Abstract
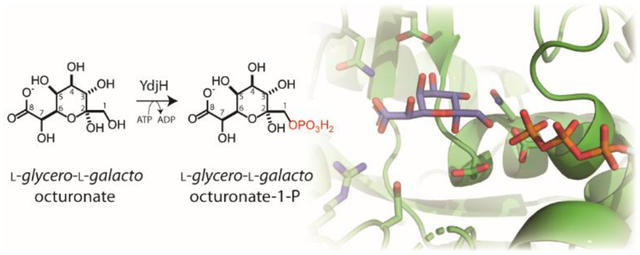
The ydj gene cluster is annotated to catalyze the catabolism of an unknown carbohydrate. Previously, YdjI, a class II aldolase, was shown to catalyze the retro-aldol cleavage of L-glycero-L-galacto-octuluronate-1-phosphate into DHAP and L-arabinuronate. In this report, the functional characterization of YdjH is presented. YdjH catalyzes the phosphorylation of 2-keto-monosaccharides at the C1 hydroxyl group with a significantly more stringent substrate profile relative to YdjI. Similar to YdjI, YdjH shows a strong preference for higher-order monosaccharides (seven to nine carbons) with a carboxylate terminus. The best substrate was determined to be L-glycero-L-galacto-octuluronate yielding L-glycero-L-galacto-octuluronate-1-phosphate with a kcat = 16 s−1 and a kcat/Km = 2.1 × 104 M−1 s−1. This is apparently the first reported example of kinase activity with eight-carbon monosaccharides. Two crystal structures of YdjH were previously solved to 2.15 Å and 1.8 Å (PDB entries: 3H49 and 3IN1). We present an analysis of the active site layout and use computational docking to identify potential key residues in the binding of L-glycero-L-galacto-octuluronate.
Graphical Abstrcat
INTRODUCTION
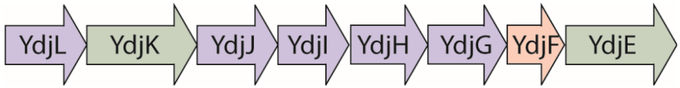
The ydj gene cluster is a putative carbohydrate degradation pathway found in Escherichia coli K12, and is highly conserved among other E. coli strains, appearing in 80% of the sequenced E. coli genomes (1). It is also found in other bacterial species including Citrobacter rodenium, Klebsiella oxytoca, Shigella flexneri, Clostridium bolteae, and Coriobacterium glomerans, among others. The ydj gene cluster contains five enzymes, two transporters, and a regulator (Figure 1).
Figure 1.
The ydj gene cluster in E. coli K12. This gene cluster includes five enzymes: YdjL, YdjJ, YdjI, YdjH, and YdjG (shown in blue); two transporters: YdjK and YdkE (shown in green); and a transcriptional regulator, YdjF (shown in red).
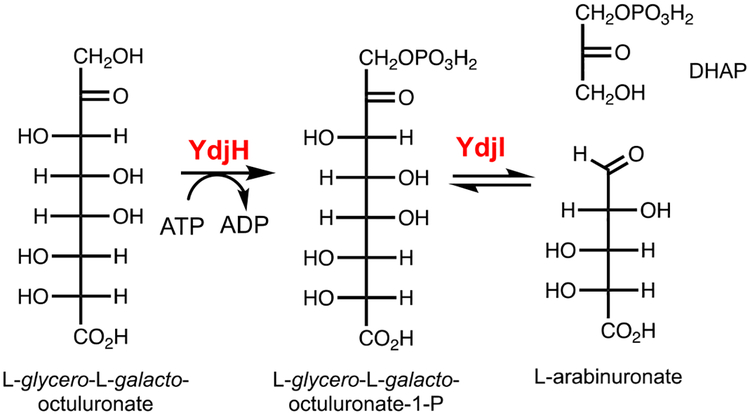
In the preceding paper, the class-II zinc dependent aldolase, YdjI, was shown to catalyze the retro-aldol cleavage of phosphorylated keto acid sugars of seven to nine carbons with a specific stereochemistry at C3, C4, C5, and C6 resembling L-galactoheptulose (1). YdjI shows a preference for substrates with a negatively charged terminus, favoring a carboxylate moiety relative to phosphate. The best aldose substrate for YdjI was L-arabinuronate when combined with DHAP to yield L-glycero-L-galacto-octuluronate-1-P (Scheme 1). The focus of this investigation is on the functional characterization of YdjH, the kinase that is expected to produce the phosphorylated product that will be the ultimate substrate for YdjI.
Scheme 1:
Reaction catalyzed by YdjI and the proposed reaction catalyzed by YdjH.
Kinases catalyze the ATP-dependent phosphorylation of specific substrates and these transformations are often essential for the catabolic degradation of carbohydrates (2–4). Most of the enzymes that phosphorylate carbohydrates are found in three families: the FGGY family (IPR000577), the ribokinase/fructokinase family (IPR002139) and the mevalonate/galactokinase family (IPR006206) (5, 6). YdjH is annotated as part of the ribokinase/fructokinase family. This family of enzymes contains ribokinase (Uniprot: P0A9J6), fructokinase (Uniprot: Q6XZ78), 6-phosphofructokinase (Uniprot: P06999), inosine kinase (Uniprot: P0AEW6) 2-dehydro-3-deoxyglucokinase (Uniprot: P37647), and fructolysine kinase (Uniprot: P45543).
In this paper, we describe the functional characterization of the substrates for YdjH and provide an analysis of how these substrates may bind in the active site. YdjH is shown to catalyze the phosphorylation at C1 of a ketose sugar with the best substrate determined to be L-glycero-L-galacto-octuluronate. Thus, the best substrate for YdjH produces the best substrate known to date for YdjI. The coordinated activities of YdjI and YdjH provide strong evidence of possible substrates for the ydj gene cluster.
MATERIALS and METHODS
Materials.
All materials used in this study were obtained from Sigma-Aldrich, Carbosynth, or GE Healthcare Bio-Sciences, unless stated otherwise. E. coli strains XL1 Blue and BL21-Gold (DE3) were obtained from New England Biolabs. NMR spectra were collected on a Bruker Avance III 400 MHz system equipped with a broadband probe and sample changer or a Bruker Avance 500 MHz system equipped with a triple resonance cyroprobe. Mass spectrometry was conducted with an MDS-Sciex 4000 Qtrap system or a Thermo Scientific Q Exactive Focus system run in negative ion mode. UV spectra were obtained using a SpectraMax340 UV-visible plate reader.
Sequence Similarity Networks.
Sequence similarity networks (SSN) were generated by submitting the InterPro number for the protein family of interest or a list of the Uniprot IDs assigned to an individual Cluster of Orthologous Genes (COG) to the EFI-EST webtool (7, 8). The initial SSN was generated with an alignment score cutoff set such that each connection (edge) represents a sequence identity of approximately 30%. Sequences that share high sequence identity (>90%) were grouped into a single node. All network layouts were created and visualized using Cytoscape 3.4 or higher (9). Within Cytoscape, more stringent SSNs were created by increasing the alignment score cutoff in small increments (usually by 5–10). This process was continued until the clusters were estimated to be isofunctional, where each cluster represents enzymes that catalyze the same reaction. Isofunctionality was determined by mapping the known functions onto the SSNs and following their movements as the alignment cutoff is increased until each cluster contains a single known catalytic activity.
Cloning of YdjH and GlpX from E. coli K12.
The gene for YdjH (Uniprot ID: P77492) was amplified from the genomic DNA of E. coli K12 with the primer pair:
5ʹ-ATCGGATTC ATGGATAATCTCGAC-3ʹ
5ʹ-ATCGTCGACTTATCCTTCGTATTCTTC-3ʹ.
Restriction sites for BamHI and SalI (underlined) were introduced into the forward and reserve primers, respectively. These sites allow for the addition of an N-terminal 6x-His-tag in the pET-30a+ expression vector. The gene for GlpX (Uniprot ID: P0A9C9) was amplified from the genomic DNA of E. coli K12 with the primer pair:
5ʹ-ATATCATATGAGACGAGAACTTGCCAT-3ʹ
5ʹ-ATATAAGCTTGAGGAGTGGCACCTGCATTTCCG −3ʹ.
Restriction sites for NdeI and HindIII (underlined) were introduced into the forward and reserve primers, respectively. These sites allow for the addition of a C-terminal 6x-His-tag in the pET-30a+ expression vector. Gene amplification, restriction digest, ligation, and plasmid isolation procedures were followed as previously described (1). Plasmids were sequenced to confirm the genes inserted into the vector.
Expression and Purification of YdjH.
The recombinant plasmid containing the gene for YdjH was transformed into E. coli BL21 (DE3) using the method of heat-shock (10). Conditions for expression and purification of the enzyme were followed as previously published with the following minor adjustments (1). During cell growth, zinc acetate was not added to the medium, as done previously for the expression of YdjI. Yields of YdjH were typically 150–200 mg from 2.0 L of cell culture. The oligomeric state of YdjH was determined by HPLC gel filtration using a Superdex 200 Increase 10/300 GL column.
Expression and Purification of GlpX.
The recombinant plasmid containing the gene for GlpX was transformed into E. coli BL21 (DE3) as described above. Conditions for expression and purification were followed as described previously with the following minor adjustments (1, 11, 12). During cell growth, 1.0 mM MnCl2 was added to the growth medium when the OD6oo = 0.1–0.2, as done previously with zinc acetate. Yields of GlpX were typically 25–50 mg from 2.0 L of cell culture.
Kinetic assays for YdjH.
The catalytic activity of YdjH was monitored using a coupled enzyme assay by following the oxidation of NADH at 340 nm at 30°C in 50 mM HEPES/K+ buffer with 100 mM KCl, pH 7.4. The coupled assay contained 2.0 mM ATP, 1.0 mM phosphoenolpyruvate (PEP), 300 μM NADH, 5.0 mM MgCl2, 1 U lactate dehydrogenase, and 1 U pyruvate kinase in a total volume of 250 μL. Assays were initiated by the addition of enzyme (150 μL) containing the above components to a well containing 100 μL of substrate. The values of kcat and kcat/Km were determined by fitting the initial velocity data to eq. 1 using GraFit 5, where v is the initial velocity of the reaction, E is the enzyme concentration, kcat is the turnover number, and Km is the Michaelis constant. The concentration of YdjH varied between 0.2 and 5 μM depending on the observed rate of product formation. Substrate concentration ranged between (0–20 mM).
| (1) |
Synthesis of Phosphorylated Carbohydrates.
Many of the potential substrates for YdjH are not commercially available, therefore they were synthesized using DHAP, an aldehyde co-substrate, and the YdjI aldolase. A list of compounds synthesized with their structures can be found in the Supporting Information (Scheme S1). These compounds include: L-galacto-heptulose-1-P, D-altro-heptulose-1-P, 6-deoxy-D-fructose-1-P, 6-deoxy-L-sorbose-1-P, L-galacto-hepturonate-1-P, L-galacto-heptulose-1,7-bisphosphate, L-glycero-L-galacto-octuluronate-1-P, D-glycero-L-galacto-octuluronate-1-P, L-glycero-D-altro-octuluronate-1-P, L-glycero-L-galacto-octuluronate-1,8-bisphospate, D-glycero-L-galacto-octuluronate-1,8-bisphospate, L-threo-L-galacto-nonulonate-1-P, and D-erythro-L-galacto-nonulonate-1-P. Phosphorylated carbohydrates were synthesized by mixing 0.14–0.28 mmol of DHAP (25–50 mg) with 0.2–0.4 mmol of the appropriate aldose, and 40 μM YdjI in 25 mM HEPES/K+ buffer with 100 mM KCl, pH 7.4 (1). Each reaction was allowed to reach equilibrium (4–16 h) and then YdjI was removed using a PALL Nanosep 10K omega centrifugation filter. The phosphorylated compounds were isolated by one of two methods; barium salt precipitation or DEAE anion exchange chromatography (1.5 × 13 cm) using a gradient of 0–500 mM ammonium bicarbonate as the elution buffer (13). Ketose-containing fractions (2.0 mL) were identified using the phenol-sulfuric acid assay (14, 15). The phosphorylated compounds eluted between 50–400 mM bicarbonate depending on the number of negative charges. The fractions containing carbohydrates were pooled and acidified (pH 2–3) with Dowex H+ resin. The resin was removed by filtration, and the filtrate bought back to pH 5–6 by the addition of ammonium bicarbonate and dried under reduced pressure. Typical yields of the phosphorylated compound were around 60–70% after purification. Purified compounds were subjected to MS as described previously and confirmed by 31P NMR spectroscopy (1).
Dephosphorylation of Selected Carbohydrates.
Phosphorylated carbohydrates, which did not require specific dephosphorylation at C1, were resuspended in 25 mM HEPES/K+ buffer with 100 mM KCl, pH 7.4 and then 10 μL of recombinant shrimp alkaline phosphatase (rSAP) was added for an incubation period of 4–16 h at 37°C. These compounds include L-galacto-heptulose-1-P, D-altro-heptulose-1-P, 6-deoxy-D-fructose-1-P, 6-deoxy-L-sorbose-1-P, L-galacto-hepturonate-1-P, L-glycer-L-galacto-octuluronate-1-P, D-glycero-L-galacto-octuluronate-1-P, L-glycero-D-altro-octuluronate-1-P, L-threo-L-galacto-nonulonate-1-P, and D-erythro-L-galacto-nonulonate-1-P. The progress of the reaction was monitored by 31P NMR spectroscopy. For compounds that needed specific dephosphorylation at C1, purified GlpX (10–20 μM) was added with 2.0 mM MgCl2 and 0.5 mM MnCl2 (11, 12). The reaction was monitored using the PiColorLock phosphate detection kit (Innova biosciences). These compounds were L-galacto-heptulose-1,7-bisphosphate, L-glycero-L-galacto-octuluronate-1,8-bisphospate, and D-glycero-L-galacto-octuluronate-1,8-bisphospate. GlpX was removed by centrifugal filtration and then the sample loaded onto a DEAE anion exchange column and purified as described above. After purification, all dephosphorylated compounds were submitted for mass analysis (Table S1) and, for compounds with remaining phosphate, were analyzed by 31P NMR spectroscopy.
Synthesis of D-Glycero-D-ido-octulose-1,8-bisphosphate.
D-Glycero-D-ido-octulose-1,8-bis-phosphate (17) cannot be synthesized using YdjI, DHAP, and D-arabinose-5-P. However, this molecule was previously synthesized using FBP aldolase (16). Briefly, D-arabinose-5-P (0.052 mmol) was mixed with DHAP (0.042 mmol) in 50 mM HEPES/K+ buffer, pH 8.5. To this solution, ~50 U (4.0 mg) of FBP aldolase (Sigma Aldrich) was added and the reaction monitored for 96 h. The aldolase was removed and D-glycero-D-ido-octulose-1,8-bis-P was purified by anion-exchange as described above. D-Glycero-D-ido-octulose-1,8-bis-P was specifically dephosphorylated at the C1 position using GlpX and purified by anion-exchange chromatography.
Synthesis of D-Fructuronate using UxaC.
D-Fructuronate (9) was synthesized using uronate isomerase (UxaC), as described previously (17–19). The gene for UxaC (Uniprot ID: P0A8G3) was amplified from the genomic DNA of E. coli K12 with the primer pair:
5ʹ- ATCCAAGCTTCGATGACTCCGTTTATGACTGAAGATTTCCTG-3ʹ
5ʹ- ATCCCTCGAGTTAGTTCAGTTCAATGGCGAAGTAGTC −3ʹ.
Restriction sites for HindIII and XhoI (underlined) were introduced into the forward and reserve primers, respectively. These sites allow for the addition of an N-terminal 6x-His-tag in the pET-30a+ expression vector. UxaC was expressed and purified as previously described (18). The overall yield was 50–75 mg/mL of purified protein from 2 L of cell culture. To test the catalytic activity of YdjH (10 μM) with D-fructuronate (9), 10 mM D-glucuronate, 5.0 mM ATP, and 5.0 mM MgCl2 were mixed in three different buffers; 50 mM borate buffer, pH 8.3, 50 mM imidazole buffer, pH 7.4, and 50 mM HEPES/K+ buffer, pH 7.4. To each reaction, 2.0 μM UxaC was added and the reaction monitored by 31P NMR spectroscopy for 24 h. These reactions were repeated and monitored using the UV-coupled assay that measures the rate of formation of ADP.
Crystal Structure and Computational Docking.
There are two three-dimensional crystal structures for YdjH (PDB entries: 3H49 and 3IN1). The 3-D structures of YdjH were compared with the following proteins: a putative ribokinase II from E. coli in complex with MgATP (PDB entry: 3IQ0), human ketohexokinase with fructose (PDB entry: 2HW1) (20), a putative phosphofructokinase from Staphylococcus aureus (PDB entry: 2JG5), 2-keto-3-deoxygluconate kinase from Sulfolobus solfataricus bound to 2-keto-3-deoxygluconate (PDB entry: 2VAR) (21), a ribokinase from E. coli bound to ribose and ADP (PDB entry: 1RDK) (22), human ribokinase (PDB entry: 2FV7), 2-keto-3-deoxygluconate kinase from Thermus thermophilus bound to 2-keto-3-deoxygluconate and ADP (PDB entry: 1V1A) and with ATP (PDB entry: 1V1B) (23), and phosphofructokinase-2 from E. coli bound with FBP and ADP, and also with F6P and ATP (PDB entry: 3UQD).
Computational docking studies were carried out with AutoDock Vina using the command-line scripts from AutoDock Tools Package provided by The Scripps Research Institute (24–26). The search space was confined to a 25 Å × 25 Å × 25 Å box, centered near residue Asp43. The box was made large enough to include most of the ATP binding site, the residues from the lid subdomain, specifically Arg108, and the loop containing Arg175. The receptor used was a modified version of YdjH with ADP and sodium bound (chain B of PDB entry: 3IN1). For the modification, the bound ADP and sodium were removed and ATP was added. The ATP was extracted from the structure of 2-keto-3-deoxygluconate kinase from T. thermophilus (PDB entry: 1V1B) (23). A total of 20 poses were provided with the best pose chosen by location to known active site residues and the known binding orientation found in other family members.
Growth of E. coli on L-glycero-L-galacto-octuluronate.
Growth of E. coli BW25113 and BW25113 ΔydjH on minimal media plates containing L-glycero-L-galacto-octuluronate as the sole carbon source was conducted following a previously published method (27). L-glycero-L-galacto-octuluronate-1-P was prepared by incubating dihydroxyacetone phosphate (0.3 mmol) with 0.35 mmol l-arabinuronic acid and 15 μM YdjI in 25 mM imidazole/HCl, pH 7.2. The reaction was monitored by 31P NMR spectroscopy and the reaction complete after 16 h. The product, L-glycero-L-galacto-octuluronate-1-P, was purified by DEAE-anion exchange as previously described and then dephosphorylated using rSAP (1). Minimal media plates prepared from M9 minimal media, 1.5% (w/v) agar, and 100 μM thiamine were prepared as described previously with a 1.0 mM carbon source (27). The five carbon sources tested were: glucose (positive control), L-glycero-L-galacto-octuluronate, dihydroxyacetone (positive control), l-arabinuronic acid, and sucrose (negative control). Plates with no added carbon source served as an additional negative control. Cells from single colonies of E. coli BW25113 and E. coli BW25113 ΔydjH were grown, washed with M9 minimal media, and serial diluted such that single colonies could be observed as described previously (27).
RESULTS
Bioinformatic Analysis of YdjH.
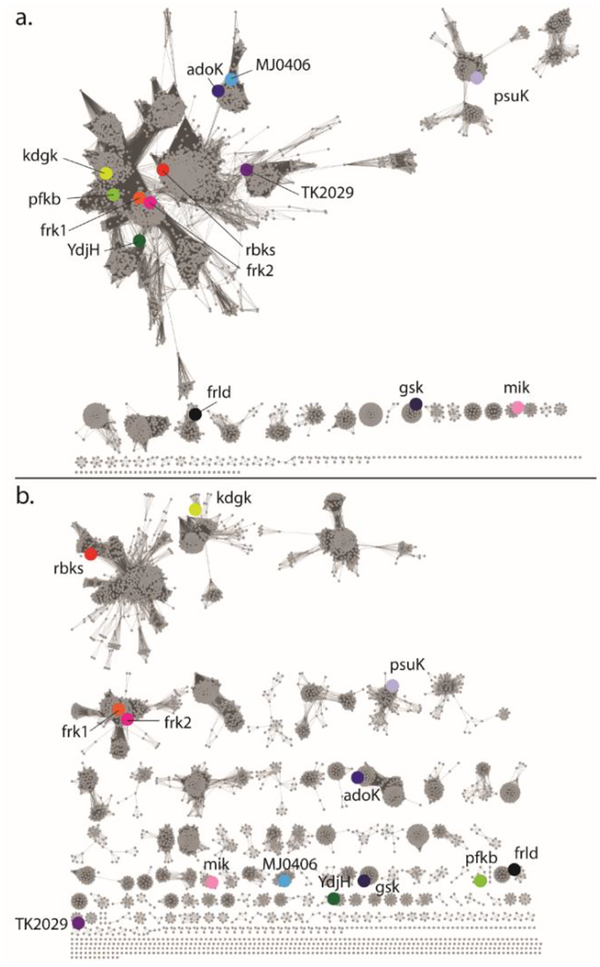
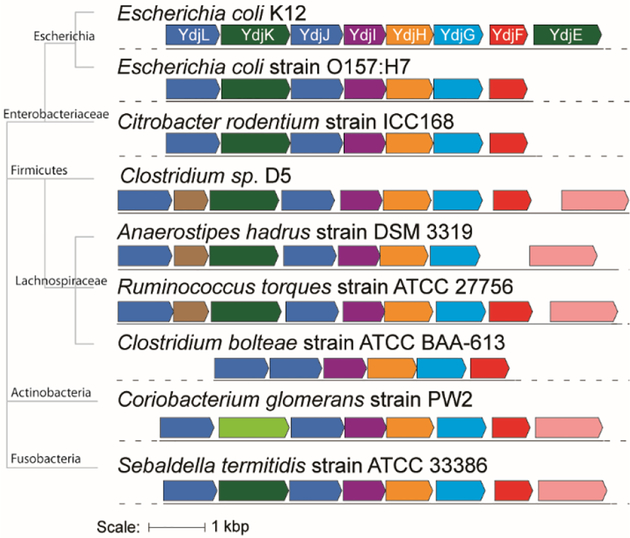
YdjH is assigned to COG0524, which is mostly comprised of members from the ribokinase/fructokinase family. As of 01/07/2019, COG0524 contains 6238 unique protein sequences with 4872 of these containing less than 90% sequence identity. The sequence similarity network (SSN) of COG524 at an alignment score cutoff of 35, or ~26% sequence identity shows YdjH in the major cluster with the enzymes for fructokinase (frk1/2), ribokinase, and 2-dehydro-3-deoxygluconokinase (Figure 2a). At an alignment score of 60, or ~35% sequence identity, YdjH is found in a small cluster containing a total of 13 nodes representing 28 unique proteins with a sequence identity of ~40% (Figure 2b). The core (10 nodes) contains homologs to YdjH from various bacterial species including E. coli O157:H7, C. bolteae, C. glomerans, Sebaldella termitidis, and Citrobacter rodentium. All of these homologs to YdjH are found in a similar genomic context as the ydj gene cluster, suggesting that all of these enzymes have the same function despite their relatively low sequence identity (Figure 3).
Figure 2.
(a) Sequence similarity network of COG0524 at an alignment score of 35. (b) Sequence similarity network of COG0524 at an alignment score of 60. At this alignment score all of the known catalytic functions have been separated into single clusters and thus the SSN is probably isofunctional. Abbreviation for enzymes are as follows: rbks-ribokinase (Uniprot: P0A9J6); kdgk-2-dehydro-3-deoxygluconokinase (Uniprot: P37647); frk1/frk2-fructokinase1/2 (Uniprot: Q6XZ78/Q0JGZ6); psuK-pseudouridine kinase (Uniprot: P30235); adoK-adenosine kinase (Uniprot: P9WID5); TK2029-ADP-dependent ribose-1-phosphate kinase from Thermococcus kodakarensis (Uniprot: Q5JDG9); mik-inositol 3-kinase (Uniprot: Q5GA22); MJ0406-nucleoside kinase from Methanocaldococcus jannaschii (Uniprot: Q57849); gsk-inosine-guanosine kinase (Uniprot: P0AEW6); pfkb-ATP-dependent 6-phosphofructokinase (Uniprot: D9TT10); and frld-fructoselysine 6-kinase (Uniprot: P45543).
Figure 3.
Genomic neighborhood of YdjH homologs from various bacterial species. Genes that share high similarity are colored the same. The phylogenetic tree shows the evolutionary relationship among the bacteria with major branch points labeled. The E. coli K12 genes are labeled using their gene names as described in Figure 1. There are three genes that are not present in the E. coli cluster. The pink gene is annotated as a sodium/dicarboxylate symporter. The light green gene is annotated as a major facilitator superfamily 1 member. The brown gene is annotated as a pentulose-5-phosphate 3-epimerase.
Screening Catalytic Activity of YdjH.
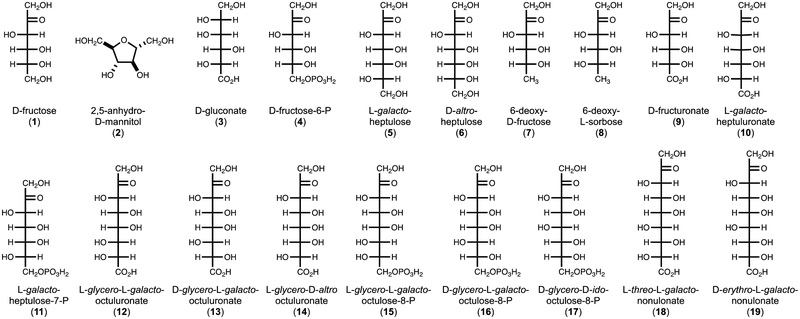
The initial substrate activity screen with 96-commercially purchased compounds (Table S2) demonstrated that YdjH catalyzes the phosphorylation of D-fructose (1), 2,5-anhydro-D-mannitol (2), D-gluconate (3), D-fructose-6-P (4), L-galacto-heptulose (5), and D-altro-heptulose (6) (Figure 4). The phosphorylated products of D-fructose, L-galacto-heptulose, and D-gluconate with YdjH were identified as D-fructose-1-P, L-galacto-heptulose-1-P, and D-gluconate-6-P, respectively, by 31P NMR spectroscopy. The steady-state kinetic constants for the phosphorylation of substrates 1-6 are presented in Table 1.
Figure 4:
Compounds tested for catalytic activity with YdjH.
Table 1.
Steady-state kinetic parameters of substrates with YdjHa
| Substrate | kcat (s−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| 1 | 0.11 ± 0.01 | 13.5 ± 2.5 | 7.8 ± 1.6 |
| 2 | 0.06 ± 0.01 | 44 ± 5 | 1.4 ± 0.2 |
| 3 | 0.025 ± 0.001 | 0.018 ± 0.001 | 1400 ± 100 |
| 4 | 0.0020 ± 0.0001 | 1.4 ± 0.3 | 1.4 ± 0.3 |
| 5 | 0.35 ± 0.01 | 14.5 ± 0.4 | 24.4 ± 0.8 |
| 6 | 0.055 ± 0.002 | 29.9 ± 1.2 | 1.8 ± 0.1 |
| 10 | 0.42 ± 0.03 | 2.2 ± 0.4 | 190 ± 40 |
| 11b | - | - | 0.3 ± 0.01 |
| 12 | 16.2 ± 0.3 | 0.77 ± 0.03 | 21000 ± 1000 |
| 13 | 0.68 ± 0.01 | 6.3 ± 0.3 | 108 ± 5 |
| 14 | 0.22 ± 0.03 | 0.54 ± 0.15 | 410 ± 120 |
| 18 | 0.34 ± 0.03 | 4.7 ± 0.5 | 72 ± 7 |
| 19 | 1 ± 0.5 | 7.0 ± 4.5 | 123 ± 5e |
Monitored using a coupled enzyme assay by following the oxidation of NADH at 30°C in 50 mM HEPES/K+ buffer with 100 mM KCl, pH 7.4.
Saturation of 11 could not be obtained, so the data was fit to a line with the slope of the line equal to kcat/Km.
Carbohydrates Identified from the Catalytic Activity of YdjI.
The initial characterization of YdjH began as a larger endeavor to identify the catalytic properties of the entire ydj gene cluster. The aldolase, YdjI, was shown to have a strong preference for extended monosaccharides of 7–9 carbons with a negatively charged terminus that is either a carboxylate or is phosphorylated (1). These longer monosaccharides were therefore tested as potential substrates for YdjH (Figure 4).
To synthesize these compounds, YdjI was combined with DHAP and the appropriate aldose sugar and the products purified by anion exchange chromatography (1). For those compounds with a single phosphate attached to C1, it was enzymatically removed with rSAP. For those compounds with a phosphate attached to both C1 and the terminal carbon, the phosphoryl group at C1 was specifically removed with GlpX (11, 12). The dephosphorylated compounds were purified by anion exchange chromatography and the molecular weight confirmed by mass spectrometry (Table S1). Two compounds, D-fructuronate (9) and D-glycero-D-ido-octulose-8-P (17) were synthesized enzymatically with UxaC and FBP aldolase, respectively (16–18). The steady-state kinetic constants for compounds found to be substrates for YdjH are summarized in Table 1. No activity (kcat < 0.001 s−1) could be detected for compounds 7, 8, 9, 15, 16, and 17. YdjH exhibits the most robust catalytic activity with L-glycero-L-galacto-octulonate (12) and preferentially phosphorylates 7–9 carbon monosaccharides, relative to those with 5 or 6 carbons. A terminal carboxylate is greatly preferred relative to those terminated with a phosphate.
Characterization of L-Glycero-L-galacto-octuluronate by NMR Spectroscopy.
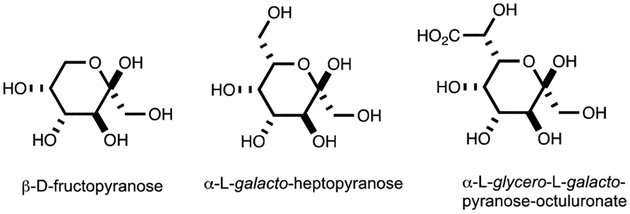
Given the high level of activity of YdjH with L-glycero-L-galacto-octuluronate (12), it was of interest to more fully characterize the conformational structure of this molecule by 13C and 1H NMR spectroscopy (Figures S1a and S1b). These spectra show that L-glycero-L-galacto-octuluronate (12) is predominately found in a single anomeric form. 2D-NMR experiments (HSQC, HMBC, and COSY) enabled the assignment of the chemical shifts for the corresponding carbons and protons (Figures S2, S3, and S4, respectively). Comparison of the 13C and 1H chemical shifts and the 1H coupling constants of L-glycero-L-galacto-octuluronate (12) to those of the corresponding chemical shifts for D-fructose (1) (28, 29) and L-galacto-heptulose (5) (30) suggests that L-glycero-L-galacto-octuluronate (12) is predominately found in the α-pyranose form (Table 2, Table S3, Table S4, and Figure 5)(31).
Table 2.
13C Chemical shifts for D-fructose, L-galacto-heptulose, and L-glycero-L-galacto-octuluronate.
| Compound | Form | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 |
|---|---|---|---|---|---|---|---|---|---|
| D-fructosea | β-pyranose (75%) | 64.9 | 98.9 | 68.6 | 70.7 | 70.2 | 64.2 | - | - |
| β-furanose (21%) | 63.7 | 102.3 | 76.4 | 75.4 | 81.5 | 63.3 | - | - | |
| α-furanose (4 %) | 63.9 | 105.2 | 83.0 | 77.0 | 82.2 | 62.1 | - | - | |
| L-galacto-heptuloseb | α-pyranose (78%) | 64.9 | 98.6 | 68.6 | 71.2 | 70.3 | 72.1 | 62.1 | - |
| α-furanose (16%) | 63.5 | 102.2 | 76.3 | 75.4 | 80.9 | 72.5 | 63.3 | - | |
| β-furanose (6%) | 63.8 | 105.1 | 82.9 | 77.0 | 81.2 | 71.6 | - | - | |
| L-glycero-L-galacto-octuluronate | α-pyranose | 63.9 | 97.8 | 67.3 | 70.6 | 69.3 | 71.6 | 72.5 | 179.0 |
Figure 5:
Anomeric forms of D-fructose, L-galacto-heptulose and L-glycero-L-galacto-octuluronate.
Analysis of Three-Dimensional Crystal Structure.
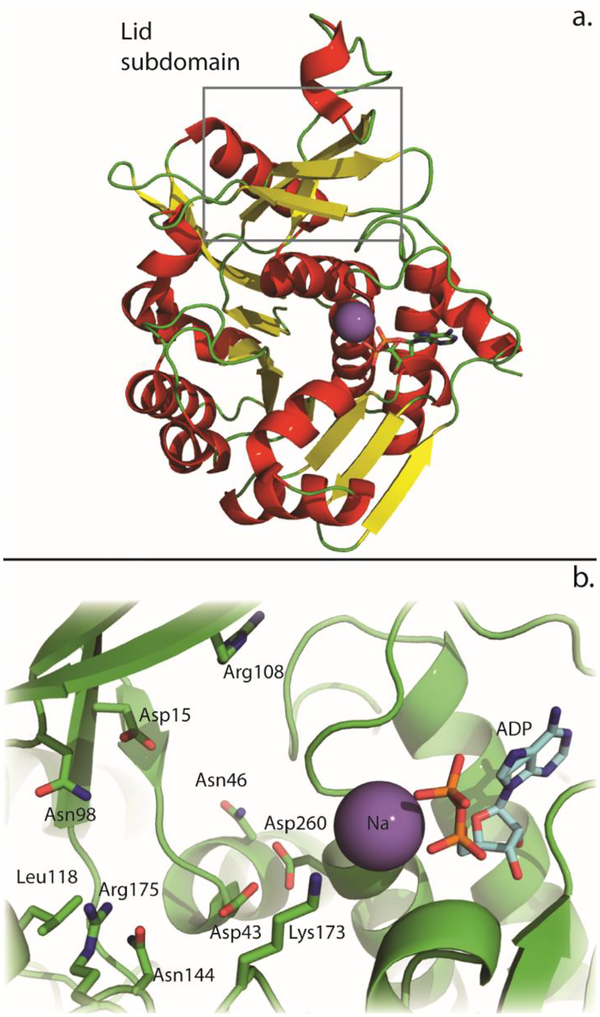
Two crystal structures for YdjH (PDB entries: 3IN1 and 3H49) were previously reported. One of these structures (PDB entry: 3IN1) contains ADP bound within the active site, while the other has nothing bound (PDB entry: 3H49). There are no major structural differences between YdjH and other members of the ribokinase/fructokinase family. Both of the YdjH structures contain two monomers in the asymmetric unit. Each monomer contains a major α/β domain with 10 α-helices positioned around an inner β-sheet (Figure 6a). The β-sheet is made up of eight parallel and one antiparallel β-strands. The lid subdomain consists of three β-strands and a small α-helix and is connected to the major α/β domain by the highly conserved GG hinge (Gly41 and Gly42) shared by all members of this enzyme family (2–4). Based on gel filtration analysis, YdjH exists as a dimer in solution, similar to the quaternary structures of other known family members (3, 4, 20–23). Both structures were solved at pH 5.6 in 200 mM ammonium acetate and sodium citrate buffer. This is different than the conditions used to test activity (50 mM HEPES/K+ buffer, pH 7.4). YdjH was assayed in 100 mM ammonium acetate, sodium citrate buffer, pH 5.6 with L-glycero-L-galacto-octuluronate. The UV-based coupled assay showed that the coupling components (pyruvate kinase, lactate dehydrogenase) displayed significant loss in activity in this buffer (>100-fold), which prevented quantification of YdjH activity. However, a 31P NMR experiment showed robust activity of YdjH at pH 5.6, where YdjH (5 μM) completely phosphorylated L-glycero-L-galacto-octuluronate (5 mM) within 30 minutes.
Figure 6:
(a) Ribbon diagram of YdjH where the α-helices are red, β-strands are yellow and the sodium appears as a purple sphere. The lid subdomain is highlighted by the box (PDB entry: 3IN1). (b) The active site of YdjH. Key residues are labeled and shown as green sticks. The Na is shown as a purple sphere and the bound ADP is shown in cyan sticks.
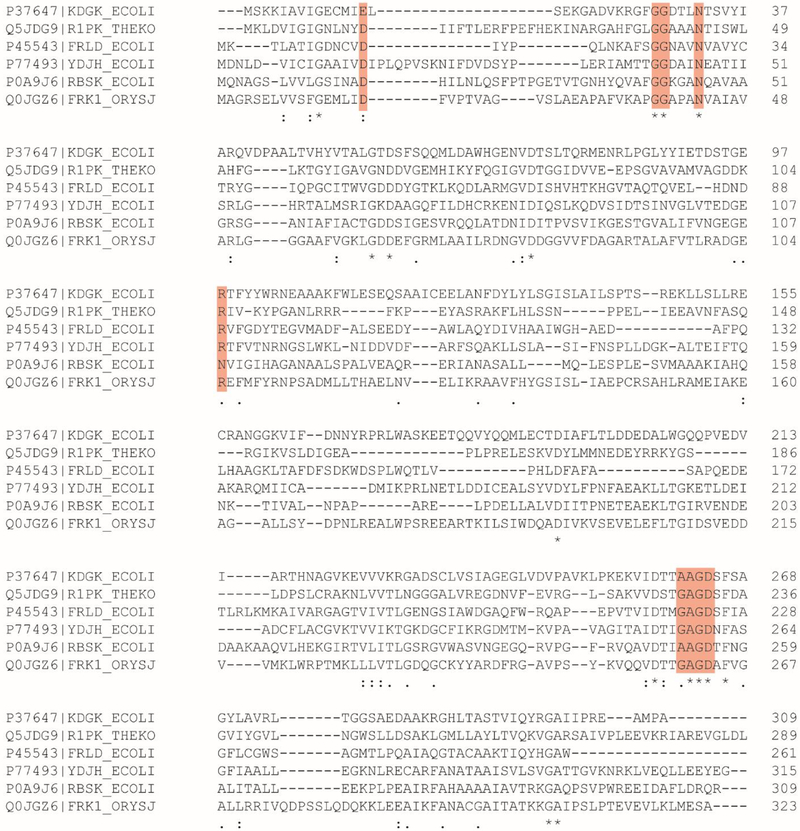
The active site is located in a deep pocket formed by the major α/β domain and the lid subdomain. The active site is defined by the strictly conserved G/AxGD motif where the conserved aspartate functions as the general base to deprotonate the hydroxyl group of the substrate, which in turn attacks the γ-phosphoryl group of ATP (3). In YdjH, this residue is Asp260 (Figure 6b). The ATP binding site is similar to others found in the ribokinase/fructokinase enzyme family whose three-dimensional structures have been determined. The 2ʹ hydroxyl group of the ribose ring is predicted to interact with Asn254 and the 3ʹ hydroxyl interacts with the backbone carbonyl of Gly232 (20–23). These residues are partially conserved among the other family members. The YdjH structure is the only one within the group with a bound sodium in the active site. The sodium interacts with the backbone carbonyl of Thr255 and the α-phosphate of the bound ADP. The sodium is positioned where the β-phosphate binds in the other structures from this enzyme family. This has apparently caused the β-phosphoryl group of ADP in YdjH to be placed in an unusual orientation. A sequence alignment among ribokinase family members reveals other well-conserved residues in the active site (Figure 7). These residues include Asp15, Asn46 and Arg108. Conserved residues specific to YdjH and close functional homologs are Arg175, Asn144, Asn98, and Asp43 (Figure S5).
Figure 7.
Sequence alignment of selected ribokinase family members. Key conserved residues and motifs as discussed in the text are highlighted in red boxes.
Binding of L-Glycero-L-galacto-octulonate in Active Site.
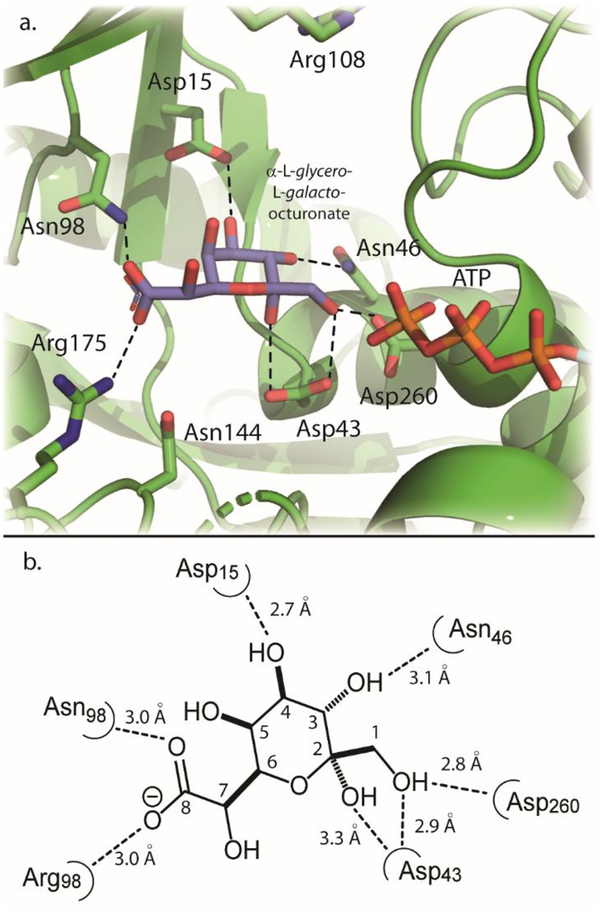
Computational docking studies were conducted to better understand how L-glycero-L-galacto-octuluronate (12) might bind in the active site of YdjH. Docking computations were conducted using AutoDock Vina with the search space confined to an area centered near Asp43, as this residue is near the center of the active site pocket (Figure S6). The α-pyranose anomer of L-glycero-L-galacto-octuluronate was docked into the active site. The receptor used for docking was chain B (PDB entry: 3IN1) of YdjH in which the bound ADP and sodium were removed. ATP was added to the YdjH receptor using the orientation of ATP found in the active site of 2-keto-3-deoxy-gluconate kinase (PDB entry: 1V1B). This specific binding mode of ATP was chosen based on an overlay of bound ATP/ADP in other ribokinase family members (Figure S7). The catalytic aspartate residue is found in approximately the same position in all observed structures (Figure S7). The docking pose for the α-pyranose anomer of L-glycero-L-galacto-octuluronate into the modified YdjH active site is shown in Figure 8a. A two-dimensional illustration with measured distances of interactions is shown in Figure 8b.
Figure 8.
Docking pose of α-L-glycero-L-galacto-octulonopyranose (blue sticks) into a modified active site of YdjH. Labeled in green sticks are key active site residues. The tripolyphosphate chain of ATP is shown in the orange and red sticks. (b) Two-dimensional illustration of the active site and interactions with docked α-L-glycero-L-galacto-octulonopyranose. Interactions are shown with the dotted lines and measured in angstroms.
These results show that the α-pyranose anomer of L-glycero-L-galacto-octuluronate fits well into the YdjH active site pocket. It is found in a position similar to the orientation of substrates bound in the structures of other ribokinase family members (Figure S8a) (20). The γ-phosphoryl group of ATP is positioned 3.9 Å from the hydroxyl group at C1. The C1 hydroxyl group is 2.8 Å away from the putative general base Asp260, and 2.9 Å from Asp43. Asp43 is predicted to interact with the hydroxyl group attached to C2. The C8 carboxylate moiety is positioned to form a salt bridge with Arg175, which is 3.0 Å away and potentially hydrogen bonds with Asn98 (3.0 Å). The conserved residues, Asp15 and Asn46, are predicted to hydrogen bond to the hydroxyl groups attached to C4 and C3, respectively. There are no predicted interactions with the hydroxyl groups at C5 or C7. Docking of the α-furanose anomer of L-glycero-L-galacto-octuluronate shows similar interactions and binding orientation as the α-pyranose anomer (Figure S8b). Docking the α-pyranose anomer of L-glycero-L-galacto-octuluronate-1-P into the YdjH receptor with ATP molecule removed, yields a slightly different binding orientation, but the phosphate group attached at C1 is found in the same position as the γ-phosphoryl group of ATP used for substrate docking (Figure S8c).
Growth of E. coli on L-glycero-L-galacto-octuluronate.
Stocks of E. coli BW25113 and BW25113 ΔydjH were serial diluted (100, 10−2, 10−4, and 10−6) and plated on M9 minimal media plates containing no carbon source, glucose (1.0 mM), dihydroxyacetone (1.0 mM), sucrose (1.0 mM), L-arabinuronic acid (1.0 mM), and L-glycero-L-galacto-octuluronate (1.0 mM). After three days (at 37°C), no colonies were visible on any plates containing no carbon source, sucrose or L-arabinuronic acid. After 24 h, small individual colonies were visible on the plates containing glucose. After three days, very small individual colonies were visible on the plates containing dihydroxyacetone, a known carbon source for E. coli (32). However, after three days, no colonies were observed on the plates containing L-glycero-L-galacto-octuluronate.
DISCUSSION
YdjH is a member of the ribose/fructokinase family of enzymes and found in the ydj gene cluster whose function in E. coli K12 is unknown. Analysis of the sequence similarity network (SSN) at a stringent alignment score shows that YdjH and its close homologs are segregated from the major cluster of enzymes (Figure 2b). The core of the cluster containing YdjH represents a set of enzymes that are all found within the same genomic context and annotated such that they resemble a catabolic pathway for the metabolism of a carbohydrate of unknown structure (Figure 3). The co-localization of the putative aldolase (YdjI) and sugar kinase (YdjH) in this gene cluster strongly suggests that the product of the kinase will be the substrate for the aldolase (YdjI).
In the preceding paper, the substrate profile of the YdjI aldolase was characterized. YdjI was shown to have a strong preference for the retro-aldol cleavage of 7 to 9 carbon ketoses where the hydroxyl group at C1 is phosphorylated and the terminal carbon is either a carboxylate or is phosphorylated (1). The best substrate identified thus far for YdjI is L-glycero-L-galacto-octuluronate-1-P. Here, the best substrate for YdjH is L-glycero-L-galacto-octuluronate (12) yielding L-glycero-L-galacto-octuluronate-1-P. Unlike the substrate profile for YdjI, YdjH shows relatively high substrate specificity for L-glycero-L-galacto-octuluronate with a kcat/Km that is 15-fold greater than the next best substrate.
Interestingly, YdjH is unable to efficiently catalyze the phosphorylation of ketoses that are phosphorylated at the terminal carbon. Since the phosphorylated product of the best substrate for the YdjH kinase is the best substrate identified to date for the YdjI aldolase, it is highly likely that the apparent physiological substrate for these two enzymes will be very similar in structure to L-glycero-L-galacto-octuluronate.
YdjH is a typical member of the ribose/fructokinase family of enzymes (20–23). YdjH contains two conserved motifs, GG and G/AxGD and other conserved key residues found within the active site. These residues include Asp260, Asn46, and Asp15. The three-dimensional structure of YdjH provides some clues as to which residues are responsible for establishment of the substrate specificity. These residues are proposed to be Arg175, Asn144, Asn98, and Asp43 as these residues are not conserved in other family members, but all four are conserved among the close homologs of YdjH (Figure S5).
Computational docking of L-glycero-L-galacto-octuluronate (12) illustrates how this substrate may bind into the active site of YdjH. L-Glycero-L-galacto-octuluronate is docked with the C1 hydroxyl positioned near the proposed catalytic base, Asp260. The C4 and C3 hydroxyl groups are predicted to have interactions with the conserved residues Asp15 and Asn46, respectively. The terminal carboxylate group likely interacts with the YdjH-specific residues Arg175 and Asn98, while Asp43 is predicted to interact with the C1 and C2 hydroxyl groups. The putative binding site for the carboxylate group is not too dissimilar from the binding site for the carboxylate group observed in the crystal structure of 2-keto-3-dehydro-D-gluconokinase (PDB entry: 1V1B) (23). This enzyme utilizes two arginine residues, Arg105 and Arg167, and a tyrosine residue, Tyr103, to coordinate the carboxylate group of a linear substrate (23). Similar interactions were observed for docking results of alternative the substrates D-glycero-L-galacto-octuluronate (13) and L-glycero-L-galacto-octulose (Figures S9a and S9b, respectively). However, L-glycero-L-galacto-octulose-8-P (15), which was shown to have no measurable activity with YdjH, is also predicted to have similar binding, although slightly twisted to accommodate the larger molecule in the active site space (Figure S9c).
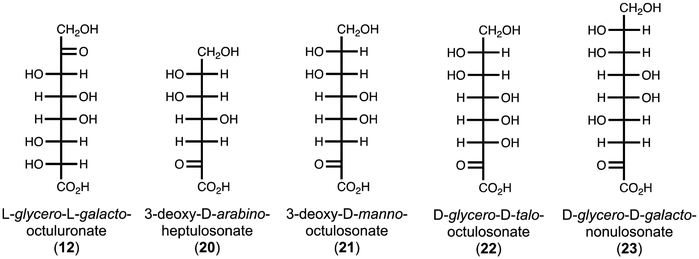
Currently, L-glycero-L-galacto-octuluronate (12) has no known natural abundance. However, there is a series of known compounds that are similar in structure (Figure 9). 3-Deoxy-D-arabino-heptulosonic acid 7-P (DAHP, 20) is an intermediate in the biosynthesis of shikimic acid, which is critical for the production of aromatic amino acids (33). 3-Deoxy-D-manno-octulosonate (KDO, 21) is a key component of lipopolysaccharide (LPS) structures of many bacterial species, including E. coli (34). KDO functions as the linker between lipid A, which is imbedded in the membrane, and the O-antigen, which is present on the surface of the bacterium. KDO-8-P is synthesized by KDO8P synthase, which utilizes PEP and D-arabinose-5-P in a condensation reaction (34). D-Glycero-D-talo-octulosonate (KO, 22) is a rare substitute for KDO in the LPS structure of some acid-stable bacteria species, including Acetobacter pasteurianus (35). 3-Deoxy-D-glycero-D-galacto-nonulosonate (KDN, 23) is a deaminated neuraminic acid derivative found on glycoproteins of vertebrates and bacteria (36). KDN has a known degradation pathway, but DAHP and KDO/KO do not (37). KDO was shown to function as a sole carbon source for growth of an E. coli mutant, but the exact pathway of degradation remains unknown (38).
Figure 9:
Known compounds similar to L-glycero-L-galacto-octuluronate.
Additional heptulose, octulose, and nonulose molecules have been previously described (39). These molecules are most commonly found in plants, but also appear in bacteria and mammals (39). The concentrations of these monosaccharides vary significantly. Octuloses, are typically found in relatively low quantities (39), but D-glycero-D-ido-octulose has been shown to constitute about 90% of the total carbohydrate in the hydrated leaves of Craterostigma plantaginum (39, 40). The catabolic pathways for these molecules are unknown with the proposed synthesis and metabolism to occur via the L-type pentose phosphate pathway (41). This catabolic pathway was developed around experimental data with D-altro-heptulose, D-glycero-D-altro-octulose and D-glycero-D-ido-octulose for both mono- and bis-phosphorylated derivatives. The L-type pentose phosphate pathway utilizes transketolase enzymes to transform these molecules into derivatives of more common monosaccharides such as D-glucose-6-P, D-fructose-6-P, or D-ribose-5-P. There are no data available for the degradation of an L-glycero-L-galacto-octulose. However, both D-glycero-L-galacto-octulose and L-glycero-L-galacto-octulose have been found in the leaves of Trifolium pretense (39).
E. coli BW25113 and the ΔydjH knockout were used to test the ability of E. coli to utilize L-glycero-L-galacto-octuluronate and L-arabinuronic acid as sole carbon sources. The in vitro results for YdjH and YdjI demonstrate that L-glycero-L-galacto-octuluronate can be phosphorylated to L-glycero-L-galacto-octuluronate-1-P and that YdjI can catalyze the retro-aldol reaction yielding DHAP and L-arabinuronate. E. coli BW25113 and the ΔydjH knockout strain were able to grow on glucose and dihydroxyacetone, and unable to grow on sucrose, as expected. Both strains failed to utilize L-arabinuronic acid or L-glycero-L-galacto-octuluronate as a sole carbon source. The lack of growth on L-glycero-L-galacto-octuluronate is most likely the result of L-glycero-L-galacto-octuluronate failing to inactivate the repressor (YdjF) such that YdjH/I gene expression can occur, and/or L-glycero-L-galacto-octuluronate is unable to be transported into the cell. The most likely transporters are the nearby gene products of ydjK and/or ydJE. These genes would be subject to the same expression problems as YdjH and YdjI if L-glycero-L-galacto-octuluronate fails to inactivate YdjF.
KDO and KO were tested as possible substrates for YdjH. However, there was no observed activity with either compound. This result was not surprising as YdjH appears to require a ketone at carbon 2. Docking of KDO and KDO-8-P into the active site suggests that these compounds will not bind well into the active site of YdjH (Figures S9d and S9e). However, it is possible that one of the three uncharacterized dehydrogenases in the ydj-gene cluster could catalyze an oxidation at C2. However, we have not observed measurable activity (oxidation or reduction) for L-glycero-L-galacto-octulonate, KDO, or KO with any of three dehydrogenases, YdjL, YdjJ, and YdjG (Huddleston et al. unpublished results. 2019). YdjJ does oxidize the penultimate carbon of two similar sugar alcohols and sugar acids to form a keto-sugar (Huddleston et al. unpublished results. 2019). While it is possible that L-glycero-L-galacto-octulonate, or a similar compound, is the intermediate for the pathway, it seems the initial substrate for this pathway remains to be identified. The lack of growth on L-glycero-L-galacto-octuluronate lends further support to this conclusion.
In summary, YdjH is a member of the ribokinase family that preferentially phosphorylates the C1 position of longer keto-sugars with a terminal carboxylate group. The best substrate was identified to be L-glycero-L-galacto-octuluronate, which has no known natural abundance. Currently, similar functional and structural characterization of the dehydrogenase enzymes, YdjJ, YdjL, and YdjG are underway to identify shared associated activities. The most likely reaction will be the formation of a keto-sugar from an acid sugar similar to the dehydrogenases described for L-gulonate and L-idonate degradation, which are found in the same COG family (COG1063) as YdjJ and YdjL (42).
Supplementary Material
Funding
This work was supported in parts by grants from the Robert A. Welch Foundation (A-840) and the National Institutes of Health (GM122825).
Footnotes
Supporting Information
Mass spectral data for synthesized compounds for YdjH activity; 1H chemical shifts and coupling constants for L-glycero-L-galacto-octuluronate; 13C and 1H spectra for L-glycero-L-galacto-octuluronate; Sequence alignment of close homologs to YdjH; Modified active site of YdjH with space of cavity near active site; Overlay of ATP/ADP molecules from ribokinase family members; Docking result comparisons of L-glycero-L-galacto-octuluronate to D-fructose binding, L-glycero-L-galacto-octuluronate furanose-form and L-glycero-L-galacto-octuluronate-1-P.
The authors declare no competing financial interest.
REFERENCES
- (1).Huddleston JP, Thoden JB, Dopkins BJ, Narindoshvili T, Fose BJ, Holden HM, and Raushel FM (2019) Structural and functional characterization of YdjI, an aldolase of unknown specificity in Escherichia coli K12, Biochemistry, (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Bork P, Sander C, and Valencia A (1993) Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases, Protein Sci, 2, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Cabrera R, Babul J, and Guixé V (2010) Ribokinase family evolution and the role of conserved residues at the active site of the PfkB subfamily representative, Pfk-2 from Escherichia coli, Arch. Biochem. Biophys, 502, 23–30. [DOI] [PubMed] [Google Scholar]
- (4).Park J, and Gupta RS (2008) Adenosine kinase and ribokinase--the RK family of proteins, Cell. Mol. Life Sci, 65, 2875–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).The UniProt Consortium. (2015) UniProt: a hub for protein information, Nucleic Acids Res, 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mitchell A, Chang H-Y, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong S-Y, Bateman A, Punta M, Attwood TK, Sigrist CJA, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, and Finn RD (2014) The InterPro protein families database: the classification resource after 15 years, Nucleic Acids Res, D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, and Whalen KL (2015) Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks, Biochim. Biophys. Acta: Proteins Proteomics, 1854, 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Galperin MY, Makarova KS, Wolf YI, and Koonin EV (2015) Expanded microbial genome coverage and improved protein family annotation in the COG database, Nucleic Acids Res, 43, D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang P-L, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, and Bader GD (2007) Integration of biological networks and gene expression data using Cytoscape, Nat. Protoc, 2, 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Inoue H, Nojima H, and Okayama H (1990) High efficiency transformation of Escherichia coli with plasmids, Gene, 96, 23–28. [DOI] [PubMed] [Google Scholar]
- (11).Donahue JL, Bownas JL, Niehaus WG, and Larson TJ (2000) Purification and characterization of glpx encoded fructose 1,6-bisphosphatase, a new enzyme of the glycerol 3-phosphate regulon of Escherichia coli, J. Bacteriol, 182, 5624–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Brown G, Singer A, Lunin VV, Proudfoot M, Skarina T, Flick R, Kochinyan S, Sanishvili R, Joachimiak A, Edwards AM, Savchenko A, and Yakunin AF (2009) Structural and biochemical characterization of the type II fructose-1,6-bisphosphatase GlpX from Escherichia coli, J. Biol. Chem, 284, 3784–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wong CH, Mazenod FP, and Whitesides GM (1983) Chemical and enzymatic syntheses of 6-deoxyhexoses. Conversion to 2,5-dimethyl-4-hydroxy-2,3-dihydrofuran-3-one (furaneol) and analogs, J. Org. Chem, 48, 3493–3497. [Google Scholar]
- (14).Masuko T, Minami A, Iwasaki N, Majima T, Nishimura S-I, and Lee YC (2005) Carbohydrate analysis by a phenol–sulfuric acid method in microplate format, Anal. Biochem, 339, 69–72. [DOI] [PubMed] [Google Scholar]
- (15).Nielsen SS (2010) Phenol-sulfuric acid method for total carbohydrates, In Food Analysis Laboratory Manual (Nielsen SS, Ed.), pp 47–53, Springer; US, Boston, MA. [Google Scholar]
- (16).Franke FP, Kapuscinski M, Macleod JK, and Williams JF (1984) A 13C-N.M.R. study of intermediates in the l-type pentose phosphate cycle, Carbohydr. Res, 125, 177–184. [DOI] [PubMed] [Google Scholar]
- (17).Ashwell G, Wahba AJ, and Hickman J (1960) Uronic acid metabolism in bacteria: I. Purification and properties of uronic acid isomer in Escherichia coli, J. Biol. Chem, 235, 1559–1565. [PubMed] [Google Scholar]
- (18).Williams L, Nguyen T, Li Y, Porter TN, and Raushel FM (2006) Uronate isomerase: a nonhydrolytic member of the amidohydrolase superfamily with an ambivalent requirement for a divalent metal ion, Biochemistry, 45, 7453–7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Nguyen TT, Fedorov AA, Williams L, Fedorov EV, Li Y, Xu C, Almo SC, and Raushel FM (2009) The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily, Biochemistry, 48, 8879–8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Trinh CH, Asipu A, Bonthron DT, and Phillips SEV (2009) Structures of alternatively spliced isoforms of human ketohexokinase, Acta. Crystallogr. D Biol. Crystallogr, 65, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Potter JA, Kerou M, Lamble HJ, Bull SD, Hough DW, Danson MJ, and Taylor GL (2008) The structure of Sulfolobus solfataricus 2-keto-3-deoxygluconate kinase, Acta. Crystallogr. D Biol. Crystallogr, 64, 1283–1287. [DOI] [PubMed] [Google Scholar]
- (22).Ng KKS, Drickamer K, and Weis WI (1996) Structural analysis of monosaccharide recognition by rat liver mannose-binding protein, J. Biol. Chem, 271, 663–674. [DOI] [PubMed] [Google Scholar]
- (23).Ohshima N, Inagaki E, Yasuike K, Takio K, and Tahirov TH (2004) Structure of Thermus thermophilus 2-keto-3-deoxygluconate kinase: evidence for recognition of an open chain substrate, J. Mol. Biol, 340, 477–489. [DOI] [PubMed] [Google Scholar]
- (24).Trott O, and Olson AJ (2009) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comp. Chem, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, and Olson AJ (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem, 30, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Seeliger D, and Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina, J. Comput.-Aided Mol. Des, 24, 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mukherjee K, Narindoshvili T, and Raushel FM (2018) Discovery of a kojibiose phosphorylase in Escherichia coli K-12, Biochemistry, 57, 2857–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Barclay T, Ginic-Markovic M, Johnston MR, Cooper P, and Petrovsky N (2012) Observation of the keto tautomer of d-fructose in D2O using 1H NMR spectroscopy, Carbohydr. Res, 347, 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kristy MR, Kevin T, Robert AL, Rensheng L, Jose GSM, Armando AUS-S, Maria ISM, Enayde de AM, Maria R. d. M., Helena TG, Modesto AC, Célio K. d. S., Andrew LT, Doug M, and Robert ES (2014) Improved extraction of soluble solids from some brazilian and north american fruits, Nat. Prod. J, 4, 201–210. [Google Scholar]
- (30).Jaseja M, Perlin AS, and Dais P (1990) Two-dimensional NMR spectral study of the tautomeric equilibria of D-fructose and related compounds, Magn. Reson. Chem, 28, 283–289. [Google Scholar]
- (31).Angyal SJ, and Tran TQ (1983) Equilibria between pyranoses and furanoses. The composition in solution and the 13C N.M.R. spectra of the heptoses and the heptuloses, Aust. J. Chem, 36, 937–946. [Google Scholar]
- (32).Jin RZ, and Lin EC (1984) An inducible phosphoenolpyruvate: dihydroxyacetone phosphotransferase system in Escherichia coli, J Gen Microbiol, 130, 83–88. [DOI] [PubMed] [Google Scholar]
- (33).Görlach J, Schmid J, and Amrhein N (1994) Abundance of transcripts specific for genes encoding enzymes of the prechorismate pathway in different organs of tomato (Lycopersicon esculentum L.) plants, Planta, 193, 216–223. [DOI] [PubMed] [Google Scholar]
- (34).Radaev S, Dastidar P, Patel M, Woodard RW, and Gatti DL (2000) Structure and mechanism of 3-deoxy-D-manno-octulosonate 8-phosphate synthase, J. Biol. Chem, 275, 9476–9484. [DOI] [PubMed] [Google Scholar]
- (35).Hashimoto M, Ozono M, Furuyashiki M, Baba R, Hashiguchi S, Suda Y, Fukase K, and Fujimoto Y (2016) Characterization of a novel D-glycero-D-talo-oct-2-ulosonic acid-substituted lipid A moiety in the lipopolysaccharide produced by the acetic acid bacterium Acetobacter pasteurianus NBRC 3283, J. Biol. Chem, 291, 21184–21194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Varki A (2008) Sialic acids in human health and disease, Trends Mol. Med, 14, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hopkins AP, Hawkhead JA, and Thomas GH (2013) Transport and catabolism of the sialic acids N-glycolylneuraminic acid and 3-keto-3-deoxy-D-glycero-D-galactonononic acid by Escherichia coli K-12, FEMS Microbiol. Lett, 347, 14–22. [DOI] [PubMed] [Google Scholar]
- (38).Nyberg K, and Ögren S (1984) A mutant of Escherichia coli capable of utilizing 3-deoxy-d-manno-2-octulosonic acid as sole carbon source, FEMS Microbiol. Lett, 23, 77–81. [Google Scholar]
- (39).Zhang Q, and Bartels D (2017) Octulose: a forgotten metabolite?, J. Exp. Bot, 68, 5689–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhang Q, Linnemann TV, Schreiber L, and Bartels D (2016) The role of transketolase and octulose in the resurrection plant Craterostigma plantagineum, J. Exp. Bot, 67, 3551–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Williams JF, and MacLeod JK (2006) The metabolic significance of octulose phosphates in the photosynthetic carbon reduction cycle in spinach, Photosynthesis Res, 90, 125–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wichelecki DJ, Vendiola JAF, Jones AM, Al-Obaidi N, Almo SC, and Gerlt JA (2014) Investigating the physiological roles of low-efficiency D-mannonate and D-gluconate dehydratases in the enolase superfamily: pathways for the catabolism of L-gulonate and L-idonate, Biochemistry, 53, 5692–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.