ABSTRACT
B7-H4, an immune suppressive member of the B7 family, is highly expressed in a wide variety of human malignancies making it an attractive immunotherapeutic target. However, the association between B7-H4 expression in the tumor microenvironment and the immune infiltrate has not been comprehensively examined. To evaluate the immune tumor microenvironment, we analyzed epithelial ovarian tumors from 28 patients using flow cytometry, immunohistochemistry, functional, and genomic analyses. We determined B7-H4 expression patterns and compared the immune infiltrates of tumors with high and low surface expression of B7-H4. Frequencies and phenotypes of tumor and immune cells were determined using multiple flow cytometry panels. Immunohistochemistry was used to analyze cellular infiltration and location. Publicly available datasets were interrogated to determine intratumoral cytokine and chemokine expression. We found that B7-H4 was predominantly expressed by tumor cells in the epithelial ovarian tumor microenvironment. Surface expression of B7-H4 on tumor cells was correlated with higher levels of infiltrating mature antigen-presenting cells. Further, expression of CXCL17, a monocyte and dendritic cell chemoattractant, correlated strongly with B7-H4 expression. T cells expressed activation markers, but T cells expressing a combination of markers associated with T cell activation/exhaustion phenotype were not prevalent. Overall, our data suggest that B7-H4 is associated with a pro-inflammatory tumor microenvironment.
KEYWORDS: B7-H4, epithelial ovarian cancer, CXCL17, immune infiltration, tumor infiltration
Introduction
Due to nonspecific symptoms and limited screening capabilities, the majority of ovarian cancer patients present with advanced disease,1 resulting in low 5-year survival at under 30% (National Cancer Institute, SEER Database). Patients with late stage epithelial ovarian cancer typically undergo a cytoreductive surgery to remove as much tumor mass as possible, coupled with platinum-based chemotherapy. High-grade serous ovarian cancers are the most common ovarian cancer histological subtype and have the worst prognosis but show high levels of sensitivity to platinum-based chemotherapy and other DNA damaging agents. Unfortunately, resistance to these therapies emerges in 80–90% of patients.2 Other treatment options include olaparib, a PARP-inhibitor,3 and bevacizumab, a VEGF-blocking antibody; however, further investigation into therapies that could provide lasting remissions is needed.
The immune system plays an important role in combating ovarian cancer. A prominent total T cell infiltration,4 CD8+ T cell infiltration,5 and a high CD8+:Treg ratio5 are predictive of positive outcomes. Further, molecular classification of high-grade ovarian cancers revealed a subset of patients with high levels of immune involvement6 and better survival outcomes compared to other high-grade subsets.6,7 However, adoptive cell transfer8,9 and immune checkpoint blockade10 have shown limited success in ovarian cancer, with lower response rates than has been seen in other malignancies such as melanoma. Understanding the ovarian immune microenvironment will help tailor immune treatments to the disease, facilitating the rational development of combination therapies that may improve treatment outcomes.
B7-H4, encoded by the VTCN1 gene, is an inhibitory member of the B7 family of immunomodulatory molecules. B7-H4 has been proposed to bind with the Semaphorin 3a/Plexin A4/Neuropilin-1 complex.11 However, Ohaegbulam et al. (2017) did not observe interactions between B7-H4 and either Semaphorin 3a or Neuropilin-112 necessitating further investigation into the interaction. Early experiments where T cells were stimulated in the presence of B7-H4-Ig demonstrated B7-H4’s role in inhibiting T cell proliferation, cytokine production, and cytotoxicity.13–15 Prior to receptor identification, B7-H4 was demonstrated to bind to a receptor induced on T cells by concanavalin A13 or TCR stimulation.14 Li et al. (2018) reported that while expression of the B7-H4 receptor steadily declined after day 12 post tumor implantation on the majority of T cells, expression was maintained on a small subset of proliferating PD-1+ T cells with cytotoxic features.16
Several studies indicate that cytokines can modulate B7-H4 expression. B7-H4 expression has been shown to be promoted by IL-6 plus IL-1017–19 or TGFβ plus IL-10,20 and decreased by GM-CSF and IL-4.17 B7-H4 protein expression has been reported to be restricted in healthy tissues and consistently elevated in many malignancies21 including ovarian cancer.22 However, the expression pattern of B7-H4 on immune cells remains inconclusive, with some groups observing B7-H4 on resting immune populations,13 others only seeing expression with stimulation,14 and some not seeing expression regardless of stimulation.23 Multiple groups have reported the presence and functional relevance of B7-H4 expression on a highly immunosuppressive population of tumor-associated macrophages.17,18
B7-H4’s inhibitory activity and enhanced expression in tumor tissues makes it an attractive target for checkpoint blockade. Further, correlation of B7-H4 with inflammatory patterns could make this molecule useful as a biomarker for defining characteristics of the tumor immune microenvironment. Therefore, our goal was to examine the ovarian carcinoma tumor microenvironment using flow cytometry, immunohistochemistry (IHC), functional, and genomic analyses to create an in-depth profile of the microenvironment associated with B7-H4 expression.
Material and methods
Tumor and blood specimens
All human tissue and bloods were obtained through protocols approved by the institutional review board (University Health Network Research Ethics Board). Surgical specimens were obtained from the UHN Biospecimen Program. Written informed consent was obtained from all donors.
Tumor digestion
Tumors were mechanically dissociated into pieces less than 1 mm in diameter and resuspended in enzymatic digestion media consisting of IMDM (Lonza) supplemented with 1 mg/mL collagenase type IV (Sigma), 10 µg/mL DNase I (Pulmozyme, Roche), 100 units/mL penicillin, 100 µg/mL streptomycin (Lonza), 10 µg/mL gentamicin sulfate (Lonza), 2 mM L-glutamine (Lonza), and 1.25 µg/mL amphotericin B. Tumor suspension was incubated for two 30 min incubations under rotation at 37°C with mechanical dissociation on the gentleMACS dissociator (Miltenyi Biotec) using the programs for soft human tumors before, in between, and after incubations. Single cell suspension was washed 3 times with wash media consisting of PBS supplemented with 10% FCS, 100 units/mL penicillin, and 100 µg/mL streptomycin (Lonza). All centrifugations were done at slow speed (1000rpm).
Ex vivo flow cytometry staining
Staining was completed at 4°C. Fc receptors were blocked with Fc block (BD Biosciences) or media supplemented with 10% human serum for 30 min prior to staining for surface expression. Cells were washed with PBS prior to staining with fixable viability dye (eBioscience) according to manufacturer’s protocol. Cells were washed with FACS buffer and fixed in 2% paraformaldehyde for 30 min. Intracellular staining for TIA-1, GzmB, and IFNγ was performed after fixation with 2% paraformaldehyde using permeabilization buffer (eBioscience) according to the manufacturer’s protocol. Information on antibodies used for staining can be found in Supplementary Table S1.
Flow cytometric gating strategies
For staining of fresh samples from ovarian tumors, markers that exhibited a negative population were gated according to patient-matched FMOs (lineage-defining markers, total expression of T cell inhibitory and activation markers). For markers that exhibited shifts in expression levels (APC inhibitory and activating markers), gMFI was normalized to control PBMCs from a hemochromatosis donor draw run in parallel according to the following formula: (gMFIsample–gMFIFMO)/(gMFIPBMCs). These measures were taken to control for variability introduced due to tumor sample processing, staining, and data acquisition being completed on different days as a result of the requirement for fresh tissue.
In vitro cell culture
T cells were cultured ex vivo in complete media consisting of IMDM supplemented with 10% human serum, 25mM HEPES (Lonza), 100 units/mL penicillin, 100 µg/mL streptomycin (Lonza), 10 µg/mL gentamicin sulfate (Lonza), 5.5 × 10−5 M β-mercaptoethanol (Gibco), and 2mM L-glutamine (Lonza). T cell marker expression was assessed following 72 h in vivo expansion. T cells were sorted out of tumor single cell suspensions using a CD3+ collection kit (Stemcell Technologies) according to the manufacturer’s instructions. CD3+ cells (2x105/96-well) were plated in complete media and stimulated with 1 µg/mL platebound anti-CD3 (clone OKT3) and 1 µg/mL soluble anti-CD28. Cells were harvested at 72 h as previously described.
Immunohistochemistry
Tumor specimens were fixed in 10% formalin solution (VWR), processed, and embedded in paraffin. Sections (4.5 µm) were dewaxed, rehydrated, and peroxidase activity was blocked with 3% hydrogen peroxide solution. In cases where two antibody clones were used to detect an antigen, three sample cases were stained with both antibody clones to ensure consistency in the results.
Antigen was retrieved with heat treatment and either 10mM sodium citrate (pH 6.0) (anti-B7-H3, anti-CD8 (clone C8/144B), antiCD3 (clone 2GV6), anti-CD20, anti-FoxP3), Tris-EDTA (pH 9.0) (anti-B7-H4, anti-CD8 (clone4B11)), or 1% pepsin (pH 2.0) (anti-CD3 (polyclonal)) prior to incubation in blocking solution. Primary antibodies used were: anti-B7-H4 (D1M8I), anti-B7-H3 (SP206), anti-CD3 (clone 2GV6 or polyclonal), anti-CD8 (clone C8/144B or 4B11), anti-FoxP3 (clone mAb22510 or 236A/E7), anti-CD20 (clone EP459Y or L26), and anti-CD68 (KP1). Slides were scanned using a Nanozoomer 2.0HT (Hamamatsu Photonics) and cell number quantification (CD3, CD8, FoxP3, CD20, CD68) and expression area quantification (B7-H4, B7-H3) was done using Halo analysis software (v2.0.1145.14).
Scoring of immune cell infiltration density
Stained slides were blinded and scored on a 5-point scale for the level of immune cell infiltration into epithelial or stromal areas in relation to range of infiltration of stained cohort according to the following scale:
1 – no positive events found on slide
2 – rare positive events observed
3 – low density of infiltration
4 – medium density of infiltration
5 – high density of infiltration
Cell lines
SK-OV-3 [SKOV-3; SKOV3] (ATCC HTB-77) and SK-BR-3 [SKBR3] (ATCC HTB-30) cell lines were cultured in McCoy’s 5A media (Gibco) supplemented with 10% FCS, 100 units/mL penicillin, and 100 µg/mL streptomycin (Lonza). OVCAR-3 [OVCAR3] (ATCC HTB-161) were cultured in RPMI-1640 (Gibco) supplemented with 20% FCS, 1mM sodium pyruvate, 0.01mg/mL bovine insulin, 100 units/mL penicillin, and 100 µg/mL streptomycin (Lonza). SK-BR-3 cells were gifted from the lab of Dr. Hal Berman, SK-OV-3 and OVCAR-3 cells were gifted from the lab of Dr. Tak Mak.
Cytokine stimulation of cell lines
Cell lines were plated in 24-well plates at 105 cells/well in complete media supplemented with cytokines (30ng/mL IL-6, 30ng/mL IL-10, 50ng/mL TGFβ, 10ng/mL IFNβ, 10ng/mL IFNα2, 10ng/mL IFNγ) for 24 h. Cell lines were plated in 96-well plates at 104 cells/well in complete media and stimulated with CXCL17 (10ng/mL, 30ng/mL, 100ng/mL, 300ng/mL for 48 h). Cells were harvested with Versene (Gibco) and stained according to the above protocol.
RNA isolation from OCT-embedded tissues
OCT-embedded tissues were sectioned using a cryotome into RNAse/DNAse-free tubes. RNA was isolated from frozen tissue sections by Trizol/chloroform extraction.
qRT-PCR
cDNA was reverse transcribed from RNA using qScript cDNA SuperMix (Quantabio) according to the manufacturer’s protocol. All qRT-PCR reactions were run using Perfecta SYBR Green FastMix with an initial 2 min 95°C incubation, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Genes were amplified with primers reported in PrimerBank24 and all primers were blasted to ensure specificity with reaction conditions used. Primer sequences used can be found in Supplementary Table S2.
Statistical analysis
Linear regressions, two-tailed Mann–Whitney U tests, and Mantel-Cox survival analyses done on flow cytometric and clinical data were calculated using GraphPad Prism Version 5.0c. One-way ANOVA with pairwise comparisons done by Tukey’s T-tests and likelihood-ratio tests to interrogate TCGA dataset (ovarian serous cystadenocarcinoma) were done through cBioPortal25,26 and with R software (R version 3.4.0). P values less than or equal to 0.05 were deemed significant (* p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001). Additional detail on statistical methodology used for ovarian cancer expression subtyping, differential analysis, and clustering can be found in the Supplementary Methods.
Results
B7-H4 is expressed by tumor cells in epithelial ovarian tumors
Interrogation of the publicly available TCGA ovarian serous cystadenocarcinoma dataset revealed that B7-H4 mRNA is differentially expressed in ovarian cancer subtypes as defined by Chen et al. (2018).27 Particularly high levels of B7-H4 were expressed in the immunoreactive/C2 and differentiated/C4 subsets, which both have high levels of infiltrating T cells (Figure 1(a)). Because the expression of B7-H4 showed notable correlations with subtypes of ovarian cancer that have higher levels of immune involvement, we sought to identify inflammatory patterns associated with B7-H4 expression. We evaluated tumor samples from primary debulking surgeries of 28 patients with epithelial ovarian cancer by flow cytometry, qRT-PCR, and IHC (Supplementary Table S3).
Figure 1.
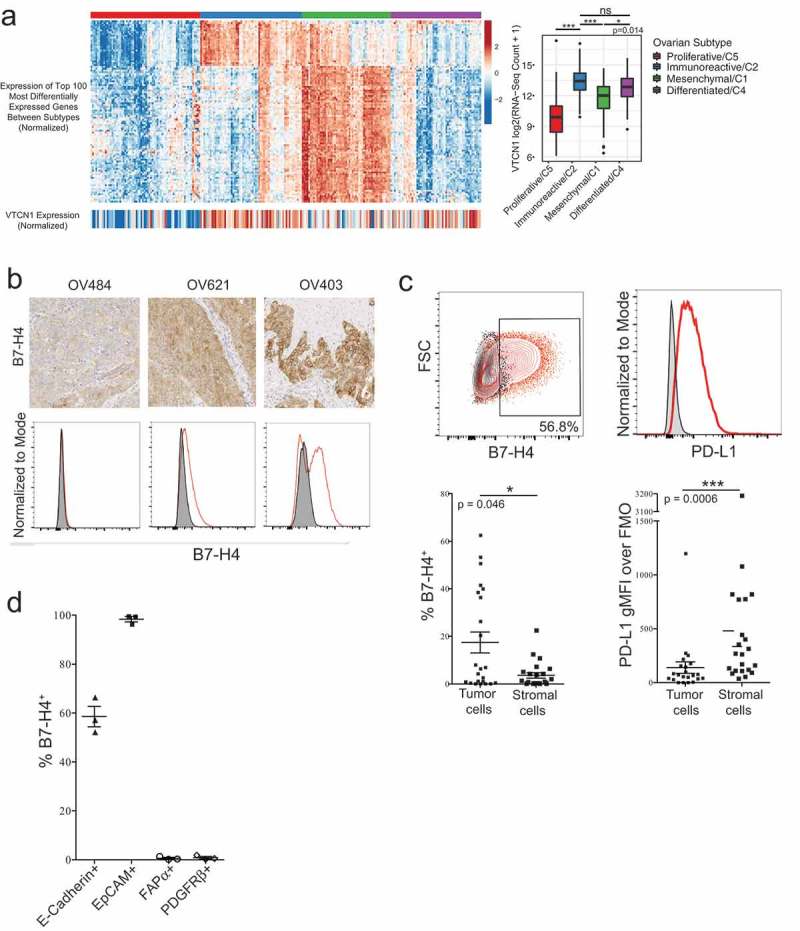
B7-H4 is expressed in the cytoplasm and on the surface of tumor cells but not stromal cells in epithelial ovarian cancer. B7-H4 mRNA expression in the TCGA ovarian serous cystadenocarcinoma dataset according to consensus molecular subtypes. Statistical significance was determined by ANOVA with Tukey’s test for pairwise comparisons (a). B7-H4 expression from three epithelial ovarian tumors assessed by both IHC and flow cytometry. Flow cytometry histograms show tumor sample (red) and matched FMO control (gray) (b). Expression of B7-H4 (p = 0.046) and PD-L1 (p = 0.0006) by the tumor and stromal compartments. Statistical significance was determined by Mann–Whitney U test (c). Expression of B7-H4 on CD45- cells expressing E-Cadherin, EpCAM, FAPα, or PDGFRβ (d).
We examined the localization of the B7-H4 protein by IHC in this cohort of samples. We observed both cytoplasmic and membrane B7-H4 staining in tumor cells but did not detect expression on stromal cells by IHC (Figure 1(b)). By distinguishing between tumor and stromal cells using the level of B7-H3 in the CD45− compartment (as described in MacGregor et al., in preparation), we found that B7-H4 was more frequently expressed by the tumor cell compartment (Figure 1(c)). When nonimmune cells were stained for B7-H4, epithelial markers (EpCAM, E-Cadherin), and stromal markers (PDGFRβ, FAPα), we observed that B7-H4 was co-expressed with EpCAM and E-Cadherin, but not FAPα or PDGFRβ (Figure 1(d)) confirming that B7-H4 expression is restricted to the tumor.
Unlike B7-H4 expression, which was confined to the tumor cell compartment in the epithelial ovarian tumors examined (Figure 1(b–d)), PD-L1 expression was more highly expressed in the stromal compartment (Figure 1(c)). The frequency of co-expression of B7-H4 and PD-L1 was low, and the total expression of B7-H4 and PD-L1 was not significantly correlated (R2 = 0.033;p = 0.41) (Supplementary Figure S1). The differences in the expression patterns of B7-H4 and PD-L1 along with the low frequencies of co-expression indicate that B7-H4 and PD-L1 expression are not co-regulated in EOC.
While B7-H4 has previously been correlated with poor survival and other negative prognostic indicators,21 B7-H4 surface expression on tumor cells was not significantly associated with overall survival or time to recurrence in our cohort of patients with EOC (Supplementary Figure S2). However, our sample size is small and further investigation into the role of B7-H4 expression in different compartments is required before firm conclusions could be made.
Tumors with high B7-H4 expression have higher proportions of infiltrating APCs
To determine if high surface expression of B7-H4 on EOC cells correlated with particular patterns of immune cell infiltration, single cell suspensions were made from epithelial ovarian tumor samples and stained for lineage-defining markers. The frequency of T and B cells within the tumor did not differ between tumors with a low or high proportion of B7-H4-expressing tumor cells (Figure 2(a,b); Supplementary Figure S3A–C). However, the frequency of APCs (CD11c+HLA-DRhigh) was higher in tumors with more B7-H4 expression (Figure 2(c)). No differences were observed in the density of total T cells (CD3+), macrophages (CD68+), or B cells (CD20+) in either the intraepithelial tumor nests or the intrastromal regions between tumors with low or high B7-H4 expression (Supplementary Figure S3D). These data indicate that B7-H4 may be associated with alterations in the EOC TME affecting the recruitment or maturation of APCs but is not associated with differences in total lymphocyte recruitment.
Figure 2.
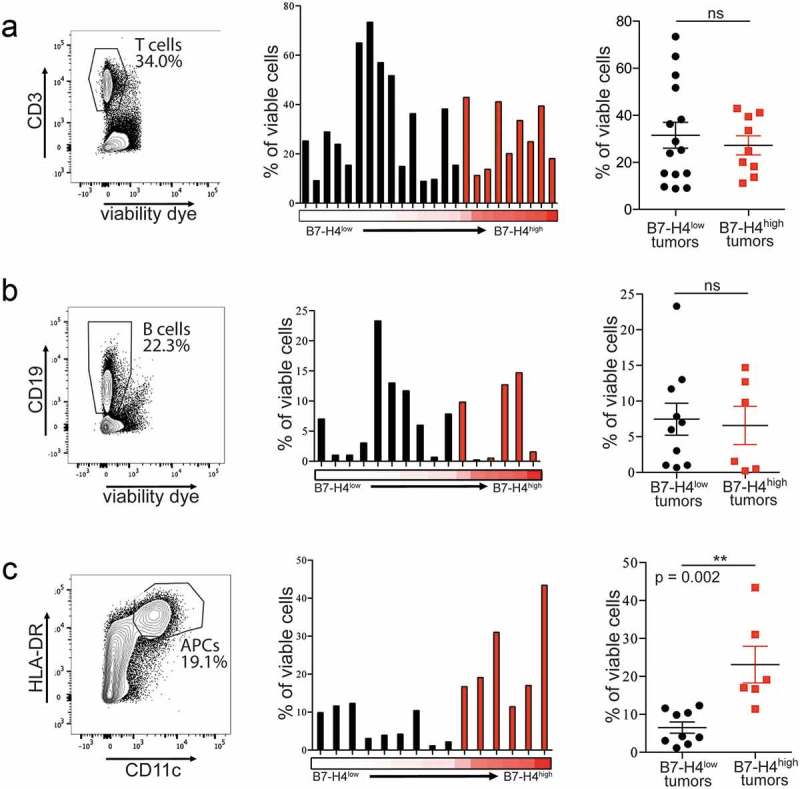
Tumors with surface expression of B7-H4 have comparable frequencies of infiltrating T and B cells, and higher frequencies of infiltrating APCs. T cell (a; n = 24), B cell (b; n = 16), and APC (c; n = 15) frequencies as percentage of viable cells from tumors expressing low (black) or high (red) levels of B7-H4 on the tumor cell surface. Spike plots represent the frequency of immune cells isolated from individual tumors with the proportion of tumor cells expressing B7-H4 represented by red heatmap below (low = 0%, high = 67%). Column plots show data from tumors expressing low (black) and high (red) levels of B7-H4 with mean ± SEM.
APCs infiltrating B7-H4-high tumors express higher levels of PD-L1 and PD-L2
APCs infiltrating EOC were stained for coinhibitory and costimulatory molecules to examine whether phenotypic differences correlated with B7-H4 surface expression on the tumor cells (Figure 3(a)). Contrary to previous reports showing high B7-H4 expression on an immunosuppressive TAM population,17,18 we found that the majority of samples exhibited undetectable (6/18) or very low (10/18) B7-H4 expression on cells from the APC compartment, with only two samples showing appreciable expression (2/18) (Supplementary Figure S4). Similarly, CLEC9a (n = 13), an activating receptor that mediates cross-presentation by myeloid DC,28–30 showed negligible or very low expression in all samples stained. Expression of B7-H4 or CLEC9a on APCs was not related to B7-H4 expression on tumor cells (Figure 3(b)). APCs from all patients expressed ICOS-L (n = 15), CD40 (n = 16), and CD86 (n = 17) at variable levels, but the variation in expression was not significantly associated with B7-H4 expression on tumor cells (Figure 3(b)). APCs from tumors expressing high levels of B7-H4 did exhibit significantly higher expression of PD-L1 (n = 15; p = 0.01 Mann–Whitney U test), and PD-L2 (n = 9; p = 0.02 Mann–Whitney U test) (Figure 3(b)) which could indicate a higher level of immune activation within the tumor.
Figure 3.
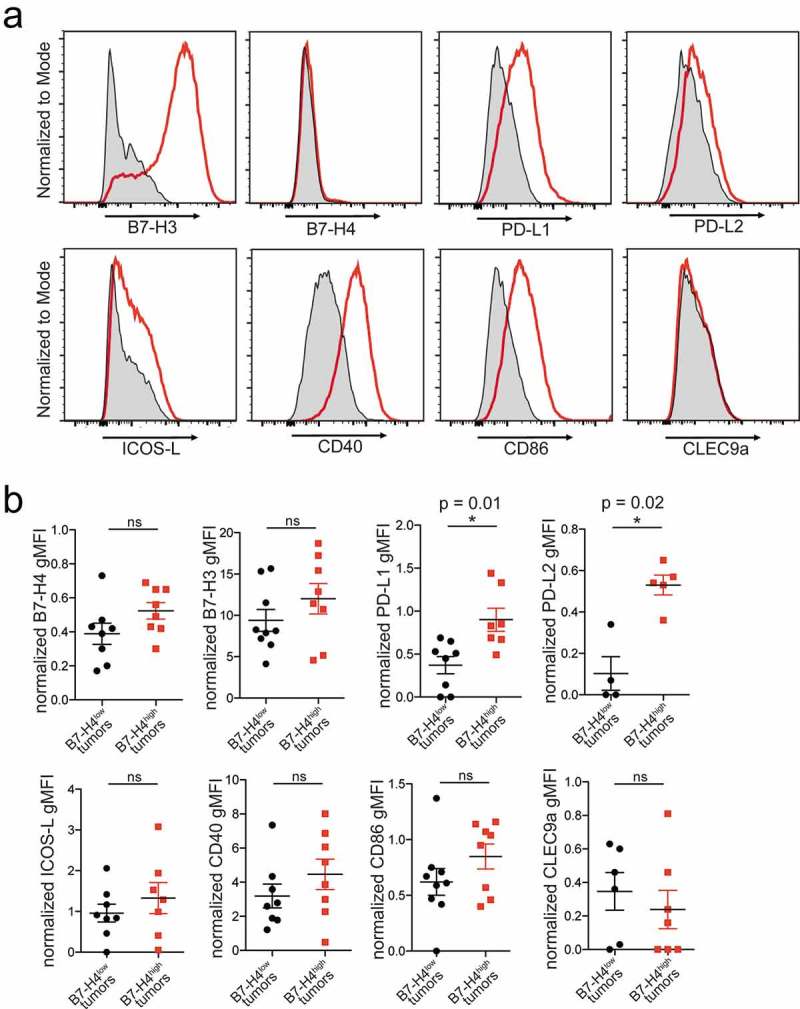
Expression of activating and inhibitory markers on APCs. APCs (CD11c+HLA-DRhigh) were stained for activating (ICOS-L (n = 15), B7-H3 (n = 17), CD40 (n = 16), CD86 (n = 17), CLEC9a (n = 13)) and inhibitory markers (B7-H4 (n = 16), B7-H3 (n = 17), PD-L1 (n = 15), PD-L2 (n = 9)). Flow cytometry histograms show tumor sample (red) and matched FMO control (gray) (a). Normalized marker expression on APCs from tumors with low (black) or high (red) proportions of tumor cells expressing B7-H4 with mean ± SEM. Significance was determined by Mann–Whitney U test (b).
Lymphocyte heterogeneity is not associated with B7-H4 expression by tumor cells
Because engagement with B7-H4 can suppress T cell activity,13–16 we analyzed the phenotype of T cells infiltrating EOC to determine whether we could observe changes in the T cell population that correlated with expression of B7-H4 on the tumor. An initial assessment of CD8 to CD4 ratios did not reveal differences in the proportions of CD8+ and CD4+ T cells isolated from tumors with different levels of surface B7-H4 expression (Figure 4(a)). We next evaluated the density of CD8+ and FoxP3+ T cell infiltration in either the epithelial tumor nests or stromal regions of the tumor. We did not find significant differences in CD8+, FoxP3+, or the ratio of CD8:FoxP3+ T cells infiltrating either region of B7-H4low or B7-H4high tumors (Figure 4(b)).
Figure 4.
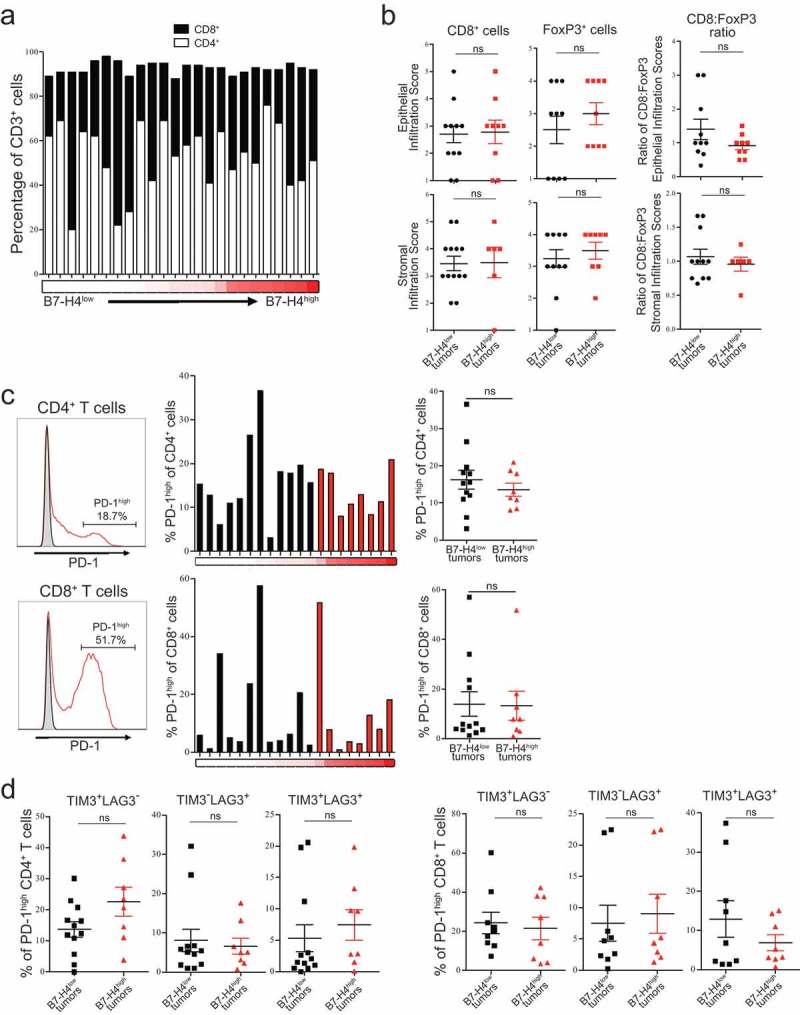
Expression of inhibitory markers on T cells infiltrating epithelial ovarian tumors. Proportions of CD4+ and CD8+ T cells isolated from tumors in order of proportion of B7-H4+ tumor cells (n = 24) (a). Infiltration scores of CD8+ and FoxP3+ cells, and the ratio of CD8+:FoxP3+ infiltration scores, from epithelial and stromal regions of tumors with low (black) and high (red) proportions of tumor cells expressing B7-H4 (b). Proportion of CD4+ and CD8+ T cells (n = 20) exhibiting high levels of PD-1 expression from tumors with low (black) and high (red) proportions of tumor cells expressing B7-H4. Flow cytometry histograms show tumor sample (red) and matched FMO control (gray) (c). Percentage of tumor cells expressing B7-H4 on the cell surface indicated by red heatmap below (low = 0%; high = 67%) (a,c). Co-expression of TIM3 and LAG3 on CD4+ and CD8+ T cells expressing high levels of PD-1. Data from tumors with low (black) or high (red) proportions of B7-H4−expressing tumor cells reported with mean ± SEM (d). Significance was determined by Mann–Whitney U test (B,C,D).
Because high levels of PD-1 expression have been shown to identify either an antigen-specific31 or exhausted T cell population,32 we analyzed the frequency of expression of this population. We did not observe differences in the proportion of CD4+ or CD8+ T cells expressing high levels of PD-1 in relation to B7-H4 expression on the tumor cells (Figure 4(c)). We further assessed the potential exhaustion of this population by analyzing the co-expression of high levels of PD-1 with TIM3 or LAG3 and did not find differences in the expression of either in relation to B7-H4 expression by the tumor cells (Figure 4(d)). Together, these data indicate that B7-H4 expression by tumor cells does not further exacerbate exhaustion by contributing to the upregulation of other exhaustion markers.
While most tumors did not have a high proportion of infiltrating T cells expressing LAG3, some tumors had T cells with marked LAG3 expression (Supplementary Figure S5). LAG3 expression was more frequently observed on the PD-1neg T cells; however, cases that had high proportions of PD-1high T cells demonstrated that LAG3 could be expressed on the PD-1high population. This data indicates that LAG3 is not selectively expressed on PD-1high T cells in EOC.
To examine the activation status of tumor-infiltrating T cells, we stained for activation and cytotoxicity markers (ICOS, TIA-1, GzmB, CD107a, GITR) and the SLAM family member 2B4 that has both activating and inhibitory functions (Figure 5(a)). While CD4+ T cells expressed ICOS and a subpopulation expressed GITR, no significant differences in expression were observed in relation to B7-H4 expression on tumor cells (Figure 5(b)). All markers except GITR were expressed at high frequencies by CD8+ T cells, but no significant differences were observed between CD8+ T cells isolated from B7-H4low and B7-H4high tumors (Figure 5(c)).
Figure 5.
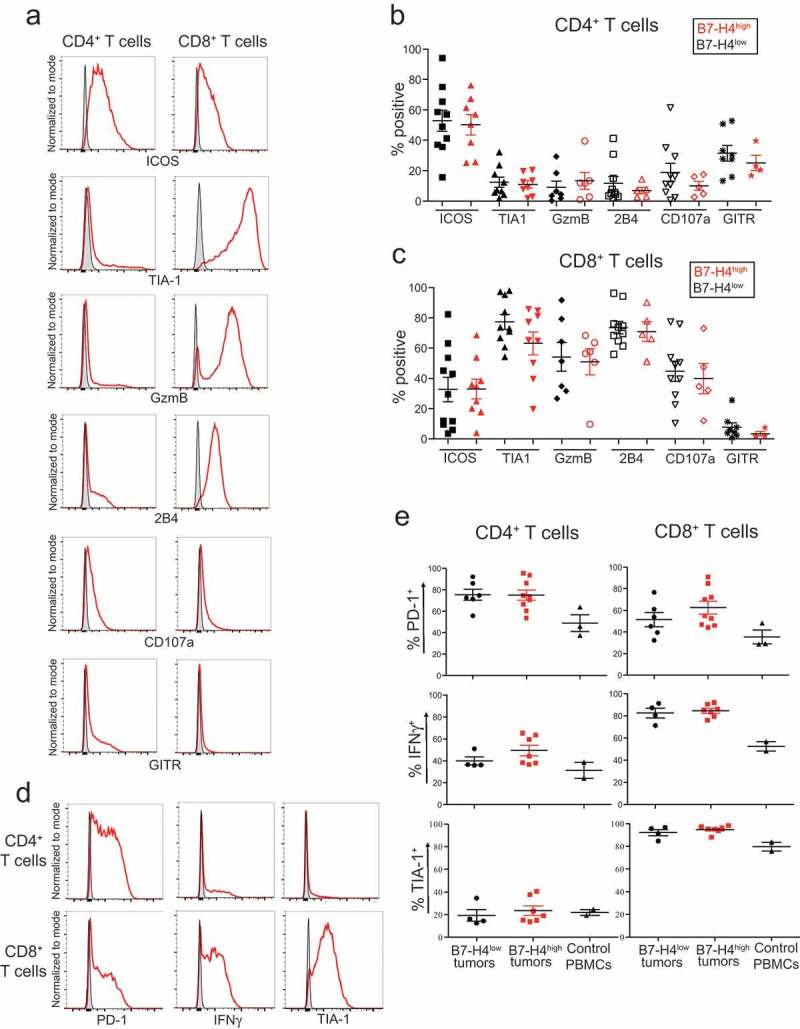
Expression of activation markers on T cells infiltrating ovarian tumors. ICOS, TIA-1, GzmB, 2B4, CD107a, and GITR staining on CD4+ and CD8+ T cells directly ex vivo (a). Summary of activation marker expression on CD4+ (b) and CD8+ (c) T cells from tumors with low (black) or high (red) proportions of B7-H4-expressing tumors cells reported with mean ± SEM. PD-1, IFNγ, and TIA-1 expression by T cells after 72 h stimulation with αCD3 and αCD28 stimulation (d). Column plots of PD-1, IFNγ, and TIA-1 expression by T cells after stimulation from tumors with low (black) or high (red) proportions of B7-H4-expressing tumors cells reported with mean ± SEM (e). Flow cytometry histograms show tumor sample (red) and matched FMO control (gray).
To investigate differences in response to stimulation, T cells isolated from epithelial ovarian tumors were stained for PD-1, TIA-1, and IFNγ after a 72 h stimulation with αCD3 and αCD28 (Figure 5(d)). In comparison to control PBMCs, tumor-infiltrating CD4+ TIL expressed higher levels of PD-1 and IFNγ, and CD8+ TIL expressed higher levels PD-1, IFNγ, and TIA-1 after stimulation; however, differences in marker expression were not correlated with B7-H4 expression by tumors (Figure 5(e)). Of note, high frequencies of CD4+ and CD8+ T cells from tumors readily produced IFNγ in response to stimulation suggesting that the T cells from these tumors are not fully functionally exhausted (Figure 5(e)). Together with the expression of PD-1, TIM3, and LAG3, these data suggest that neither the phenotype nor functionality of T cells infiltrating EOC is indelibly changed by the presence of B7-H4 in the TME.
We evaluated the phenotype of B cells infiltrating EOC using several markers to determine whether this population has changes in activation markers that correlate with expression of B7-H4 on tumor cells. The proportion of B cells present in the tumor varied widely (Figure 2(b)). CD19+ B cells co-expressing both CD20 and CD22 did not vary significantly in relation to B7-H4 expression by tumor cells (Supplementary Figure S6A,B). B cells expressing markers for naive (IgD+), activated (BTLA+), or memory (CD27+) populations (Supplementary Figure S6A) did not vary significantly in relation to B7-H4 expression by the tumor cells (Supplementary Figure S6B).
Cytokines previously shown to regulate B7-H4 expression do not alter B7-H4 expression on ovarian and breast cancer cell lines in vitro
Because B7-H4 expression has been shown to be regulated by IL-6, IL-10, TGFβ, and IFNγ on various cell lines,17–20,33 we looked at the expression of these cytokines in relation to B7-H4 mRNA expression in the TCGA dataset of mRNA expression from serous ovarian adenocarcinomas. IL6 (Spearman Rho = 0.14; p = 0.017), IL10 (Spearman Rho = 0.19; p = 0.00071), TGFB1 (Spearman Rho = 0.11; p = 6 x 10−2), and IFNG (Spearman Rho = 0.25; p = 1.3 x 10−5) all showed weak but significant positive correlations with expression of B7-H4 mRNA (Supplementary Figure S7A).
To determine whether expression of these cytokines could be responsible for eliciting B7-H4 expression on tumor cells we analyzed the effect of cytokine stimulation on cell lines SK-BR-3, OVCAR-3, and SK-OV-3. Cell lines were stimulated for 24 h with cytokines previously shown to elicit B7-H4 expression (TGFβ, IL-6, IL-10, IFNγ) as well as type I interferon cytokines because of their ability to upregulate B7 family member PD-L1.34 Cells were stained for class I HLA to confirm responsiveness to interferon treatments. SK-BR-3 and OVCAR3 cells expressed low surface levels of B7-H4, but SK-OV-3 cells did not express B7-H4 on the cell surface; however, all cell lines expressed some level of intracellular B7-H4, with SK-BR-3 showing the highest level of expression (Supplementary Figure S7B). Cytokine stimulation failed to increase surface or intracellular expression of B7-H4 in any cell lines, and some treatments even reduced B7-H4 expression slightly (Supplementary Figure S7B).
The low Spearman Rho of the correlations between B7-H4 mRNA and the expression of the different cytokines, along with the failure of these cytokines to induce B7-H4 expression alone, suggest that other factors are required in conjunction with cytokine stimulation to elicit B7-H4 expression.
B7-H4 mRNA expression correlates with different chemokine milieus in tumors
To gain insights into the biology relating to B7-H4, we examined the TCGA dataset of ovarian serous cystadenocarcinoma samples to identify genes that correlated with B7-H4 mRNA expression. B7-H4 mRNA expression positively correlated with multiple monocyte-attracting chemokines including CCL2, CCL4, CCL5, CCL8, and CXCL17, as well as the T cell chemoattractants CXCL10 and CXCL11 (Supplementary Figure S8). In addition, ACKR2 expression strongly correlated with B7-H4 mRNA expression (Figure 6(a,b)). ACKR2, atypical chemokine receptor 2, promiscuously binds almost all inflammatory CC-type chemokines leading to their internalization and degradation.35–37 Together, these expression data indicate that B7-H4 mRNA expression correlates with increased regulation of CC-type chemokines, which would selectively increase the impact of the CXCL-type chemokines expressed.
Figure 6.
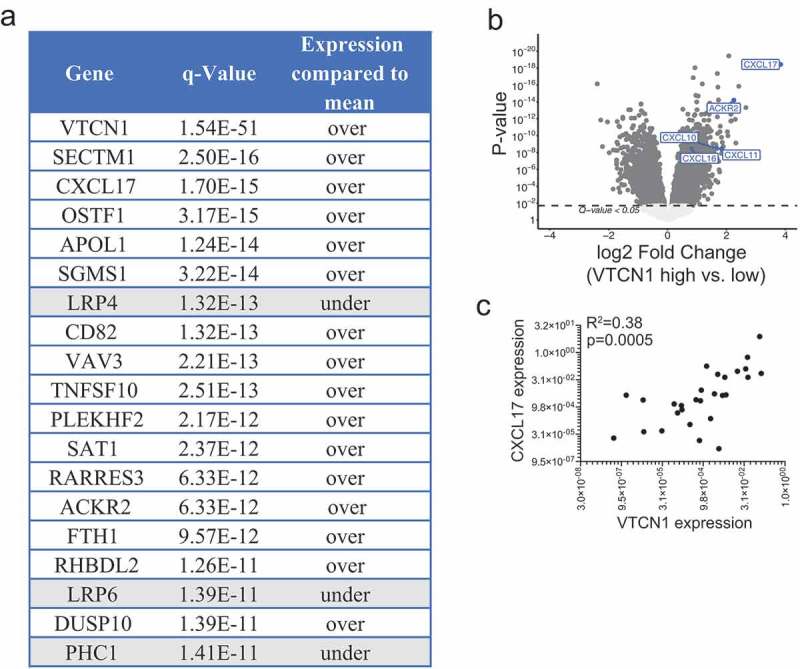
B7-H4 mRNA expression by tumor cells strongly correlates with CXCL17 mRNA expression. Most significant differentially regulated genes in ovarian serous cystadenocarcinoma tumors (TCGA dataset) enriched for B7-H4 mRNA expression (a,b). CXCL17 mRNA expression in relation to B7-H4 mRNA expression in our cohort of epithelial ovarian tumors. Significance was determined by linear regression (c).
B7-H4 mRNA expression strongly correlates with CXCL17 expression
In the TCGA ovarian serous cystadenocarcinoma dataset, B7-H4 mRNA expression strongly correlated with expression of CXCL17. CXCL17, an angiogenic chemokine that has been reported to selectively attract cells of the myeloid-lineage,38–42 exhibited a high level of enrichment in B7-H4 mRNA-enriched tumors (Figure 6(a–c)). CXCL17 was the only chemokine or chemokine receptor in the top 250 most significantly differentially expressed genes and showed a highly significant positive correlation with B7-H4 mRNA expression (Spearman Rho = 0.53; p = 2.2 x 10−16) (Supplementary Figure S8).
We confirmed higher levels of CXCL17 mRNA expression in relation to B7-H4 mRNA expression in our cohort of ovarian tumors (R2 = 0.38; p = 0.0005) (Figure 6(c)). To address the possibility that CXCL17 could be regulating B7-H4 expression by tumor cells, we stimulated cell lines with CXCL17 and assessed B7-H4 expression (Supplementary Figure S9). We did not observe differences in B7-H4 in relation to stimulation with CXCL17. These data demonstrate a correlation between CXCL17 and B7-H4 expression but suggest that CXCL17 is not directly inducing B7-H4 expression.
CXCL17 has been reported to induce VEGF expression43 suggesting that tumors high in CXCL17 may express higher levels of VEGF. Additionally, serum levels of B7-H4 and VEGF were positively correlated in patients with renal cell carcinoma.44 To see if B7-H4 expression was positively correlated with VEGF expression in ovarian cancer we interrogated the TCGA dataset; however, there was no significant association between B7-H4 mRNA and VEGFA expression (Spearman Rho = 0.01; p = 8.4 x 10−1) (Supplementary Figure S7A). These data suggest that, unlike in other contexts, expression of VEGF may not be positively correlated with B7-H4 expression in the EOC TME.
Discussion
In this study, we evaluated differences in frequency and phenotype of infiltrating immune populations in ovarian tumor samples that differentially expressed B7-H4. Our goal was to determine whether B7-H4 expression was associated with a particular inflammatory phenotype.
One of our aims was to distinguish the impact of membrane-bound B7-H4 from total expression of B7-H4. Because B7-H4 can be expressed in multiple cellular compartments and may carry out different biological functions depending on location of expression, we used flow cytometry and immunohistochemistry to quantify cell surface and total expression, respectively. Significant B7-H4 expression on tumor cell surface (36.0%; 9/25 tumors) was less frequent than B7-H4 expression in cytoplasm (85.3%; 29/34 tumors). Thus far, most reports of B7-H4 in human cancer have not differentiated between location of expression; however, this will be relevant when designing therapeutic modalities, as monoclonal blocking antibodies and CAR T cells would bind surface B7-H4 (and soluble forms), but would be unable to target tumors with intracellular expression.
Many previous reports have found a negative correlation between B7-H4 expression and survival outcomes, but differences in clinical impact of B7-H4 expression in different cellular locations have rarely been assessed.45 Therefore, instead of quantifying total B7-H4 expression we analyzed cell-surface B7-H4 expression in EOC by flow cytometry and compared patient survival. Under these conditions, an unexpected trend toward improved survival in patients with B7-H4high tumors was observed; however, the differences did not reach significance (Supplementary Figure S2). This is in agreement with our previous study in mouse models suggesting that B7-H4 expression by the tumor is important for anti-tumor immunity.33 This difference from the reported association between B7-H4 expression and worse survival outcomes could be due to our analysis focusing on surface expression of B7-H4, uncoupling the protein’s roles in immune cell inhibition through the binding of an unknown receptor13–15 with its potential cell-intrinsic roles in tumor cells46–48 but further investigation and a larger sample size is warranted.
Another aim was to evaluate if membrane-bound B7-H4 had an impact on the phenotype of T cells infiltrating tumors. We did not find any differences in T cell frequency (Figure 2(a)), phenotype (Figure 4; Figure 5(a–c)), or response to αCD3/αCD28 stimulation (Figure 5(d,e)) in relation to the level of B7-H4 expressed on the surface of tumor cells. These data indicate that B7-H4 expression in the tumor microenvironment does not exert detectable inhibitory effects on the bulk tumor-infiltrating T cell population. However, the expression of the B7-H4 receptor could not be evaluated.
While B7-H4 expression was not associated with differences in frequencies of infiltrating T or B cells, B7-H4 expression correlated with higher frequencies of APC infiltration in tumors (Figure 2(c)). Previous reports have found high expression of B7-H4 on tumor-infiltrating macrophages in ovarian cancer17 and glioma;18 however, we found no or low expression of B7-H4 on APCs (Figure 3; Supplementary Figure S4). This could be due to differences in tumor types18 or in antibody clones used.17
We examined the expression of multiple other activating and inhibitory molecules by APCs, including ICOS-L, B7-H3, PD-L1, PD-L2, CD40, CD86, and CLEC9a (Figure 3). APCs from B7-H4high tumors expressed significantly higher levels of PD-L1 and PD-L2 than APCs from B7-H4low tumors indicating that the microenvironment associated with high B7-H4 expression on tumors correlates with high expression of other B7 family members.
Because IFNγ has been shown to induce CXCL1741 and B7-H4 in a mouse model of breast cancer,33 we hypothesized that B7-H4 expression could be indicative of elevated levels of IFNγ expression. We found a weak positive correlation between IFNG and B7-H4 mRNA expression in the TCGA dataset of ovarian serous cystadenocarcinomas (Supplementary Figure S7A). However, when we stimulated breast and ovarian cancer cell lines in vitro with IFNγ we did not see any upregulation of B7-H4 expression (Supplementary Figure S7B). Together with reports that B7-H4 expression does not correlate with Th1 signature in endometrial tumors49 or T cell infiltration in lung cancer,50 these data indicate that B7-H4 expression is not induced by IFNγ.
Because, to our knowledge, there are not yet any studies assessing the co-expression of B7-H4 and PD-L1 in ovarian cancer, we looked at their expression in the TME of epithelial ovarian tumors. Previous studies have reported that B7-H4 and PD-L1 are both expressed by the tumor cell compartment in gastric cancer and NSCLC; however, while their expressions were positively correlated in gastric cancer51 they were rarely co-expressed by the same tumors in NSCLC.52 We found that B7-H4 was exclusively expressed by tumor cells (Figure 1(b–d)), but PD-L1 was more commonly expressed by the stromal compartment (Figure 1(c)). Further, we did not find any correlation between the level of PD-L1 expressed and the proportion of B7-H4+ cells in a tumor (Supplementary Figure S1). These data indicate differences in the biology of these two B7 family members, suggesting that therapeutics recognizing B7-H4 will target different cell types than those recognizing PD-L1 in epithelial ovarian cancer.
B7-H4 expression in the serum has been associated with higher serum VEGF in renal cell carcinoma.44 We interrogated the TCGA ovarian serous cystadenocarcinoma dataset to determine whether B7-H4 mRNA was correlated with VEGFA in ovarian cancer and found a weak positive correlation (Supplementary Figure S7A). However, we found a strong positive correlation between B7-H4 mRNA and CXCL17 (Figure 6), a chemokine whose expression has been correlated with VEGF and increased tumor growth.43 CXCL17 is able to attract monocytes, DCs, 38,39 macrophages,40 and MDSCs.41,42 In addition to CXCL17, expression of the CC-type chemokines CCL2, CCL4, CCL5, and CCL8 were positively associated with B7-H4 mRNA expression (Supplementary Figure S8). However, due to ACKR2’s ability to bind and internalize CC-type chemokines35 the CC-type chemokines might be selectively depleted, diminishing their chemoattractant activity. This scavenging receptor is one way to regulate the chemokine milieu in tumors, apart from differential expression of chemokines. This finding provides a molecular explanation for the increased frequency of infiltrating APCs and is consistent with the previously reported positive correlation between serum levels of B7-H4 and VEGF44 as CXCL17 has been reported to induce VEGF.
In summary, our data support a correlation between B7-H4 and CXCL17 expression resulting in an accumulation of mature APCs in B7-H4-expressing tumors. B7-H4 expression was not associated with PD-L1 expression in the tumor, and unlike PD-L1 its expression was not regulated by IFNγ. Furthermore, B7-H4 expression did not correlate with T cells expressing multiple inhibitory ligands including PD-1, TIM3, and LAG3. Taken together this suggests that B7-H4 is not associated with an inhibitory microenvironment.
Funding Statement
This work was supported by CIHR Foundation Award (CIHR-FDN 143220) and the CIHR China/Canada Team Grant (CCM-104887) awarded to PSO.
Acknowledgments
The authors would like to thank N.K. Grimshaw, A.R. Elford, C.S. Lo, J.Y. Yam, M. Fyrsta, J. Nie, and P.H. Yen for their technical expertise and support. We wholeheartedly thank the patients who have made this research possible by donating tumor tissue.
Author Contributions
Conception and design: H.L. MacGregor, S.Q. Crome, C. Robert-Tissot, P.S. Ohashi
Development of Methodology: H.L. MacGregor, L.T. Nguyen
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): P.A. Shaw, M.Q. Bernardini
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): H.L. MacGregor, C. Garcia-Batres, A. Sayad, H.K. Berman
Writing, review, and/or revision of the manuscript: H.L. MacGregor, A. Toker, C. Robert-Tissot, P.S. Ohashi
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): L.T. Nguyen, S.R. Katz, M.Q. Bernardini
Study supervision: P.S. Ohashi
Other (conducted experiments): H.L. MacGregor, C. Garcia-Batres, A. Elia
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:1–13. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 2.Bowtell DD, Böhm S, Ahmed AA, Aspuria P-J, Bast RC, Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15:668–679. doi: 10.1038/nrc4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim G, Ison G, McKee AE, Zhang H, Tang S, Gwise T, Sridhara R, Lee E, Tzou A, Philip R, et al. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res. 2015;21:4257–4261. doi: 10.1158/1078-0432.CCR-15-0887. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 5.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, Jungbluth AA, Frosina D, Gnjatic S, Ambrosone C, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci. 2005;102:18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 7.Helland Å, Anglesio MS, George J, Cowin PA, Johnstone CN, House CM, Sheppard KE, Etemadmoghadam D, Melnyk N, Rustgi AK, et al. Deregulation of MYCN, LIN28B and LET7 in a molecular subtype of aggressive high-grade serous ovarian cancers. PLoS One. 2011;6:1–9. doi: 10.1371/journal.pone.0018064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita K, Ikarashi H, Takakuwa K, Kodama S, Tokunaga A, Takahashi T, Tanaka K. Prolonged disease-free period in patients with advanced epithelial ovarian cancer after adoptive transfer of tumor-infiltrating lymphocytes. Clin Cancer Res. 1995;1:501–507. [PubMed] [Google Scholar]
- 9.Aoki Y, Takakuwa K, Kodama S, Tanaka K, Takahashi M, Tokunaga A, Takahashi T. Use of adoptive transfer of tumor-infiltrating lymphocytes alone or in combination with cisplatin-containing chemotherapy in patients with epithelial ovarian cancer. Cancer Res. 1991;51:1934–1939. [PubMed] [Google Scholar]
- 10.Ring KL, Pakish J, Jazaeri AA.. Immune checkpoint inhibitors in the treatment of gynecologic malignancies. Cancer J. 2016;22:101–107. doi: 10.1097/PPO.0000000000000179. [DOI] [PubMed] [Google Scholar]
- 11.Podojil JR, Chiang M-Y, Ifergan I, Copeland R, Liu LN, Maloveste S, Langermann S, Liebenson D, Balabanov R, Chi H, et al. B7-H4 modulates regulatory CD4+ T cell induction and function via ligation of a Semaphorin 3a/Plexin A4/Neuropilin-1 complex. J Immunol. 2018;201:897–907. doi: 10.4049/jimmunol.1700811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohaegbulam KC, Liu W, Jeon H, Almo SC, Zang X. Tumor-expressed immune checkpoint B7x promotes cancer progression and antigen-specific CD8 T cell exhaustion and suppressive innate immune cells. Oncotarget. 2017;8:82740–82753. doi: 10.18632/oncotarget.21098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasad DVR, Richards S, Mai XM, Dong C. B7S1, a novel B7 family member that negatively regulates T cell activation. Immunity. 2003;18:863–873. doi: 10.1016/S1074-7613(03)00147-X. [DOI] [PubMed] [Google Scholar]
- 14.Sica GL, Choi IH, Zhu G, Tamada K, Wang SD, Tamura H, Chapoval AI, Flies DB, Bajorath J, Chen L. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity. 2003;18:849–861. doi: 10.1016/S1074-7613(03)00152-3. [DOI] [PubMed] [Google Scholar]
- 15.Zang X, Loke P, Kim J, Murphy K, Waitz R, Allison JP. B7x: a widely expressed B7 family member that inhibits T cell activation. Proc Natl Acad Sci U S A. 2003;100:10388–10392. doi: 10.1073/pnas.1434299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Lee Y, Li Y, Jiang Y, Lu H, Zang W, Zhao X, Liu L, Chen Y, Tan H, et al. Co-inhibitory molecule B7 superfamily member 1 expressed by tumor-infiltrating myeloid cells induces dysfunction of anti-tumor CD8+ T cells. Immunity. 2018; 48: 773-786. doi:10.1016/j.immuni.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Kryczek I, Zou L, Rodriguez P, Zhu G, Wei S, Mottram P, Brumlik M, Cheng P, Curiel T, Myers L, et al. B7-H4 expression identifies a novel suppressive macrophage population in human ovarian carcinoma. J Exp Med. 2006;203:871–881. doi: 10.1084/jem.20050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Y, Ye H, Qi Z, Mo L, Yue Q, Baral A, Hoon DSB, Vera JC, Heiss JD, Chen CC, et al. B7-H4(B7x)-mediated cross-talk between glioma-initiating cells and macrophages via the IL6/JAK/STAT3 pathway lead to poor prognosis in glioma patients. Clin Cancer Res. 2016;22:2778–2790. doi: 10.1158/1078-0432.CCR-15-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kryczek I, Wei S, Zhu G, Myers L, Mottram P, Cheng P, Chen L, Coukos G, Zou W. Relationship between B7-H4, regulatory T cells, and patient outcome in human ovarian carcinoma. Cancer Res. 2007;67:8900–8905. doi: 10.1158/0008-5472.CAN-07-1866. [DOI] [PubMed] [Google Scholar]
- 20.Cao Q, Wang Y, Zheng D, Sun Y, Wang Y, Lee VWS, Zheng G, Tan TK, Ince J, Alexander SI, et al. IL-10/TGF-b–modified macrophages induce regulatory T cells and protect against adriamycin nephrosis. J Am Soc Nephrol. 2010;21:933–942. doi: 10.1681/ASN.2009060592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacGregor HL, Ohashi PS. Molecular pathways: Evaluating the potential for B7-H4 as an immunoregulatory target. Clin Cancer Res. 2017;23:2934-2941. doi: 10.1158/1078-0432.CCR-15-2440. [DOI] [PubMed] [Google Scholar]
- 22.Tringler B, Liu W, Corral L, Torkko KC, Enomoto T, Davidson S, Lucia MS, Heinz DE, Papkoff J, Shroyer KR. B7-H4 overexpression in ovarian tumors. Gynecol Oncol. 2006;100:44–52. doi: 10.1016/j.ygyno.2005.08.060. [DOI] [PubMed] [Google Scholar]
- 23.Lee JS, Scandiuzzi L, Ray A, Wei J, Hofmeyer KA, Abadi YM, Loke P, Lin J, Yuan J, Serreze DV, et al. B7x in the periphery abrogates pancreas-specific damage mediated by self-reactive CD8 T cells. J Immunol. 2012;189:4165–4174. doi: 10.4049/jimmunol.1201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: A PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:1144–1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the {cBioPortal.}. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen GM, Kannan L, Geistlinger L, Kofia V, Safikhani Z, Gendoo DMA, Parmigiani G, Birrer M, Haibe-Kains B, Waldron L. Consensus on molecular subtypes of high-grade serous ovarian carcinoma. Clin Cancer Res. 2018;24:5037–5047. doi:10.1158/1078-0432.CCR-18-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreibelt G, Klinkenberg LJJ, Cruz LJ, Tacken PJ, Tel J, Kreutz M, Adema GJ, Brown GD, Figdor CG, de Vries IJ. The C-type lectin receptor CLEC9A mediates antigen uptake and (cross-)presentation by human blood BDCA3+ myeloid dendritic cells. Blood. 2012;119:2284–2292. doi: 10.1182/blood-2011-08-373944. [DOI] [PubMed] [Google Scholar]
- 29.Zelenay S, Keller AM, Whitney PG, Schraml BU, Deddouche S, Rogers NC, Schulz O, Sancho D, Reis e Sousa C. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J Clin Invest. 2012;122:1615–1627. doi: 10.1172/JCI60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iborra S, Izquierdo HM, Martínez-López M, Blanco-Menéndez N, Reis E Sousa C, Sancho D. The DC receptor DNGR-1 mediates cross-priming of CTLs during vaccinia virus infection in mice. J Clin Invest. 2012;122:1628–1643. doi: 10.1172/JCI60660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, Wunderlich JR, Mixon A, Farid S, Dudley ME, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahbar R, Lin A, Ghazarian M, Yau H-L, Paramathas S, Lang PA, Schildknecht A, Elford AR, Garcia-Batres C, Martin B, et al. B7-H4 expression by nonhematopoietic cells in the tumor microenvironment promotes antitumor immunity. Cancer Immunol Res. 2015;3:184–195. doi: 10.1158/2326-6066.CIR-14-0113. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonecchi R, Garlanda C, Mantovani A, Riva F. Cytokine decoy and. scavenger receptors as key regulators of immunity and inflammation. Cytokine. 201. 6;87:37–45. doi: 10.1016/j.cyto.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nibbs RJ, Wylie SM, Yang J, Landau NR, Graham GJ. Cloning and characterization of a novel promiscuous human beta-chemokine receptor D6. J Biol Chem. 1997;272:32078–32083. doi: 10.1074/jbc.272.51.32078. [DOI] [PubMed] [Google Scholar]
- 37.Bonini JA, Martin SK, Dralyuk F, Roe MW, Philipson LH, Steiner DF. Cloning, expression, and chromosomal mapping of a novel human CC-chemokine receptor (CCR10) that displays high-affinity binding for MCP-1 and MCP-3. DNA Cell Biol. 1997;16:1249–1256. doi: 10.1089/dna.1997.16.1249. [DOI] [PubMed] [Google Scholar]
- 38.Pisabarro MT, Leung B, Kwong M, Corpuz R, Frantz GD, Chiang N, Vandlen R, Diehl LJ, Skelton N, Kim HS, et al. Cutting edge: novel human dendritic cell- and monocyte-attracting chemokine-like protein identified by fold recognition methods. J Immunol. 2006;176:2069–2073. doi: 10.4049/jimmunol.176.4.2069. [DOI] [PubMed] [Google Scholar]
- 39.Hiraoka N, Yamazaki-Itoh R, Ino Y, Mizuguchi Y, Yamada T, Hirohashi S, Kanai Y. CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology. 2011;140:310–321. doi: 10.1053/j.gastro.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 40.Burkhardt AM, Maravillas-Montero JL, Carnevale CD, Vilches-Cisneros N, Flores JP, Hevezi PA, Zlotnik A. CXCL17 is a major chemotactic factor for lung macrophages. J Immunol. 2014;193:1468–1474. doi: 10.4049/jimmunol.1400551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oka T, Sugaya M, Takahashi N, Takahashi T, Shibata S, Miyagaki T, Asano Y, Sato S. CXCL17 attenuates imiquimod-induced psoriasis-like skin inflammation by recruiting myeloid-derived suppressor cells and regulatory T cells. J Immunol. 2017;198:3897–3908. doi: 10.4049/jimmunol.1601607. [DOI] [PubMed] [Google Scholar]
- 42.Matsui A, Yokoo H, Negishi Y, Endo-Takahashi Y, Chun NAL, Kadouchi I, Suzuki R, Maruyama K, Aramaki Y, Semba K, et al. CXCL17 expression by tumor cells recruits CD11b+GR1highF4/80- cells and promotes tumor progression. PLoS One. 2012;7:1–11. doi:10.1371/journal.pone.0044080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinstein EJ, Head R, Griggs DW, Sun D, Evans RJ, Swearingen ML, Westlin MM, Mazzarella R. VCC-1, a novel chemokine, promotes tumor growth. Biochem Biophys Res Commun. 2006;350:74–81. doi: 10.1016/j.bbrc.2006.08.194. [DOI] [PubMed] [Google Scholar]
- 44.Fukuda T, Kamai T, Masuda A, Nukui A, Abe H, Arai K, Yoshida K-I. Higher preoperative serum levels of PD-L1 and B7-H4 are associated with invasive and metastatic potential and predictable for poor response to VEGF-targeted therapy and unfavorable prognosis of renal cell carcinoma. Cancer Med. 2016;5:1810–1820. doi: 10.1002/cam4.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyatake T, Tringler B, Liu W, Liu SH, Papkoff J, Enomoto T, Torkko KC, Dehn DL, Swisher A, Shroyer KR. B7-H4 (DD-O110) is overexpressed in high risk uterine endometrioid adenocarcinomas and inversely correlated with tumor T-cell infiltration. Gynecol Oncol. 2007;106:119–127. doi: 10.1016/j.ygyno.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Cai L, Zhang G, Shen Y, Huang J. B7-H4 promotes tumor growth and metastatic progression in lung cancer by impacting cell proliferation and survival. Oncotarget. 2017;8:18861–18871. doi:10.18632/oncotarget.14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Wu H, Lu D, Li G, Sun C, Song H, Li J, Zhai T, Huang L, Hou C, et al. The costimulatory molecule B7-H4 promote tumor progression and cell proliferation through translocating into nucleus. Oncogene. 2013;32: 5347-5358. doi:10.1038/onc.2012.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Wang L, Wang W, Zhao L, Shan B. B7-H4 facilitates proliferation of esophageal squamous cell carcinoma cells through promoting interleukin-6/signal transducer and activator of transcription 3 pathway activation. Cancer Sci. 2016;107:944–954. doi: 10.1111/cas.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bregar A, Deshpande A, Grange C, Zi T, Stall J, Hirsch H, Reeves J, Sathyanarayanan S, Growdon WB, Rueda BR. Characterization of immune regulatory molecules B7-H4 and PD-L1 in low and high grade endometrial tumors. Gynecol Oncol. 2017;145:446–452. doi: 10.1016/j.ygyno.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Schalper KA, Carvajal-Hausdorf D, McLaughlin J, Altan M, Velcheti V, Gaule P, Sanmamed MF, Chen L, Herbst RS, Rimm DL. Differential expression and significance of PD-L1, IDO-1 and B7-H4 in human lung cancer. Clin Cancer Res. 201. 7;23:370–378. doi:10.1158/1078-0432.CCR-16-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geng Y, Wang H, Lu C, Li Q, Xu B, Jiang J, Wu C. Expression of costimulatory molecules B7-H1, B7-H4 and Foxp3+ Tregs in gastric cancer and its clinical significance. Int J Clin Oncol. 2015;20:273–281. doi: 10.1007/s10147-014-0701-7. [DOI] [PubMed] [Google Scholar]
- 52.Altan M, Pelekanou V, Schalper KA, Toki M, Gaule P, Syrigos K, Herbst RS, Rimm DL. B7-H3 expression in NSCLC and its association with B7-H4, PD-L1 and tumor-infiltrating lymphocytes. Clin Cancer Res. 2017;23:5202–5209. doi: 10.1158/1078-0432.CCR-16-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


