Abstract
The Salt Overly Sensitive (SOS) pathway in Arabidopsis thaliana plays important roles in maintaining appropriate ion homeostasis in the cytoplasm and regulating plant tolerance to salinity. However, little is known about the details regarding SOS family genes in the tuber mustard crop (Brassica juncea var. tumida). Here, 12 BjSOS family genes were identified in the B. juncea var. tumida genome including two homologous genes of SOS1, one and three homologs of SOS2 and SOS3, two homologs of SOS4, two homologs of SOS5 and two homologs of SOS6, respectively. The results of conserved motif analysis showed that these SOS homologs contained similar protein structures. By analyzing the cis-elements in the promoters of those BjSOS genes, several hormone- and stress-related cis-elements were found. The results of gene expression analysis showed that the homologous genes were induced by abiotic stress and pathogen. These findings indicate that BjSOS genes play crucial roles in the plant response to biotic and abiotic stresses. This study provides valuable information for further investigations of BjSOS genes in tuber mustard.
Introduction
Tuber mustard, Brassica juncea var. tumida (AABB, 2n = 36), which belongs to Brassicaceae, is an allotetraploid species that was produced from a natural cross between B. rapa (AA, 2n = 20) and B. nigra (BB, 2n = 16), followed by chromosome doubling [1]. It is an important vegetable in China and some Southeast Asian countries, as it serves as the raw material for Fuling mustard and is also famous for its special flavor and nutritional value. However, during growth and development, tuber mustard frequently suffers from abiotic and biotic stresses, such as salinity and pathogens, resulting in the inhibition of plant growth and huge economic loss. Therefore, determining the mechanisms underlying its resistance to salt stress will be helpful for improving the production of this vegetable.
The Salt Overly Sensitive (SOS) signaling pathway plays important roles in the plant response to salt stress and has three components: SOS1, SOS2 and SOS3. SOS1 encodes a Na+/H+ anti-transport protein in the cell membrane, which transports excess Na+ from the cytoplasm to the extracellular region [2]; SOS2 encodes a serine/threonine protein kinase; and SOS3 encodes a Ca2+ binding protein [3, 4]. Under salt stress, the concentration of Ca2+ in the cytoplasm immediately increases and is perceived by SOS3. SOS3 further activates SOS2 protein kinase by combining with Ca2+. SOS2 can interact with SOS3 and forms the SOS2-SOS3 complex to regulate the expression of SOS1 by phosphorylation. The transport activity of SOS1 is activated by SOS2-SOS3, and excess Na+ is discharged to alleviate the toxic effects of Na+ on cells [4–6]. Besides that, AtSOS4, AtSOS5 and AtSOS6 were also identified using a root-bending assay for the regulation of ion homeostasis and cell expansion under salt stress [7–10]. AtSOS4 encodes a pyridoxal kinase which is involved in the biosynthesis of vitamin B6 and regulates Na+ and K+ homeostasis [7, 8]. AtSOS5 encodes a putative cell surface adhesion protein and is required for normal cell expansion [9]. The root tips of sos5 mutant swell and root growth is arrested [9]. AtSOS6 encodes a cellulose synthase-like protein, AtCSLD5 [10]. The sos6-1 mutant shows hypersensitive to salt stress and osmotic stress, and accumulates high level of reactive oxygen species (ROS) [10].
The Arabidopsis mutants sos1, sos2 and sos3 are hypersensitive to Na+ and Li+, indicating that SOS proteins are involved in the regulation of plant tolerance to salinity [11]. Transgenic Arabidopsis seedlings overexpressing SOS1 show enhanced tolerance to salt stress and less Na+ content compared to wild-type plants under NaCl treatment [12]. The lateral root development of the sos3-1 mutant shows increased sensitivity even at low salt concentration, confirming that the SOS signaling pathway also modulates organ development in response to salt stress [13]. Overexpression of B. juncea SOS3 (BjSOS3) in the Arabidopsis mutant sos3 complements the sos3 mutant phenotype and transgenic plants exhibit enhanced tolerance to salinity, indicating that BjSOS3 has conserved function with AtSOS3 in regulating plant resistance to salt stress [13, 14]. However, the role of the SOS gene family in tuber mustard remains mostly unknown. Therefore, we identified the SOSs genes and elucidated their putative role for better understanding of SOS signaling pathway in tuber mustard (Brassica juncea var. tumida).
In this study, we identified 12 SOS family genes in the B. juncea var. tumida genome. Based on the analysis of phylogenic relationship, gene structures, protein motifs, and promoter cis-elements, similar gene characteristics were found between BjSOS and AtSOS. In addition, we analyzed the transcript levels of BjSOS family genes under biotic and abiotic stresses, including NaCl, ABA, low temperature, and the pathogen Plasmodiophora Brassicae. The results showed that BjSOS genes were induced by abiotic stresses and pathogen in tuber mustard. The findings indicate that SOS family genes play crucial roles in the plant response to biotic and abiotic stresses. The findings not only are helpful for further understanding of the SOS signaling pathway but also provide clues about the defense responses of tuber mustard against different stresses.
Materials and methods
Materials and growth conditions
The tuber mustard cultivar Yong’an was used in this study. The seeds were surface sterilized and plated on MS medium (Sigma-Aldrich, St. Louis, MO, USA) with 1% sucrose and 8 g/L agar (Sigma-Aldrich, St. Louis, MO, USA) and then cultivated in a growth room at 22°C and 6000 lx under long-day conditions (16 h light/8 h dark). To analyze the gene expression patterns of BjSOS genes under abiotic stresses, 1-week-old seedlings grown on MS medium were treated with 50 μM ABA, 200 mM NaCl and low temperature (4°C) for the indicated time points. To analyze the gene expression patterns of BjSOS genes under biotic stress, 2-week-old seedlings were irrigated with P. brassicae suspension liquid (OD600 = 0.07) for 0, 0.25, 0.5, 1, 3, 5, 7, and 9 days.
Bioinformatics analysis
The gene sequences of AtSOS1, AtSOS2, AtSOS3, AtSOS4, AtSOS5 and AtSOS6 and their homologous genes in tuber mustard were searched in the Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), TAIR (http://www.arabidopsis.org/) and Brassica databases (http://brassicadb.org/brad/). The protein sequences were aligned by ClustalX 1.83 [15], and a phylogenic tree was constructed using the neighbor-joining method with bootstrap values of 1000 by MEGA5 [16]. Gene structure analysis was performed using online software (http://gsds.cbi.pku.edu.cn/). The regions located 2 kb upstream of the BjSOS coding sequences were used as the promoter sequences, and the promoter cis-element analysis was performed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace) online software. Protein domain analysis was done using SMART (http://smart.embl-heidelberg.de/) and ExPASy (http://prosite.expasy.org/prosite.html) online analysis tools.
Gene expression analysis
Total RNA was extracted from tuber mustard seedlings that had been subjected to NaCl, ABA, low temperature, and pathogen treatment using TRIzol reagent (Invitrogen, Carlsbad, CA, USA; Catalog No. 15596026). The RNA samples were used for cDNA synthesis using the cDNA synthesis Supermix with gDNA remover kit (Transgen Biotech, China; Catalog No. AT301) following the manufacturer’s instructions. qRT-PCR was carried out using SYBR Green qPCR Supermix (Invitrogen, Carlsbad, CA, USA; Catalog No. 4309155). The transcript abundance was calculated by the comparative CT (cycle threshold) method, and BjActin3 was used as the internal control. The qRT-PCR experiments were carried out three times with three replicates each. The primers used in this study were listed in S1 Table.
Statistical analysis
All data were analyzed using SigmaPlot 10.0 (Systat Software, Inc., Chicago, IL) and SPSS 16.0 software. The averages and standard deviations of all results were calculated, and for multiple groups of samples, the one-way ANOVA followed by the Dunnett test was used. The statically significant treatments were marked with ‘***’ (P<0.001), ‘**’ (P<0.01) and ‘*’ (P<0.05).
Results
Genome-wide identification and characterization of SOS homologs in B. juncea var. tumida
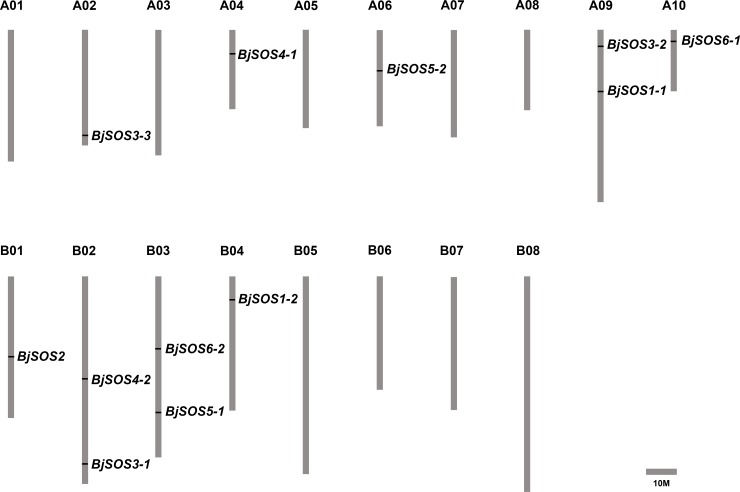
Twelve genes as homologs of SOS genes were identified in B. juncea var. tumida genome through BLASTP in Brassica database using six AtSOS protein sequences as references (Table 1). The gene lengths ranged from 1191 bp to 5965 bp with 1–23 exons in each sequence. The protein lengths of these twelve SOS homologs ranged from 180 (BjSOS3-3) to 1192 (BjSOS6-2) amino acid (aa) residues. The relative molecular weights of those proteins varied from 20.59 kD (BjSOS3-3) to 133.61 kD (BjSOS6-2), and the isoelectric point (PI) ranged from 4.90 to 9.25 (Table 1). The twelve BjSOS genes were distributed in 9 of the 18 chromosomes of B. juncea var. tumida. Each of the chromosomes A02, A04, A06, A10, B01, and B04 contained one gene, and each of the chromosomes A09, B02 and B03 contained two genes (Fig 1).
Table 1. The SOS family members in Brassica juncea var. Tumida.
| Group | Gene name | Locus | Sequence ID | Exon | Genomics (bp) | CDS (bp) | Protein (aa) | pI | MW (kD) |
|---|---|---|---|---|---|---|---|---|---|
| SOS1 | BjSOS1-1 | A09 | BjuA002024 | 23 | 5789 | 3030 | 1009 | 6.82 | 111.85 |
| BjSOS1-2 | B04 | BjuB027801 | 23 | 5965 | 3321 | 1106 | 6.21 | 122.13 | |
| SOS2 | BjSOS2 | B01 | BjuB024452 | 12 | 2716 | 1221 | 406 | 9.25 | 46.16 |
| SOS3 | BjSOS3-1 | B02 | BjuB038085 | 8 | 1418 | 660 | 219 | 5.04 | 25.35 |
| BjSOS3-2 | A09 | BjuA046788 | 8 | 1492 | 657 | 218 | 4.90 | 25.13 | |
| BjSOS3-3 | A02 | BjuA033133 | 7 | 1191 | 543 | 180 | 5.08 | 20.59 | |
| SOS4 | BjSOS4-1 | A04 | BjuA015607 | 12 | 2615 | 930 | 309 | 5.34 | 34.16 |
| BjSOS4-2 | B02 | BjuB048173 | 12 | 2627 | 930 | 309 | 5.11 | 34.06 | |
| SOS5 | BjSOS5-1 | B03 | BjuB019581 | 1 | 1269 | 1272 | 423 | 5.68 | 44.16 |
| BjSOS5-2 | A06 | BjuA022634 | 1 | 1260 | 1263 | 420 | 5.52 | 44.22 | |
| SOS6 | BjSOS6-1 | A10 | BjuA037683 | 3 | 3697 | 3534 | 1177 | 8.21 | 132.02 |
| BjSOS6-2 | B03 | BjuB043857 | 3 | 3784 | 3579 | 1192 | 7.85 | 133.61 |
pI: Isoelectric point; MW: molecular weight.
Fig 1. The distribution of BjSOS in Brassica juncea var. tumida chromosomes.
Twelve identified SOS homologs genes were mapped to the 9 of 18 chromosomes. The chromosome name is at the top of each bar. The scale of the chromosome is in millions of bases (Mb).
Phylogenic analysis and gene structures of SOS family genes
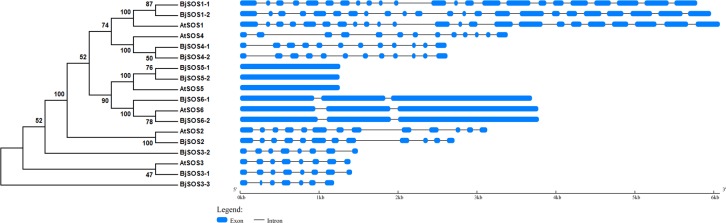
To analyze the evolutionary relationships between BjSOSs and AtSOSs, a phylogenetic tree was constructed using MEGA5 software with the neighbor-joining method. According to the phylogenic tree, twelve BjSOS genes with six AtSOS genes were identified and clustered into six clades. The first clade was AtSOS1 and two homologs BjSOS1-1 and BjSOS1-2; the second clade was AtSOS4 and two homologs BjSOS4-1 and BjSOS4-2; the third clade was AtSOS5 and its two homologs BjSOS5-1 and BjSOS5-2; the fourth clade was AtSOS6 and its two homologs BjSOS6-1 and BjSOS6-2; the fifth clade was AtSOS2 and its homolog BjSOS2; and the last clade was AtSOS3 and its homologs BjSOS3-1, BjSOS3-2 and BjSOS3-3 (Fig 2). SOS family genes in the same subfamilies may have similar functions. To understand their gene structures, we analyzed the gene exon-introns using the GSDS2.0 online server. According to the results, AtSOS1, BjSOS1-1 and BjSOS1-2 all had 23 exons and 22 introns; AtSOS2 and BjSOS2 contained 13 and 12 exons, respectively; AtSOS3, BjSOS3-1 and BjSOS3-2 all contained eight exons with the exception of BjSOS3-3 (7 exons); AtSOS4, BjSOS4-1 and BjSOS4-2 contained 13 and 12 exons, respectively; AtSOS5, BjSOS5-1 and BjSOS5-2 all contained one exon; AtSOS6, BjSOS6-1 and BjSOS6-2 all contained 3 exons (Fig 2). These results indicated that the homologs clustered into the same subfamily had similar gene structures and might have conserved functions.
Fig 2. The phylogenic tree and gene structures of SOS family genes.
The phylogenic tree was built using the neighbor-joining (NJ) method and the exon-intron structure of SOS homologs was drawn according to their phylogenic relationships. The blue boxes and gray lines denoted exons and introns, respectively.
Protein sequence alignment and conserved motif analysis of SOS homologs
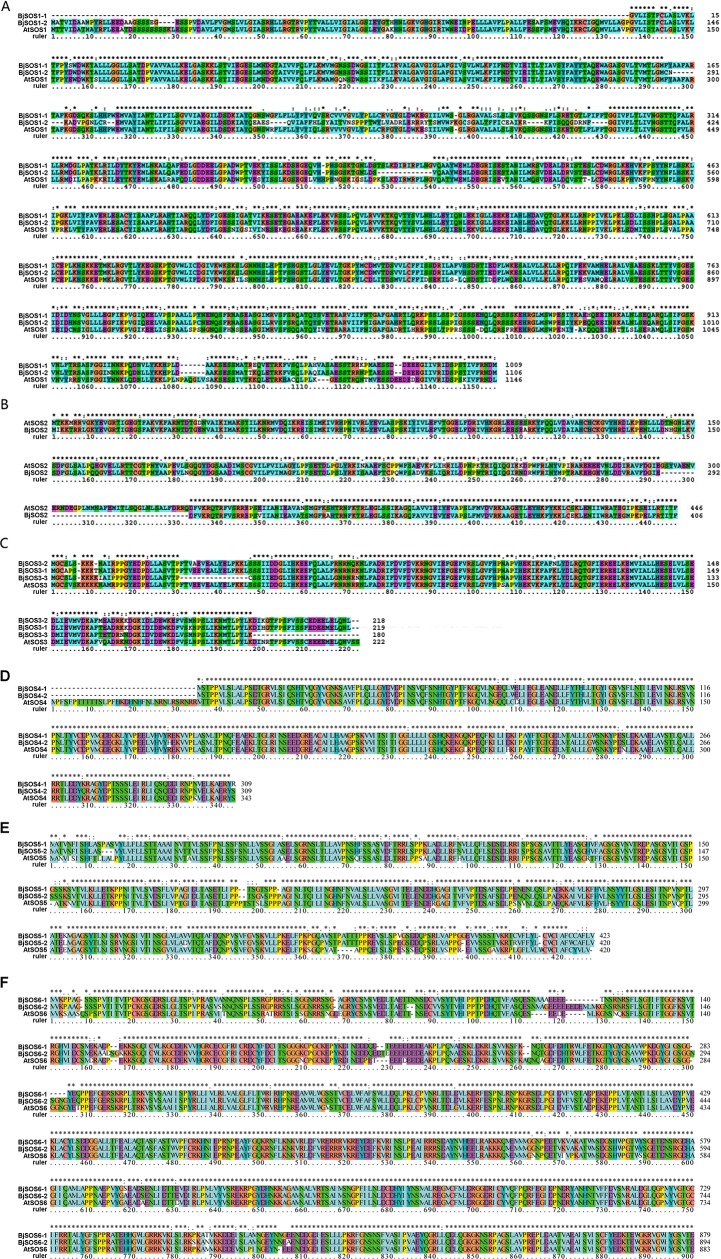
The SOS homologous protein sequences were aligned by Clustalx and the results showed that homologs clustered into the same subfamily had conserved protein sequences (Fig 3). BjSOS1-1 and BjSOS1-2 shared 73.11% and 75.74% sequence identity, respectively, with AtSOS1; AtSOS2 and BjSOS2 shared 81.39% sequence identity; AtSOS3 and its homologs BjSOS3-1, BjSOS3-2, and BjSOS3-3 shared 89.19%, 88.29%, and 72.07% sequence identity, respectively; BjSOS4-1 and BjSOS4-2 shared 91% and 90% sequence identity with AtSOS4, respectively; BjSOS5-1 and BjSOS5-2 shared 74% and 73% sequence identity with AtSOS5, respectively; and BjSOS6-1 and BjSOS6-2 shared 88% and 87% sequence identity with AtSOS6, respectively (Fig 3).
Fig 3. The protein sequence alignment of SOS homologs.
The protein sequences were aligned by Clustalx. A. The protein sequence alignment of AtSOS1 and its homologs. B. The protein sequence alignment of AtSOS2 and its homologs. C. The protein sequence alignment of AtSOS3 and its homologs. D. The protein sequence alignment of AtSOS4 and its homologs. E. The protein sequence alignment of AtSOS5 and its homologs. F. The protein sequence alignment of AtSOS6 and its homologs.
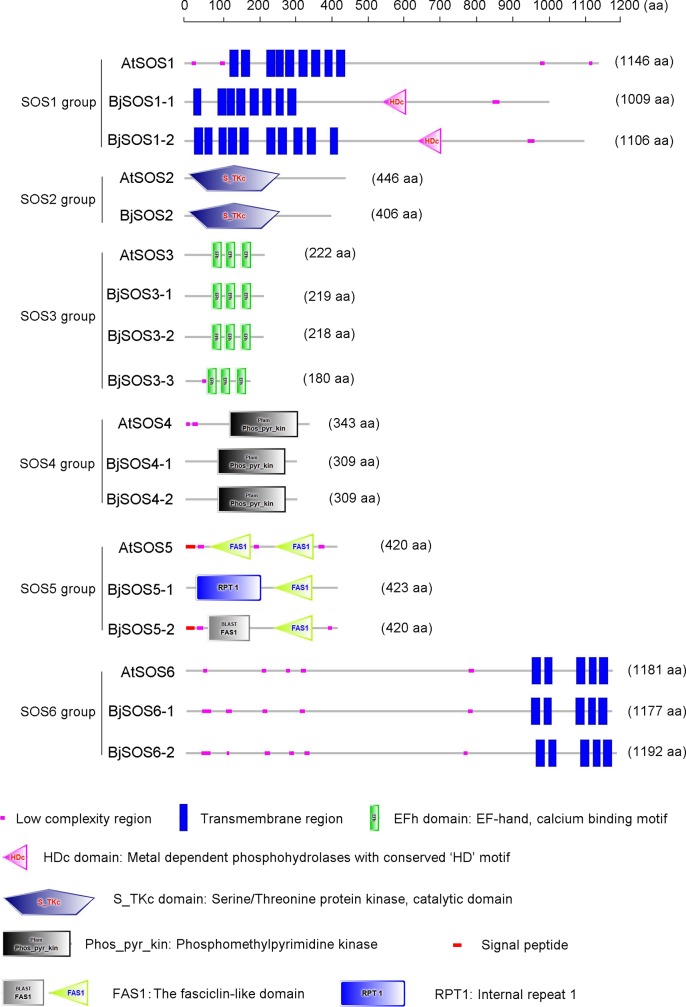
Protein conserved motif analysis was conducted using the SMART and ExPASy online analysis tools. The results showed that AtSOS1 and its homologs BjSOS1-1 and BjSOS1-2 all contained 8–10 transmembrane regions; both BjSOS1-1 and BjSOS1-2 contained 1 HDc domain; both AtSOS2 and BjSOS2 contained the S_TKc domain, which was a serine/threonine protein kinase catalytic domain; AtSOS3 and its homologs BjSOS3-1, BjSOS3-2, and BjSOS3-3 all contained the EFh domain, which was calcium Ca2+- binding motif; AtSOS4, BjSOS4-1 and BjSOS4-2 all contained Phos_pyr_kin domain, which was a phosphomethylpyrimidine kinase domain; AtSOS5, BjSOS5-1 and BjSOS5-2 all contained FAS1 domain, which was the fasciclin-like domain; AtSOS6, BjSOS6-1 and BjSOS6-2 all contained 5 transmembrane regions (Fig 4). The results indicated that the BjSOS homologs might have conserved functions in response to abiotic stress.
Fig 4. The conserved motifs of the SOS homologs proteins.
These motifs were identified using the online software of SMART, and colored boxes indicated conserved motifs and gray lines represent non-conserved sequences.
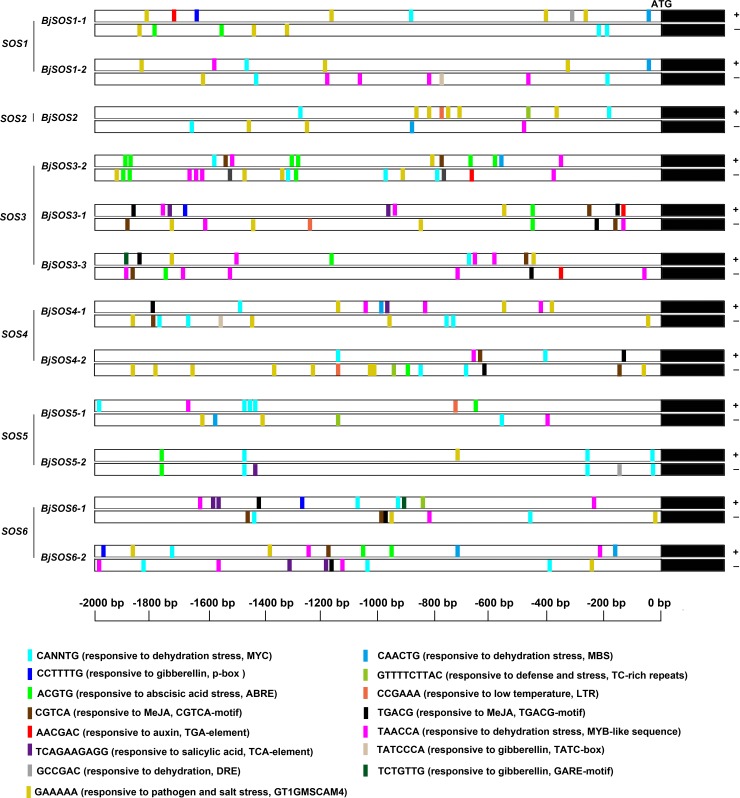
Promoter cis-acting regulatory elements prediction of SOS homologs
To further understand the potential roles of SOS homologs and how their gene expression is regulated, we chose the 2000 bp DNA fragment upstream of the ATG start codon as the promoter sequences and performed promoter cis-element analysis using PlantCARE and PLACE online software. According to the results, the promoters of all BjSOS genes, except BjSOS2, contained at least one hormone-related elements such as the ABRE (ACGTG, responsive to abscisic acid stress) [17], p-box (CCTTTTG, responsive to gibberellin) [18], and CGTCA motif (CGTCA, responsive to MeJA) [19] (Fig 5 and S2 Table). In addition, the promoters of the SOS homologs contained at least eight stressed-related elements such as the DRE (GCCGAC, responsive to dehydration) [20], MBS (CAACTG, responsive to dehydration stress) [21], TC-rich repeats (GTTTTCTTAC, responsive to defense and stress) [22], LTR (CCGAAA, responsive to low temperature) [23], and GT1GMSCAM4 motif (GAAAAA, responsive to pathogen and salt stress) [24] (Fig 5 and S2 Table). Together, the promoters of SOS homologs contained diverse cis-elements responsive to ABA, auxin, GA, SA, and abiotic stresses, indicating that the genes expression of SOS homologous were regulated by hormone and abiotic stresses, and they might play a role in regulating tuber mustard response to hormone and stresses.
Fig 5. The promoter cis-elements analysis of SOS homologs genes.
The 2 kb DNA fragments upstream of the ATG staring code of SOS homologs genes were analyzed using online analysis software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace).
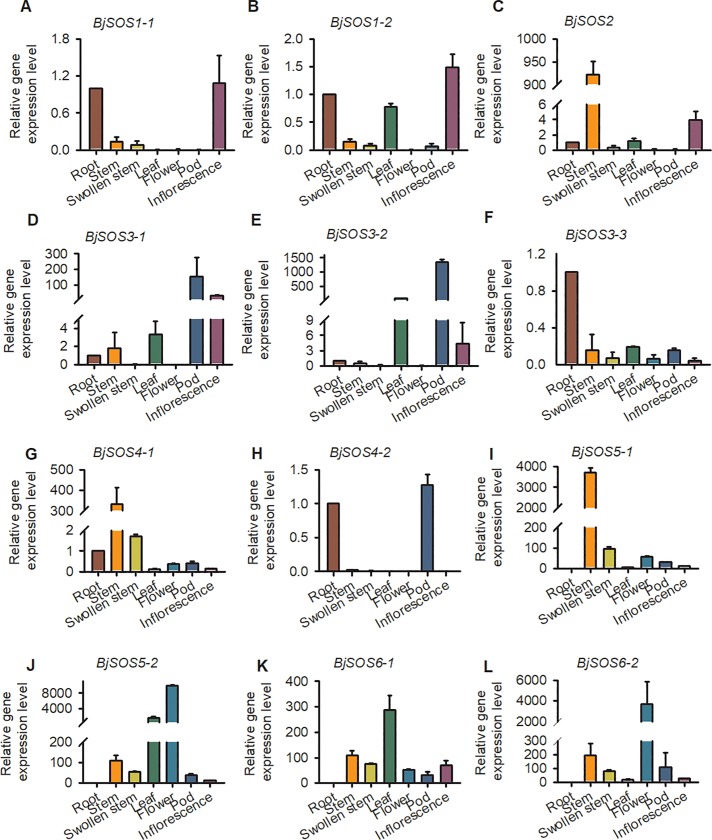
Tissue specific expression pattern analysis of SOS homologs genes
To investigate the tissue specific expression patterns of SOS homologs, we analyzed the gene expression levels at different growth stages and tissues (root, stem, swollen stem, leaf, pod and inflorescence) using qRT-PCR. The results showed that the SOS1 homolog BjSOS1-2 was highly expressed in the root, leaf, and inflorescence. In contrast, the expression level of another homolog BjSOS1-1 was very low, with nearly no expression in pod; BjSOS2 was highly expressed in the stem; the SOS3 homologs BjSOS3-1 and BjSOS3-2 were highly expressed in the leaf, pod, and inflorescence, whereas there was very low BjSOS3-3 expression in B. juncea var. tumida (Fig 6). BjSOS4-1 and BjSOS5-1 was highly expressed in stem; BjSOS5-2 was highly expressed in leaf and flower; and the expression level of BjSOS6-1 and BjSOS6-2 were high in almost all the tissues (Fig 6). According to the results, the expression levels of the SOS homologs varied in different tissues and organs, indicating that they may play different roles in different tissues. In addition, the different expression patterns of the same gene in different tissues and organs suggest that the expression pattern of the genes was existence of space-time specificity.
Fig 6. Expression levels of SOS homologs genes in different tissues.
Tissue specific expression pattern of SOS homologs genes were analyzed by qPCR. Data were normalized to the expression level of BjActin3. The values are means ± standard error. Three independent biological repeats were performed.
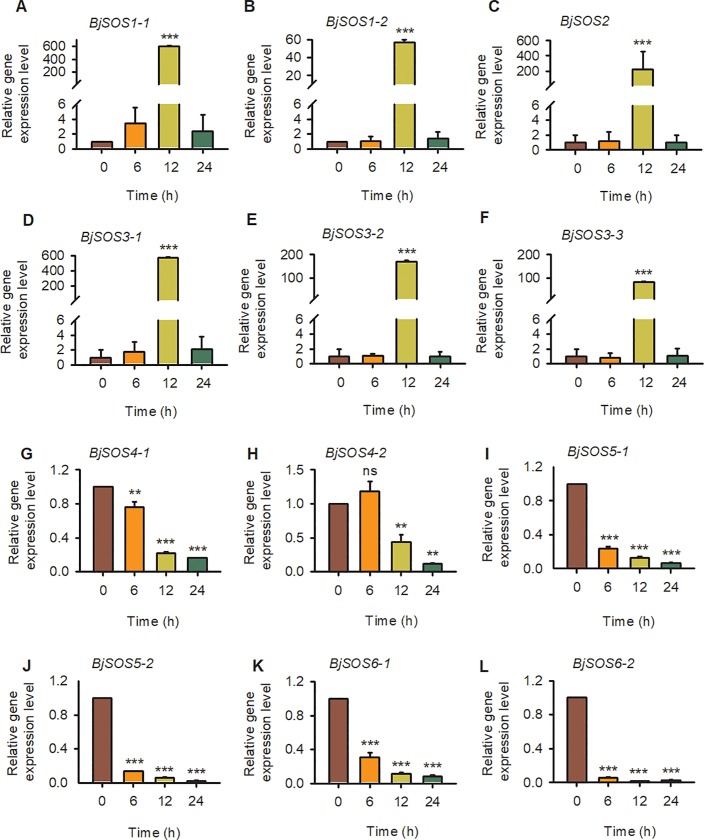
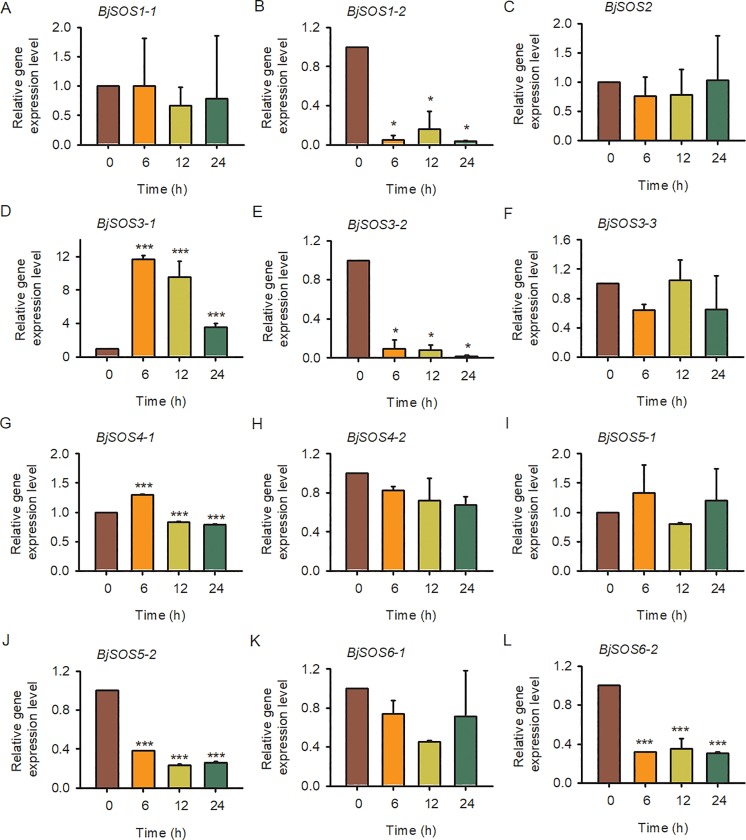
Gene expression levels of SOS homologs in B. juncea var. tumida under abiotic and biotic stresses
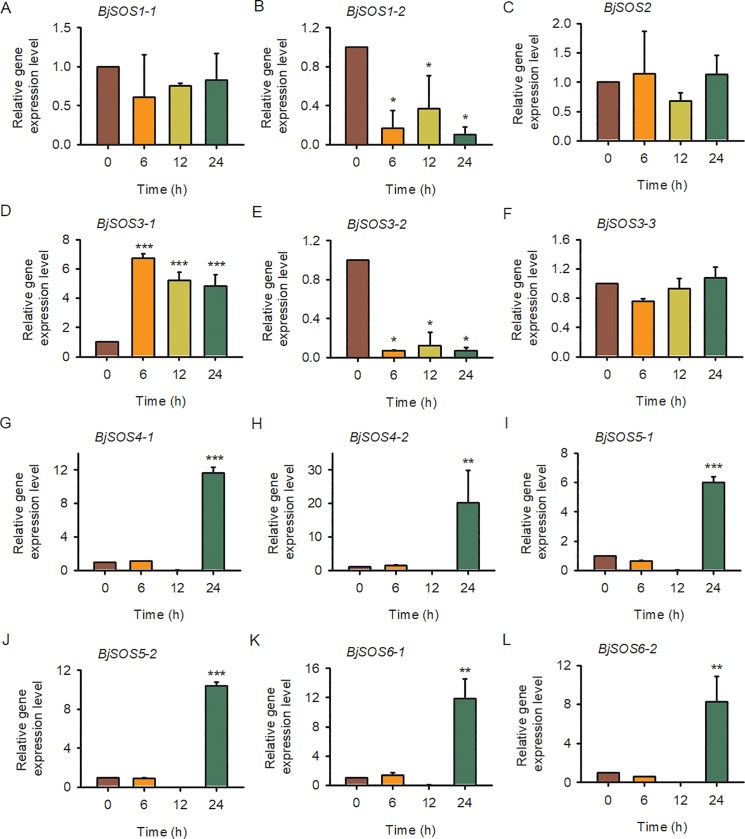
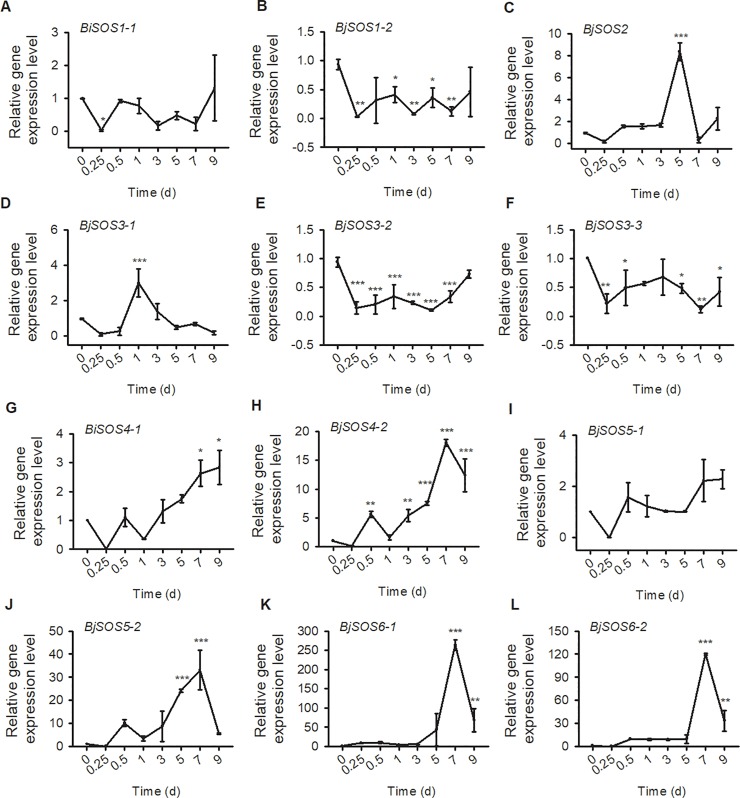
To further explore the SOS homolog expression levels in tuber mustard under biotic and abiotic stresses, qRT-PCR was performed using tuber mustard seedlings treated with 200 mM NaCl, 50 μM ABA, low temperature (4°C) and the pathogen P. brassicae at different time points. Under NaCl treatment, all of the BjSOS1-1, BjSOS1-2, BjSOS2, BjSOS3-1, BjSOS3-2 and BjSOS3-3 genes were induced by salt stress, especially at 12 h after NaCl treatment, and then decreased to normal level at 24 h; and all of the BjSOS4-1, BjSOS4-2, BjSOS5-1, BjSOS5-2, BjSOS6-1 and BjSOS6-2 genes were repressed by salt stress, indicating that these SOS homologs play important roles in the plant response to salt stress (Fig 7). The expression levels of BjSOS3-1 and BjSOS4-1 were significantly induced after ABA treatment. In contrast, the gene expression level of BjSOS1-2, BjSOS3-2, BjSOS5-2 and BjSOS6-2 were downregulated under ABA treatment, suggesting that BjSOS1-2, BjSOS3-1, BjSOS3-2, BjSOS4-1, BjSOS5-2 and BjSOS6-2 might be involved in the ABA signaling pathway (Fig 8). Under low temperature stress condition, the gene expression level of BjSOS3-1, BjSOS4-1, BjSOS4-2, BjSOS5-1, BjSOS5-2, BjSOS6-1 and BjSOS6-2 were significantly induced and the transcript levels of BjSOS1-2 and BjSOS3-2 were downregulated. However, there was no obvious expression difference at 0, 6, 12 and 24 h after 4°C treatment of BjSOS1-1, BjSOS2, and BjSOS3-3, indicating that BjSOS1-2, BjSOS3-1, BjSOS3-2, BjSOS4-1, BjSOS4-2, BjSOS5-1, BjSOS5-2, BjSOS6-1 and BjSOS6-2 regulated the tuber mustard response to low temperature stress (Fig 9). Under pathogen stress, we treated the tuber mustard seedlings with P. brassicae for 0, 0.25, 0.5, 1, 3, 5, 7, and 9 days, and the qRT-PCR results showed that BjSOS3-1 was induced by pathogen on day 1; the SOS2 homolog BjSOS2 was upregulated by pathogen, especially on day 5, and then downregulated at later time points; BjSOS4-2, BjSOS5-2, BjSOS6-1 and BjSOS6-2 were up-regulated by pathogen, especially on day 7, suggesting that BjSOS3-1, BjSOS2, BjSOS4-2, BjSOS5-2, BjSOS6-1 and BjSOS6-2 may play crucial roles in biotic tolerance (Fig 10). Taken together, the expression patterns of SOS homologous genes changed under salt, ABA, 4°C, and pathogen treatment, indicating that these genes in tuber mustard might be important candidates for regulating plant tolerance to biotic and abiotic stresses.
Fig 7. The expression patterns of SOS homologs genes under salt stress.
Total RNA was extracted from tuber mustard seedlings treated with 200 mM NaCl at the indicated time points. Data were normalized to the expression level of BjActin3. The values are means ± standard error. Three independent biological repeats were performed.
Fig 8. The expression patterns of SOS homologs genes under ABA treatment.
Total RNA was extracted from tuber mustard seedlings treated with 50 μM ABA or not at the indicated time points. Data were normalized to the expression level of BjActin3. The values are means ± standard error. Three independent biological repeats were performed.
Fig 9. The expression patterns of SOS homologs genes under low tempreture treatment.
Total RNA was extracted from tuber mustard seedlings treated with 4°C or not at the indicated time points. Data were normalized to the expression level of BjActin3. The values are means ± standard error. Three independent biological repeats were performed.
Fig 10. The expression patterns of SOS homologs genes under pathogen treatment.
Total RNA was extracted from tuber mustard seedlings treated with Plasmodiophora Brassicae at the indicated time points. Data were normalized to the expression level of BjActin3. The values are means ± standard error. Three independent biological repeats were performed.
Discussion
The SOS signaling pathway plays important roles in the plant response to salt stress; however, the identity and expression patterns of B. juncea var. tumida SOS genes are unknown. Here, a total of twelve BjSOS genes were identified and located in 9 of 18 chromosomes (Fig 1). There was high sequence identity between BjSOSs and their relative homologs in A. thaliana, and they shared the same protein motifs (Fig 4). The expression patterns of BjSOS genes indicated that they might play specific roles in different tissues and stress conditions. These results shed light on the roles of BjSOS genes in regulating plant growth and response to abiotic and biotic stresses, which will be helpful for improving the production of tuber mustard.
In A. thaliana, six SOS genes SOS1, SOS2, SOS3, SOS4, SOS5 and SOS6 were identified [2–5, 7–10]. According to our results, twelve SOS homologs were found in the genome of B. juncea var. tumida. There were more members of the SOS genes in the B. juncea var. tumida genome than in A. thaliana, most likely because B. juncea var. tumida is an allotetraploid species that resulted from the hybridization between B. rapa and B. nigra following with genome duplication [1]. The comparable homologous gene number in the A sub-genome and B sub-genome indicated the B. juncea var. tumida genome experienced co-linearity gene duplication. However, the homologous genes of AtSOS2 were lost or not duplicated in B. juncea var. tumida, suggesting that these homologous genes may have had functional redundancy or divarication during the evolutionary process. The loss of genes during the genome duplication event has also frequently occurred in other Brassica species, such as chitinase gene family in B. rapa [25].
The cis-elements and functional characterization of the promoters of SOS genes have been identified in many species such as Salicornia brachiate, B. juncea, and A. thaliana [26–28]. In this study, we analyzed the promoter cis-elements of SOS homologs in B. juncea var. tumida, and found that all of the promoters contained diverse cis-elements responsive to plant hormones (ABA, auxin, GA, and SA) and abiotic stresses (drought, cold, and salt stresses) (Fig 5). These promoter cis-elements of SOS genes in B. juncea var. tumida are in accordance with previous studies in other species [26–28], indicating that the expression patterns of SOS homologs were regulated by hormone and abiotic stresses, and that these homologs might play roles in regulating the tuber mustard response to hormone and abiotic stresses.
To date, although the expression patterns of SOS family genes have been determined in other species, such as wheat, Arabidopsis and Brassica [12, 14, 27–30], no detailed studies on the expression of BjSOS genes were found. Here, tissue specific expression pattern analysis results revealed that majority of BjSOS genes expressed in various tissues, such as root, stem, pod, leaf and flower (Fig 6). Notably, some genes highly expressed in root, stem and leaf, pointing to the important roles of BjSOS genes in those tissues and the diverse biological functions of different BjSOS genes in B. juncea var. tumida. In Arabidopsis, AtSOS1 promoter-driven GUS expression was mainly found in root, inflorescence and leaf [31], similar tissue specific expression patterns were also found for BjSOS1 genes, suggesting that SOS1 genes played conserved functions in Arabisopsis and tuber mustard (Fig 6). In Arabidopsis, AtSOS4 expressed ubiquitously in all organs, in contrast, BjSOS4-1 mainly expressed in stem, while almost no expression of BjSOS4-2 could be detected in stem, leaf and flower (Fig 6), indicating the different roles of BjSOS4 genes in the regulation of tuber mustard growth and development [7, 8]. The expression patterns of BjSOS genes under different stress treatments were examined by qPCR and found that they were differentially expressed after different stress treatments. According to the results of gene expression pattern analysis, all of the SOS homologs were significantly response to salt stress; BjSOS3-1 was induced by ABA and low temperature; BjSOS4-1, BjSOS4-2, BjSOS5-1, BjSOS5-2, BjSOS6-1 and BjSOS6-2 were induced by low temperature, BjSOS2, BjSOS3-1, BjSOS4-2, BjSOS5-2, BjSOS6-1 and BjSOS6-2 were significantly induced by treatment with the pathogen P. brassicae, indicating that BjSOS genes might regulate the tuber mustard response to abiotic and biotic stresses (Figs 7–10). In rice, OsSOS1 was highly induced in root after 15 h salt stress treatment comparing with non-treated plant [32], in accordance with this, both the expression levels of BjSOS1-1 and BjSOS1-2 were significantly induced in root by salt treatment at 12 h (Fig 7), and the expression induction was also found for PabSOS1 Populus [33]. Besides that, AtSOS1 was significantly induced by NaCl but not ABA and cold [2], similar expression patterns were also found for BjSOS1-1 and BjSOS1-2 (Figs 7–9), those results suggested that the functions of SOS1 genes are conserved in the regulation of plant response to salt stress in different species. Although abiotic and biotic stresses responsive cis-elements could be found in the promoters of some BjSOS genes, the expressions of those genes were not induced by ABA, low temperature or pathogen. The event of gene expression level not in agreement with the promoter analysis of cis-element also frequently exists in other species, such as BnPYLs in B. napus and BjuTIR1/AFBs in B. juncea var. tumida [34–35].
In conclusion, our study identified twelve BjSOS genes in tuber mustard and analyzed their transcript levels under the biotic and abiotic stresses. The results suggest that SOS family genes might potentially be utilized for improving the tolerance of B. juncea var. tumida to biotic and abiotic stresses.
Supporting information
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by The National Natural Science Foundation of China (Program Nos. 31701451 and 31770407), Zhaoming Cai and Diandong Wang; Chongqing technology innovation and application demonstration project (social livelihood) (Program No. cstc2018jscx-msyb0152), Changman Li; Chongqing Basic Research and Frontier Exploration Project (Program No. cstc2018jcyjAX0628), the Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN201801434), Chunhong Cheng; and Yangtze Normal University Startup Foundation for Dr. Scientific Research (Program Nos. 2017KYQD151 and 2017KYQD152), Zhaoming Cai and Chunhong Cheng. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B, et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. 10.1038/ng.3657 [DOI] [PubMed] [Google Scholar]
- 2.Shi HZ, Ishitani M, Kim CS, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA. 2000, 97, 6896–6901. 10.1073/pnas.120170197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA. 2000, 97, 3730–3734. 10.1073/pnas.060034197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong D, Guo Y, Schumaker KS, Zhu JK. The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol. 2004, 134, 919–926. 10.1104/pp.103.037440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA. 2002, 99, 8436–8441. 10.1073/pnas.122224699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu JK. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [DOI] [PubMed] [Google Scholar]
- 7.Shi H, Xiong L, Stevenson B, Lu T, Zhu JK. The Arabidopsis salt overly sensitive 4 Mutants Uncover a Critical Role for Vitamin B6 in Plant Salt Tolerance. Plant Cell. 2002, 14, 575–588. 10.1105/tpc.010417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi H, Zhu JK. SOS4, A Pyridoxal Kinase Gene, Is Required for Root Hair Development in Arabidopsis. Plant Physiol. 2002, 129, 585–593. 10.1104/pp.001982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK. The Arabidopsis SOS5 Locus Encodes a Putative Cell Surface Adhesion Protein and Is Required for Normal Cell Expansion. Plant cell. 2003, 15, 19–32. 10.1105/tpc.007872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu JH, Lee BH, Dellinger M, Cui XP, Zhang CQ, Wu S, et al. A cellulose synthase‐like protein is required for osmotic stress tolerance in Arabidopsis. The Plant Journal. 2010, 63, 128–140. 10.1111/j.1365-313X.2010.04227.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu JK, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis evidence for a critical role of potassium nutrition. Plant Cell. 1998, 10, 1181–1191. 10.1105/tpc.10.7.1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H, Lee BH, Wu SJ, Zhu JK. Overexpression of a plasmamembrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 81–85. 10.1038/nbt766 [DOI] [PubMed] [Google Scholar]
- 13.Nutan KK, Kumar G, Singla-Pareek SL, Pareek A. A salt overly sensitive pathway member from Brassica juncea BjSOS3 can functionally complement ΔAtsos3 in Arabidopsis. Curr. Genomics. 2017, 19, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar G, Purty RS, Sharma MP, Singla-Pareek SL, Pareek A. Physiological response among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. Plant Physiol. 2009, 166, 507–520. [DOI] [PubMed] [Google Scholar]
- 15.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, et al. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 2006, 60, 51–68. 10.1007/s11103-005-2418-5 [DOI] [PubMed] [Google Scholar]
- 18.Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P. A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol. 2002, 130, 111–119. 10.1104/pp.005561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Despres C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, et al. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell. 2003, 15, 2181–2191. 10.1105/tpc.012849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker SS, Wilhelm KS, Thomashow MF. The 5'-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 1994, 24, 701–713. 10.1007/bf00029852 [DOI] [PubMed] [Google Scholar]
- 21.Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 1993, 5, 1529–1539. 10.1105/tpc.5.11.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakravarthy S, Tuori RP, DAscenzo MD, Fobert PR, Despres C, Martin GB. The tomato transcription factor Pti4 regulates defence-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 2003, 15, 3033–3050. 10.1105/tpc.017574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn MA, White AJ, Vural S, Hughes MA. Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.). Plant Mol. Biol. 1998, 38, 551–564. 10.1023/a:1006098132352 [DOI] [PubMed] [Google Scholar]
- 24.Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004, 135, 2150–2161. 10.1104/pp.104.041442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Piao Y, Liu Y, Li X, Piao Z. Genome-wide identification and expression analysis of chitinase gene family in, Brassica rapa, reveals its role in clubroot resistance. Plant Sci. 2018, 270, 257–267. 10.1016/j.plantsci.2018.02.017 [DOI] [PubMed] [Google Scholar]
- 26.Goyal E, Singh RS, Kanika K. Isolation and functional characterization of Salt overly sensitive 1 (SOS1) gene promoter from Salicornia brachiata. Biol. Plantarum. 2013, 57, 465–473. [Google Scholar]
- 27.Kaur C, Kumar G, Kaur S, Ansari MW, Pareek A, Sopory SK, et al. Molecular cloning and characterization of salt overly sensitive gene promoter from Brassica juncea (BjSOS2). Mol. Biol. Rep. 2015, 42, 1139–1148. 10.1007/s11033-015-3851-4 [DOI] [PubMed] [Google Scholar]
- 28.Feki K, Brini F, Amar SB, Saibi W, Masmoudi K. Comparative functional analysis of two wheat Na+/H+ antiporter SOS1 promoters in Arabidopsis thaliana under various stress conditions. J. Appl. Genet. 2015, 56, 15–26. 10.1007/s13353-014-0228-7 [DOI] [PubMed] [Google Scholar]
- 29.Sathee L, Sairam RK, Chinnusamy V, et al. Differential transcript abundance of salt overly sensitive (SOS) pathway genes is a determinant of salinity stress tolerance of wheat. Acta Physiol. Plant. 2015, 37, 169. [Google Scholar]
- 30.Chakraborty K, Sairam RK, Bhattacharya RC. Differential expression of salt overly sensitive pathway genes determines salinity stress tolerance in Brassica genotypes. Plant Physiol. Biochem. 2012, 51, 90–101. 10.1016/j.plaphy.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 31.Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. The Plant Cell. 2002, 14, 465–477. 10.1105/tpc.010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, et al. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. 10.1104/pp.106.092635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang RJ, Liu H, Bao Y, Lv QD, Yang L, Zhang HX. The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol. Biol. 2010, 74, 367–380. 10.1007/s11103-010-9680-x [DOI] [PubMed] [Google Scholar]
- 34.Di F, Jian H, Wang T, Chen X, Ding Y, Du H, et al. Genome-wide analysis of the PYL gene family and identification of PYL genes that respond to abiotic stress in Brassica napus. Genes. 2018, 9, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Z, Zeng DE, Liao J, Cheng C, Sahito ZA, Xiang M, et al. Genome-wide analysis of auxin receptor family genes in Brassica juncea var. tumida. Genes. 2019, 10, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.