ABSTRACT
Post-transcriptional non-template additions of nucleotides to 3′-ends of RNAs play important roles in the stability and function of RNA molecules. Although tRNA nucleotidyltransferase (CCA-adding enzyme) is known to add CCA trinucleotides to 3′-ends of tRNAs, whether other RNA species can be endogenous substrates of CCA-adding enzyme has not been widely explored yet. Herein, we used YAMAT-seq to identify non-tRNA substrates of CCA-adding enzyme. YAMAT-seq captures RNA species that form secondary structures with 4-nt protruding 3′-ends of the sequence 5′-NCCA-3′, which is the hallmark structure of RNAs that are generated by CCA-adding enzyme. By executing YAMAT-seq for human breast cancer cells and mining the sequence data, we identified novel candidate substrates of CCA-adding enzyme. These included fourteen ‘CCA-RNAs’ that only contain CCA as non-genomic sequences, and eleven ‘NCCA-RNAs’ that contain CCA and other nucleotides as non-genomic sequences. All newly-identified (N)CCA-RNAs were derived from the mitochondrial genome and were localized in mitochondria. Knockdown of CCA-adding enzyme severely reduced the expression levels of (N)CCA-RNAs, suggesting that the CCA-adding enzyme-catalyzed CCA additions stabilize the expression of (N)CCA-RNAs. Furthermore, expression levels of (N)CCA-RNAs were severely reduced by various cellular treatments, including UV irradiation, amino acid starvation, inhibition of mitochondrial respiratory complexes, and inhibition of the cell cycle. These results revealed a novel CCA-mediated regulatory pathway for the expression of mitochondrial non-coding RNAs.
KEYWORDS: tRNA, CCA-adding enzyme, TRNT1, YAMAT-seq, Mitochondria
Introduction
After transcription, newly-synthesized primary RNAs undergo various maturation steps prior to becoming functional molecules. Post-transcriptional non-template additions of nucleotides to 3′-ends of RNAs represent a major maturation step that regulates the stability and function of RNA molecules. For example, co-transcriptional addition of poly(A) tails to mRNAs by poly(A) polymerases regulate the stability of mRNAs and facilitates their translation, and this is a nearly universal phenomena among eukaryotic cells [1,2]. Uridylation of precursor molecules is an essential maturation step for U6 small nuclear RNA (snRNA) [3,4], and uridylation or adenylation of microRNAs (miRNAs) and their precursors regulates their stability [5–7]. Analysis of non-template nucleotide additions of RNAs are therefore necessary to the understanding biology of RNA molecules. Yet, whereas RNA sequencing (RNA-seq) has become a ubiquitous tool in biomedical research, RNAs containing such additional 3′-terminal sequences are expected to be infrequently recognized and characterized in RNA-seq data, because the presence of non-genomic sequences reduces their chances of being mapped on the genome. Considering the relative obscurity of these RNAs, cells may express many yet-unidentified RNA molecules with post-transcriptionally-added non-genomic sequences on their 3′-ends.
Transfer RNAs (tRNAs) are universally expressed non-coding RNAs (ncRNAs) that play central roles in translation machinery by converting information of mRNA codons into amino acids. In their clover-leaf secondary structures, tRNAs invariably bear 4-nucleotide (nt) protruding 3′-terminal sequences comprising 5′-CCA-3′ trinucleotides and a preceding discriminator nucleotide. In most organisms, the 3′-CCA end is not genetically encoded and is post-transcriptionally added by tRNA nucleotidyltransferase (CCA-adding enzyme) [8,9]. This enzyme not only synthesizes de novo CCA ends of tRNAs but also is involved in the maintenance and quality control of tRNAs [10]. CCA-adding enzyme adds CCA sequences to 1-nt protruding 3′-ends of stem-structured RNAs, which forms protruding 5′-NCCA-3′ sequences [11,12]. In addition to tRNAs, other ncRNAs can be substrates of CCA-adding enzyme. For example, in humans, MALAT1-associated small cytoplasmic RNA (mascRNA), which is a tRNA-like cytoplasmic RNA generated from the locus of MALAT1 ncRNA, contains post-transcriptionally-added CCA sequences [13] that are generated by CCA-adding enzyme [14]. Non-tRNA molecules with 3′-terminal non-genomic CCA sequences have also been found in plant mitochondria [15] and plastids [16]. In vitro experiments further suggest that non-tRNA sequences can be substrates of CCA-adding enzymes [17–19]. However, endogenous non-tRNA substrates of CCA-adding enzyme have not yet been widely explored.
We previously developed a Y-shaped Adapter-ligated MAture TRNA sequencing (YAMAT-seq) that efficiently and conveniently allows high-throughput sequencing of tRNAs [20]. In YAMAT-seq, a Y-shaped adapter is specifically ligated to 5′-ends and to the 4-nt protruding 3′-ends of tRNAs by T4 RNA Ligase 2 (Rnl2), and the ligation products are amplified and sequenced. Theoretically, the Y-shaped adapter should be ligated not only to tRNAs but also to any RNA species with a secondary structure that includes a 4-nt protruding 3′-end of the sequence 5′-NCCA-3′. Because such protruding 3′-ends are the hallmark structure of RNAs that are generated by CCA-adding enzyme, we reasoned that YAMAT-seq can be used to identify endogenous non-tRNA substrates of CCA-adding enzyme. Herein, we report our execution of YAMAT-seq for human breast cancer cells and subsequent mining of sequence data, leading to identification of novel mitochondrial 3′-CCA-containing ncRNAs that are regulated by CCA-adding enzyme and are responsive to various cellular treatments.
Results and discussion
Sequences identified by YAMAT-seq contain non-tRNA species bearing non-genomic 3′-terminal CCA sequences
YAMAT-seq was designed to selectively capture RNAs with secondary structures that bear protruding 3′-ends of the sequence 5′-NCCA-3′, and it’s specificity, quantitative ability, and broad applicability have been demonstrated previously [20]. To investigate endogenous, non-tRNA substrates of CCA-adding enzyme, we applied YAMAT-seq to total RNA isolated from MCF-7 breast cancer cells. As observed previously [20], the method specifically amplified abundant cDNA bands of 170–230 base pairs (bp) (Fig. 1A). Illumina next-generation sequencing of these bands yielded approximately 28 million raw reads, and, as expected, a high percentage (~80%) of these were mapped to tRNA sequences. Because maximum read lengths of sequencing were 100 nt and the YAMAT-seq reads contain 13-nt 5′-Y-adapter sequences, the maximum read lengths of the extracted RNAs was 87 nt. We focused our attention on reads that were derived from non-tRNA regions of the genome. To extract potential substrates of CCA-adding enzyme, non-tRNA reads were filtered by 1) the presence of non-genomic 3′-terminal CCA sequences and 2) the formation of secondary structures with 4-nt protruding 3′-ends of the sequence 5′-NCCA-3′ (Fig. 1B).
Figure 1.
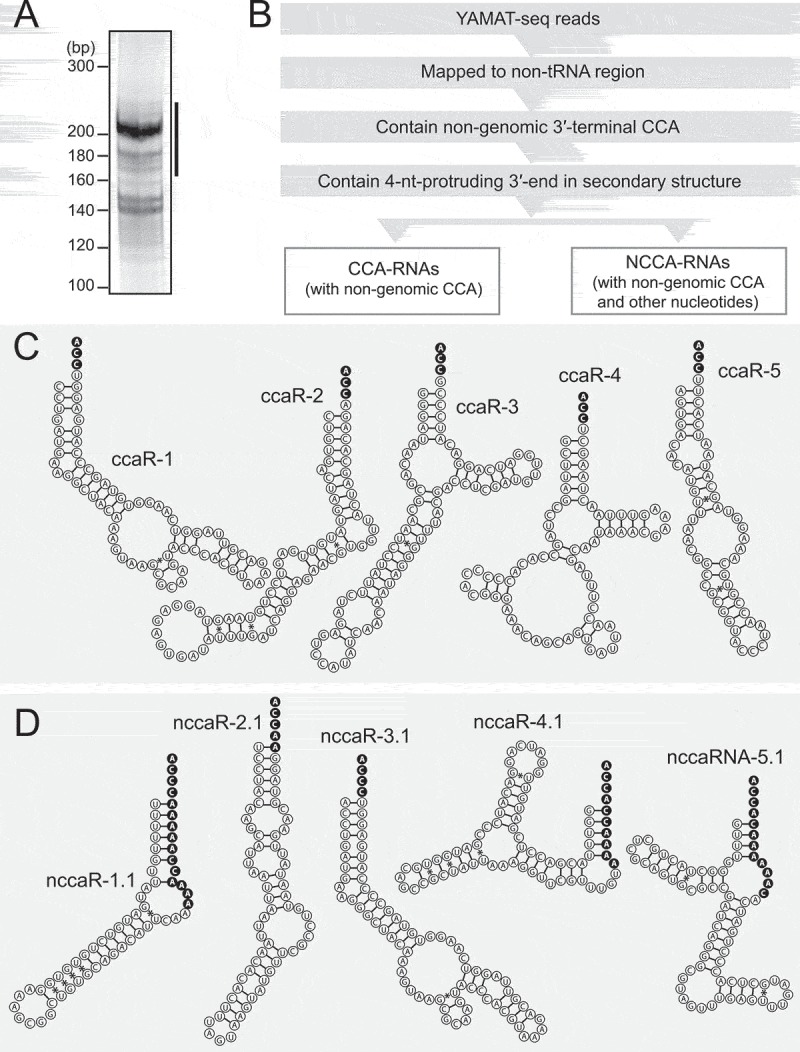
YAMAT-seq identified non-tRNA species containing non-genomic 3′-terminal CCA sequences.
(A) YAMAT-seq amplified cDNAs from MCF-7 cells were electrophoresed in 8% native polyacrylamide gel. The region designated with a line was subjected to gel-purification and Illumina sequencing.(B) Bioinformatics procedures to extract sequences of CCA-RNAs and NCCA-RNAs.(C, D) Sequences and secondary structures of the top 5 CCA-RNAs (C) and NCCA-RNAs (D).
The obtained reads were then divided into 1) CCA-RNAs that only contain CCA as non-genomic sequences and 2) NCCA-RNAs that contain CCA and other nucleotides (mostly A) as non-genomic sequences (Fig. 1C, D and S1A-B). A previously-reported mascRNA [13] was the most abundantly-detected CCA-RNA, but all other 14 CCA-RNAs and 11 NCCA-RNAs were identified as novel candidate endogenous ncRNA substrates of CCA-adding enzyme (Table S1). The majority of the identified (N)CCA-RNAs (ccaR-1–11, ccaR-13, ccaR-14, nccaR-1–8, and nccaR-10) were also observed in previously-reported YAMAT-seq data from MCF-7 cells [20], confirming the reproducibility of the expression of (N)CCA-RNAs. Because many RNA post-transcriptional modifications impair reverse transcription [21] and thereby prevent identification of heavily-modified RNAs in sequencing data, there is a possibility that some endogenous (N)CCA-RNAs remain uncaptured by our YAMAT-seq approach and subsequent strict filtering based on the prediction of secondary structures.
Many tRNAs carry A at the discriminator position [22], and accordingly, in vitro experiments showed that human CCA-adding enzyme has a preference for substrates with A at that position [19]. This preference was also observed among the newly-identified CCA-RNAs, of which half carry A adjacent to CCA sequences, and 43% carry U in this position. Among NCCA-RNAs, consecutive As are the most commonly observed in non-genomic sequences that could be generated by successive actions of poly(A) polymerase and CCA-adding enzyme, as shown in Fig. S2. More than half of the NCCA-RNAs have variations in their 5′-ends and/or in their 3′-terminal non-genomic sequences (Table S1), suggesting that, during biogenesis of NCCA-RNAs, interplay between poly(A) polymerase, CCA-adding enzyme, and nucleases that cleave precursor molecules, is not uniformly regulated.
CCA-RNAs are derived from the mitochondrial genome and are localized in mitochondria
All of the newly-identified CCA-RNAs and NCCA-RNAs are derived from the mitochondrial genome (Fig. 2). Regions encoding rRNAs are the most-frequently involved in generating (N)CCA-RNAs, and 10 CCA-RNAs and 4 NCCA-RNAs are derived from 16S or 12S rRNAs. This may reflect the abundant expression of 16S and 12S rRNAs in mitochondria. The two (N)CCA-RNAs ccaR-6 and nccaR-9 are derived from mRNAs for CO II and CO I, respectively, and the remaining 3 CCA-RNAs and 6 CCA-RNAs are generated from non-coding regions of the mitochondrial genome. Regarding the top 5 abundant (N)CCA-RNAs, ccaR-1 and ccaR-3 are derived from 12S and 16S rRNA, respectively; ccaR-2 is derived from an anti-sense strand of ND4 mRNA; and nccaR-1 and nccaR-2 originate from the non-coding regions close to the light strand promoter (LSP) of mitochondria. The generation of (N)CCA-RNAs might influence the expression of their substrate RNAs or of the RNAs anti-sense to them. The generation of NCCA-RNAs from the regions close to the LSP might affect mitochondrial transcription. It remains unclear why (N)CCA-RNAs are exclusively identified in the mitochondrial genome but not in the nuclear genome. Given that cytoplasmic tRNAs are much more abundant than mitochondrial tRNAs, the relative abundance of non-tRNA substrates of CCA-adding enzyme may be higher in mitochondria than in the cytoplasm.
Figure 2.
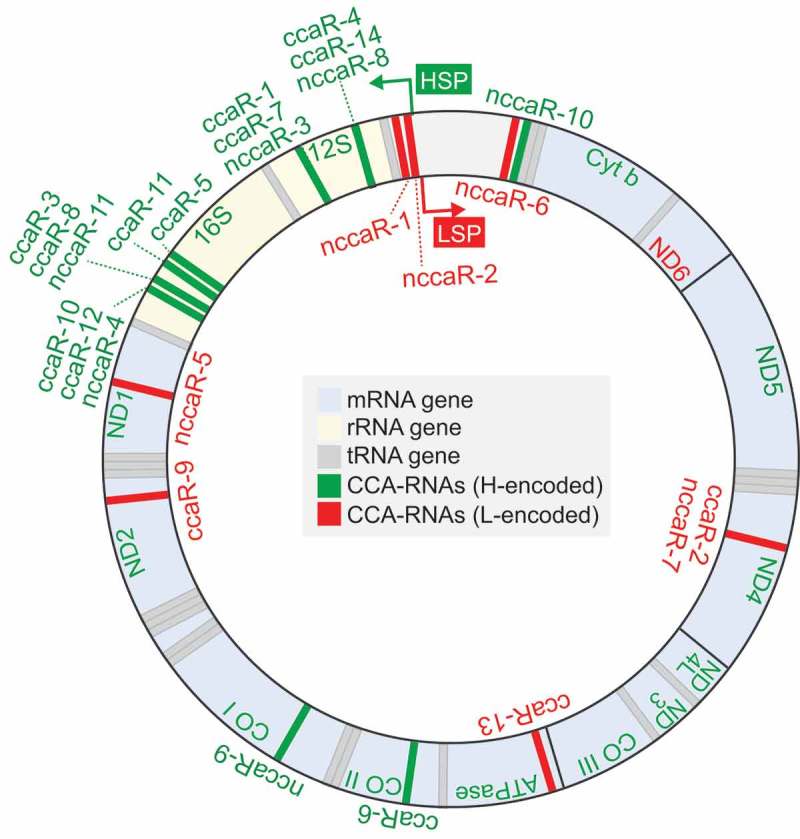
Mitochondrial genome regions that generate CCA-RNAs.
To confirm expression in cells, three (N)CCA-RNAs were selected for northern blot using total RNAs from cultured human cell lines. To confirm that the signals are specifically derived from targeted RNAs, two different probes were designed for each RNA (Fig. S3). Both probes detected ccaR-2 and nccaR-2 as clear bands (Fig. 3). Although ccaR-2 was expressed in all five examined cell lines, nccaR-2 was detected only in 293T, MCF-7, and LNCaP cells, and not in HeLa or MCF10A cells, suggesting that (N)CCA-RNA expressions are not always ubiquitous and can differ between (N)CCA-RNA species and/or cell lines. The most clearly-observed nccaR-1 band corresponded with a precursor RNA that lacks non-genomic nucleotide additions (the starting RNA species depicted in Fig. S2), and smeared bands in the upper region were likely nccaR-1. These smeared bands may indicate the co-existence of many nccaR-1 variants, as observed in sequencing data (Table S1).
Figure 3.
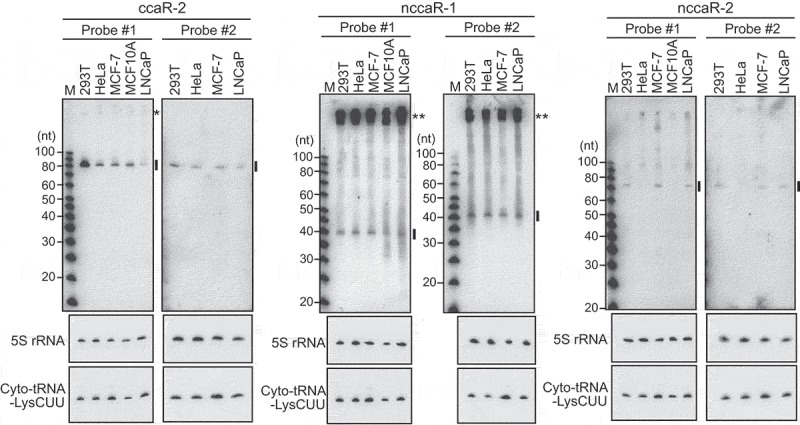
Experimental validation of the expression of CCA-RNAs.
Total RNAs from the indicated human cultured cell lines were subjected to northern blot using the probes shown in Fig. S3. 5S rRNA and cytoplasmic tRNALysCUU were analyzed as controls. Detected (N)CCA-RNAs are indicated by lines. In the detection of ccaR-2 using probe #1, we observed a non-specific band (indicated by a single asterisk) that was derived from a cytoplasmic RNA (Fig. 4) and was not regulated by CCA-adding enzyme (Fig. 5). In contrast, detection of nccaR-1 using both probes #1 and #2 identified potential precursor molecules (indicated by double asterisks) that were present in mitochondria (Fig. 4). Given its size, the clearly observed nccaR-1 band (indicated by a line) may correspond with RNAs that lack nucleotide additions, and nucleotide-added sequences are likely represented by the upper smeared bands.
To determine the localizations of (N)CCA-RNAs, cytoplasmic and mitochondrial fractions from HeLa cells were analyzed using northern blot. Whereas cytoplasmic and mitochondrial tRNAs were detected in respective fractions, both of the examined (N)CCA-RNAs were observed in the mitochondrial fraction (Fig. 4), indicating that (N)CCA-RNAs are produced from the mitochondrial genome and the generated RNAs remain inside mitochondria. The mitochondrial expressions of (N)CCA-RNAs suggest that mitochondrial enzymes are responsible for their biogenesis. Only one CCA-adding enzyme gene (TRNT-1) is encoded on the human genome [23,24]. As is the case of yeast CCA-adding enzyme, TRNT1 mRNA is thought to have two alternative translation starting site [24,25], and one of the two generated proteins, containing N-terminal mitochondrial targeting signal, is imported into mitochondria and would be involved in the biogenesis of (N)CCA-RNAs. Mitochondrial poly(A) polymerase [26,27] would be additionally required for the biogenesis of NCCA-RNAs.
Figure 4.
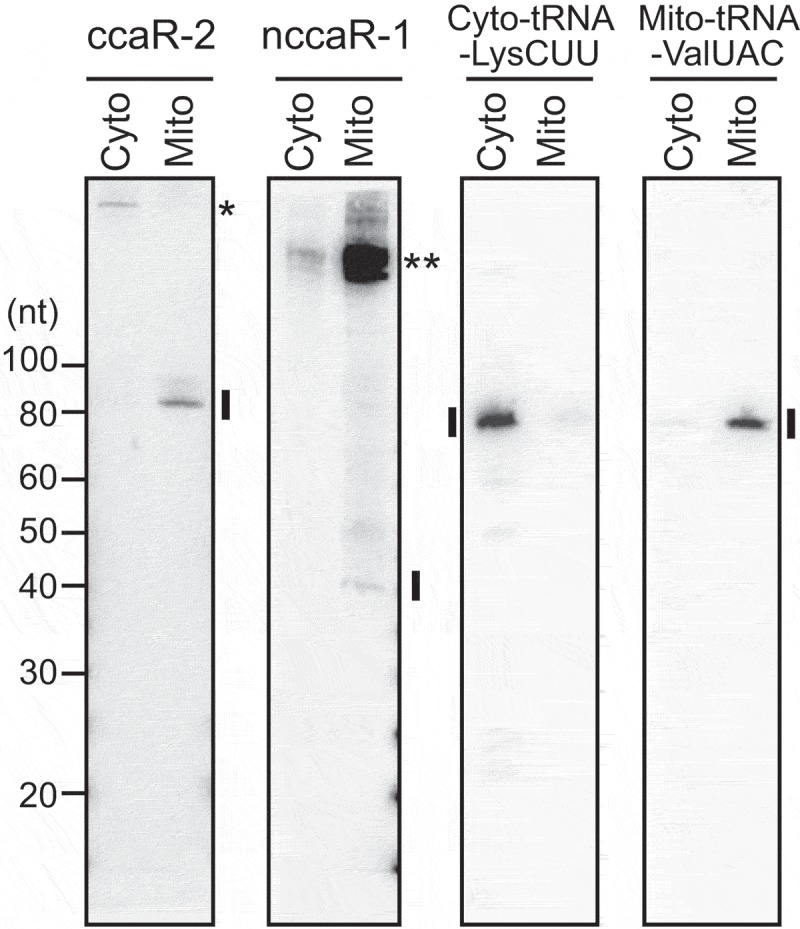
Mitochondrial localization of CCA-RNAs.
Cytoplasmic and mitochondrial fractions of HeLa cells were subjected to northern blot for (N)CCA-RNAs. Cytoplasmic tRNALysCUU and mitochondrial tRNAValUAC were analyzed as control cytosolic and mitochondrial RNAs, respectively. A single asterisk: a non-specific cytoplasmic band; double asterisks: the bands of potential precursor RNAs of nccaR-1.
CCA-adding enzyme stabilizes the expression of CCA-RNAs
To elucidate the functional significance of CCA addition in (N)CCA-RNAs, we performed RNAi knockdown of the human CCA-adding enzyme (TRNT1) in HeLa cells, which reduced the expression levels of TRNT1 mRNA to approximately 25% (Fig. 5A). (N)CCA-RNAs and other control RNAs were then quantitated using northern blot. TRNT1 knockdown did not affect the expression of unrelated RNAs, such as microRNA, snRNAs, and rRNA, but it reduced the expression levels of cytoplasmic and mitochondrial tRNAs (Fig. 5B, S4A). The reduced tRNA levels following TRNT1 depletion, as reported previously[28], confirmed that the knockdown efficiency was sufficient to observe effects on TRNT1 substrate RNAs. Under the conditions of TRNT1 knockdown, we observed a severe reduction in expression levels of both examined (N)CCA-RNAs, suggesting that CCA-adding enzyme stabilizes the expression of (N)CCA-RNAs by adding 3′-terminal CCA sequences. Although addition of short poly(A) sequences or two tandem CCA sequences (CCACCA) has been shown to favor degradation of RNAs [14,29,30], two CCA sequences flanking several A residues stabilize nccaR-1, but the associated mechanism remains to be characterized. In the case of tRNAs, CCA addition enables aminoacylation and interaction with elongation factor, which stabilizes tRNAs. (N)CCA-RNAs may also interact with partner proteins for stabilization, and these interactions may require CCA addition by CCA-adding enzyme.
Figure 5.
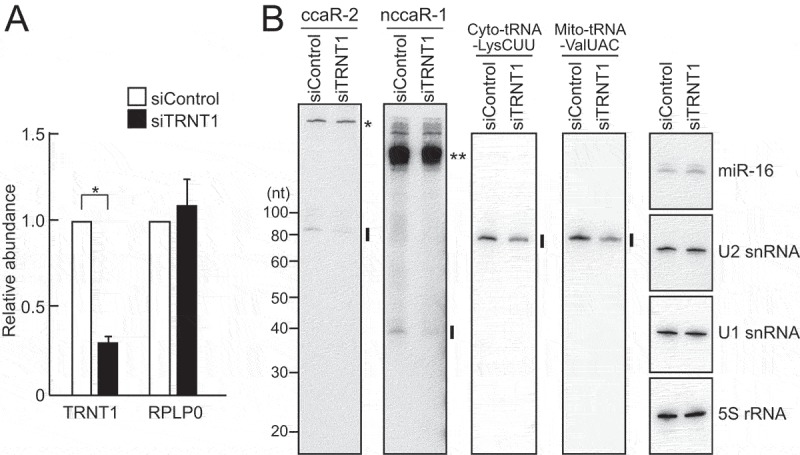
The CCA-adding enzyme TRNT1 stabilizes the expression of CCA-RNAs.
(A) HeLa cells were transfected with control siRNA or siRNA targeting TRNT1. TRNT1 mRNA expression was efficiently knocked down by the targeted siRNA.(B) Total RNAs from control and TRNT1 KD cells were used for northern blots of (N)CCA-RNAs. Cytoplasmic tRNALysCUU and mitochondrial tRNAValUAC or other ncRNAs miR-16, U1/U2 snRNAs, and 5S rRNA were analyzed as control RNAs that are regulated, or not regulated, by TRNT1, respectively. Band quantifications are shown in Fig. S4A. A single asterisk: a non-specific cytoplasmic band; double asterisks: the bands of potential precursor RNAs of nccaR-1.
The expressions of (N)CCA-RNAs are responsive to various cellular treatments
To investigate modes of regulation for (N)CCA-RNA expressions, HeLa cells were subjected to UV irradiation, amino acid starvation, inhibition of mitochondrial respiratory complexes, or inhibition of the cell cycle. In expression analyses of (N)CCA-RNAs, a control microRNA was unaffected by any of the treatments, but the expressions of (N)CCA-RNAs were severely reduced by all of the treatments (Fig. 6, S4B). Because mitochondrial tRNA was not affected (Fig. 6, S4B), the observed reductions would not result from decreased levels or activities of CCA-adding enzyme. The reduction of (N)CCA-RNAs upon inhibition of respiratory complexes may simply be due to decreased levels of precursor RNAs, because levels of potential precursors of nccaR-1 were also reduced, as observed in the upper region of the blot. However, UV irradiation, amino acid starvation, and inhibition of the cell cycle severely reduced the expression levels of nccaR-1 without affecting precursor band intensities, suggesting that these reductions are caused by molecular mechanisms that are specific to (N)CCA-RNA molecules.
Figure 6.
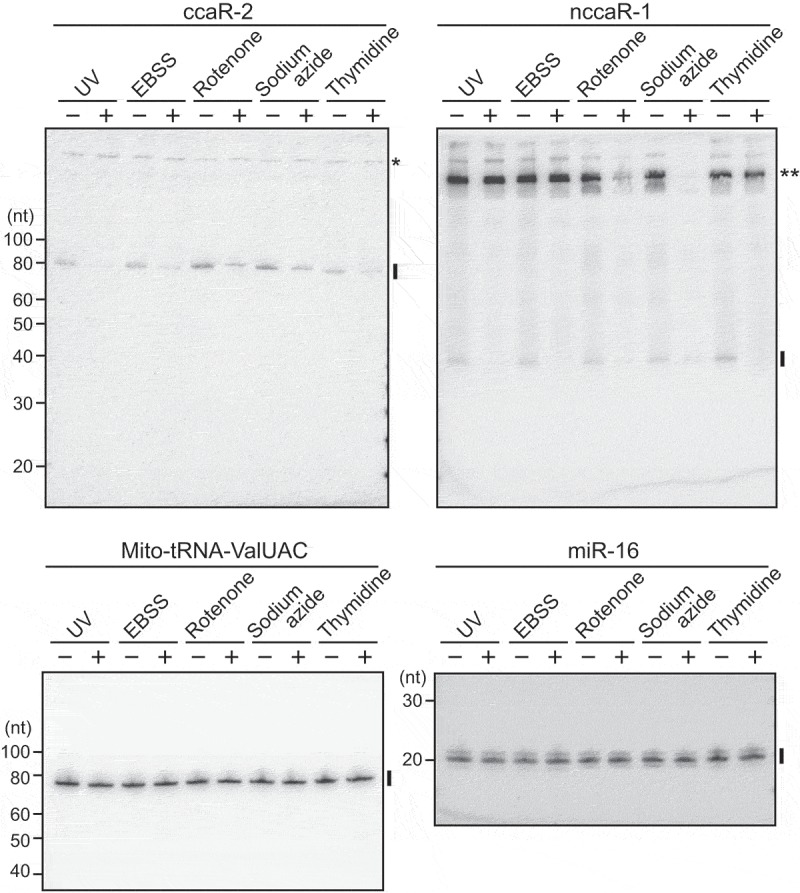
The expression of CCA-RNAs are responsive to cellular treatments.
HeLa cells were subjected to UV irradiation (UV), starvation cultured with EBSS, inhibition of mitochondrial respiratory complex I (rotenone) or IV (sodium azide), or inhibition of cell cycle (thymidine block), and the expression of CCA-RNAs were analyzed using northern blots. Mitochondrial tRNAValUAC and miR-16 were analyzed as control RNAs whose expressions were unaffected by the treatments. Band quantifications are shown in Fig. S4B. A single asterisk: a non-specific cytoplasmic band; double asterisks: the bands of potential precursor RNAs of nccaR-1.
Conclusion
Using YAMAT-seq, we identified novel substrate ncRNAs of CCA-adding enzyme. Other than previously-reported cytoplasmic mascRNA, all of the identified CCA-RNAs and the NCCA-RNAs were derived from the mitochondrial genome and their mitochondrial localizations were experimentally confirmed. CCA addition by CCA-adding enzyme stabilizes the expression of (N)CCA-RNAs, and the expressions of (N)CCA-RNAs are responsive to various cellular treatments. These results revealed a novel regulatory pathway for the stabilization of mitochondrial ncRNAs.
Material and methods
Cell culture
Cells were cultured as described previously [20,31]. Briefly, MCF-7 cells were cultured in MEM (Corning) containing 10% FBS, 1 mM sodium pyruvate (Corning), 1 × nonessential amino acids solution (Life Technologies), and 10 μg/ml insulin (Sigma). MCF10A cells were cultured in DMEM F12 (Corning) containing 10% FBS, 10 μg/ml insulin (Sigma), 20 ng/ml epidermal growth factor (Pepro Tech), 1 mg/ml hydrocortisone (Sigma), and 100 ng/ml cholera toxin (Sigma). HeLa and 293T cells were cultured in DMEM (Corning) containing 10% FBS. LNCaP cells were cultured in improved MEM (Corning) containing 5% FBS.
YAMAT-seq
YAMAT-seq was performed as described previously[20]. Briefly, total RNA was extracted from MCF-7 cells using TRIsure (Bioline) and then incubated at 37°C for 40 min in 20 mM Tris-HCl (pH 9.0) for deacylation, followed by ethanol precipitation. Deacylated RNAs were mixed with Y-shaped adapters, were incubated at 90°C for 2 min, and were then subjected to annealing by incubating at 37°C for 15 min in an annealing buffer composed of 5 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, and 10 mM MgCl2. To ligate Y-shaped adapters with their annealed RNAs, they were incubated with T4 RNA Ligase 2 (New England Biolabs) at 37°C for 1 h, followed by overnight incubation at 4°C. We then amplified cDNAs as described previously[20]. Amplified cDNA products were electrophoresed on 8% native polyacrylamide gels, gel-purified, and sequenced (100 nt single-read) using an Illumina HiSeq 2500 system by the Next-Generation Sequencing Core at University of Pennsylvania.
Bioinformatics analyses
YAMAT-seq sequence reads from MCF-7 cells (raw read numbers: 28,148,240) are publically available at NCBI’s Sequence Read Archive via accession No. SRR9102952. We performed quality trimming and adapter cutting using cutadapt[32] to remove the 5′-adapter (ACTGGATACTGGN) and the 3′-adapter (GTATCCAGTTGGAATTCTCGGGTGCCAAGG), and used a similar previously described method[20] to align reads to the tRNA-reference set. To be more conservative, we used a larger number of tRNAs and did not disqualify tRNA-mapped reads that were better mapped to genomic locations outside of the tRNA-reference set. Our tRNA-reference set comprised 1,132 tRNA-reference genes that included 613 nuclear-encoded cytoplasmic tRNAs (listed in gtRNAdb[33]) from the GRCh37 assembly, 22 known mitochondrial tRNAs from tRNAdb[34], and 497 tRNA lookalikes[35]. As in previous studies, we also removed introns and added 3′-terminal CCA sequences to the tRNA-reference set. SHRiMP2[36] was used to map reads to this reference set, allowing for non-unique mappings with a 10% mismatch rate and equal penalties for each mismatch and gap extension. Subsequently, we discarded all reads that were mapped to this tRNA-reference set. We then sub-selected reads that were 60–87 nt in length and were encoded on the genome. The remaining reads were further filtered according to the presence of 3′-terminal CCA sequences that are not encoded on the genome. After discarding RNA sequences with single reads only, the secondary structures of selected reads were analyzed using mfold[37] and RNAs that formed stem structures with 4 nt-protruding 3′-ends were selected as potential RNAs with CCA sequences that are added by CCA-adding enzyme. These reads were subclassified into CCA-RNAs or NCCA-RNAs which contained non-genomically-encoded CCA of CCA plus other nucleotides, respectively, at their 3′-ends (Table S1).
Northern blot
Northern blot was performed as described previously [31,38] with 10 µg of total RNA per lane and the antisense probes shown in Table S2. Typhoon 9400 (GE Healthcare) was used to visualize and quantitate phosphor autoradiography.
Cell fractionation
Cytoplasmic and mitochondrial fractions were prepared from HeLa cells as described previously[38]. Briefly, about 500 µl of cell pellets were mixed with 5 ml of fractionation buffer containing 0.25 M sucrose, 20 mM HEPES (pH 7.5), 3 mM EDTA, and 1 mM PMSF (Sigma), and were homogenized on ice using a daunce tissue grinder (Wheaton). Homogenates were then made up to 10 ml and were centrifuged twice at 600 g for 15 min to pellet cell debris and nuclei. Supernatants were further centrifuged at 15,000 g for 30 min, and the resulting supernatants were collected as cytoplasmic fractions. Residual pellets were resuspended in fractionation buffer and were centrifuged again at 15,000 g for 15 min to yield mitochondrial fractions as pellets.
RNAi knockdown of TRNT1
RNAi knockdown was performed as described previously[31]. To knockdown human TRNT1, Stealth RNA interference duplexes (Invitrogen) were used as described previously[28], and ON-TARGETplus (#D-001810-02-05, Dharmacon) was used as a control siRNA. Briefly, HeLa cells were transfected with 50 nM of each siRNA using a reverse-transfection method with RNAiMAX (Invitrogen). Two more transfections were repeated over 3 and 6 d after the first transfection, and the cells were harvested at 3 d after the final transfection (9 d after the first transfection). The efficiency of RNAi knockdown was verified by quantifying TRNT1 mRNA using real-time RT-qPCR with the primers shown in Table S3. GAPDH mRNA was quantified as a control.
Cellular treatments
HeLa cells were UV irradiated as described previously[39]. Briefly, PBS-washed cells were exposed to 40 J/m[2] of UV using CL-1000 UV Crosslinker (Analytik Jena). Cells were then incubated in fresh medium for 4 h. Starvation conditions were produced as described previously [40,41]. Specifically, PBS-washed HeLa cells were cultured in Earle’s balanced salt solution (EBSS, Sigma) for 4 h. To inhibit mitochondrial electron transport as described previously [42–45], HeLa cells were incubated in medium containing either 50 µM rotenone (Complex I inhibitor) or 10 µM sodium azide (Complex IV inhibitor) for 24 h. To induce cell cycle arrest, a double thymidine block was performed as described previously[31]. HeLa cells were cultured with 2.5 mM thymidine (Sigma) for 16 h, were washed in PBS, and were then incubated in normal medium for 9 h. Finally, cells were incubated again with 2.5 mM thymidine for 16 h [10].
Funding Statement
This work was supported by National Institute of General Medical Sciences [GM106047]; National Institute of Allergy and Infectious Diseases[AI130496];the American Cancer Society [RSG-17-059-01-RMC]; The W.W. Smith Charitable Trust [C1608].
Acknowledgments
We are grateful to Dr. Dmitry Temiakov (Thomas Jefferson University) and the members of Kirino lab for helpful discussions. This study was supported in part by the National Institutes of Health Grant (GM106047 and AI130496, to YK), American Cancer Society Research Scholar Grant (RSG-17-059-01-RMC, to YK), and The W.W. Smith Charitable Trust Grant (C1608, to YK).
Author contributions
YK, PL, and SH designed the project with contributions from all co-authors. MS performed YAMAT-seq and generated the sequence data. PL designed and implemented the pipeline to process YAMAT-seq reads to identify non-tRNA reads with CCA sequences, followed by further selections of (N)CCA-RNAs by KP, MS, and SH. KP finalized (N)CCA-RNA data and performed experimental works to confirm their cellular expression and their regulation by CCA-adding enzyme and by cellular treatments. All authors discussed the results, and KP and YK wrote the paper with contributions from MS, PL, and IR. All authors have read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- [1].Sachs A. The role of poly(A) in the translation and stability of mRNA. Curr Opin Cell Biol. 1990;2:1092–1098. [DOI] [PubMed] [Google Scholar]
- [2].Dreyfus M, Regnier P. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. [DOI] [PubMed] [Google Scholar]
- [3].Lund E, Dahlberg JE. Cyclic 2ʹ,3ʹ-phosphates and nontemplated nucleotides at the 3ʹ end of spliceosomal U6 small nuclear RNA’s. Science. 1992;255:327–330. [DOI] [PubMed] [Google Scholar]
- [4].Trippe R, Sandrock B, Benecke BJ. A highly specific terminal uridylyl transferase modifies the 3ʹ-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. [DOI] [PubMed] [Google Scholar]
- [6].Heo I, Joo C, Kim YK, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. [DOI] [PubMed] [Google Scholar]
- [7].Heo I, Joo C, Cho J, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. [DOI] [PubMed] [Google Scholar]
- [8].Weiner AM. tRNA maturation: RNA polymerization without a nucleic acid template. Curr Biol. 2004;14:R883–5. [DOI] [PubMed] [Google Scholar]
- [9].Xiong Y, Steitz TA. A story with a good ending: tRNA 3ʹ-end maturation by CCA-adding enzymes. Curr Opin Struct Biol. 2006;16:12–17. [DOI] [PubMed] [Google Scholar]
- [10].Betat H, Morl M. The CCA-adding enzyme: A central scrutinizer in tRNA quality control. Bioessays. 2015;37:975–982. [DOI] [PubMed] [Google Scholar]
- [11].Tomita K, Fukai S, Ishitani R, et al. Structural basis for template-independent RNA polymerization. Nature. 2004;430:700–704. [DOI] [PubMed] [Google Scholar]
- [12].Xiong Y, Steitz TA. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004;430:640–645. [DOI] [PubMed] [Google Scholar]
- [13].Wilusz JE, Freier SM, Spector DL. 3ʹ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wilusz JE, Whipple JM, Phizicky EM, et al. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Williams MA, Johzuka Y, Mulligan RM. Addition of non-genomically encoded nucleotides to the 3ʹ-terminus of maize mitochondrial mRNAs: truncated rps12 mRNAs frequently terminate with CCA. Nucleic Acids Res. 2000;28:4444–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zandueta-Criado A, Bock R. Surprising features of plastid ndhD transcripts: addition of non-encoded nucleotides and polysome association of mRNAs with an unedited start codon. Nucleic Acids Res. 2004;32:542–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hegg LA, Kou M, Thurlow DL. Recognition of the tRNA-like structure in tobacco mosaic viral RNA by ATP/CTP: tRNAnucleotidyltransferases from Escherichia coli and Saccharomyces cerevisiae. J Biol Chem. 1990;265:17441–17445. [PubMed] [Google Scholar]
- [18].Cho HD, Tomita K, Suzuki T, et al. U2 small nuclear RNA is a substrate for the CCA-adding enzyme (tRNA nucleotidyltransferase). J Biol Chem. 2002;277:3447–3455. [DOI] [PubMed] [Google Scholar]
- [19].Wende S, Bonin S, Gotze O, et al. The identity of the discriminator base has an impact on CCA addition. Nucleic Acids Res. 2015;43:5617–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shigematsu M, Honda S, Loher P, et al. YAMAT-seq: an efficient method for high-throughput sequencing of mature transfer RNAs. Nucleic Acids Res. 2017;45:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kellner S, Burhenne J, Helm M. Detection of RNA modifications. RNA Biol. 2010;7:237–247. [DOI] [PubMed] [Google Scholar]
- [22].Limmer S, Hofmann HP, Ott G, et al. The 3ʹ-terminal end (NCCA) of tRNA determines the structure and stability of the aminoacyl acceptor stem. Proc Natl Acad Sci U S A. 1993;90:6199–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Levinger L, Morl M, Florentz C. Mitochondrial tRNA 3ʹ end metabolism and human disease. Nucleic Acids Res. 2004;32:5430–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Reichert AS, Thurlow DL, Morl M. A eubacterial origin for the human tRNA nucleotidyltransferase? Biol Chem. 2001;382:1431–1438. [DOI] [PubMed] [Google Scholar]
- [25].Nagaike T, Suzuki T, Tomari Y, et al. Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J Biol Chem. 2001;276:40041–40049. [DOI] [PubMed] [Google Scholar]
- [26].Chang JH, Tong L. Mitochondrial poly(A) polymerase and polyadenylation. Biochim Biophys Acta. 2012;1819:992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tomecki R, Dmochowska A, Gewartowski K, et al. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sasarman F, Thiffault I, Weraarpachai W, et al. The 3ʹ addition of CCA to mitochondrial tRNASer(AGY) is specifically impaired in patients with mutations in the tRNA nucleotidyl transferase TRNT1. Hum Mol Genet. 2015;24:2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Slomovic S, Fremder E, Staals RH, et al. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc Natl Acad Sci U S A. 2010;107:7407–7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Carpousis AJ, Vanzo NF, Raynal LC. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. [DOI] [PubMed] [Google Scholar]
- [31].Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112:E3816–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- [33].Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Juhling F, Morl M, Hartmann RK, et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Telonis AG, Loher P, Kirino Y, et al. Nuclear and mitochondrial tRNA-lookalikes in the human genome. Front Genet. 2014;5:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].David M, Dzamba M, Lister D, et al. SHRiMP2: sensitive yet practical SHort read mapping. Bioinformatics. 2011;27:1011–1012. [DOI] [PubMed] [Google Scholar]
- [37].Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Honda S, Kirino Y, Maragkakis M, et al. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. Rna. 2013;19:1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sharma A, Kaur M, Kar A, et al. Ultraviolet radiation stress triggers the down-regulation of essential replication factor Mcm10. J Biol Chem. 2010;285:8352–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nazio F, Carinci M, Valacca C, et al. Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J Cell Biol. 2016;215:841–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ni HM, Bockus A, Wozniak AL, et al. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7:188–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen Y, McMillan-Ward E, Kong J, et al. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007;120:4155–4166. [DOI] [PubMed] [Google Scholar]
- [43].Hayashi Y, Yoshida M, Yamato M, et al. Reverse of age-dependent memory impairment and mitochondrial DNA damage in microglia by an overexpression of human mitochondrial transcription factor a in mice. J Neurosci. 2008;28:8624–8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Schwoebel ED, Ho TH, Moore MS. The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J Cell Biol. 2002;157:963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Maro B, Bornens M. Reorganization of HeLa cell cytoskeleton induced by an uncoupler of oxidative phosphorylation. Nature. 1982;295:334–336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


