Abstract
Background:
The enzyme beta-secretase 1 (BACE1) and its antisense transcript (BACE1-AS) have been implicated in the pathogenesis of Alzheimer's disease. Moreover, several lines of evidence point to their contribution in tumorigenesis.
Methods:
In the present study, we evaluated expression of BACE1 mRNA (BACE1) and BACE1-AS in 54 breast cancer tissues and 54 adjacent non-cancerous tissues (ANCTs) from the same patients using quantitative real-time PCR.
Results:
BACE1 was significantly down-regulated in tumoral tissues compared with ANCTs, while BACE1- AS expression was not significantly different between tumoral tissues and ANCTs. The Bayesian Multilevel model showed a significant difference in BACE1 expression between stage 1 and 2 cancers after age-effect adjustments. BACE1-AS expression was significantly greater in ER-positive than in ER-negative samples (P=0.01). BACE1 and BACE1-AS expression were not correlated with patient ages in any sample sets.
Conclusion:
Significant correlations were detected between expression of these genes in both tumoral tissues and ANCTs. The current study provides evidence for differential BACE1 expression in breast tissues and suggests further assessment of the role of BACE1 in the pathogenesis of cancer.
Key Words: BACE1, BACE1-AS, Breast Cancer, Lncrna
Introduction
The enzyme beta-secretase 1 (BACE1) is involved in the primary step of proteolytic cleavage of amyloid precursor protein (APP), which is completed by the subsequent function of gammasecretase. Accumulation of the resultant amyloid beta (Abeta) peptide in neuritic plaque of Alzheimer's brain has underscored the role of BACE1 in the pathogenesis of this disorder and has been proposed as a therapeutic target in Alzheimer's disease (AD) (1). A long non-coding RNA (lncRNA) transcribed from the BACE1 gene (BACE1) antisense (AS) strand regulates its expression via a feed-forward loop. The observed up-regulation of BACE1-AS in human AD brains compared with matched controls provided further evidence for the participation of BACE1 and BACE1-AS in the pathogenesis of AD (2). The significance of BACE1 in the pathogenesis of cancer has also been the focus of other researchers. The presence of APP in the endothelium during vessel synthesis implies its participation in angiogenesis. Moreover, BACE1 inhibitors have been shown to decrease endothelial cell proliferation and inhibit neoangiogenesis both in vitro and in vivo (3). Consistent with these studies, two independent studies have reported elevated APP levels in breast cancer cells (4, 5). Considering the regulatory role of BACE1-AS in BACE1 expression and subsequent APP production (2) on one hand, and the detected high APP abundance in breast cancer cells (4) on the other, we hypothesized that BACE1 and BACE1- AS expression in breast cancer tissues might differ from that in corresponding adjacent non-cancerous tissues (ANCTs). Further evidence for our hypothesis were provided by a growing number of studies reporting the role of lncRNAs in breast cancer (6-8) and the observed up-regulation of the lncRNA BACE1-AS by several cell stressors (9), which might be present in the cancer microenvironment as well. Consequently, we performed the present study to evaluate BACE1 and BACE1-AS expression in invasive ductal carcinoma samples and their paired ANCTs.
Materials and Methods
Patients
In the present study, we recruited 54 patients with invasive ductal carcinoma of breast who were hospitalized in Sina and Farmanieh hospitals (Tehran, Iran) for surgical removal of breast mass. The diagnosis of cancer was confirmed through histopathological examination. Hormone receptor status and other relevant information were extracted from medical records. All patients signed written informed-consent forms. The study protocol was approved by the local ethical committee of Shahid Beheshti University of Medical Sciences. Tumoral tissues and ANCTs were excised and transferred in liquid nitrogen to the Medical Genetics Laboratory for further assessments.
RNA extraction, cDNA synthesis, and expression analysis
RNA was extracted and cDNA synthesized using TRIzol™ Reagent (Invitrogen, Carlsbad, CA, USA) and a RevertAid First Strand cDNA Synthesis Kit (TaKaRa, Japan), respectively, according to company protocols. Relative BACE1 and BACE1-AS expression were compared between tumoral tissues and ANCTs using TaqMan Fast Universal PCR Master Mix (Applied Biosystems) in a Rotor Gene 6000 Corbett Real-Time PCR System. Expression was normalized to the hypoxanthine-guanine phosphoribosyl transferase gene (HPRT). The primer sequences are shown in Table 1.
Table 1.
Nucleotide sequences of primers and probes used in the study.
| Gene name | Primer and probe sequences | Product length |
|---|---|---|
| HPRT1 | F: AGCCTAAGATGAGAGTTC | 88 |
| R: CACAGAACTAGAACATTGATA | ||
| FAM -CATCTGGAGTCCTATTGACATCGC- TAMRA | ||
| BACE1 | F: CCAAGACGACTGTTACAA | 79 |
| R: GAAGCCCTCCATGATAAC | ||
| FAM-TTGCCATCTCACAGTCATCCAC-TAMRA | ||
| BACE1-AS | F: GACACTGTACCATCTCTTTTACCC | 113 |
| R: CACCACCAACCTTCGTTTGC | ||
| FAM - AGTCCACTCACGGAGGAGGTCGCC -TAMRA |
Statistical methods
Statistics were analyzed using R software version 3.0.5 and SPSS 18 (Chicago, IL, USA). The Spearman correlation and Bayesian Multilevel model were used. Kruschke's Bayesian estimation was used to assess the significance of mean expression differences between tumoral tissues and ANCTs. A student-t prior distribution was assumed for parameters with 200,000 iteration and 5000 burn-outs. The 95% highest density interval (HDI) values were calculated based on Bayesian approach. The difference between relative mean gene expression values in patient categories was evaluated using Tukey's honest significance test. CT values were corrected for the efficiencies of each primer set. The pairwise correlations between relative BACE1 and BACE1-AS transcripts levels in each set of samples were calculated using the regression model.
Results
Patient demographic and clinical features
Patient demographic and clinical features are shown in Table 2.
Table 2.
Patient demographic and clinical features.
| Variables | Values |
|---|---|
| Age (years) (mean± SD) | 51.79 ± 13.54 (29-81) |
| Menarche age (years) (mean± SD) | 13 ± 1.65 (10-18) |
| Menopause age (years) (mean± SD) | 44.91 ± 14.91 (38-60) |
| First pregnancy age (years) (mean± SD) | 18.04 ± 8.36 (14-32) |
| Breast feeding duration (months) (mean± SD) | 41.62 ± 34.1 (3-120) |
| Positive family history for other cancers (%) | 17% |
| Cancer stage (%) | |
| I | 30.8 |
| II | 28.8 |
| III | 30.8 |
| IV | 9.6 |
| Overall grade (%) | |
| I | 17 |
| II | 49 |
| III | 34 |
| Mitotic rate (%) | |
| I | 45.2 |
| II | 42.9 |
| III | 11.9 |
| Tumor size (%) | |
| <2 cm | 32 |
| ≥2 cm, <5 cm | 66 |
| ≥5 cm | 2 |
| Estrogen receptor (%) | |
| Positive | 87.8 |
| Negative | 12.2 |
| Progesterone receptor (%) | |
| Positive | 77.1 |
| Negative | 22.9 |
| Her2/neu expression (%) | |
| Positive | 25 |
| Negative | 75 |
| Ki67 expression (%) | |
| Positive | 100 |
| Negative | 0 |
BACE1 and BACE1-AS expression in tumoral tissues and ANCTs
BACE1 expression was significantly less in tumoral tissues than in ANCTs; however , BACE1-AS expression was not significantly different in the two tissue types (Fig. 1).
Fig. 1.
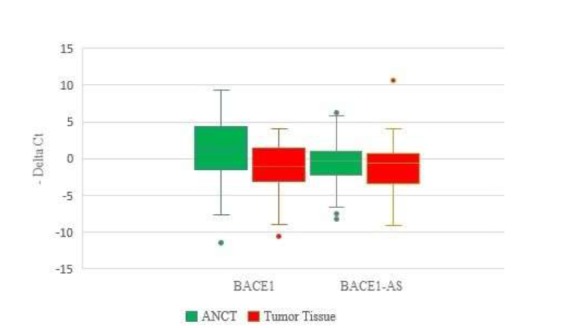
Relative BACE1 and BACE1-AS expression in tumoral tissues and ANCTs.
The Bayesian t test results comparing BACE1 and BACE1-AS relative expression in tumoral tissues and ANCTs are shown in Table 3.
Table 3.
Bayesian t test to compare relative gene expression between two paired groups (a: Tumor-ANCTs, b: computed from frequentist method, c: 95% highest density interval (HDI)
| Gene | Posterior mean | Relative Expression differencea | SD | Effect Size | P valueb | 95% HDIc | |
|---|---|---|---|---|---|---|---|
| Tumoral tissues | ANCTs | ||||||
| BACE1 | -0.44±0.68 | 1.02±0.67 | -1.637 | 0.48 | -0.487 | 0.001 | [-2.58, -0.71] |
| BACE1-AS | 0.42±0.43 | 0.63±0.5 | -0.3562 | 0.51 | 0.094 | 0.499 | [-1.37, 0.62] |
Association between expression levels and patients' clinical data
We assessed associations between expression levels and cancer stages using the Bayesian Multilevel model after age-effect adjustments and found higher BACE1 expression in stage 1 compared with stage 2 cancers in a way that expression in stage 2 cancers were 1.81 unit higher than stage 2 cancers (Table 4); however, BACE1-AS expression was not significantly different between the various stages (Table 5).
Table 4.
Bayesian Multilevel results of association between BACE1 expression and stage with age-effect adjustments (Stage 1= Reference group).
| BACE1 expression | Estimate | SE | P value | 95% Credible Interval |
|---|---|---|---|---|
| Stage2 | 1.81 | 1.25 | 0.035 | [0.65, 4.28] |
| Stage 3 | 0.38 | 1.28 | 0.141 | [-2.14, 2.96] |
| Stage 4 | -0.29 | 1.77 | 0.8 | [-3.68, 3.24] |
| Age | -0.03 | 0.04 | 0.272 | [-0.11, 0.04] |
Table 5.
Bayesian Multilevel results of association between BACE1-AS expression and stage with age-effect adjustments (Stage 1= Reference group).
| BACE1-AS expression | Estimate | SE | P value | 95% Credible Interval |
|---|---|---|---|---|
| Stage2 | 0.46 | 1.47 | 0.347 | [-2.43, 3.34] |
| Stage 3 | 0.33 | 1.42 | 0.543 | [-2.43, 3.17] |
| Stage 4 | -2.14 | 2.1 | 0.482 | [-6.28, 2.04] |
| Age | -0.06 | 0.05 | 0.248 | [-0.15, 0.03] |
We also assessed correlations between expression levels in each sample set and patients' ages. BACE1 and BACE1-AS expression were not correlated with patients' ages in any sample sets (Table 6).
Table 6.
Spearman correlation between gene expression in each sample set and patients' ages.
| BACE1 | BACE1-AS | ||
|---|---|---|---|
| Group | Tumoral tissues | -0.252 | 0.059 |
| ANCTs | -0.164 | 0.107 |
We also evaluated associations between relative gene expression and clinicopathological data (Table 7). BACE1-AS expression was significantly greater in estrogen receptor (ER)-positive than in ERnegative samples (P=0.01). No significant gene expression differences were found between patient subgroups.
Table 7.
Association between BACE1 and BACE1-AS transcript levels and tumor characteristics. Mean ± standard deviation values of Efficiency ^CT reference gene-Efficiency ^CT target gene are presented.
| BACE1 | P value | BACE1-AS | P value | |
|---|---|---|---|---|
| Age | ||||
| <55 years old vs. ≥55 years old | 3.06 (1.42) vs. 91.45 (288.38) | 0.35 | 327.39 (1.54) vs. 489.87 (2.15) | 0.78 |
| ER Status | ||||
| ER (+) vs. ER (-) | 2.39 (1.26) vs. 484.36 (1.17) | 0.71 | 55.67 (282.95) vs. 1.47 (3.61) | 0.01 |
| PR Status | ||||
| PR (+) vs. PR (-) | 571 (2.98) vs. 7.699 (2.45) | 0.08 | 63.68 (304.84) vs. 809.02 (2.66) | 0.09 |
| HER2 Status | ||||
| HER2 (+) vs. HER2 (-) | 6.81 (2.36) vs. 666.5 (3.04) | 0.12 | 1.73 (1.97) vs. 312.08 (1.49) | 0.47 |
| Tumor Grade | ||||
| Grade 1 vs. 2 | 336.89 (419.44) vs. 10.6 (37.95) | 0.9 | 56.93 (45.06) vs. 1.4 (1.84) | 0.95 |
| Grade 1 vs. 3 | 336.89 (419.44) vs. 6.42 (2.05) | 0.47 | 56.93 (45.06) vs. 1.27 (3.14) | 0.28 |
| Grade 2 vs. 3 | 10.6 (37.95) vs. 6.42 (2.05) | 0.24 | 1.4 (1.84) vs. 1.27 (3.14) | 0.09 |
Correlation between BACE1 and BACE1-AS expression in tumors and ANCTs
Significant correlation was found between RACE1 and BACE1-AS expression in both tumors and ANCTs (Fig. 2A and 2B).
Fig. 2.
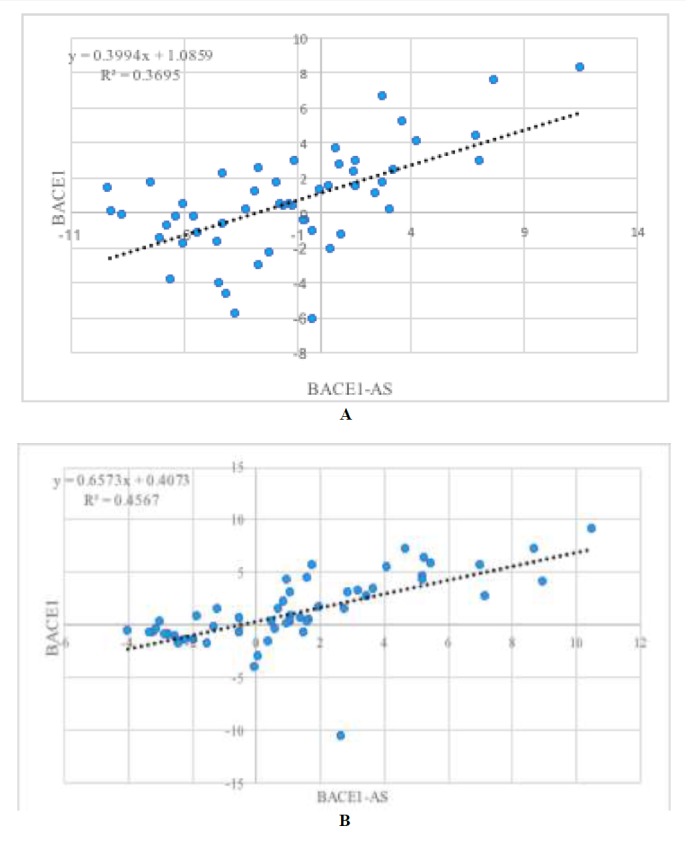
Correlation between BACE1 and BACE1-AS expression in tumors (A) and ANCTs (B).
Discussion
In the present study we evaluated BACE1 and BACE1-AS expression in invasive ductal carcinoma of breast and found significantly less BACE1 in tumors than in ANCTs. BACE1 catalyzes the primary step of APP production (1), which has been shown to be elevated in some human malignancies and to enhance cell proliferation (10). In breast cancer cell lines, APP expression was up-regulated by dihydrotestosterone and its expression was associated with cell proliferation. In addition, APP protein abundance has been suggested as a prognostic factor in ER-positive breast cancer patients (4). In addition, increased APP levels have been shown to increase tumorigenicity and invasiveness of aggressive breast cancer cells (5). However, BACE1 activity and abundance have not yet been assessed in breast cancer samples. Several lines of evidences have highlighted the role of BACE1 in the pathogenesis of other cancer types. BACE1 inhibitors have been suggested as putative therapeutic agents in some human malignancies as they suppressed the growth and angiogenesis of human glioblastoma and lung adenocarcinoma tumors in a xenograft animal model (3). BACE1 is also involved in the photolytic cleavage of the insulin receptor (IR). The soluble IR fragment (IRsol) generated by this enzymatic action is increased in the plasma of hepatic cancer patients (11). Our study showed different BACE1 expression patterns in breast cancer tissues than were seen in other malignancy types. A possible explanation for this discrepancy is the presence of a tissue-specific function for BACE1. On the other hand, as previous studies have shown up-regulation of APP in breast cancer tissues, we hypothesize that the observed downregulation of BACE1 in our study might be compensated by activation of or over-expression of gamma-secretase to accomplish the final step of APP production. Simultaneous analysis of BACE, gamma-secretase, and APP levels in a larger cohort of breast cancer patients might help clarify the role of these proteins in the pathogenesis of breast cancer. The similar level of BACE1-AS in umoral tissues and ANCTs, despite the down-regulation of BACE1 in tumoral tissues as revealed by our study, might imply a defect in regulation of BACE1-AS expression or in the feed-forward loop between these two transcripts in tumoral tissues. The latter possibility is also reflected in the observed stronger correlation between these two transcripts in ANCTs than in tumoral tissues, as demonstrated by R2 values. Future studies are needed to assess the interaction between these two transcripts in cancerous and normal tissues.
The Bayesian Multilevel model results showed a significant difference in BACE1 expression between stages 1 and 2 cancers after age-effect adjustments. Such a finding might imply a putative stage-specific signature for this lncRNA or its involvement in certain stages of cancer development, which should be assessed in larger patient cohorts.
Previous animal studies have shown agedependent differential expression of BACE splice variants in brain tissues (12). Further evidence for age-dependent regulation of BACE1 in brain tissue included the observed down-regulation of miR-186 as a negative regulator of BACE1 in aged brains (13); however, we found no correlations between BACE1 or BACE1-AS transcript levels in breast tissues and patient age. Such inconsistencies might be due to the presence of tissue-specific regulatory mechanisms for BACE1 expression.
Finally, we found greater BACE1-AS expression in ER-positive than in ER-negative samples. Previous studies have shown an association between BACE1 activity and brain estrogen reduction both in female AD patients and animal models (14). Moreover, estrogen treatment significantly decreased BACE1 protein levels in an ER-dependent manner (15). The observed association between BACE1-AS expression and ER status suggest an extra level of complexity in estrogen-regulated BACE production.
In conclusion, we demonstrated down-regulation of BACE1 in invasive breast cancer samples in association with some patient clinicopathologies. Future studies are needed to elaborate the role of BACE1 in breast cancer pathogenesis.
Acknowledgements
The current study was supported by a grant from Shahrekord University of Medical Sciences.
The authors declare they have no conflict of interest.
References
- 1.John V. Human beta-secretase (BACE) and BACE inhibitors: progress report. Current Topics in Medicinal Chemistry. 2006;6(6):569–78. doi: 10.2174/156802606776743084. [DOI] [PubMed] [Google Scholar]
- 2.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of betasecretase. NatureMedicine. 2008;14(7):723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paris D, Quadros A, Patel N, DelleDonne A, Humphrey J, Mullan M, et al. Inhibition of angiogenesis and tumor growth by beta and gamma-secretase inhibitors. European Journal of Pharmacology. 2005;514(1):1–15. doi: 10.1016/j.ejphar.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 4.Takagi K, Ito S, Miyazaki T, Miki Y, Shibahara Y, Ishida T, et al. Amyloid precursor protein in human breast cancer: an androgen-induced gene associated with cell proliferation. Cancer Science. 2013;104(11):1532–8. doi: 10.1111/cas.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim S, Yoo BK, Kim HS, Gilmore HL, Lee Y, Lee HP, et al. Amyloid-beta precursor protein promotes cell proliferation and motility of advanced breast cancer. BMC Cancer. 2014;14:928. doi: 10.1186/1471-2407-14-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaghoobi H, Azizi H, Oskooei VK, Taheri M, Ghafouri-Fard S. Assessment of expression of interferon gamma (IFN-G) gene and its antisense (IFNG-AS1) in breast cancer. World Journal of Surgical Oncology. 2018;16(1):211. doi: 10.1186/s12957-018-1508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esfahani ZT, Dashti S, Taheri M, Kholghi-Oskooei V, Arsang-Jang S, Ghafouri-Fard S. Expression of brain-derived neurotrophic factor (BDNF) and its naturally occurring antisense in breast cancer samples. Meta Gene. 2019;19:69–73. [Google Scholar]
- 8.Taheri M, Omrani MD, Ghafouri-Fard S. Long non-coding RNAs expression in renal cell carcinoma. Journal of Biology and Today's World. 2017;6(12):240–7. [Google Scholar]
- 9.Xiong DD, Li ZY, Liang L, He RQ, Ma FC, Luo DZ, et al. The LncRNA NEAT1 Accelerates Lung Adenocarcinoma Deterioration and Binds to Mir- 193a-3p as a Competitive Endogenous RNA. Cellular Physiology and Biochemistry : International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2018;48(3):905–18. doi: 10.1159/000491958. [DOI] [PubMed] [Google Scholar]
- 10.Hansel DE, Rahman A, Wehner S, Herzog V, Yeo CJ, Maitra A. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Research. 2003;63(21):7032–7. [PubMed] [Google Scholar]
- 11.Meakin PJ, Mezzapesa A, Benabou E, Haas ME, Bonardo B, Grino M, et al. The beta secretase BACE1 regulates the expression of insulin receptor in the liver. Nature Communication. 2018;9 doi: 10.1038/s41467-018-03755-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zohar O, Pick CG, Cavallaro S, Chapman J, Katzav A, Milman A, et al. Age-dependent differential expression of BACE splice variants in brain regions of tg2576 mice. Neurobiology of Aging. 2005;26(8):1167–75. doi: 10.1016/j.neurobiolaging.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Yoon H, Chung DE, Brown JL, Belmonte KC, Kim J. miR-186 is decreased in aged brain and suppresses BACE1 expression. Journal of Neurochemistry. 2016;137(3):436–45. doi: 10.1111/jnc.13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue X, Lu M, Lancaster T, Cao P, Honda S, Staufenbiel M. Brain estrogen deficiency accelerates Abeta plaque formation in an Alzheimer's disease animal model. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):19198–203. doi: 10.1073/pnas.0505203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan M, Li XY, Jiang W, Huang Y, Li JD, Wang ZM. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Experimental and Therapeutic Medice. 2013;5(4):1143–6. doi: 10.3892/etm.2013.933. [DOI] [PMC free article] [PubMed] [Google Scholar]


