ABSTRACT
Ewing´s Sarcoma Family of Tumors (ESFT) are clinically aggressive bone and soft tissue tumors in children and young adults. Analysis of the immune tumor microenvironment (TME) provides insight into tumor evolution and novel treatment options. So far, the scarcity of immune cells in ESFT has hindered a comprehensive analysis of rare subtypes. We determined the relative fraction of 22 immune cell types using 197 microarray gene expression datasets of primary ESFT tumor samples by using CIBERSORT, a deconvolution algorithm enumerating infiltrating leucocytes in bulk tumor tissue. The most abundant cells were macrophages (mean 43% of total tumor-infiltrating leukocytes, TILs), predominantly immunosuppressive M2 type macrophages, followed by T cells (mean 23% of TILs). Increased neutrophils, albeit at low number, were associated with a poor overall survival (OS) (p = .038) and increased M2 macrophages predicted a shorter event-free survival (EFS) (p = .033). High frequency of T cells and activated NK cells correlated with prolonged OS (p = .044 and p = .007, respectively). A small patient population (9/32) with combined low infiltrating M2 macrophages, low neutrophils, and high total T cells was identified with favorable outcome. This finding was confirmed in a validation cohort of patients with follow up (11/38). When comparing the immune TME with expression of known stemness genes, hypoxia-inducible factor 1 α (HIF1α) correlated with high abundance of macrophages and neutrophils and decreased T cell levels. The immune TME in ESFTs shows a distinct composition including rare immune cell subsets that in part may be due to expression of HIF1α.
KEYWORDS: CIBERSORT, tumor-infiltrating immune cells, tumor microenvironment, Ewing´s Sarcoma Family of Tumors, HIF1α
1. Introduction
The Ewing´s Sarcoma Family of Tumors (ESFT) includes the previously as primitive neuroectodermal tumor of bone, Ewing sarcoma, and Askin tumor defined soft tissue and bone tumors with an EWS/ETS balanced translocation, giving rise to oncogenic chimeric proteins, the most common being the EWS-FLI1 t(11;22)(q24;q12) (85%) with type 1 (exon 7 of EWS to exon 6 of FLI1) and type 2 (exon 7 of EWS to exon 5 of FLI1).1 In another 10-15% of cases, alternative translocations are found such as t(21;22)(22;12) resulting in EWS-ERG (ETS-related gene) fusion.2 More complex molecular aberrations are found in a minority of cases. Histologically, they belong to the group of “small round blue cell tumors” composed of small scattered tumor cells with high nucleus/cytoplasm ratio, finely dispersed chromatin arranged in sheets with occasional rosettes and varying degree of neuroectodermal differentiation and areas of necrosis. Treatment comprises local surgery, radiotherapy and polychemotherapy, and emerging novel agents are being tested in patients in the relapsed and metastatic stage.3 ESFTs are clinically aggressive tumors with a survival of 70–80% for patients with standard risk and localized disease and around 30% for those with metastatic disease, often to the lungs and bones.4 The recent Euro-Ewing 99 clinical trial showed 3-year overall survival (OS) rates of 72–78% and 3-year event-free survival (EFS) rates of 57–69 %.5 The corresponding 8-year OS ranged from 56–65% and the 8-year EFS ranged from 47–61 %.5 The Children´s Oncology Group reported 5-year OS rates of 77–83% and 5-year EFS rates of 65–73 %.6 Patients with relapsed ESFT have a dismal prognosis once metastasized.3 Thus, the continued search for biomarkers and novel therapeutic targets is urgently needed.7
The exact cell of origin in ESFT is unclear. However, recent evidence suggests that these tumors may arise from a mesenchymal stem cell (MSC) locked into a stemness phenotype through oncoprotein driven overexpression of enhancer of zeste homolog2 (EZH2).8–12
So far, a comprehensive immune tumor microenvironment (TME) characterization has been lacking. ESFT primary tumors contain varying numbers of tumor-infiltrating lymphocytes (TILs),13,14 with macrophages and T cells being far less abundant than in other malignant bone tumors,15 rendering a comprehensive quantitative characterization of immune subsets in tissue by standard immunohistochemistry (IHC) impossible. We try to fill this gap by using CIBERSORT,16 a retrospective in silico analysis that allows immune cell profiling by deconvolution of gene expression microarray data. It reconstructs the type and relative quantity of immune cell subsets in bulk tumor tissue using an expression matrix derived from gene expression data of 22 known immune cells from the peripheral blood using 547 characteristic marker genes.16 It has the advantage of being able to detect even rare and functionally distinct immune cell types (e.g. mast cells, γδ T cells, memory B cells, regulatory T cells/Tregs, etc.). This method has been successfully validated by flow cytometry and used to determine the infiltration of immune cells in various other malignant tumors (e.g. breast cancer and colon cancer).17,18
ESFT is a prime example of an embryonic tumor displaying stemness features, both morphologically through its “small round blue cell” morphology, phenotypically and through epigenetic reprogramming via EZH2.11,12
We therefore investigated whether the expression of previously published stemness genes19 might influence the composition of the immune TME found in ESFT.
2. Materials and methods
2.1. ESFT datasets and CIBERSORT analysis
Gene expression microarray data of 197 primary ESFT tumor samples from Gene Expression Omnibus (GEO)20 datasets GSE1825,10 GSE37371,21 GSE34620,22,23 GSE17679,24 and GSE1575712 were analyzed using CIBERSORT.16 Affymetrix HG-U133A (GEO accession number GPL96) and Affymetrix HG-U133 Plus 2.0 (GEO accession number GPL570) platform data was selected, other datasets were excluded from this study because the leucocyte signature comparison matrix was validated on the above platforms.16 Seventy of 197 patients had follow up data including OS and EFS. Thirty-two patients from GSE1767924 were included in the training, 38 patients from GSE3462022 in the validation cohort for Kaplan-Meier analysis.
Patients’ characteristics and microarray datasets used in this study are shown in Table 1.
Table 1.
Patient characteristics and microarray datasets.
| age (in years) |
sex |
no. of patients with survival information |
no. of patients with pretreatment |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEO ID | no. of primary ESFT | EWS translocated | platform | < 10 | 10 to 18 | >18 | female | male | OS data | EFS data | yes | no |
| GSE1825 | 5 (2.5%) | 5/5 | U133A | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| GSE37371 | 39 (19.8%) | NA – data not availible | U133A/U133Plus2 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| GSE34620 | 117 (59.4%) | 117/117 | U133Plus2 | 30/117 | 63/117 | 24/117 | 51/117 | 66/117 | 38/117 | 39/117 | NA | NA |
| GSE17679 | 32 (16.2%) | 32/32 | U133Plus2 | 4/32 | 15/32 | 13/32 | 11/32 | 21/32 | 32/32 | 32/32 | 5/32 | 27/32 |
| GSE15757 | 4 (2%) | NA – data not availible | U133A | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| TOTAL | 197 (100%) | |||||||||||
GEO, gene expression omnibus; OS, overall survival; EFS, event-free survival; NA, not applicable.
Gene expression microarray profiles of 197 primary ESFT tumor samples from Gene Expression Omnibus (GEO)20 datasets GSE1825, 10 GSE37371, 21 GSE34620, 22,23 GSE1767924 and GSE15757;12 total numbers of primary ESFTs within Gene Series Expression (GSE) and the percentage within the entire cohort. Age of patient at onset of disease in years. A total of 70 patients had complete OS and EFS survival data, 32 patients (GSE17679) (Figures 2 and 3) were used as a training cohort and 38 patients (GSE34620) (supplementary Figures 2 and 3) were used as a validation cohort.
2.2. Microarray preprocessing
Affymetrix array data was downloaded as CEL files (raw data) from GEO20 and probes were aggregated to HUGO gene symbols. All microarray studies were normalized according to the “Robust Multi-array Average” (RMA) method25 prior to analysis using the “affy” package in Bioconductor and R (R Foundation for Statistical Computing).
2.3. Assessment of immune infiltration by CIBERSORT
We used CIBERSORT to examine the relative fractions of 22 infiltrating immune cell types in each tumor tissue, using the LM22 signature matrix with 1,000 permutations (other parameters were left at default values). The LM22 matrix includes naïve and memory B cells, plasma cells, seven T cell types (CD8 T cells, naïve CD4 T cells, resting memory CD4 T cells, activated memory CD4 T cells, follicular helper T cells, regulatory T cells, γδ T cells), resting and activated natural killer (NK) cells, monocytes, macrophages (M0 macrophages, M1 macrophages, M2 macrophages), resting and activated dendritic cells (DC), resting and activated mast cells, eosinophils and neutrophils .16 The sum of all evaluated immune cell type fractions equals one for each tumor sample, hence all estimates are relative to total leukocyte content.
2.4. Statistical analysis
Statistical analysis was performed using the software package IBM SPSS (Chicago, IL) Statistics for Windows (version 24). Mean value comparisons were performed with the Mann-Whitney-U and Kruskal–Wallis test depending on the number of compared groups. The Benjamini-Hochberg (BH) procedure was used to correct for multiple testing errors with a false discovery rate of 0.05, where indicated.26 If not otherwise specified in the figure legends, data are presented as box plots with horizontal bars representing the median.
Survival analyses were performed using the Kaplan–Meier method and the log-rank test. Optimal cutoff points of cell abundance expressed as the percentage of a specific cell fraction within all immune cells were set at the point with the most significant (log-rank test) separation using the web-based tool “cutoff Finder”.27 Due to the small number of patients with survival data (training cohort: n = 32, validation cohort: n = 38), only exploratory data analysis within these subgroups was performed (no further multiple testing error analysis due to small cohort size). Univariate and multivariate analysis were performed using Cox regression analysis. Multivariate Cox regression analysis was run backwards with p(in) = 0.05 and p(out) = 0.1. Hazard ratios (HR) and their 95% confidence intervals (95% CI) were calculated.
P-values less than 0.05 were considered statistically significant.
3. Results
3.1. Immune cell composition in ESFT
The predominant immune cell type in ESFT determined by CIBERSORT were macrophages (43% of all leucocytes) with immunosuppressive M2 macrophages being the predominant population. The second most prominent cell fraction were T cells (mean: 23%) (follicular helper T cells > CD4 memory T cells > CD8 T cells > regulatory T cells > CD4 naïve T cells > γδ T cells). Interestingly, B cells (8%) and plasma cells (5%) as well as neutrophils (3%) and NK cells (6%) represented small immune cell subsets (Figure 1).
Figure 1.
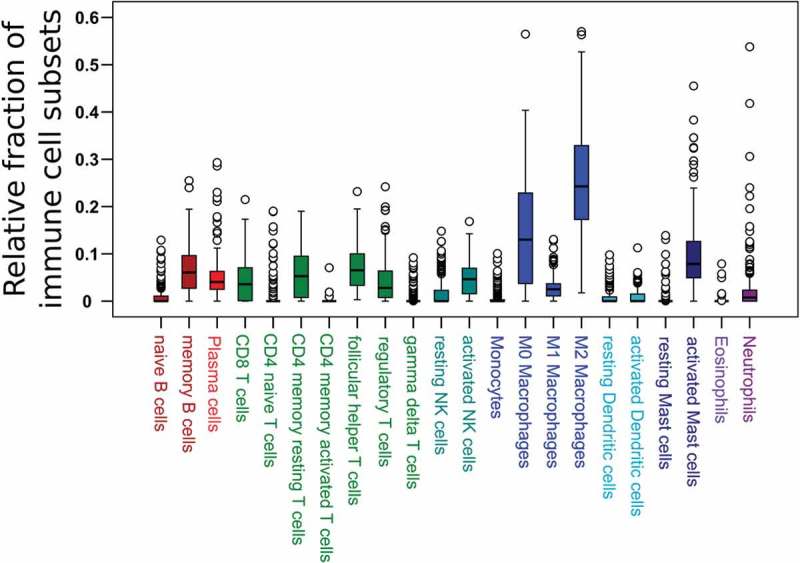
Immune cell distribution in 197 ESFT primary tumors (CIBERSORT).
Relative proportion of all tumor-infiltrating leucocytes of 22 immune cell subsets in ESFT.
3.2. No effect of neoadjuvant therapy and age of onset on immune subsets in ESFT
By subgroup analysis, pretreated tumors (n = 5) were compared to tumors of ESFT that were not treated with neoadjuvant chemotherapy (n = 27) (GSE17679) (Table 1). Although the absolute number of infiltrating leukocyte content is not given by CIBERSORT analysis, neoadjuvant treatment had no significant impact on relative immune cell subsets nor on PD-L1 expression (data not shown).
The age of disease onset was available in 149 ESFT cases. This subgroup was divided into group 1 (0 to 10 years of age), group 2 (10 to 18 years) and group 3 (>18 years). No significant differences between age groups were found (Kruskal–Wallis test) (data not shown).
3.3. Prognostic role of tumor-infiltrating immune cells
The distribution of relative immune cell fractions in ESFT (Figure 1) was correlated with OS and EFS in a training cohort of 32 patients with clinical follow-up data (GSE17679) and validated in an independent cohort of 38 patients (GSE34620). Infiltrating neutrophils correlated with shortened OS (median OS not reached vs. median OS of 20.7 months, 95% CI 11.9–29.5 months, p = .038, Figure 2(a)), whereas activated NK cells were associated with a prolonged OS (median OS not reached vs. median OS of 20.7 months, 95% CI 12.1–29.3 months, p = .007, Figure 2(b)) by Kaplan-Meier analysis. Higher levels of M2 macrophages were associated with shorter EFS (median EFS of 47 months vs. median EFS of 15.3 months, 95% CI 8.6–22.0 months, p = .033, Figure 2(d)) and memory B cells also correlated with shorter EFS (median EFS of 28.6 months, 95% CI 0.8–56.4 months, vs. median EFS of 11.7 months, 95% CI 7.5–15.9 months, p = .024, Figure 2(e)). When all T cell fractions were summed, high frequency of total T cells was associated with longer OS and EFS (median OS not reached vs. median OS of 21.3 months, 95% CI 12.1–30.5 months, p = .044, Figure 2(c); median EFS of 47 months vs. median EFS of 14 months, 95% CI 6.9–21.1 months, p = .032, Figure 2(f)). The combination of low M2 macrophages, low neutrophils and high T cells (“T cell predominant”) identified a small patient cohort (9/32 patients) with favorable outcome for both, OS (median OS not reached, p = .014, Figure 3(a)) and EFS (median EFS: 47 months, 95% CI not determined, p = .005, Figure 3(b)). The subgroup with high M2 macrophages, high neutrophils and low T cells (“M2-neutrophil predominant”) showed the shortest OS and EFS (median OS: 15.6 months, 95% CI 3.2–28 months; median EFS: 11 months, 95% CI 8.7–13.3 months). All other combinations of M2 macrophages, neutrophils and T cells (“mixed”) did not reach the median OS and had a median EFS of 19.4 months (95% CI 0–39 months).
Figure 2.
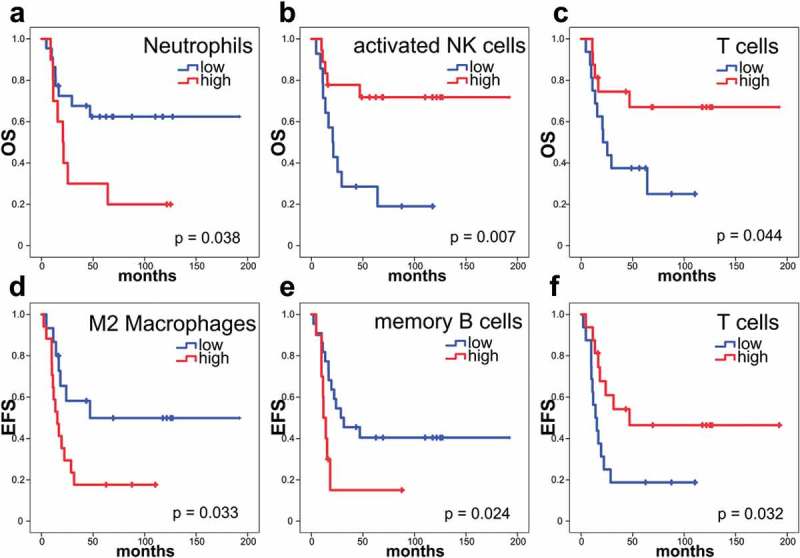
Survival in patients with ESFT is dependent on the abundance of immune cell types.
Kaplan-Meier analyses showing OS and EFS with tumors with low (blue) and high (red) frequency of specific immune cells determined by CIBERSORT, as indicated: (a, b, c) OS analysis. (d, e, f) EFS analysis. Thirty-two patients within the training cohort (GSE17679) were included in this survival analysis. (a) Low frequency of neutrophils in ESFT (<0.2546% of total immune cells) were associated with a good prognosis (median OS not reached), high frequency of neutrophils (>0.2546% of total immune cells) with an estimated median survival of only 20.7 months (95% CI 11.9–29.5 months) (p = .038). (b) High frequency of activated NK cells (>3.771% of total immune cells) were associated with a good prognosis (median OS not reached), low frequency of activated NK cells (<3.771% of total immune cells) with an estimated median survival of only 20.7 months (95% CI 12.1–29.3 months) (p = .007). (c) High frequency of total T cells (>22.43% of total immune cells) were associated with a good prognosis (median OS not reached), low frequency of total T cells (<22.43% of total immune cells) with an estimated median survival of only 21.3 months (95% CI 12.1–30.5 months) (p = .044). (d) Whereas patients ESFTs with a low frequency of M2 macrophages (<24.83% of total immune cells) had a median EFS of 47 months, high frequency of M2 macrophages (>24.83% of total immune cells) were associated with an estimated median survival of only 15.3 months (95% CI 8.6–22.0 months) (p = .033). (e) Low frequency of memory B cells (<8.655% of total immune cells) were associated with a median EFS of 28.6 months (95% CI 0.8–56.4 months), high frequency of memory B cells (>8.655% of total immune cells) with an estimated median survival of only 11.7 months (95% CI 7.5–15.9 months) (p = .024). (f) High frequency of total T cells (>22.43% of total immune cells) were associated with a median EFS of 47 months, low frequency of total T cells (>22.43% of total immune cells) with an estimated median survival of only 14 months (95% CI 6.9–21.1 months) (p = .032). Log-rank test.
Figure 3.
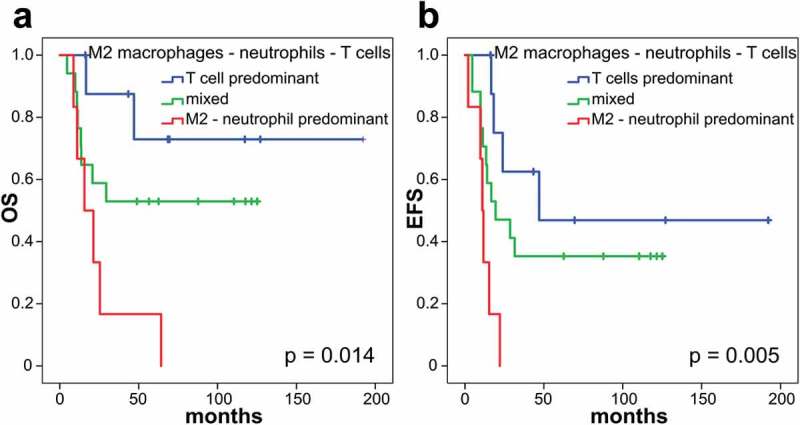
Combining M2 macrophage, neutrophil and T cell frequency predicts prognosis in primary ESFT.
Thirty-two patients within the training cohort (GSE17679) were included in this survival analysis. Blue curve (9/32 patients): low M2, low neutrophils and high T cells (“T cell predominant”) were associated with a good prognosis (median OS not reached; median EFS: 47 months, 95% CI not determined), red curve (6/32 patients): high M2, high neutrophils and low T cells (“M2-neutrophil predominant”) were associated with a dismal prognosis (median OS: 15.6 months, 95% CI 3.2–28 months; median EFS: 11 months, 95% CI 8.7–13.3 months) and green curve (17/32 patients): any other combination of M2 macrophages, neutrophils and T cells (“mixed”) with intermediate risk (median OS not reached, median EFS: 19.4 months, 95% CI 0–39 months). (a) Overall survival analysis, p = .014. (b) Event-free survival analysis, p = .005. Log-rank test.
For univariate and multivariate comparative analysis binary variables were used (Table 3).
Table 3.
Univariate and multivariate analysis of immune cell subsets and survival of 32 ESFT patients within the training cohort (GSE17679). All variables with a p-value of less than 0.05 by Kaplan-Meier analysis were analyzed using univariate and multivariate Cox regression analysis.
| A |
Overall survival |
|||
|---|---|---|---|---|
| Parameter | Category | HR | 95% CI | p |
| Univariate | ||||
| Neutrophils | low vs. high | 2.733 | 1.018–7.337 | .046 |
| Activated NK cells | low vs. high | 0.252 | 0.086–0.736 | .012 |
| T cells | low vs. high | 0.350 | 0.121–1.016 | .053 |
| M2-neutrophil-T cell | T cell/mixed vs.M2-neutro | 3.755 | 1.341–10.51 | .012 |
| Multivariate | ||||
| Neutrophils | low vs. high | 1.427 | 0.300–6.790 | .655 |
| Activated NK cells | low vs. high | 0.252 | 0.086–0.736 | .012 |
| M2-neutrophil-T cell |
T cell/mixed vs.M2-neutro |
2.178 |
0.703–6.743 |
.177 |
| B |
Event-free survival |
|||
| Parameter |
Category |
HR |
95% CI |
p |
| Univariate | ||||
| M2 macrophages | low vs. high | 2.603 | 1.043–6.492 | .040 |
| Memory B cells | low vs. high | 2.787 | 1.100–7.059 | .031 |
| T cells | low vs. high | 0.391 | 0.161–0.952 | .039 |
| M2-neutrophil-T cell | T cell/mixed vs.M2-neutro | 4.380 | 1.579–12.15 | .005 |
| Multivariate | ||||
| M2 macrophages | low vs. high | 1.813 | 0.654–5.025 | .253 |
| Memory B cells | low vs. high | 2.250 | 0.841–6.020 | .106 |
| T cells | low vs. high | 1.118 | 0.212–5.908 | .895 |
| M2-neutrophil-T cell | T cell/mixed vs.M2-neutro | 4.380 | 1.579–12.15 | .005 |
(A) Univariate Cox regression of overall survival (OS) analysis showed that neutrophils (p = .046), activated NK cells (p = .012) and a combination of M2 macrophages, neutrophils and total T cells (p = .012) correlated with OS. By multivariate Cox regression analysis, high abundance of activated NK cells (p = .012) showed a prolonged OS.
(B) Univariate Cox regression of event-free survival analysis (EFS) showed that M2 macrophages (p = .04), memory B cells (p = .031), T cells (p = .039) and a combination of M2 macrophages, neutrophils and total T cells (p = .005) were associated with EFS. By multivariate Cox regression analysis, a combination of M2 macrophages, neutrophils and total T cells (p = .005) correlated best with EFS.
Multivariate Cox regression analysis confirmed that activated NK cells (p = .012) and the combination of M2 macrophages, neutrophils and T cells (p = .005) were the most significant and independent prognostic factors for OS and EFS, respectively (Table 3).
Kaplan-Meier analysis of 38 patients (GSE34620) within the validation cohort confirmed the negative prognostic value of neutrophils and positive prognostic value of T cells by both OS and EFS (p = .029 and p = .026 for neutrophils; p = .005 and p = .02 for T cells; supplementary Figure 1). When M2 macrophages, neutrophils, and T cells were combined, a subgroup with favorable OS and EFS was confirmed in the validation cohort (p = .026 and p = .012; supplementary Figure 2). The prognostic value of the combination of M2 macrophages, neutrophils, and T cells was also confirmed by multivariate analysis in the validation cohort (p = .019 and p = .014; supplementary table 1).
3.4. Gene expression of published stemness genes and checkpoint molecules in ESFT
Out of 18 previously published stemness-related genes19 Myc, HIF1α and EZH2 showed the highest gene expression in ESFT with mean expression levels of 10.1, 9.9 and 9.6 (log2 Affymerix RMA intensity values) respectively (Figure 4(a)). Next, we analyzed checkpoint molecule expression in our cohort of ESFT cases. PD-L1 gene expression and most other molecules were low to undetectable in ESFTs (Figure 4(b)). No difference in PD-L1 expression was observed in tumors with low vs. high total T cell infiltrates (data not shown).
Figure 4.
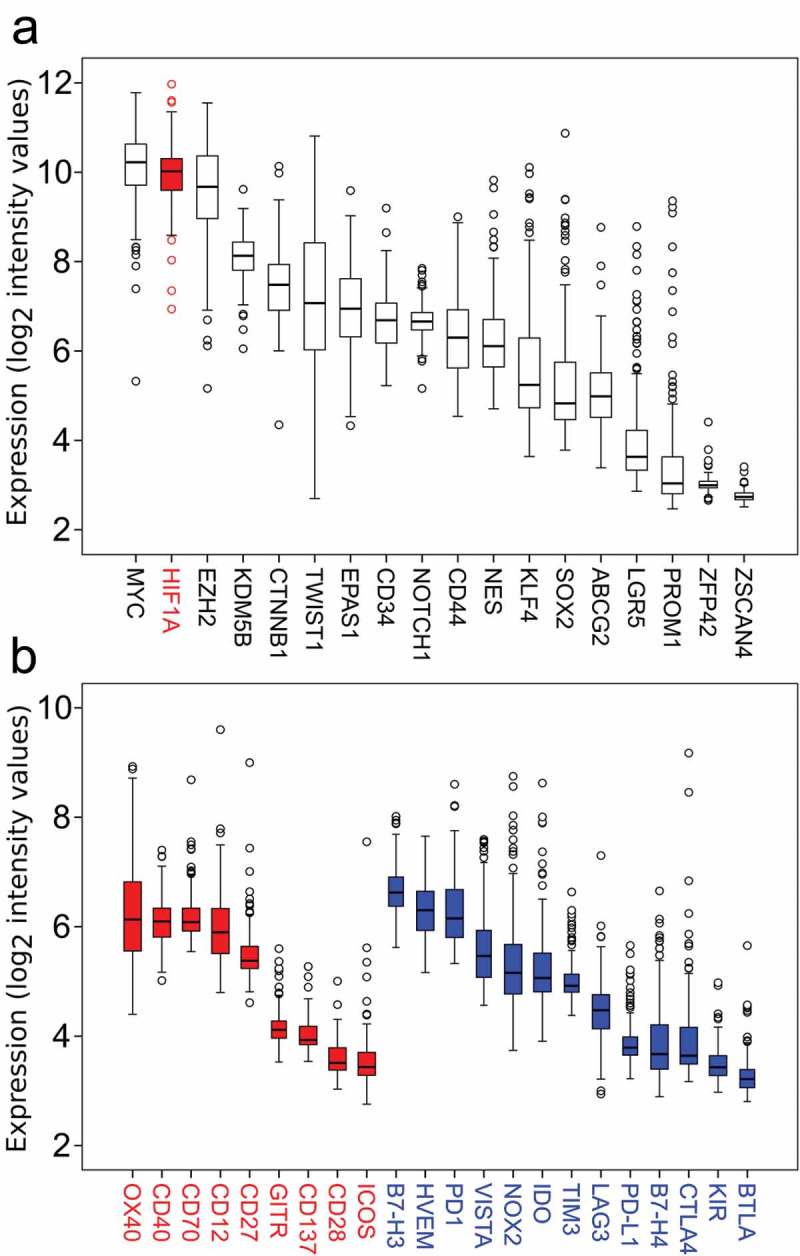
Gene expression of hallmark stemness genes and checkpoint molecules in 197 human ESFT samples.
(a) Myc, HIF1α, and EZH2 are highly expressed compared to other stemness genes in ESFT. (b) Co-stimulatory (red, left) and co-inhibitory checkpoint molecules (blue, right) show a low (<7 log2 intensity value) expression in ESFTs.
3.5. Correlation between HIF1α expression and immune cell subsets
When expression of known stemness genes was compared with the fraction of immune cell types generated by CIBERSORT, HIF1α showed a correlation with 12 immune cell populations (Table 2). We therefore divided the cohort into a low (< median) and high (> median) HIF1α expression group. The HIF1α high expression group showed significantly more macrophages, especially M2 macrophages (p < .001, Figure 5(b)) and neutrophils (p< .01, Figure 5(c)), while several T cell populations including CD8 positive T cells, follicular helper T cells, and Tregs were significantly reduced in the high HIF1α group (Table 2). When all T cell populations were summed, total T cells were significantly reduced in the high HIF1α expression group (p < .001, Figure 5(a)). The mean of relative immune cell subset in the low and high HIF1α group is shown in Table 2.
Table 2.
Relative proportions of immune cell subsets in low and high HIF1α expression subgroups of ESFT. The mean relative fractions of immune cell subsets in 197 ESFT tumors in subgroups with low and high HIF1α expression. Significant differences after correcting for multiple testing error using the Benjamini-Hochberg (BH) procedure with a false discovery rate of 0.05 are indicated in bold. *p < .05, **p < .01, ***p < .001 (Mann-Whitney-U-test).
| low HIF1α | high HIF1α | p | BH corrected p value | |
|---|---|---|---|---|
| Naive B cells | 0.009 | 0.01 | .211 | .253 |
| Memory B cells | 0.073 | 0.058 | .022* | .044* |
| Plasma cells | 0.056 | 0.047 | .001** | .003** |
| CD8 T cells | 0.06 | 0.03 | <.001*** | .003** |
| CD4 naive T cells | 0.011 | 0.01 | .252 | .275 |
| CD4 memory resting T cells | 0.052 | 0.063 | .080 | .113 |
| CD4 memory activated T cells | 0 | 0.001 | .077 | .113 |
| Follicular helper T cells | 0.081 | 0.056 | <.001*** | .003** |
| Regulatory T cells | 0.058 | 0.025 | <.001*** | .003** |
| Gamma delta T cells | 0.004 | 0.008 | .109 | .145 |
| Total T cells | 0.265 | 0.194 | <.001*** | .003** |
| Resting NK cells | 0.017 | 0.016 | .78 | .78 |
| Activated NK cells | 0.05 | 0.042 | .057 | .105 |
| Monocytes | 0.008 | 0.006 | .199 | .251 |
| M0 macrophages | 0.128 | 0.158 | .067 | .113 |
| M1 macrophages | 0.03 | 0.028 | .515 | .537 |
| M2 macrophages | 0.222 | 0.288 | <.001*** | .003** |
| Total macrophages | 0.38 | 0.474 | <.001*** | .003** |
| Resting DC | 0.011 | 0.005 | .001** | .003** |
| Activated DC | 0.013 | 0.007 | .021* | .044* |
| Resting mast cells | 0.003 | 0.007 | .006** | .014* |
| Activated mast cells | 0.095 | 0.096 | .073 | .113 |
| Eosinophils | 0 | 0.001 | .239 | .273 |
| Neutrophils | 0.019 | 0.036 | .005** | .013* |
Figure 5.
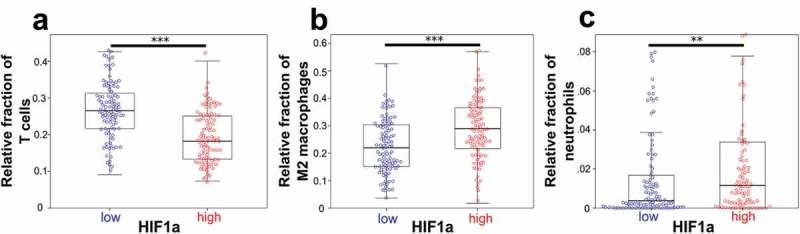
HIF1α correlates with total T cells, M2 macrophages, and neutrophils.
(a) T cells were significantly reduced in the high HIF1α expression group (mean: 26.5% vs. 19.4%). (b) M2 macrophages were significantly higher in the high HIF1α expression group (mean: 22.2% vs. 28.8%). (A) Neutrophils were significantly more abundant in the high HIF1α expression group (mean: 1.9% vs. 3.6%). The entire cohort of 197 ESFT was included. Box plots. **p < .01, ***p < .001 (Mann-Whitney-U-test).
As HIF1α positively correlated with dismal prognostic immune cell subsets like M2 macrophages and neutrophils and negatively correlated with good prognostic T cells, we further asked, whether HIF1α also was associated with a shortened survival. A trend was observed toward poor prognosis by OS and EFS, respectively, by Kaplan–Meier analysis of the training cohort (p = .071, p = .091, respectively). This correlation was confirmed by Kaplan-Meier analysis in the validation cohort. Here, high HIF1α expression was also associated with a shortened OS (p = .043).
4. Discussion
ESFTs represent a group of highly aggressive bone and soft tissue tumors in children and young adults. Clinically, ESFTs rapidly metastasize with 25% of patients presenting with metastatic disease initially.4,28 The relapse rate has been lowered in patients with localized disease due to intensified chemotherapy protocols and combination with autologous stem-cell therapy.5 While therapy has improved for patients with localized disease, novel-targeted therapies are urgently required for patients with relapsed and metastatic disease.
Our retrospective gene expression analysis shows low levels of PD-L1 transcripts in ESFTs (Figure 4), confirming previous reports demonstrating PD-L1 protein expression determined by IHC in only 0-19% of ESFT tumor samples depending on type of tissue, antibody used, and whether patients had been treated prior to biopsy.13,29,30
Immunoprofiling via CIBERSORT of ESFT gene expression microarrays confirms and extends prior findings of a TME rich in macrophages31 and devoid of checkpoint molecules such as PD-L1.30 Our results provide a road map by which tumor-infiltrating immune cells can be used as biomarkers and possible novel immunotherapeutic targets.
As this is an exploratory study, the CIBERSORT results will need to be compared with other methods like single-cell RNA sequencing, which would allow a more detailed analysis of the immune cell infiltrates. Moreover, larger clinical cohorts and longitudinal studies correlating molecular as well as immune TME data will be required to confirm the findings. In addition, other methods assessing absolute immune cell numbers and phenotypic data using flow cytometry and immunohistochemistry as well as algorithms assessing absolute cell counts will need to confirm the above findings.
Previously, unsupervised clustering of gene expression data identified gene signatures that may have come from non-neoplastic tumor-associated stromal cells.32 Here, we can show that at least a part may be derived from immune cells. It appears that M2 type macrophages and neutrophils are associated with a poor prognosis, whereas T cells and even relatively few activated NK cells predict a good prognosis in ESFT patients.
Embryonal tumors such as ESFT bear similarities morphologically and by gene expression with stem cells, thus we were interested to determine, which stemness genes were upregulated in ESFT and whether relative immune subsets in our cohort correlated with stemness. Interestingly, out of 18 known hallmark genes of stemness19 examined, HIF1α emerged as an upregulated hallmark stemness gene that also shows significant correlation with immune cell composition (Table 2). In ESFT with high HIF1α expression high numbers of macrophages and neutrophils and low numbers of adaptive immune cells were observed fitting other observations of a relative lack of adaptive immune cells and high frequency of innate immune cells in tumors with a hypoxic TME (Table 2, Figure 5).
Increased HIF1α protein expression has previously been observed by IHC studies in ESFT.33,34 HIF1α directly binds and regulates EWS-FLI-1 protein33,35,36 and is induced within hypoxic/necrotic areas in ESFT.33
EZH2 is highly upregulated in ESFT in our analysis (Figure 4), in line with previous observations that EZH2 maintains a stemness expression signature through epigenetic regulation in ESFT.12,37 It is upregulated through the oncogenic EWS/FLI1 fusion protein.8 Recently, it was shown that EZH2 inhibition together with chimeric antigen receptor (CAR) T cell therapy is an interesting novel immunotherapeutic treatment approach in EWST, as it leads to upregulation of the immune target ganglioside GD2.38
Similar to adult tumor types, the relative contribution of total T cells correlates directly with a better OS and EFS (training cohort: p = .044, p = .032, respectively; validation cohort: p = .005, p = .02, respectively) in ESFT. Previously, it has been demonstrated that HLA-G, a non-classical, immune-inhibitory MHC class 1 molecule is expressed in tumor cells and/or lymphocytes in ESFT primary tumor samples.39,40
The finding that activated NK cells confer a prolonged OS, is encouraging regarding current efforts to use chimeric antigen receptor-expressing NK cells in patients with ESFT therapeutically.41
Our finding of macrophages contributing the major immune cell subset in ESFT is consistent with IHC analyses31 and appears to be a general feature in pediatric cancers.42 Tumor-associated macrophages are known to promote cancer cell proliferation, immunosuppression and angiogenesis supporting metastasis and progression and through differentiation to M2 type macrophages expressing anti-inflammatory cytokines (IL-10, TGFβ) that have an inhibitory effect on cytotoxic CD8 + T cells.43 Interestingly, in the peripheral blood monocytosis and other changes in blood parameters are more commonly observed in ESFT patients with a poor prognosis,44 a phenomenon that supports the systemic changes of immune cells and cytokines in ESFT patients. Our findings may encourage efforts to target macrophages in ESFT, as currently tested for other types of malignancies using macrophage ablation or restoring their immunostimulatory potential.45–47
By combining low number of neutrophils, low M2 macrophages and high T cells in ESFT tumor samples we retrospectively identified a small patient subpopulation (9/32 patients) with a good prognosis (Figure 3). This was confirmed in a validation cohort (11/38 patients, supplementary Figure 2). Future clinical studies will need to determine whether the frequency of neutrophils, M2 macrophages and T cells in ESFTs may improve prognostication in this tumor entity and whether these cells can be targeted in a therapeutically favorable way.43
In chronic inflammatory conditions, neutrophils and monocytes/macrophages interact directly and enable the host to efficiently defend against and eliminate foreign pathogens as a first line of defense and during regeneration and repair.48 In the immune TME, neutrophils and macrophages can exert protumoral functions, enhancing tumor cell invasion and metastasis, angiogenesis, and extracellular matrix remodeling, while inhibiting antitumoral immune surveillance.49 Recently, neutrophil plasticity in the TME has been revisited with PMN-MDSCs being identified as immunosuppressive neutrophil suppressor cells50 and neutrophil extracellular traps containing chromatin and neutrophil proteins have been identified in ESFT tumor samples.51 Clearly, additional functional and phenotypic experiments are necessary to look more into the subtypes of neutrophils detected by CIBERSORT in ESFT and to characterize the contribution of each of these subsets further.
The connection of HIF1α overexpression and elevated innate immune cells (macrophages and neutrophils), but decreased adaptive immune cells such as T cells and plasma cells (Table 2) in ESFT is interesting, because HIF1α is a major transcription factor responsible for triggering tumor progression. In response to changes in oxygen, tension HIF1α regulates angiogenesis and tumor growth.52,53 In hypoxic tumors such as ESFT, hypoxia-driven formation of reactive oxygen species (ROS) destabilizes Prolyl Hydroxylases causing stabilization of HIF1α. Expression of the EWS-FLI1 oncoprotein in ESFT highjacks the developmental transcription factor SOX6, leading to constitutively elevated ROS levels and therapeutic vulnerability (synthetic lethality) to Elesclomol.54
Our findings are in line with previous hypotheses that HIF1α promotes tumor progression through suppressive myeloid and T cell populations and by creating a metabolically hostile environment for immune effector cells.33,55 Reprogramming of the tumor metabolic program into glycolysis in part via HIF1α results in “metabolic competition” between cancer cells and T cells, which may explain the paucity of T cells in ESFT.56 Alternatively, low levels of T cells in ESFT may occur because mutation frequencies are low in ESFT, similar to other pediatric tumors,57 thus providing few neoantigens. A lack of available neoantigens to induce effective T cell responses are frequently the cause of a poor adaptive immune response.
Currently, several novel avenues of immunotherapeutic treatment are being explored for ESFT patients in the relapsed or metastatic stage; however, most have shown limited success or are in early clinical trials.58 Anti-insulin like growth factor receptor-1 (IGFR1) therapy may provide therapeutic benefit for a selected group of patients.59 Here, characterization of the immune TME in clinical trials using CIBERSORT may be of interest to identify predictive biomarkers.
Of interest, most checkpoint molecules that we assessed were expressed at only low levels (Figure 4(b)), with somewhat elevated expression of OX40 (Figure 4(b)). While these in silico data need to be confirmed, OX40 may be an interesting immunotherapeutic target in otherwise “cold” EWST tumors,60 as co-stimulatory anti-OX40 antibodies are already tested in other advanced malignancies.61
Interestingly, HIF1α is responsible for regeneration and tissue repair.62 This may explain why immune cells involved in reparative processes such as phagocytes move in and adaptive immune cells that are necessary to fight infection move out in hypoxic tumors such as ESFT. HIF1α mediated hypoxia could play a role in an increase of myeloid-derived suppressor cells in the immune TME of ESFT63 and polarization toward M2 macrophages.64 Therefore, targeting macrophages through specific ablation or repolarization7,45 either alone or in combination with other treatment modalities could be an interesting novel treatment strategy in the era of personalized and TME adapted medicine. Additional in vitro and in vivo studies will need to address which soluble factors are responsible for M2 macrophage and neutrophil migration into the tumor and retention.
Funding Statement
This work was supported by the Bonn NeuroImmunology (BonnNi) program funded by the Else Kröner-Fresenius-Stiftung under Grant Q-611.1754 provided to David Stahl and Ines Gütgemann.
Acknowledgments
We thank Dr. Stefan Schönberger, Department of Pediatric Hematology and Oncology, University Children’s Hospital Bonn, for helpful discussions.
Conflict of interest
The authors report no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G.. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:1–10. Cited in: PMID: 1522903. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 2.Sorensen PH, Triche TJ. Gene fusions encoding chimaeric transcription factors in solid tumours. Semin Cancer Biol. 1996;7:3–14. Cited in: PMID: 8695764. doi: 10.1006/scbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 3.Lawlor ER, Sorensen PH. Twenty years on: what do we really know about Ewing Sarcoma and what is the path forward? Crit Rev Oncog. 2015;20:155–171. doi: 10.1615/CritRevOncog.2015013553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Álava E, Kovar H, Sorensen PH, Delattre O, Dirksen U. Ewing sarcoma. Nat Rev Dis Primers. 2018;4:5 Cited in: PMID: 29977059. doi: 10.1038/s41572-018-0003-x. [DOI] [PubMed] [Google Scholar]
- 5.Whelan J, Le Deley M-C, Dirksen U, Le Teuff G, Brennan B, Gaspar N, Hawkins DS, Amler S, Bauer S, Bielack S, et al. High-dose chemotherapy and blood autologous stem-cell rescue compared with standard chemotherapy in localized high-risk Ewing Sarcoma: results of Euro-E.W.I.N.G.99 and Ewing-2008. JCO. 2018;36:3110–3119. doi: 10.1200/JCO.2018.78.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Womer RB, West DC, Krailo MD, Dickman PS, Pawel BR, Grier HE, Marcus K, Sailer S, Healey JH, Dormans JP, et al. Randomized Controlled Trial of Interval-Compressed Chemotherapy for the Treatment of Localized Ewing Sarcoma: A Report From the Children’s Oncology Group. JCO. 2012;30:4148–4154. doi: 10.1200/JCO.2011.41.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdach SEG, Westhoff M-A, Steinhauser MF, Debatin K-M. Precision medicine in pediatric oncology. Mol Cell Pediatr. 2018;5:83. doi: 10.1186/s40348-018-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richter GHS, Plehm S, Fasan A, Rössler S, Unland R, Bennani-Baiti IM, Hotfilder M, Löwel D, von Luettichau I, Mossbrugger I, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A 2009;106:5324–5329. Cited in: PMID: 19289832. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ewing J. Classics in oncology. diffuse endothelioma of bone. James Ewing. Proceedings of the New York pathological society, 1921. CA Cancer J Clin. 1972;22:95–98. Cited in: PMID: 4622125. doi: 10.3322/canjclin.22.2.95. [DOI] [PubMed] [Google Scholar]
- 10.Staege MS, Hutter C, Neumann I, Foja S, Hattenhorst UE, Hansen G, Afar D, Burdach SEG. DNA microarrays reveal relationship of Ewing family tumors to both endothelial and fetal neural crest-derived cells and define novel targets. Cancer Res. 2004;64:8213–8221. Cited in: PMID: 15548687. doi: 10.1158/0008-5472.CAN-03-4059. [DOI] [PubMed] [Google Scholar]
- 11.Suvà M-L, Riggi N, Stehle J-C, Baumer K, Tercier S, Joseph J-M, Suvà D, Clément V, Provero P, Cironi L, et al. Identification of cancer stem cells in Ewing’s sarcoma. Cancer Res 2009;69:1776–1781. Cited in: PMID: 19208848. doi: 10.1158/0008-5472.CAN-08-2242. [DOI] [PubMed] [Google Scholar]
- 12.Burdach S, Plehm S, Unland R, Dirksen U, Borkhardt A, Staege MS, Müller-Tidow C, Richter GHS. Epigenetic maintenance of stemness and malignancy in peripheral neuroectodermal tumors by EZH2. Cell Cycle. 2009;8:1991–1996. Cited in: PMID: 19502792. doi: 10.4161/cc.8.13.8929. [DOI] [PubMed] [Google Scholar]
- 13.Paydas S, Bagir EK, Deveci MA, Gonlusen G. Clinical and prognostic significance of PD-1 and PD-L1 expression in sarcomas. Med Oncol. 2016;33:93 Cited in: PMID: 27421997. doi: 10.1007/s12032-016-0807-z. [DOI] [PubMed] [Google Scholar]
- 14.Berghuis D, Santos SJ, Baelde HJ, Taminiau AH, Egeler RM, Schilham MW, Hogendoorn PC, Lankester AC. Pro-inflammatory chemokine-chemokine receptor interactions within the Ewing sarcoma microenvironment determine CD8(+) T-lymphocyte infiltration and affect tumour progression. J Pathol. 2011;223:347–357. Cited in: PMID: 21171080. doi: 10.1002/path.2819. [DOI] [PubMed] [Google Scholar]
- 15.Inagaki Y, Hookway E, Williams KA, Hassan AB, Oppermann U, Tanaka Y, Soilleux E, Athanasou NA. Dendritic and mast cell involvement in the inflammatory response to primary malignant bone tumours. Clin Sarcoma Res. 2016;6:13 Cited in: PMID: 27482375. doi: 10.1186/s13569-016-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. Cited in: PMID: 25822800. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali HR, Chlon L, Pharoah PDP, Markowetz F, Caldas C. Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study. PLoS Med. 2016;13:e1002194 Cited in: PMID: 27959923. doi: 10.1371/journal.pmed.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong Y, Wang K, Zhou H, Peng L, You W, Fu Z. Profiles of immune infiltration in colorectal cancer and their clinical significant: A gene expression-based study. Cancer Med. 2018;7:4496–4508. Cited in: PMID: 30117315. doi: 10.1002/cam4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J, Omberg L, Gevaert O, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 2018;173:338–354.e15. Cited in: PMID: 29625051. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau W-C, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 2005;33:D562–6. Cited in: PMID: 15608262. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart E, Goshorn R, Bradley C, Griffiths LM, Benavente C, Twarog NR, Miller GM, Caufield W, Freeman BB, Bahrami A, et al. Targeting the DNA repair pathway in Ewing sarcoma. Cell Rep 2014;9:829–841. Cited in: PMID: 25437539. doi: 10.1016/j.celrep.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postel-Vinay S, Véron AS, Tirode F, Pierron G, Reynaud S, Kovar H, Oberlin O, Lapouble E, Ballet S, Lucchesi C, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet 2012;44:323–327. Cited in: PMID: 22327514. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 23.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, Zhang Z, Lapouble E, Grossetête-Lalami S, Rusch M, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 2014;4:1342–1353. Cited in: PMID: 25223734. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savola S, Klami A, Myllykangas S, Manara C, Scotlandi K, Picci P, Knuutila S, Vakkila J. High expression of complement component 5 (C5) at tumor site associates with superior survival in Ewing’s Sarcoma family of tumour patients. ISRN Oncol. 2011;2011:168712 Cited in: PMID: 22084725. doi: 10.5402/2011/168712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. Cited in: PMID: 12925520. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 27.Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862 Cited in: PMID: 23251644. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JA, Kim DH, Cho J, Lim JS, Koh J-S, Yoo JY, Kim MS, Kong C-B, Song WS, Cho WH, et al. Treatment outcome of Korean patients with localized Ewing sarcoma family of tumors: a single institution experience. Jpn J Clin Oncol 2011;41:776–782. Cited in: PMID: 21415003. doi: 10.1093/jjco/hyr033. [DOI] [PubMed] [Google Scholar]
- 29.Machado I, López-Guerrero JA, Scotlandi K, Picci P, Llombart-Bosch A. Immunohistochemical analysis and prognostic significance of PD-L1, PD-1, and CD8+ tumor-infiltrating lymphocytes in Ewing’s sarcoma family of tumors (ESFT). Virchows Arch. 2018;472:815–824. Cited in: PMID: 29445891. doi: 10.1007/s00428-018-2316-2. [DOI] [PubMed] [Google Scholar]
- 30.Spurny C, Kailayangiri S, Jamitzky S, Altvater B, Wardelmann E, Dirksen U, Hardes J, Hartmann W, Rossig C. Programmed cell death ligand 1 (PD-L1) expression is not a predominant feature in Ewing sarcomas. Pediatr Blood Cancer 2018;65:e26719 Cited in: PMID: 28868758. doi: 10.1002/pbc.26719. [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara T, Fukushi J-I, Yamamoto S, Matsumoto Y, Setsu N, Oda Y, Yamada H, Okada S, Watari K, Ono M, et al. Macrophage infiltration predicts a poor prognosis for human ewing sarcoma. Am J Pathol 2011;179:1157–1170. Cited in: PMID: 21771572. doi: 10.1016/j.ajpath.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volchenboum SL, Andrade J, Huang L, Barkauskas DA, Krailo M, Womer RB, Ranft A, Potratz J, Dirksen U, Triche TJ, et al. Gene expression profiling of Ewing Sarcoma tumors reveals the prognostic importance of tumor-stromal interactions: a report from the children’s oncology group. J Pathol Clin Res 2015;1:83–94. Cited in: PMID: 26052443. doi: 10.1002/cjp2.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knowles HJ, Schaefer K-L, Dirksen U, Athanasou NA. Hypoxia and hypoglycaemia in Ewing’s sarcoma and osteosarcoma: regulation and phenotypic effects of Hypoxia-Inducible Factor. BMC Cancer. 2010;10:372 Cited in: PMID: 20637078. doi: 10.1186/1471-2407-10-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Schaft DWJ, Hillen F, Pauwels P, Kirschmann DA, Castermans K, Egbrink MGAO, Tran MGB, Sciot R, Hauben E, Hogendoorn PCW, et al. Tumor cell plasticity in Ewing sarcoma, an alternative circulatory system stimulated by hypoxia. Cancer Res 2005;65:11520–11528. Cited in: PMID: 16357161. doi: 10.1158/0008-5472.CAN-05-2468. [DOI] [PubMed] [Google Scholar]
- 35.Hameiri-Grossman M, Porat-Klein A, Yaniv I, Ash S, Cohen IJ, Kodman Y, Haklai R, Elad-Sfadia G, Kloog Y, Chepurko E, et al. The association between let-7, RAS and HIF-1α in Ewing Sarcoma tumor growth. Oncotarget 2015;6:33834–33848. Cited in: PMID: 26393682. doi: 10.18632/oncotarget.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aryee DNT, Niedan S, Kauer M, Schwentner R, Bennani-Baiti IM, Ban J, Muehlbacher K, Kreppel M, Walker RL, Meltzer P, et al. Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of Ewing’s sarcoma cells in vitro. Cancer Res 2010;70:4015–4023. Cited in: PMID: 20442286. doi: 10.1158/0008-5472.CAN-09-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. Cited in: PMID: 17060944. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 38.Kailayangiri S, Altvater B, Lesch S, Balbach S, Göttlich C, Kühnemundt J, Mikesch J-H, Schelhaas S, Jamitzky S, Meltzer J, et al. EZH2 inhibition in Ewing Sarcoma upregulates GD2 expression for targeting with gene-modified T cells. Mol Ther. 2019;27:933–946. doi: 10.1016/j.ymthe.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spurny C, Kailayangiri S, Altvater B, Jamitzky S, Hartmann W, Wardelmann E, Ranft A, Dirksen U, Amler S, Hardes J, et al. T cell infiltration into Ewing sarcomas is associated with local expression of immune-inhibitory HLA-G. Oncotarget 2018;9:6536–6549. Cited in: PMID: 29464090. doi: 10.18632/oncotarget.23815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rizzo R, Lanzoni G, Stignani M, Campioni D, Alviano F, Ricci F, Tazzari PL, Melchiorri L, Scalinci SZ, Cuneo A, et al. A simple method for identifying bone marrow mesenchymal stromal cells with a high immunosuppressive potential. Cytotherapy 2011;13:523–527. Cited in: PMID: 21171826. doi: 10.3109/14653249.2010.542460. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C, Oberoi P, Oelsner S, Waldmann A, Lindner A, Tonn T, Wels WS. Chimeric antigen receptor-engineered NK-92 cells: an off-the-shelf cellular therapeutic for targeted elimination of cancer cells and induction of protective antitumor immunity. Front Immunol. 2017;8:533 Cited in: PMID: 28572802. doi: 10.3389/fimmu.2017.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, Zhou X, Li Y, Rusch MC, Easton J, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018;555:371–376. Cited in: PMID: 29489755. doi: 10.1038/nature25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genard G, Lucas S, Michiels C. Reprogramming of tumor-associated macrophages with anticancer therapies: radiotherapy versus chemo- and immunotherapies. Front Immunol. 2017;8:828 Cited in: PMID: 28769933. doi: 10.3389/fimmu.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Przybyl J, Kozak K, Kosela H, Falkowski S, Switaj T, Lugowska I, Szumera-Cieckiewicz A, Ptaszynski K, Grygalewicz B, Chechlinska M, et al. Gene expression profiling of peripheral blood cells: new insights into Ewing sarcoma biology and clinical applications. Med Oncol 2014;31:109 Cited in: PMID: 25008066. doi: 10.1007/s12032-014-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv Drug Deliv Rev. 2017;114:206–221. doi: 10.1016/j.addr.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Georgoudaki A-M, Prokopec KE, Boura VF, Hellqvist E, Sohn S, Östling J, Dahan R, Harris RA, Rantalainen M, Klevebring D, et al. Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 2016;15:2000–2011. doi: 10.1016/j.celrep.2016.04.084. [DOI] [PubMed] [Google Scholar]
- 47.Movahedi K, Laoui D, Gysemans C, Baeten M, Stange G, van Den Bossche J, Mack M, Pipeleers D, In’t Veld P, de Baetselier P, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70:5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 48.Prame Kumar K, Nicholls AJ, Wong CHY. Partners in crime: neutrophils and monocytes/macrophages in inflammation and disease. Cell Tissue Res. 2018;371:551–565. Cited in: PMID: 29387942. doi: 10.1007/s00441-017-2753-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Bae J-S. Tumor-Associated macrophages and neutrophils in tumor microenvironment. Mediators Inflamm. 2016;2016:6058147 Cited in: PMID: 26966341. doi: 10.1155/2016/6058147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giese MA, Hind LE, Huttenlocher A. Neutrophil plasticity in the tumor microenvironment. Blood. 2019;133:2159–2167. Cited in: PMID: 30898857. doi: 10.1182/blood-2018-11-844548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger-Achituv S, Brinkmann V, Abed UA, Kühn LI, Ben-Ezra J, Elhasid R, Zychlinsky A. A proposed role for neutrophil extracellular traps in cancer immunoediting. Front Immunol. 2013;4:48 Cited in: PMID: 23508552. doi: 10.3389/fimmu.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. Cited in: PMID: 21376230. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Ratcliffe PJ, Pugh CW, Maxwell PH. Targeting tumors through the HIF system. Nat Med. 2000;6:1315–1316. Cited in: PMID: 11100107. doi: 10.1038/82113. [DOI] [PubMed] [Google Scholar]
- 54.Marchetto A, Ohmura S, Orth MF, Li J, Wehweck FS, Knott MML, Stein S, Saucier D, Arrigoni C, Gerke JS, et al. Oncogenic hijacking if a developmental transcription factor evokes therapeutic vulnerability for ROS-induction in Ewing sarcoma. BioRxiv. 2019. Epub ahead of print. doi: 10.1101/578666. [DOI] [Google Scholar]
- 55.Chouaib S, Noman MZ, Kosmatopoulos K, Curran MA. Hypoxic stress: obstacles and opportunities for innovative immunotherapy of cancer. Oncogene. 2017;36:439–445. Cited in: PMID: 27345407. doi: 10.1038/onc.2016.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petrova V, Annicchiarico-Petruzzelli M, Melino G, Amelio I. The hypoxic tumour microenvironment. Oncogenesis. 2018;7:10 Cited in: PMID: 29362402. doi: 10.1038/s41389-017-0011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, Johann PD, Balasubramanian GP, Segura-Wang M, Brabetz S, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321–327. Cited in: PMID: 29489754. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 58.Xu J, Xie L, Sun X, Dong S, Tang X, Guo W. Management of recurrent or refractory Ewing sarcoma: A systematic review of phase II clinical trials in the last 15 years. Oncol Lett. 2019. Epub ahead of print. doi: 10.3892/ol.2019.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pappo AS, Patel SR, Crowley J, Reinke DK, Kuenkele K-P, Chawla SP, Toner GC, Maki RG, Meyers PA, Chugh R, et al. R1507, a monoclonal antibody to the insulin-like growth factor 1 receptor, in patients with recurrent or refractory Ewing sarcoma family of tumors: results of a phase II Sarcoma Alliance for Research through Collaboration study. J Clin Oncol 2011;29:4541–4547. Cited in: PMID: 22025149. doi: 10.1200/JCO.2010.34.0000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reuter D, Staege MS, Kühnöl CD, Föll J. Immunostimulation by OX40 ligand transgenic Ewing Sarcoma cells. Front Oncol. 2015;5:1451. doi: 10.3389/fonc.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Curti BD, Kovacsovics-Bankowski M, Morris N, Walker E, Chisholm L, Floyd K, Walker J, Gonzalez I, Meeuwsen T, Fox BA, et al. OX40 is a potent immune-stimulating target in late-stage cancer patients. Cancer Res. 2013;73:7189–7198. doi: 10.1158/0008-5472.CAN-12-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Strehin I, Bedelbaeva K, Gourevitch D, Clark L, Leferovich J, Messersmith PB, Heber-Katz E. Drug-induced regeneration in adult mice. Sci Transl Med. 2015;7:290ra92 Cited in: PMID: 26041709. doi: 10.1126/scitranslmed.3010228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simpson KD, Templeton DJ, Cross JV. Macrophage migration inhibitory factor promotes tumor growth and metastasis by inducing myeloid-derived suppressor cells in the tumor microenvironment. J Immunol. 2012;189:5533–5540. Cited in: PMID: 23125418. doi: 10.4049/jimmunol.1201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barsoum IB, Smallwood CA, Siemens DR, Graham CH. A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014;74:665–674. Cited in: PMID: 24336068. doi: 10.1158/0008-5472.CAN-13-0992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


