flattening of the cyclam ring, decreasing the σ-bonding interaction with the metal center and facilitating CO release, which was found to be the rate-limiting step.66–68 A buffer molecule that binds to [Ni(cyclam)]2+ in the axial position may mimic these electrode interactions, perturbing the conformational equilibria and changing the reactivity.
Weakened differential interaction between carbon dioxide and [Ni(cyclam)]2+ in the presence of HEPES
The distinct catalytic behavior of [Ni(cyclam)]2+ in the series of buffers studied here might have been attributed to enhanced binding affinity of CO2 to the metal center in the presence of different buffering agents. However, the experiments presented here indicate that there is no significant difference in CO2 binding constants for [Ni(cyclam)]+ between HEPES, phosphate, and TEOA buffers, despite dramatic changes in activity, overpotential, and selectivity. For this reason, it can be concluded that the binding of CO2 is not the sole factor responsible for the observed high selectivity reported in photoassays using TEOA as the buffering agent. However, the shifts in potential upon increasing CO2 concentration are less pronounced for the neutral buffering agent (HEPES) when compared to the charged buffers. This affects the curvature of the data, which is not fit well to a model that only includes CO2 binding to Ni[cyclam]+, and necessitates use of a model that includes a stronger interaction between CO2 and the divalent state of the metal in the fit (Figure S20). We consider two possible explanations for these differences. On one hand, the increased hydrophobicity of HEPES could stabilize a carbonate- or bicarbonate-bound [Ni(cyclam)]2+ species, which would inhibit reduction of the metal center. Alternatively, the HEPES molecule could stabilize a catalytically incompetent Ni[cyclam]2+-CO2 adduct. Both effects would explain the apparent observed binding of CO2 to Ni[cyclam]2+ as well as the low catalytic currents observed in HEPES at a given overpotential. While the electrocatalytic activity is largely unaffected by the pKa of the buffering component when analyzing current values at 100 mV more negative than the onset potential (Figure S29), HEPES displays a markedly lower catalytic current value compared to the other buffering agents when measured at a constant overpotential (Figure S30). This difference may also be explained by slow proton transfer from HEPES to either [Ni(cyclam)]+ or a CO2-bound species due to steric hindrance, as HEPES is one of the largest buffers investigated in this work.
Experimental design considerations gleaned from buffer dependence studies
One of the most striking observations in this study is the substantially divergent reactivity observed in the different buffers as well as between the electrochemical and photochemical experiments. While it has previously been reported that selectivity and activity for CO2 reduction can vary between the two types of assays,18,57,69 the different trends observed across the buffering systems investigated suggest the mechanisms of catalysis, or the rate-determining process(es), may differ between the two experiment types. Photochemically, [Ni(cyclam)]2+ is the most active in imidazole, Tris, bicarbonate, and phosphate, though the latter demonstrates high H2 evolution as well. However, [Ni(cyclam)]2+ in bicarbonate exhibits lower electrochemical currents at the catalytic onset potential, suggesting the absolute activity in a given buffer for one technique cannot be directly correlated to another. This observation suggests that a new metric may need to be established for photocatalytic assays that is different from those used in electrocatalytic assays to reflect differences in catalytic performance between analysis procedures.
No direct correlation is seen between the turnover frequency and the overpotential in the different buffers, contradicting standard Tafel-like behavior and indicating that the initial reduction event may not be the rate-determining step. This deviation is suggested to derive largely from outer sphere interactions between the catalyst, substrate, and the buffer in at least one protonation state, interactions that have been noted to play important roles in natural enzyme systems as well as other catalysts for CO2 reduction.70–75 This observation provides a guide for identifying key molecular components necessary for selective and efficient catalysis, which depend on the reaction conditions and lifetimes of different intermediates. For example, a buffer with multiple protonatable sites might support a proton “wire” between solvent and the active site, leading to rapid substrate protonation and higher turnover rates. This is a common technique employed in natural enzyme systems, where amino acid residues are carefully placed to deliver protons to the active site in a controlled manner,2,70,76,77 and has recently been modeled through the use of encapsulation of a catalyst within a polymer matrix.78 To probe these interactions, detailed, high-resolution structural information along with a proton inventory study would be advantageous. On the other hand, if studying the proton reduction mechanism of [Ni(cyclam)]2+, a buffer that is active for H2 production but does not bind directly to the metal center, such as HEPES or PIPES, is preferred.
The vastly different activity observed for a single catalyst across this small subset of buffering systems must be considered when comparing catalytic systems. It is highly possible that a catalyst may give a wide range of results in terms of both selectivity and activity under different reaction conditions depending on the interaction of the compound with solvent, substrate, or the buffer molecules.29,79–81 It is thus critical to consider benchmarking a new compound against prior literature results under identical reaction conditions (buffer, pH, temperature, ionic strength), as the buffer should be treated as an active medium that can participate in and modulate catalysis.
The best buffers provide an outer coordination sphere that mimics those in natural CO2-reducing enzymes
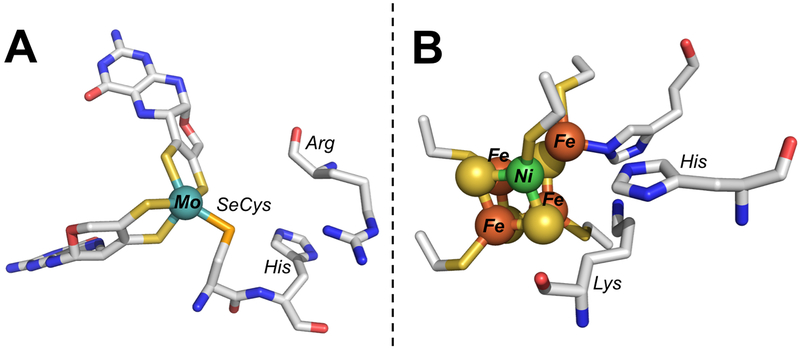
The gold standards for catalyst design are naturally occurring enzymes that reduce CO2, such as formate dehydrogenase (FDH) and carbon monoxide dehydrogenase (CODH). These enzymes are extremely selective for CO2 reduction over H2 evolution, operate reversibly at the thermodynamic potentials, and exhibit high turnover rates in solution-phase assays and electrochemically.82–86 While the primary coordination environments of the active-site metals in these classes of enzyme differ considerably (Figure 6), examining the secondary coordination spheres shows a number of similarities. Importantly, the active sites of both CODH and FDH contain strictly conserved histidine residues.2 The Mo/W FDHs also have conserved arginine residues, while CODH has a nearby lysine. It is proposed that the arginine acts to stabilize formate while the histidine transfers protons to and from the carbon atom in FDH.2,73,74,87 Similarly, histidine is invoked for proton transfer in CODH, and the positively charged lysine may stabilize a bound carboxylate ligand.88 The secondary sphere interactions prime the system for water loss and concomitant CO formation, which occurs on the order of 10 s−1.2,10,89,90
Figure 6.
Structures of the catalytic active sites of (A) formate dehydrogenase (PDB 1FDO) and (B) carbon monoxide dehydrogenase (PDB 1MJG) shown with conserved, cationic secondary sphere residues.
Given the coordination environments in FDH and CODH, it is unsurprising that cationic buffers are both the most active and most selective for CO2 reduction to CO by [Ni(cyclam)]2+; these compounds closely mimic the functional groups that are conserved in the secondary spheres of the native enzyme systems. This is consistent with previous reports, which have shown that not only can [Ni(cyclam)]2+ interact with pyridine-like structures,91 the activity and selectivity are enhanced by pyridine or imidazole binding.20,22 This has also been observed in other catalytic systems using an imidazolium functional group.92 The larger catalytic overpotential in imidazole can be understood by the modest preferential binding of imidazole to the NiII state of the catalyst, stabilizing the divalent oxidation state and decreasing the reduction potential; steric constraints of the protein fold prevent this inhibitory, direct binding from occurring in the native enzyme.73,93 Once [Ni(cyclam)]2+ is reduced, the imidazole buffer may functionally mimic the His residues in CODH or FDH, protonating only the bound substrate rather than the metal center directly. In this way, the surrounding buffer molecules could serve as a less-constrained, pseudo-secondary coordination sphere. The importance of outer sphere interactions is increasingly being recognized as critical for catalysis in natural and engineered systems. Many secondary sphere residues are highly conserved across organisms, with specific amino acids precisely placed in order to stabilize intermediates and disperse extra charge, often through extensive hydrogen bonding networks.71,94–97 Similar interactions have been implemented in a great number of synthetic catalysts, for which ligands that contain a well-structured secondary sphere enhance catalysis.40,72,98–103 However, the ligand syntheses are often challenging and expensive, limiting practical application of these compounds. Instead, taking advantage of the buffer to provide a rudimentary secondary sphere, albeit one that is unstructured and disordered, may overcome some of these limitations and provide an understanding of how to generate effective, complete systems for CO2 reduction.
Conclusions
The activity of [Ni(cyclam)]2+ towards CO2 reduction in aqueous solution varies dramatically depending on the method of providing electrons and the identity of the buffer. The onset potentials for electrocatalysis in a suite of eight buffers studied here span a range of 80 mV, with the lowest overpotential of 0.69 V observed in bicarbonate. Photochemical carbon monoxide production spans two orders of magnitude, with CO selectivity relative to H2 production ranging from 20 – 100%. The cationic buffers were found to be the most effective at promoting high levels of CO2 reduction due to electrostatic effects, though size also dictates accessibility of protons that can bind to the catalyst. These observations are discussed in the context of the secondary coordination environments of native CO2-reducing enzymes. Taken together, this study shows that simply changing the buffering component can significantly modulate the catalytic activity, and the buffer cannot be treated as a spectator species. Thus, the nature of the buffer-catalyst interactions must be carefully considered for characterizing catalytic activity and benchmarking new molecular compounds.
Supplementary Material
Acknowledgements
This work was supported by the Department of Energy Office of Science with an Early Career Award to HSS (DE-SC0018020). CRS acknowledges past support by an NIH Chemistry-Biology Interface Training Program Fellowship (GM08512) and a Presidential Fellowship from The Ohio State University Graduate School. We would like to acknowledge Dr. Anastasia Manesis, Shelby Behnke, and all other members of the Shafaat lab for helpful discussions and advice.
Footnotes
Electronic Supplementary Information (ESI) available: Electrochemical and photochemical assay control experiments.
Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Because a dependence on imidazole lot was observed, all experiments reported here used imidazole from the same Alfa Aesar lot.
References
- 1.Bell AT, Gates BC, Ray D and Thompson MR, Basic research needs: catalysis for energy, Pacific Northwest National Laboratory (PNNL), Richland, WA (US), 2008. [Google Scholar]
- 2.Appel AM, Bercaw JE, Bocarsly AB, Dobbek H, DuBois DL, Dupuis M, Ferry JG, Fujita E, Hille R, Kenis PJA, Kerfeld CA, Morris RH, Peden CHF, Portis AR, Ragsdale SW, Rauchfuss TB, Reek JNH, Seefeldt LC, Thauer RK and Waldrop GL, Chem. Rev, 2013, 113, 6621–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arakawa H, Aresta M, Armor JN, Barteau MA, Beckman EJ, Bell AT, Bercaw JE, Creutz C, Dinjus E, Dixon DA, Domen K, DuBois DL, Eckert J, Fujita E, Gibson DH, Goddard WA, Goodman DW, Keller J, Kubas GJ, Kung HH, Lyons JE, Manzer LE, Marks TJ, Morokuma K, Nicholas KM, Periana R, Que L, Rostrup-Nielson J, Sachtler WMH, Schmidt LD, Sen A, Somorjai GA, Stair PC, Stults BR and Tumas W, Chem. Rev, 2001, 101, 953–996. [DOI] [PubMed] [Google Scholar]
- 4.Ramanathan V and Feng Y, Atmos. Environ, 2009, 43, 37–50. [Google Scholar]
- 5.Benson EE, Kubiak CP, Sathrum AJ and Smieja JM, Chem. Soc. Rev, 2009, 38, 89–99. [DOI] [PubMed] [Google Scholar]
- 6.Dibenedetto A, Angelini A and Stufano P, J. Chem. Technol. Biotechnol, 2014, 89, 334–353. [Google Scholar]
- 7.Najafabadi AT, Int. J. Energy Res, 2013, 37, 485–499. [Google Scholar]
- 8.Takeda H, Cometto C, Ishitani O and Robert M, ACS Catal, 2017, 7, 70–88. [Google Scholar]
- 9.Dry ME, Catal. Today, 2002, 71, 227–241. [Google Scholar]
- 10.Can M, Armstrong FA and Ragsdale SW, Chem. Rev, 2014, 114, 4149–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maia LB, Moura I and Moura JJG, Inorganica Chim. Acta, 2017, 455, Part 2, 350–363. [Google Scholar]
- 12.Francke R, Schille B and Roemelt M, Chem. Rev, 2018, 118, 4631–4701. [DOI] [PubMed] [Google Scholar]
- 13.Windle CD and Perutz RN, Coord. Chem. Rev, 2012, 256, 2562–2570. [Google Scholar]
- 14.Taheri A, Thompson EJ, Fettinger JC and Berben LA, ACS Catal, 2015, 5, 7140–7151. [Google Scholar]
- 15.Beley M, Collin J-P, Ruppert R and Sauvage J-P, J. Chem. Soc. Chem. Commun, 1984, 0, 1315–1316. [DOI] [PubMed] [Google Scholar]
- 16.Balazs GB and Anson FC, J. Electroanal. Chem, 1993, 361, 149–157. [Google Scholar]
- 17.Beley M, Collin JP, Ruppert R and Sauvage JP, J. Am. Chem. Soc, 1986, 108, 7461–7467. [DOI] [PubMed] [Google Scholar]
- 18.Schneider J, Jia H, Kobiro K, Cabelli DE, Muckerman JT and Fujita E, Energy Environ. Sci, 2012, 5, 9502–9510. [Google Scholar]
- 19.Froehlich JD and Kubiak CP, Inorg. Chem, 2012, 51, 3932–3934. [DOI] [PubMed] [Google Scholar]
- 20.Saravanakumar D, Song J, Jung N, Jirimali H and Shin W, ChemSusChem, 2012, 5, 634–636. [DOI] [PubMed] [Google Scholar]
- 21.Kuehnel MF, Sahm CD, Neri G, Lee JR, Orchard KL, Cowan AJ and Reisner E, Chem. Sci, 2018, 9, 2501–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider CR and Shafaat HS, Chem Commun, 2016, 52, 9889–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behnke SL, Manesis AC and Shafaat HS, Dalton Trans, 2018, 47, 15206–15216. [DOI] [PubMed] [Google Scholar]
- 24.Dyson PJ and Jessop PG, Catal. Sci. Technol, 2016, 6, 3302–3316. [Google Scholar]
- 25.Rigsby ML, Wasylenko DJ, Pegis ML and Mayer JM, J. Am. Chem. Soc, 2015, 137, 4296–4299. [DOI] [PubMed] [Google Scholar]
- 26.Natali M, Badetti E, Deponti E, Gamberoni M, Scaramuzzo FA, Sartorel A and Zonta C, Dalton Trans, 2016, 45, 14764–14773. [DOI] [PubMed] [Google Scholar]
- 27.Sinha S and Warren JJ, Inorg. Chem, 2018, 57, 12650–12656. [DOI] [PubMed] [Google Scholar]
- 28.Rosen BA, Salehi-Khojin A, Thorson MR, Zhu W, Whipple DT, Kenis PJA and Masel RI, Science, 2011, 334, 643–644. [DOI] [PubMed] [Google Scholar]
- 29.Costentin C, Passard G and Savéant J-M, J. Am. Chem. Soc, 2015, 137, 5461–5467. [DOI] [PubMed] [Google Scholar]
- 30.Grills DC, Matsubara Y, Kuwahara Y, Golisz SR, Kurtz DA and Mello BA, J. Phys. Chem. Lett, 2014, 5, 2033–2038. [DOI] [PubMed] [Google Scholar]
- 31.Ceballos BM, Tsay C and Yang JY, Chem. Commun, 2017, 53, 7405–7408. [DOI] [PubMed] [Google Scholar]
- 32.Ryu J, Wuttig A and Surendranath Y, Angew. Chem. Int. Ed, 2018, 57, 9300–9304. [DOI] [PubMed] [Google Scholar]
- 33.Good NE, Arch. Biochem. Biophys, 1962, 96, 653–661. [DOI] [PubMed] [Google Scholar]
- 34.Good NE, Winget GD, Winter W, Connolly TN, Izawa S and Singh RMM, Biochemistry, 1966, 5, 467–477. [DOI] [PubMed] [Google Scholar]
- 35.Grady JK, Chasteen ND and Harris DC, Anal. Biochem, 1988, 173, 111–115. [DOI] [PubMed] [Google Scholar]
- 36.Krishnamoorthy CR and Nakon R, J. Coord. Chem, 1991, 23, 233–243. [Google Scholar]
- 37.Mash HE, Chin Y-P, Sigg L, Hari R and Xue H, Anal. Chem, 2003, 75, 671–677. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Huang Y, Ye W and Li Y, Adv. Sci, 2017, 4, 1700194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Call A, Cibian M, Yamamoto K, Nakazono T, Yamauchi K and Sakai K, ACS Catal, DOI: 10.1021/acscatal.8b04975. [DOI] [Google Scholar]
- 40.Dutta A, DuBois DL, Roberts JAS and Shaw WJ, Proc. Natl. Acad. Sci, 2014, 111, 16286–16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neri G, Aldous IM, Walsh JJ, Hardwick LJ and Cowan AJ, Chem. Sci, 2016, 7, 1521–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoi VS, Sun Y, Long JR and Chang CJ, Chem. Soc. Rev, 2013, 42, 2388–2400. [DOI] [PubMed] [Google Scholar]
- 43.Koshiba K, Yamauchi K and Sakai K, Angew. Chem, 2017, 129, 4311–4315. [DOI] [PubMed] [Google Scholar]
- 44.Bosnich B, Tobe ML and Webb GA, Inorg. Chem, 1965, 4, 1109–1112. [Google Scholar]
- 45.Froehlich JD and Kubiak CP, J. Am. Chem. Soc, 2015, 137, 3565–3573. [DOI] [PubMed] [Google Scholar]
- 46.Jung HJ, Bang H and Suh MP, Notes, 2001, 22, 523. [Google Scholar]
- 47.Fourmond V, Anal. Chem, 2016, 88, 5050–5052. [DOI] [PubMed] [Google Scholar]
- 48.Pierce DT, Hatfield TL, Billo EJ and Ping Y, Inorg. Chem, 1997, 36, 2950–2955. [DOI] [PubMed] [Google Scholar]
- 49.Wilhelm E, Battino R and Wilcock RJ, Chem. Rev, 1977, 77, 219–262. [Google Scholar]
- 50.Sommer DJ, Vaughn MD and Ghirlanda G, Chem Commun, 2014, 50, 15852–15855. [DOI] [PubMed] [Google Scholar]
- 51.Bard AJ and Faulkner LR, Electrochemical Methods: Fundamentals and Applications, 2nd Edition, Wiley Global Education, 2000. [Google Scholar]
- 52.Savéant J-M, Chem. Rev, 2008, 108, 2348–2378. [DOI] [PubMed] [Google Scholar]
- 53.Khusnutdinova D, Wadsworth BL, Flores M, Beiler AM, Reyes Cruz EA, Zenkov Y and Moore GF, ACS Catal, 2018, 8, 9888–9898. [Google Scholar]
- 54.Savéant J-M and Vianello E, Electrochimica Acta, 1965, 10, 905–920. [Google Scholar]
- 55.Kaeffer N, Morozan A, Fize J, Martinez E, Guetaz L and Artero V, ACS Catal, 2016, 6, 3727–3737. [Google Scholar]
- 56.Herrero C, Quaranta A, El Ghachtouli S, Vauzeilles B, Leibl W and Aukauloo A, Phys. Chem. Chem. Phys, 2014, 16, 12067. [DOI] [PubMed] [Google Scholar]
- 57.Kumar B, Llorente M, Froehlich J, Dang T, Sathrum A and Kubiak CP, Annu. Rev. Phys. Chem, 2012, 63, 541–569. [DOI] [PubMed] [Google Scholar]
- 58.Slater JW and Shafaat HS, J. Phys. Chem. Lett, 2015, 6, 3731–3736. [DOI] [PubMed] [Google Scholar]
- 59.Stevenson MJ, Marguet SC, Schneider CR and Shafaat HS, ChemSusChem, 2017, 10, 4424–4429. [DOI] [PubMed] [Google Scholar]
- 60.Craig CA, Spreer LO, Otvos JW and Calvin M, J. Phys. Chem, 1990, 94, 7957–7960. [Google Scholar]
- 61.Kimura E, Wada S, Shionoya M and Okazaki Y, Inorg. Chem, 1994, 33, 770–778. [Google Scholar]
- 62.Joseph Billo E, Connolly PJ, Sardella DJ, Jasinski JP and Butcher RJ, Inorganica Chim. Acta, 1995, 230, 19–28. [Google Scholar]
- 63.Connolly PJ and Billo EJ, Inorg. Chem, 1987, 26, 322–3226. [Google Scholar]
- 64.Balazs GB and Anson FC, J. Electroanal. Chem, 1992, 322, 325–345. [Google Scholar]
- 65.Fujihira M, Hirata Y and Suga K, J. Electroanal. Chem. Interfacial Electrochem, 1990, 292, 199–215. [Google Scholar]
- 66.Wu Y, Rudshteyn B, Zhanaidarova A, Froehlich JD, Ding W, Kubiak CP and Batista VS, ACS Catal, 2017, 5282–5288. [Google Scholar]
- 67.Smieja JM and Kubiak CP, Inorg. Chem, 2010, 49, 9283–9289. [DOI] [PubMed] [Google Scholar]
- 68.Song J, Klein EL, Neese F and Ye S, Inorg. Chem, 2014, 53, 7500–7507. [DOI] [PubMed] [Google Scholar]
- 69.Grant JL, Goswami K, Spreer LO, Otvos JW and Calvin M, J. Chem. Soc. Dalton Trans, 1987, 2105–2109. [Google Scholar]
- 70.Drennan CL, Heo J, Sintchak MD, Schreiter E and Ludden PW, Proc. Natl. Acad. Sci, 2001, 98, 11973–11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nichols EM and Chang CJ, Organometallics, 2019, 38, 1213–1218. [Google Scholar]
- 72.Chapovetsky A, Welborn M, Luna JM, Haiges R, Miller TF and Marinescu SC, ACS Cent. Sci, 2018, 4, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC and Sun PD, Science, 1997, 275, 1305–1308. [DOI] [PubMed] [Google Scholar]
- 74.Mota CS, Rivas MG, Brondino CD, Moura I, Moura JJG, González PJ and Cerqueira NMFSA, JBIC J. Biol. Inorg. Chem, 2011, 16, 1255–1268. [DOI] [PubMed] [Google Scholar]
- 75.Dobbek H, Svetlitchnyi V, Gremer L, Huber R and Meyer O, Science, 2001, 293, 1281–1285. [DOI] [PubMed] [Google Scholar]
- 76.Seravalli J and Ragsdale SW, Biochemistry, 2008, 47, 6770–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim EJ, Feng J, Bramlett MR and Lindahl PA, Biochemistry, 2004, 43, 5728–5734. [DOI] [PubMed] [Google Scholar]
- 78.Liu Y and McCrory CCL, Nat. Commun, 2019, 10, 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Artero V and Savéant JM, Energy Environ. Sci, 2014, 7, 3808–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Costentin C and Savéant J-M, J. Am. Chem. Soc, 2018, 140, 16669–16675. [DOI] [PubMed] [Google Scholar]
- 81.Pegis ML, Wise CF, Koronkiewicz B and Mayer JM, J. Am. Chem. Soc, 2017, 139, 11000–11003. [DOI] [PubMed] [Google Scholar]
- 82.Bassegoda A, Madden C, Wakerley DW, Reisner E and Hirst J, J. Am. Chem. Soc, 2014, 136, 15473–15476. [DOI] [PubMed] [Google Scholar]
- 83.Hille R, Trends Biochem. Sci, 2002, 27, 360–367. [DOI] [PubMed] [Google Scholar]
- 84.Parkin A, Seravalli J, Vincent KA, Ragsdale SW and Armstrong FA, J. Am. Chem. Soc, 2007, 129, 10328–10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reda T, Plugge CM, Abram NJ and Hirst J, Proc. Natl. Acad. Sci, 2008, 105, 10654–10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang VC-C, Islam STA, Can M, Ragsdale SW and Armstrong FA, J. Phys. Chem. B, 2015, 119, 13690–13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raaijmakers HCA and Romão MJ, JBIC J. Biol. Inorg. Chem, 2006, 11, 849–854. [DOI] [PubMed] [Google Scholar]
- 88.Jeoung J-H and Dobbek H, Science, 2007, 318, 1461–1464. [DOI] [PubMed] [Google Scholar]
- 89.Heo J, Staples CR and Ludden PW, Biochemistry, 2001, 40, 7604–7611. [DOI] [PubMed] [Google Scholar]
- 90.Kumar M, Lu W-P and Ragsdale SW, Biochemistry, 1994, 33, 9769–9777. [DOI] [PubMed] [Google Scholar]
- 91.El Ghachtouli S, Cadiou C, Déchamps-Olivier I, Chuburu F, Aplincourt M, Patinec V, Le Baccon M, Handel H and Roisnel T, New J. Chem, 2006, 30, 392. [Google Scholar]
- 92.Sung S, Li X, Wolf LM, Meeder JR, Bhuvanesh NS, Grice KA, Panetier JA and Nippe M, J. Am. Chem. Soc, 2019, 141, 6569–6582. [DOI] [PubMed] [Google Scholar]
- 93.Doukov TI, Iverson TM, Seravalli J, Ragsdale SW and Drennan CL, Science, 2002, 298, 567–572. [DOI] [PubMed] [Google Scholar]
- 94.Bowman SEJ and Bren KL, Inorg. Chem, 2010, 49, 7890–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lancaster KM, in Molecular Electronic Structures of Transition Metal Complexes I, eds. Mingos DMP, Day P and Dahl JP, Springer Berlin Heidelberg, 2011, pp. 119–153. [Google Scholar]
- 96.Spreitzer RJ and Salvucci ME, Annu. Rev. Plant Biol, 2002, 53, 449–475. [DOI] [PubMed] [Google Scholar]
- 97.Volbeda A and Fontecilla-Camps JC, Coord. Chem. Rev, 2005, 249, 1609–1619. [Google Scholar]
- 98.Borovik AS, Acc. Chem. Res, 2005, 38, 54–61. [DOI] [PubMed] [Google Scholar]
- 99.Dutta A, Lense S, Hou J, Engelhard MH, Roberts JAS and Shaw WJ, J. Am. Chem. Soc, 2013, 135, 18490–18496. [DOI] [PubMed] [Google Scholar]
- 100.Ginovska-Pangovska B, Dutta A, Reback ML, Linehan JC and Shaw WJ, Acc. Chem. Res, 2014, 47, 2621–2630. [DOI] [PubMed] [Google Scholar]
- 101.Lewis JC, ACS Catal, 2013, 3, 2954–2975. [Google Scholar]
- 102.Shaw WJ, Catal. Rev, 2012, 54, 489–550. [Google Scholar]
- 103.Shearer J, Neupane KP and Callan PE, Inorg. Chem, 2009, 48, 10560–10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.