ABSTRACT
During replication of long terminal repeat (LTR)-retrotransposons, their proteins and genome (g) RNA assemble into virus-like particles (VLPs) that are not infectious but functionally related to retroviral virions. Both virions and VLPs contain gRNA in a dimeric form, but contrary to retroviruses, little is known about how gRNA dimerization and packaging occurs in LTR-retrotransposons. The LTR-retrotransposon Ty1 from Saccharomyces cerevisiae is an informative model for studying LTR-retrotransposon and retrovirus replication. Using structural, mutational and functional analyses, we explored dimerization of Ty1 genomic RNA. We provide direct evidence that interactions of self-complementary PAL1 and PAL2 palindromic sequences localized within the 5′UTR are essential for Ty1 gRNA dimer formation. Mutations disrupting PAL1-PAL2 complementarity restricted RNA dimerization in vitro and Ty1 mobility in vivo. Although dimer formation and mobility of these mutants was inhibited, our work suggests that Ty1 RNA can dimerize via alternative contact points. In contrast to previous studies, we cannot confirm a role for PAL3, tRNAiMet as well as recently proposed initial kissing-loop interactions in dimer formation. Our data also supports the critical role of Ty1 Gag in RNA dimerization. Mature Ty1 Gag binds in the proximity of sequences involved in RNA dimerization and tRNAiMet annealing, but the 5′ pseudoknot in Ty1 RNA may constitute a preferred Gag-binding site. Taken together, these results expand our understanding of genome dimerization and packaging strategies utilized by LTR-retroelements.
KEYWORDS: LTR-retrotransposons, RNA dimerization, Ty1, Gag, RNA structure
Introduction
Retrotransposons are widespread in most eukaryotic genomes, and active elements are present in fungi, plants, insects and mammals, including humans. They influence the architecture and evolution of genomes, and can impact the expression of genes [1]. Furthermore, expression of retrotransposon RNAs and proteins, and in some cases the production of VLPs, accompanies various types of cancer and autoimmune diseases in humans [2,3], and enhances synaptic plasticity in Drosophila and mammals [4,5]. The packaging of gRNA into virions or VLPs is critical for propagation of infectious and endogenous LTR-retroelements. Extensive studies with retroviruses indicate that gRNA packaging is coupled with dimerization, which preludes gRNA encapsidation into virions and ensures the proper packaging specificity [6–9]. A variety of structure-based mapping strategies have been used to reveal the molecular mechanisms and structural determinants of gRNA dimerization in retroviruses [10–12]. In general, dimeric retroviral RNA is stabilized by base-pairing between cis-acting palindromic sequences located near the 5′-end of the gRNA [10,13]. The nucleocapsid (NC) domain of Gag is responsible for recognition of specific dimerization and packaging signals in viral RNA, and then Gag or mature NC nucleic acid chaperone activity promotes the structural rearrangements of gRNA and interactions between RNA partners [8,10,14–16]. In addition, Gag participates in selective packaging of cellular tRNA into virions and facilitates its annealing with the complementary primer-binding site (PBS) in viral gRNA [17–19].
Much less is known about gRNA dimerization and packaging during propagation of endogenous retroelements. The Ty1 retrotransposon of S. cerevisiae is a well-studied member of the widely disseminated Ty1/Copia family of the Pseudoviridae [20]. Ty1 shares many critical features with endogenous and infectious LTR-retroelements [21]. For example, Ty1 contains partially overlapping GAG and POL genes flanked by LTRs at the 5′ and 3′ termini. GAG encodes the capsid protein of VLPs, and POL encodes enzymes required for replication (reverse transcriptase, protease, integrase). Analogous to other retroelements, Ty1 replicates via an RNA intermediate and full-length Ty1 gRNA plays dual role in replication [22]. It is translated into Ty1 proteins and utilized as a genome that is packaged into VLPs, and reverse transcribed to form a DNA copy that is integrated into the host genome [21,23,24]. Assembly of VLPs from Ty1 or the yeast Metavirus Ty3 occurs in specific cytoplasmic foci (termed retrosomes or T-bodies) where element-encoded proteins and gRNA colocalize during an early stage of retrotransposition [25–30]. Ty1 VLPs are mainly composed of Gag and contain Ty1-encoded enzymes and gRNA in a dimeric form [24]. During or soon after VLP assembly, the precursor Ty1 Gag polyprotein (Gag-p49) undergoes a single C-terminal cleavage that results in mature Gag-p45 (Fig. 1b) [31–34]. Despite the lack of sequence homology, Ty1 Gag is functionally related to retroviral Gag proteins as it comprises the capsid of VLPs and mediates critical RNA transactions during the retrotransposon replication cycle [21,28]. Direct interactions between Gag and gRNA are required for effective Ty1 retrotransposition and mutations in GAG can disrupt retrosome nucleation and VLP assembly [26,30,35–37]. Based on in vitro studies of truncated derivatives of Ty1 Gag, the RNA binding and nucleic acid chaperone region has been mapped to the C-terminus and contains three clusters of basic amino acid residues (Fig. 1b) [38,39].
Figure 1.
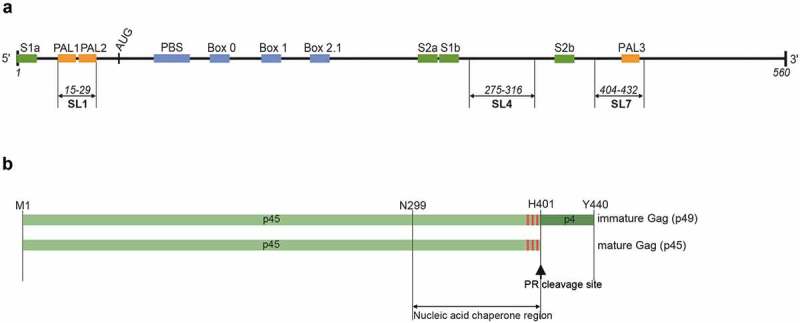
Organization of mini Ty1 RNA and Ty1 Gag. (a) Mini Ty1 RNA (+1–560) contains the primer binding site (PBS), three short boxes (Box 0, Box 1, Box 2.1; blue) that anneal with cellular tRNAiMet, and the palindromic sequences PAL1, PAL2, PAL3 (orange). The strands forming the stems of the pseudoknot (S1 and S2) are marked with green. (b). Schematic representation of Ty1 Gag: p49 (immature) and p45 (mature) with amino acid coordinates. The red boxes represent stretches of basic amino acids present in the nucleic acid chaperone region.
Mutational and functional analyses identified a ~ 380 nt region at the 5′-terminus of the gRNA that is required for Ty1 retrotransposition [40,41]. The 5′-terminus of Ty1 gRNA contains cis-acting sequences required for packaging, reverse transcription, translation initiation and genome cyclization [42–45]. In VLPs, this region forms a long-range pseudoknot that is needed for efficient retrotransposition [46,47]. However, the sequences essential for Ty1 gRNA dimerization and packaging have not been precisely defined. An early study, based on in vitro dimerization assays with a synthetic peptide from a predicted chaperone domain of Ty1 Gag, suggested a role for tRNAiMet in Ty1 RNA dimerization and proposed a model for dimer formation mediated by interaction of two tRNAiMet molecules bound to the PBS sequences from two gRNAs [38]. More recently, SHAPE (selective 2′-hydroxyl acylation analysed by primer extension) [48] analysis of Ty1 gRNA in vitro and in virio identified the changes in the flexibility of nucleotides contiguous to the three palindromic PAL sequences from the 5′ terminus [46]. By analogy to retroviruses and in agreement with SHAPE reactivity profiles, PAL1, PAL2 and PAL3 sequences were suggested as potential contact sites for dimer formation. Two palindromes (PAL1 and PAL2) are located in 5′UTR, while PAL3 is located in the Gag open reading frame (Fig. 1a). However, the role of PALs in Ty1 RNA dimerization was not analysed. Recent work also suggests that two SL1a – SL3a kissing-loop interactions (SL1 – SL4 according to our notation, Fig. 1a) participate in Ty1 RNA dimerization [49]. Mutations disturbing the complementarity between SL1 and SL4 decreased Ty1 RNA stability in vivo and significantly inhibited Ty1 mobility, but the direct contact between these hairpins in dimerized RNA was not shown. The SL1 – SL4 kissing-loop complex is mutually exclusive with the SHAPE-based model of dimeric Ty1 RNA, as SL1 is formed by an intramolecular PAL1 and PAL2 interaction [46].
Here, we have taken a multifaceted approach to characterize the nucleotide sequences involved in Ty1 RNA dimerization and define features of monomeric and dimeric states of Ty1 RNA. The importance of selected sequences was assessed by their impact on RNA dimerization and structure, and Ty1 mobility. Our results indicate that similar to retroviruses dimeric Ty1 gRNA is maintained by interactions of self-complementary palindromic sequences within the 5′UTR. We show that intermolecular PAL1 – PAL2 interactions are important for in vitro dimerization of Ty1 RNA and Ty1 mobility in vivo. However mutations that disrupt PAL1 – PAL2 complementarity partially inhibit Ty1 RNA functionality and Ty1 RNA apparently dimerizes via alternative contact points. Our data support the critical role of Ty1 Gag in RNA dimerization. Interestingly, dimerization induced by Gag does not require initiator tRNAiMet annealing. Although mature Ty1 Gag binds in the proximity of sequences involved in RNA dimerization and tRNAiMet annealing, the 5′ pseudoknot may serve as a major Gag-binding site. We propose that this interaction could be essential for Ty1 RNA stability during the process of retrotransposition.
Materials and methods
Genetic techniques, media, strains, and plasmids
Standard yeast genetic and microbiological procedures were used in this work [50]. Ty1 mobility was determined in the Ty1-less S. paradoxus strain DG3582 (MATα his3-Δ200hisG trp1 ura3) [39]. The helper-Ty1 and mini-Ty1his3-AI plasmids were kindly provided by M. Joan Curcio. The Ty1 helper plasmid (pEIB, 2μ TRP1) contains nucleotides 241–5561 of Ty1-H3 [42]. The mini-Ty1his3AI plasmid (pJC994, 2μ URA3) was constructed by deleting the HpaI-SnaBI fragment of pGTy1his3AI-[∆1] (818–5463 nt of Ty1-H3 DNA) [51]. Plasmids PAL1, PAL2, PAL3, and IL3 were derived from pJC994 by site-directed mutagenesis using the QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) following the manufacturer’s protocol (for specific oligonucleotide sequences see Supplementary Table S1). Plasmid DNA was purified using PureLink Quick Plasmid Miniprep Kit (Invitrogen) and mutations were verified by DNA sequencing. The multicopy (2μ) TRP1-based pEIB plasmid was introduced into DG3582 by transformation to generate DG4148. Multicopy (2μ) URA3-based plasmids pJC994, PAL1, PAL2, PAL3, and IL3 were transformed individually into DG4148 to generate strains DG4151-DG4155, respectively, containing the helper and mini-Ty1his3-AI plasmids. Both Ty1 helper and mini-Ty1his3-AI element transcription is under the control of the GAL1 promoter.
Ty1his3-AI mobility
Ty1 movement was monitored using the selectable indicator gene his3-AI [52]. A single colony of each strain was grown in SC-Ura-Trp 2% raffinose broth for 16 hrs at 30°C on a tube roller. Cultures diluted 25-fold into SC-Ura-Trp 2% galactose broth in quadruplicate were grown at 22°C for 2 days to induce Ty1 expression from the GAL1 promoter. Cells were centrifuged, suspended in water and dilutions plated on SC-Ura-Trp 2% glucose and SC-Ura-Trp-His 2% glucose plates. Following incubation for 5 days at 30°C, the frequency of Ty1his3-AI mobility was calculated by dividing the number of Ura+ Trp+ His+ colonies by the number of Ura+ Trp+ colonies. Mobility analysis for wild type and the mutants were performed together, repeated at least twice, and representative data are shown. Note that Ty1HIS3 insertions usually occur by retrotransposition following splicing of the artificial intron. Since His+ cells can also result from recombination of Ty1HIS3 cDNA with Ty1 elements or solo-LTRs [53,54], the term Ty1 ‘mobility’ is used to describe both types of insertion.
Cloning, expression and purification of Ty1 Gag-p45
Recombinant Gag-p45 was expressed from pGEX-p45 and purified as described previously [55]. Briefly, the Gag-p45-GST fusion protein was purified from Escherichia coli BL21(DE3)pLysS strain (Invitrogen) by affinity chromatography using Glutathione Sepharose (GE Healthcare). The GST tag was removed by thrombin cleavage and Gag-p45 was eluted using a high – salt wash buffer containing 1 M NaCl, concentrated by centrifugal filtration and stored at −80°C.
DNA and RNA substrates
The template for transcription of unmodified yeast tRNAiMet was generated by PCR, and RNA was synthesized using T7-MEGAshortscript (Invitrogen). RNA was purified by denaturing gel electrophoresis (8 M urea) in 1 x TBE buffer, eluted from the gel matrix and concentrated by ethanol precipitation. The template for in vitro transcription of mini Ty1 RNA (560 nt) was generated by PCR amplification of sequences corresponding to the RNA nt +1–560 from pBDG433 using a forward primer F-miniRNA containing an SP6 promoter sequence followed by 5′ Ty1 RNA sequence and a reverse primer R-miniRNA (Supplementary Table S2). DNA templates for PAL1, PAL2 and ∆S1a mini Ty1 RNA mutants were obtained using forward primers introducing desired mutations and primer R-mini RNA. Templates for the PAL3, SL4 and IL3 mutants were generated by PCR-driven overlap extension. All transcripts were synthesized using SP6-MEGAscript (Invitrogen) and purified using Direct-zol RNA MiniPrep Kit (Zymo Research). The quality of transcripts was monitored by high-resolution agarose-gel electrophoresis in the presence of formaldehyde. Mini Ty1RNA and Ty1 RNA mutants were 3′-end labeled with [α-32P] pCp using T4 RNA ligase (Fermentas) and purified on NucAway Spin Columns (Invitrogen Ambion). Purified RNAs were stored at −20°C.
Ty1 RNA dimerization
32P-labeled Ty1 RNA (1 pmol) was refolded in buffer containing 40 mM Tris–HCl pH 8.0, 130 mM KCl, 0.1 mM EDTA by heating at 95°C for 3 minutes, slowly cooled to 60°C, placed on ice for 1 minute, and then incubated at 37°C for 30 minutes following the addition of MgCl2 to 2 mM. For the dimerization assays with tRNAiMet, tRNAiMet was folded separately in equivalent conditions and Ty1 RNA was combined with unlabeled tRNAiMet at 1:2 molar ratio before addition of Ty1 Gag-p45. RNAs were incubated with increasing concentrations of protein at 37°C for 30 minutes. All reactions were quenched by incubation with 1% (w/v) SDS at room temperature for 5 minutes. The samples were phenol/chloroform extracted and 10 μl of aqueous phase was mixed with 2 μl of 50% glycerol. RNA was resolved on a 1% agarose gel in 0.5 x TB at room temperature or TBM (TB with 0.2 mM MgCl2) running buffer at 4°C. Gels were autoradiographed and quantified by phosphorimaging using FLA-5100 phosphorimager with MultiGaugeV 3.0 software (FujiFilm). The obtained data were analysed using Origin software (OriginLab). In all cases, at least three independent experiments were performed and the data presented are representative of the whole.
Ty1 RNA structure probing
RNAs (8 pmol) were refolded in 20 µl of renaturation buffer (10 mM Tris-HCl [pH 8.0], 100 mM KCl, and 0.1 mM EDTA) by heating for 3 minutes at 95°C, slow cooling to 4°C, then adding 100 µl of water and 30 μl of 5× folding buffer (final concentration: 40 mM Tris-HCl [pH 8.0], 130 mM KCl, 0.2 mM EDTA, and 4 mM MgCl2), followed by incubation for 25 minutes at 37°C. The RNA sample was divided into two tubes and treated with 8 µl of NMIA in DMSO ([+], final concentration 1 mM) or DMSO alone (−), and the modification reaction was carried for 50 minutes at 37°C. RNA was recovered by ethanol precipitation and resuspended in 10 μl of water.
Detecting Gag-induced changes in RNA structure
Ty1 RNA (8 pmol) and tRNAiMet (16 pmols) were folded as described for dimerization assays. Subsequently, 240 pmols of Gag-p45 (8 μl in the protein buffer containing 50 mM Tris–HCl, pH 8.0, 1 M NaCl, 10 mM β-mercaptoethanol, 2.5 mM DTT) or equal volume of protein buffer were added to 72 μl of RNA mixture. The reaction was incubated at 37°C for 30 minutes and quenched by incubation with 1% SDS at room temperature for 5 minutes followed by phenol/chloroform extraction. Each reaction was divided into two separate tubes and treated with 8 μl of NMIA in DMSO [(+), 1.5mM NMIA, final concentration] or DMSO alone (-). The modification reactions were carried out for 50 minutes at 24°C. RNA was recovered by ethanol precipitation and was resuspended in 10 μl of water. Detection of 2′-O-adducts and data processing were performed as described below. The contribution of Ty1 RNA dimer to the ensemble SHAPE reactivity profile(s) was calculated according to:
Where RE is the overall reactivity obtained experimentally, RM, RD are the reactivities for monomer and dimer and pM, pD, are the per cent contribution from monomer/dimer to total RNA. To reduce bias produced by extreme values, calculated reactivity values greater than 1 were set equal to 1 and negative values were set to 0 [56].
Hydroxyl radical footprinting
RNA samples (5 pmol) were refolded by heating at 95°C for 1 minute followed by incubation for 25 minutes at 37°C in folding buffer (40 mM Tris–HCl, pH 8.0, 130 mM KCl, 0.5 mM EDTA and 5 mM MgCl2). Folded mini Ty1 RNA samples were diluted 20-fold with 20 mM Tris–HCl, pH 8.0 prior to adding Ty1 Gag-p45. Subsequently, 50 or 100 pmol of Gag-p45 in a total volume of 6 μl was added to a 70 μl reaction. As a control for non-specific cleavage, buffer was added instead of protein. RNA/protein complexes were formed by incubating 20 minutes at 0°C. To initiate footprinting reactions, 1.5 μl of 2.5 mM (NH4)Fe(SO4)2, 50 mM sodium ascorbate, 1.5% H2O2 and 2.75 mM EDTA were applied on the wall of the tube followed by centrifugation [57]. Reactions were incubated for 15 seconds at 24°C and quenched by addition of thiourea and EDTA to final concentrations of 20 mM and 40 mM, respectively. RNA was purified using Direct-zol RNA MiniPrep Kit (Zymo Research).
Primer extension reactions and data processing
A total of 10 pmols of fluorescently labeled primer PR3 [5′- TCAGGTGATGGAGTGCTCAG-3′] was added to 4 pmols of RNA [Cy5 (+) and Cy5.5 (−)], and 12 μl of primer-template was incubated at 95°C for 3 minutes, 37°C for 10 minutes and 55°C for 2 minutes, then reverse transcribed as described previously [46]. Sequencing ladders were prepared using primers labeled with WellRed D2 (ddA) or LicorIRD-800 (ddT) and a Thermo Sequenase Cycle Sequencing kit (Affymetrix) according to the manufacturer’s protocol. Samples and sequencing ladders were purified using ZR DNA Sequencing Clean-up Kit (Zymo Research) and analysed on a GenomeLab GeXP Analysis System (Beckman-Coulter). Electropherograms were processed using SHAPEfinder software [58], normalized as described previously [18] and converted into nucleotide reactivity tables [59]. All reactivity data used in analysis were averaged from at least three independent experiments. Tables with HR footprinting and SHAPE data are provided in Supplementary Data set 2.
RNA structure prediction
Secondary structure models were generated using RNAstructure software with default parameters [60]. RNAComposer was used for prediction of RNA 3D structural models (http://rnacomposer.cs.put.poznan.pl/). This is a fully automated RNA structure modeling server that predicts RNA 3D structures based on sequence and secondary structure [61–63].
Results
Requirements for in vitro dimerization of mini Ty1 RNA
Similar to retroviral RNAs, full-length Ty1 gRNA (5900 nt) dimerizes inefficiently in vitro, even in the presence of recombinant Gag-p45 (Fig. S1). To analyse Ty1 RNA dimerization and further define nucleotide sequences and structural motifs required for this process, we used a mini Ty1 RNA that encompasses +1–560 nucleotides of Ty1 gRNA and contains sequences suggested previously as candidates for mediating Ty1 gRNA dimerization, packaging into VLPs, and undergoing reverse transcription and integration (Fig. 1a) [22,38,41,46,49]. To determine if mature Ty1 Gag-p45 displays properties similar to retroviral Gag polyproteins, we purified recombinant Gag-p45 from E. coli and examined whether Gag-p45 enhanced mini Ty1 RNA dimerization in vitro. 32P-labeled mini Ty1 RNA was incubated with the increasing amounts of the Gag-p45 and dimeric RNA formation was assayed by electrophoresis under TB and TBM (includes MgCl2) running conditions after protein removal. This experimental approach allows one to distinguish ‘loose dimers’, which are mediated by kissing loops from ‘tight dimers’, which may be held together by extended intermolecular duplexes [64–67]. Kissing loop complexes require Mg2+ for stabilization and are only detectable by electrophoresis using running buffers containing Mg2+ that prevent electrophoresis-dependent dissociation. In contrast, tight dimers are stable under mild denaturing conditions (TB or TBE running buffers) in the absence of Mg2+.
Mini Ty1 RNA formed dimers in the presence of Gag-p45 (Fig. 2), whereas the RNA migrated as a single band corresponding to the monomeric form under TBM and TB electrophoresis conditions in the absence of Gag-p45. Incubation of mini Ty1 RNA with increasing amounts of Gag-p45 resulted in significant conversion toward the dimeric state. At a 3 µM concentration of Gag-p45 (1:23 protein to nt molar ratio), ~45% of mini Ty1 RNA was present in a dimeric state. Dimerization assays were performed in the presence (Fig. 2a) or absence of tRNAiMet (Fig. 2b) to directly assess whether tRNAiMet – tRNAiMet interactions act as a ‘bridge’ in Ty1 RNA dimerization, as suggested by earlier work [38]. The inclusion of tRNAiMet did not alter the monomer – dimer ratio, indicating that similar to retroviruses, dimeric Ty1 RNA is stabilized by direct base pairing of complementary cis-acting sequences. A similar efficiency of dimerization was obtained under TB or TBM running conditions, showing that mature Gag promotes formation of tight mini Ty1 RNA dimers. However, two discrete bands migrating slower than the major dimer were visible in the presence of Mg2+ (Fig. 2, bottom panel TBM gels). These bands may contain the higher-order multimers of mini Ty1 RNA or transient forms of dimeric RNA that are unstable during electrophoresis when the running buffer lacks Mg2+.
Figure 2.
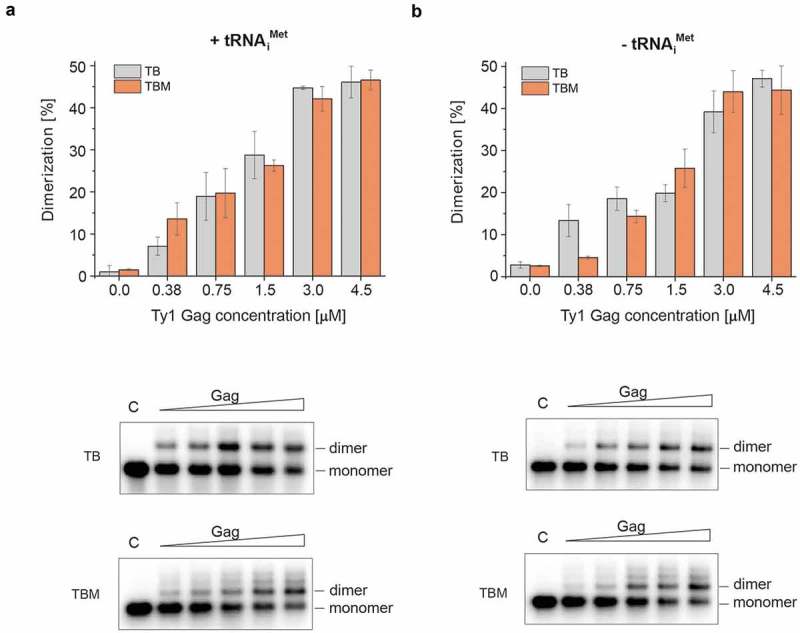
Gag-mediated dimerization of mini Ty1 RNA in the presence (a) and absence (b) of tRNAiMet analysed in the presence or absence of Mg2+. The graphs represent dimerized RNA (%) with increasing concentrations of Gag-p45 (0; 0.375; 0.75; 1.5; 3; 4.5 μM). Samples were analysed by agarose gel electrophoresis in TBM (2 mM MgCl2, orange) and TB (no MgCl2, grey). Representative analyses of mini Ty1 RNA dimerization are presented at the bottom. Lanes denoted C represent control samples that lack protein.
Gag binding sites on mini Ty1 RNA
Hydroxyl radical (HR) footprinting was used to map Gag-p45 binding sites within mini Ty1 RNA. This method offers an advantage over SHAPE analysis for detecting sequences protected by protein binding because HR cleavage of the RNA backbone occurs independently of RNA secondary structure [68]. Ribonucleoprotein complexes were formed on ice to minimize Gag-induced structural changes of mini Ty1 RNA and maintain the monomeric state of the RNA (Fig. S2). Overall reactivity profiles for mini Ty1 RNA in the presence or absence of recombinant Gag-p45 were compared to reveal protected nucleotide sequences (Fig. 3a). HR footprinting data were correlated with the SHAPE-based secondary structure model of the mini Ty1 RNA monomer (Figs. 3b & S3). The strongest and statistically significant decrease in HR cleavage was observed in sequences forming the Ty1 pseudoknot, raising the possibility that this is a preferred region for Gag-p45 binding. We also observed smaller but statistically significant protection in three regions located downstream of PBS and in the apical part of the SL1 hairpin that is formed by intramolecular base pairing between PAL1 and PAL2 palindromes (Figs. 3b & S3). Analysis of the composition of protected sites revealed that the content of C, A and U residues in the protected stretches of nucleotides were similar, while the number of G residues was significantly lower (Fig. 3c). These results suggest a lack of Ty1 Gag preference for particular nucleotide base(s), whereas retroviral Gag binding sites contain a high purine content (A and/or G) [18,69].
Figure 3.
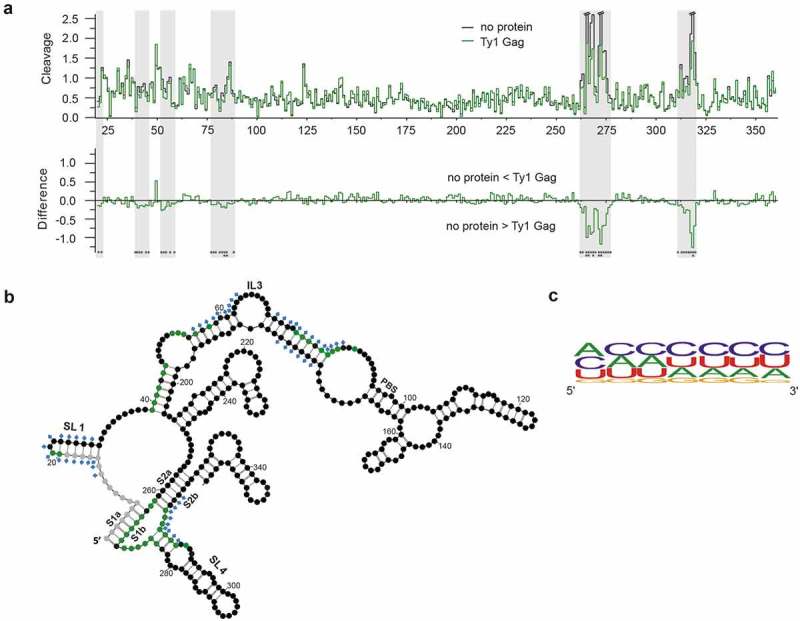
Gag-p45 binding sites in mini Ty1 RNA. (A) Hydroxyl radical (HR) cleavage and difference plot analyses of protein free mini Ty1 RNA in comparison with RNA probed in the presence of Gag-p45. Regions showing statistically significant decrease HR reactivity over several nucleotides are indicated by gray stripes (absolute cleavage decrease of 0.1 or greater and a p-value <0.05). Sites of decreased HR cleavage upon Gag-p45 binding are indicated by negative peaks. Asterisks below the plot correspond to statistical significance (Student’s t-test). (B) 2D structure model of +1–358 region of mini Ty1 RNA with the positions protected from HR cleavage in the presence of Gag-p45 coloured with green (absolute cleavage decrease of 0.1 or greater and a p-value <0.05). Not analysed nucleotides are marked with gray. Nucleotide positions which most likely correspond to protein binding sites within the Ty1 genomic RNA in VLPs are marked with blue diamonds [46]. Tables with HR footprinting data are provided in Supplementary Data set 2. (C) Frequency of nucleotide occurrence within Gag-p45 binding sites and their vicinities represented as a logo (https://weblogo.berkeley.edu/).
SHAPE analysis of wild type mini Ty1 RNA in monomeric and dimeric states
To further explore Ty1 RNA dimerization, we analysed the structure of the Gag-induced mini Ty1 RNA dimers by SHAPE [48,70]. In addition to mini Ty1 RNA, tRNAiMet was also present in the reaction mixtures as dimerization and primer binding are two critical RNA-RNA hybridization processes occurring during Ty1 VLPs assembly. When Gag-p45 was not added to the reaction, dimers were not formed and tRNAiMet was not annealed. Therefore, this reaction was used to monitor the monomeric state prior tRNAiMet annealing (thereafter referred to as monomer). Addition of Gag-p45 induced both dimerization of mini Ty1 RNA and tRNAiMet annealing (thereafter referred to as dimer). Prior to SHAPE analysis, protein was gently removed by phenol-chloroform extraction. Since the maximal efficiency of dimerization was ~ 50%, the mini Ty1 RNA population was a mixture of monomers and dimers after incubation with Gag-p45. To address this problem, we applied a mathematical method that allows one to determine the secondary structures of individual RNAs in a mixture of the conformers. This method was used successfully to analyse RNA folding intermediates of the HIV-2 Rev-responsive element [56]. Accordingly, the reactivity value of each nucleotide obtained for the mixture of two RNA states is a sum of reactivity values of both states, weighted according to the fractional contribution of each to the total RNA population. Structural differences were detected both by comparing monomer and dimer SHAPE reactivity profiles and by subtracting individual nucleotide reactivities for the monomer from values of dimer to yield a difference plot (Fig. 4a).
Figure 4.
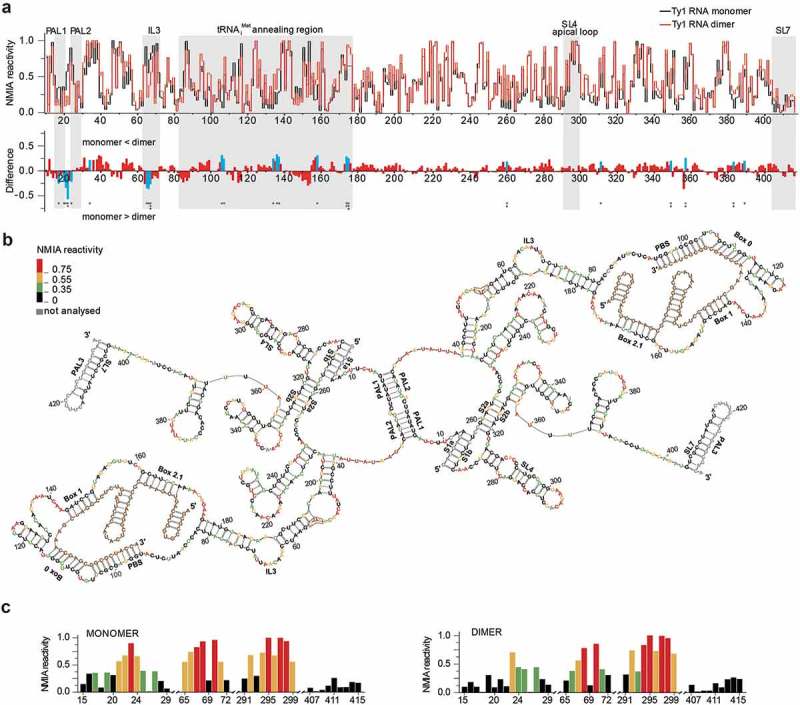
SHAPE analysis of mini Ty1 RNA. (a) Comparison of SHAPE reactivities for mini Ty1 RNA in monomeric and dimeric states. At the top is the step plot of NMIA reactivity for monomeric (black) and dimeric RNA (red). At the bottom is the difference plot calculated by subtracting the monomer intensities from those of the dimer. Negative values indicate nucleotides that are less flexible in the dimer. Regions of the interest are indicated by gray stripes. Columns corresponding to nucleotides that exhibit statistically significant differential reactivity (10% of highest SHAPE reactivity differences and a p-value <0.05 using the Student’s t-test) are coloured in blue. Asterisks below the plot correspond to statistical significance. (b) +1–433 region of mini Ty1 RNA in the dimeric form. Nucleotide residue reactivity to NMIA are scaled by colour (see insert on the left). The tRNAiMet is shown in brown. (c) Colour-coded histograms of NMIA reactivity for mini Ty1 RNA in monomeric and dimeric states. Tables with SHAPE data are provided in Supplementary Data set 2.
The SHAPE profiles for mini Ty1 RNA monomer and dimer are similar, indicating that Gag-induced transition from the monomer to dimer state does not cause extensive refolding of the RNA (Fig. 4a). Despite this overall similarity, we observed statistically significant reactivity differences that reveal important local changes in RNA conformation. NMIA reactivity alterations in the region +82–176 can be attributed to the formation of a mini Ty1 RNA/tRNAiMet complex in the presence of Gag-p45. These changes correspond well with NMIA reactivity profiles characteristic for Ty1 RNA/tRNAiMet complex described earlier [46]. The tRNAiMet binding sequences constitute a specific border in SHAPE reactivity (Fig. 4a). In the downstream region, stretches of small positive peaks are revealed by difference plots, while upstream from the PBS, stretches of both positive and negative peaks are detected. Most of positive peaks correspond to increases in local nucleotide reactivity resulting from a structure destabilizing activity of Gag-p45 that is similar to the nucleic acid chaperone activity of retroviral Gag polyproteins [28,38,39]. The stretches of negative peaks are attributable to intermolecular interactions critical for dimer formation. The strongest and statistically significant reactivity decrease was observed in the 21UCU23 sequence linking PAL1 and PAL2. In the monomer, nucleotides 21UCU23 were highly reactive since they comprise apical loop of the SL1 hairpin formed by intramolecular base-pairing of PAL1 and PAL2 (Figs. 4c & S2). PAL1 and PAL2 palindromic sequences were unreactive in both monomer and dimer states, therefore, it is challenging to prove that they constitute contact sites in the dimer. Importantly, the observed reactivity decrease in 21UCU23 is independent from tRNAiMet annealing (Fig. S4) and supports a Gag-induced structural transition that may correspond to the SL1 hairpins forming an intermolecular extended duplex with a UCU/UCU distortion (Fig. 4b). Alternatively, reactivity changes in SL1 apical part may arise from intermolecular kissing loop interactions between SL1 and SL4 that were recently proposed to help initiate Ty1 RNA dimerization [49]. Since the SL4 apical loop was highly reactive in both monomeric and dimeric states (Figs. 4c & S2), these kissing loop interactions are unlikely to be present in dimeric mini Ty1 RNA. Similar to PAL1 and PAL2, PAL3 was also unreactive in the monomeric and dimeric states. This palindrome is present in the stem of SL7 (Figs. 4c & S2) and its engagement in intermolecular base-pairing is anticipated to disturb the structure of SL7 and increase reactivity in the complementary strand [46]. However, we did not observe SHAPE reactivity changes in the SL7 stem (Fig. 4c) that would support PAL3-PAL3 interactions in the Ty1 RNA dimer.
In addition to the change of 21UCU23 reactivity in dimeric mini Ty1 RNA, a reproduceable statistically relevant decrease in reactivity was detected for nucleotides in the third internal loop – IL3 (nt 63–71) (Fig. 4a). This asymmetric internal loop contains a short 67AAUU70 palindromic sequence that could serve as an additional dimerization site (Fig. 4b). However, despite the reactivity decrease in IL3, nucleotides A67, A68 and U70 in the palindrome remained reactive to NMIA in the dimer (Figs. 4b, c), suggesting this interaction is less stable or transient in nature. In support of the validity of our approach, a similar change in the IL3 reactivity pattern was detected between full-length monomeric Ty1 RNA probed in vitro and in dimeric Ty1 RNA probed within VLPs [46].
3D models of PAL1-PAL2 interactions in monomer and dimer
Our data suggest the SHAPE reactivity changes in the 21UCU23 sequence linking PAL1 and PAL2 result from a Gag-induced monomer to dimer transition. To extend and validate these findings, we generated 3D structure models of SL1 and the PAL1-PAL2 extended duplex (Fig. 5). Models were generated using RNAComposer, which is an advanced tool for modeling atomic-resolution 3D models of RNAs based on their sequence and secondary structures [61,62]. Predicted structural models correspond well with the NMIA reactivity changes observed in the linking sequence. In monomeric Ty1 RNA, nucleotides 21UCU23 constitute the apical loop of SL1 and were highly reactive to NMIA. The 3D structure model of PAL1-PAL2 extended duplex suggests formation of non-canonical C-U intermolecular base-pairs involving U21 and C22 residues. Non-canonical C-U base pairs were identified in several biologically important RNAs and RNA duplexes containing the C-U base pairs can be considered similar to typical A-RNA helices [71,72]. The preservation of continuous stacking contributes significantly to the stability of RNA folds and minimizes the exposure of hydrophobic base surfaces to polar solvent. This can explain the lack of U21 and C22 NMIA modification in dimeric mini Ty1 RNA. Moreover, U23 residues were predicted to be extruded from the duplex with the base pointing towards the solvent. Such looped out nucleotides would allow the overall duplex geometry to remain close to the regular A-form. In agreement with the 3D structure model, U23 residues were readily modified by NMIA in dimeric mini Ty1 RNA.
Figure 5.
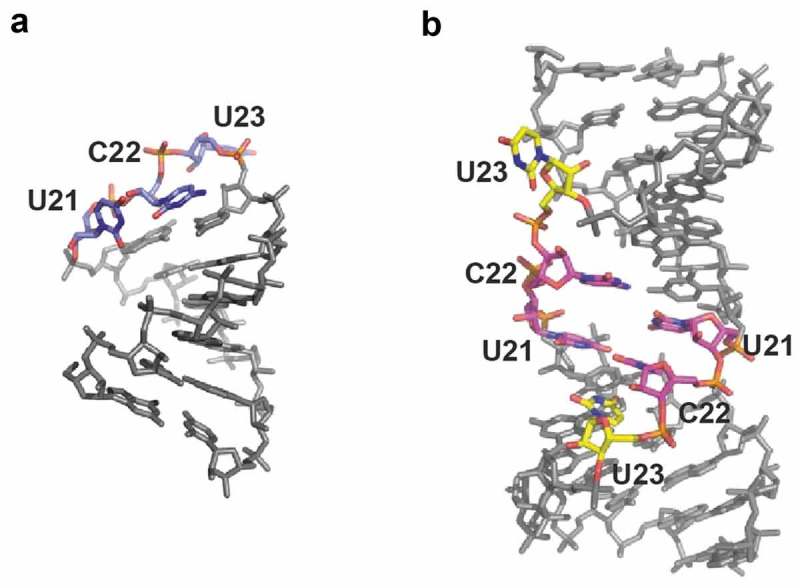
3D structure models of SL1 (a) and the PAL1-PAL2 intermolecular duplex (b), characteristic for monomeric and dimeric RNA, respectively. In SL1, nucleotides 21UCU23 are marked with blue. In duplex U21 and C22 are marked with magenta, U23 are marked with yellow.
Mutational analysis of cis-acting sequences involved in mini Ty1 RNA dimerization in vitro
To further define the role of candidate sequences in Ty1 RNA dimerization, we analysed in vitro dimerization of mini Ty1 RNA mutants with nucleotide substitutions in the postulated contact sites (Table 1). Our data reveal that substitution of PAL1 or PAL2 by non-palindromic sequences (PAL1 mutant and PAL2 mutant, respectively) results in a two-fold decrease in dimerization when compared to wild type mini Ty1 RNA (Figs. 6a & S5). Investigation of a mini Ty1 RNA mutant with a single nucleotide substitution (U16C) in PAL1 additionally confirmed the role of this palindrome in Ty1 RNA dimerization (Fig. S6). The U16C RNA mutant dimerized less efficiently than wild type but as expected, the inhibition was weaker than observed for the PAL1 non-palindromic mutant. U16C disturbs the U16-A28 base pair and thereby might destabilize the PAL1-PAL2 intermolecular interaction. However, mutations in SL1 hairpin could also impact the potential SL1-SL4 interactions [49] and thus, the observed inhibitory effects might result from disturbance of an initial kissing loop complex. Despite the lack of support for this possibility from our SHAPE analysis, we also assayed dimerization of a mini Ty1 RNA mutant with substitutions in the SL4 apical loop (SL4 mutant) that disturb SL1-SL4 complementarity and impairs Ty1 retrotransposition [49]. We observed that mutations in SL4 altered monomer to dimer ratio only slightly, indicating that the SL1-SL4 kissing complex is not required for dimerization of mini Ty1 RNA in vitro (Figs. 6b & S5).
Table 1.
Ty1 RNA mutants used in this study.
| Mutant | Description of introduced mutations | Sequence of wild type mini Ty1 RNA and mutants | |
|---|---|---|---|
| U16C mutant | Single nucleotide substitution in PAL1 sequence | 15GUAUAU20 | → GCAUAU |
| PAL1 mutant | Substitution of PAL1 sequence with non palindromic sequence | 13UAGUAUA19 | → CGCCCGC |
| PAL2 mutant | Substitution of PAL2 sequence with non palindromic sequence | 24GUAUAC29 | → CGCCCG |
| PAL3 mutant | Substitution of PAL3 sequence with non palindromic sequence | 423CCUGGG428 | → AAGAGG |
| IL3 mutant | Substitution of internal loop 3 sequence | 65ACAAUUAU72 | → CCAACCAU |
| SL4 mutant | Substitution of SL4 apical loop sequence | 291CCACAGAAU299 | → CCUCUCUAA |
| ∆S1a mutant | Deletion of 5 nt within S1a pseudoknot stem | 1GAGGA5 | → – – – – – |
Figure 6.
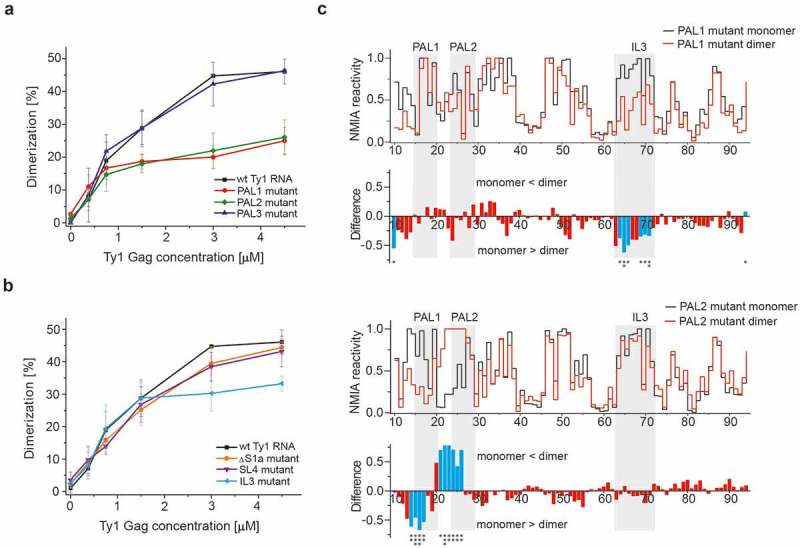
Identification of cis-acting sequences important for mini Ty1 RNA dimerization. (a) Dimerization assays for wild type mini Ty1 RNA and PAL1, PAL2, PAL3 mutants. (b) Dimerization assays for wild type mini Ty1 RNA and ∆S1a, SL4 and IL3 mutants. The graphs represent averaged measurements from three independent experiments. (c) SHAPE analysis of PAL1 and PAL2 RNA mutants in monomeric and dimeric states. For clarity, the plots show the +10–90 region of mini Ty1 RNA mutants. At the top is the step plot of NMIA reactivity for monomeric (black) and dimeric (red) RNA. At the bottom is the difference plot calculated by subtracting the monomer intensities from those of the dimer. Negative values indicate nucleotides that are less flexible in the dimer. Columns corresponding to nucleotides that exhibit statistically significant differential reactivity (10% of highest SHAPE reactivity differences and a p-value <0.05 using the Student’s t-test) are coloured in blue. Asterisks below the plot correspond to statistical significance. Tables with SHAPE data are provided in Supplementary Data set 2.
Taken together, our results suggest that PAL1-PAL2 intermolecular interactions play an important role in Ty1 RNA dimerization. However, dimerization of RNA containing PAL1 or PAL2 mutations was reduced but not abolished completely, suggesting another sequence could facilitate dimerization in addition to PAL1 and PAL2. Although SHAPE analysis of Ty1 RNA in VLPs raises the possibility that a PAL3–PAL3 interaction influences dimer formation [46], a non-palindromic PAL3 mutant displayed a dimerization efficiency comparable to that of wild type mini Ty1 RNA (Fig. 6a), suggesting that PAL3 is dispensable for dimerization in vitro. We also analysed dimerization of mini Ty1 RNA with substitutions in IL3 (IL3 mutant) because NMIA reactivity of dimeric RNA decreased in the IL3 internal loop. The IL3 mutant dimerized similar to wild type mini Ty1 RNA at lower Gag-p45 concentration, but less efficiently at higher Gag-p45 concentration (Figs. 6b & S5). However, the decrease in IL3 RNA mutant dimerization was weaker then that observed for the PAL1 or PAL2 non-palindromic RNA mutants.
SHAPE analysis of monomeric and dimeric mini Ty1 RNA containing non-palindromic mutations
To exclude mutation-induced RNA misfolding that could indirectly affect dimerization, we performed SHAPE analysis of non-palindromic RNA mutants in monomer form. The obtained reactivity profiles of PAL1, PAL2 and PAL3 non-palindromic RNA mutants resembled closely that of monomeric wild type mini Ty1 RNA and the majority of statistically significant reactivity changes were found only for the mutated sequences and adjacent nucleotides (Fig. 7a–c). These results confirm that the observed perturbations in dimerization are not caused by extensive alterations in the secondary structure of the mutated RNAs extending beyond the altered sequence. Interestingly, the IL3 RNA mutant displayed decreased NMIA reactivity in residues of the SL1 apical loop (Fig. 7d). Therefore, the IL3 mutation may influence the architecture of the 5′ terminus of Ty1 RNA and indirect effect on mini Ty1 RNA dimerization in vitro is also possible.
Figure 7.
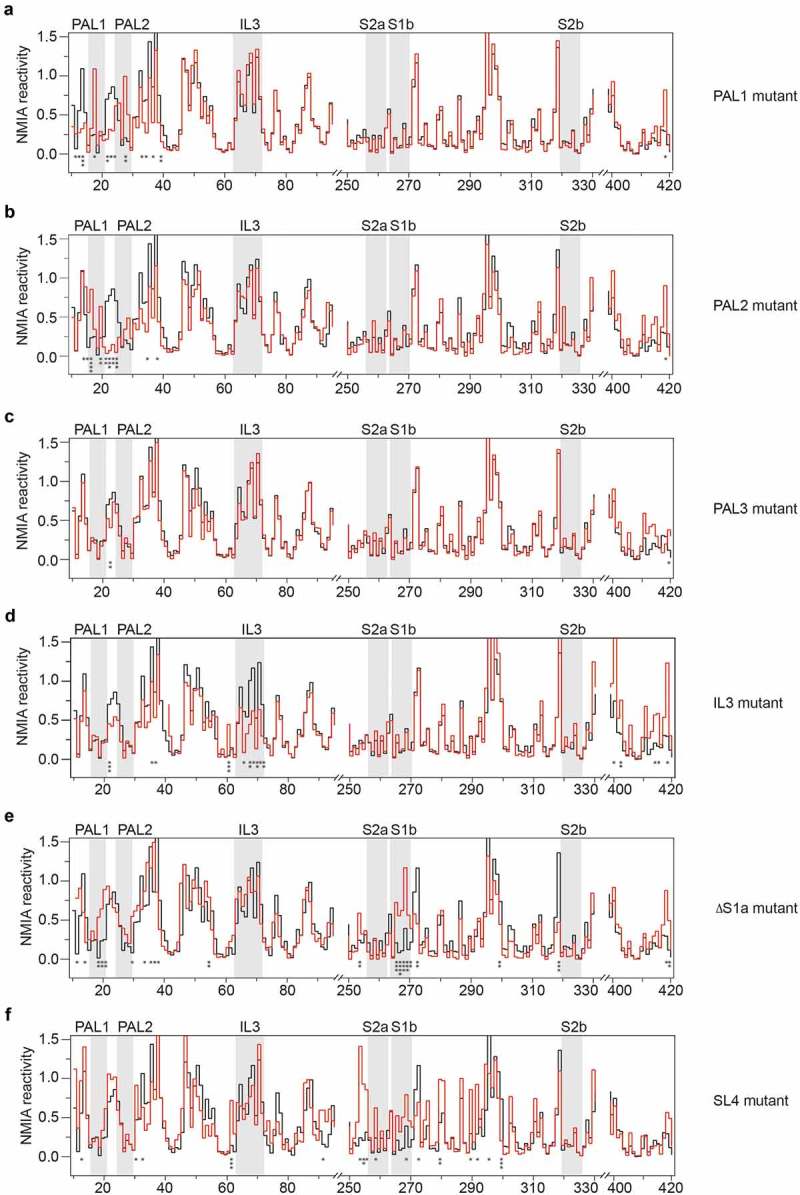
SHAPE analysis of the secondary structure of mini Ty1 RNA mutants in monomeric form. Step plots of NMIA reactivity of PAL1 (a), PAL2 (b), PAL3 (c), IL3 (d), ∆S1a (e) and SL4 (f) mutants. For clarity, the plots show the selected regions of mini Ty1 RNA mutants. Each mutant (red line) is plotted against the wild type mini Ty1 RNA (black line) for comparison. Nucleotides within the PAL1, PAL2, IL3 and stems (S1b, S2a, S2b) of the pseudoknot are indicated by gray stripes. Nucleotides that exhibit statistically significant differential reactivity (10% of highest SHAPE reactivity differences and a p-value <0.05 using the Student’s t-test) are marked with asterisks below the plot. Tables with SHAPE data are provided in Supplementary Data set 2.
We also characterized PAL1 and PAL2 non-palindromic RNA mutants in the dimeric form by SHAPE as described above (Fig. 6c). Similar to wild type mini Ty1 RNA, differences in SHAPE reactivity between monomeric and dimeric RNAs were limited to the regions (i) +82–176 (indicating tRNA annealing) and (ii) upstream from the PBS. For the PAL1 mutant, a stretch of statistically relevant high negative peaks was observed in the region corresponding to IL3. This loss of reactivity in the dimeric state suggests that the PAL1 mutant dimerizes via an interaction between palindromic sequences located in the IL3. The decrease in SHAPE reactivity in this region was also observed for wild type mini Ty1 RNA dimer but was less evident when PAL1-PAL2 intermolecular interactions were present (Fig. 4a). This effect was not detected for the PAL2 mutant, where a surprisingly strong and statistically significant reactivity decrease was identified in PAL1 sequence, suggesting intermolecular PAL1-PAL1 interaction in dimeric PAL2 RNA mutant. Taken together, our observations suggest that alternate dimerization contacts are utilized when the wild-type dimerization sites are mutated.
Role of the 5′-pseudoknot in Ty1 RNA dimerization
The 5′-terminus of Ty1 RNA in VLPs is stabilized by a long-range pseudoknot structure with two helical regions: S1 and S2 [46]. This tertiary motif provides an important biological function as mutations disrupting its conformation interfere with Ty1 retrotransposition [41,47,49]. This pseudoknot is not present in the full-length Ty1 RNA in vitro, but is stable in the mini Ty1 RNA monomer [47]. Our structural analysis reveals that the dimerization does not change the reactivity pattern in the sequence forming the S1 and S2 stems of the pseudoknot (Fig. 4b), and this critical motif is also present in dimeric mini Ty1 RNA. To explore the relationship between pseudoknot formation and dimerization, we analysed the structure and dimerization of a mutant with a defect in S1 stem of the pseudoknot (ΔS1a mutant) (Figs. 6b & 7e). Deletion of 5 nts from the S1a strand caused a statistically relevant increase in NMIA reactivity in the S1b strand, but not in the S2 stem, confirming that two helical regions of pseudoknot fold independently (Fig. 7e) [47]. Interestingly, statistically significant increased NMIA reactivity was also detected for residues A19, U20 and U21, suggesting destabilization of the upper part of SL1. This SL1 destabilization did not greatly influence dimerization as the ΔS1a mutant when compared with wild type mini Ty1 RNA (Fig. 6b).
The pseudoknot stems are linked by the sequence constituting SL4 hairpin. Although our findings do not support a role for SL4 in dimerization, a strong retrotransposition defect was previously reported for a mini Ty1 RNA mutant with identical substitutions in SL4 apical loop [49]. To further explain the role of SL4 in Ty1 retrotransposition, we performed SHAPE analysis of the SL4 RNA mutant in the monomer state (Fig. 7f). It revealed that substitutions in the SL4 apical loop disrupt the pseudoknot as increased NMIA reactivity was detected for residues in S1b and in direct proximity of S2a. Similar to the ΔS1a mutant, dimerization of the SL4 mutant was moderately weaker than that of wild type mini Ty1 RNA (Fig. 6b), additionally confirming a lack of direct correlation between the disruption of pseudoknot and Ty1 RNA dimerization. However, our data show the importance of SL4 for pseudoknot architecture, which might explain the retrotransposition defect induced by mutations in SL4 apical loop [49].
Involvement of dimerization sequences in Ty1 retrotransposition
To determine if sequences involved in dimerization of Ty1 RNA affect retrotransposition, we utilized a mini-Ty1his3-AI element containing the same sequences as those in our analyses in vitro, and that can be mobilized by Ty1 proteins supplied by a helper element (Fig. 8a) [41,49,54]. The helper Ty1 RNA expressed from pEIB lacks the PBS and 3′ U3 and R sequences, which should block its involvement in reverse transcription. Unlike previous studies, Ty1 mobility assays were performed in a yeast strain that lacks Ty1 elements [73,74] (C. Bergman and D. J. Garfinkel, personal communication), thereby minimizing the effects of Ty1 products such as the p22 restriction factor that may complicate interpretation of the results [75]. Here, the PAL1 and PAL2 mutations decreased Ty1 mobility about 2-fold whereas the PAL3 and IL3 mutations did not affect mobility (Fig. 8b). The Ty1 mobility assays correspond well with results obtained in vitro and support our proposition of Ty1 RNA dimerization via alternative RNA-RNA contact points when PAL1-PAL2 complementarity is disrupted. Unchanged mobility of the IL3 mutant suggests that the IL3 palindrome is not required when PAL1-PAL2 can interact. Like retroviruses [76,77], alternative dimerization sites appear to be biologically active. Therefore, comparing in vivo replication and in vitro RNA dimerization confirms that in vitro assays reflect the biological relevance of dimerization sequences.
Figure 8.
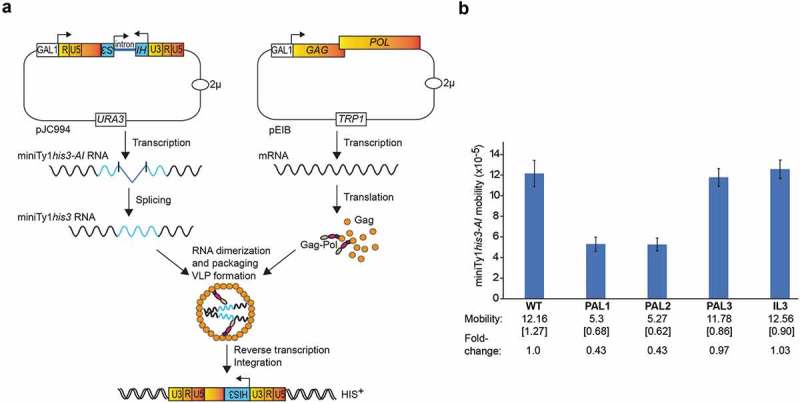
Involvement of dimerization sequences in Ty1 mobility. (a) Mini Ty1his3-AI mobility assay using two-plasmid system. Expression of the mini Ty1his3-AI element (pJC994) and Ty1 helper (pEIB), is driven from the GAL1 promoter in a Ty1-less S. paradoxus strain. The Ty1 helper element contains GAG and POL ORFs. The absence of 3′ LTR and silent substitutions in PBS preclude this element from being used as a template for reverse transcription. The mini Ty1his3-AI element contains the first 575 nucleotides, as well as the last 357 nucleotides of Ty1 gRNA and the his3-AI retrotransposition indicator gene. After splicing, miniTy1his3 RNA is reverse transcribed and cDNA is integrated into the host genome. Cells containing spliced miniTy1HIS3 element give rise to His+ colonies. (b) Effect of PAL1, PAL2, PAL3 and IL3 mutations on mini Ty1his3-AI mobility. Cells expressing the Ty1 helper plasmid pEIB and mini Ty1his3-AI plasmids containing wild type sequences (WT) or PAL1, PAL2, PAL3 or IL3 mutations were analysed for mini Ty1his3-AI mobility as described in the Materials and Methods. The average frequency of mini Ty1his3-AI mobility is shown with standard deviations in brackets, and fold-change in mobility relative to WT.
Discussion
Understanding how RNA dimerization occurs and its critical role in propagation has been well-studied in retroviruses [8,10], but has not been extensively analysed for other retroelements. Here, we used a combination of mutational and structural analyses to develop a model for RNA dimerization of the yeast LTR-retrotransposon Ty1 that builds on earlier work where we characterized the structure of Ty1 RNA in three distinct biological states (in virio, ex virio and in vitro) using SHAPE [46]. This analysis suggested PAL1, PAL2, and PAL3 might be involved in forming dimeric RNA if Ty1 followed a retroviral paradigm that involves base-pairing between palindromic sequences from two RNAs [46]. Full-length Ty1 RNA exists as monomer in vitro while it forms a dimer in VLPs [24,46]. In monomeric Ty1 RNA PAL1 and PAL2 form the stem of SL1, while PAL3 is a part of SL7 stem [46]. Like other classical RNA probing methods, SHAPE cannot discriminate intra- from intermolecular base-pairing, therefore, PAL sequences were unreactive in all analysed biological states and PAL’s engagement in dimerization was based on changes in reactivity of nucleotides adjacent to PAL sequences. Our current model was refined by analysing the effects of mutations in PAL sequences and additional potential contact sites on Ty1 RNA structure, Gag-induced dimerization in vitro and Ty1 mobility. By demonstrating that mutations in PAL1 and PAL2, designed to impair dimerization, do not cause major RNA secondary structure refolding while diminishing the ability of RNA to form dimers (Figs. 6a & 7a, b), we provide evidence for PAL1 and PAL2 involvement in Ty1 RNA intermolecular interactions. Furthermore, comparative SHAPE analyses of wild type mini Ty1 RNA monomeric and dimeric states (Fig. 4) together with 3D models of PAL1 and PAL2 intra- and inter-molecular interactions (Fig. 5) have allowed us to assign reactivity alterations in the sequence linking PAL1 and PAL2 to dimer formation. These alterations are also observed in VLPs [46], which validates and extends our understanding of how Ty1 RNA dimers form. Also, the present work is the first to directly implicate PAL1 and PAL2 sequences in Ty1 RNA dimerization, and the proposed model clearly explains the inhibitory effect of mutations in PAL1 or PAL2 on Ty1 mobility in yeast (Fig. 8). The biological relevance of the proposed dimerization sequences is supported by analysis of conserved nucleotides within the Ty1 RNA 5′ terminus indicating high preservation of PAL1 and PAL2 palindromes among Saccharomyces Ty1 elements [49].
Interestingly, we do not detect reactivity changes in the SL7 hairpin harboring PAL3 that were proposed to result from PAL3–PAL3 intermolecular base pairing in VLPs [46]. Although our analyses support the idea that PAL3 is not required for Ty1 RNA dimerization, we cannot exclude the possibility that PAL3–PAL3 interactions occur under specific conditions in VLPs. However, even if PAL3-PAL3 interactions occur in VLPs, they are not critical for Ty1 replication as the PAL3 mutations do not affect mobility (Fig. 8). Our data initially suggested that the IL3 palindrome might be a contact site that stabilizes the dimer in addition to PAL1-PAL2 duplex formation. Additional dimerization sequences have been identified in retroviral RNAs, and they can support stability of the RNA dimer or serve as alternative contact sites when primary dimerization sites are altered [10]. For example, wild type MMTV RNA dimerizes via PAL II as a major point of contact, while deletion of PAL II induces dimerization via the palindromic sequence located within the PBS (PBS-PAL), which constitutes additional point of contact in wild type MMTV RNA dimer [78]. Repression of the HIV-2 RNA dimerization initiation site (DIS) by antisense oligonucleotides also promotes tight dimer formation by interaction between 10-nt long PAL sequence located upstream from DIS [64,79].Ty1 RNA with a mutated PAL1 dimerizes via IL3-IL3 interaction, and IL3 mutations negatively affect dimerization in vitro (Fig. 6). However, despite the reactivity decrease in IL3, the IL3 palindrome is still reactive toward NMIA in the wild type mini Ty1 RNA dimer (Fig. 4) and in full-length Ty1 RNA probed in virio [46]. Also, IL3 mutations do not affect Ty1 mobility (Fig. 8b). Taken together, our results indicate that IL3 serves as alternative contact point in Ty1 RNA dimer when PAL1-PAL2 intermolecular interaction is disrupted. Although, we cannot exclude that under some conditions IL3-IL3 interaction additionally stabilizes the dimer of wild type Ty1 RNA, this interaction is transient and not critical for Ty1 replication.
Ty1 RNA dimers resemble retroviral dimers in that they undergo stabilization during maturation of the VLPs, resulting from proteolytic processing of Gag-p49 to Gag-p45 as well as the Gag-Pol precursor [24]. A recent study proposed that initial Ty1 RNA dimers are maintained by two intermolecular kissing loop interactions between the SL1 and SL4 hairpins [49].These kissing loop interactions are not detected in full-length Ty1 RNA in VLPs [46] or in the mini Ty1 RNA dimer induced by Gag-p45 (Fig. 4). The RNA dimers in VLPs and induced in vitro by Gag-p45 represent the mature dimer stabilized by the intermolecular PAL1-PAL2 extended duplex that is mutually exclusive with the SL1-SL4 kissing loop. Retroviral transcripts can form loose dimers stabilized by kissing loop interactions even in the absence of proteins [64,66,80]. Therefore, spontaneously-formed kissing loop dimers of Ty1 RNA could be selected for packaging by the Gag-p49 precursor and then converted to mature dimer stabilized by PAL1-PAL2 interactions. However, Ty1 RNA dimers are not detected in the absence of protein (Fig. 2) and mutational disruption of the SL1–SL4 kissing loop interactions impacts mini Ty1 RNA dimerization in vitro only modestly, if any (Fig. 6b). Our results do not support SL1–SL4 intermolecular interactions, but they confirm the significance of SL4 in Ty1 retrotransposition. We propose that while SL4 is not involved in dimerization, it is critical for global architecture of Ty1 RNA including folding of the biologically important pseudoknot.
Our work re-examines an early study where it was proposed that unlike retroviruses, Ty1 dimeric RNA results from base paring between two tRNAiMet molecules bound to PBS sequences that initiate reverse transcription [38]. This model is based on dimerization assays with a 103-residue synthetic peptide from the C-terminal region of Ty1 Gag that possesses RNA chaperone activity (Fig. 1b). Here we show that Ty1 RNA dimerization induced by mature Gag-p45 does not require tRNAiMet sequences (Fig. 2), and that the retrotransposon RNA dimers resemble those of retroviruses in that they are maintained via cis-acting RNA sequences as described above (Fig. 4b). The lack of tRNA involvement in Ty RNA dimer formation is also supported by data showing that dimerization of Ty3 retrotransposon gRNA is not reduced in mutants that fail to package tRNAiMet, and deletion of the PBS does not alter gRNA packaging into Ty3 VLPs [81–83].
Assembly of Ty1 VLPs and effective retrotransposition requires direct interactions between Gag and gRNA [26,28,30], but specific nucleotide sequences recognized by Ty1 Gag have not been defined. Recently, we identified sites within mini Ty1 RNA that are recognized in vitro by a protein comprising the C-terminal half of Ty1 Gag called the CTR [39]. The N-terminal part of Gag does not bind RNA, but is known to play an important role in VLP structure as residues from this region face the outside of the particle [37]. Gag-p45 binding with Ty1 RNA is significantly less salt-dependent than that of CTR [55], raising the possibility that Gag-p45 has a higher non-electrostatic RNA-binding component than the CTR, and therefore, may be more specific. We used hydroxyl radical footprinting to show that Gag-p45 recognizes the same nucleotide sequences in Ty1 RNA that we previously identified as CTR binding sites [39]. This observation indicates that although the N-terminal part of Gag-p45 influences the salt-dependent protein binding to Ty1 RNA it does not change the selective recognition of nucleotide sequences in Ty1 RNA in vitro.
A majority of RNA binding sites for Gag-p45 identified in vitro may also bind Gag or other proteins in VLPs [46]. In support of this idea, Gag-p45 binds close to RNA sequences critical for Ty1 dimerization as well as to sequences involved in cellular tRNAiMet packaging and initiation of reverse transcription (Fig. 3). Interestingly, the most prominent Gag-p45 binding site in vitro contains sequences forming the pseudoknot. This motif is functionally important as mutations that destabilize the S1 stem inhibit reverse transcription [47], and mutations in S2 stem decrease RNA stability [49]. Surprisingly, the pseudoknot is not required for packaging since mutations in S1 or S2 do not influence the level of Ty1 RNA in VLPs [47]. Although it is not known whether LTR-retrotransposon gRNA dimerization is a precondition for packaging or occurs in VLPs, we show that pseudoknot disruption by deletions in the S1 stem or by substitutions in SL4 have only moderate effects on Ty1 RNA dimerization, suggesting that the link between the pseudoknot and dimerization may be indirect. Indeed, pseudoknot destabilization might impact the overall Ty1 RNA structure and interactions with Gag at other steps during retrotransposition. Checkley et al. showed that Gag is required for Ty1 RNA stability, efficient nuclear export and localization into cytoplasmic foci [30]. Although, it remains to be determined if Ty1 Gag recognizes pseudoknot architecture or is involved in its folding process in vivo, we propose that sequences involved in pseudoknot formation constitute a critical Gag binding site and these interactions are essential for Ty1 RNA stability and trafficking in the cell. Further work will be required to determine whether Ty1 RNA contains high affinity binding sites for Gag-p45 that are involved in selective packaging and support VLPs assembly, as has been demonstrated for several retroviral Gag proteins [8,16].
Funding Statement
This work was sponsored by National Science Centre Poland [2016/22/E/NZ3/00426 to K.P.W. and 2016/23/B/ST6/03931] and the National Institutes of Health [GM095622 and GM124216, to D.J.G]. This publication was also supported by the Polish Ministry of Science and Higher Education, under the KNOW programme, and the grants for young researches and Ph.D. students [to J.G.]. Funding for open access charge was provided by the National Science Centre Poland [2016/22/E/NZ3/00426 to K.P.W.].
Acknowledgments
We thank M. J. Curcio for plasmids pEIB and pJC994 and A. Kiliszek for help preparing Fig. 5. We also thank members of K.P.W. and D.J.G. laboratories for fruitful discussion and critical reading of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Huang CR, Burns KH, Boeke JD.. Active transposition in genomes. Annu Rev Genet. 2012;46:651–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Contreras-Galindo R, Kaplan MH, Dube D, et al. Human endogenous retrovirus type K (HERV-K) particles package and transmit HERV-K-related sequences. J Virol. 2015;89:7187–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mager DL, Stoye JP. Mammalian endogenous retroviruses. Microbiol Spectr. 2015;3:1–20 [DOI] [PubMed] [Google Scholar]
- [4].Ashley J, Cordy B, Lucia D, et al. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell. 2018;172:262–274 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pastuzyn ED, Day CE, Kearns RB, et al The neuronal gene arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell. 2018;172:275–288 e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Johnson SF, Telesnitsky A. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog. 2010;6:e1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kuzembayeva M, Dilley K, Sardo L, et al. Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology. 2014;454–455:362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Comas-Garcia M, Davis SR, Rein A. On the selective packaging of genomic RNA by HIV-1. Viruses-Basel. 2016;8:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].D’Souza V, Summers MF. How retroviruses select their genomes. Nature Rev Microbiol. 2005;3:643–655. [DOI] [PubMed] [Google Scholar]
- [10].Dubois N, Marquet R, Paillart JC, et al. Retroviral RNA dimerization: from structure to functions. Front Microbiol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jayaraman D, Kenyon JC. New windows into retroviral RNA structures. Retrovirology. 2018;15:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Keane SC, Summers MF. NMR studies of the structure and function of the HIV-1 5-leader. Viruses-Basel. 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011;410:609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Abd El-Wahab EW, Smyth RP, Mailler E, et al. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat Commun. 2014;5:4304. [DOI] [PubMed] [Google Scholar]
- [15].Mailler E, Bernacchi S, Marquet R, et al. The life-cycle of the HIV-1 Gag-RNA complex. Viruses-Basel. 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Olson ED, Musier-Forsyth K. Retroviral Gag protein-RNA interactions: implications for specific genomic RNA packaging and virion assembly. Semin Cell Dev Biol. 2018;8:129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guo F, Saadatmand J, Niu MJ, et al. Roles of Gag and NCp7 in facilitating tRNA(3)(Lys) annealing to viral RNA in human immunodeficiency virus type 1. J Virol. 2009;83:8099–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pachulska-Wieczorek K, Blaszczyk L, Biesiada M, et al. The matrix domain contributes to the nucleic acid chaperone activity of HIV-2 Gag. Retrovirology. 2016;13:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kleiman L, Jones CP, Musier-Forsyth K. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010;584:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Peterson-Burch BD, Voytas DF. Genes of the pseudoviridae (Ty1/copia retrotransposons). Mol Biol Evol. 2002;19:1832–1845. [DOI] [PubMed] [Google Scholar]
- [21].Curcio MJ, Lutz S, Lesage P. The Ty1 LTR-retrotransposon of budding yeast. Microbiol Spectr. 2015;3:1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Boeke JD, Garfinkel DJ, Styles CA, et al. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. [DOI] [PubMed] [Google Scholar]
- [23].Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. [DOI] [PubMed] [Google Scholar]
- [24].Feng YX, Moore SP, Garfinkel DJ, et al. The genomic RNA in Ty1 virus-like particles is dimeric. J Virol. 2000;74:10819–10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dutko JA, Kenny AE, Gamache ER, et al. 5ʹ to 3ʹ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J Virol. 2010;84:5052–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Malagon F, Jensen TH. T-body formation precedes virus-like particle maturation in S. cerevisiae. RNA Biol. 2011;8:184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Beliakova-Bethell N, Beckham C, Giddings TH Jr., et al. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pachulska-Wieczorek K, Le Grice SF, Purzycka KJ. Determinants of genomic RNA encapsidation in the Saccharomyces cerevisiae long terminal repeat retrotransposons Ty1 and Ty3. Viruses. 2016;8:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sandmeyer S, Patterson K, Bilanchone V. Ty3, a position-specific retrotransposon in budding yeast. Microbiol Spectr. 2015;3:1–29 [DOI] [PubMed] [Google Scholar]
- [30].Checkley MA, Mitchell JA, Eizenstat LD, et al. Ty1 gag enhances the stability and nuclear export of Ty1 mRNA. Traffic. 2013;14:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Garfinkel DJ, Hedge AM, Youngren SD, et al. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65:4573–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Garfinkel DJ, Mastrangelo MF, Sanders NJ, et al. Transposon tagging using Ty elements in yeast. Genetics. 1988;120:95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kingsman AJ, Mellor J, Adams S, et al. The genetic organization of the yeast Ty element. J Cell Sci Suppl. 1987;7:155–167. [DOI] [PubMed] [Google Scholar]
- [34].Muller F, Bruhl KH, Freidel K, et al. Processing of TY1 proteins and formation of Ty1 virus-like particles in Saccharomyces cerevisiae. Mol Gen Genet. 1987;207:421–429. [DOI] [PubMed] [Google Scholar]
- [35].Tucker JM, Larango ME, Wachsmuth LP, et al. The Ty1 retrotransposon restriction factor p22 targets Gag. PLoS Genet. 2015;11:e1005571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Martin-Rendon E, Hurd DW, Marfany G, et al. Identification of proteolytic cleavage sites within the gag-analogue protein of Ty1 virus-like particles. Mol Microbiol. 1996;22:1035–1043. [DOI] [PubMed] [Google Scholar]
- [37].Brookman JL, Stott AJ, Cheeseman PJ, et al. Analysis of Tya protein regions necessary for formation of the Ty1 virus-like particle structure. Virology. 1995;212:69–76. [DOI] [PubMed] [Google Scholar]
- [38].Cristofari G, Ficheux D, Darlix JL. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem. 2000;275:19210–19217. [DOI] [PubMed] [Google Scholar]
- [39].Nishida Y, Pachulska-Wieczorek K, Blaszczyk L, et al. Ty1 retrovirus-like element Gag contains overlapping restriction factor and nucleic acid chaperone functions. Nucleic Acids Res. 2015;43:7414–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xu H, Boeke JD. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol Cell Biol. 1990;10:2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bolton EC, Coombes C, Eby Y, et al. Identification and characterization of critical cis-acting sequences within the yeast Ty1 retrotransposon. RNA. 2005;11:308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Boeke JD, Eichinger D, Castrillon D, et al. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol Cell Biol. 1988;8:1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chapman KB, Bystrom AS, Boeke JD. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc Natl Acad Sci U S A. 1992;89:3236–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Friant S, Heyman T, Bystrom AS, et al. Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA(iMet) primer are required for initiation of reverse transcription in vivo. Mol Cell Biol. 1998;18:799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cristofari G, Bampi C, Wilhelm M, et al. A 5ʹ-3ʹ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. Embo J. 2002;21:4368–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Purzycka KJ, Legiewicz M, Matsuda E, et al. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res. 2013;41:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Huang Q, Purzycka KJ, Lusvarghi S, et al. Retrotransposon Ty1 RNA contains a 5ʹ-terminal long-range pseudoknot required for efficient reverse transcription. RNA. 2013;19:320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Merino EJ, Wilkinson KA, Coughlan JL, et al. RNA structure analysis at single nucleotide resolution by selective 2ʹ-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc. 2005;127:4223–4231. [DOI] [PubMed] [Google Scholar]
- [49].Gamache ER, Doh JH, Ritz J, et al. Structure-function model for kissing loop interactions that initiate dimerization of Ty1 RNA. Viruses. 2017;9:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Guthrie CF, Fink GR. Guide to yeast genetics and molecular biology. Meth Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- [51].Scholes DT, Banerjee M, Bowen B, et al. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Curcio MJ, Garfinkel DJ. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci U S A. 1991;88:936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Melamed C, Nevo Y, Kupiec M. Involvement of cDNA in homologous recombination between Ty elements in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sharon G, Burkett TJ, Garfinkel DJ. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Blaszczyk L, Biesiada M, Saha A, et al. Structure of Ty1 internally initiated RNA influences restriction factor expression. Viruses. 2017;9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lusvarghi S, Sztuba-Solinska J, Purzycka KJ, et al. The HIV-2 Rev-response element: determining secondary structure and defining folding intermediates. Nucleic Acids Res. 2013;41:6637–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Somarowthu S, Legiewicz M, Keating KS, et al. Visualizing the ai5gamma group IIB intron. Nucleic Acids Res. 2014;42:1947–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Vasa SM, Guex N, Wilkinson KA, et al. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. Rna. 2008;14:1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. Bmc Bioinformatics. 2010;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Antczak M, Popenda M, Zok T, et al. New functionality of RNAComposer: an application to shape the axis of miR160 precursor structure. Acta Biochim Pol. 2016;63:737–744. [DOI] [PubMed] [Google Scholar]
- [62].Popenda M, Szachniuk M, Antczak M, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Purzycka KJ, Popenda M, Szachniuk M, et al. Automated 3D RNA structure prediction using the RNAComposer method for riboswitches. Methods Enzymol. 2015;553:3–34. [DOI] [PubMed] [Google Scholar]
- [64].Purzycka KJ, Pachulska-Wieczorek K, Adamiak RW. The in vitro loose dimer structure and rearrangements of the HIV-2 leader RNA. Nucleic Acids Res. 2011;39:7234–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Berkhout B, Ooms M, Beerens N, et al. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J Biol Chem. 2002;277:19967–19975. [DOI] [PubMed] [Google Scholar]
- [66].Tran T, Liu Y, Marchant J, et al Conserved determinants of lentiviral genome dimerization. Retrovirology. 2015;12:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. [DOI] [PubMed] [Google Scholar]
- [68].Nilsen TW. Mapping RNA-protein interactions using hydroxyl-radical footprinting. Cold Spring Harb Protoc. 2014;2014:1333–1336. [DOI] [PubMed] [Google Scholar]
- [69].Kutluay SB, Zang T, Blanco-Melo D, et al. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159:1096–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wilkinson KA, Gorelick RJ, Vasa SM, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Rypniewski W, Banaszak K, Kulinski T, et al. Watson-Crick-like pairs in CCUG repeats: evidence for tautomeric shifts or protonation. RNA. 2016;22:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Blaszczyk L, Rypniewski W, Kiliszek A. Structures of RNA repeats associated with neurological diseases. Wiley Interdiscip Rev RNA. 2017;8:e1412. [DOI] [PubMed] [Google Scholar]
- [73].Moore SP, Liti G, Stefanisko KM, et al. Analysis of a Ty1-less variant of Saccharomyces paradoxus: the gain and loss of Ty1 elements. Yeast. 2004;21:649–660. [DOI] [PubMed] [Google Scholar]
- [74].Garfinkel DJ, Nyswaner K, Wang J, et al. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics. 2003;165:83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Saha A, Mitchell JA, Nishida Y, et al. A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J Virol. 2015;89:3922–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Laughrea M, Shen N, Jette L, et al. Variant effects of non-native kissing-loop hairpin palindromes on HIV replication and HIV RNA dimerization: role of stem-loop B in HIV replication and HIV RNA dimerization. Biochemistry. 1999;38:226–234. [DOI] [PubMed] [Google Scholar]
- [77].Paillart JC, Berthoux L, Ottmann M, et al. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Aktar SJ, Vivet-Boudou V, Ali LM, et al. Structural basis of genomic RNA (gRNA) dimerization and packaging determinants of mouse mammary tumor virus (MMTV). Retrovirology. 2014;11:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lanchy JM, Ivanovitch JD, Lodmell JS. A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA. RNA. 2003;9:1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Huthoff H, Berkhout B. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry. 2002;41:10439–10445. [DOI] [PubMed] [Google Scholar]
- [81].Clemens K, Bilanchone V, Beliakova-Bethell N, et al. Sequence requirements for localization and packaging of Ty3 retroelement RNA. Virus Res. 2013;171:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Nymark-McMahon MH, Beliakova-Bethell NS, Darlix JL, et al. Ty3 integrase is required for initiation of reverse transcription. J Virol. 2002;76:2804–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Larsen LS, Zhang M, Beliakova-Bethell N, et al Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J Virol. 2007;81:6957–6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


