ABSTRACT
Immune checkpoints are intensively investigated as targets in cancer therapy. T-cell immunoreceptor with immunoglobulin (Ig) and ITIM domains (TIGIT) and its ligand poliovirus receptor (PVR) are recently emerging as novel promising targets in immunotherapy. Here, we show that high expression of PVR represents an independent prognostic marker being associated with poor outcome for breast cancer patients. Furthermore, PVR mRNA, as well as protein expression, is associated with more aggressive breast cancer subtypes such as HER2 positive and triple-negative breast cancer. In vitro, blocking TIGIT or PVR resulted in enhanced immune cell-mediated lysis of breast cancer cell lines SKBR-3, MDA-MB-231, MDA-MB-468, and BT549 and additionally increased the cytotoxic effects of a bispecific T cell engager BiTE® antibody construct targeting EGFR. Taken together, our data identify the immune checkpoint factor PVR as a novel prognostic marker in breast cancer and indicate that blocking the TIGIT-PVR axis might represent a novel therapeutic option for the treatment of breast cancer patients.
KEYWORDS: TIGIT, PVR, immunotherapy, breast cancer
Introduction
Escape of neoplastic cells from destruction mediated by the immune system represents one of the hallmarks of cancer.1 One mechanism of immune escape is the induction of an exhausted phenotype in effector lymphocytes and thereby prevention of an efficient tumor rejection.2,3 The inhibition of T-cell activation is regulated by several receptor/ligand systems involved in checkpoint control of T-cell effector functions and the concept of inhibiting these immune checkpoints to reactivate the cytotoxic phenotype has revolutionized the field of cancer immunotherapy. Recently, antibodies blocking the negative immune checkpoint molecules cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1) or PD-1 ligand 1 (PD-L1) were approved by the FDA for the treatment of various cancer entities due to remarkable survival benefits.4–6 But only a subset of cancer patients benefits from the treatment with blocking antibodies against the PD-1, PD-L1 or CTLA-4 immune checkpoint axis.7 Therefore, much effort is being directed to the therapeutic evaluation of additional immune checkpoints.8
T cell immunoreceptor with Ig and ITIM domains (TIGIT) and its ligand PVR (poliovirus receptor, CD155) have recently emerged as promising targets in immunotherapy.9 TIGIT is a type I transmembrane protein with an Ig-like variable extracellular domain expressed on activated and memory T cells, regulatory T cells, as well as natural killer (NK) and natural killer T cells (NKT).10,11
Initially, PVR has been identified as entry mediator for the poliovirus.12 Furthermore, as nectin-like molecule PVR participates in cell-cell adhesion via heterophilic and homophilic trans interactions in adherence junctions.13 In the immunologic context, on the one hand, PVR binds to the co-stimulatory receptor DNAX accessory molecule-1 (DNAM-1) resulting in an activated immune response. But, on the other hand, PVR’s binding affinity to the inhibitory receptor TIGIT is much higher and therefore TIGIT outcompetes the positive receptor DNAM-1 for its ligand PVR.14,15 Upon binding of PVR to TIGIT, TIGIT suppresses the immune response through its cytosolic immunoglobulin tail tyrosine-like phosphorylation motif and immunoreceptor tyrosine-based inhibitory motif.16 High expression of PVR could be associated with a poor survival in several cancer entities and the blockade of the TIGIT/PVR axis resulted in anti-tumor activity in a number of preclinical models.17–23 Based on those promising observations, the clinical evaluation of blocking TIGIT was initiated (NCT02794571, NCT03119428, NCT02913313).
Breast cancer (BC) was always considered to be a less immunogenic entity. Nevertheless, BC is a heterogeneous disease comprising several molecular subtypes and among them, triple-negative breast cancer (TNBC) has been shown to be the most sensitive to immune therapeutic intervention.24–27 But so far, only one phase III study in first-line treatment of triple-negative metastatic breast cancer showed a benefit for the checkpoint inhibitor atezolizumab.28 Based on these data, atezolizumab was recently approved by the FDA for TNBC patients. The majority of other clinical trials in pretreated patients have reported low response rates.29,30
In the present investigation, we explored the therapeutic potential of blocking the immune checkpoint receptor TIGIT or its ligand PVR in breast cancer. We show that a high expression of PVR is associated with a poor outcome for breast cancer patients. Blocking of TIGIT or PVR resulted in enhanced immune cell-mediated lysis of breast cancer cells and could additionally augment the cytotoxic effects of a bispecific T cell engager (BiTE®) antibody construct targeting EGFR.
Results
PVR is associated with the triple-negative breast cancer molecular subtype and poor survival in breast cancer patients
PVR mRNA levels were evaluated in 197 tumor samples of breast cancer patients using microarray data. Four different probesets (32699_s_at, 214443_at, 214444_s_at and 212662_at) corresponding to PVR were analyzed. Additionally, the mean value of all four probesets was calculated and included in further analyses. As mentioned in the Material and Methods section, for each probeset and the mean value, the cohort was divided into quartiles (Q) of similar size, representing low, moderate-low, moderate-high and high PVR levels. Correlations between PVR mRNA levels and clinicopathological factors showed a significant association of high PVR expression with higher grading (in 2 of 4 probesets), ER- (in all probesets and the mean value), PR-negativity (in 2 of 4 probesets and the mean value) and nodal involvement (2 of 4 probesets), whereas no correlation between PVR and stage, or HER2 status was found (supplementary table 1). Further, PVR mRNA levels from all probesets and the mean values were significantly associated with shorter overall survival (OS) (supplementary table 1), while only probesets 32699_s_at, 212662_at and the mean value correlated significantly with shorter recurrence-free interval (RFI) using cox regressions with the continuous data (p < .001 and p < .001, and p = .017, respectively; supplementary table 1). Here, in multivariate Cox regression analyses including clinical stage, nodal involvement, and molecular subtype, PVR remained an independent prognostic factor for overall survival (in 3 of 4 probesets and the mean value) and recurrence-free survival (in 2 of 4 probesets and the mean value). The mean PVR level could be also identified as an independent prognostic marker for the overall survival and recurrence-free interval in Cox regression analyses using quartiles (p = .004; HR 1.426, 95% CI 1.11–1.82 for OS and p = .037; HR 1.251; 95% CI 1.01–1.54 for RFI).
For probeset 32699_s_at, which showed the highest correlation with the PVR protein level as described in the next section, additional analyses were carried out and are represented in Figure 1. Here, cases with PVR levels lower than the median (Q1 and Q2) compared to those in Q3 and Q4 behaved similarly in survival analysis and were therefore combined. PVR mRNA levels were significantly higher in ER and PR negative (Figure 1(a, b); p < .001 and p = .01, respectively), tumors with overexpression of HER2 (Figure 1(c); p = .04) and poor histological differentiation (grading) (Figure 1(e), p = .002). Consequently, HER2 positive and triple-negative tumors, which represent the most aggressive BC subtypes, showed higher PVR mRNA levels in comparison with the luminal subtypes (Figure 1(d); p = .001). Kaplan-Meier analyses revealed a significant association of high PVR mRNA levels with shorter overall survival (p = .003) and shorter recurrence-free interval (p < .001) for the entire cohort (Figure 1(f,g), respectively). Further, in a multivariate Cox regression including clinical stage, nodal involvement, and molecular subtype, PVR remained an independent prognostic factor for recurrence-free survival (p = .004; HR 2.040; 95% CI 1.26–3.3) and overall survival (p = .033, HR 1.822, 95% CI 1.05–3.161).
Figure 1.
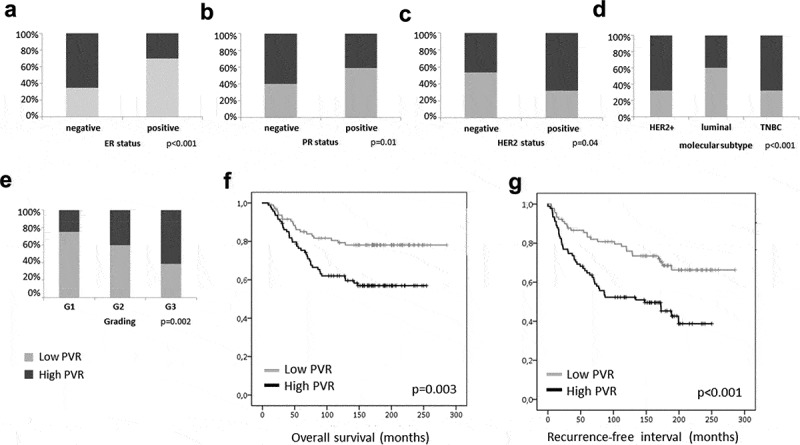
PVR mRNA levels in breast cancer tumor tissue. Correlation analyses of probeset 32699_s_at showed a significant association of high PVR mRNA levels with ER- and PR-negativity (a,b) and HER2 status (c). Furthermore, HER2 positive and triple-negative tumors representing the most aggressive breast cancer subtypes showed higher PVR mRNA levels in comparison with the luminal subtypes (d) and high PVR mRNA levels were associated with a higher grading (e). Kaplan-Meier analysis and Log-rank tests showing a significant correlation of high PVR mRNA levels with shorter overall survival (f) and recurrence-free interval (g); PVR levels < median are shown in light gray, PVR levels > median are shown in dark gray.
The negative impact of a high PVR expression could be verified using two independent and publicly available breast cancer patient cohorts (Gene Expression Omnibus datasets GSE1456 and GSE42568, n = 156 and n = 104 patients, respectively) accessible at the PROGgeneV2 prognostic database.31 When analyzing low vs. high PVR expressors by separating the cohort using the median expression value as cutoff, high PVR expression levels were associated with a poorer overall survival as well as recurrence-free interval in both datasets (GSE1456: p = .02 for OS and p = .009 for RFI; GSE42568: p = .002 for OS and p = .008 for RFI; see supplemental figure S1 for Kaplan-Meier survival curves).
PVR mRNA levels correlate with protein levels in breast cancer samples
In order to test for the correlation of PVR mRNA and protein level, we analyzed PVR protein expression in a subgroup of breast cancer samples (n = 30) included in the microarray cohort using western blot analysis. Expression of PVR in the AML cell line MV4-11 was used as a reference and set to 100%.32 In the PVR-specific Western blot of the breast cancer samples, we detected a band of varying intensity with a mean expression level after densitometry of 23.6% and a range from 0% to 128.4% (Figure 2(a)). A positive correlation between PVR mRNA level and protein expression was found, with a Spearman correlation coefficient of 0.488 (p = .006) for probeset 32699_s_at. Similar to the results obtained from the microarray data, we observed a higher PVR protein expression in samples with higher grading (p = .334), ER- and PR-negativity (p = .008 and p = .02, respectively) as well as in HER2-positive and TNBC subtypes compared with luminal breast cancer samples (p = .012) (Figure 2(b–d)).
Figure 2.
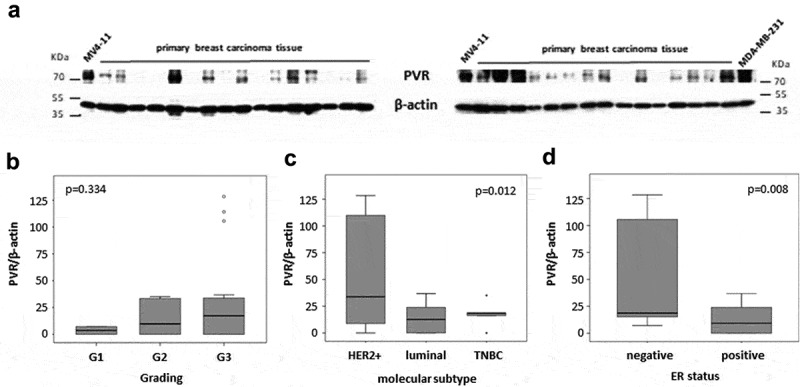
PVR protein levels in breast cancer samples. (a) Western blot results showing PVR expression in 30 breast cancer samples. Protein extract from the cell line MV4-11 was included in each gel as an internal control and β-actin was used as a loading control. (b–d) Box plots showing PVR protein expression in G1 – G3 breast cancer samples (b), HER2 positive, luminal and TNBC (c) as well in tumors with negative vs. positive ER status (d). Expression values are relative to the MV4-11 control which was set as 100%.
Immune checkpoint blockade of the TIGIT-PVR axis increases the specific lysis of breast cancer cell lines and augments the cytotoxic effects of an EGFR-specific BiTE® antibody construct
We analyzed eight different breast cancer cell lines for the expression of PVR. All analyzed cell lines showed cell surface expression of PVR on their cellular membrane. The observed varying median fluorescence intensity of PVR staining compared to the isotype control suggests differences in the protein expression intensity on different breast cancer cell lines (Figure 3). Moreover, PVR expression is high across cell lines of several different breast cancer subtypes, including luminal A (MCF-7 and T47D), HER2 positive (AU565, HCC1954, and SKBR-3), triple-negative basal B (BT549 and MDA-MB-231) and triple-negative basal A (MDA-MB-468).33
Figure 3.
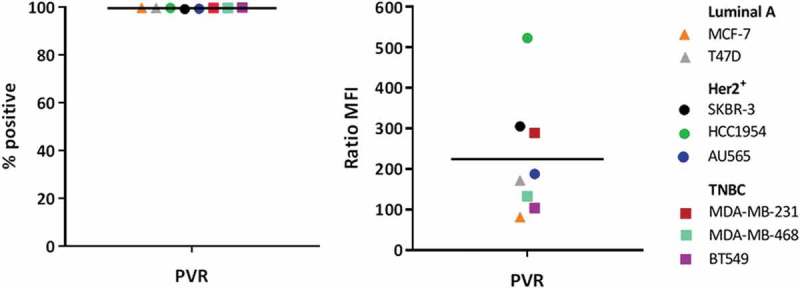
Breast cancer cell lines show a high expression of PVR across several different subtypes. All analyzed cell lines are positive for PVR expression. The expression intensities vary between cell lines as depicted by the median fluorescence intensity (MFI) ratio but nevertheless all cell lines show very high PVR expression intensities.
Next, we investigated whether interference with the TIGIT-PVR axis could be utilized as a novel therapeutic approach for the treatment of breast cancer. Therefore, we evaluated the impact of antibody blockade of PVR on breast cancer cells or the corresponding receptor TIGIT on allogenic immune cells isolated from healthy donor peripheral blood mononuclear cells (HD-PBMCs) in in vitro cytotoxicity assays. The specific lysis of breast cancer cells by immune cells was normalized to the respective sample with an isotype control (IgG1 or IgG2a for anti-PVR or anti-TIGIT antibody, respectively). Additionally, we examined whether the lysis of breast cancer cells induced by a BiTE® antibody construct, which is a bispecific T-cell recruiting antibody binding to EGFR and CD3, could be enhanced by blocking PVR or TIGIT. A BiTE® antibody construct specific for an irrelevant target antigen served as control. The cytotoxic effect of immune cells was non-significantly, but measurably enhanced by the addition of blocking PVR or TIGIT antibodies in the triple-negative cell lines MDA-MB-231 (basal B), MDA-MB-468 (basal A) and BT549 (basal B) as well as for the HER2-positive cell line SKBR-3. Furthermore, the cytotoxic effect of the EGFR-specific BiTE® antibody construct was clearly increased by the addition of PVR or TIGIT blocking antibodies for all analyzed cell lines (Figure 4). Cytokine-induced killer cells (CIKs) are ex vivo stimulated natural killer-like T lymphocytes that represent a promising option as adoptive immunotherapy to fight cancer.34 In analogy to the assays using HD-PBMCs as effector cells, we generated CIKs from HD-PBMCs and analyzed the impact of blocking the TIGIT-PVR axis using the cell lines MDA-MB-231, BT549 and SKBR-3. Here, we also observed an enhanced immune cell-mediated lysis of breast cancer cells by the addition of blocking PVR and TIGIT antibodies alone or in combination with an EGFR-specific BiTE® antibody construct (Figure 5).
Figure 4.
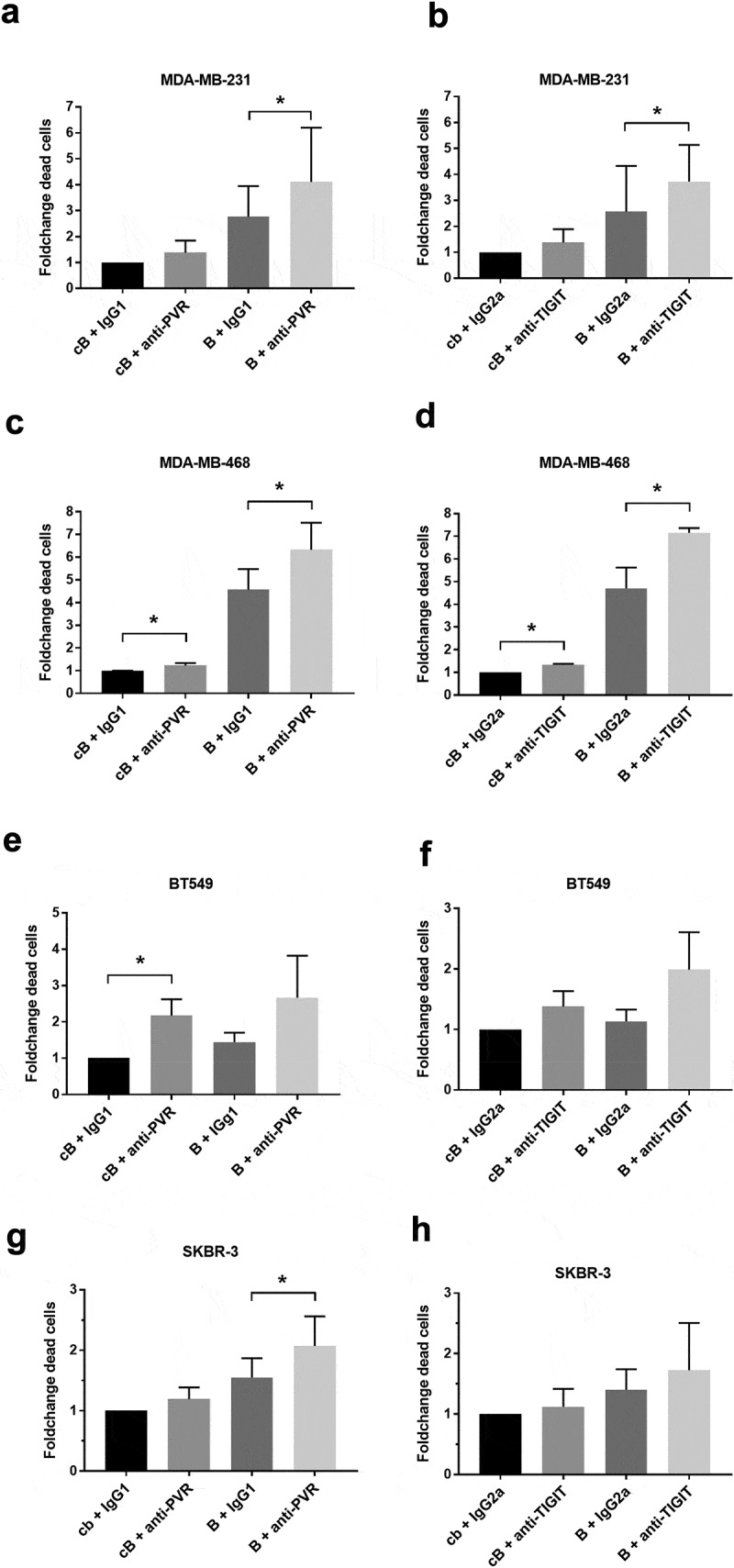
Blocking of the TIGIT-PVR axis increased the lysis of breast cancer cell lines by HD-PBMCs alone or in combination with an EGFR-specific BiTE® antibody construct. The specific lysis for the triple-negative breast cancer cell lines MDA-MB-231 ((a,b); n = 6), MDA-MB-468 ((c,d); n = 4/3) and BT549 ((e,f); n = 3) as well as for the HER2-positive cell line SKBR-3 ((g,h); n = 4) was measured after 24 h of incubation with healthy donor peripheral mononuclear cells in presence or absence of blocking antibodies against PVR on breast cancer cells or TIGIT on immune cells and the EGFR-specific BiTE® antibody construct PL-38112 (b). As controls for the blocking and bispecific antibodies, respective isotype controls and a non-binding control BiTE® (cB) were used. Results are depicted as the mean ± SD fold changes of dead target cells, relative to the sample with control BiTE® and blocking antibody isotype control. For statistical analysis paired T-tests were performed (*p ≤ 0.05). cB = control BiTE® antibody construct, B = EGFR BiTE® antibody contruct, IgG1 = IgG1 isotype control antibody, IgG2a = IgG2a isotype control antibody.
Figure 5.
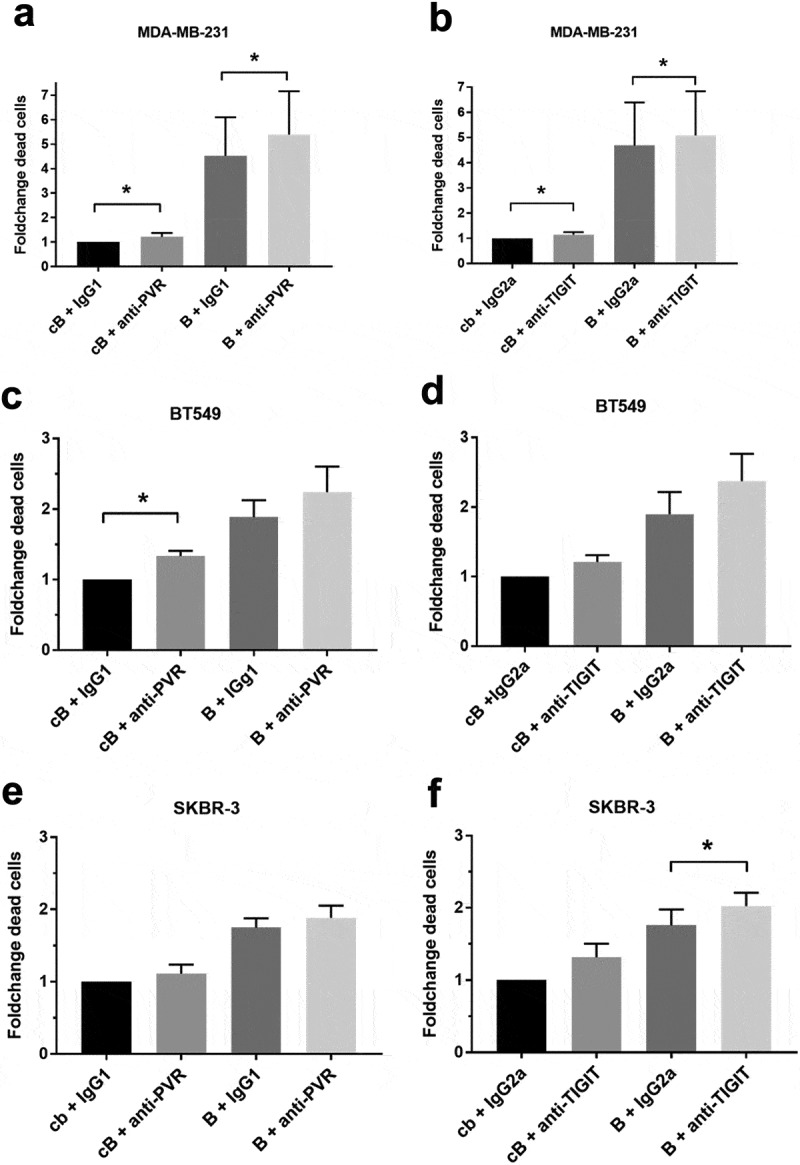
Blocking of the TIGIT-PVR axis increased the lysis of breast cancer cell lines by CIKs alone or in combination with an EGFR-specific BiTE® antibody construct. As described in Figure 4, we analyzed the effects of blocking PVR on the triple-negative breast cancer luminal B cell line MDA-MB-231 ((a,b), n = 6) and BT549 ((c,d), n = 3) as well as on the HER2-positive cell line SKBR-3 ((e,f); n = 3) or TIGIT on cytokine-induced killer cells (CIKs) produced from healthy donors. Results are depicted as the mean ± SD fold changes (FC) of dead target cells, relative to the sample with control BiTE® and blocking antibody isotype control. For statistical analysis paired T-tests were performed (*p ≤ 0.05). cB = control BiTE® antibody construct, B = EGFR BiTE® antibody contruct, IgG1 = IgG1 isotype control antibody, IgG2a = IgG2a isotype control antibody.
To further characterize the cytotoxic potential of immune cells after blocking the TIGIT-PVR axis, we analyzed the granzyme B levels in the supernatant of killing assays using HD-PBMCs and the cell lines SKBR-3, MDA-MB-231 and MDA-MB-468 using an enzyme-linked immunosorbent assay (ELISA). A clear increase in granzyme B secretion could be demonstrated for the combination of the PVR and the TIGIT blocking antibodies, respectively, with the EGFR-specific BiTE® antibody construct compared to control antibodies (Figure 6). We also analyzed the granzyme B secretion of CIKs in killing assays using the cell line SKBR-3. Using CIKs as effector cells, an increase of granzyme B concentration in the supernatant by blocking PVR or TIGIT compared to the isotype controls could be detected in the control BiTE® setting, which was further enhanced with the EGFR-CD3-BiTE® antibody construct (supplemental figure S2).
Figure 6.
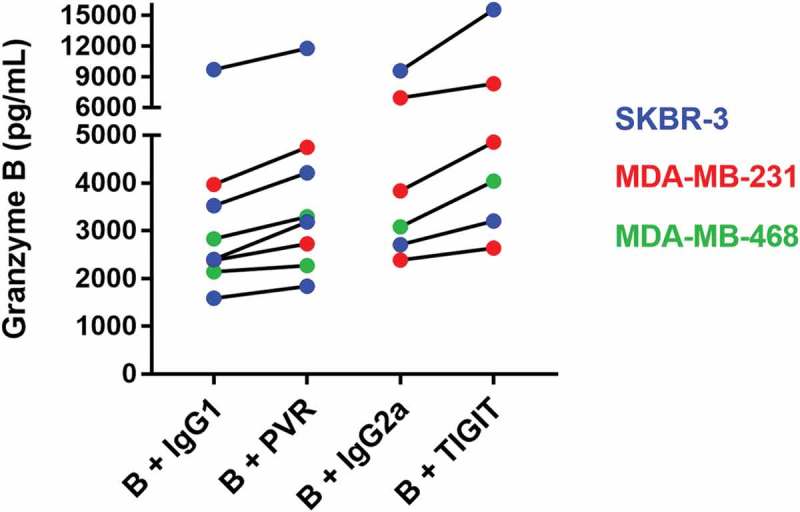
The blocking of the TIGIT-PVR axis resulted in increased granzyme B secretion of immune cells. Breast cancer cell lines SKBR-3, MDA-MB-231, and MDA-MB-468 were incubated with healthy donor peripheral mononuclear cells in the presence or absence of PVR and TIGIT blocking antibodies and the EGFR-specific BiTE® antibody construct. The supernatants of these cytotoxicity assays were harvested after 24 h. The increase of granzyme B concentration measured by ELISA between isotype control antibody and blocking antibody from several different experiments is visualized. B = EGFR BiTE® antibody contruct, IgG1 = IgG1 isotype control antibody, IgG2a = IgG2a isotype control antibody.
Discussion
Although the overall survival has increased during the last decades, breast cancer remains one of the leading causes of death as the survival rates for metastatic breast cancer are still low with a median 5-year and 10-year survival of only 27% and 13%, respectively.35,36 Therefore, numerous novel therapeutic approaches including immunotherapy are investigated to improve the patients’ outcome. Only a few phase I and phase II trials with immune checkpoint inhibitors targeting PD-1, PD-L1 or CTLA-4 have been conducted so far in metastatic breast cancer. The overall response rates were only moderate, but as observed in other cancer entities, those patients who responded had durable remissions.37–39 These observations could indicate that different immune checkpoint axes other than PD-1 or CTLA-4 may be more relevant and therefore better targets in breast cancer.40 Our present study provides strong evidence that the PVR-TIGIT axis may represent a promising therapeutic target for breast cancer patients.
We could show that high mRNA levels of the TIGIT ligand PVR were significantly associated with a poor overall and recurrence-free survival in a cohort of 197 breast cancer patients analyzed by microarray gene expression. Furthermore, high PVR expression was related to more aggressive breast cancer subtypes as high PVR levels were especially observed in HER2 positive and triple-negative breast cancer patients. To our knowledge, this is the first study proving that high PVR mRNA levels represent an independent negative prognostic marker in a large cohort of breast cancer patients. Furthermore, we could verify our findings using the mRNA expression data of two public available independent breast cancer patient cohorts.31 In two recently published studies, the authors analyzed the PVR protein expression by immunohistochemistry in large cohorts of breast cancer patients. In both studies, high PVR expression was associated with poor patients’ outcome41,42 supporting our findings. Moreover, these observations are in line with the correlation between PVR mRNA and protein levels that we found in our patient cohort. Furthermore, Johnston and colleagues attempted to determine the expression of TIGIT on tumor-associated T cells using Cancer Genome Atlas Networks. They found a strong correlation between TIGIT and CD3ϵ in several solid tumor entities including breast cancer.17 Compared to normal matched tissue samples, the TIGIT:CD3ϵ expression ratio was significantly increased in the majority of analyzed tumor entities indicating that TIGIT is upregulated by T cells in a broad range of solid tumors.17 Conversely, another PVR binding T-cell antigen DNAM-1 is the main costimulatory counterpart of TIGIT and often downregulated on cancer patients’ immune cells.21,22,43,44 Due to high TIGIT and low DNAM-1 expression in cancer and the fact that TIGIT has a significantly higher binding affinity for PVR compared to DNAM-1,10,45 it seems reasonable to assume that high expression of PVR on tumor cells is associated with an immunosuppressive phenotype and consequently a poor prognosis.
To test the therapeutic potential of blocking the TIGIT-PVR axis, we performed in vitro cytotoxicity assays using breast cancer cell lines and allogeneic immune cells. The blockade of TIGIT on immune effector cells or PVR on the cancer cells using blocking antibodies enhanced the cytotoxic effects. For the majority of analyzed cell lines, the effects of blocking TIGIT or PVR alone were only modest. However, the effects were more pronounced when combining TIGIT or PVR inhibition with an EGFR targeting BiTE® antibody construct. Furthermore, this enhanced cytotoxicity of immune cells was accompanied by increased release of granzyme B confirming a functional response in immune effector cells.
In support of our findings, Xu et al. recently published that the addition of anti-PVR antibodies to trastuzumab enhanced the cytotoxicity of NK cells against HER2-positive breast cancer cell lines in vitro.23
Bispecific T-cell engagers (BiTE®) are antibodies possessing binding sites for CD3 on T cells and for tumor antigens, bringing neoplastic cells and T cells in close contact to induce the cytolytic action. BiTE® antibody constructs are investigated in a wide variety of solid and hematopoietic malignancies with Blinatumomab, a CD19-specific BiTE® antibody construct, representing the most advanced member in this class being FDA and EMA approved for the treatment of acute lymphoblastic leukemia.46 EGFR overexpression is observed in all subtypes of breast cancer but especially pronounced in triple-negative breast cancer. Hence, EGFR was evaluated as a therapeutic target in several studies using monoclonal antibodies such as cetuximab and panitumumab. However, so far results have been disappointing.47–51 A recently published study revealed that PD-L1 expression on tumor cells contributed to the resistance against EGFR tyrosine kinase inhibitors among patients with non-small cell lung cancer.52 Up-regulation of immune checkpoint ligands on cancer cells as a resistance mechanism has been described in several studies including up-regulation of PD-L1 in response to treatment with Blinatumomab in acute lymphoblastic leukemia.53–56 In another study, up-regulation of PD-L1 was observed in colorectal cancer patients treated with chemotherapy.56 Xia et al. described an activating EGFR receptor mutation as an acquired resistance mechanism to the PD-1 antibody nivolumab in a lung cancer patient.57 These data underline the rationale to combine immune checkpoint inhibitors with other targeted therapies such as bispecific T-cell engagers to evade resistance mechanisms and enhance the therapeutic efficacy.
The combination of blocking the TIGIT-PVR axis with other immune therapeutic approaches was analyzed in a number of studies. Chauvin et al. observed enhanced proliferation, cytokine production and degranulation of CD8+ tumor-infiltrating lymphocytes from melanoma patients upon combined blockade of TIGIT and PD-1.21 In a different study, the inhibition of both TIGIT and PD-1 resulted in significantly reduced tumor growth in a murine breast as well as a colon cancer model. Furthermore, the authors observed the induction of a protective antigen-specific immune response, as mice in which the tumor had completely been cleared by combined antibody treatment, were resistant to a re-challenge.17 In a Wilms tumor protein-1 (WT1)-targeted dendritic cell vaccination study in patients with gastric cancer, upregulation of TIGIT and other immune checkpoint molecules such as PD-1 and Tim3 on cytotoxic T cells was observed. Indeed, the TIGIT+PD-1+Tim3+ population represented the most dysfunctional subset of WT1-specific CTLs and the triple blockade of TIGIT, PD-1 and Tim3 enhanced the proliferation and cytokine production of CTLs in vitro.58 In a different study, the addition of a blocking anti-PVR antibody to a co-culture of melanoma cells and antigen-specific cytotoxic T cells led to increased interferon-gamma production by the immune cells.20
In our present study, the cytotoxicity assays were performed in an allogeneic setting with breast cancer cell lines and healthy donor-derived immune cells. This approach differs from the situation in patients due to the lack of tumor-primed specific immune cells, on the one hand, as well as exhausted immune cells due to tumor-mediated immunosuppressive effects, on the other hand. Further studies should, therefore, include the TIGIT blockade in an autologous setting using primary breast cancer cells and the corresponding tumor-infiltrating lymphocytes.
Taken together, our preclinical data identify the immune checkpoint molecule PVR as a novel prognostic marker in breast cancer and indicate that blocking the TIGIT-PVR axis alone or in combination with, e.g. BiTE® antibody constructs might represent a novel promising therapeutic option for the treatment of breast cancer patients that should be further explored.
Patients and methods
Patient cohort and microarray data
We analyzed the PVR mRNA levels using microarray data (Affymetrix HG-U133A) from a cohort of 197 mammary carcinoma patients enrolled at the department of Gynecology at the University Medical Center Hamburg-Eppendorf between 1991 and 2002. The clinical and histological characteristics of this cohort as well as the technical details have been described elsewhere.59
Informed consent for the scientific use of tissue materials, which was approved by the local ethics committees (Ethik-Kommission der Ärztekammer Hamburg, #OB/V/03) was obtained from all patients. The study was performed in accordance with the principles of the declaration of Helsinki and REMARK criteria.60
The mRNA data of five probesets (32699_s_at, 214443_at, 214444_s_at, 216283_s_at, and 212662_at) corresponding to PVR were available. Only probeset 216283_s_at showed low mRNA levels in our samples and was excluded from further analysis. Additionally, the mean value of the four probesets mentioned was calculated and included in further analyses. According to the PVR mRNA values of each probeset and the mean value, the cohort was divided into quartiles of similar size, representing low, moderate-low, moderate-high and high PVR levels. Correlations between PVR mRNA levels (quartiles) and clinicopathological factors such as histological grading, stage, lymph node involvement, estrogen, and progesterone receptor status (ER, PR) were statistically examined by χ2-tests. Correlation between PVR mRNA levels (continuous data) and overall survival or progression-free interval was calculated using Cox regression analyses. Multivariate Cox regression analysis (with continuous data or quartiles) including the clinical stage, nodal involvement, and molecular subtype was performed for all probe sets and the PVR mean value. Here, a backward analysis with stepwise removal of insignificant terms was used. Additionally, the correlation between PVR mRNA levels and overall survival or progression-free interval was analyzed by Kaplan-Meier analysis and Log-Rank-Tests as well. Probability values less than 0.05 were regarded as statistically significant. All statistical analyses were conducted using SPSS software Version 23 (SPSS Inc., Chicago, IL, USA). The PROGgeneV2 database was used to verify our findings.31 We chose only breast cancer patient cohorts for our analysis for which overall survival, as well as recurrence-free interval data, was available. Furthermore, we have excluded datasets with restricted cohorts such as those with only nodal negative or ER-positive patients or with less than 100 patients. The median PVR expression was chosen as cutoff for the division into PVR low vs. high expressors.
Cell lines and cell culture
All breast cancer cell lines were cultured at 37°C and 5% CO2 in a humidified atmosphere and were tested for mycoplasma contamination (MycoAlert, Lonza) regularly. MDA-MB-231, MDA-MB-468, BT549, and MCF-7 were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS Superior, Biochrom), SKBR-3 were cultured in McCoy’s 5A (PromoCell) supplemented with 20% FBS and all other cell lines used in this study were cultured in RPMI 1640 (Gibco) supplemented with 10% FBS.
Western blot analysis
Thirty breast cancer samples included in the microarray analysis were analyzed by western blotting. The characteristics of this cohort are given in supplementary table S2. Western blot was performed as described previously.61,62 Briefly, 20 µg protein lysate of each sample was loaded per well. The protein lysate from the cell line MV4-11 was used as a reference and internal control in all blots.18 After electrophoresis and blotting to PVDF membranes, PVR was detected using the monoclonal anti-PVR antibody L95.63 Equal loading was verified by immunoblotting with anti-β-actin antibody (sc-47778, Santa Cruz Biotechnology). Horseradish peroxidase-conjugated rabbit anti-mouse antibody (Cell Signaling) was used as the secondary antibody. After visualization by a chemiluminescence reagent (SuperSignal West Pico kit, Pierce) band intensities were quantified by densitometry (Imaging Densitometer GS-700, BioRad) and calculated as percent intensity of the specific control sample.
Flow cytometry
To examine the expression of PVR on breast cancer cell lines, 0.2 × 106 cells were incubated with an APC-conjugated anti-PVR antibody (Clone SKII.4, BioLegend) or the corresponding isotype control. Flow cytometric data were acquired using a BD FACSCalibur (BD Biosciences) and analyzed using FlowingSoftware (Version 2.5.1). The median fluorescence intensity (MFI) ratio (MFI target divided by MFI isotype control) was calculated as a measure of expression intensity.
Cytotoxicity assays
To analyze the specific lysis of breast cancer cells, target cells were labeled with 70 nM CellTracker™ Green CMFDA Dye (Thermo Fisher). Healthy donor peripheral blood mononuclear cells (HD-PBMCs) were isolated using density-gradient centrifugation of buffy coats from cytomegalovirus seronegative, anonymous donors kindly provided from the blood bank of the University Medical Center Hamburg-Eppendorf (Germany). For the generation of cytokine-induced killer cells (CIKs), PBMCs were stimulated for 7 days with IFN-y (1000 U/mL; Peprotech), IL-2 (600 U/mL, Peprotech) and anti-CD3 antibody (5 ng/mL, clone OKT3; Biolegend). Labeled target cells were mixed with HD-PBMCs or CIKs at 1 × 106 cells/ml in a ratio of 6:1 or 2:1, respectively, in the cell culture medium used to cultivate the target cell line. Two hundred microliter of cell suspension was plated in triplicates in 96-well plates and incubated for 24 h with antibodies against PVR (4 µg/ml, clone D171, NeoMarkers) or TIGIT (50 µg/ml, clone #A15153G, Biolegend) in the presence of the EGFR-specific BiTE® antibody construct PL-38112 (AMGEN Inc.). The EGFR BiTE® antibody construct concentrations of 0.1 ng/ml for the cell lines MDA-MB-231, MDA-MB-468 and BT549 or 1.0 ng/ml for the cell line SKBR-3 were selected based on pilot experiments defining BiTE® antibody construct concentrations eliciting suboptimal cytotoxicity in order to be able to demonstrate additional effects of combined treatment. As controls, we used IgG1 or IgG2a isotype control antibodies for the PVR and TIGIT antibodies, respectively, and a non-binding control BiTE® antibody construct at equal concentrations, respectively. Assessment of specific lysis of target cells was performed after 24 h of incubation by measuring the 7AAD (BD Biosciences) staining of gated CMFDA positive cells in flow cytometry. All experiments were performed at least three times.
Granzyme B ELISA
To determine the degree of immune cell activation, the granzyme B concentration in supernatants of cytotoxicity assays was measured using the human granzyme B DuoSet ELISA (R&D Systems), according to manufacturer’s instructions.
Funding Statement
This work was funded in part by the Medical Faculty of the University of Hamburg, Germany and by Amgen Inc.
Acknowledgments
We would like to thank the members of the Center for Transfusion Medicine (University Medical Center Hamburg-Eppendorf, Germany) for providing buffy coats from healthy donors. Furthermore, we thank Dr. Michael Linnebacher (University of Rostock, Germany) for his support regarding the generation of CIK cells and Pauline E. Schneeberger for the establishment of the CIK cultures in our lab. We also gratefully thank Dr. Daniela Pende (IRCCS AOU San Martino-IST, Genoa, Italy) for provision of the anti-PVR L95 antibody.
Disclosure of Potential Conflicts of Interest
CB: honoraria from Merck KGaA, Sanofi, Roche, Bayer, Bristol-Myers Squibb, Servier/Pfizer and AstraZeneca, participated in advisory boards of Lilly/ImClone, Merck Serono, Sanofi, Mundipharma, Bayer Schering Pharma, Hexal, Merck Sharp Dohme, GSO and AOK Health, support for meeting attendance from Merck Serono, Sanofi, Pfizer and Bristol-Myers Squibb. WF: participated in advisory boards of Amgen, Pfizer, Novartis, Jazz Pharmaceuticals and Ariad/Incyte, research funding from Amgen, support for meeting attendance from Amgen, Jazz Pharmaceuticals, Daiichi Sanchyo Oncology and Servier, support in medical writing from Amgen, Pfizer, and Abbvie. VM: speaker honoraria from Amgen, Astra Zeneca, Celgene, Daiichi-Sankyo, Eisai, Pfizer, Novartis, Roche and Teva, consultancy honoraria from Genomic Health, Hexal, Roche, Pierre Fabre, Amgen, Novartis, MSD, Daiichi-Sankyo and Eisai, Lilly, Tesaro and Nektar, research support from Novartis, Roche, Seattle Genetics and Genentech. RK: employment by Amgen Research (Munich) GmbH and stock ownership of Amgen Inc.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Hanahan D, Weinberg RA.. Hallmarks of cancer: the next generation. Cell. 2011;144(5):1–10. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R.. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 3.Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, Rosenberg SA. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114(8):1537–1544. doi: 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti–PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callahan MK, Wolchok JD. At the bedside: CTLA-4- and PD-1-blocking antibodies in cancer immunotherapy. J Leukoc Biol. 2013;94(1):41–53. doi: 10.1189/jlb.1212631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burugu S, Dancsok AR, Nielsen TO. Emerging targets in cancer immunotherapy. Semin Cancer Biol. 2018;52:39–52. doi: 10.1016/j.semcancer.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Dougall WC, Kurtulus S, Smyth MJ, Anderson AC. TIGIT and CD96: new checkpoint receptor targets for cancer immunotherapy. Immunol Rev. 2017;276(1):112–120. doi: 10.1111/imr.12518. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10(1):48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 11.Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, Levine Z, Beiman M, Dassa L, Achdout H, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci. 2009;106(42):17858–17863. doi: 10.1073/pnas.0903474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nobis P, Zibirre R, Meyer G, Kuhne J, Warnecke G, Koch G. Production of a monoclonal antibody against an epitope on HeLa cells that is the functional poliovirus binding site. J Gen Virol. 1985;66(12):2563–2569. doi: 10.1099/0022-1317-66-12-2563. [DOI] [PubMed] [Google Scholar]
- 13.Sakisaka T, Takai Y. Biology and pathology of nectins and nectin-like molecules. Curr Opin Cell Biol. 2004;16(5):513–521. doi: 10.1016/j.ceb.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Pauken KE, Wherry EJ. TIGIT and CD226: tipping the balance between costimulatory and coinhibitory molecules to augment the cancer immunotherapy toolkit. Cancer Cell. 2014;26(6):785–787. doi: 10.1016/j.ccell.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 15.Lozano E, Dominguez-Villar M, Kuchroo V, Hafler DA. The TIGIT/CD226 axis regulates human T cell function. J Immunol. 2012;188(8):3869–3875. doi: 10.4049/jimmunol.1103627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Xia P, Du Y, Liu S, Huang G, Chen J, Zhang H, Hou N, Cheng X, Zhou L, et al. T-cell immunoglobulin and ITIM domain (TIGIT) Receptor/Poliovirus Receptor (PVR) ligand engagement suppresses interferon-γ production of natural killer cells via β-arrestin 2-mediated negative signaling. J Biol Chem. 2014;289(25):17647–17657. doi: 10.1074/jbc.M114.572420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8+T cell effector function. Cancer Cell. 2014;26(6):923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Stamm H, Klingler F, Grossjohann E-M, Muschhammer J, Vettorazzi E, Heuser M, Mock U, Thol F, Vohwinkel G, Latuske E, et al. Immune checkpoints PVR and PVRL2 are prognostic markers in AML and their blockade represents a new therapeutic option. Oncogene. 2018;37:39. doi: 10.1038/s41388-018-0288-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurtulus S, Sakuishi K, Ngiow S, Joller N, Tan DJ, Teng MWL, Smyth MJ, Kuchroo VK, Anderson AC. TIGIT predominantly regulates the immune response via regulatory T cells. J Clin Invest. 2015;125(11):4053–4062. doi: 10.1172/JCI81187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Inozume T, Yaguchi T, Furuta J, Harada K, Kawakami Y, Shimada S. Melanoma cells control antimelanoma CTL responses via interaction between TIGIT and CD155 in the effector phase. J Invest Dermatol. 2016;136(1):255–263. doi: 10.1038/JID.2015.404. [DOI] [PubMed] [Google Scholar]
- 21.Chauvin J-M, Pagliano O, Fourcade J, Sun Z, Wang H, Sander C, Kirkwood JM, Chen TT, Maurer M, Korman AJ, et al. TIGIT and PD-1 impair tumor antigen–specific CD8+ T cells in melanoma patients. J Clin Invest. 2015;125(5):2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong Y, Zhu L, Schell TD, Zhang J, Claxton DF, Ehmann WC, Rybka WB, George MR, Zeng H, Zheng H. T-Cell Immunoglobulin and ITIM domain (TIGIT) associates with CD8+ T-cell exhaustion and poor clinical outcome in AML patients. Clin Cancer Res. 2016;22(12):3057–3066. doi: 10.1158/1078-0432.CCR-15-2626. [DOI] [PubMed] [Google Scholar]
- 23.Xu F, Sunderland A, Zhou Y, Schulick RD, Edil BH, Zhu Y. Blockade of CD112R and TIGIT signaling sensitizes human natural killer cell functions. Cancer Immunol Immunother. 2017;66(10):1367–1375. doi: 10.1007/s00262-017-2031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curigliano G. Gyneco-oncological genomics and emerging biomarkers for cancer treatment with immune-checkpoint inhibitors. Semin Cancer Biol. 2018;52:253–258. doi: 10.1016/j.semcancer.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Li M, Jiang Z, Wang X. A comprehensive immunologic portrait of triple-negative breast cancer. Transl Oncol. 2018;11(2):311–329. doi: 10.1016/j.tranon.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, Kellokumpu-Lehtinen P-L, Bono P, Kataja V, Desmedt C, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol Off J Eur Soc Med Oncol. 2014;25(8):1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 27.Smyth MJ, Ngiow SF, Ribas A, Teng MWL. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13(3):143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 28.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Diéras V, Hegg R, Im S-A, Shaw Wright G, et al. Atezolizumab and Nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379(22):2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 29.Rugo HS, Delord J-P, Im S-A, Ott PA, Piha-Paul SA, Bedard PL, Sachdev J, Le Tourneau C, van Brummelen EMJ, Varga A, et al. Safety and antitumor activity of pembrolizumab in patients with estrogen receptor-positive/human epidermal growth factor receptor 2-negative advanced breast cancer. Clin Cancer Res. 2018;24(12):2804–2811. doi: 10.1158/1078-0432.CCR-17-3452. [DOI] [PubMed] [Google Scholar]
- 30.Adams S, Diamond JR, Hamilton E, Pohlmann PR, Tolaney SM, Chang C-W, Zhang W, Iizuka K, Foster PG, Molinero L, et al. Atezolizumab Plus nab-paclitaxel in the treatment of metastatic triple-negative breast cancer with 2-year survival follow-up: a phase 1b clinical trial. JAMA Oncol. 2018;5(3):334. doi: 10.1001/jamaoncol.2018.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14(1):970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamm H, Klingler F, Grossjohann E-M, Muschhammer J, Vettorazzi E, Heuser M, Mock U, Thol F, Vohwinkel G, Latuske E, et al. Immune checkpoints PVR and PVRL2 are prognostic markers in AML and their blockade represents a new therapeutic option. Oncogene. 2018. May:1. doi: 10.1038/s41388-018-0288-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai X, Cheng H, Bai Z, Li J. Breast cancer cell line classification and Its relevance with breast tumor subtyping. J Cancer. 2017;8(16):3131–3141. doi: 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao X, Mi Y, Guo N, Xu H, Xu L, Gou X, Jin W. Cytokine-induced killer cells as pharmacological tools for cancer immunotherapy. Front Immunol. 2017;8:774. doi: 10.3389/fimmu.2017.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eng LG, Dawood S, Sopik V, Haaland B, Tan PS, Bhoo-Pathy N, Warner E, Iqbal J, Narod SA, Dent R. Ten-year survival in women with primary stage IV breast cancer. Breast Cancer Res Treat. 2016;160(1):145–152. doi: 10.1007/s10549-016-3974-x. [DOI] [PubMed] [Google Scholar]
- 36.Newman LA. Epidemiology of locally advanced breast cancer. Semin Radiat Oncol. 2009;19(4):195–203. doi: 10.1016/j.semradonc.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Nanda R, Chow LQM, Dees EC, Berger R, Gupta S, Geva R, Pusztai L, Pathiraja K, Aktan G, Cheng JD, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J Clin Oncol. 2016;34(21):2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams S, Loi S, Toppmeyer D, Cescon DW, De Laurentiis M, Nanda R, Winer EP, Mukai H, Tamura K, Armstrong A, et al. Phase 2 study of pembrolizumab as first-line therapy for PD-L1–positive metastatic triple-negative breast cancer (mTNBC): preliminary data from KEYNOTE-086 cohort B. J Clin Oncol. 2017;35(15_suppl):1088. doi: 10.1200/JCO.2017.35.15_suppl.1088. [DOI] [Google Scholar]
- 39.Rugo HS, Delord J-P, Im S-A, Ott PA, Piha-Paul SA, Bedard PL, Sachev J, Le Tourneau C, van Brummelen EMJ, Varga A, et al. Safety and antitumor activity of pembrolizumab in patients with estrogen receptor‒positive/human epidermal growth factor receptor 2‒negative advanced breast cancer. Clin Cancer Res. 2018. March 20:clincanres.3452.2017. doi: 10.1158/1078-0432.CCR-17-3452. [DOI] [PubMed] [Google Scholar]
- 40.Chrétien S, Zerdes I, Bergh J, Matikas A, Foukakis T. Beyond PD-1/PD-L1 inhibition: what the future holds for breast cancer immunotherapy. Cancers (Basel). 2019;11(5):628. doi: 10.3390/cancers11050628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Triki H, Charfi S, Bouzidi L, Ben Kridis W, Daoud J, Chaabane K, Sellami-Boudawara T, Rebai A, Cherif B. CD155 expression in human breast cancer: clinical significance and relevance to natural killer cell infiltration. Life Sci. 2019;231:116543. doi: 10.1016/j.lfs.2019.116543. [DOI] [PubMed] [Google Scholar]
- 42.Yong H, Cheng R, Li X, Gao G, Jiang X, Cheng H, Zhou X, Zhao W. CD155 expression and its prognostic value in postoperative patients with breast cancer. Biomed Pharmacother. 2019;115:108884. doi: 10.1016/j.biopha.2019.108884. [DOI] [PubMed] [Google Scholar]
- 43.Carlsten M, Norell H, Bryceson YT, Poschke I, Schedvins K, Ljunggren H-G, Kiessling R, Malmberg K-J. Primary human tumor cells expressing CD155 impair tumor targeting by down-regulating DNAM-1 on NK cells. J Immunol. 2009;183(8):4921–4930. doi: 10.4049/jimmunol.0901226. [DOI] [PubMed] [Google Scholar]
- 44.Mamessier E, Sylvain A, Thibult M-L, Houvenaeghel G, Jacquemier J, Castellano R, Gonçalves A, André P, Romagné F, Thibault G, et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest. 2011;121(9):3609–3622. doi: 10.1172/JCI45816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tahara-Hanaoka S, Shibuya K, Onoda Y, Zhang H, Yamazaki S, Miyamoto A, Honda S-I, Lanier LL, Shibuya A. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int Immunol. 2004;16(4):533–538. doi: 10.1093/intimm/dxh059. [DOI] [PubMed] [Google Scholar]
- 46.Zugmaier G, Klinger M, Schmidt M, Subklewe M. Clinical overview of anti-CD19 BiTE® and ex vivo data from anti-CD33 BiTE® as examples for retargeting T cells in hematologic malignancies. Mol Immunol. 2015;67(2):58–66. doi: 10.1016/j.molimm.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 47.Dickler MN, Cobleigh MA, Miller KD, Klein PM, Winer EP. Efficacy and safety of erlotinib in patients with locally advanced or metastatic breast cancer. Breast Cancer Res Treat. 2009;115(1):115–121. doi: 10.1007/s10549-008-0055-9. [DOI] [PubMed] [Google Scholar]
- 48.Dickler MN, Rugo HS, Eberle CA, Brogi E, Caravelli JF, Panageas KS, Boyd J, Yeh B, Lake DE, Dang CT, et al. A phase II trial of erlotinib in combination with bevacizumab in patients with metastatic breast cancer. Clin Cancer Res. 2008;14(23):7878–7883. doi: 10.1158/1078-0432.CCR-08-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Minckwitz G, Jonat W, Fasching P, Du Bois A, Kleeberg U, Lück H-J, Kettner E, Hilfrich J, Eiermann W, Torode J, et al. A multicentre phase II study on gefitinib in taxane- and anthracycline-pretreated metastatic breast cancer. Breast Cancer Res Treat. 2005;89(2):165–172. doi: 10.1007/s10549-004-1720-2. [DOI] [PubMed] [Google Scholar]
- 50.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A, et al. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol. 2012;30(21):2615–2623. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yardley DA, Ward PJ, Daniel BR, Eakle JF, Lamar RE, Lane CM, Hainsworth JD. Panitumumab, gemcitabine, and carboplatin as treatment for women with metastatic triple-negative breast cancer: a Sarah Cannon Research Institute Phase II Trial. Clin Breast Cancer. 2016;16(5):349–355. doi: 10.1016/j.clbc.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Su S, Dong Z-Y, Xie Z, Yan L-X, Li Y-F, Su J, Liu S-Y, Yin K, Chen R-L, Huang S-M, et al. Strong programmed death ligand 1 expression predicts poor response and de novo resistance to EGFR tyrosine kinase inhibitors among NSCLC patients with EGFR mutation. J Thorac Oncol. 2018;13(11):1668–1675. doi: 10.1016/j.jtho.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 53.Köhnke T, Krupka C, Tischer J, Knösel T, Subklewe M. Increase of PD-L1 expressing B-precursor ALL cells in a patient resistant to the CD19/CD3-bispecific T cell engager antibody blinatumomab. J Hematol Oncol. 2015;8(1):111. doi: 10.1186/s13045-015-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feucht J, Kayser S, Gorodezki D, Hamieh M, Döring M, Blaeschke F, Schlegel P, Bösmüller H, Quintanilla-Fend L, Ebinger M, et al. T-cell responses against CD19+ pediatric acute lymphoblastic leukemia mediated by bispecific T-cell engager (BiTE) are regulated contrarily by PD-L1 and CD80/CD86 on leukemic blasts. Oncotarget. 2016;7(47):76902–76919. doi: 10.18632/oncotarget.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krupka C, Kufer P, Kischel R, Zugmaier G, Lichtenegger FS, Köhnke T, Vick B, Jeremias I, Metzeler KH, Altmann T, et al. Blockade of the PD-1/PD-L1 axis augments lysis of AML cells by the CD33/CD3 BiTE antibody construct AMG 330: reversing a T-cell-induced immune escape mechanism. Leukemia. 2015;(August):1–8. doi: 10.1038/leu.2015.214. [DOI] [PubMed] [Google Scholar]
- 56.Dosset M, Vargas TR, Lagrange A, Boidot R, Végran F, Roussey A, Chalmin F, Dondaine L, Paul C, Lauret Marie-Joseph E, et al. PD-1/PD-L1 pathway: an adaptive immune resistance mechanism to immunogenic chemotherapy in colorectal cancer. Oncoimmunology. 2018;7(6):e1433981. doi: 10.1080/2162402X.2018.1433981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia C, Zeng F, Zhang Y. EGFR exon 21 L858R as an acquired resistance mechanism to nivolumab in a lung cancer patient originally driver gene-negative. Thorac Cancer. 2019. February 27. doi: 10.1111/1759-7714.13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu X, Liu J, Cui P, Liu T, Piao C, Xu X, Zhang Q, Xiao M, Liu X, Wang Y, et al. Co-inhibition of TIGIT, PD1, and Tim3 reverses dysfunction of Wilms tumor protein-1 (WT1)-specific CD8+ T lymphocytes after dendritic cell vaccination in gastric cancer. Am J Cancer Res. 2018;8(8):1564–1575. [PMC free article] [PubMed] [Google Scholar]
- 59.Milde-Langosch K, Karn T, Schmidt M, Zu Eulenburg C, Oliveira-Ferrer L, Wirtz RM, Schumacher U, Witzel I, Schütze D, Müller V. Prognostic relevance of glycosylation-associated genes in breast cancer. Breast Cancer Res Treat. 2014;145(2):295–305. doi: 10.1007/s10549-014-2949-z. [DOI] [PubMed] [Google Scholar]
- 60.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM; Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics . REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. 2005;93(4):387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eckhoff K, Flurschütz R, Trillsch F, Mahner S, Jänicke F, Milde-Langosch K. The prognostic significance of Jun transcription factors in ovarian cancer. J Cancer Res Clin Oncol. 2013;139(10):1673–1680. doi: 10.1007/s00432-013-1489-y. [DOI] [PubMed] [Google Scholar]
- 62.Mahner S, Baasch C, Schwarz J, Hein S, Wölber L, Jänicke F, Milde-Langosch K. C-Fos expression is a molecular predictor of progression and survival in epithelial ovarian carcinoma. Br J Cancer. 2008;99(8):1269–1275. doi: 10.1038/sj.bjc.6604650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, Cantoni C, Grassi J, Marcenaro S, Reymond N, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198(4):557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


