Abstract
Mineral dust-induced gene (mdig) encodes a member of the evolutionarily conserved JmjC family proteins that play fundamental roles in regulating chromatin-based processes as well as transcription of the genes in eukaryotic cells. This gene is also named as myc-induced nuclear antigen 53 (MINA), nucleolar protein 52 (NO52) and ribosomal oxygenase 2 (RIOX2). Increased expression of mdig had been noted in a number of human cancers, esp. lung cancer. Emerging evidence suggests that the oncogenic activity of mdig is most likely achieved through its regulation on the demethylation of histone proteins, despite it lacks the structural identities of the demethylases. Here, we discuss the latest discoveries on the characteristics of the mdig protein and its roles in a wide variety of normal and carcinogenic processes. We will also provide perspectives on how mdig is involved in the maintenance and differentiation of the embryonic stem cells, somatic stem cells and cancer stem cells.
Keywords: Mdig, Cancer, Epigenetics, Stem cells, Methylation
1. Introduction
Cancer is a major public health concern worldwide and the second leading cause of death in many countries, among which lung cancer is the number one malignant-related deaths [1]. In United States, lung cancer kills more Americans than any other types of malignancy. In 2018, the estimated number of new lung cancer (including bronchus cancer) cases in US is 234,030 and the estimated number of lung cancer deaths in US is 154,050, which is large than the combination of the cancer deaths from colon cancer, breast cancer and prostate cancer. The incidence rate of lung cancer is declining in men from a high of 102 per 100,000 in 1984 to 73 in 2018, whereas the rate in women has been increasing at the same period, from 39 to 53 per 100,000. Thus, the burden has grown steadily in recent decades due to a rising incidence of female lung cancer patients. The death rate of lung cancer is the highest among all races of US male population. Except for Hispanic females in which the death rate of lung cancer is the second highest, the death rate of lung cancer is also the highest among all other races of US female population. Owing to the lack of reliable biomarkers and symptoms in the early stages, most cases of lung cancer are discovered at an advanced stage. Nearly 60% of patients die within a year after diagnosis and 80% die within five years. Despite immense progress in diagnosis and molecular-targeting therapy of the lung cancer, the improvement in the survival rate over the last three decades is modest. The present 5-year survival rate of 18% is only marginally better than the survival rate of about 10% in the 1970s. Thus, an effort in revealing the new mechanisms of this deadly disease is urgently required. There are two main categories of lung cancer with diverse, histologically distinct pulmonary neoplasms: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) [2]. About 87% of lung cancer patients are diagnosed with NSCLC, including squamous cell carcinoma (˜35%), adenocarcinoma (˜40%) and large-cell carcinoma (˜10%). The squamous cell lung cancer usually begins in one of the large airway tubes (bronchi). This type of lung cancer mainly occurs among elderly male or female with a history of tobacco smoking. The adenocarcinoma, a type of lung cancer more common in women who are either non-smokers or former smokers, mostly appears near the outside surface of the lung with a likelihood of spreading to lymph nodes and other organs. The large cell lung cancer is generally originated from bronchial epithelial cells accompanied by extensive bleeding and tissue damage. This type of lung cancer, especially those with neuroendocrine features, is commonly associated with the quick spread of the tumor to the brain or other organs at the earlier stage. Based on the size of the tumor and the status of metastasis, the lung cancer can be roughly classified into four different stages. The stage I lung cancer is much localized with a tumor size less than 3 cm without notable lymph node metastasis. The tumor size of stage II lung cancer is larger than 3 cm. Sub-stages of stage II lung cancer are made based on the metastatic status (IIA, limited lymph node metastasis; IIB, chest wall or diaphragm metastasis). The lung cancers at stages III and IV are generally those tumors with larger size and much severe metastasis into the other side of the lung and the distant organs.
The most prominent factors associated with lung cancer include tobacco smoking and exposure to environmental or occupational hazards. Other etiological factors have also been linked to lung cancer, albeit at a lower frequency, such as tuberculosis (TB), chronic obstructive pulmonary disease (COPD) and marijuana smoking [2]. It is believed that the majority of lung cancers are caused by tobacco smoking. Epidemiological studies suggest that the longer a person has been smoking and the more cigarettes per day smoked, the greater the lung cancer risk will be. Such risk is also presented for ex-smokers and passive smokers. More than 4000 chemicals and toxic metals have been identified in tobacco smoke, many of which are well known carcinogens that cause acute damage to the lung cells and chronic alterations of the intracellular signaling and genomic DNA in these cells. Some tobacco carcinogens, including polycyclic aromatic hydrocarbons (PAH), mainly the benzo[a]pyrene (BaP) and derivatives, and nitrosamines, e.g. 4-(methylnitrosamino)-l-(3-pyridyl)-l-butanone (NNK), Nʹ-nitrosonornicotine (NNN), can directly induce tumorigenesis in experimental animals, possibly through the formation of DNA adducts that cause genetic mutations. Mechanistically, these carcinogens are also potent in the activation of a wide range of intracellular signaling pathways, such as EGF receptor, MAP kinases, PI3K-Akt, NF-κB, and cell cycle checkpoint cascades.
There is a disturbing trend of more and more non-smoking men and women with the lung cancer in recent years, indicating factors other than tobacco smoke must play a considerable role in the development of the lung cancer. Indirect evidence supporting this notion is the highest incidence rates of lung cancer among US black males who have similar tobacco smoking rate with that of the white American men [3]. Although the second-hand or so-called passive tobacco smoking has been suspected, mounting evidence suggests that exposures to environmental and occupational hazards are attributable to the lung cancer incidence among non-smokers and smokers. The risk of lung cancer is higher in workers who are exposed to mineral dusts including asbestos, silica and some other metal-containing particles. Asbestos is a group of minerals that occurs in underground deposits as fibers and is used in certain industries including building construction, fire-proofing and auto mechanics. Asbestos fibers tend to break easily into particles that can float in the air. Inhalation of asbestos increases the risk of both squamous cell carcinoma and adenocarcinoma. The hypothesis of the carcinogenic effect of silica particles has been supported by a relatively large body of epidemiologic literatures that formed the basis to classify silica as a group I carcinogen by the International Agency for Research on Cancer (IARC) [4]. Silica exposure occurs in a mixed environment such as ore mining, ceramics, pottery, or brick manufacturing, where exposure may be to two or more polymorphs of crystalline silica. Many epidemiologic studies indicated that the increased lung cancer risks in people who exposed to silica could not be explained by confounding factors, such as cigarette smoking or other exposures. These epidemiologic data were supported by a number of animal studies that showed silica inhalation induces lung tumor in rats. Other environmental factors linked to the lung cancer incidence include metropolitan air pollution and radiation [5]. One study suggested that people living in the most heavily polluted metropolitan areas have a 12% increased risk of lung cancer than people in the least polluted areas. It was believed that fuel exhaust and sulfates are comparable to the effects of second-hand smoke from a cigarette [6].
Certain genetic abnormalities or polymorphisms have been found to be associated with a statistically greater risk of lung cancer development in both smoker and non-smoker [7]. It is widely acknowledged that tobacco smoking is a dominant risk factor for lung cancer. However, only a limited percentage (˜11%) of tobacco smokers develop lung cancer. Clearly genetic signature for a given individual determines the outcome of the gene-environment interaction through affecting the expression and function of the oncogenes, tumor suppressors, checkpoint proteins, and enzymes responsible for the metabolism of carcinogens. A number of genome-wide loss-of-heterozygosity studies have identified activating mutations in K-Ras, c-myc, EGFR, cyclin D1, telomerase, Bcl2, and mutations that compromise the function of p53, p16INK/p14ARF, fragile histidine triad (FHIT), RASFF1A, SEMA3B, and Rb-pathway in lung cancer patients. In addition, chromosomal comparative genomic hybridization and high-resolution genomic profiles of lung cancer revealed a high number of recurrent chromosomal aberrations, particularly amplifications and deletions in certain chromosome regions, including 1q36, 3q11.2–12.3, 3q25–27, 5p13–14, etc., where some oncogenes are localized [8]. Furthermore, a various case-control and cohort studies indicated presence of a rare autosomal dominant gene predisposing to lung cancer in members of the family with lung cancer history [9].
2. Epigenetics in cancer
In addition to genetic aberrations, cancer has also been viewed as a disease that is initiated by epigenetic changes in which the gene expression is altered without genomic abnormalities. The most common epigenetic changes occur on the methylation levels of DNA and histone proteins. Many epigenetic studies of lung cancer focused on hypermethylation of DNA in the promoter region of tumor suppressors, such as adenomatous polyposis coli (APC), retinoic acid receptor-2, H-cadherin, FHIT, and RASSF1A [10], whereas investigations on the alterations of histone methylation in lung cancer are just beginning. The status of histone methylation, especially on the N-terminal tails of histones H3 and H4, determines the accessibility of genes in the condensed packed chromatin structure. The lysine residues in histones H3 and H4 can be modified by mono-, di- and tri-methylation. It has been generally viewed that tri-methylation of lysines 4, 36 and 79 of histone H3 is associated with an active transcription of the genes in the de-condensed chromatin (euchromatin) region. In contrast, tri-methylation of lysines 9 and 27 of histone H3 and lysine 20 of histone H4 is refractory for gene expression due to the formation of the highly condensed heterochromatin architecture [11]. A number of histone methyltransferases have been identified, among which human SUV39hl is the best characterized methyltransferase responsible for the di- and tri-methylation of lysine 9 on histone H3.
Methylations on histone proteins were traditionally regarded as permanent marks and irreversible [12]. Since the first discovery of histone lysine specific demethylases-1 (LSD1) in 2004 [13], now we know that histone methylation is a dynamic process. The histone proteins can be methylated under the control of a variety of histone methyltransferases [11]. Meanwhile, these methylations on histone proteins can also be removed by histone demethylases. In 2006, several groups independently identified a JmjC family of histone demethylases that demethylate di- or tri-methylated lysines 9 and 36 on histone H3 [14–19]. Although histone methylation has been considered as a key event for gene transcription, it is plausible to speculate that such modification can also regulate DNA replication, recombination and damage repair. Biochemically, additions of methyl groups to lysines or arginines are unable to change the charges that affect the chromatin structure due to the nature of small molecular weight of methyl groups [20]. It is very likely, thus, methylations on the side chains of lysine or arginine residues create binding code for regulatory proteins. In other word, methylations on histone proteins will influence the accessibility of the chromatin by chromatin binding proteins, transcription factors, DNA repairing complexes, and DNA replication machineries [21].
There is no compelling evidence suggesting that lung cancer or other types of cancer develop by purely aberrations in histone methylation or the signaling pathways that regulate histone methylation. However, perturbations in histone methylation have been observed in some types of human cancer, such as colorectal carcinomas and hepatocellular cancer [22,23]. In experimental animal, mice with gene deficiency of SUV39, a methyltransferase catalyzing di- or tri-methylation of lysine 9 on histone H3, suffer dramatic genome instability and develop B-cell lymphomas due to a substantial loss of lysine 9 methylation on histone H3 [24]. Reduced expression of RIZ1, another methyltransferase for lysine 9 on histone H3, has been frequently observed in lung cancer, breast cancer, hepatocellular carcinoma, colon cancer, neuroblastoma and melanoma [25]. An increased lung cancer risk has been recently reported in people carrying 1624 G > C polymorphism of SUV39h2 [26]. Since this polymorphism occurs in the 3ʹ-UTR region, it is very likely that this polymorphism may reduce the mRNA stability or translational potential of SUV39h2 and therefore, compromises the tri-methylation of lysine 9 on histone H3.
3. Mdig gene and cancer
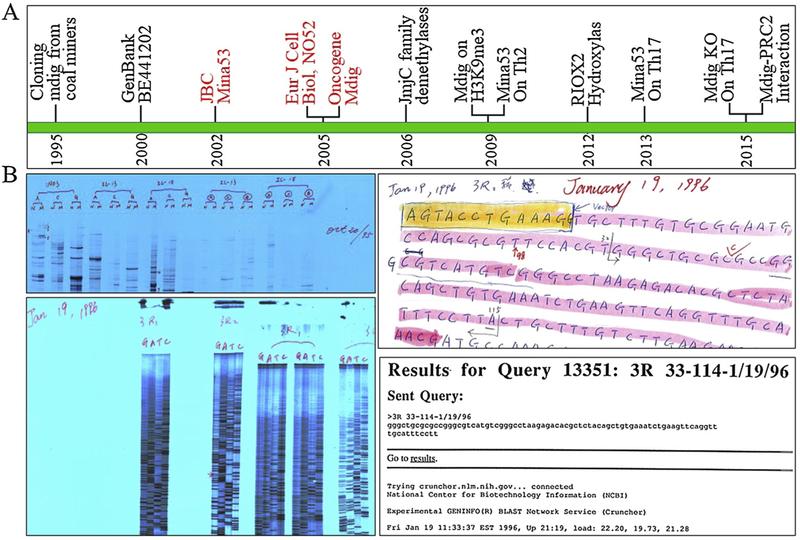
Mdig gene was first identified in later 1990s through differentiation display RT-PCR (DDRT-PCR) using mRNAs from alveolar macrophages of the people who had suffered from chronic lung diseases due to exposure to mineral dust in mining industries (Fig. 1A), but the partial DNA sequence was not deposited into the GenBank until year 2000 (GenBank BE441202) [27]. Similar to the cases of many other genes, the original identification of mdig was unintended. During the course of studying inflammatory cytokines in occupational lung diseases, we applied DDRT-PCR to investigate mRNA expression profiles and failed to achieve meaningful or reproducible results after several attempts. As a last-gasp in experiments, we replaced the random upstream primers of DDRT-PCR with gene specific primers for iNOS, IL-13 and IL-15, and detected several unique cDNA bands in the reactions containing iNOS primer (Fig. 1B, up left). After further cloning and DNA sequencing, we discovered that the last eight nucleotides at the 3′-terminus of the iNOS primer marches the sequence of mdig from 163 to 170 (GenBank AY302110) (Fig. 2). Accordingly, under the less stringent DDRT-PCR condition, iNOS specific primer we used was able to serve as a bait to capture mdig mRNAs in DDRT-PCR.
Fig. 1.
Discover history of mdig. A. Chronicle of the key events in studying mdig/MINA/NO52/RIOX2 from 1995 to 2015; B. Historical images of the first differentiation display RT-PCR (upper left), DNA sequencing (bottom left), Reading recorder of DNA sequencing (upper right), and GenBank query (bottom right) of the mdig gene. In differentiation display RT-PCR panel, N and M depict total RNAs from alveolar macrophages of normal control subjects (N) and coal miners (M), respectively; A, C and G are the down-stream PCR primers, H-T11A, H-T11C and H-T11 G, provided by DDRT-PCR kit (GenHunter Corporation, Nashville, TN, USA); iNOS, IL-13 and IL-15 represent up-stream PCR primers derived from iNOS, IL-13 and IL-15, respectively. The numbers marked on these bands represent cDNA fragments from DDRT-PCR for cloning and DNA sequencing. In DNA sequencing panel, 3R1 and 3R2 indicate using right (upstream) primer for sequencing of the 3rd cDNA fragment from DDRT-PCR, which led to the discovery of mdig.
Fig. 2.
Diagram shows partial base pairing between iNos PCR primer and the mdig mRNA.
In 2002, Tsuneoka et al [28] reported a myc-induced nuclear antigen, mina53 that is 100% homologous to mdig identified from human alveolar macrophages, in human glioblastoma cell line T98 G cells with myc overexpression. In addition to human cells, mdig was also found as a nucleolar protein in Xenopus laevis and named NO52 by Schmidt-Zachmann and colleagues [29]. All of these studies pointed out the presence of a conserved JmjC domain in the encoded protein by mdig. The functional significance of this JmjC domain was unknown until year 2006 when several groups demonstrated a histone demethylase activity of some JmjC family proteins [15,17,30]. For the mdig protein, indeed, a histone demethylase-like activity was detected in the cells subjected to mdig silencing or overexpression [31]. The mdig gene maps on the region of chromosome 3q12.1-q12.2 with a gene length about 28 kb. This gene contains 10 exons and 2 or 3 alternative exons that transcribe mRNA containing a single open reading frame of 465 amino acids with a molecular weight of 52,800.2 kDa and pI 6.23. The gene locus of mdig is located in one of several chromosome instability regions involved in amplifications and deletions in human lung cancer. In A549 cell line that is derived from human lung cancer, the expression of mdig can be enhanced by the treatment of the cells with silica particles. A number of human lung cancer cell lines exhibit a higher basal expression of mdig [32]. Silencing expression by RNA interference using mdig siRNA resulted in a significant inhibition of cell proliferation compared to the cells transfected with control siRNA. Furthermore, mdig silencing sensitized the cytotoxic response to silica particles in A549 cells. However, it was unclear at that time how the gene product of mdig regulated the signals for cell growth or proliferation.
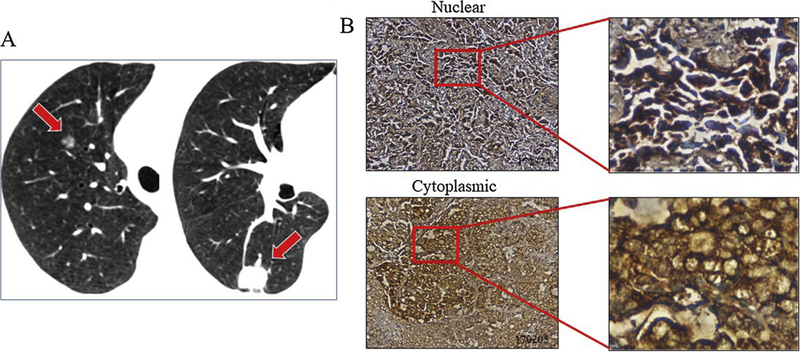
The pro-proliferative function of mdig for the cells implicates a potential oncogenic role of mdig in lung cancer and other cancers. By screening the expression pattern of mdig in more than 150 cases of human lung cancers, we found that more than 90% of human lung cancers showed an increased expression of mdig relative to the case-matched normal lung tissues [31]. Most recently, we also noted a unique pattern of mdig expression in human synchronous multiple primary lung cancer (SMPLC), a T3 or T4 stage lung cancer as classified by the latest, the 7th version of TNM classification. Among 34 cases of SMPLC, 33 cases showed a clear staining of mdig expression in the cancer tissues. In contrast, mdig was not detected in the adjacent normal lung tissues (Fig. 3). Another interesting finding in this SMPLC study is that a considerable number of cancer cells exhibited cytosolic, rather than nuclear localization of mdig protein (Fig. 3). The cytoplasmic localization of mdig was previously noted in the bronchial epithelial cells with stable overexpression of the mdig gene or the cells treated with arsenic [33]. Thus, it is possible that mdig may not only serve as an epigenetic regulator in the nuclei for the configuration of the chromatin, but also regulate the functions of some cytosolic proteins.
Fig. 3.
Higher rate of mdig expression in human synchronous multiple primary lung cancer (SMPLC). A. Computed tomography (CT) scan shows SMPLC in one individual. Red block arrows indicate position of tumors in the lung; B. Nuclear and cytoplasmic localization of mdig protein in cancer cells of the SMPLC samples.
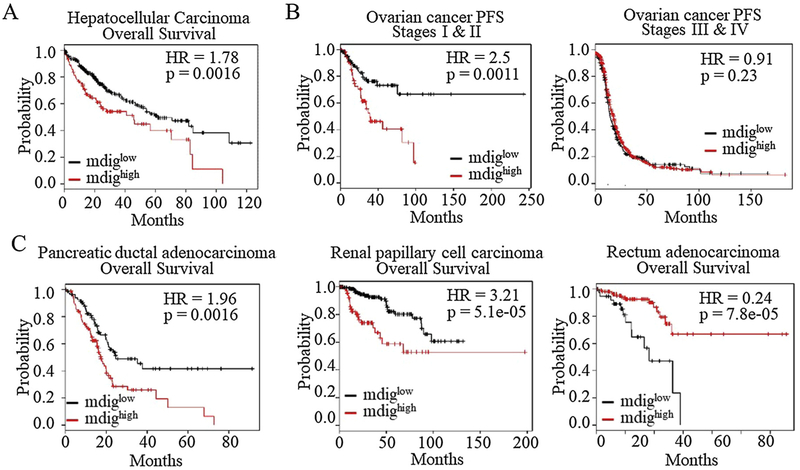
Increased expression of mdig had also been observed in breast cancer [34], colon cancer [35], esophageal squamous cell carcinoma [36], gingival squamous cell carcinoma [37], lymphoma [38], renal cell carcinoma [39], glioblastoma [40], gastric cancer [41], hepatocellular carcinoma [42], cholangiocarcinoma [43], multiple myeloma [44], pancreatic cancer [45], etc. In some of these cancers, the level of mdig is correlated with the overall survival of the patients. In general, higher mdig expression in cancers is associated with a poorer overall survival or poorer progress free survival, such as squamous lung cancer and breast cancer without lymph node metastasis [34,46]. In human hepatocellular carcinoma, higher mdig expression also indicated a poorer overall survival (Fig. 4A). Intriguingly, increased expression of mdig predicted a better survival of the breast cancer patients who had lymph node or distal organ metastasis [34]. A similar phenomenon occurs in ovarian cancer. Higher mdig expression predicts a poorer progress free survival (PFS) when the cancer stages are not specified. However, if the cancers were stratified according to the disease stages, higher mdig expression only predicted poorer survival of the stages I and II cancer patients, but not the stages III and IV patients (Fig. 4B). These data suggested that mdig may play dual roles in some cancers. In earlier stage cancers, mdig is oncogenic for the growth of the tumors. In later stages, in contrast, mdig may be tumor suppressive by limiting the metastatic potentials of the cancer cells.
Fig. 4.
Prognostic value of mdig in human cancer. A. Higher expression of mdig predicts poorer overall survival of hepatocellular carcinoma; B. Higher expression of mdig predicts poorer progress free survival (PFS) of the stages I and II, but not the stages III and IV ovarian cancer. C. Association of higher mdig expression with poorer survival of the patients with pancreatic ductal adenocarcinoma (left) and renal papillary cell carcinoma (middle). Higher mdig level indicated better survival of the patients with rectum adenocarcinoma (right). The patient survival information is derived from Kaplan Meier-plotter (kmplot.com).
The inverse relationship between mdig level and patient survival is also evident for pancreatic cancer, kidney cancer, cervical cancer, head-neck squamous cell carcinoma, sarcoma, thymoma, thyroid carcinoma, and uterine corpus endometrial carcinoma (Fig. 4C and data not shown). For the rectum adenocarcinoma, intriguingly, the higher expression of mdig is associated with a highly significant better survival of the cancer patients (Fig. 4C, right panel), suggesting that mdig may play protective role for the patients or delay the progression of the rectum cancer.
4. Mdig in protein hydroxylation and demethylation
The mdig protein contains a conserved JmjC domain from amino acids 128 to 271. The JmjC domain had been recently identified as a signature motif of the JmjC family histone demethylases [11]. It is very likely, therefore, that mdig may promote cell growth or cell cycle transition by affecting the methylation status of the histone proteins. Based on the similarity of the amino acid sequence of the JmjC domain in mdig and the other known histone demethylases, we originally believed that mdig may be able to regulate the level of histone H3 lysine 9 trimethylation (H3K9me3) [31]. Several lines of evidence supported this assumption. First, an inverse relationship between the levels of mdig and H3K9me3 was clearly observed in human lung cancer samples [31]. Secondly, overexpression or siRNA silencing of mdig in lung epithelial cells reduced or up-regulated the level of H3K9me3. Lastly, demethylase activity, although it was moderate, was detected in a test tube reaction of the immunoprecipitated mdig protein and the H3K9me3 peptide [47]. Possible involvement of mdig in H3K9me3 demethylation was also evidenced in glioblastoma cells and Human hepatocellular carcinoma (HCC), respectively. Silencing mdig by shRNAs not only increased the level of H3K9me3 but also inhibited expression of some cell cycle regulatory proteins, such as cyclin B, cyclin D1, CDK1, CDK2, etc [40]. In human HCC, studies by Huo et al demonstrated that about 70% of human HCC showed an increased expression of mdig. In HCC cell lines, inhibition of mdig by siRNAs elevated the level of H3K9me3 substantially [42].
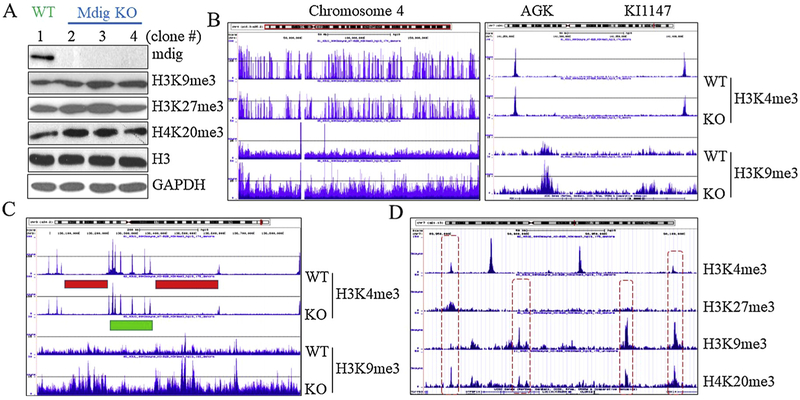
Using CRISPR-Cas9 gene editing approach, our recent data provided direct evidence showing that mdig is highly capable of regulating the level of H3K9me3, H3K27me3 and H4K20me3, the three major repressive histone trimethylation markers, in the human bronchial epithelial cell line, BEAS-2B cells, A549 cells, and triple negative breast cancer cell line, MDA-MB-231 cells. Knockout (KO) of mdig caused a pronounced increase of H3K9me3, H3K27me3 and H4K20me3 (Fig. 5A), along with increased protein levels of EZH2 and RBBP4, two key components of the PRC2 complex that catalyzes H3K27me3 (data not shown). Chromosomal immunoprecipitation-sequencing (ChIP-seq) revealed a substantial enrichment of H3K9me3 in the KO cells (Fig. 5B, the entire chromosome 4 region). Except Y chromosome, gain of H3K9me3 can be visualized in all chromosomes in the mdig KO cells. Distinguishing from the sharp peaks of H3K4me3 that exclusively located at the transcription start sites of genes as demonstrated by ChIP-seq, the enriched peaks of H3K9me3 in the KO cells are mostly in the gene body and intergenic regions (Fig. 5B, right panel, and Fig. 5C). The peaks of H3K27me3 is either co-localized with H3K4me3, or in the gene body and intergenic regions. Despite there are considerable number of genes with an enrichment of both H3K9me3 and H3K27me3 following mdig KO, most peaks of these two histone markers are not overlapped on the individual genes (Fig. 5D). In contrast, many peaks of H4K20me3 and H3K9me3 are overlapped (Fig. 5D). These effects of mdig on the repressive histone trimethylation markers, nevertheless, are not surprising, because we had previously shown a direct protein-protein interaction between mdig and CBX3, CBX5, RBBP4, and RBBP7 [48]. Both CBX3 and CBX5 can recognize and bind to H3K9me3. RBBP4 and RBBP7 are regulatory subunit of the PRC2 complex [48]. Interaction of mdig with RBBP4 and RBBP7 may compromise the methyltransferase activity of the PRC2 complex. Knockout of mdig, accordingly, leads to an enhanced activity of the PRC2, and the subsequent elevation of H3K27me3.
Fig. 5.
Mdig promotes histone demethylation. A. Knockout (KO) of mdig by CRISPR-Cas9 approach increases the levels of H3K9me3, H3K27me3 and H4K20me3 in BEAS-2B cells; B. Knockout of mdig enriches H3K9me3 in the entire chromosome region of chromosome 4 (left panel). Right panel shows a typical different location of H3K4me3 and H3K9me3 in a given gene. C. Enrichment of H3K9me3 in the mdig KO cells mostly occurred in the gene poor region on the chromosome as indicated by red bars. Green bar depict gene rich region as indicated by a cluster of H3K4me3 peaks. D. A representative region of chromosome showing distributions of H3K4me3, H3K27me3, H3K9me3, and H4K20me3. KO: Knockout of mdig by CRISPR-Cas9 gene editing; WT: cell colonies from CRISPR-Cas9-transfected cells without knockout of mdig gene.
In human breast cancer cell line MDA-MB-231 cells, there is a similar level of enrichment of H4K20me3 and H3K9me3 following CRISPR-Cas9-mediated knockout of mdig (data not shown). H4K20me3 is another heterochromatin marker for somatic cells and embryonic stem cells [49]. In human fibroblasts, increasing in H4K20me3 enhances chromatin compaction and induces quiescence of the cells. Reducing the level of H4K20me3 by silencing the methyltransferase Suv4-20h1 (KMT5B) and Suv4-20h2 (KMT5C) that are responsible in catalyzing H4K20me3 from H4K20me1, on the other hand, promoted cell cycle entry into the S-phase [50]. Meanwhile, H4K20me3 is considered as an important epigenetic marker in maintaining the pluripotent status of the embryonic stem cells (ESCs). Loss of H4K20me3 will de-repress the expression of the cell lineage specific genes, leading to the differentiation of the ESCs [51]. An increase of H4K20me3 following the knockout of mdig, thus, may lead to growth inhibition of the somatic cells where mdig expression is inhibited by siRNA silencing or knockout through CRISPR-Cas9 approach. This provides a new explanation for our previous observations showing decreasing of the S-phase cells by mdig silencing [27].
Despite containing the signature motif of the demethylases, the JmjC domain in the mdig protein, and the increased H3K9me3, H3K27me3 and H4K20me3 following mdig silencing in several reports and our recent ChIP-seq experiments following mdig knockout by CRISPR-Cas9 gene editing, questions remain on whether mdig is truly a histone demethylase. Unlike other demethylases, mdig protein does not possess other known chromatin-binding domains, such as PHD, Tudor, F-box, etc. Structural analysis revealed that the C-terminus of the JmjC domain in mdig is linked to helical dimerization and carboxy-terminal “winged helix” domains that are required for the hydroxylase activity towards ribosomal protein L27A but not for the demethylase activity for di- or tri-methylated lysine residues in histone proteins [52]. It is plausible, thus, to speculate that if mdig contributes to demethylation of the histone proteins, it requires interaction with other proteins or cofactors that either modulate the structure of mdig protein or regulate the recruitment/assemble of other histone demethylase or methyltransferase complexes. Indeed, our data from protein pull-down in combination of proteomics analysis had unraveled physical interaction of mdig with several chromatin-binding proteins, DNA repair and replication proteins, and proteins involved in RNA splicing [48].
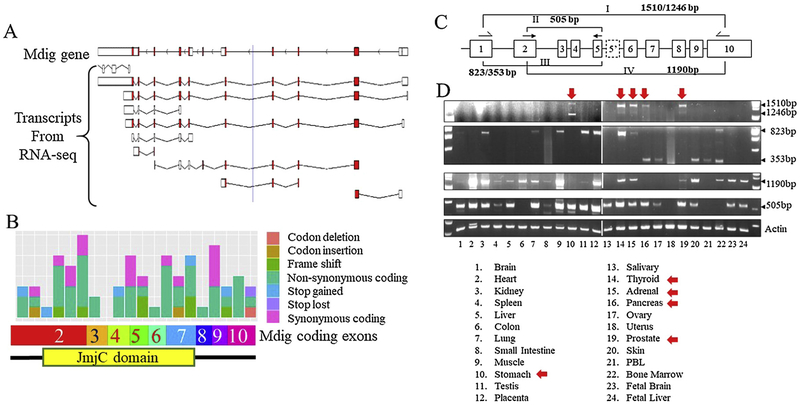
The complexity of mdig in histone demethylation is further intensified by the presence of multiple alternatively spliced mdig transcripts in cancer cells. As earlier as in 2005, we had first discovered an alternatively spliced mdig mRNA in one of the lung cancer cell lines [27]. This alternatively spliced mdig transcript was resulted from splicing skipping of exon 2 and insertion of an alternative exon 5, exon 5′, between exons 5 and 6, which encodes a mdig protein without the JmjC domain. In fact, RNA-seq data had identified at least 10 different alternatively spliced mdig mRNAs (Fig. 6A). Furthermore, higher frequent mutations in the JmjC domain had been identified in a number of human cancers (Fig. 6B). All of these alternative splicing and mutations, without questions, will influence the possible enzymatic activity of mdig on protein hydroxylation or demethylation.
Fig. 6.
Alternative splicing and mutations of mdig genes. A. Multiple transcripts of mdig identified by RNA-seq (grch37.ensembl.org); B. Known mutation sites and mutation types of mdig genes in human cancers (https://cancer.sanger.ac.uk/cosmic/); C. Schematic diagram of the RT-PCR strategy to detect alternatively spliced mdig mRNAs; D. Tissue scanning of mdig mRNA expression by RT-PCR. Red arrows indicate the tissues expressing the full-length mdig mRNA.
The alternative splicing of mdig mRNA is also common in normal human tissues. By using different sets of PCR primers to scan mdig expression among 24 different human tissues (Fig. 6C), we had found that the full length mdig mRNA could be detected in the stomach, thyroid, adrenal, pancreas, and prostate only, but not other normal human tissues (Fig. 6D). The stomach also expressed an alternatively spliced mdig mRNA without the exon 2 region but contained an alternative exon 5′, as what we had previously reported in human lung cancer cell line H441 cells [27]. When the PCR primer set that amplifies the mdig mRNA corresponding to the region from exon 1 to exon 5, the canonical 823bp fragment could be detected in kidney, lung, muscle, testis, placenta, thyroid, adrenal, and bone marrow, whereas the alternatively spliced 353bp mdig mRNA resulting from skipping of exon 2 was observed in pancreas, ovary, skin, PBL, and bone marrow (Fig. 6D). These data are consistent to the RNA-seq findings showing mdig transcripts lacking of the upstream exons, down-stream exons, or both (Fig. 6A).
A mutual regulation between histone lysine methylation and DNA methylation had long been established [53]. Although different methyltransferases and demethylases are involved in DNA and histone methylation, respectively, these two systems are highly interrelated and depended on each other for epigenetic modifications. Emerging evidence suggests that some histone methyltransferases bind to the methylated DNA for the propagation of histone lysine methylation during the formation of heterochromatin. Conversely, some histone methyltransferases responsible for H3K9me3 are able to recruit DNA methyltransferase (DNMT3a/b) to the heterochromatin region for DNA methylation. In addition, there are observations indicating a direct binding of DNMT3a/b to H3K9me3 [53]. In a test tube reaction, we noted demethylation of the methylated DNA fragments following incubation with the immunoprecipitated mdig protein. In cellular experiments using human noncancerous and cancerous breast cell lines, a significant increase in the level of DNA methylation, 5-methyl cytosine, was detected among the cells where mdig was silenced by siRNAs, indicating that mdig might be capable of reducing the level of DNA methylation [54]. In MDA-MB-231 breast cancer cell line, mdig knockout by CRISPR-Cas9 approach caused down-regulation of Tet1 and Tet2, two DNA demethylases, and pronounced increase of DNMT2, an enzyme originally attributed to DNA methylation, but later classified as a tRNA methyltransferase for the methylation of cytosine 38 in the anticodon loop of aspartic acid tRNA [tRNA(Asp)] [55]. Accordingly, these data suggest that mdig can either directly act as a DNA demethylase or indirectly regulate DNA demethylation through Tet family DNA demethylases.
5. Mdig in stem cells
All stem cells, including embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), cell lineage specific stem cells (somatic stem cells), and cancer stem cells (CSCs), have a unique epigenetic program differing from differentiated cells for the maintenance of the stemness [56]. A number of epigenetic writers and erasers play a central role in establishing the stem cell specific epigenetic profiles on the genome. Methyltransferases and acetyltransferases are the writers of epigenetics, whereas demethylases and deacetylases have been viewed as erasers of the epigenetics. The activities of both writers and erasers are regulated by the metabolites from either glycolysis in cytoplasm or the tricarboxylic acid (TCA) cycle in mitochondria. The glycolysis pathway facilitates S-adenosylmethionine (SAM) production through serine/glycine and tetrahydrofolate metabolism to provide methyl donors for DNA and histone methylation. α-ketoglutarate (α-KG), one of the key metabolites of TCA cycle on the other hand, is an essential cofactor for the activity of the JmjC family histone demethylases and Tet family DNA demethylases [57].
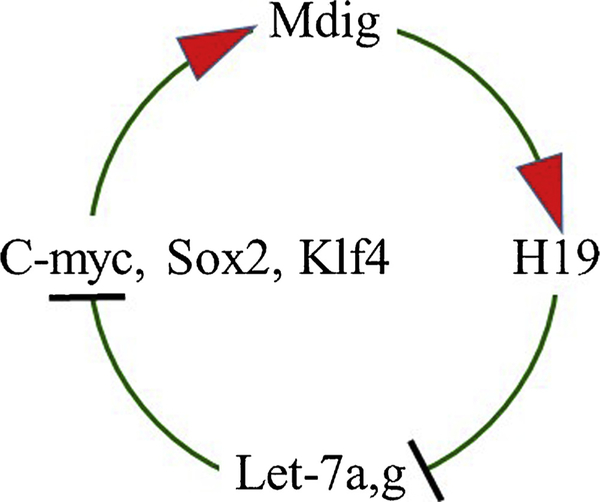
Currently, there is no direct evidence linking mdig to the pluripotency, self-renewal and differentiation of the stem cells. Our gene knockout study in mice failed to produce homozygous, but only the heterozygous mdig knockout mice, indicating pivotal role of mdig in the processes of embryonic development [58]. Mdig was previously identified as a target gene of c-myc [28], one of the four Yamanaka factors that are essential for the stemness of the ESCs, iPSCs, somatic stem cells, and CSCs [59]. In our overexpression and siRNA-silencing experiments, we noted that mdig and c-myc are mutually regulated. Overexpression of mdig caused a substantial increase in c-myc expression in bronchial epithelial cells [47]. In the same study, we also noted that mdig is able to promote expression of H19, a paternally imprinted but maternally expressed oncofetal gene transcribing a large intergenic non-coding RNA (lincRNA), through reducing H3K9me3 in the imprint control region and CTCF binding region [47]. A number of studies implicated important roles of H19 in ESCs, somatic stem cells and CSCs [60]. H19 has also been recognized as an endogenous antagonist of the tumor suppressive miRNAs let-7a and let-7 g. Indeed, overexpression of mdig repressed the levels of both let7a and let-7 g, but not let-7c. Our additional tests further demonstrated that both let-7a and let-7 g are highly capable of inhibiting the expression of c-myc, and to a lesser degree of inhibition on Sox2 and Klf4, two other stem cell transcription factors. These data, therefore, suggest that mdig may participate in the regulatory circuit among H19, let-7 s and c-myc for the stem cells (Fig. 7).
Fig. 7.
Forward feedback circuit of mdig and c-myc. Oncogene c-myc induces mdig transcription. Mdig, in turn, causes de-repression of H19 that can antagonize Let-7a and Let-7 g. Both Let-7a and Let-7 g are tumor suppressive miRNAs that target c-myc, Sox2 and Klf4.
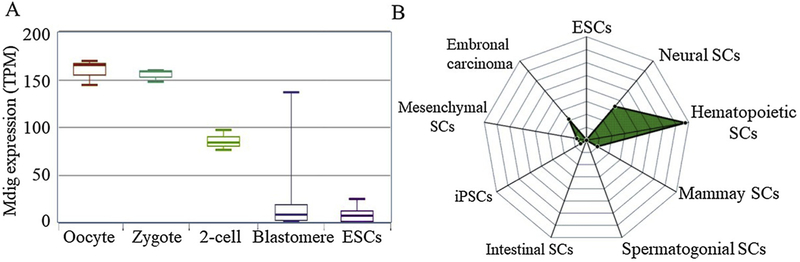
Based on the available datasets of single-cell RNA-seq from 174 cell groups, it indicates that mdig is highly expressed in human oocyte, zygote and preimplantation blastomere. However, mdig expression appears to be lost in human ESCs (Fig. 8A). The levels of mdig are also much lower in several established human iPSCs. Thus, loss of mdig may be important for the self-renewal and pluripotency of the ESCs or iPSCs. Expression of mdig, in turn, may induce differentiation of the ESCs or iPSCs. If this is true, the question is how mdig affects the stemness of these cells. The answer may be from the observed effect of mdig on these heterochromatin markers, including H3K9me3, H3K27me3 and H4k20me3. In ESCs or iPSCs, the expression of the differentiation genes must be repressed by H3K9me3, H3K27me3 and H4K20me3 through the formation of heterochromatin [51]. Expression of mdig reduces the levels of these repressive chromatin markers, leading to de-repression of the differentiation genes and the differentiation of the ESCs or iPSCs.
Fig. 8.
Possible involvement of mdig in regulating the stemness of stem cells. A. Expression levels of mdig in oocyte, zygote, 2-cell blastomere, early blastomere, and ESCs. Data are derived from scRNASeqDB (https://bioinfo.uth.edu); B. RNA-seq data indicated that knockout of mdig through CRISPR-Cas9 approach resulted in down-regulation of the stemness genes featured in hematopoietic stem cells and neural stem cells as analyzed by StemChecker (stemchecker.sysbiolab.eu).
In histone methylation profiling of the bronchial epithelial cell line BEAS-2B and breast cancer cell line MDA-MB-231 following mdig knockout through CRISPR-Cas9 gene editing, we indeed noted an up-regulation of H3K9me3, H3K27me3 and H4K20me3 (Fig. 5). Therefore, if the ESCs or iPCs are subjected to an enforced expression of mdig, differentiation will occur in these stem cells. However, above speculation may be not suitable for somatic stem cells, progenitor cells or cancer stem cells (CSCs) due to the required partial differentiation for cell lineage commitment. Unlike the ESCs or iPSCs that are pluripotent, somatic stem cells, progenitor cells and CSCs are multipotent or oligopotent. Some genes associated with cell or tissue lineage development are primed for expression or partially expressed, which requires removal of the repressive histone methylation markers, including H3K9me3, H3K27me3 and H4K20me3. Because of the demethylase-like activity of mdig, it is plausible to hypothesize that mdig is required for functional specification and lineage commitment of the somatic stem cells or progenitor cells. This hypothesis may be also true for the CSCs. Hierarchically, CSCs are roughly equal to the somatic stem cells that are multipotent or oligopotent, but not pluripotent. This is the reason why CSCs of breast cancer can only form breast cancer cells with different functional characteristics, and CSCs of glioblastoma can only generate glioblastoma cells with diverse cell surface markers or phenotypes. Although it is unknown in other types of cancers, our recent data from human breast cancer indicated that mdig is important for the tumor initiation or tumor growth of the earlier stage breast cancers. In later stages of breast cancer, in contrast, mdig expression might be beneficial for the inhibition of migration, invasion and metastasis of the cancer cells [54]. CSCs have been viewed as tumor-initiating cells that are capable of self-renewal and giving rise to more differentiated progenies of tumor cells. Certainly, expression of mdig, therefore, is critical for the stemness maintenance of the CSCs.
An additional indication supporting the contribution of mdig to the stem cells is from our ongoing knockout study of mdig through CRISPR-Cas9 approach and RNA-seq. In human bronchial epithelial cell line BEAS-2B, mdig knockout resulted in the down regulation of 883 genes. Among the most significantly down-regulated 63 genes, 41 genes had been previously reported as stem cell-related genes. Gene signature analysis indicated that these down-regulated stem cell-related genes are mostly associated with the hematopoietic stem cells, neural stem cells, or mesenchymal stem cells (Fig. 8B), but not the ESCs or iPSCs, further suggesting a possible requirement of mdig in somatic stem cells or CSCs.
6. Summary
It is more than 20 years since the first discovery of mdig from the coal miners’ alveolar macrophages. Considerable progress had been made in the past years in our understanding on how mdig is associated with human cancers, esp. the human lung cancer and breast cancer. Debates continue, however, on whether mdig is a protein hydroxylase or histone demethylase, or both. The current information on the structure and function of mdig is exclusively derived from studying the full-length mdig protein. The biological function and mechanism of action of the alternatively spliced and mutated mdig was uncharted at the present. Unequivocal structural evidence suggests that mdig is a ribosomal protein hydroxylase. Yet how this hydroxylation of ribosomal protein impacts the protein translational machinery and whether there are other proteins can be hydroxylated by mdig remain to be further investigated. Studying the potential role of mdig in stem cells and CSCs is just beginning. Based on the diminished expression of mdig in ESCs and iPSCs, mdig may be not required for the self-renewal of the stem cells, but rather act at the boundary of stemness and differentiation. This effect of mdig should be necessary not only for the initial phase of differentiation but also for the expansion of cell populations required for tissue regeneration or tumorigenesis, if the stem cells are CSCs. Certainly, extensive studies on the characteristic of mdig are much needed for our better understanding on the role of mdig in normal development and cancers.
Acknowledgements
The research work in Chen’s laboratory was/is supported by NIH R01 ES017217, ES020137, ES028263, and P30 ES020957 to FC. JS was supported by Natural Science Foundation of Liaoning Province 20,170,541,015.
References
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2018, CA Cancer J. Clin 68 (1) (2018) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].Dela Cruz CS, Tanoue LT, Matthay RA, Lung cancer: epidemiology, etiology, and prevention, Clin. Chest Med 32 (4) (2011) 605–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gallagher CM, Goovaerts P, Jacquez GM, Hao Y, Jemal A, Meliker JR, Racial disparities in lung cancer mortality in U.S. Congressional districts, 1990–2001, Spat. Spatiotemporal Epidemiol 1 (1) (2009) 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brown T, Silica exposure, smoking, silicosis and lung cancer–complex interactions, Occup. Med. (Lond) 59 (2) (2009) 89–95. [DOI] [PubMed] [Google Scholar]
- [5].Tango T, Effect of air pollution on lung cancer: a Poisson regression model based on vital statistics, Environ. Health Perspect 102 (Suppl. 8) (1994) 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Raaschou-Nielsen O, Andersen ZJ, Beelen R, Samoli E, Stafoggia M, Weinmayr G, Hoffmann B, Fischer P, Nieuwenhuijsen MJ, Brunekreef B, Xun WW, Katsouyanni K, Dimakopoulou K, Sommar J, Forsberg B, Modig L, Oudin A, Oftedal B, Schwarze PE, Nafstad P, De Faire U, Pedersen NL, Ostenson CG, Fratiglioni L, Penell J, Korek M, Pershagen G, Eriksen KT, Sorensen M, Tjonneland A, Ellermann T, Eeftens M, Peeters PH, Meliefste K, Wang M, Bueno-de-Mesquita B, Key TJ, de Hoogh K, Concin H, Nagel G, Vilier A, Grioni S, Krogh V, Tsai MY, Ricceri F, Sacerdote C, Galassi C, Migliore E, Ranzi A, Cesaroni G, Badaloni C, Forastiere F, Tamayo I, Amiano P, Dorronsoro M, Trichopoulou A, Bamia C, Vineis P, Hoek G, Air pollution and lung cancer incidence in 17 European cohorts: prospective analyses from the European Study of Cohorts for Air Pollution Effects (ESCAPE), Lancet Oncol 14 (9) (2013) 813–822. [DOI] [PubMed] [Google Scholar]
- [7].Kanwal M, Ding XJ, Cao Y, Familial risk for lung cancer, Oncol. Lett 13 (2) (2017) 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, Martin ES, Yang Z, Ji H, Chin L, Depinho RA, High-resolution genomic profiles of human lung cancer, Proc Natl Acad Sci U S A 102 (27) (2005) 9625–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schwartz AG, Ruckdeschel JC, Familial lung cancer: genetic susceptibility and relationship to chronic obstructive pulmonary disease, Am. J. Respir. Crit. Care Med 173 (1) (2006) 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Langevin SM, Kratzke RA, Kelsey KT, Epigenetics of lung cancer, Transl. Res 165 (1) (2015) 74–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Greer EL, Shi Y, Histone methylation: a dynamic mark in health, disease and inheritance, Nat. Rev. Genet 13 (5) (2012) 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Byvoet P, Shepherd GR, Hardin JM, Noland BJ, The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells, Arch. Biochem. Biophys 148 (2) (1972) 558–567. [DOI] [PubMed] [Google Scholar]
- [13].Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y, Histone demethylation mediated by the nuclear amine oxidase homolog LSD1, Cell 119 (7) (2004) 941–953. [DOI] [PubMed] [Google Scholar]
- [14].Klose RJ, Kallin EM, Zhang Y, JmjC-domain-containing proteins and histone demethylation, Nat. Rev. Genet 7 (9) (2006) 715–727. [DOI] [PubMed] [Google Scholar]
- [15].Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, Hagman J, Hansen K, Shi Y, Zhang G, Structural insights into histone demethylation by JMJD2 family members, Cell 125 (4) (2006) 691–702. [DOI] [PubMed] [Google Scholar]
- [16].Schneider J, Shilatifard A, Histone demethylation by hydroxylation: chemistry in action, ACS Chem. Biol 1 (2) (2006) 75–81. [DOI] [PubMed] [Google Scholar]
- [17].Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y, Histone demethylation by a family of JmjC domain-containing proteins, Nature 439 (7078) (2006) 811–816. [DOI] [PubMed] [Google Scholar]
- [18].Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y, Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases, Cell 125 (3) (2006) 467–481. [DOI] [PubMed] [Google Scholar]
- [19].Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y, JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor, Cell 125 (3) (2006) 483–495. [DOI] [PubMed] [Google Scholar]
- [20].Bannister AJ, Kouzarides T, Regulation of chromatin by histone modifications, Cell Res 21 (3) (2011) 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hunt CR, Ramnarain D, Horikoshi N, Iyengar P, Pandita RK, Shay JW, Pandita TK, Histone modifications and DNA double-strand break repair after exposure to ionizing radiations, Radiat. Res 179 (4) (2013) 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vaiopoulos AG, Athanasoula K, Papavassiliou AG, Epigenetic modifications in colorectal cancer: molecular insights and therapeutic challenges, Biochim. Biophys. Acta 1842 (7) (2014) 971–980. [DOI] [PubMed] [Google Scholar]
- [23].Nishida N, Kudo M, Clinical significance of epigenetic alterations in human hepatocellular carcinoma and its association with genetic mutations, Dig. Dis 34 (6) (2016) 708–713. [DOI] [PubMed] [Google Scholar]
- [24].Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T, Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability, Cell 107 (3) (2001) 323–337. [DOI] [PubMed] [Google Scholar]
- [25].Steele-Perkins G, Fang W, Yang XH, Van Gele M, Carling T, Gu J, Buyse IM, Fletcher JA, Liu J, Bronson R, Chadwick RB, de la Chapelle A, Zhang X, Speleman F, Huang S, Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily, Genes Dev 15 (17) (2001) 2250–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yoon KA, Hwangbo B, Kim IJ, Park S, Kim HS, Kee HJ, Lee JE, Jang YK, Park JG, Lee JS, Novel polymorphisms in the SUV39H2 histone methyltransferase and the risk of lung cancer, Carcinogenesis 27 (11) (2006) 2217–2222. [DOI] [PubMed] [Google Scholar]
- [27].Zhang Y, Lu Y, Yuan BZ, Castranova V, Shi X, Stauffer JL, Demers LM, Chen F, The Human mineral dust-induced gene, mdig, is a cell growth regulating gene associated with lung cancer, Oncogene 24 (31) (2005) 4873–4882. [DOI] [PubMed] [Google Scholar]
- [28].Tsuneoka M, Koda Y, Soejima M, Teye K, Kimura H, A novel myc target gene, mina53, that is involved in cell proliferation, J. Biol. Chem 277 (38) (2002) 35450–35459. [DOI] [PubMed] [Google Scholar]
- [29].Eilbracht J, Kneissel S, Hofmann A, Schmidt-Zachmann MS, Protein NO52–a constitutive nucleolar component sharing high sequence homologies to protein NO66, Eur. J. Cell Biol 84 (2–3) (2005) 279–294. [DOI] [PubMed] [Google Scholar]
- [30].Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM, Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A, Science 312 (5774) (2006) 748–751. [DOI] [PubMed] [Google Scholar]
- [31].Lu Y, Chang Q, Zhang Y, Beezhold K, Rojanasakul Y, Zhao H, Castranova V, Shi X, Chen F, Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3, Cell Cycle 8 (13) (2009) 2101–2109. [DOI] [PubMed] [Google Scholar]
- [32].Thakur C, Chen F, Current understanding of mdig/MINA in human cancers, Genes Cancer 6 (7–8) (2015) 288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yu M, Sun J, Thakur C, Chen B, Lu Y, Zhao H, Chen F, Paradoxical roles of mineral dust induced gene on cell proliferation and migration/invasion, PLoS One 9 (2) (2014) e87998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Thakur C, Lu Y, Sun J, Yu M, Chen B, Chen F, Increased expression of mdig predicts poorer survival of the breast cancer patients, Gene 535 (2) (2014) 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Teye K, Tsuneoka M, Arima N, Koda Y, Nakamura Y, Ueta Y, Shirouzu K, Kimura H, Increased expression of a Myc target gene Mina53 in human colon cancer, Am. J. Pathol 164 (1) (2004) 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tsuneoka M, Fujita H, Arima N, Teye K, Okamura T, Inutsuka H, Koda Y, Shirouzu K, Kimura H, Mina53 as a potential prognostic factor for esophageal squamous cell carcinoma, Clin. Cancer Res 10 (21) (2004) 7347–7356. [DOI] [PubMed] [Google Scholar]
- [37].Kuratomi K, Yano H, Tsuneoka M, Sakamoto K, Kusukawa J, Kojiro M, Immunohistochemical expression of Mina53 and Ki67 proteins in human primary gingival squamous cell carcinoma, Kurume Med. J 53 (3–4) (2006) 71–78. [DOI] [PubMed] [Google Scholar]
- [38].Teye K, Arima N, Nakamura Y, Sakamoto K, Sueoka E, Kimura H, Tsuneoka M, Expression of Myc target gene mina53 in subtypes of human lymphoma, Oncol. Rep 18 (4) (2007) 841–848. [PubMed] [Google Scholar]
- [39].Ishizaki H, Yano H, Tsuneoka M, Ogasawara S, Akiba J, Nishida N, Kojiro S, Fukahori S, Moriya F, Matsuoka K, Kojiro M, Overexpression of the myc target gene Mina53 in advanced renal cell carcinoma, Pathol. Int 57 (10) (2007) 672–680. [DOI] [PubMed] [Google Scholar]
- [40].Huang MY, Xuan F, Liu W, Cui HJ, MINA controls proliferation and tumorigenesis of glioblastoma by epigenetically regulating cyclins and CDKs via H3K9me3 demethylation, Oncogene 36 (3) (2017) 387–396. [DOI] [PubMed] [Google Scholar]
- [41].Zhang Q, Hu CM, Yuan YS, He CH, Zhao Q, Liu NZ, Expression of Mina53 and its significance in gastric carcinoma, Int. J. Biol. Markers 23 (2) (2008) 83–88. [DOI] [PubMed] [Google Scholar]
- [42].Huo Q, Ge C, Tian H, Sun J, Cui M, Li H, Zhao F, Chen T, Xie H, Cui Y, Yao M, Li J, Dysfunction of IKZF1/MYC/MDIG axis contributes to liver cancer progression through regulating H3K9me3/p21 activity, Cell Death Dis 8 (5) (2017) e2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tan XP, Zhang Q, Dong WG, Lei XW, Yang ZR, Upregulated expression of Mina53 in cholangiocarcinoma and its clinical significance, Oncol. Lett 3 (5) (2012) 1037–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wu K, Li L, Thakur C, Lu Y, Zhang X, Yi Z, Chen F, Proteomic Characterization of the World Trade Center dust-activated mdig and c-myc signaling circuit linked to multiple myeloma, Sci. Rep 6 (2016) 36305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kumar SA, Thakur C, Li L, Cui H, Chen F, Pathological and prognostic role of mdig in pancreatic cancer, Genes Cancer 8 (7–8) (2017) 650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sun J, Yu M, Lu Y, Thakur C, Chen B, Qiu P, Zhao H, Chen F, Carcinogenic metalloid arsenic induces expression of mdig oncogene through JNK and STAT3 activation, Cancer Lett 346 (2) (2014) 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen B, Yu M, Chang Q, Lu Y, Thakur C, Ma D, Yi Z, Chen F, Mdig de-represses H19 large intergenic non-coding RNA (lincRNA) by down-regulating H3K9me3 and heterochromatin, Oncotarget 4 (9) (2013) 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang W, Lu Y, Stemmer PM, Zhang X, Bi Y, Yi Z, Chen F, The proteomic investigation reveals interaction of mdig protein with the machinery of DNA double-strand break repair, Oncotarget 6 (29) (2015) 28269–28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pannetier M, Julien E, Schotta G, Tardat M, Sardet C, Jenuwein T, Feil R, PR-SET7 and SUV4-20H regulate H4 lysine-20 methylation at imprinting control regions in the mouse, EMBO Rep 9 (10) (2008) 998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, Coller HA, H4K20 methylation regulates quiescence and chromatin compaction, Mol. Biol. Cell 24 (19) (2013) 3025–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kidder BL, Hu G, Cui K, Zhao K, SMYD5 regulates H4K20me3-marked heterochromatin to safeguard ES cell self-renewal and prevent spurious differentiation, Epigenetics Chromatin 10 (2017) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chowdhury R, Sekirnik R, Brissett NC, Krojer T, Ho CH, Ng SS, Clifton IJ, Ge W, Kershaw NJ, Fox GC, Muniz JRC, Vollmar M, Phillips C, Pilka ES, Kavanagh KL, von Delft F, Oppermann U, McDonough MA, Doherty AJ, Schofield CJ, Ribosomal oxygenases are structurally conserved from prokaryotes to humans, Nature 510 (7505) (2014) 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rose NR, Klose RJ, Understanding the relationship between DNA methylation and histone lysine methylation, Biochim. Biophys. Acta 1839 (12) (2014) 1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Thakur C, Chen B, Li L, Zhang Q, Yang Z-Q, Chen F, Loss of mdig expression enhances DNA and histone methylation and metastasis of the aggressive breast cancer, Signal Transduct. Target. Ther 3 (2018) 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH, Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2, Science 311 (5759) (2006) 395–398. [DOI] [PubMed] [Google Scholar]
- [56].Wainwright EN, Scaffidi P, Epigenetics and Cancer Stem Cells: Unleashing, Hijacking, and Restricting Cellular Plasticity, Trends Cancer 3 (5) (2017) 372–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Reid MA, Dai Z, Locasale JW, The impact of cellular metabolism on chromatin dynamics and epigenetics, Nat. Cell Biol 19 (11) (2017) 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Thakur C, Wolfarth M, Sun J, Zhang Y, Lu Y, Battelli L, Porter DW, Chen F, Oncoprotein mdig contributes to silica-induced pulmonary fibrosis by altering balance between Th17 and Treg T cells, Oncotarget 6 (6) (2015) 3722–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liu X, Huang J, Chen T, Wang Y, Xin S, Li J, Pei G, Kang J, Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells, Cell Res 18 (12) (2008) 1177–1189. [DOI] [PubMed] [Google Scholar]
- [60].Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, Kang ZJ, Fan WJ, Zhu LL, Li M, Cui B, Zheng FM, Wang HJ, Lam EW, Wang B, Xu J, Liu Q, H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance, Cell Death Dis 8 (1) (2017) e2569. [DOI] [PMC free article] [PubMed] [Google Scholar]