Abstract
Introduction
Long non-coding RNAs (LncRNAs) have been demonstrated to play a vital role in human carcinogenesis. HOXA cluster antisense RNA 2 (HOXA-AS2), a 1048-bp lncRNA located between the HOXA3 and HOXA4 genes, is identified as an oncogene in several malignancies, including glioma. However, the biological functions of HOXA-AS2 and its underlying molecular mechanisms in glioma progression remain to be investigated.
Method
The expression of HOXA-AS2 and RND3 mRNA was determined using qRT-PCR analysis. The protein level of RND3 and EZH2 was measured by Western blot analysis. The biological function of HOXA-AS2 or RND3 in glioma was detected by CCK-8 assay, colony formation assays, transwell assay, and flow cytometry. Dual-luciferase reporter, RIP, RNA-protein pull down and ChIP assays were performed to explore the molecular mechanism of HOXA-AS2 in glioma. The effect of HOXA-AS2 in vivo was examined using xenograft tumor assay.
Results
HOXA-AS2 expression was increased in glioma tissues and cells. High HOXA-AS2 expression was associated with larger tumor size and advanced pathological stage. Functionally, knockdown of HOXA-AS2 suppressed cell proliferation and invasion, and promoted apoptosis. Mechanically, HOXA-AS2 epigenetically inhibited RND3 transcription by binding to EZH2. Moreover, overexpression of RND3 exerted similar tumor-suppressive effects to the depletion of HOXA-AS2. Furthermore, the anti-cancer effects induced by si-HOXA-AS2 were greatly reversed by silencing of RND3. Finally, knockdown of HOXA-AS2 impaired tumor growth in vivo possibly via increasing RND3 expression.
Conclusion
Taken together, HOXA-AS2 recruits EZH2 to the promoter region of RND3 and inhibits its expression, thereby facilitating glioma progression. Our findings provide a prospective therapeutic strategy for glioma intervention.
Keywords: glioma, lncRNA, HOXA-AS2, RND3, EZH2
Introduction
Glioma is demonstrated as the most common and aggressive primary brain tumor in central nervous system.1 Glioma is divided into low-grade glioma (WHO grade I and II) and high-grade glioma (WHO grade III and IV) according to the histopathological features.2 Until now, surgical resection followed by radiotherapy or chemotherapy is still the standard therapy for glioma.3 Due to high proliferative and invasive ability of glioma, the prognosis remains unsatisfactory.4 Therefore, it is in an urgent need to gain insight into the pathogenic mechanism of glioma for developing more effective molecular targets.
Long non-coding RNAs (lncRNAs) are identified as a class of transcripts with a minimum length of 200 bases and limited protein-coding potential. In recent years, lncRNAs have become a hot topic for its important participation in various human diseases, including cancers.5 LncRNAs are able to affect cell growth, apoptosis, metastasis and angiogenesis in malignant tumors through regulating miRNAs, chromatin remodeling, and histone protein modification.6,7 Enhancer of zeste homolog 2 (EZH2), a core catalytic component of polycomb repressive complex 2 (PRC2), serves as a histone methyltransferase to induce histone H3 lysine 27 trimethylation (H3K27me3) and silence transcription.8 A growing number of evidence reveals that many lncRNAs recruit EZH2 to the promoter of target genes and affect a wide range of cancer-related cellular processes.9 For example, lncRNA CASC9 facilitated esophageal squamous cell carcinoma growth by suppressing PDCD4 expression via EZH2.10 LncRNA AWPPH enhanced cell proliferation, autophagy, and migration, while decreased apoptosis in bladder cancer by repressing SMAD4 through binding to EZH2.11 LncRNA FOXC2-AS1 aggravated non-small-cell lung cancer (NSCLC) progression by epigenetic silencing of p15 expression via interaction with EZH2.12 HOXA cluster antisense RNA 2 (HOXA-AS2), a 1048-bp lncRNA located between the human HOXA3 and HOXA4 genes in the HOXA cluster, functions as an oncogene in multiple malignancies, such as breast cancer, gastric cancer, gallbladder carcinoma, hepatocellular carcinoma and pancreatic cancer, via different mechanisms.13 For instance, Xie et al found that up-regulation of HOXA-AS2 promoted tumorigenesis in gastric cancer partly by epigenetically silencing P21, PLK3 and DDIT3 transcription via binding with EZH2.14 Jiang et al reported that overexpression of HOXA-AS2 contributed to cell proliferation, migration, and invasion in papillary thyroid cancer by inducing HOXA3 expression via binding to miR-15a-5p.15 As previously reported, HOXA-AS2 expression was up-regulated in glioma tissues and cells, and knockdown of HOXA-AS2 suppressed malignant glioma behaviors and vasculogenic mimicry formation through regulating miR-373/EGFR axis.16 However, the possible action mechanisms of HOXA-AS2 in glioma progression still needs to be elaborated.
In the present study, it was found that HOXA-AS2 expression was increased in glioma tumor tissues and cells. High HOXA-AS2 expression was associated with tumor size and WHO grade. Furthermore, HOXA-AS2 promoted proliferation and invasion, and lowered apoptosis of glioma cells possibly by epigenetically suppressing RND3 expression via binding with EZH2. These data highlighted the potential of HOXA-AS2 as a therapy target for glioma patients.
Materials And Methods
Clinical Tissue Samples
A total of 50 pairs of glioma tissues and adjacent normal brain tissues were obtained from patients who underwent surgical resection at the First Affiliated Hospital of Zhengzhou University between Jan 2015 and Jun 2016. The adjacent normal brain tissues were taken from at least 1 cm away from the edge of the lesions and contained no obvious tumor cells. Histopathological characteristics were evaluated by two independent pathologists. The detailed patient information was provided in Table 1. No patients received local or systemic treatment before surgery. The collected specimens were snap-frozen in liquid nitrogen and stored at −80°C. This study was approved by the Research Ethic Committee of our hospital in accordance with the Declaration of Helsinki Principle. All written informed consents were signed by the participants.
Table 1.
Correlation Of HOXA-AS2 Expression And Clinicopathological Characteristics In Glioma
| Variable | HOXA-AS2 Expression | P value | |
|---|---|---|---|
| Low | High | ||
| Age | 0.390 | ||
| <50 | 9 | 12 | |
| ≥50 | 16 | 13 | |
| Gender | 0.774 | ||
| Male | 11 | 10 | |
| Female | 14 | 15 | |
| Tumor size | 0.047* | ||
| <5 cm | 15 | 8 | |
| ≥5 cm | 10 | 17 | |
| WHO grade | 0.024* | ||
| I–II | 16 | 8 | |
| III–IV | 9 | 17 | |
| Tumor position | 0.777 | ||
| Frontal | 13 | 12 | |
| Temporal | 12 | 13 | |
Notes: Low/high by the sample mean. Pearson χ2 test. *P < 0.05 was considered statistically significant.
Cell Culture
Human glioma cell lines (A172, SHG44, LN229, U87, U251) and normal human astrocytes (NHA) were purchased from the Chinese Academy of Sciences (Shanghai; China). All cells were maintained in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere with 5% CO2.
Vector Construction And Cell Transfection
Small interference RNAs (siRNAs) oligos targeting HOXA-AS2 (si-HOXA-AS2), EZH2 (si-EZH2), or RND3 (si-RND3), and respective non-targeting sequences (si-NC) were purchased from GenePharma (Shanghai, China). Interference sequences were listed as below: si-NC, 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ; si-HOXA-AS2, 5ʹ-GAGUUCAGCUCAAGUUGAACAUACA-3ʹ; si-EZH2, 5ʹ-GTGCCCTTGTGTGATAGCACAA-3ʹ; si-RND3, 5ʹ-CCCUGAUUCGGAUGCUGUGCUGAUU-3ʹ. To achieve the overexpression of HOXA-AS2 or RND3, the full length sequences of HOXA-AS2 or RND3 were amplified from human cDNA and cloned into pcDNA3.1 vector (Invitrogen). When cells reached at approximately 80% confluency, transfection was performed using Lipofectamine 2000 reagent (Invitrogen). After 48 h, transfection efficiency was examined using qRT-PCR.
RNA Isolation And qRT-PCR
Total RNA was extracted from glioma tissues and cells using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instruction. Total RNA (1 μg) was used for cDNA synthesis using PrimeScript RT Reagent Kit (Takara, Dalian, China). qRT-PCR reaction was carried out using SYBR Select Master Mix (Applied Biosystems, Foster City, CA, USA) on a StepOnePlus Real-time PCR system (Applied Biosystems). Primer sequences for PCR were listed as follows: HOXA-AS2, GTTCAGCTCAAGTTGAACATA (Forward) and AAACCTTGTAGATAGCTTGAG (Reverse); RND3, CTATGACCAGGGGGCAAATA (Forward) and TCTTCGCTTTGTCCTTTCGT (Reverse); GAPDH, ACCACAGTCCATGCCATCAC (Forward) and TCCACCACCCTGTTGCTGTA (Reverse); U6, CTCGCTTCGGCAGCACA (Forward) and AACGCTTCACGAATTTGCGT (Reverse). The 2−ΔΔCt method was used for determining the relative expressions of HOXA-AS2 and RND3 mRNA. GAPDH was used as an internal reference for qPCR normalization.
Cell Viability Assay
Glioma cell viability was detected using the Cell Counting Kit-8 (CCK-8; Beyotime, Beijing, China) in accordance with the manual. Briefly, si-HOXA-AS2-transfected U87 and U251 cells or pcDNA3.1-transfected LN229 cells were seeded into 96-well plates at a density of 2 × 103 cells/well. At 24, 48, 72, and 96 h, 10 μl of CCK-8 solution was added into each well and incubated for another 4 h. The optical absorbance of each well was measured at a wavelength of 450 nm.
Colony Formation Assay
Si-HOXA-AS2-transfected U87 and U251 cells or pcDNA3.1-transfected LN229 cells were inoculated in 6-well plates and incubated in proper medium for 12 days. The culture medium was changed every 4 days. Following fixed with 96% ethanol and stained with 0.1% crystal violet, the colonies were counted manually under a light microscope.
Cell Invasion Assay
Transwell assay inserts (Millipore, Billerica, MA, USA) with a Matrigel-coated membrane were used for evaluating the invasive ability. Si-HOXA-AS2-transfected U87 and U251 cells or pcDNA3.1-transfected LN229 cells were plated into the upper chamber with 200 μL serum-free medium. The lower chambers were filled with 600 μL complete medium with 10% FBS. After incubation at 37°C for 24 h, the invaded cells on the lower surface of the transwell membrane were fixed with methanol and stained with crystal violet. The stained cells were counted from at least 5 random fields.
Apoptosis Analysis
Si-HOXA-AS2-transfected U87 and U251 cells or pcDNA3.1-transfected LN229 cells were plated into 6-well plates and cultured for 48 h. Then, cells were washed with PBS, stained with the FITC-Annexin V Apoptosis Detection Kit (BD Biosciences, Franklin Lakes, NJ, USA), and subjected to a FACSCalibur flow cytometer (BD Biosciences) for apoptosis detection.
Western Blot Analysis
Total protein was isolated from harvested glioma tissues and cells using RIPA buffer (Beyotime, Shanghai, China). Protein extractions were separated on a 10% SDS-PAGE gel, and then further incubated overnight with primary antibodies against RND3 (Cell Signaling Technology, Boston, MA, USA), EZH2 Cell Signaling Technology) and GAPDH (Cell Signaling Technology). Protein signals were visualized using ECL system (Millipore, Bedford, MA, USA). GAPDH was used as an endogenous control.
Subcellular Fractionation Location
The Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA) was used to separate the nuclear and cytosolic fractions in accordance with the manufacturer’s instruction. U6 was used as nuclear control, and GAPDH as cytoplasm control.
Dual-Luciferase Reporter Assay
To detect the effect of HOXA-AS2 or EZH2 on RND3 transcription, U87 and U251 cells were co-transfected with the RND3 promoter reporter vector (Genechem, Shanghai, China) and si-HOXA-AS2 or si-EZH2. At 48 h after transfection, the luciferase activity was determined by the Dual-Lucierase Reporter Assay System (Promega, Madison, WI, USA). Renilla luciferase vector was used as the endogenous control to normalize the Firefly luciferase activity.
RIP Analysis
RIP experiments were conducted using Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) following the manufacturer’s procotol. The antibodies against EZH2 and IgG were obtained from Cell Signaling Technology. The co-precipitated RNAs were subjected to qRT-PCR analysis for detecting the level of HOXA-AS2.
RNA-Protein Pull Down Assays
HOXA-AS2 transcripts were transcribed with TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA) in vitro. Then, purified RNAs were labeled with biotin using Pierce RNA 3ʹ Desthiobiotinylation Kit (Thermo Fisher Scientific). RNA-protein pull-down experiments were performed using a Pierce Magnetic RNA-Protein Pull down Kit (Thermo Fisher Scientific). Briefly, biotinylated transcripts were mixed and incubated with cell lystates, followed by addition of magnetic beads and further incubation. After washing the beads, the eluted proteins were analyzed by Western blot.
Chromatin Immunoprecipitation (ChIP)
U87 and U251 cells were treated with 1% formaldehyde to acquire protein-DNA crosslinks. Cell lysates were sonicated to obtain chromatin fragments of 200–300 bp. Then, the chromatin was immunoprecipitated with EZH2 (Cell Signaling Technology), H3K27me3 (Millipore) or IgG (Millipore) antibody. The IgG was used as a negative control. Precipitated chromatin DNA was purified and detected by qRT-PCR. The primer sequences for RND3 promoter were 5ʹ-GCTTCCTATGTTTTATCACTG-3ʹ (Forward) and 5ʹ-CTGATTTGAACTACAATCCC-3ʹ (Reverse).
Xenografts In Nude Mice
All animal experiment procedures were approved by the Ethic Committee and performed according to the guidelines of Animal Care and Use Committee at the First Affiliated Hospital of Zhengzhou University. U87 cells (7×106) stably transfected with sh-NC or sh-HOXA-AS2 were subcutaneously injected into the right flanks of male BALB/c athymic nude mice (4–6 weeks old). Tumor sizes were measured every 4 days and tumor volumes were calculated using the following formula: V (mm3) = (L × W2)/2. At 31 days after inoculation, the mice were euthanized and tumor weights were examined. Then, the excised tumor masses were subjected to qRT-PCR and Western blot assays.
Statistical Analysis
Quantitative data were represented as mean ± standard deviation (mean ± SD). Statistical difference between groups was evaluated by one-way ANOVA or Student’s t-test using GraphPad Prism (GraphPad Software, San Diego, CA, USA). The Chi-square test was used to determine the association between HOXA-AS2 levels and clinicopathological features. Pearson’s correlation analyses were performed to detect the correlation of HOXA-AS2 and RND3 levels in glioma tissues. P < 0.05 denotes a significant difference.
Results
HOXA-AS2 Expression Was Up-Regulated In Glioma Tissues And Cells
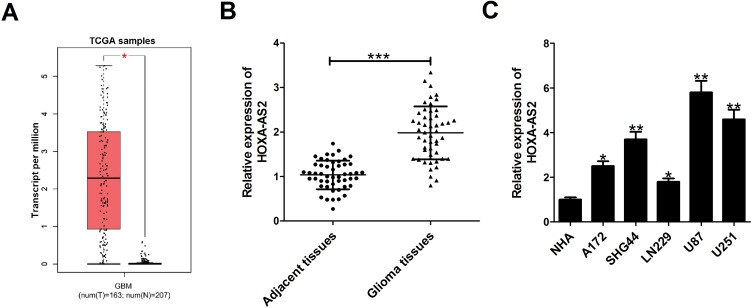
Firstly, the expression difference of HOXA-AS2 in 163 glioma tissues and 207 normal tissues was analyzed based on TCGA database from GEPIA (http://gepia.cancer-pku.cn/detail.php?gene=&clicktag=boxplot). Results showed a significant increase of HOXA-AS2 expression in tumor tissues (Figure 1A). Then, the expression level of HOXA-AS2 in 50 cases of glioma tissues and adjacent normal tissues was detected. A significant up-regulation of HOXA-AS2 expression was observed in glioma tumor tissues in comparison to matched non-cancerous tissues (Figure 1B). According to the clinicopathologic characteristics of 50 glioma patients presented in Table 1, high HOXA-AS2 expression was positively associated with larger tumor size (P=0.047) and advanced tumor stage (P=0.024). Also, the expression levels of HOXA-AS2 in five glioma cell lines (A172, SHG44, LN229, U87, U251) and normal human astrocytes (NHA) were measured. As expected, HOXA-AS2 expression was higher in glioma cells than that in NHA (Figure 1C). These data suggested that HOXA-AS2 may be a facilitator in glioma progression.
Figure 1.
HOXA-AS2 expression is up-regulated in glioma tissues and cells. (A) HOXA-AS2 expression in glioma tissues (n=163) and normal tissues (n=207) was compared using the GEPIA tool (http://gepia.cancer-pku.cn/detail.php?gene=&clicktag=boxplot). (B) qRT-PCR analysis of HOXA-AS2 expression in tumor tissues and adjacent normal tissues from 50 cases of glioma patients. (C) qRT-PCR analysis of HOXA-AS2 expression in glioma cells (A172, SHG44, LN229, U87, U251) and normal human astrocytes (NHA). Data are shown as mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Knockdown Of HOXA-AS2 Suppressed Proliferation And Invasion, And Promoted Apoptosis In Glioma Cells
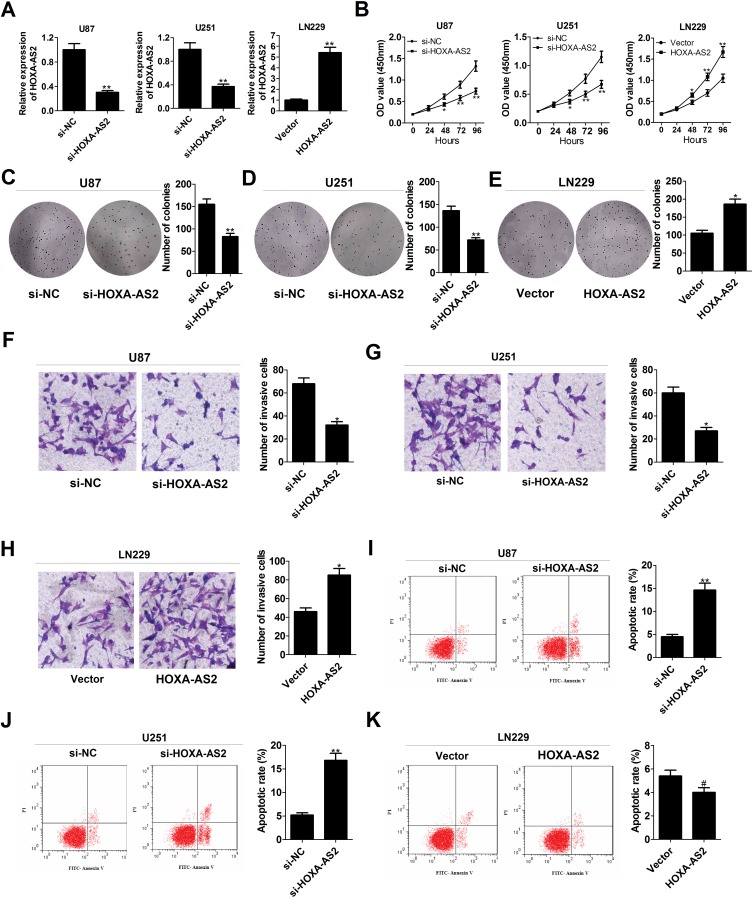
To explore the effects of HOXA-AS2 on the biological behaviors of glioma, HOXA-AS2 expression was down-regulated in U87 and U251 cells by transfection with specific siRNA, and was also up-regulated in LN229 cells by transfection with HOXA-AS2-overexpression vector (Figure 2A). CCK-8 assays demonstrated that knockdown of HOXA-AS2 greatly repressed the growth of U87 and U251 cells, while overexpression of HOXA-AS2 facilitated LN229 cell growth (Figure 2B). Colony forming assays revealed that depletion of HOXA-AS2 resulted in a significant decrease of clonogenic capacities in U87 and U251 cells (Figure 2C and D), while enforced expression of HOXA-AS2 increased the colony number of LN229 cells (Figure 2E). Transwell assays manifested a suppression of invasive ability in si-HOXA-AS2-transfected U87 and U251 cells (Figure 2F and G), and a promotion of invasiveness in HOXA-AS2-transfected LN229 cells (Figure 2H). Flow cytometry assays illuminated that the apoptotic rate was obviously increased in U87 and U251 cells following the silencing of HOXA-AS2 (Figure 2I and J). However, there was no significant alteration of apoptosis in LN229 cells transfected with HOXA-AS2 when compared to vector group (Figure 2K). To conclude, down-regulation of HOXA-AS2 inhibited glioma progression in vitro.
Figure 2.
Knockdown of HOXA-AS2 decreases growth, suppresses invasion, and accelerates apoptosis in glioma cells. U87 and U251 were transfected with si-NC or si-HOXA-AS2, and LN229 cells were transfected with control vector or pcDNA-HOXA-AS2. (A) qRT-PCR analysis was used to determine the transfection efficiency of U87, U251 and LN229 cells. (B) CCK-8 analysis was performed to detect the effects of HOXA-AS2 knockdown or overexpression on cell proliferation. (C–E) Colony forming ability was examined in transfected U87, U251 and LN229 cells. (F–H) Transwell invasion assay was carried out to measure the effects of HOXA-AS2 knockdown or overexpression on cell invasion. (I–K) The effects of HOXA-AS2 knockdown or overexpression on apoptosis were evaluated by flow cytometry. Data are shown as mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, #P > 0.05.
HOXA-AS2 Epigenetically Repressed RND3 Expression By Binding With EZH2
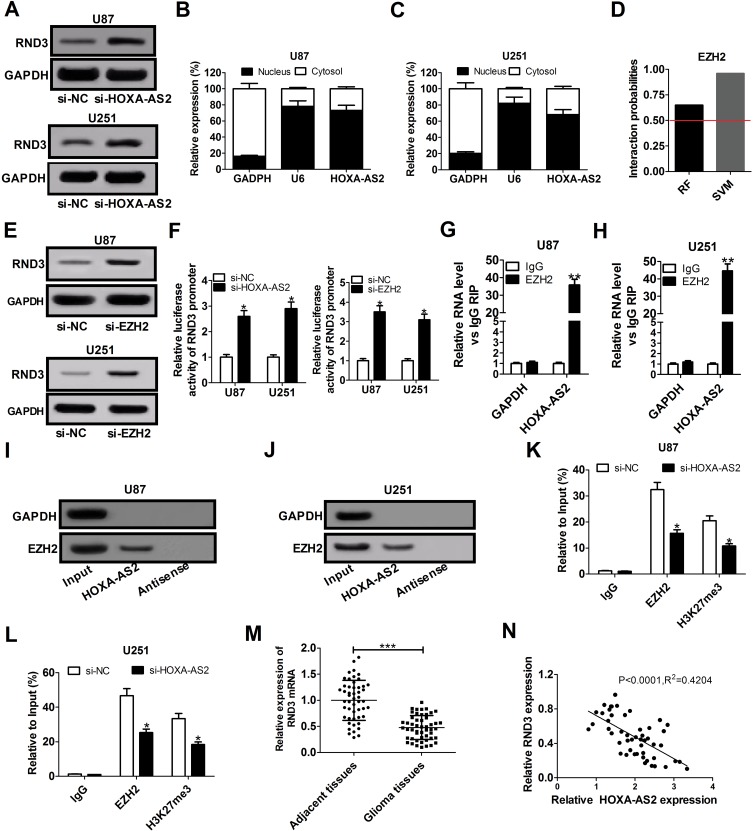
Up to 20% of human lncRNAs are reported to be physically associated with Polycomb Repressive Complex 2 (PRC2).17 HOXA-AS2 was previously elucidated to function as an oncogene in colorectal cancer by epigenetically repressing p21 and KLF2 transcription via binding to EZH2 and LSD1.18 RND3, a tumor suppressor in glioma, was discovered to be decreased by EZH2 via enhancing the level of H3K27me3 at its promoter region in preeclampsia.19,20 Hence, we hypothesized that HOXA-AS2 could regulate RND3 expression via EZH2 in glioma. To validate this idea, the effects of HOXA-AS2 depletion on RND3 expression were detected in glioma cells by Western blot analysis. The data indicated that knockdown of HOXA-AS2 led to a dramatic rise of RND3 protein level in both U87 and U251 cells (Figure 3A). Then, subcellular fractionation assays were conducted to evaluate the distribution of HOXA-AS2 in the nuclear and cytosolic fractions of glioma cells. Results showed that HOXA-AS2 was mainly distributed in the nucleus of U87 and U251 cells (Figure 3B and C), suggesting its important regulatory functions at transcriptional level. RNA-protein interaction prediction website (http://pridb.gdcb.iastate.edu/RPISeq/) showed that HOXA-AS2-EZH2 interaction score was 0.65 using RF classifier, and 0.96 using SVM classifier (Figure 3D), indicating the interaction possibility between HOXA-AS2 and EZH2. Moreover, RND3 expression was prominently promoted in U87 and U251 cells after transfection with EZH2 siRNA (Figure 3E), implying that EZH2 was able to affect RND3 expression. Luciferase reporter experiments further showed that knockdown of HOXA-AS2 or EZH2 resulted in an increase of the luciferase activity of RND3 promoter in U87 and U251 cells (Figure 3F), highlighting that HOXA-AS2 or EZH2 could modulate RND3 expression at the transcriptional level.
Figure 3.
HOXA-AS2 represses RND3 expression by recruiting EZH2 to its promoter region. (A) Western blot assay of RND3 protein level in U87 and U251 cells after transfection with si-NC or si-HOXA-AS2. (B and C) qRT-PCR assay was used to examine the distribution of HOXA-AS2 in cytoplasm and nucleus of U87 and U251 cells, with GAPDH as a cytoplasm marker and U6 as a nucleus marker. (D) RNA-Protein interaction prediction website (http://pridb.gdcb.iastate.edu/RPISeq/) was applied to evaluate the interaction probability between HOXA-AS2 and EZH2. Predictions with probabilities > 0.5 are regarded as positive. RPISeq predictions are based on Random Forest (RF) or Support Vector Machine (SVM). (E) Western blot assay was used to assess the effects of EZH2 knockdown on RND3 protein expression in U87 and U251 cells. (F) The influence of si-HOX1-AS2 or si-EZH2 on the luciferase activity of RND3 promoter reporter was determined in U87 and U251 cells. (G and H) RIP assays with anti-EZH2 or anti-IgG from U87 and U251 cell extracts, and then the coprecipitated RNA was subjected to qRT-PCR for HOXA-AS2. The fold enrichment of HOXA-AS2 in EZH2 RIP is relative to its matched IgG control. (I and J) The RNA pull-down assays revealed the binding between HOXA-AS2 and EZH2 in U87 and U251 cells. GAPDH is presented as the control. (K and L) ChIP of EZH2 occupancy and H3K27me3 binding in the RND3 promoter region in U87 and U251 cells transfected with si-NC or si-HOXA-AS2. IgG was used as a negative control. (M) qRT-PCR analysis of RND3 mRNA expression in tumor tissues and adjacent non-cancerous tissues from 50 glioma patients. (N) The correlation analysis of HOXA-AS2 and RND3 expression in glioma tissues. Data are shown as mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Subsequently, RIP assays displayed a substantial enrichment of HOXA-AS2 level in the anti-EZH2 RIP fraction compared with the IgG fraction in U87 and U251 cells (Figure 3G and H). RNA pull down assays exhibited that EZH2 in the nuclear extract fraction of U87 and U251 cells was pulled down by labeled HOXA-AS2 (Figure 3I and J). These data suggested a specific interaction between HOXA-AS2 and EZH2 in glioma cells. Additionally, the results of ChIP assays manifested that EZH2 could bind to RND3 promoter region and induce H3K27 trimethylation, and knockdown of HOXA-AS2 reduced the enrichment of EZH2 and H3K27me3 in the promoter RND3 region (Figure 3K and L). As we might expect, RND3 expression was found to be decreased in tumor tissues of glioma patients compared to adjacent normal tissues (Figure 3M). Furthermore, RND3 expression was inversely correlated with HOXA-AS2 expression in glioma patients (Figure 3N). All these data made us draw a conclusion that HOXA-AS2 negatively regulated RND3 expression by recruiting EZH2 to its promoter region and increasing the H2K27me3 enrichment of its promoter in glioma.
Up-Regulation Of RND3 Inhibited Proliferation And Invasion, And Induced Apoptosis In Glioma Cells
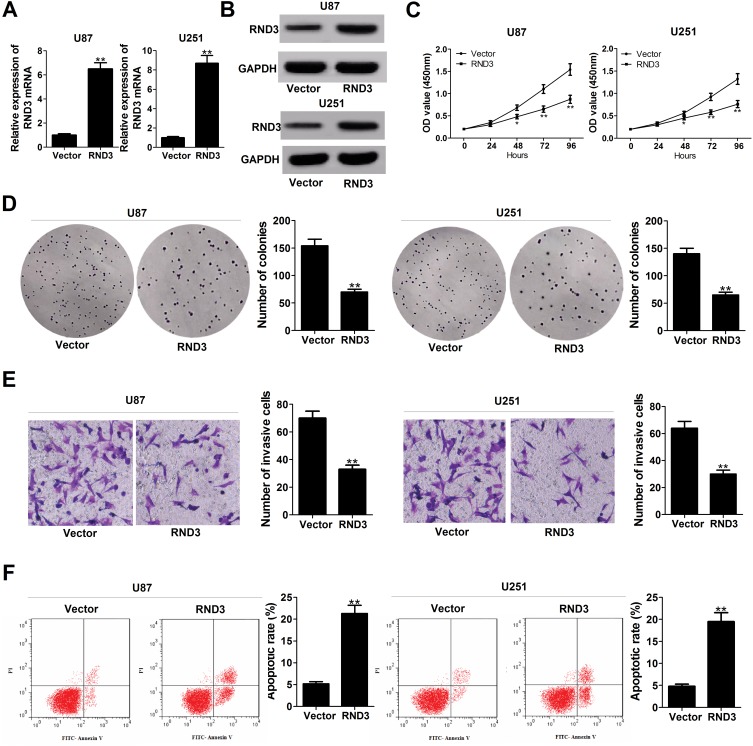
To clarify the biological functions of RND3 in glioma, RND3-overexpression vector (RND3) was introduced into glioma cells. A substantial up-regulation of RND3 expression at both mRNA and protein levels was observed in RND3-transfected U87 and U251 cells (Figure 4A and B). Function assays elucidated a suppressive effect of RND3 overexpression on U87 and U251 cell proliferation (Figure 4C). Also, the number of colonies was evidently reduced in U87 and U251 cells following transfection with RND3 (Figure 4D). Moreover, exogenous expression of RND3 led to an apparent suppression of invasiveness in U87 and U251 cells (Figure 4E). Furthermore, apoptotic rate was significantly increased by overexpression of RND3 in U87 and U251 cells (Figure 4F). Together, overexpression of RND3 could exert an inhibitory role in glioma progression in vitro.
Figure 4.
Enforced expression of RND3 inhibits proliferation, hinders invasion and enhances apoptosis glioma cells. U87 and U251 cells were transfected with pcDNA empty vector or pcDNA-RND3. (A and B) RND3 expression at mRNA and protein levels was determined in transfected cells. (C–F) Effects of RND3 overexpression on cell proliferation (C), colony forming ability (D), invasiveness (E), and apoptosis (F) were detected. Data are shown as mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01.
Down-Regulation Of HOXA-AS2 Repressed The Malignant Process Of Glioma By Regulating RND3
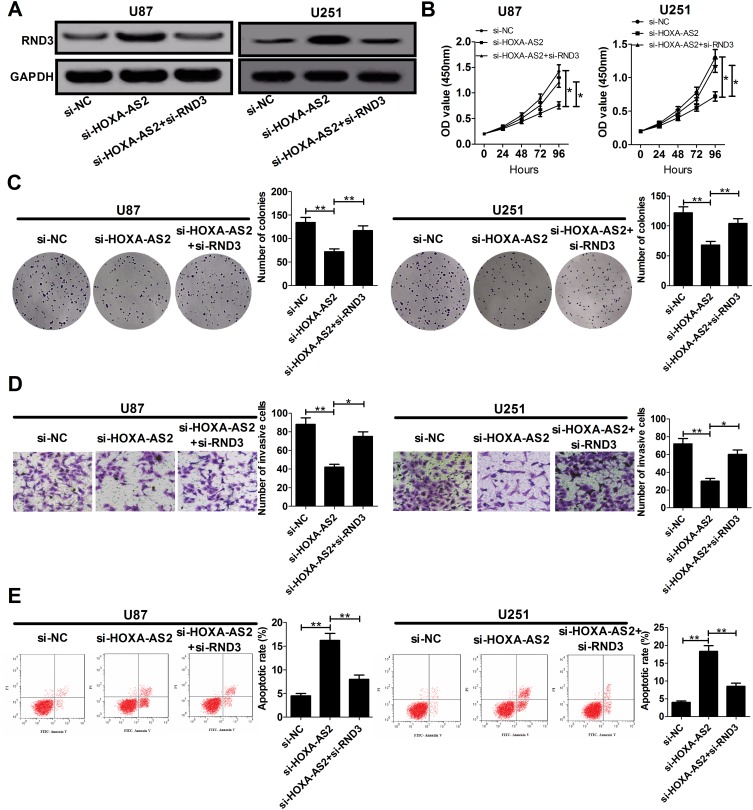
In order to address whether RND3 was involved in HOXA-AS2-mediated growth promotion, U87 and U251 cells were co-transfected with si-HOXA-AS2 and si-RND3. As exhibited in Figure 5A, si-HOXA-AS2-elicited enhancement of RND3 protein expression in U87 and U251 cells was remarkably reversed by transfection with si-RND3. The anti-proliferative effects induced by HOXA-AS2 knockdown were abated by suppression of RND3 (Figure 5B and C). Moreover, si-HOXA-AS2-mediated suppression of invasion was greatly counteracted following inhibition of RND3 (Figure 5D). Also, silencing of RND3 partly abrogated si-HOXA-AS2-induced apoptosis (Figure 5E). Collectively, HOXA-AS2 facilitated glioma development through negatively modulating RND3 expression.
Figure 5.
The anti-cancer effects mediated by HOXA-AS2 knockdown are partly reversed by down-regulation of RND3. U87 and U251 cells were transfected with si-NC, si-HOXA-AS2, or co-transfected with si-HOXA-AS2 and si-RND3, followed by the examination of RND3 protein level (A), cell proliferation (B), colony formation (C), invasiveness (D), and apoptosis (E). Data are shown as mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01.
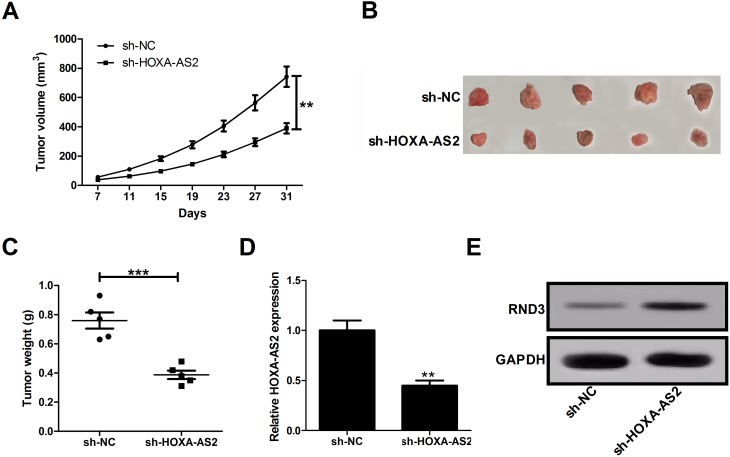
Depletion Of HOXA-AS2 Hindered Tumor Growth In Vivo
To further determine whether HOXA-AS2 affects glioma cell growth in vivo, sh-HOXA-AS2- or sh-NC-transfected U87 cells were injected into the nude mice. Results showed that down-regulation of HOXA-AS2 slowed down the tumor growth when compared with sh-NC group (Figure 6A). At 31 days after inoculation, the average tumor weight was lower in mice derived from sh-HOXA-AS2-transfected cells than that from sh-NC-transfected cells (Figure 6B and C). Moreover, knockdown of HOXA-AS2 resulted in a decrease of HOXA-AS2 expression (Figure 6D), and an increase of RND3 protein level (Figure 6E) compared to control sh-NC group. These data suggested that silencing of HOXA-AS2 inhibited tumorigenesis possibly via promoting RND3 expression in glioma.
Figure 6.
Depletion of HOXA-AS2 slows down glioma growth in vivo. The male nude mice were subcutaneously injected with sh-NC- or sh-HOXA-AS2-transfected U87 cells. (A) Tumor sizes were measured and tumor volumes were calculated. (B) Representative images of resected tumor masses. (C) Tumor weights were detected at 31 days after cell inoculation. (D and E) qRT-PCR analysis of HOXA-AS2 expression and Western blot analysis of RND3 expression in removed tumor tissues. Data are shown as mean ± SD of 3 independent experiments. **P < 0.01, ***P < 0.001.
Discussion
It is well known that lncRNAs drive the malignant phenotypes of human cancers via interacting with several cellular macromolecules, such as DNA, protein, and RNA.21 Increasing lncRNAs are found to be associated with the initiation, differentiation, progression, recurrence and stem-like characteristics in glioma, implying the potential of lncRNAs as diagnostic biomarkers and therapeutic targets.22,23 HOXA-AS2 was previously found to be increased in glioma tissues and cells, and depletion of HOXA-AS2 inhibited the malignant progression of glioma by decreasing EGFR expression via miR-373.16 However, the regulatory mechanisms of HOXA-AS2 in the nucleus in glioma remain unkonwn.
In the present study, HOXA-AS2 expression was confirmed to be up-regulated in glioma tissues and cells. Moreover, high HOXA-AS2 expression was positively associated with larger tumor size and advanced tumor grade. Subsequent functional assays manifested that knockdown of HOXA-AS2 repressed proliferation and invasion, and induced apoptosis in U87 and U251 cells, while overexpression of HOXA-AS2 exerted the contrary effects in LN229 cells, suggesting the carcinogenicity of HOXA-AS2 in glioma in vitro. In agreement with our results, HOXA-AS2 was demonstrated as an oncogene in bladder cancer, acute myeloid leukemia, and NSCLC.24–26
Enhancer of zeste homolog 2 (EZH2), a core subunit of the PRC2 complex, is a master regulator of transcription in human cancers, and functions as an oncogene in various malignancies.27,28 Mounting evidence reports that lncRNAs affect the downstream target genes through binding to the PRC2, and PRC2-mediated epigenetic regulation plays an important part in tumorigenesis.29 For instance, LINC00511 functioned as an oncogene partially by epigenetically silencing of p57 expression via binding with EZH2.30 LncRNA FOXD2-AS1 contributed to gastric cancer progression by inhibiting EphB3 expression through direct interaction with EZH2 and LSD1.31 SNHG14 functioned as a facilitator in cell proliferation and metastasis of colorectal cancer through targeting EZH2-regulated EPHA7.32 RND3, a tumor suppressor in glioma, was found to be suppressed by HOXA11-AS, via recruiting EZH2 and LSD1 protein and thereby inducing H3K27me3 in preeclampsia.19 Therefore, whether HOXA-AS2 could affect RND3 expression through interaction with EZH2 were further discussed. The results found that down-regulation of HOXA-AS2 or EZH2 resulted in an increase of RND3 protein level in glioma cells. Moreover, HOXA-AS2 was mainly located in the nuclear fractions in glioma cells. Luciferase reporter experiments proved the transcriptional regulation of HOXA-AS2 or EZH2 on RND3 expression. The follow-up RIP and RNA-protein pull down assays further verified that HOXA-AS2 could directly bind to EZH2. ChIP assays disclosed that EZH2 could directly bind to the promoter of RND3 in glioma cells, and down-regulation of HOXA-AS2 reduced the binding ability of EZH2 to the promoter region of RND3 and repressed H3K27 trimethylation. All these data indicated that HOXA-AS2 inhibited RND3 expression partially by recruiting EZH2 to its promoter region in glioma. Consistent with our findings, Lian et al documented that HOXA-AS2 exhibits oncogenic activity through binding to EZH2 and LSD1 in pancreatic cancer.33 Ding et al discovered that HOXA-AS2 facilitated tumorigenesis in colorectal cancer by recruiting EZH2 and LSD1 to the promoter regions of p21 and KLF2 to repress their transcription.18
RND3, an atypical member of the Rho GTPase family in that it lacks detectable GTPase activity, plays a vital role in arresting cell cycle, inhibiting cell growth, and facilitating apoptosis and differentiation.34 RND3 may serve as a either a suppressor or a promoter in carcinogenesis depending on different tumor types.35 Liu et al uncovered that RND3 expression was reduced in human glioblastoma, enforced expression of RND3 inhibited glioblastoma cell proliferation and glioblastoma tumor growth in mice through inactivating Notch signaling.36 Liu et al delineated that RND3 protein level was down-regulated in human glioblastoma tissues, and overexpression of RND3 impeded cell migration and invasion activity by promoting Snail 1 protein degradation.37 A recent research demonstrated that RND3 promoted GBM cell apoptosis by inactivation of NF-κB signaling through binding to p65 protein and inducing its ubiquitination.38 In this study, RND3 expression was validated to be reduced and negatively correlated with HOXA-AS2 expression in glioma tissues. Gain-of-function experiments showed that up-regulation of RND3 suppressed proliferation and invasion, and enhanced apoptosis of glioma cells. Further restoration experiments suggested that silencing of RND3 partly reversed HOXA-AS2-knockdown-mediated growth suppression. Finally, knockdown of HOXA-AS2 was confirmed to inhibit tumor growth in vivo possibly by increasing RND3 expression.
Conclusion
In conclusion, HOXA-AS2 facilitated glioma progression partly by epigenetically suppressing RND3 expression via binding with EZH2. Our finding highlights the promising prospect of HOXA-AS2 as a molecular target for glioma therapy. Nevertheless, the other potential regulatory mechanisms of HOXA-AS2 involved in glioma progression still needs to be further identified.
Supplementary Materials
The original images for all relevant western blots have been provided (Supplementary file).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
- 1.Demuth T, Berens ME. Molecular mechanisms of glioma cell migration and invasion. J Neurooncol. 2004;70(2):217–228. doi: 10.1007/s11060-004-2751-6 [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 3.Ghotme KA, Barreto GE, Echeverria V, et al. Gliomas: new Perspectives in Diagnosis, Treatment and Prognosis. Curr Top Med Chem. 2017;17(12):1438–1447. doi: 10.2174/1568026617666170103162639 [DOI] [PubMed] [Google Scholar]
- 4.Wen PY, Reardon DA. Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12(2):69–70. doi: 10.1038/nrneurol.2015.242 [DOI] [PubMed] [Google Scholar]
- 5.Saha P, Verma S, Pathak RU, Mishra RK. Long Noncoding RNAs in Mammalian Development and Diseases. Adv Exp Med Biol. 2017;1008:155–198. [DOI] [PubMed] [Google Scholar]
- 6.Bhat SA, Ahmad SM, Mumtaz PT, et al. Long non-coding RNAs: mechanism of action and functional utility. Noncoding RNA Res. 2016;1(1):43–50. doi: 10.1016/j.ncrna.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camacho CV, Choudhari R, Gadad SS. Long Noncoding RNAs and Cancer, an Overview. Steroids. 2018;133:93–95. doi: 10.1016/j.steroids.2017.12.012 [DOI] [PubMed] [Google Scholar]
- 8.Cao R, Wang L, Wang H, et al. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- 9.Bian EB, Li J, Xie YS, Zong G, Li J, Zhao B. LncRNAs: new players in gliomas, with special emphasis on the interaction of lncRNAs With EZH2. J Cell Physiol. 2015;230(3):496–503. doi: 10.1002/jcp.24549 [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Hu L, Liang Y, et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16(1):150. doi: 10.1186/s12943-017-0715-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu F, Zhang X, Yu Q, et al. LncRNA AWPPH inhibits SMAD4 via EZH2 to regulate bladder cancer progression. J Cell Biochem. 2018;119(6):4496–4505. doi: 10.1002/jcb.v119.6 [DOI] [PubMed] [Google Scholar]
- 12.Sun Z, He C, Xiao M, et al. LncRNA FOXC2 antisense transcript accelerates non-small-cell lung cancer tumorigenesis via silencing p15. Am J Transl Res. 2019;11(7):4552–4560. [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Su Z, Lu S, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233. doi: 10.1016/j.cca.2018.07.004 [DOI] [PubMed] [Google Scholar]
- 14.Xie M, Sun M, Zhu YN, et al. Long noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression. Oncotarget. 2015;6(32):33587–33601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang L, Wu Z, Meng X, Chu X, Huang H, Xu C. LncRNA HOXA-AS2 facilitates tumorigenesis and progression of papillary thyroid cancer by modulating the miR-15a-5p/HOXA3 axis. Hum Gene Ther. 2019;30(5):618–631. doi: 10.1089/hum.2018.109 [DOI] [PubMed] [Google Scholar]
- 16.Gao Y, Yu H, Liu Y, et al. Long non-coding RNA HOXA-AS2 regulates malignant glioma behaviors and vasculogenic mimicry formation via the MiR-373/EGFR axis. Cell Physiol Biochem. 2018;45(1):131–147. doi: 10.1159/000486253 [DOI] [PubMed] [Google Scholar]
- 17.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding J, Xie M, Lian Y, et al. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis. 2017;6(1):e288. doi: 10.1038/oncsis.2016.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Wu D, Liu J, et al. Downregulated lncRNA HOXA11-AS affects trophoblast cell proliferation and migration by regulating RND3 and HOXA7 expression in PE. Mol Ther Nucleic Acids. 2018;12:195–206. doi: 10.1016/j.omtn.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y, Ge Z, Zhang E, et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017;8(10):e3104. doi: 10.1038/cddis.2017.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rynkeviciene R, Simiene J, Strainiene E, et al. Non-coding RNAs in glioma. Cancers. 2019;11(1):17. doi: 10.3390/cancers11010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vecera M, Sana J, Lipina R, Smrcka M, Slaby O. Long non-coding RNAs in gliomas: from molecular pathology to diagnostic biomarkers and therapeutic targets. Int J Mol Sci. 2018;19(9):E2754. doi: 10.3390/ijms19092754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Wu D, Chen J, et al. Long non-coding RNA HOXA-AS2 promotes the migration, invasion and stemness of bladder cancer via regulating miR-125b/Smad2 axis. Exp Cell Res. 2019;375(1):1–10. doi: 10.1016/j.yexcr.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 25.Dong X, Fang Z, Yu M, et al. Knockdown of long noncoding RNA HOXA-AS2 suppresses chemoresistance of acute myeloid leukemia via the miR-520c-3p/S100A4 axis. Cell Physiol Biochem. 2018;51(2):886–896. doi: 10.1159/000495387 [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Lin X, Zhou S, Zhang P, Shao G, Yang Z. Long noncoding RNA HOXA-AS2 promotes non-small cell lung cancer progression by regulating miR-520a-3p. Biosci Rep. 2019;39(5):BSR20190283. doi: 10.1042/BSR20190283 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–134. doi: 10.1038/nm.4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamagishi M, Uchimaru K. Targeting EZH2 in cancer therapy. Curr Opin Oncol. 2017;29(5):375–381. doi: 10.1097/CCO.0000000000000390 [DOI] [PubMed] [Google Scholar]
- 29.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015;21(12):2007–2022. doi: 10.1261/rna.053918.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol Ther Nucleic Acids. 2016;5(11):e385. doi: 10.1038/mtna.2016.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu TP, Wang WY, Ma P, et al. Upregulation of the long noncoding RNA FOXD2-AS1 promotes carcinogenesis by epigenetically silencing EphB3 through EZH2 and LSD1, and predicts poor prognosis in gastric cancer. Oncogene. 2018;37(36):5020–5036. doi: 10.1038/s41388-018-0308-y [DOI] [PubMed] [Google Scholar]
- 32.Di W, Weinan X, Xin L, et al. Long noncoding RNA SNHG14 facilitates colorectal cancer metastasis through targeting EZH2-regulated EPHA7. Cell Death Dis. 2019;10(7):514. doi: 10.1038/s41419-019-1707-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lian Y, Li Z, Fan Y, et al. The lncRNA-HOXA-AS2/EZH2/LSD1 oncogene complex promotes cell proliferation in pancreatic cancer. Am J Transl Res. 2017;9(12):5496–5506. [PMC free article] [PubMed] [Google Scholar]
- 34.Jie W, Andrade KC, Lin X, Yang X, Yue X, Chang J. Pathophysiological Functions of Rnd3/RhoE. Compr Physiol. 2015;6(1):169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paysan L, Piquet L, Saltel F, Moreau V. Rnd3 in cancer: a review of the evidence for tumor promoter or suppressor. Mol Cancer Res. 2016;14(11):1033–1044. doi: 10.1158/1541-7786.MCR-16-0164 [DOI] [PubMed] [Google Scholar]
- 36.Liu B, Lin X, Yang X, et al. Downregulation of RND3/RhoE in glioblastoma patients promotes tumorigenesis through augmentation of notch transcriptional complex activity. Cancer Med. 2015;4(9):1404–1416. doi: 10.1002/cam4.2015.4.issue-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu B, Dong H, Lin X, et al. RND3 promotes Snail 1 protein degradation and inhibits glioblastoma cell migration and invasion. Oncotarget. 2016;7(50):82411–82423. doi: 10.18632/oncotarget.12396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Q, Dong H, Li Y, et al. Small GTPase RHOE/RND3, a new critical regulator of NF-κB signalling in glioblastoma multiforme? Cell Prolif. 2019;22:e12665. [DOI] [PMC free article] [PubMed] [Google Scholar]