Abstract
Hepatocellular carcinoma (HCC) is one of the primary causes of cancer-associated deaths worldwide. Current treatment methods include surgical resection, chemotherapy and radiotherapy; however the curative rate remains low, thus novel treatments are required. The aim of the present study was to investigate the role of targeting protein for Xenopus kinesin-like protein 2 (TPX2) in the growth of HCC and its underlying molecular mechanism. Immunohistochemistry staining, reverse transcription-quantitative (RT-q)PCR and western blotting were used to detect the expression of TPX2 mRNA and protein in liver cancer tissue samples, adjacent normal liver tissue samples, and the HCC cell lines Huh7, Hep3B, PLC/PRF/5 and MHCC97-H. The recombinant plasmid pMagic4.1-shRNA-TPX2 was constructed and transfected into Huh7 and Hep3B HCC cells to silence TPX2 expression. The proliferation, apoptosis, migration and invasion of Huh7 cells and Hep3B cells were evaluated before and after TPX2 silencing. The mRNA and protein expression levels of multiple signaling pathway-associated genes were detected by RT-qPCR and western blotting. The expression levels of TPX2 mRNA and protein were significantly higher in HCC tissue samples compared with adjacent normal liver tissue sample. TPX2 mRNA and protein expression levels were detected in the different HCC cell lines. The recombinant plasmid pMagic4.1-shRNA-TPX2 was successfully transfected into Huh7 and Hep3B cells, resulting in TPX2 silencing. TPX2 knockdown significantly reduced cell proliferation, cell migration and cell invasion of Huh7 and Hep3B cells, whilst also increasing the rate of apoptosis in these cells. Following TPX2 silencing, the expression levels of PI3K, phospho-AKT, Bcl-2, c-Myc and Cyclin D1 were significantly decreased, whereas the expression levels of P21 and P27 were significantly increased. In conclusion, TPX2 may suppress the growth of HCC by regulating the PI3K/AKT signaling pathway and thus, TPX2 may be a potential target for the treatment of liver cancer.
Keywords: hepatocellular carcinoma, targeting protein for Xenopus kinesin-like protein 2, proliferation, apoptosis, PI3K/AKT signaling pathway
Introduction
Liver cancer is the fourth most common malignant tumor in the world and the third leading cause of cancer-associated death (1). China has a high incidence of chronic hepatitis B, and persistent infection and replication of hepatitis B virus causes liver fibrosis and cirrhosis or possibly liver cancer as the diseases progresses (2). A recent analysis reported that 45.69% of liver cancer deaths were due to HBV infection, so the incidence and mortality of hepatocellular carcinoma (HCC) is high as a result (3). Studies have found that due to an increase in the incidence of non-alcoholic fatty liver disease and chronic hepatitis C, the incidence of HCC in western countries is also increasing annually (4,5). According to the statistics, ~600,000 individuals are diagnosed with HCC every year worldwide, and ~500,000 individuals die of HCC-associated diseases (1,6). Therefore, it is of great significance to explore the pathogenesis of liver cancer and develop new therapeutic targets for the treatment of liver cancer.
Targeting protein for Xenopus kinesin-like protein 2 (TPX2) is a nuclear proliferation microtubule-associated protein that can regulate spindle formation and stabilize spindle microtubules by promoting chromatin microtubule nucleation (7). TPX2 is the activating protein of Aurora A and can localize with Aurora A in mitotic spindle microtubules (8). However, upregulation of TPX2 can cause centrosome amplification and lead to DNA polyploidy (9). Previous studies have found that TPX2 is upregulated in a wide range of malignant tumors, including esophageal cancer (10), colon cancer (11,12), breast cancer (13), cervical cancer (14,15), ovarian cancer (16), bladder carcinoma (17) and medullary thyroid carcinoma (18). To the best of our knowledge, there are no studies on the association between TPX2 and the occurrence and development of HCC. The role of TPX2 in liver cancer progression and its potential molecular mechanism are unclear.
In the present study, the mechanism by which TPX2 participated in the development of HCC was investigated. RNA interference was used to silence TPX2 expression in HCC cells and changes in the molecular biological behavior of HCC cells were observed. Additionally, the molecular mechanism underlying TPX2-mediated regulation of growth of HCC cells was elucidated.
Materials and methods
Reagents
DMEM, FBS and trypsin were purchased from HyClone (GE Healthcare Life Sciences). The empty vector, pMagic4.1 with a construct for green fluorescent protein was purchased from Clontech Laboratories, Inc., and the recombinant the plasmids pMagic4.1-shRNA-TPX2 and pMagic4.1-shRNA-NC were constructed in our laboratory and verified by PCR, endonuclease cleavage and sequencing in our previous study (19). Lipofectamine™ 2000 and TRIzol® were purchased from Invitrogen (Thermo Fisher Scientific, Inc). Cell Counting Kit-8 (CCK-8) solution was purchased from Dojindo Molecular Technologies, Inc. The Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) kit was purchased from Beijing Solarbio Science & Technology Co., Ltd. Transwell chambers were purchased from BD Biosciences. DMSO was obtained from Sigma-Aldrich (Merck KGaA). The reverse transcription kit was purchased from Fermentas (Thermo Fisher Scientific, Inc.). PCR primers were synthesized by Generay Biotech Co. Ltd. Total protein extraction kit was purchased from Sangon Biotech Co. Ltd. Primary antibodies against TPX2 (D2R5C) (cat. no. 12245), PI3K (cat. no. 4249), phospho (p)-AKT (Ser 473) (cat. no. 4060), AKT (cat. no. 4685), P21 (cat. no. 2947), Bcl-2 (cat. no. 15071), p38 MAPK (cat. no. 8690), p-p38 MAPK (Thr180/Tyr182) (cat. no. 4511), STAT3 (cat. no. 12640), p-STAT3 (Tyr705) (cat. no. 9145) and β-actin (cat. no. 4970) were purchased from Cell Signaling Technology. Anti-TPX2 (cat. no. ab32795) primary antibody for immunohistochemical staining was purchased from Abcam. Primary antibodies against Smad2/3 (cat. no. sc-398844) and p-Smad2/3 (Ser 423/425) (cat. no. sc-11769) were purchased from Santa Cruz Biotechnology, Inc. Horseradish peroxidase-conjugated secondary antibodies were purchased from OriGene Technologies, Inc.
Clinical specimen
A total of 14 HCC tissue samples and adjacent normal liver tissue samples (distance from tumor >5 cm) from patients who underwent surgical resection were collected at Jiading District Central Hospital of Shanghai University of Medicine & Health Sciences (Shanghai, China) between May 2016 and March 2018. The 14 HCC cases included 10 males and 4 females aged 42-79 (55.86±2.81) years. None of the patients had received chemotherapy or radiotherapy prior to radical resection of liver cancer. The present study was approved by the Human Ethics Committee at Jiading District Central Hospital of Shanghai University of Medicine & Health Sciences and prior written consent was obtained from all patients.
Cell culture
The human liver cancer cell lines Huh7, Hep3B, PLC/PRF/5 and MHCC97-H were obtained from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. All cells were maintained in DMEM containing 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. The cells were routinely plated at a density of 1x105 cells/ml in 6-well plates and incubated in a humidified incubator at 37°C with 95% air and 5% CO2.
Reverse transcription-quantitative (RT-q)PCR
RT-qPCR was performed on all tissue samples and cells. Total RNA was extracted from frozen tissue samples and cells using TRIzol®, and 1 µg total RNA was used as a template for the synthesis of the first-strand cDNA synthesis using a reverse transcription kit (Fermentas; Thermo Fisher Scientific, Inc.). The reverse transcription temperature protocol used was 37°C for 15 min and 85°C for 30 sec. TPX2, PI3K, AKT, P21, Bcl-2, c-Myc, P27, BCL2L1 and Cyclin D1 primers were designed using Primer Premier version 5.0 (Premier Biosoft International) and synthesized by Generay Biotech Co. Ltd. The sequences of the primers are presented in Table I. A Takara quantitative kit with TB Green Premix Ex Taq (Takara Bio, Inc.) was used for quantitative PCR. The thermocycling conditions were: Initial denaturation at 95°C for 5 min; followed by 40 cycles of 95°C for 30 sec and 60°C for 30 sec. Relative expression of TPX2, PI3K, AKT, P21, Bcl-2, c-Myc, P27, BCL2L1 and Cyclin D1 mRNA was calculated using the 2–ΔΔCq (20) method and β-actin was used as the internal reference.
Table I.
Primer sequences used for PCR.
| Gene | Forward sequence, 5'-3' | Reverse sequence, 5'-3' |
|---|---|---|
| TPX2 | ACCTTGCCCTACTAAGATT | AATGTGGCACAGGTTGAGC |
| PI3K | TGGCCTTAGCTCTTAGCCAAACAC | ATTGGAACACGGCCTTTGACA |
| AKT C | TGTGCCTATGCTGCCCAT | CAGTGCGATGTCGTGGAGG |
| P21 | GACCTGTCACTGTCTTGTAC | CTCTCATTCAACCGCCTAG |
| Bcl-2 | GGATAACGGAGGCTGGGATGC | GACTTCACTTGTGGCCCAGAT |
| C-Myc | TGTGTTACGGTCGCGTCTTT | AACAGCTCGGTCACCATCTC |
| Cyclin D1 | CCAGACCCACGTTTCTTTGC | ATCCCTAGAAACACCACGGC |
| P27 | TGGAAAGCGGTCTGCAAGTG | TCACTGTCACATTCAGGGGC |
| BCL2L1 | TCCCCATGGCAGCAGTAAAG | TCCACAAAAGTATCCTGTTCAAAGC |
| β-actin | AAGGTGACAGCAGTCGGTT | TGTGTGGACTTGGGAGAGG |
Immunohistochemistry
Immunohistochemical staining was performed on paraffin-embedded HCC tissue samples to examine the level of TPX2 protein as described previously (13). Briefly, the HCC tissue samples were fixed in 4% paraformaldehyde for 48 h at room temperature, dehydrated in a graded series of ethanol (50, 75, 85, 95 and 100%), embedded in paraffin and sectioned into 4 µm thick slices. Xylene and a graded series of ethanol (100, 95, 85 and 75%) were used to dewax and hydrate the samples, respectively, followed by 30 min of antigen retrieval in Tris-EDTA (pH 9.0) in a 720 W microwave. Subsequently, the sections were blocked in 3% H2O2 for 10 min and incubated with anti-TPX2 (1:400) primary antibody at room temperature for 2 h, washed with TBS-Tween, and incubated with the horseradish peroxidase-linked anti-goat immunoglobulin G secondary antibody (1:1,500; Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. Following incubation with the antibodies, a diaminobenzidine substrate kit (Vector Laboratories, Inc.) was used to visualize bound antibodies. Hematoxylin was used to stain the cell nuclei at room temperature for 4 min. The tissues were observed and imaged under an inverted light microscope at a x200 and x400 magnification (Olympus Corporation) and evaluated by a pathologist blinded to the patient's information. The staining score was assessed as described previously (13). Expression grading was stratified according to the final score as follows: 0-3, low TPX2 expression and 4-7, high TPX2 expression (13).
Western blotting
Western blot analysis of all tissue samples and cells were performed according to the manufacturers' protocols. Briefly, total proteins were extracted from tissue samples or cells using RIPA lysis buffer (Cell Signaling Technology, Inc.) and protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). A total of 40 mg/lane of samples were loaded on a 10% SDS gel, resolved using SDS-PAGE and transferred to nitrocellulose membranes. Subsequently, membranes were blocked in 5% non-fat milk for 1 h at room temperature and incubated with anti-TPX2 (1:1,000), anti-PI3K (1:1,000), anti-p-AKT (1:2,000), anti-AKT (1:1,000), anti-P21 (1:2,000), anti-Bcl-2 (1:1,000), anti-Smad2/3 (1:1,000), anti-p-Smad2/3 (1:1,000), anti-p38 MAPK (1:1,000), anti-p-p38 MAPK (1:1,000), anti-STAT3 (1:1,000), anti-p-STAT3 (1:2,000) or anti-β-actin (1:3,000) primary antibody overnight at 4°C, followed by incubation with horseradish peroxidase-conjugated anti-rabbit and anti-mouse immunoglobulin G secondary antibodies at a dilution of 1:5,000 (cat. no. ab6721 and ab205719, respectively; Abcam) at room temperature for 2 h. The signal was visualized using enhanced chemiluminescence reagent (Beyotime Institute of Biotechnology) according to manufacturer's protocol. The densities of the protein bands were quantified using ImageJ 1.8.0 software (National Institutes of Health). Expression of β-actin antibody was used as the internal control.
TPX2 short hairpin (sh)RNA and cell transfection
Huh7 or Hep3B cells were divided into three groups: i) untrans-fected control group (Ctrl); ii) pMagic4.1-shRNA-negative control plasmid transfected group (shRNA-NC); and iii) pMagic4.1-shRNA-TPX2 plasmid transfected group (shRNA-TPX2). The TPX2 shRNA sequence and the detailed procedures of the transfection are described in our previous publication (19). Green fluorescence was observed under a fluorescence microscope (magnification, x100) after plasmid transfection for 48 h. The TPX2 mRNA and protein levels in Huh7 and Hep3B cells were detected using RT-qPCR and western blot analysis.
Cell proliferation assay
Huh7 or Hep3B cells were plated at a density of 5x103 cells/well into 96-well plates following transfection. After 0, 24, 48 or 72 h of culture, 10 ml CCK-8 solution was added to each well and further incubated at 37°C for 1 h. Absorbance was measured at 450 nm using a micro-plate reader.
Cell apoptosis assay
Huh7 or Hep3B cells in each group were trypsinized and collected by centrifugation at 37°C for 5 min at a speed of 1,000 x g. Cells were washed twice with PBS, and 1x105 cells were re-suspended in 500 ml binding buffer and incubated with Annexin V-FITC/PI dual stain for 15 min at room temperature. A FACSCalibur flow cytometer (BD Biosciences) was used to detect the apoptotic rate of cells. BD CellQuest™ Pro software version 5.1 (BD Biosciences) was used to analyze the data.
Transwell migration and invasion assays
Cell migration and invasion assays were performed using Transwell chambers with (invasion) or without (migration) Matrigel according to the manufacturer's protocol. A total of 2x105 Huh7 or Hep3B cells were plated into the upper chamber of the insert in serum-free DMEM. DMEM containing 10% FBS was added to the lower chamber. The Huh7 or Hep3B cells remaining on the insert's top layer were removed with a cotton swab after 48 h of incubation. The cells which had migrated or invaded to the lower surface of the membrane were stained with crystal violet for 30 min at room temperature. The cells on the lower surface of the membrane were imaged under an inverted light microscope at x50 magnification (Olympus Corporation), and cells in 5 fields of view were counted to estimate cell migration/invasion. Each experiment was performed three times independently.
Statistical analysis
Data are presented as the mean ± standard deviation of at least three repeats. Differences in the expression levels of TPX2 in HCC tissue samples were evaluated using a Pearson χ2 test or Fisher's exact test. Other experimental data were evaluated using a one-way ANOVA with a post-hoc least significant difference test, using SPSS version 19.0 (IBM, Corp.). P<0.05 was considered to indicate a statistically significant difference.
Results
TPX2 expression is upregulated in HCC tissues and human hepatoma cell lines
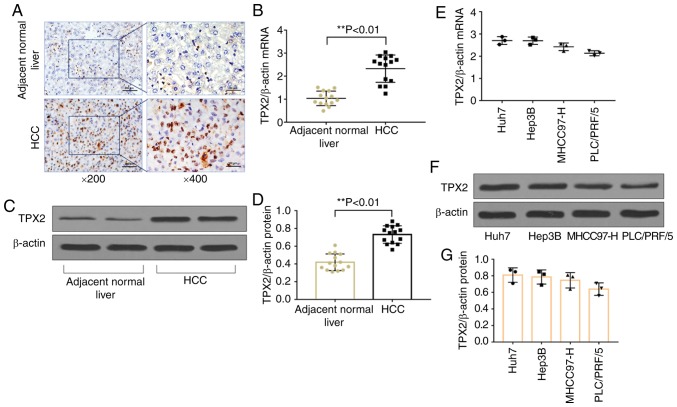
Immunohistochemistry staining, RT-qPCR and western blot analysis demonstrated that the protein (P<0.01) and mRNA (P<0.01) expression levels of TPX2 were significantly increased in HCC tissues compared with adjacent normal liver tissues (Fig. 1A-D). The correlation between TPX2 expression and various clinicopathological characteristics are shown in Table II. TPX2 expression was correlated with tumor differentiation (P=0.017) and clinical Tumor-Node-Metastasis stage (21) (P=0.016), suggesting that TPX2 may be associated with carcinogenesis and progression of HCC. Therefore, TPX2 mRNA and protein expression levels were assessed in various human hepatoma cell lines, including Huh7, Hep3B, PLC/PRF/5 and MHCC97-H. TPX2 mRNA and protein expression levels were detected in all human liver cancer cell lines (Fig. 1E-G). Huh7 and Hep3B cell lines had the highest levels of expression of TPX2 and were thus used for TPX2 knockdown and subsequent functional experiments.
Figure 1.
TPX2 expression levels in HCC tissues and liver cancer cell lines. (A) Representative immunohistochemical stain of TPX2 expression in patients' tissues. (B) mRNA expression levels of TPX2 in HCC tissues and the adjacent normal liver tissue. **P<0.01. (C) Representative western blot of TPX2 protein expression levels in HCC tissues and the adjacent normal liver tissue. (D) Densitometry analysis of TPX2 protein expression levels in (C). **P<0.01. (E) mRNA and (F) protein expression levels in the liver cancer cell lines, Huh7, Hep3B, MHCC97-H, PLC/PRF/5. (G) Densitometric analysis of TPX2 protein expression levels in (F). Data are presented as the mean ± standard deviation. HCC, hepatocellular carcinoma; TPX2, targeting protein for Xenopus kinesin-like protein 2.
Table II.
Association between TPX2 expression and clinicopathological features in patients with HCC.
| Characteristics | No. of patients | Low TPX2 expression | High TPX2 expression | P-value |
|---|---|---|---|---|
| Sex | 0.852 | |||
| Male | 10 | 3 | 7 | |
| Female | 4 | 1 | 3 | |
| Age, years | 0.597 | |||
| ≤50 | 5 | 1 | 4 | |
| >50 | 9 | 3 | 6 | |
| Tumor size, cm | 0.481 | |||
| ≤2 | 5 | 2 | 3 | |
| >2 | 9 | 2 | 7 | |
| Differentiation | 0.017a | |||
| Well | 1 | 1 | 0 | |
| Moderate | 5 | 3 | 2 | |
| Poor | 8 | 0 | 8 | |
| TNM stage | 0.016a | |||
| I | 1 | 1 | 0 | |
| II | 4 | 3 | 1 | |
| III | 8 | 0 | 8 | |
| IV | 1 | 0 | 1 |
P<0.05. TPX2, targeting protein for Xenopus kinesin-like protein 2; HCC, hepatocellular carcinoma; TNM, Tumor-Node-Metastasis.
TPX2 shRNA plasmid transfection and downregulation of TPX2
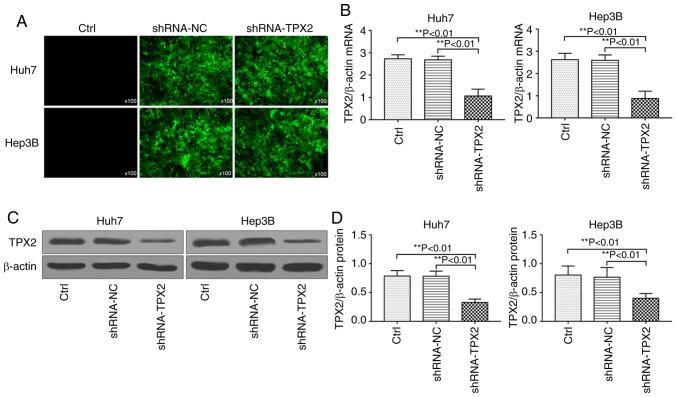
The pMagic4.1-shRNA-TPX2 plasmid or pMagic4.1-shRNA-NC plasmid were transiently transfected into Huh7 and Hep3B cells. After 48 h, cells were observed under a fluorescence microscope. As shown in Fig. 2A, the cells which were successfully transfected with pMagic4.1-shRNA-TPX2 or pMagic4.1-shRNA-NC plasmid showed green fluorescence. RT-qPCR and western blot analysis demonstrated that the mRNA and protein expression levels of TPX2 in Huh7 or Hep3B cells were significantly downregulated (P<0.01; Fig. 2B-D).
Figure 2.
TPX2 expression is downregulated in cells transfected with pMagic4.1-shRNA-TPX2. (A) Green fluorescence was observed following transfection. Magnification, x100. (B) mRNA and (C) protein expression levels of TPX2 in Huh7 and Hep3B transfected with the shRNA-TPX2 vector. (D) Densitometry analysis of TPX2 protein expression levels in (C). **P<0.01. Data are presented as the mean ± standard deviation. TPX2, targeting protein for Xenopus kinesin-like protein 2; sh, short hairpin; Ctrl, untransfected control; NC, negative control plasmid.
TPX2 silencing inhibits the proliferation of Huh7 or Hep3B cells
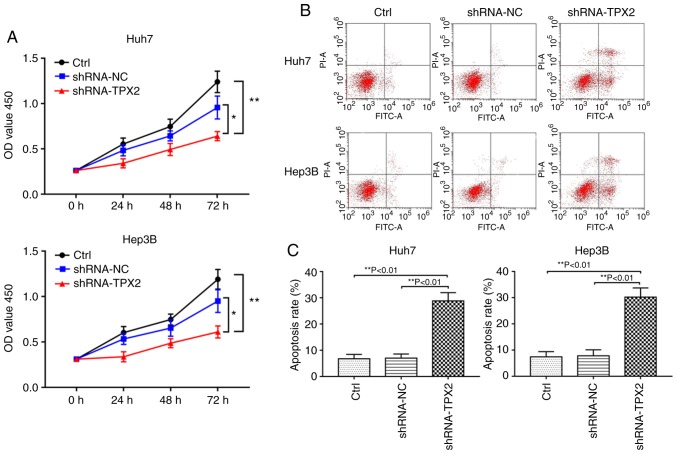
A cell viability assay was used to investigate whether TPX2 silencing affected proliferation of Huh7 and Hep3B cells. As shown in Fig. 3A, the optical density (OD) values of the shRNA-TPX2 group was significantly lower compared with the shRNA-NC group and Ctrl group (P<0.05 and P<0.01, respectively). There was no significant difference in the OD value between the Ctrl group and shRNA-NC group (P>0.05).
Figure 3.
Effects of TPX2 silencing on the proliferation and apoptosis of Huh7 and Hep3B cells. (A) A Cell Counting Kit-8 assay was used to detect the proliferation of Huh7 and Hep3B cells. *P<0.05, **P<0.01. (B) Representative dot plots of apoptosis in Huh7 and Hep3B cells. (C) Quantitative analysis of apoptosis in Huh7 and Hep3B cells transfected with an shRNA-TPX2 or shRNA-NC plasmid. **P<0.01. Data are presented as the mean ± standard deviation. TPX2, targeting protein for Xenopus kinesin-like protein 2; sh, short hairpin; Ctrl, untransfected control; NC, negative control plasmid.
Downregulation of TPX2 increases apoptosis of Huh7 or Hep3B cells
Flow cytometry showed that Huh7 or Hep3B cell apoptosis were significantly increased when TPX2 was knocked down. The apoptotic rate of the shRNA-TPX2 group was significantly higher compared with the Ctrl group and the shRNA-NC group (P<0.01). There was no significant difference in the apoptotic rate between the Ctrl group and the shRNA-NC group (P>0.05; Fig. 3B and C).
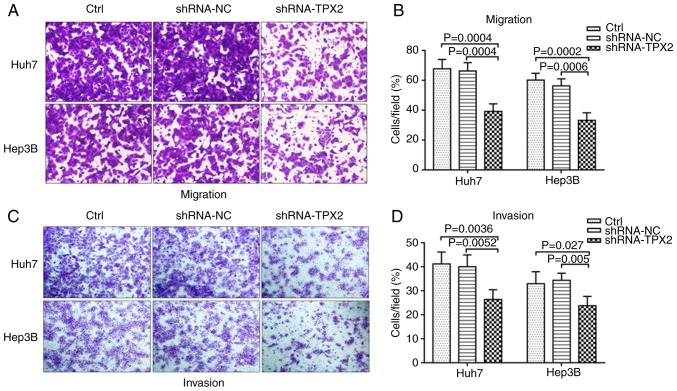
TPX2 knockdown suppresses the migration and invasion of Huh7 and Hep3B cells
The effect of silencing TPX2 on the migration and invasion of Huh7 and Hep3B cells was assessed using Transwell migration and invasion assays. As shown in Fig. 4A and B, the migratory capacity of Huh7 and Hep3B cells in the shRNA-TPX2 group was significantly reduced compared with the Ctrl group and shRNA-NC group (both P<0.01). The knockdown of TPX2 in Huh7 and Hep3B cells resulted in decreased invasion compared with the shRNA-NC group and Ctrl group (P<0.05 or P<0.01; Fig. 4C and D). There was no significant difference in the migratory and invasive capacities between the Ctrl group and the shRNA-NC group (P>0.05).
Figure 4.
Effect of TPX2 silencing on the migration and invasion of Huh7 and Hep3B cells. (A and B) Effect of TPX2 knockdown on cell migration and (C and D) invasion of Huh7 and Hep3B cells was examined using a Transwell assay. Data are presented as the mean ± standard deviation. TPX2, targeting protein for Xenopus kinesin-like protein 2; sh, short hairpin; Ctrl, untransfected control; NC, negative control plasmid.
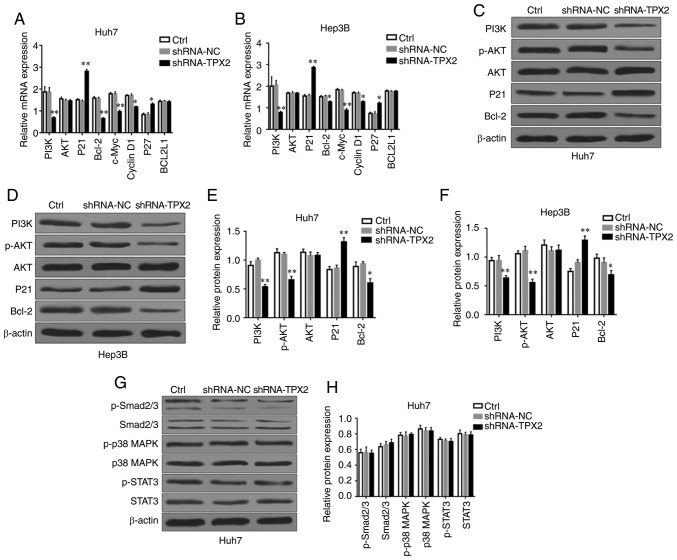
TPX2 silencing suppresses the PI3K/AKT signaling pathway
The development and progression of HCC are associated with multiple signaling pathways, such as the PI3K/AKT, MAPK/P38, JAK2/STAT3, TGF-β/Smad and NF-κB signaling pathways (22-26). Previous studies have found that TPX2 can regulate the proliferation and apoptosis of a number of different types of malignant tumors via the PI3K/AKT signaling pathway (13,16). In the present study, the potential effects of TPX2 silencing on the expression levels of PI3K/AKT signaling pathway-associated factors in Huh7 or Hep3B cells were assessed. As shown in Fig. 5A and B, PI3K, Bcl-2, c-Myc and Cyclin D1 mRNA expression levels in Huh7 or Hep3B cells were significantly decreased in the shRNA-TPX2 group compared with the Ctrl group and shRNA-NC group (P<0.05, P<0.01), whereas P21 and P27 mRNA expression levels in the shRNA-TPX2 group was significantly upregulated (P<0.05, P<0.01). There were no significant differences in the BCL2L1 mRNA expression levels among the shRNA-TPX2, Ctrl and shRNA-NC groups (P>0.05). Compared with the Ctrl group and shRNA-NC group, PI3K, p-AKT and Bcl-2 protein expression levels were significantly downregulated in the shRNA-TPX2 group (P<0.05 or P<0.01), whereas P21 protein expression levels in the shRNA-TPX2 group were significantly increased (P<0.01; Fig. 5C-F). There was no significant difference in the AKT mRNA and protein expression levels among the shRNA-TPX2, Ctrl or shRNA-NC groups (P>0.05). Additionally, there was no significant difference in the expression levels of PI3K/AKT signaling pathway-associated factors between the Ctrl group and the shRNA-NC group (P>0.05). To determine the association between TPX2 and other signaling pathways in HCC, Smad2/3, p-Smad2/3, p38 MAPK, p-p38 MAPK, STAT3 and p-STAT3 protein expression levels were examined in the Huh7 cells following silencing of TPX2. There were no significant differences in the expression levels of TGFβ/Smad, MAPK/p38 and JAK2/STAT3 signaling pathway-associated factors among the Ctrl group, shRNA-NC group and shRNA-TPX2 group (P>0.05; Fig. 5G and H). These results confirmed that TPX2 activated the PI3K/AKT signaling pathway in liver cancer.
Figure 5.
Effect of TPX2 knockdown on multiple signaling pathways in Huh7 and Hep3B cells. mRNA expression levels of PI3K, AKT, P21, Bcl-2, c-Myc, Cyclin D1, P27 and BCL2L1 in (A) Huh7 and (B) Hep3B cells. *P<0.05, **P<0.01. Western blots of PI3K, p-AKT, AKT, P21 and Bcl-2 expression in (C) Huh7 and (D) Hep3B cells. Quantitative analysis of protein expression levels of PI3K, p-AKT, AKT, P21 and Bcl-2 in (E) Huh7 and (F) Hep3B cells. *P<0.05, **P<0.01. (G) Western blots of p-Smad2/3, Smad2/3, p-p38 MAPK, p38 MAPK, p-STAT3 and STAT3 expression in Huh7 cells. β-actin was used as the loading control. (H) Quantitative analysis of p-Smad2/3, Smad2/3, p-p38 MAPK, p38 MAPK, p-STAT3 and STAT3 expression in Huh7 cells. Data are presented as the mean ± standard deviation. TPX2, targeting protein for Xenopus kinesin-like protein 2; sh, short hairpin; Ctrl, untransfected control; NC, negative control plasmid.
Discussion
According to the latest statistics from Global Cancer 2018, liver cancer is one of the most common malignant tumors and ranks 4th in incidence and 3rd in mortality rates of all types of cancer (1). Over the past few decades, with the development of radical hepatectomy and liver transplantation, ~40% of patients with liver cancer have been cured (27,28), but ~60% of patients with liver cancer are still unable to receive effective treatment due to advanced stages of liver cancer at initial diagnosis, poor economic status or a lack of livers for transplants (29). In the majority of the patients who undergo radical hepatectomy or liver transplantation, the tumor may exhibit recurrence or may have metastasized (30,31). Although several biomarkers have been thought to be associated with the occurrence and development of liver cancer (32,33), the majority of these markers have not proven beneficial for diagnosing or treating patients with liver cancer. Recently, several novel molecular inhibitors were approved for the treatment of advanced liver cancer, but the overall survival rate of patients has not improved significantly, and the efficacy of these drugs are not promising (34,35). Therefore, understanding the molecular mechanisms underlying the occurrence of liver cancer may assist in the development of new therapeutic strategies and targets for treating liver cancer.
TPX2, a microtubule-associated protein located on human chromosome 20q 11.2, serves an important role in regulating mitotic spindles and chromosome segregation (36,37). TPX2 is a downstream effector of the small GTPase Ran, which is involved in spindle formation (38). Overexpression of TPX2 can induce centrosome amplification, lead to DNA polyploidy and induce tumor formation (36). A number of studies have confirmed that TPX2 is closely associated with the occurrence of tumors. Overexpression of TPX2 promotes the occurrence and development of esophageal cancer, colon cancer, breast cancer, cervical cancer, ovarian cancer, bladder carcinoma and medullary thyroid carcinoma (10-18). In the present study it was also demonstrated that the expression of TPX2 in human HCC tissues was significantly upregulated compared with the adjacent normal tissues. The expression of TPX2 in the normal liver cell line, LO2, and compared with the different HCC cell lines Huh7, Hep3B, PLC/PRF/5 and MHCC97-H. The expression levels of TPX2 in LO2 cells were significantly lower compared with the different HCC cell lines; however, the data from LO2 cells were removed as their identity could not be verified using STR profiling. As such, the fact that the data in the HCC cells were not compared to a normal liver cell line is a limitation of the present study. Additionally, silencing TPX2 gene expression using RNA interference, significantly reduced proliferation, migration and invasion of Huh7 and Hep3B cells, whilst increasing apoptosis. These results suggest that TPX2 may improve the viability of HCC cells and inhibit cell apoptosis.
Previous studies have demonstrated that the PI3K/AKT/P21 signaling pathway serves an important role in the occurrence and development of malignant tumors (39-42). The activation of AKT is closely associated with cell proliferation, survival, migration and invasion of tumors (43). Inhibiting the activation of AKT and promoting the expression of P21 inhibits the proliferation of tumor cells, promotes cell apoptosis and decreases tumor progression (44,45). In the present study, expression of TPX2 was demonstrated to be associated with phosphorylation of AKT. Following silencing of TPX2, the expression levels of p-AKT were significantly decreased, suggesting that TPX2 may promote the activation of AKT and PI3K/AKT signal transduction. P21 is an inhibitor of cyclin-dependent kinase and an important cell cycle regulator in the PI3K/AKT signaling pathway. Previous studies have shown that P21 and P27 can inhibit cell cycle progression and promote apoptosis, behaviors crucial for tumorigenesis (46,47). The present study also found that the expression levels of P21 and P27 were significantly increased when TPX2 expression was knocked down. Bcl-2, c-Myc, BCL2L1 and Cyclin D1 are tumor-associated regulatory factors that are significantly upregulated during tumorigenesis (48,49). TPX2 knockdown significantly decreased the expression levels of Bcl-2, c-Myc and Cyclin D1. Together, the present study demonstrated that downregulation of TPX2 inhibited PI3K/AKT signal transduction, suppressed cell proliferation and promoted cell apoptosis, and thus may prevent the occurrence and development of HCC.
In summary, expression of TPX2 in human HCC was significantly upregulated. Targeted silencing of TPX2 reduced cell viability, abrogated cell cycle progress and promoted apoptosis of HCC cells by inhibiting the PI3K/AKT signal transduction pathway. Based on the results of the present study, TPX2 and its downstream effectors may be potential targets for the diagnosis and treatment of liver cancer and provide novel avenues for successful treatment of malignant tumors.
Acknowledgements
The authors would like to thank Dr Xia Gan, Dr Li-Hong Gan, Dr Fei Chen, Dr Li Zheng, Dr Ya-Qing Huang, and Dr Ling Yao (Department of Gastroenterology, Third Affiliated Hospital of Nanchang University) for their help.
Funding
The present study was supported by a grant from the Scientific Research Project of Health system in Jiading District of Shanghai (Shanghai, China; grant no. 2015-KY-04).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
DH, JJ and LL designed the experiments. DH, YZ and SL performed the experiments. JJ contributed to the analysis of the data and wrote the manuscript. LL corrected the manuscript. All authors approved the final of the version manuscript.
Ethics approval and consent to participate
The present study was approved by the Human Ethics Committee at Jiading District Central Hospital of Shanghai University of Medicine & Health Sciences (Shanghai, China) and prior written consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Dandri M, Petersen J. Mechanism of hepatitis B virus persistence in hepatocytes and its carcinogenic potential. Clin Infect Dis. 2016;62(Suppl 4):S281–S288. doi: 10.1093/cid/ciw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M, Wang Y, Feng X, Wang R, Wang Y, Zeng H, Qi J, Zhao H, Li N, Cai J, Qu C. Contribution of hepatitis B virus and hepatitis C virus to liver cancer in China north areas: Experience of the Chinese National Cancer Center. Int J Infect Dis. 2017;65:15–21. doi: 10.1016/j.ijid.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 4.de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62:1190–1200. doi: 10.1002/hep.27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Younossi ZM, Otgonsuren M, Henry L, Venkatesan C, Mishra A, Erario M, Hunt S. Association of nonalcoholic fatty liver disease (NAFLD) with hepatocellular carcinoma (HCC) in the United States from 2004 to 2009. Hepatology. 2015;62:1723–1730. doi: 10.1002/hep.28123. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Jiang Y, Yuan H, Fang Q, Cai N, Suo C, Jin L, Zhang T, Chen X. The trends in incidence of primary liver cancer caused by specific etiologies: Results from the Global Burden of Disease Study 2016 and implications for liver cancer prevention. J Hepatol. 2019;70:674–683. doi: 10.1016/j.jhep.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Gruss OJ, Vernos I. The mechanism of spindle assembly: Functions of Ran and its target TPX2. J Cell Biol. 2004;166:949–955. doi: 10.1083/jcb.200312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rennie YK, McIntyre PJ, Akindele T, Bayliss R, Jamieson AG. A TPX2 proteomimetic has enhanced affinity for Aurora-A due to hydrocarbon stapling of a Helix. ACS Chem Biol. 2016;11:3383–3390. doi: 10.1021/acschembio.6b00727. [DOI] [PubMed] [Google Scholar]
- 9.Pascreau G, Eckerdt F, Lewellyn AL, Prigent C, Maller JL. Phosphorylation of p53 is regulated by TPX2-Aurora A in xenopus oocytes. J Biol Chem. 2009;284:5497–5505. doi: 10.1074/jbc.M805959200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu HC, Zhang Y, Wang XL, Qin WS, Liu YH, Zhang L, Zhu CL. Upregulation of the TPX2 gene is associated with enhanced tumor malignance of esophageal squamous cell carcinoma. Biomed Pharmacother. 2013;67:751–755. doi: 10.1016/j.biopha.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi Y, Sheridan P, Niida A, Sawada G, Uchi R, Mizuno H, Kurashige J, Sugimachi K, Sasaki S, Shimada Y, et al. The AURKA/TPX2 axis drives colon tumorigenesis cooperatively with MYC. Ann Oncol. 2015;26:935–942. doi: 10.1093/annonc/mdv034. [DOI] [PubMed] [Google Scholar]
- 12.Wei P, Zhang N, Xu Y, Li X, Shi D, Wang Y, Li D, Cai S. TPX2 is a novel prognostic marker for the growth and metastasis of colon cancer. J Transl Med. 2013;11:313. doi: 10.1186/1479-5876-11-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, Zhang H, Zhang G, Zhong A, Ma Q, Kai J, Tong Y, Xie S, Wang Y, Zheng H, et al. Targeting TPX2 suppresses proliferation and promotes apoptosis via repression of the PI3k/AKT/P21 signaling pathway and activation of p53 pathway in breast cancer. Biochem Biophys Res Commun. 2018;507:74–82. doi: 10.1016/j.bbrc.2018.10.164. [DOI] [PubMed] [Google Scholar]
- 14.Jiang P, Shen K, Wang X, Song H, Yue Y, Liu T. TPX2 regulates tumor growth in human cervical carcinoma cells. Mol Med Rep. 2014;9:2347–2351. doi: 10.3892/mmr.2014.2106. [DOI] [PubMed] [Google Scholar]
- 15.Chang H, Wang J, Tian Y, Xu J, Gou X, Cheng J. The TPX2 gene is a promising diagnostic and therapeutic target for cervical cancer. Oncol Rep. 2012;27:1353–1359. doi: 10.3892/or.2012.1668. [DOI] [PubMed] [Google Scholar]
- 16.Tian Y, Liu LL, Guo DM, Wang Y, Zha WH, Li Y, Wu FJ. TPX2 gene silencing inhibits cell proliferation and promotes apoptosis through negative regulation of AKT signaling pathway in ovarian cancer. J Cell Biochem. 2018;119:7540–7555. doi: 10.1002/jcb.27065. [DOI] [PubMed] [Google Scholar]
- 17.Yan L, Li Q, Yang J, Qiao B. TPX2-p53-GLIPR1 regulatory circuitry in cell proliferation, invasion, and tumor growth of bladder cancer. J Cell Biochem. 2018;119:1791–1803. doi: 10.1002/jcb.26340. [DOI] [PubMed] [Google Scholar]
- 18.Yang X, Liu G, Xiao H, Yu F, Xiang X, Lu Y, Li W, Liu X, Li S, Shi Y. TPX2 overexpression in medullary thyroid carcinoma mediates TT cell proliferation. Pathol Oncol Res. 2014;20:641–648. doi: 10.1007/s12253-014-9743-4. [DOI] [PubMed] [Google Scholar]
- 19.Jian J, Huang Y, Liu LZ, Li SX, Deng F. TPX2 gene-silencing inhibits the proliferation and invasion of human colon cancer SW480 cells. TUMOR. 2016;36:628–634. [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Llovet JM, Bruix J, Fuster J, Castells A, Garcia-Valdecasas JC, Grande L, Franca A, Brú C, Navasa M, Ayuso MC, et al. Liver transplantation for small hepatocellular carcinoma: The tumor-node-metastasis classification does not have prognostic power. Hepatology. 1998;27:1572–1577. doi: 10.1002/hep.510270616. [DOI] [PubMed] [Google Scholar]
- 22.Xue S, Zhou Y, Zhang J, Xiang Z, Liu Y, Miao T, Liu G, Liu B, Liu X, Shen L, et al. Anemoside B4 exerts anti-cancer effect by inducing apoptosis and autophagy through inhibiton of PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Am J Transl Res. 2019;11:2580–2589. [PMC free article] [PubMed] [Google Scholar]
- 23.Feng PC, Ke XF, Kuang HL, Pan LL, Ye Q, Wu JB. BMP2 secretion from hepatocellular carcinoma cell HepG2 enhances angiogenesis and tumor growth in endothelial cells via activation of the MAPK/p38 signaling pathway. Stem Cell Res Ther. 2019;10:237. doi: 10.1186/s13287-019-1301-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Li SJ, Sui MH, Sun ZX, Zhang WW. LncRNA 00152 promotes the development of hepatocellular carcinoma by activating JAK2/STAT3 pathway. Eur Rev Med Pharmacol Sci. 2019;23:1038–1046. doi: 10.26355/eurrev_201902_16991. [DOI] [PubMed] [Google Scholar]
- 25.Zuo J, Ma H, Cai H, Wu Y, Jiang W, Yu L. An inhibitory role of NEK6 in TGFβ/Smad signaling pathway. BMB Rep. 2015;48:473–478. doi: 10.5483/BMBRep.2015.48.8.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Yang X, Li L, Yang G, Ouyang X, Xiang J, Zhang T, Min X. LASS2 inhibits proliferation and induces apoptosis in HepG2 cells by affecting mitochondrial dynamics, the cell cycle and the nuclear factor-κB pathways. Oncol Rep. 2019;41:3005–3014. doi: 10.3892/or.2019.7058. [DOI] [PubMed] [Google Scholar]
- 27.Thelen A, Benckert C, Tautenhahn HM, Hau HM, Bartels M, Linnemann J, Bertolini J, Moche M, Wittekind C, Jonas S. Liver resection for hepatocellular carcinoma in patients without cirrhosis. Br J Surg. 2013;100:130–137. doi: 10.1002/bjs.8962. [DOI] [PubMed] [Google Scholar]
- 28.Rhu J, Kim JM, Choi GS, Kwon CHD, Joh JW. Continuing five or more locoregional therapies before living donor salvage liver transplantation for hepatocellular carcinoma is related to poor recurrence-free survival. Ann Surg Treat Res. 2018;95:152–160. doi: 10.4174/astr.2018.95.3.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson TN, Zarrinpar A. Hepatocyte transplantation: Past efforts, current technology, and future expansion of therapeutic potential. J Surg Res. 2018;226:48–55. doi: 10.1016/j.jss.2018.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Famularo S, Di Sandro S, Giani A, Lauterio A, Sandini M, De Carlis R, Buscemi V, Uggeri F, Romano F, Gianotti L, De Carlis L. Recurrence patterns after anatomic or paren-chyma-sparing liver resection for hepatocarcinoma in a western population of cirrhotic patients. Ann Surg Oncol. 2018;25:3974–3981. doi: 10.1245/s10434-018-6730-0. [DOI] [PubMed] [Google Scholar]
- 31.Meischl T, Rasoul-Rockenschaub S, Györi G, Sieghart W, Reiberger T, Trauner M, Soliman T, Berlakovich G, Pinter M. C-reactive protein is an independent predictor for hepatocellular carcinoma recurrence after liver transplantation. PLoS One. 2019;14:e0216677. doi: 10.1371/journal.pone.0216677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scaggiante B, Kazemi M, Pozzato G, Dapas B, Farra R, Grassi M, Zanconati F, Grassi G. Novel hepatocellular carcinoma molecules with prognostic and therapeutic potentials. World J Gastroenterol. 2014;20:1268–1288. doi: 10.3748/wjg.v20.i5.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao B, Li S, Tan Z, Ma L, Liu J. ACTG1 and TLR3 are biomarkers for alcohol-associated hepatocellular carcinoma. Oncol Lett. 2019;17:1714–1722. doi: 10.3892/ol.2018.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Augello G, Emma MR, Cusimano A, Azzolina A, Mongiovì S, Puleio R, Cassata G, Gulino A, Belmonte B, Gramignoli R, et al. Targeting HSP90 with the small molecule inhibitor AUY922 (luminespib) as a treatment strategy against hepatocellular carcinoma. Int J Cancer. 2019;144:2613–2624. doi: 10.1002/ijc.31963. [DOI] [PubMed] [Google Scholar]
- 35.Pan W, Luo Q, Yan X, Yuan L, Yi H, Zhang L, Li B, Zhang Y, Sun J, Qiu MZ, Yang DJ. A novel SMAC mimetic APG-1387 exhibits dual antitumor effect on HBV-positive hepatocellular carcinoma with high expression of cIAP2 by inducing apoptosis and enhancing innate anti-tumor immunity. Biochem Pharmacol. 2018;154:127–135. doi: 10.1016/j.bcp.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Neumayer G, Belzil C, Gruss OJ, Nguyen MD. TPX2: Of spindle assembly, DNA damage response, and cancer. Cell Mol Life Sci. 2014;71:3027–3047. doi: 10.1007/s00018-014-1582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moss DK, Wilde A, Lane JD. Dynamic release of nuclear RanGTP triggers TPX2-dependent microtubule assembly during the apoptotic execution phase. J Cell Sci. 2009;122:644–655. doi: 10.1242/jcs.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Pan YZ, Cheung M, Cao M, Yu C, Chen L, Zhan L, He ZW, Sun CY. LAMB3 mediates apoptotic, proliferative, invasive, and metastatic behaviors in pancreatic cancer by regulating the PI3K/Akt signaling pathway. Cell Death Dis. 2019;10:230. doi: 10.1038/s41419-019-1320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, Cao FL, Qu LL, Wang ZM, Liu XY. MEG3 promotes liver cancer by activating PI3K/AKT pathway through regulating AP1G1. Eur Rev Med Pharmacol Sci. 2019;23:1459–1467. doi: 10.26355/eurrev_201902_17103. [DOI] [PubMed] [Google Scholar]
- 41.Cen D, Huang H, Yang L, Guo K, Zhang J. Long noncoding RNA STXBP5-AS1 inhibits cell proliferation, migration, and invasion through inhibiting the PI3K/AKT signaling pathway in gastric cancer cells. Onco Targets Ther. 2019;12:1929–1936. doi: 10.2147/OTT.S194463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yun WK, Hu YM, Zhao CB, Yu DY, Tang JB. HCP5 promotes colon cancer development by activating AP1G1 via PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2019;23:2786–2793. doi: 10.26355/eurrev_201904_17553. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Zhang Q, Wu Q, Cui Y, Zhu H, Fang M, Zhou X, Sun Z, Yu J. Interleukin-22 secreted by cancer-associated fibroblasts regulates the proliferation and metastasis of lung cancer cells via the PI3K-Akt-mTOR signaling pathway. Am J Transl Res. 2019;11:4077–4088. [PMC free article] [PubMed] [Google Scholar]
- 44.Chen T, Gu C, Xue C, Yang T, Zhong Y, Liu S, Nie Y, Yang H. LncRNA-uc002mbe.2 interacting with hnRNPA2B1 mediates AKT deactivation and p21 up-regulation induced by trichostatin in liver cancer cells. Front Pharmacol. 2017;8:669. doi: 10.3389/fphar.2017.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen T, Huang H, Zhou Y, Geng L, Shen T, Yin S, Zhou L, Zheng S. HJURP promotes hepatocellular carcinoma proliferation by destabilizing p21 via the MAPK/ERK1/2 and AKT/GSK3β signaling pathways. J Exp Clin Cancer Res. 2018;37:193. doi: 10.1186/s13046-018-0866-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Liu Y, Duan J, Yan H, Zhang J, Zhang H, Fan Q, Luo F, Yan G, Qiao K, Liu J. Hippocalcin-like 1 suppresses hepatocellular carcinoma progression by promoting p21(Waf/Cip1) stabilization by activating the ERK1/2-MAPK pathway. Hepatology. 2016;63:880–897. doi: 10.1002/hep.28395. [DOI] [PubMed] [Google Scholar]
- 47.Ohkoshi S, Yano M, Matsuda Y. Oncogenic role of p21 in hepatocarcinogenesis suggests a new treatment strategy. World J Gastroenterol. 2015;21:12150–12156. doi: 10.3748/wjg.v21.i42.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J, Ren Y, Zhang L, Kong X, Wang T, Shi Y, Bu R. Knocking-down of CREPT prohibits the progression of oral squamous cell carcinoma and suppresses cyclin D1 and c-Myc expression. PLoS One. 2017;12:e0174309. doi: 10.1371/journal.pone.0174309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Fang L, Zhang J, Li G, Ma M, Li C, Lyu J, Meng QH. Blockage of Glyoxalase I inhibits colorectal tumorigenesis and tumor growth via upregulation of STAT1, p53, and Bax and downregulation of c-Myc and Bcl-2. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18030570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.