ABSTRACT
Programmed death-ligand 1 (PD-L1) is a crucial target for lung cancer immunotherapy. In lung cancer patients with high PD-L1 expression, blocking or reducing its expression can inhibit tumor growth. PD-L1 is regulated by signaling pathways, transcription factors and epigenetic factors, such as the GSK3β/β-catenin pathway, P53 protein and EMT. In our previous study, succinate dehydrogenase 5 (SDH5) was reported to regulate ZEB1 expression, induce EMT and lead to lung cancer metastasis via the GSK3β/β-catenin pathway. It is possible that SDH5 is involved in the mechanisms of PD-L1 regulation.In the present study, we observed a negative correlation between the expression of PD-L1 and SDH5 in vivo and in vitro. The examination of patient tissues also confirmed our results. Furthermore, we also found that SDH5 could reverse PD-L1 expression by the GSK3β/β-catenin/ZEB1 pathways. All these results reveal that SDH5 regulates PD-L1 expression and suggest that SDH5 can be used as a marker to predict tumor immune micro-states and provide guidance for clinical immunotherapy.
KEYWORDS: Lung cancer, PD-L1, SDH5, EMT, immunotherapy
Introduction
The latest advances in cancer immunotherapies have played an essential role in the landscape of cancer treatment. Immune checkpoint blockade, especially programmed death 1 (PD-1)/programmed death ligand 1 (PD-L1) blockade, is one of the most effective immunotherapies.1,2 As an immune checkpoint molecule, PD-L1 on cancer cells is bound to its receptor, PD-1, on T cells to suppress their function and leads to immune suppression.3 The evidence that antibodies blocking the PD-1/PD-L1 interaction reactivate cytotoxic T cells to eradicate cancer cells has demonstrated a clear survival benefit as a single agent or in combination compared with standard chemotherapy in both treatment-naïve patients and patients previously treated for advanced non-small-cell lung cancer (NSCLC).4 Due to successful treatment outcomes, using anti-PD-1/PD-L1 antibodies has been a main topic in the oncology field. Recent studies have revealed that PD-1 blockade has also proven clinical benefits as a first-line therapy in NSCLC.5–7
Unfortunately, only a quarter of select patients with NSCLC respond to PD-1 blockade by nivolumab.8 PD-L1 protein expression assessed by immunochemistry (IHC) has been applied as a biomarker to select NSCLC patients for pembrolizumab therapy.9 Therefore, understanding the relative regulatory mechanisms of PD-L1 becomes important for establishing effective therapies. Studies have shown that PD-L1 expression is regulated by signaling pathways, transcription factors and epigenetic factors, such as the GSK-3β/β-catenin pathway, P53 protein, and EMT.10–13
Succinate dehydrogenase 5 (SDH5), also known as SDHAF2, plays an essential role in the assembly of succinate dehydrogenase and is a component of the tricarboxylic acid (TCA) cycle. SDH5 has been reported to contribute to the development of several kinds of cancers and is mutated in paraganglioma and gastrointestinal stromal tumors.14,15 Previously, we demonstrated that the loss of SDH5 could augment the expression of ZEB1, induce EMT and lead to lung cancer metastasis via the glycogen synthase kinase 3β/β-catenin pathway.16 Additionally, studies have shown that ZEB1 can increase PD-L1 expression by activating miRNA-200 in NSCLC, resulting in the dysfunction of CD8 T cells, which couples with epithelial-mesenchymal transition (EMT) to increase metastasis.17 These studies raised the possibility that SDH5 is involved in the mechanisms of PD-L1 regulation. To the best of our knowledge, no study has focused on the functions of SDH5 in the regulation of PD-L1.
In the present study, we found that SDH5 mRNA can be detected by qRT-PCR in the tumor tissues of patients and has a negative association with the expression of PD-L1. Our data further show that SDH5 exerts its PD-L1 downregulating effect by preventing β-catenin nuclear translocation and then affecting the expression of ZEB1 in large experimental cohorts. Moreover, consistent with the in vivo data, PD-L1 was higher in SDH5-/- mice than in SDH5+/+ mice. The results of this study demonstrate that the loss of SDH5 protein is a crucial oncogenic mechanism of PD-L1 expression in NSCLC. To the best of our knowledge, this is the first evidence showing that the connection between SDH5 and PD-L1 in NSCLC can be used as a potential biomarker to predict the expression of PD-L1. Moreover, we highlight the therapeutic potential of targeting SDH5 for cancer treatment either as a monotherapy or in combination with PD-L1-targeted immunotherapies.
Materials and methods
Cell lines and clinical specimens
A549, H460, HCC-827, and NCI-1975 cells were maintained in RPMI1640 culture medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco). H292 cells were maintained in DMEM (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco). Cells were maintained at 37°C in a humidified incubator containing 5% CO2 Human lung cancer and paracancerous lung tissue samples were obtained from Wuhan Union Hospital. All samples were anonymized. All protocols using human samples were reviewed and approved by the Ethical Committee of Huazhong University of Science and Technology. Written informed consent was obtained from all patients.
Regents, plasmids, and antibodies
Plasmids for SDH5 were obtained from Sigma. For cDNA transfection, cells (5 x 105 cells/well) were seeded into a 6-well plate (Costar) at 70–80% confluence before transfection. Transfection was carried out using Lipofectamine PLUS (Invitrogen) according to the manufacturer’s instructions. Wnt-CM and L-CM were collected according to the directions from the ATCC and were added to the cells for 24 h. The following primary antibodies were used: anti-SDH5 (Cell Signaling Technology,(USA), anti-PD-L1 (Cell Signaling Technology, (USA), anti-ZEB1 (Sigma), anti-β-catenin (Cell Signaling Technology, USA), anti-GSK-3 (Sigma), anti-phospho-GSK-3 (Ser-9) (Sigma), and anti-GAPDH (Abcam, UK).
siRNA transfection
An siRNA targeting SDH5 was purchased from Santa Cruz Biotechnology (sc-96879). Cells were transfected with 50 nM siRNA. GenMute transfection reagent (SL100568, SignaGen Laboratories, China) was used according to the manufacturer’s instructions. Silencer select siRNAs against Zeb1 and β-catenin were purchased from Thermo Fisher Scientific (Zeb1: 3ʹCAUAGUGGUUGCUUCAGGATTUCCUGAAGCAACCACUAUGTT and β-catenin: 3ʹACGCUGCAUAAUCUCCUGTTCAGGAGAUUAUGCAGCGUTT).
Western blot analyzes
Cultured cells were washed twice with ice-cold PBS, and total protein extraction was performed using RIPA lysis buffer containing 1 mmol/L phenylmethanesulfonyl fluoride (PMSF) together with protease and phosphatase inhibitors. Cytosolic protein, nuclear protein and membrane protein extraction was performed according to the manufacturer’s protocol. The protein concentration was then determined by the Bradford method using bovine serum albumin (BSA, 0.1 mg/mL) as the standard. Equal concentrations (30 mg) of total protein was subjected to 10% polyacrylamide SDS gel electrophoresis and transferred to PVDF membranes. The membranes were blocked with 5% skim milk in Tris-buffered saline containing Tween (TBST) buffer (10 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, and 0.1% Tween-20) for 2 h. Protein expression was detected using primary antibodies incubated overnight at 4°C, followed by incubation with secondary antibodies for 1 h at RT. After the membranes were washed with TBST buffer 3 times, the proteins were visualized with enhanced chemiluminescence reagent.
Flow cytometry
A total of 1 × 106 human GC cells were harvested and incubated with APC-conjugated PD-L1 or its isotype control mouse IgG (eBiosience, Inc., San Diego, CA, USA) at 4°C for 30 minutes, washed twice, and subjected to flow cytometry analyzes using FACSDiva Software (BD Bioscience, Franklin Lakes, NJ, USA).
Fluorescence confocal microscopy
Cells were transfected with the control or SDH5 siRNA for 48 h. Then, the cells were fixed in 4% formaldehyde and subjected to indirect immunofluorescence microscopy with anti-β-catenin. The fluorescein isothiocyanate (FITC)-conjugated anti-IgG was purchased from Molecular Probes. Confocal immunofluorescence microscopy (Olympus) was performed using an Olympus confocal microscope according to the manufacturer’s protocol (×60 magnification).
Luciferase reporter gene assay
For the reporter gene assay, cells seeded into 24-well plates were transfected with β-catenin, a firefly luciferase reporter gene construct (TOP or FOP; 200 ng), and 1 ng of pRL-SV40 Renilla luciferase (as an internal control). Cell extracts were prepared 24 h after transfection, and luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega).
Quantitative RT-PCR
Total RNA was extracted using Trizol Reagent (Invitrogen). SYBR® Premix Ex TaqTM II (Takara Bio, Japan) was used to reverse-transcribe total RNA into cDNA according to the manufacturer’s instructions. The amplification was performed using SYBR Green Mastermix (Takara Bio, Japan) with a Step One Plus Real-Time PCR system (Applied Biosystems). The primer sequences are as follows: SDH5 forward primer 5ʹ-GACTTCGTCGCTGATGCTTG-3ʹ and reverse primer 5ʹ-GTTGGGCTGTCACCTCTGTA-3ʹ; PD-L1 forward primer 5ʹ- GCTGCACTAATTGTCTATTGGG and reverse primer 5ʹ- CACAGTAATTCGCTTGTAGTCG; and GAPDH forward primer 5ʹ-ACCACAGTCCATGCCATCAC-3ʹ and reverse primer 5ʹ-TCCACCACCCTGTTGCTGTA-3ʹ. Triplicate runs of each sample were normalized to GAPDH mRNA to determine relative expression.
Histology and immunohistochemical staining
Tumors were removed, weighed, fixed in 5% formalin, and prepared for histological analyzes. Consecutive tumor sections were stained with H&E, SDH5, and PD-L1. Immunohistochemical staining was carried out using an ABC staining kit (Santa Cruz Biotechnology) and a secondary biotinylated antibody to mouse IgG (Invitrogen). Lung cancer patient tissues were washed with PBS, inflated, and fixed with 10% buffered formalin. The sections were paraffin-embedded, cut into 5-m sections, and stained with routine H&E.
Animal experiments
The animal experiments were reviewed and approved by the Ethical Committee of HUST. For the orthotopic mouse model bearing lung cancer, the lungs of male nude mice (6–8 weeks of age) were exposed and injected with 5 × 105 cells suspended in 200 µl of phosphate-buffered saline (PBS). One week after injection, surgical staples were removed. Then, the indicated proteins were measured by western blot analyzes. The SDH5 knockout mouse model was generated according to a previously reported KO strategy.16
Analyzes of the TCGA data
Level 3 data were downloaded directly from the TCGA portal and utilized in subsequent analyzes. For lung adenocarcinomas included in the TCGA cohort, experimental procedures regarding RNA extraction from tumors, mRNA library preparation, sequencing (on the Illumina HiSeq platform), quality control, and subsequent data processing for the quantification of gene expression were performed as previously reported.18 The gene expression cutoff value was chosen as the tertile value over the entire dataset to ensure that all analyzes of each gene were based on the same cutoff value. Analyses were performed in R (version 3.0.1) (http:///www.r-project.org/). All tests were two-sided and considered significant at the 0.05 level.
Statistical analysis
R language (v. 3.4.3), SPSS software (v. 22.0), and GraphPad Prism (v. 7.0) for Windows were used for statistical analyses and generating figures. All error bars in graphical data represent the mean ± S.E. Spearman’s rank correlation was used to analyze the association between SDH5 and PD-L1 expression. Student’s two-tailed t test was used to determine the statistical relevance between groups, and p < .05 was considered significant.
Results
Correlation between SDH5 expression and PD-L1 in TCGA samples from patients with NSCLC
To verify the correlation between SDH5 and PD-L1 (CD274) expression, we compared the mRNA expression levels of SDH5 and PD-L1 in TCGA samples from patients with NSCLC and found a significant negative correlation between SDH5 and PD-L1 in lung adenocarcinoma (LUAD) and squamous cell lung carcinoma (LUSC). (Figure 1(a)). This result suggests that the regulation of PD-L1 by SDH5 may be at the transcriptional level. As a crucial immune checkpoint, PD-L1 binds to PD-1 to suppress the function of T cells and leads to immune escape. To further explore whether the high SDH5 expression-induced decrease in PD-L1 expression would eventually influence the function of T cells, we next assessed the relationship between SDH5 expression and the T-effector and IFN-γ-associated gene signature, which has previously been associated with activated T cells, immune cytolytic activity, and IFN-γ release.19 An integrated heatmap depicts the expression levels of T-effector and IFN-γ-associated gene signatures in tumors with low SDH5 expression compared to those with high SDH5 expression (Figure 1(b)). We identified a significant increase in the expression of both T-effector and IFN-γ-associated genes in LUSC patients with high SDH5, while there were no differences in LUAD, indicating preexisting immunity within tumor tissues with high SDH5 expression. Quantitative analysis of four key genes (GZMA, GZMB, CXCL10 and IFN-γ) in the T-effector and IFN-γ gene signature based on SDH5 expression levels was also consistent with the integrated heatmap (Figure 1(c)).
Figure 1.
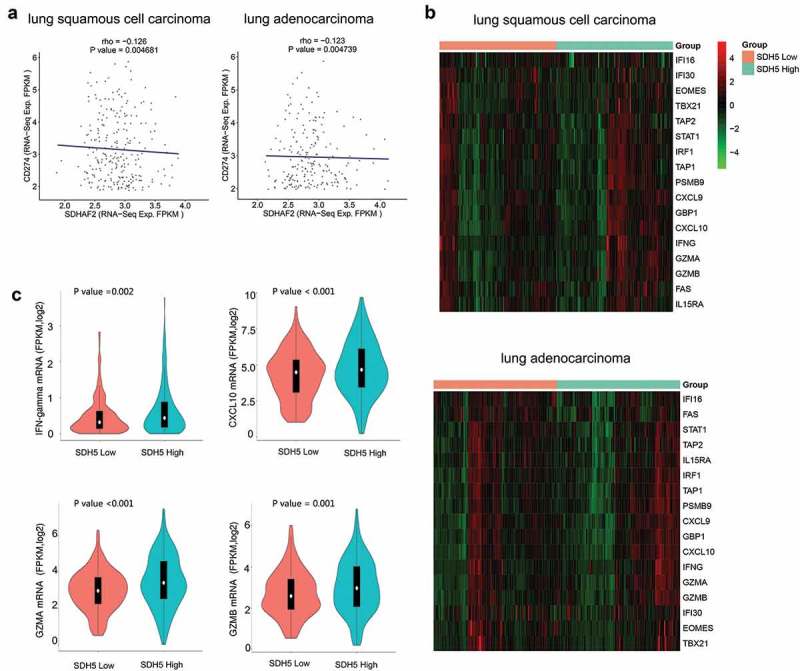
Correlation between SDH5 expression and PD-L1 in TCGA samples.
(a). The mRNA expression (FPKM log2) correlation between SDH5 and PD-L1 expression in LUAD and LUSC. (r = −0.126, P < .05 in LUSC and r = −0.123, p < .05 in LUAD). (b). Quantitative analysis of four key genes (IFN-γ, CXCL10, GZMA and GZMB) in the T-effector and IFN-γ gene signature based on the expression of SDH5 in LUAD and LUSC. (c). Heatmap depicting the mRNA expression levels of the T-effector and interferon-γ (IFN-γ)-associated gene signature in LUAD and LUSC between the SDH5-low and SDH5-high groups.
SDH5 inhibited PD-L1 expression in NSCLC cell lines
Next, we began to verify the regulation of PD-L1 by SDH5 at the cellular level in vitro. We transfected a small interfering RNA (siRNA) targeting SDH5 or an empty vector into H292, HCC827 and H1975 cells, which have high expression levels of SDH5 protein. The western blot results showed that the knockdown of SDH5 caused notably upregulated expression of PD-L1 (Figure 2 A,H292, B, HCC827, C,NCI1975). Next, we performed RT-PCR to verify whether the upregulation occurred at the transcriptional mRNA level and found that the transcriptional level of PD-L1 was significantly upregulated in several SDH5-knockdown cells (Figure 2(d–f)). In addition, flow cytometry results showed that the knockdown of SDH5 expression increased the amount of PD-L1 expression on the cell membrane (Figure 2(g–i)). To summarize these results, we hypothesized that the inhibition of SDH5 can upregulate the expression level of PD-L1 at the transcriptional level and eventually affect the final immune status of the tumor.
Figure 2.

SDH5 inhibited PD-L1 expression in vitro.
A small interfering RNA (siRNA) targeting SDH5 or an empty vector was transfected into H292, HCC827 and H1975 cells, and cell lysates were blotted with SDH5 and PD-L1 antibodies. GAPDH was used as a loading control. (a–c). Total PD-L1 mRNA was measured by RT-PCR. Knockdown of SDH5 expression significantly increased PD-L1 expression at the mRNA level in H292 (d), HCC827 (e) and H1975 (f) cells; **P < .01; ***P < .001 by unpaired Student’s t-test. (d–f) The expression of PD-L1 on the cell membrane was measured by flow cytometry (FCM) with a PD-L1 antibody.
SDH5 knockdown up-regulates the expression of PD-L1 by activating ZEB1
Epithelial-mesenchymal transition (EMT) is a process in which epithelial cells acquire mesenchymal features. In cancer, EMT is associated with tumor initiation, invasion, metastasis, and resistance to therapy.20,21 Our previous studies have shown that SDH5 plays a key role in the regulation of EMT by regulating the GSK-3β-β-catenin signaling pathway and that the inhibition of SDH5 induces upregulation of the EMT transcription factor ZEB1 protein.
The basic function of ZEB1 is to activate EMT. Chen L. et al. demonstrated that ZEB1 regulates the transcription of PD-L1 via miRNA-200.17 Therefore, we hypothesized that SDH5 inhibits the expression level of PD-L1 by inhibiting the transcription level of ZEB1. To prove this hypothesis, we first examined whether the inhibitory effect of SDH5 could be reversed by overexpressing ZEB1. In SDH5-transfected cells, increasing amounts of ZEB1 cDNA increased the level of PD-L1 protein expression (Figure 3(a), H460 and B, A549). Next, ZEB1-siRNA and SDH5-siRNA were transfected or cotransfected at the same time into HCC827 and H1975 cells, and we found that the upregulation of PD-L1 protein induced by SDH5 knockdown could be reversed by ZEB1 knockdown (Figure 3(c,d)). Flow cytometry results showed that the expression of PD-L1 membrane protein also exhibited this trend (Figure 3(e,f)).
Figure 3.
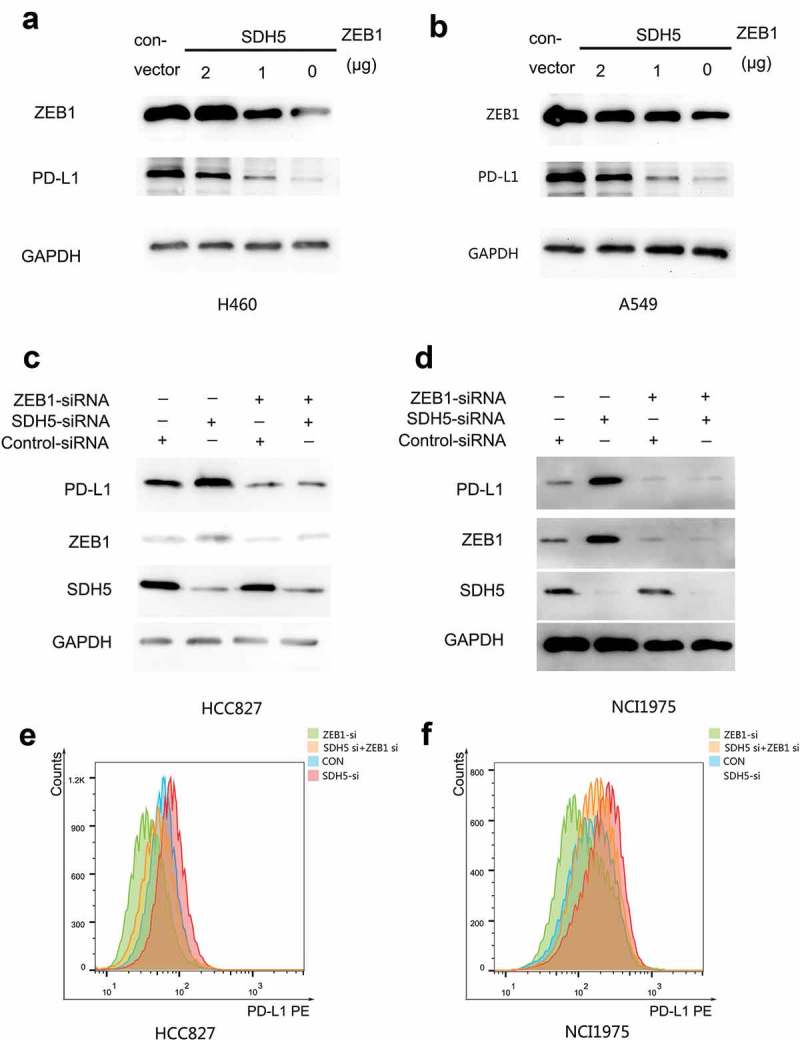
Loss of SDH5 triggers PD-L1 expression by activating ZEB1.
(a–b), ZEB1 overexpression reverses the SDH5-mediated PD-L1 decrease. Increasing amounts of ZEB1 and SDH5 cDNA were cotransfected into H460 (a) or A549 (b) cells for 24 h. Total cell lysates were probed with antibodies against ZEB1 and PD-L1. (c–d), ZEB1 and SDH5 siRNAs were transfected or cotransfected into HCC827 and H1975 cells. PD-L1, ZEB1 and SDH5 proteins were measured by Western blot analysis. (e-f). Flow cytometry (FCM) assay showing the same trend as that of western blot analysis in HCC827 (e) and H1975 (f) cells.
Furthermore, we found that the mRNA expression of SDH5 is not only associated with ZEB1 but is also negatively correlated with the mRNA expression of multiple EMT transcription factors, such as ZEB2 (Figure 4(a,b)). Linear regression analysis showed a significant negative correlation between SDH5, ZEB1 and ZEB2 in lung adenocarcinoma and lung squamous cell carcinoma (Figure 4(c)). At the same time, we found that the mRNA level of ZEB1 is not only positively correlated with the mRNA level of PD-L1 but is also positively correlated with various immune checkpoints, such as PD-L2, TIM3 and CTLA-4 (Figure 4(d,e)). However, this trend is more significant in lung squamous cell carcinoma.
Figure 4.
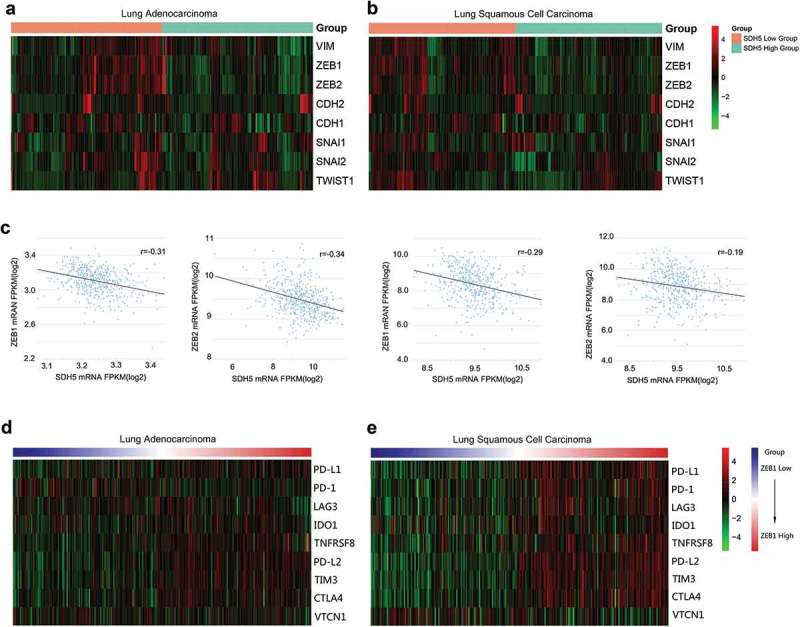
Correlation between the expression of SDH5, EMT transcription factors and selected immune inhibitory checkpoints.
(a–b). Heatmap representation of the relative mRNA expression levels of select EMT transcription factors based on the expression of SDH5 in LUAD and LUSC. (c). Quantitative analysis of two key genes, ZEB1 and ZEB2, based on the expression of SDH5 in LUAD and LUSC. (d–e). Heatmap representation of the relative mRNA expression levels of select immune inhibitory checkpoints based on the expression of ZEB1 in LUAD and LUSC.
SDH5 knockdown up-regulates the expression of PD-L1 by increasing the transcription level of β-catenin
To understand the possible mechanism by which SDH5 inhibits PD-L1, we examined the effect of SDH5 on the GSK3β/β-catenin signaling pathway. In the canonical Wnt pathway, GSK3β/β-catenin degradation is inhibited, leading to the accumulation of β-catenin in the nucleus and the transactivation of β-catenin/T cell factor (TCF) target genes.22 Thus, the hallmark of β-catenin signaling in both normal and neoplastic tissues is nuclear translocation. By knocking down endogenous SDH5 with an siRNA, we observed the accumulation of cytoplasmic β-catenin, the nuclear translocation of β-catenin, and reduced membrane-associated β-catenin (Figure 5(a,b)). SDH5 activates GSK-3β and antagonizes the Wnt-mediated GSK-3β-β-catenin signaling pathway, which is a key inducer of EMT during embryonic development and cancer progression. We tested whether manipulating SDH5 levels in various cell lines could modulate PD-L1 through the Wnt-induced GSK-3β/β- catenin signaling pathway. Whereas Wnt only slightly elicited EMT (Figure 5(c), left two lanes) in HCC827 cells, it significantly increased EMT when endogenous SDH5 was knocked down by SDH5-siRNA (Figure 5(c), right two lanes). In contrast, restoring SDH5 expression in H460 cells (SDH5-negative cells) prevented Wnt-induced EMT. Consistent with these results, β-catenin/TCF transcriptional activity was increased upon the knockdown of SDH5 (Figure 5(e–f)).
Figure 5.
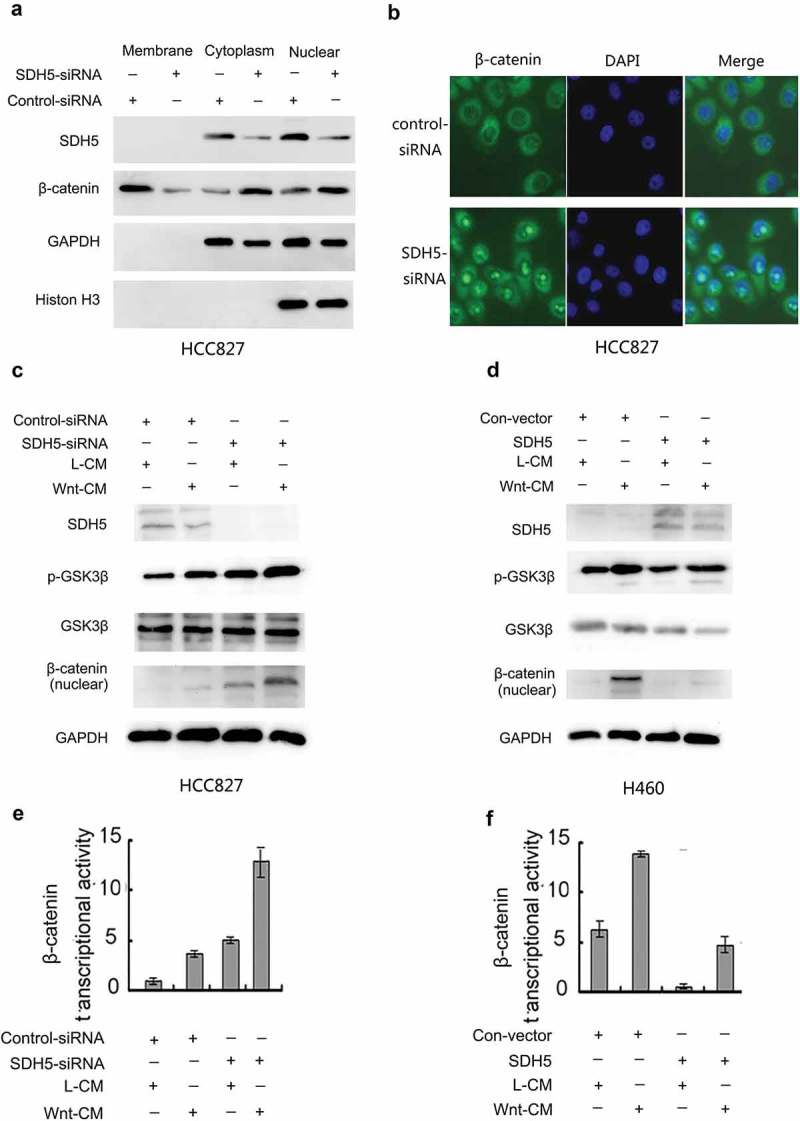
SDH5 antagonizes the Wnt-mediated GSK3β/β-catenin signaling pathway.
(a–b), SDH5 prevents the nuclear translocation of β-catenin. HCC827 cells were transfected with a control siRNA or SDH5-siRNA. The subcellular localization of β-catenin was visualized by Western blot analysis (a) and confocal microscopy (magnification, *500)(b). (c) Knockdown of SDH5 inactivates GSK-3β, promotes β-catenin activity, and enhances the Wnt-induced GSK-3/β-catenin signaling pathway in HCC827 cells. Cells were cotransfected with the control siRNA or SDH5-siRNA and TOP or FOP. Then, the cells were treated with L- or Wnt-CM. (c), Cell lysates were subjected to Western blot analysis. (d), In contrast, restoring SDH5 expression in H460 cells (SDH5-negative) prevented the Wnt-induced GSK-3/β-catenin signaling pathway. (e–f). Relative luciferase expression was measured as described above. Asterisks indicate statistically significant differences in cells transfected with the control vector versus those transfected with SDH5. Error bars, S.E.
To demonstrate whether SDH5 upregulates ZEB1 by inducing β-catenin transcription activity levels and ultimately affecting PD-L1, we knocked down β-catenin and SDH5 at the same time and found that the upregulation of PD-L1 and ZEB1 protein induced by SDH5 knockdown was abolished (Figure 6c, HCC827 B, NCI-1975). Next, we found that in SDH5-transfected cells, increasing amounts of β-catenin cDNA increased the level of PD-L1 protein expression (Figure 6(c), H460, D, A549). β-Catenin/TCF transcriptional activity was measured by the Luciferase Reporter Gene Assay (Figure 6(e-f)). Taken together, these results led us to hypothesize that the loss of SDH5 could increase PD-L1 expression by activating the GSK3β/β-catenin/ZEB1 signaling pathway.
Figure 6.
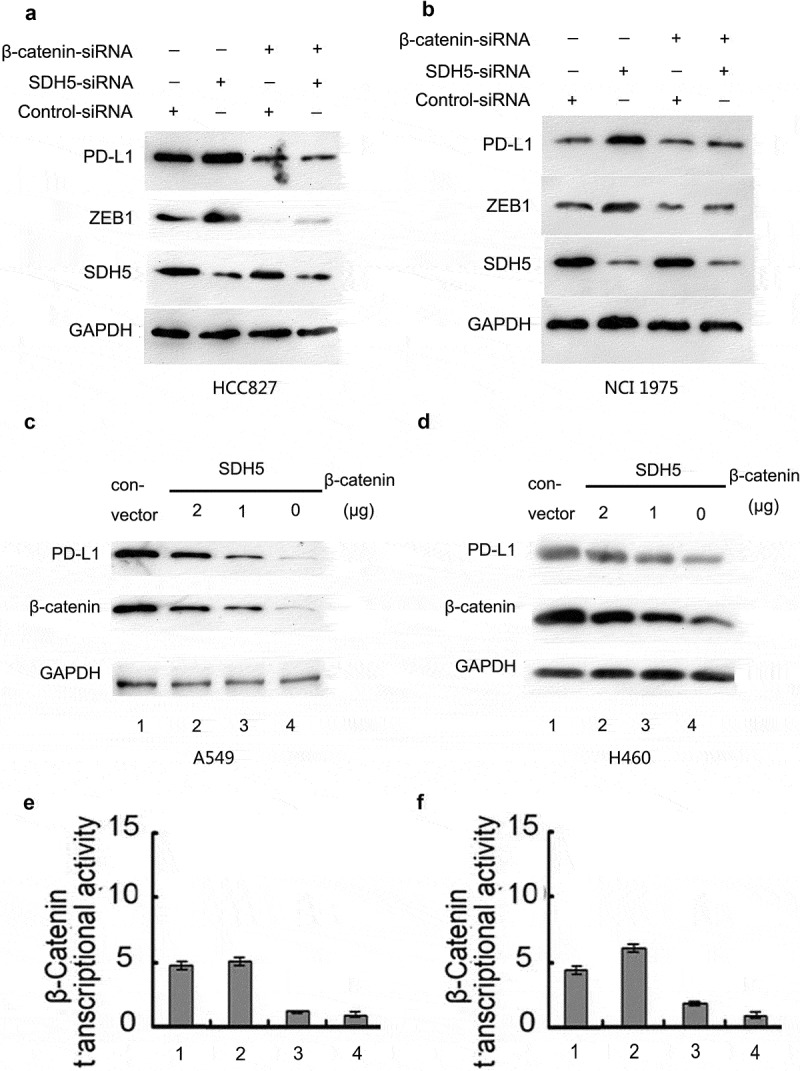
SDH5 knockdown upregulates the expression of PD-L1 by increasing the transcription level of β-catenin.
(a–b). Simultaneous knockdown of β-catenin and SDH5 abrogated the SDH5 knockdown-induced increase in PD-L1 protein in HCC827 (a) and H1975 (b) cells. (c–d). In SDH5-transfected cells, increasing amounts of β-catenin cDNA increased the level of PD-L1 protein expression in A549 (c) and H460 (d) cells. (e–f). Relative luciferase expression of β-catenin was measured as described above.
SDH5 regulates PD-L1 in SDH5-knockout mice and mice bearing SDH5-KD tumors
To further determine the impact of SDH5 on phenotypic changes in normal lung tissue, SDH5-knockout (KO; SDH5-/-) mice were employed. The strategy for targeted deletion of the mouse SDH5 gene is outlined in our previous study.16 This strategy effectively disrupts the coding regions of SDH5 that are required for its activity. Moreover, after homologous recombination, it can introduce a translation stop codon in-frame, preventing translation downstream of the SDH5 sequence. In vitro data showed that PD-L1 expression was higher in SDH5-/-mice than in SDH5+/+ mice (Figure 7(a)), suggesting that PD-L1 expression is elevated in SDH5-/- mice. Additionally, lung epithelial cells in SDH5-/- mice exhibit elevated ZEB1 and p-GSK3β expression (Figures 7(a) and 6(b)).
Figure 7.

SDH5 regulates PD-L1 in SDH5-knockout mice and mice bearing KD tumors.
(a). PD-L1, p-GSK-3β and ZEB1 protein expression levels in lung homogenates from WT and SDH5-knockout mice. (b). Expression of SDH5 and PD-L1 in paraffin-embedded lung tissue sections was determined by IHC(magnification, x100). H&E staining was used to identify pathology (magnification, x100). C. PD-L1, p-GSK-3β, and ZEB1 protein expression levels in KD (SDH5-knockdown) versus con-expressing HCC827 cells using an orthotopic mouse model.
To verify whether SDH5 can regulate PD-L1 in SDH5-KD tumors in vivo, we examined the PD-L1 protein expression level in KD mice (SDH5-knockdown cells) versus control mice (con-expressing HCC827 cells) using an orthotopic mouse model. The results showed that mice bearing KD (SDH5-knockdown) cells express higher levels of PD-L1 than control mice. Furthermore, SDH5-knockdown mice exhibited increased ZEB1 and PD-L1 expression (Figure 7(c)).
SDH5 expression regulates PD-L1 in lung cancer patients
To examine the relationship between SDH5 and the PD-L1 expression, we performed an analytical, observational, open, and retrospective study (ChiCTR1900022067). A total of 15 lung cancer specimens were retrieved by experienced surgical pathologists from Wuhan Union Hospital using computed tomography (CT)-guided needle biopsy (Figure 8(a)). The size of the tumor is between 3 to 10 cm.We examined the relationship between SDH5 expression and PD-L1 in lung cancer patients. Specimens from human lung cancer patients were examined by positron emission tomography/computed tomography (Figure 8(a)). Based on the TNM staging system for lung cancer, we selected patients with stage IV tumors. Detailed information for the chosen patients is listed in Table 1. A loss of SDH5, as well as increased p-GSK-3β, ZEB1 and PD-L1 levels, was clearly detected in tissues from lung cancer patients with low SDH5 expression (Figure 8(b)). There was a significant inverse correlation between the levels of SDH5 and PD-L1 in all of the samples tested. H&E staining verified lung cancer patient pathology (Figure 8(c)). Taken together, our human in vivo results are consistent with our in vitro results from various cancer cell lines.
Figure 8.

SDH5 in primary cancer lung cancer is correlated with the expression of PD-L1.
Loss of SDH5 expression correlates with changes in PD-L1 and EMT marker expression in clinical specimens from lung cancer patients. (a). Representative images of the position of CT-guided percutaneous needle biopsy of lung cancer. (b). Expression levels of SDH5, PD-L1, β-catenin, p-GSK-3β (Ser-9) and ZEB1 in normal (n = 3) and lung cancer (n = 3) tissues were determined by Western blot analysis. Densitometry was used to determine relative protein levels, and all proteins were normalized to the levels of β-actin. (c). Elevated mesenchymal markers and PD-L1 in lung cancer tissues. H&E staining was used to identify lung cancer patients’ pathology (magnification, x100).
Table 1.
Detailed patient information.
| Patient number | age | gender | stage | ps score | PD = L1 | SDH5 RQ |
|---|---|---|---|---|---|---|
| 1 | 65 | male | IV | 1 | >20 | <0.01 |
| 2 | 66 | female | IV | 1 | >20 | 0.05 |
| 3 | 77 | female | IV | 0 | >20 | <0.01 |
| 4 | 78 | male | IV | 0 | >20 | <0.01 |
| 5 | 31 | male | IV | 1 | >20 | 0.05 |
| 6 | 54 | female | IV | 1 | >20 | <0.01 |
| 7 | 55 | female | IV | 0 | >20 | <0.01 |
| 8 | 67 | female | IV | 1 | >20 | <0.01 |
| 9 | 55 | male | IV | 0 | <1 | 0.13 |
| 10 | 57 | female | IV | 2 | <1 | 0.16 |
| 11 | 65 | female | IV | 1 | <1 | 0.71 |
| 12 | 61 | male | IV | 2 | <1 | 0.13 |
| 13 | 45 | female | IV | 1 | <1 | 0.12 |
| 14 | 57 | female | IV | 1 | <1 | 0.57 |
| 15 | 78 | female | IV | 0 | <1 | 1 |
Discussion
In this study, we hypothesized that SDH5 might be associated with PD-L1 expression in NSCLC and found that the high expression of tumor PD-L1 was frequently identified in SDH5-deficient tumors in large experimental and validation cohorts. The current study provides evidence indicating that SDH5 can reverse the suppressive immuno-microenvironment in NSCLC and that the underlying mechanisms might involve PD-L1 and the GSK3β/β-catenin/ZEB1 signaling pathways. In addition, the TCGA analysis showed significant differences in the expression levels of T-effector and IFN-γ-associated gene signatures in tumors with low SDH5 expression compared with those with high SDH5 expression. Furthermore, we found a negative correlation between the expression status of SDH5 and PD-L1 in NSCLC patients, indicating preexisting immunity within SDH5-high tumors.
Succinate dehydrogenase 5 (SDH5), also known as SDHAF2, is required for the flavination of succinate dehydrogenase, which has been reported to contribute to the development of several kinds of cancers. Our previous study demonstrated that SDH5 depletion can facilitate EMT, leading to lung cancer metastasis. However, the mechanism by which SDH5 influences tumors in immune evasion is poorly understood. In this study, our findings provide a new role for SDH5 and suggest that SDH5 modulates the tumor immune response by regulating PD-L1.
Many previous studies have shown that EMT-related pathways and transcription factors are involved in the regulation of PD-L1.23,24 It has been reported that ZEB1 can increase PD-L1 expression by activating miRNA-200 in NSCLC and results in a dysfunction of CD8+ T cells, which couples with epithelial-mesenchymal transition (EMT) to increase metastasis. In autochthonous mouse melanoma models, Spranger S. et al. demonstrated that tumor-intrinsic active β-catenin signaling results in T cell exclusion and resistance to anti-PD-L1/anti-CTLA-4 monoclonal antibody therapy. Therefore, the activation of EMT signaling seems to be a crucial oncogenic mechanism that upregulates PD-L1 in various cancer types. In the present study, we suggest that SDH5 induces β-catenin entry into the nucleus and upregulates the EMT transcription factor ZEB1 by phosphorylating GSK3β. Furthermore, upregulated ZEB1 increases the transcriptional activity of PD-L1 and increases the expression of PD-L1 protein.
Moreover, in the current study, we found that high SDH5 expression inhibits the expression of multiple EMT-related transcription factors, such as ZEB1, from the TCGA data. Many previous studies have proven that high ZEB1 expression can increase the expression of various immune inhibitory checkpoints. We also analyzed the TCGA data and found that SDH5 negatively correlated with the T-effector interferon-γ (IFN-γ)-associated gene. Consistent with the in vitro data, our orthotopic lung cancer animal model (mice bearing SDH5-knockdown cells_ displayed a dramatic increase in the expression of PD-L1.
Moreover, western blot analysis showed that PD-L1 expression was higher in SDH5-knockout mice than in SDH5 wild type mice. We further examined the relationship between SDH5 expression and PD-L1 status in lung cancer patients. Immunohistochemistry of lung cancer tissue samples from patients revealed that the expression of PD-L1 was negatively related to the expression of SDH5, which is consistent with the TCGA results. Taken together, these data indicate that SDH5 is a key regulator of PD-L1 and may affect the tumor immune microenvironment by inhibiting EMT levels.
In particular, we reported for the first time that SDH5 could downregulate the expression of PD-L1 by inactivating the GSK3β/β-catenin/ZEB1 signaling pathways and eventually affected the TME in NSCLC. Although multiple studies have focused on PD-L1 expression and its immunosuppressive effect in NSCLC, we are the first to report its expression and relation to SDH5 in NSCLC patients, indicating that SDH5 is involved in the regulation of the tumor immune microenvironment and tumor immune escape.
Programmed cell death ligand 1 (PD-L1) expression has recently been proposed as a predictive biomarker for the response to PD-1 blockade immunotherapy. However, there are still many treatment responses beyond the explanation of these factors. It has been proposed that immunotherapy combined with targeted therapy might effectively improve clinical outcomes.25–27 Liu L. el at. showed that the combination of trametinib with immunomodulators targeting PD-1, PD-L1, or CTLA-4 was more efficacious than any single agent.28 These studies remind us that decreasing PD-L1 expression through signaling inhibition may be combined with anti-PD-L1 or anti-PD-1 therapy, which may increase treatment efficacy by controlling the PD-L1/PD-1 axis at very low levels.
Our study indicated that SDH5 could act as a tumor suppressor by inhibiting EMT and reversing the suppressive immuno-microenvironment of NSCLC. Our findings suggest that future work exploring the tumor SDH5 status as a predictive factor for PD-L1 status and cancer immunotherapy may be needed. Our data also highlight the therapeutic potential of targeting SDH5 for cancer treatment either as a monotherapy or in combination with PD-L1-targeted immunotherapies. These results would improve our understanding of the oncogenic mechanism involved in PD-L1 regulation in NSCLC. They could also help us find optimal candidates for immunotherapy.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Sznol M, Chen L.. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer–response. Clin Cancer Res. 2013;19(19):1. doi: 10.1158/1078-0432.CCR-13-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21(2):137–12. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5(12):1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 4.Tartour E, Zitvogel L. Lung cancer: potential targets for immunotherapy. Lancet Respir Med. 2013;1(7):551–563. doi: 10.1016/S2213-2600(13)70159-0. [DOI] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bondarenko I, Luft A, Serwatowski P, Barlesi F, Chacko R, Sebastian M, Neal J, Lu H, Cuillerot J-M, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line treatment in stage IIIB/IV non-small-cell lung cancer: results from a randomized, double-blind, multicenter phase II study. J Clin Oncol. 2012;30(17):2046–2054. doi: 10.1200/JCO.2011.38.4032. [DOI] [PubMed] [Google Scholar]
- 6.Hellmann MD, Rizvi NA, Goldman JW, Gettinger SN, Borghaei H, Brahmer JR, Ready NE, Gerber DE, Chow LQ, Juergens RA, et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): results of an open-label, phase 1, multicohort study. Lancet Oncol. 2017;18(1):31–41. doi: 10.1016/S1470-2045(16)30624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui R, Garon EB, Goldman JW, Leighl NB, Hellmann MD, Patnaik A, Gandhi L, Eder JP, Ahn M-J, Horn L, et al. Pembrolizumab as first-line therapy for patients with PD-L1-positive advanced non-small cell lung cancer: a phase 1 trial. Ann Oncol. 2017;28(4):874–881. doi: 10.1093/annonc/mdx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WEE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373(2):123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li CW, Lim SO, Xia W, Lee -H-H, Chan L-C, Kuo C-W, Khoo K-H, Chang -S-S, Cha J-H, Kim T, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 12.Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst. 2016;108:1. doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noman MZ, Janji B, Abdou A, Hasmim M, Terry S, Tan TZ, Mami-Chouaib F, Thiery JP, Chouaib S. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology. 2017;6(1):e1263412. doi: 10.1080/2162402X.2016.1263412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley J-P, Kunst H, Devilee P, Cremers CWRJ, Schiffman JD, Bentz BG, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325(5944):1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starker LF, Delgado-Verdugo A, Udelsman R, Bjorklund P, Carling T. Expression and somatic mutations of SDHAF2 (SDH5), a novel endocrine tumor suppressor gene in parathyroid tumors of primary hyperparathyroidism. Endocrine. 2010;38(3):397–401. doi: 10.1007/s12020-010-9399-0. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Gao L, Zhang H, Wang D, Wang M, Zhu J, Pang C, Wang C. Succinate dehydrogenase 5 (SDH5) regulates glycogen synthase kinase 3beta-beta-catenin-mediated lung cancer metastasis. J Biol Chem. 2013;288(41):29965–29973. doi: 10.1074/jbc.M113.450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Gibbons DL, Goswami S, Wuestner S, Hess O. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research N Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Saci A, Szabo PM, Chasalow SD, Castillo-Martin M, Domingo-Domenech J, Siefker-Radtke A, Sharma P, Sfakianos JP, Gong Y, et al. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun. 2018;9(1):3503. doi: 10.1038/s41467-018-05992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv YF, Dai H, Yan GN, Meng G, Zhang X, Guo QN. Downregulation of tumor suppressing STF cDNA 3 promotes epithelial-mesenchymal transition and tumor metastasis of osteosarcoma by the Wnt/GSK-3beta/beta-catenin/Snail signaling pathway. Cancer Lett. 2016;373(2):164–173. doi: 10.1016/j.canlet.2016.01.046. [DOI] [PubMed] [Google Scholar]
- 23.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, Skoulidis F, Parra ER, Rodriguez-Canales J, Wistuba II, et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res. 2016;22(3):609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asgarova A, Asgarov K, Godet Y, Jin G, Su J, Chou JW, Hoadley KA, Print C, Knowlton N, Black MA, et al. PD-L1 expression is regulated by both DNA methylation and NF-kB during EMT signaling in non-small cell lung carcinoma. Oncoimmunology. 2018;7(5):e1423170. doi: 10.1080/2162402X.2018.1490854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stagg J, Loi S, Divisekera U, et al. Anti-ErbB-2 mAb therapy requires type I and II interferons and synergizes with anti-PD-1 or anti-CD137 mAb therapy. Proc Natl Acad Sci U S A. 2011;108(17):7142–7147. doi: 10.1073/pnas.1016569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahnel PS, Thaler S, Antunes E, Huber C, Theobald M, Schuler M. Targeting AKT signaling sensitizes cancer to cellular immunotherapy. Cancer Res. 2008;68(10):3899–3906. doi: 10.1158/0008-5472.CAN-07-6286. [DOI] [PubMed] [Google Scholar]
- 27.Li B, VanRoey M, Wang C, Chen TH, Korman A, Jooss K. Anti-programmed death-1 synergizes with granulocyte macrophage colony-stimulating factor–secreting tumor cell immunotherapy providing therapeutic benefit to mice with established tumors. Clin Cancer Res. 2009;15(5):1623–1634. doi: 10.1158/1078-0432.CCR-08-1825. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, Mayes PA, Eastman S, Shi H, Yadavilli S, Zhang T, Yang J, Seestaller-Wehr L, Zhang S-Y, Hopson C, et al. The BRAF and MEK inhibitors dabrafenib and trametinib: effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res. 2015;21(7):1639–1651. doi: 10.1158/1078-0432.CCR-14-2339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


