Abstract
Introduction
Primary sarcomatoid carcinoma of the lung (PSC) is a rare subtype of non-small cell lung cancer, which has a bad prognosis and lacks biomarkers for its diagnosis and prognosis. Recent studies suggested that KDM6B (lysine demethylase 6B), also known as Jumonji domain-containing protein D3 (JMJD3), plays an oncogenic role in various human cancers. However, abnormalities of JMJD3 in sarcomatoid carcinoma of the lung and its clinical prognostic significance have not been determined. Therefore, the present study aimed to ascertain the relationship between JMJD3 and PSC.
Materials and methods
In this study, immunohistochemistry (IHC) was performed to examine the expression of JMJD3 in a tissue microarray (TMA) containing 96 cases of PSC.
Result
Overexpression of JMJD3 was observed in nuclei of the PSC cells. Further analyses indicated that the overexpression of JMJD3 was significantly associated with tumor size, pN stage, and clinical stage. By univariate survival analysis, positive expression of JMJD3 was significantly correlated with shortened patient survival. More importantly, multivariate analysis identified JMJD3 as an independent prognostic factor for sarcomatoid carcinoma of the lung.
Conclusion
These findings provide evidence that JMJD3 protein levels, as examined by IHC, may act as a novel prognostic biomarker for patients with primary sarcomatoid carcinoma of the lung.
Keywords: primary sarcomatoid carcinoma of the lung, JMJD3, IHC, prognosis
Introduction
Primary sarcomatoid carcinoma of the lung (PSC) is an unusual subtype of non-small cell lung cancer, representing only 0.1% to 0.4% of lung cancer.1 In accordance with the latest World Health Organization classification of lung cancer, PSC is a rare subtype of non-small cell lung cancer with poor histological differentiation. It contains sarcoma or sarcoma-like components, with spindle cell carcinoma or giant cell carcinoma, and pleomorphic carcinoma.2 Its outcome is significantly worse than other subtypes of non-small cell lung cancer.3 Its rarity means that there has been little research on sarcomatoid carcinoma. It is imperative to find diagnostic or prognostic biomarkers and to identify potential therapeutic targets for PSC.
Histone covalent modifications consist of acetylation, phosphorylation, methylation, and ubiquitination.4,5 The most common modifications are acetylation and methylation, which play vital roles in differentiation and disease.6 These modified histones can directly or indirectly recruit transcription factors to specific chromatin regions, thus leading to a change in the chromatin structure and function, which further lead to gene expression or repression. Histone methylation and demethylation are catalyzed by special enzymes, such as peptidyl arginine deiminase 4 (PADI4), lysine-specific demethylase 1 (LSD1), and Jumonji C (JmjC) domain.7–9 Jumonji domain-containing protein D3 (JMJD3), also known as lysine-specific demethylase 6B (KDM6B), is a member of JmjC family. JMJD3 and UTX (ubiquitously transcribed X-chromosome tetratricopeptide repeat protein) demethylate H3K27.10 JMJD3 and UTX specifically decrease H3K27 dimethylation (H3K27me2) and H3K27 trimethylation (H3K27me3): H3K27m3 is associated with an inactive gene while H3K27m is associated with an active gene.11,12 JMJD3 plays an important role in differentiation, inflammation, and cancer: overexpression of JMJD3 was found in liver cancer, multiple myeloma, and colorectal cancer.13–15 These findings suggested that JMJD3 may be a new biomarker for cancer prognosis or could serve as a therapeutic target. However, there has been no study of the association of JMJD3 with lung cancer, especially PSC. In the present study, we investigated the expression of JMJD3 in PSC and the relationship between JMJD3 and the prognosis of PSC.
Methods
Ethics Statement
This research was authorized by the ethical committee of the Sun Yat-sen University Cancer Center. Informed consent (written or verbal) was waived by the ethics committee. All the specimens analyzed were anonymized.
Patients And Tissue Specimens
In our research, a total of 96 patients diagnosed with primary sarcomatoid carcinomas at our institution between February 2000 and December 2015 were enrolled in this study. Patients were included if they underwent surgery and had pathologically confirmed primary sarcomatoid carcinomas. The diagnosis was determined according to the morphology of HE staining and subsequent biomarkers IHC analysis. The diagnosis of PSC was further reviewed by two independent pathologists with consistency according to the 2015 WHO classification of lung tumor,2 and the differential diagnosis from sarcomatoid mesothelioma was mainly made by the biomarkers of CK5/6, D2-40, calretinin, and WT1 based on the WHO guideline.16 Patients were excluded if (1) they were diagnosed with primary sarcomatoid carcinomas with other malignancies; (2) they received radiation or chemotherapy before surgery; (3) the clinical and pathological characteristics of the patients could not be assessed. The data were obtained from the medical records, pathology reports, and hospital follow-up records. Patients without full follow-up information or who died for other reasons were excluded. The TNM stage of PSC was based on the 7th edition of the American Joint Committee on Cancer (AJCC) staging system. The study included 83 men and 13 women. Among the patients, 56% were diagnosed at stage III or IV, and the others were at early stages I and II. All the clinicopathological parameters are shown in Table 1. The tissue microarray (TAM) was built according to a previously described method.17
Table 1.
Correlation Between The Clinicopathologic Variables And JMJD3 In LSC
| Variable | JMJD3 Expression | |||
|---|---|---|---|---|
| All Cases | Low | High | P valuea | |
| Age (years)b | 0.486 | |||
| ≤ 57 | 45 | 26 (57.8%) | 19 (42.2%) | |
| > 57 | 51 | 33 (64.7%) | 18 (35.3%) | |
| Gender | 0.218 | |||
| Male | 83 | 49 (59.0%) | 34 (41.0%) | |
| Female | 13 | 10 (76.9%) | 3 (23.1%) | |
| Tumor sizec | 0.022 | |||
| ≤ 4.5 | 58 | 41 (70.7%) | 17 (29.3%) | |
| > 4.5 | 38 | 18 (47.4%) | 20 (52.6%) | |
| Smoking | 0.716 | |||
| Yes | 72 | 45 (62.5%) | 27 (37.5%) | |
| No | 24 | 14 (58.3%) | 10 (41.7%) | |
| pT stage | 0.937 | |||
| T1+T2 | 54 | 33 (61.1%) | 21 (38.9%) | |
| T3+T4 | 42 | 26 (61.9%) | 16 (38.1%) | |
| pN stage | 0.006 | |||
| N0 | 48 | 36 (75.0%) | 12 (25.0%) | |
| N1+N2+N3 | 48 | 23 (47.9%) | 25 (52.1%) | |
| pM stage | 0.146 | |||
| M0 | 88 | 56 (63.6%) | 32 (36.4%) | |
| M1 | 8 | 3 (37.5%) | 5 (62.5%) | |
| Clinical stage | 0.009 | |||
| I-II | 42 | 32 (76.2%) | 10 (23.8%) | |
| III-IV | 54 | 27 (50.0%) | 27 (50.0%) | |
| Therapyd | 0.250 | |||
| Therapy1 | 73 | 48 (65.8%) | 25 (34.2%) | |
| Therapy2 | 18 | 8 (44.4%) | 10 (55.6%) | |
| Therapy3 | 5 | 3 (60.0%) | 2 (40.0%) | |
| Relapse | 0.270 | |||
| Yes | 30 | 16 (53.3%) | 14 (46.7%) | |
| No | 66 | 43 (65.2%) | 23 (34.8%) | |
Notes: aChi-square test, bMean age, cMean tumor size; dTherapy: Therapy1 is lobectomy/bilobectomy and lymphadenectomy, Therapy2 is lobectomy/bilobectomy; Therapy3 is lobectomy/bilobectomy and radiotherapy and chemoradiotherapy.
Immunohistochemistry (IHC)
We performed IHC for JMJD3 in the PSC samples using the standard EnVision procedure.17 The TMA was deparaffinized in xylene and then rehydrated in a graded series of alcohol. The slides were then immersed in 3% hydrogen peroxide for 15 mins to block endogenous peroxidase activity, and then pressure cooked in EDTA buffer (pH = 8.0) for 2.5 mins to retrieve the antigen. Subsequently, the slides were incubated in 3.5% bovine serum albumin (BSA) for 10 mins to block nonspecific binding. Subsequently, the slides were incubated with rabbit anti-JMJD3 polyclonal antibodies (Abcam38113, 1:300 dilution) overnight at 4°C in a moist chamber. A secondary antibody (Envision, Dako, K5007) was then incubated with the slides for 30 mins at 37°C, and 3,3-diaminobenzidine (DAB) was used as the chromogenic substrate. Nuclear counterstaining was performed with Mayer’s hematoxylin. Finally, the sections were mounted with a cover glass and air-dried in a fumigation hood. Normal rabbit IgG was used as the negative control.
IHC Evaluation
The protein expression of JMJD3 in PSC was evaluated under a microscope by two independent pathologists who were blinded to the clinicopathological parameters. If the two evaluations of one sample were different, a third pathologist would participate to determine the final evaluation. The detailed evaluation was operated according to a previously described method.18 JMJD3 was mainly observed in the nucleus, and the positively stained cells were brown or yellow. The stain intensity was described as 0, negative; 1, weak; 2, moderate; and 3, strong. The percentage of positively stained cells was evaluated in 5% increments from 0% to 100%. The final score was obtained by evaluating the percent of positive cells and the positive cell intensity and then multiplying them together (score of 0–300).
Statistical Analysis
We carried out the statistical analysis using SPSS software (version 21.0, SPSS, Chicago, IL, USA). A two-tailed p value of <0.05 was considered to statistically significant in all cases. The cut-off point of JMJD3 expression in PSC was determined using receiver operating characteristic (ROC) analysis. The relationship between JMJD3 and patients with PSC clinicopathological parameters was assessed using the Chi-squared test. Univariate analysis was performed using the Kaplan–Meier method. Cox-proportional hazards regression analysis was employed to identify the independent prognostic factors. All data in this research were uploaded to Research Data Deposit public platform (www.researchdata.org.cn) of Sun Yat-sen University Cancer Center for future reference (No. RDDB2019000670).
Results
JMJD3 Expression In Sarcomatoid Carcinoma Of The Lung
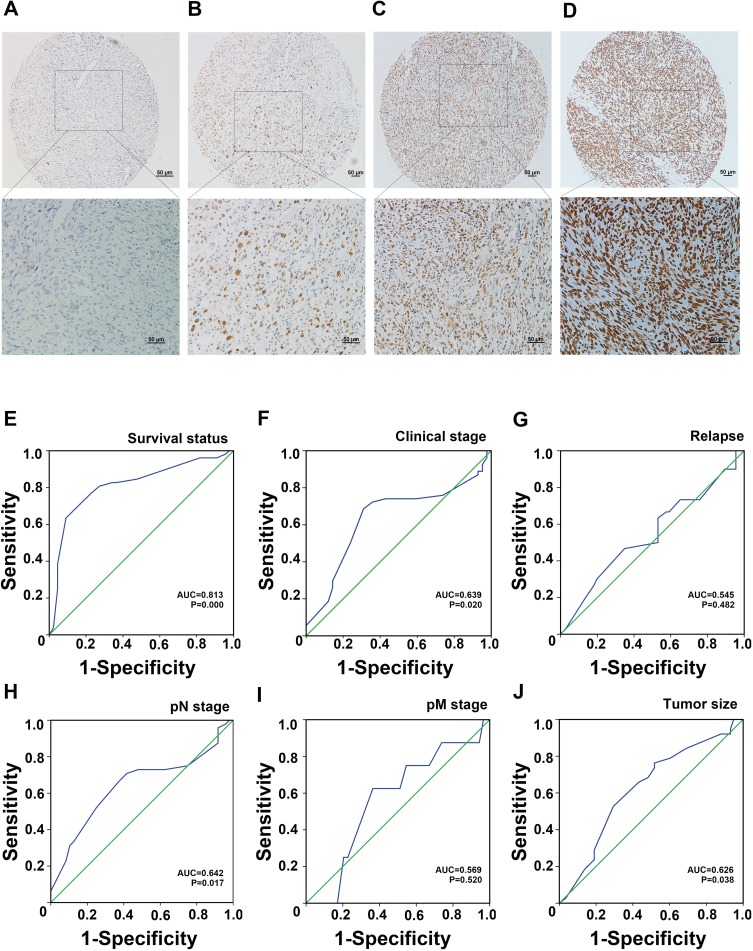
According to the IHC results, JMJD3 was primarily located in the nuclei in PSC. The detailed expression is shown in Figure 1A–D.
Figure 1.
(A–D) Expression of JMJD3 in PSC. (A) Negative expression; (B) Low expression; (C) Moderate expression; (D) Strong expression. (E–J) ROC curve analysis for JMJD3 expression in NPC. (E) Survival status; (F) Clinical stage; (G) Relapse; (H) pN stage; (I) pM stage; (J) Tumor size.
Cut-Off Point For JMJD3 Expression
We carried out ROC curve analysis to determine the cut-off point for JMJD3. The value with the highest value of specificity plus sensitivity was selected as the cut-off value.19 Therefore, we chose the survival status as a state variable. The result of ROC curve analysis showed that the cut-off point for JMJD3 status in PSC was 190. The details of the ROC curve analysis are shown in Figure 1E–J.
Association Of JMJD3 With PSC Clinicopathological Features
Chi-square analysis of the IHC results showed that JMJD3 correlated with tumor size (P = 0.022), pN stage (P = 0.006), and clinical stage (P = 0.009); however, there was no significant correlation between JMJD3 and the other clinicopathological features, such as patient age, gender, smoking, tumor stage, pM stage, and relapse. The details are shown in Table 1.
Relationship Between The Clinicopathological Characteristics, JMJD3 Expression, And Patient Survival
To identify the prognostic utility of JMJD3 in PSC, after median follow-up time of 17.5 months and 52 deaths, and we analyzed the effect of the clinicopathological characteristics on the survival of PSC. The statistical analysis showed that tumor size (P = 0.020), pT stage (P = 0.037), pN stage (P = 0.000), pM stage (P = 0.001), clinical stage (P = 0.000), and JMJD3 (P = 0.000) had a significant effect on the survival of patients with PSC (Table 2). The overall survival (OS) and disease-free survival (DFS) in patients with low JMJD3 and high JMJD3 levels were significantly different. Kaplan–Meier survival analysis showed that the mean survival time of the patients with low JMJD3 was significantly longer than that of the patients with high JMJD3 (60.0 months vs 11.1 months, P = 0.000; Table 3). Among the 96 patients, those with low JMJD3 levels had a 3-year mean survival rate of 61.0%, whereas patients with high JMJD3 had a 3-year mean survival rate of 6.0%; the five-year survival rates in patients with low JMJD3 and high JMJD3 were 56.0% and 0.0%, respectively (Table 3).
Table 2.
Univariate Analysis Of Clinicopathologic Variables In 96 Patients With LSC (Log Rank test)
| Variable | All Cases | Mean Survival (Months) | Median Survival (Months) | P valuea |
|---|---|---|---|---|
| Age (years)b | 0.530 | |||
| ≤ 57 | 45 | 47.9 | 31.0 | |
| > 57 | 51 | 54.8 | 18.0 | |
| Gender | 0.377 | |||
| Female | 13 | 38.9 | 41.0 | |
| Male | 83 | 54.9 | 24.0 | |
| Smoking | 0.104 | |||
| Yes | 72 | 59.5 | 31.0 | |
| No | 24 | 25.4 | 14.0 | |
| Tumor sizec | 0.020 | |||
| ≤ 4.5cm | 58 | 65.6 | 48.0 | |
| > 4.5cm | 38 | 36.0 | 13.0 | |
| pT stage | 0.037 | |||
| T1+T2 | 54 | 62.9 | 43.0 | |
| T3+T4 | 42 | 43.8 | 14.0 | |
| pN stage | 0.000 | |||
| N0 | 48 | 75.3 | NR | |
| N1+N2+N3 | 48 | 33.0 | 14.0 | |
| pM stage | 0.001 | |||
| M0 | 88 | 59.9 | 31.0 | |
| M1 | 8 | 12.1 | 8.0 | |
| Therapyd | 0.092 | |||
| Therapy1 | 73 | 62.6 | 43.0 | |
| Therapy2 | 18 | 26.9 | 15.0 | |
| Therapy3 | 5 | 19.0 | 24.0 | |
| Relapse | 0.647 | |||
| Yes | 30 | 35.0 | 24.0 | |
| No | 66 | 57.6 | 41.0 | |
| Clinical stage | 0.000 | |||
| I-II | 42 | 92.6 | NR | |
| III-IV | 54 | 20.0 | 11.0 | |
| JMJD3 | 0.000 | |||
| Low expression | 59 | 82.9 | NR | |
| High expression | 37 | 14.7 | 11.0 |
Notes: aChi-square test, bMean age, cMean tumor size; dTherapy: Therapy1 is lobectomy/bilobectomy and lymphadenectomy, Therapy2 is lobectomy/bilobectomy; Therapy3 Pneumonectomy and radiotherapy and chemoradiotherapy.
Table 3.
The Expression Of JMJD3 For Three-Year Survival Rate And Five-Year Survival Rate
| JMJD3 Expression | Median Survival Time (Months) | Three-Year Survival Rate (%) | Five-Year Survival Rate (%) |
|---|---|---|---|
| Low | 60.0 | 61.0 | 56.0 |
| High | 11.1 | 6.0 | 0.0 |
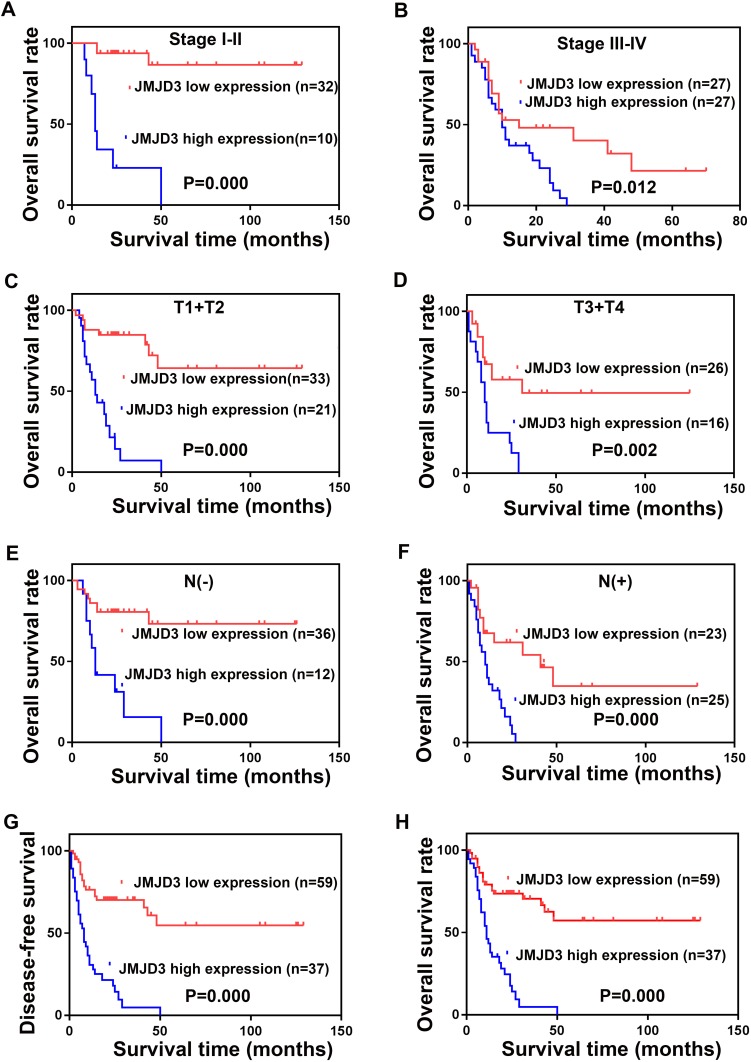
To explore the exact influence of JMJD3 on PSC, we analyzed the survival of patients with different JMJD3 levels in different stages. The results revealed that JMJD3 was a prognostic factor in patients with stage I–II (P = 0.000), stage III–IV (P = 0.012), T1+T2 (P = 0.000), T3+T4 (P = 0.002), N (-) (P = 0.000) and N (+) (P = 0.000). The details are shown in Figure 2.
Figure 2.
Survival analysis of JMJD3 in PSC patients. Clinical stage I–II (A) Clinical stage III–IV (B), T1+T2 (C), T3+T4 (D), N(−) (E), N(+) (F). The disease-free survival of all the patients (G) and the overall survival (H).
Independent Prognostic Factors Of PSC: Multivariate Cox Regression Analysis
The factors that had a significant influence in the univariate analysis were analyzed by Cox regression analysis. The results showed that a low level of JMJD3 was an independent prognostic factor associated with good overall survival (hazard ration (HR): 3.482; 95% confidence interval (CI): 1.858–6.525, P = 0.000; Table 4). Meanwhile, we also found that clinical stage (P = 0.000) was an independent prognostic factor for patients with PSC.
Table 4.
Multivariate Analysis Of Different Prognostic Factors In 96 Patients With LSC
| Variable | Hazards Ratio | 95% CI | P value |
|---|---|---|---|
| Tumor size (> 4.5cm vs ≤ 4.5cm) | 1.180 | 0.668–2.084 | 0.568 |
| Clinical stage (III-IV vs I-II) | 3.881 | 1.891–7.963 | 0.000 |
| JMJD3 (High vs Low) | 3.482 | 1.858–6.525 | 0.000 |
Discussion
Lung cancer is the most common cancer worldwide.20 Research into lung cancer has made considerable achievements; however, there has been little study of PSC, a rare subtype of lung cancer. Most cases of advanced NSCLC receive the same chemotherapy without consideration of their histological subtype.21 However, scientists are becoming increasingly aware that precise molecular biomarkers are required to identify accurately the subtype of PSC, rather than simply classifying them histologically and delivering the same therapy regimen.22 Thus, there is an urgent need for prognostic molecular biomarkers that can predict tumor recurrence.
Histone methylation plays a vital role in the regulation of gene transcription, cell cycle, and differentiation, and its dysfunction is closely related to tumorigenesis.23,24 Histone demethylases can distinguish the site and methylation state of histones and display particular activities.25 The JmjC domain-containing JMJD3 is an H3K27-specific demethylase that can demethylate H3K27me2/3.11
In the present study, we estimated the protein level of JMJD3 in 96 primary sarcomatoid carcinomas of the lung using IHC. Chi-squared analysis revealed a significant correlation between JMJD3 expression and tumor size, lymph node, and clinical stage. Univariate Kaplan–Meier analysis indicated that the JMJD3 protein level had a significant effect on patient survival. Cox multivariate analysis showed that JMJD3 was an independent prognostic factor for PSC. Taken together, our results revealed that the JMJD3 protein level is a reliable biomarker for the prognosis of PSC.
JMJD3 can act via NF-κB and BMP signaling in melanoma to promote clonogenicity, self-renewal, and transendothelial migration.26 In liver cancer, JMJD3 modulated H3K27me3 in the SLUG gene promoter, thereby inducing epithelial–mesenchymal transition (EMT), stemness, and metastasis.13 A recent study shows that JMJD3 was increased in NSCLC, the prognosis of patients with high expression of JMJD3 was significantly worse than that of low expression. JMJD3 might promote the invasion and migration of NSCLC by regulating EMT signaling pathway, all these studies support our results and give us a hint that maybe JMJD3 also plays as an oncogene by promoting tumor metastasis in PSC.27 JMJD3-mediated H3K27 methylation mutation could cause tumorigenesis; therefore, drugs or antibody targeting JMJD3 may represent a novel therapy for cancer. Pediatric brainstem gliomas often harbor an oncogenic K27M mutation of histone H3.3. GSKJ4, a pharmacological inhibitor of JMJD3, demonstrated potent antitumor efficacy by increasing cellular H3K27 methylation and is an effective therapeutic drug to treat K27M-expressing brainstem glioma,28 Ntziachristos observed the same phenomenon that GSKJ4 affects T-cell acute lymphoblastic leukemia (T-ALL) growth by targeting JMJD3 activity.29
However, another study showed that JMJD3 could inhibit cell proliferation and was not a good predictor of poor prognosis in colorectal cancer.15 There is little knowledge of JMJD3’s function in tumors, and it may influence the intersections of many pathways.30 Understanding the mechanism of JMJD3’s activity in cancer or other diseases is difficult because JMJD3’s target is H3K27, which is spread throughout the genome; therefore, the particular effect and mechanism of JMJD3 in cancer might vary. Besides, our study only included 96 cases of PSC, this is not a very big cohort, and thus our conclusion drawn from this study will be limited to be used extensively.
Conclusion
In our study, we found that JMJD3 was overexpressed in PSC which was consistent with the previous researches about JMJD3 in other cancers. We proposed that JMJD3 could be used as a biomarker for prognosis in PSC.
Acknowledgments
This work was supported by the National Key R&D Program of China (No. 2017YFC1309000) and Open Project of State Key Laboratory of Respiratory Disease of China (No. SKLRD2016OP004). The funding sources have no role in designing the experiment, the collection and analysis of data or the writing of manuscript.
Abbreviations
NSCLC, non-small cell lung cancer; PSC, primary sarcomatoid carcinoma of the lung; JMJD3, Jumonji domain-containing protein D3; IHC, immunohistochemistry.
Disclosure
All authors declared that there are no conflicts of interest.
References
- 1.Pelosi G, Sonzogni A, De Pas T, et al. Review article: pulmonary sarcomatoid carcinomas: a practical overview. Int J Surg Pathol. 2010;18(2):103–120. doi: 10.1177/1066896908330049 [DOI] [PubMed] [Google Scholar]
- 2.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thoracic Oncol. 2015;10(9):1243–1260. doi: 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 3.Yendamuri S, Caty L, Pine M, et al. Outcomes of sarcomatoid carcinoma of the lung: a Surveillance, Epidemiology, and End Results Database analysis. Surgery. 2012;152(3):397–402. doi: 10.1016/j.surg.2012.05.007 [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- 5.Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–1140. doi: 10.1101/gad.1652908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuthbert GL, Daujat S, Snowden AW, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118(5):545–553. doi: 10.1016/j.cell.2004.08.020 [DOI] [PubMed] [Google Scholar]
- 8.Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–816. doi: 10.1038/nature04433 [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- 10.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131(1):29–32. doi: 10.1016/j.cell.2007.09.026 [DOI] [PubMed] [Google Scholar]
- 11.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104(47):18439–18444. doi: 10.1073/pnas.0707292104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17(10):850–857. doi: 10.1038/cr.2007.83 [DOI] [PubMed] [Google Scholar]
- 13.Tang B, Qi G, Tang F, et al. Aberrant JMJD3 expression upregulates slug to promote migration, invasion, and stem cell-like behaviors in hepatocellular carcinoma. Cancer Res. 2016;76(22):6520–6532. doi: 10.1158/0008-5472.CAN-15-3029 [DOI] [PubMed] [Google Scholar]
- 14.Ohguchi H, Harada T, Sagawa M, et al. KDM6B modulates MAPK pathway mediating multiple myeloma cell growth and survival. Leukemia. 2017;31(12):2661–2669. doi: 10.1038/leu.2017.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokunaga R, Sakamoto Y, Nakagawa S, et al. The prognostic significance of histone lysine demethylase JMJD3/KDM6B in colorectal cancer. Ann Surg Oncol. 2016;23(2):678–685. doi: 10.1245/s10434-015-4879-3 [DOI] [PubMed] [Google Scholar]
- 16.Marchevsky AM, LeStang N, Hiroshima K, et al. The differential diagnosis between pleural sarcomatoid mesothelioma and spindle cell/pleomorphic (sarcomatoid) carcinomas of the lung: evidence-based guidelines from the International Mesothelioma Panel and the MESOPATH National Reference Center. Hum Pathol. 2017;67:160–168. doi: 10.1016/j.humpath.2017.07.015 [DOI] [PubMed] [Google Scholar]
- 17.Yang GF, He WP, Cai MY, et al. Intensive expression of Bmi-1 is a new independent predictor of poor outcome in patients with ovarian carcinoma. BMC Cancer. 2010;10:133. doi: 10.1186/1471-2407-10-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo RZ, Cai PQ, Li M, et al. Decreased expression of PTPN12 correlates with tumor recurrence and poor survival of patients with hepatocellular carcinoma. PLoS One. 2014;9(1):e85592. doi: 10.1371/journal.pone.0085592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai MY, Zhang B, He WP, et al. Decreased expression of PinX1 protein is correlated with tumor development and is a new independent poor prognostic factor in ovarian carcinoma. Cancer Sci. 2010;101(6):1543–1549. doi: 10.1111/j.1349-7006.2010.01560.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 21.Azzoli CG, Baker S Jr, Temin S, et al. American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol. 2009;27(36):6251–6266. doi: 10.1200/JCO.2009.23.5622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thoracic Oncol. 2013;8(7):823–859. doi: 10.1097/JTO.0b013e318290868f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. doi: 10.1016/j.molcel.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79(8 Suppl):1514–1519. doi: 10.1902/jop.2008.080172 [DOI] [PubMed] [Google Scholar]
- 25.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8(4):307–318. doi: 10.1038/nrm2143 [DOI] [PubMed] [Google Scholar]
- 26.Park WY, Hong BJ, Lee J, Choi C, Kim MY. H3K27 demethylase JMJD3 employs the NF-kappaB and BMP signaling pathways to modulate the tumor microenvironment and promote melanoma progression and metastasis. Cancer Res. 2016;76(1):161–170. doi: 10.1158/0008-5472.CAN-15-0536 [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Hua PY, Jin CY, Li JD, Zhang GX, Wang B. JMJD3 enhances invasiveness and migratory capacity of non-small cell lung cancer cell via activating EMT signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(11):4784–4792. doi: 10.26355/eurrev_201906_18063 [DOI] [PubMed] [Google Scholar]
- 28.Hashizume R, Andor N, Ihara Y, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20(12):1394–1396. doi: 10.1038/nm.3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ntziachristos P, Tsirigos A, Welstead GG, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514(7523):513–517. doi: 10.1038/nature13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrigue PM, Najbauer J, Barciszewski J. Histone demethylase JMJD3 at the intersection of cellular senescence and cancer. Biochim Biophys Acta. 2016;1865(2):237–244. doi: 10.1016/j.bbcan.2016.03.002 [DOI] [PubMed] [Google Scholar]