Abstract
Significant biases of dinucleotide composition in many RNA viruses including influenza A virus have been reported in recent years. Previous studies have showed that a codon-usage-altered influenza mutant with elevated CpG usage is attenuated in mammalian in vitro and in vivo models. However, the relationship between dinucleotide preference and codon usage bias is not entirely clear and changes in dinucleotide usage of influenza virus during evolution at segment level are yet to be investigated. In this study, a Monte Carlo type method was applied to identify under-represented or over-represented dinucleotide motifs, among different segments and different groups, in influenza viral sequences. After excluding the potential biases caused by codon usage and amino acid sequences, CpG and UpA were found under-represented in all viral segments from all groups, whereas UpG and CpA were found over-represented. We further explored the temporal changes of usage of these dinucleotides. Our analyses revealed significant decrease of CpG frequency in Segments 1, 3, 4, and 5 in seasonal H1 virus after its re-emergence in humans in 1977. Such temporal variations were mainly contributed by the dinucleotide changes at the codon positions 3-1 and 2-3 where silent mutations played a major role. The depletions of CpG and UpA through silent mutations consequently led to over-representations of UpG and CpA. We also found that dinucleotide preference directly results in significant synonymous codon usage bias. Our study helps to provide details on understanding the evolutionary history of influenza virus and selection pressures that shape the virus genome.
Keywords: influenza, dinucleotide usage, codon usage, evolution
1. Introduction
Dinucleotide composition is commonly used as a genomic signature for differentiating various microbial and viral metagenomes, as this species-specific property captures the major variations between different organisms (Karlin and Burge 1995; Wang et al. 2005; Willner, Thurber, and Rohwer 2009). An unbiased dinucleotide composition is expected to match the frequency predicted from the nucleotide composition. Interestingly, frequencies of some dinucleotide motifs, such as CpG (C-phosphate-G) and UpA are extremely biased from the expected in many RNA viruses, such as influenza A virus (IAV) (Greenbaum et al. 2008; Cheng et al. 2013; Di Giallonardo et al. 2017). Such biased dinucleotide composition may be the consequence of two major factors, intrinsic characteristics of the virus and mutational pressure from the host. The intrinsic properties of the virus can be due to constraints of specific protein-coding capacity (i.e. amino acid sequence conservation) and/or significant codon usage/nucleotide composition bias of the viral genome. The amino acid sequence constraint, however, is unlikely to be the key factor driving the dinucleotide bias of IAV, as similar dinucleotide bias has been commonly found across different genes. Codon usage biases of IAV from different host origins can be very different. Previous work done by us and others have further demonstrated that different IAV subtypes have different selection pressures on their codon usage patterns (Greenbaum et al. 2008; Wong et al. 2010). Uneven usage of synonymous codons might potentially result in biased dinucleotide composition, but the relationship between synonymous codon preference and dinucleotide usage has remained largely unknown. It is possible that they are just two confounding variables in the influenza viral genome.
The mutational pressure from the host is suggested to have a significant role in selecting certain dinucleotides motifs, such as CpG (Greenbaum et al. 2008; Di Giallonardo et al. 2017). Debates on the underlying reasons concerning CpG suppression in RNA viruses have been going on for over thirty years (Shpaer and Mullins 1990; Karlin, Doerfler, and Cardon 1994; Atkinson et al. 2014; Tulloch et al. 2014; Gaunt et al. 2016; Fros et al. 2017). Early explanations focused on the cytosine DNA methylation of the host, especially for retroviruses (van der Kuyl and Berkhout 2012), but this hypothesis fails to fit for other RNA viruses which are free from DNA intermediates throughout their whole life cycles. In recent years, an increasing number of researchers have found evidence linking the dinucleotide usage bias in RNA viruses and the host immune responses triggered by these dinucleotide motifs. The dinucleotide frequency of CpG or UpA usage was found to significantly influence the replication ability of echovirus 7 and IAV (Atkinson et al. 2014; Gaunt et al. 2016). Modifying dinucleotide extremes in IAV resulted in changes of pathogenicity and host response to infection (Gaunt et al. 2016). It is also believed that the CpG and UpA abundance in such RNA viruses can modulate the host immune response through currently uncharacterized pattern recognition receptors (Greenbaum et al. 2008; Atkinson et al. 2014; Takata et al. 2017).
Conventional dinucleotide researches on IAV have been focused on the full-length whole genome rather than individual segments. To achieve a more accurate estimation of dinucleotide bias in each segment, we analyzed 159,028 influenza viral sequences in this study. We identified CpG/UpA and CpA/UpG highly under- and over-represented, respectively, in IAV genome. Our analysis suggested possible interactive linkage among these four dinucleotides. We also found that dinucleotide selection, such as CpG avoidance, greatly biased synonymous codon usage.
2. Materials and methods
2.1 Sequence data
Available full-length protein-coding region sequences of influenza virus were collected through Influenza Research Database (https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi, accessed 13 Sep 2017). The sequences were grouped according to their hosts and subtypes. For all the eight genes (PB2, PB1, PA, HA, NP, NA, M, and NS), sequences were allocated to five different groups, i.e. (1) human seasonal H1 viruses; (2) human seasonal H3 viruses; (3) human seasonal H2N2 viruses; (4) human pandemic H1N1(2009) viruses; (5) avian influenza virus. For the avian HA and NA sequences, we primarily focused only on subtypes that are relevant to seasonal H1 and H3 influenza viruses (i.e. avian H1, H3, N1, and N2). The downloaded raw data were in cDNA-sense FASTA format and they were filtered according to the following data cleaning processes: (1) filtering sequences without date information; (2) filtering sequences with unidentifiable nucleotides (other than ‘a, t, c, g’); and (3) filtering sequences with unusual length (>110 or <90% of the expected length). We also excluded the human seasonal H1 viruses isolated in 1976, as it is believed that these viruses are likely to be of swine influenza origin (Zimmer and Burke 2009). The final dataset comprised a total of 159,028 protein-coding sequence regions.
2.2 Dinucleotide measurement
A conventional way to evaluate the dinucleotide patterns is to use the dinucleotide odds ratio (Burge, Campbell, and Karlin 1992; Karlin and Burge 1995; Karlin and Mrázek 1997; Willner, Thurber, and Rohwer 2009; Cheng et al. 2013). The dinucleotide odds ratio can be deduced from the ratio of the observed and expected dinucleotide usage, where the expected dinucleotide usage assumes random combinations of nucleotides. This approach assumes that the forming of dinucleotides is a random process. However, the dinucleotide usage can be biased by the amino acid sequences encoded by these genes and the codon usage bias of these genes. To adjust for these potential biases, we applied a Monte Carlo type method in our study to evaluate the level of representation of different dinucleotides, similar to the method described in a previous study (Greenbaum et al. 2008). The Monte Carlo procedure reshuffles the synonymous codons in a gene, while keeping the codon usage bias and the gene’s amino acid structure unchanged (see Supplementary Methods for further explanation). The occurrences of dinucleotides formed at the codon-codon junctions in both the observed sequences and the simulated sequences were compared. Focusing on this subset of the total dinucleotides in these sequences can eliminate the effects from gene function maintained by conserved amino acid structure, or the codon usage preference, and thus reflect the genuine dinucleotide usage bias caused by host adaption.
For every collected sequence in our study, 1,000 randomized equivalent Monte Carlo sequences were generated to evaluate the expected sequence space. P-values are calculated as the proportion of times that the dinucleotide count in simulated sequences is greater/less than the count in the real sequence, for each possible dinucleotide combination, to evaluate the level of over-representation/under-representation. Besides P-value, the index is proposed to quantify the degree of over- and under-representation of different dinucleotides for every real sequence. compares the observed dinucleotide usage in real sequences with the expected (average) dinucleotide usage from Monte Carlo sequences.
2.3 Time series and consensus sequences
The time series of the dinucleotide odds ratio was plotted for different groups of viruses to explore the dynamic variations. The yearly consensus sequences of different segments from a specific virus group were deduced from the ‘Seqinr’ (Charif and Lobry 2007) package in R and the sequence alignment was done by MAFFT (Katoh and Standley 2013). The single nucleotide polymorphisms (SNPs) and their related dinucleotide usage dynamics were identified by comparing the consensus sequences at two consecutive time points. Dinucleotide frequency was calculated as the number of occurrences of specific dinucleotide in a genomic sequence divided by the nucleic acid length of the sequence.
2.4 Statistical analysis
The association between time and dinucleotide usage was tested by Pearson’s product moment correlation. The global validation of linear models assumptions were performed by ‘gvlma’ (Peña and Slate, 2006) package in R. The Kruskal−Wallis rank sum test was applied in our study to estimate whether the odds ratios of different dinucleotide motifs have similar distribution. This test is a non-parametric equivalent to the one-way analysis of variance.
The paired Mann−Whitney U test was performed to measure the difference between dinucleotide pairs. This test is also known as ‘two-sample Wilcoxon signed test’ which is a nonparametric test used to determine whether two dependent samples were selected from populations having the same distribution.
3. Results
3.1 CpG and UpA were found substantially under-represented among segments and subtypes, whereas UpG and CpA were found over-represented
The IAV sequences are classified by their hosts and subtypes, of which eight viral segments were analyzed individually. We first estimated the relative abundance of all sixteen dinucleotide motifs in the studied sequences using the above-mentioned Monte Carlo type method (see Section 2 and Supplementary Methods).
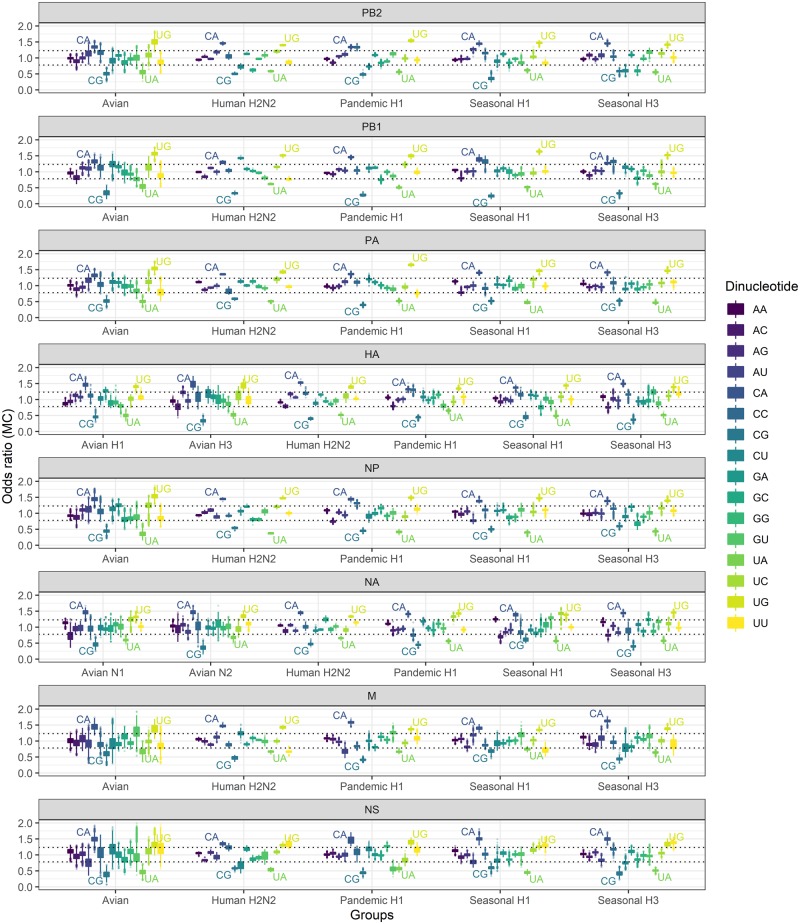
By comparing the dinucleotide composition from the observed sequences to their randomized equivalents, we obtained the Monte Carlo odds ratios of these dinucleotide sequences for each sequence. Distributions of the odds ratios are shown in the boxplot, separated by genome segments and virus groups into forty-two sets (Fig. 1). Following the statistical theory and data experience from Karlin (Karlin and Cardon 1994; Karlin and Burge 1995), an odds ratio >1.23 is considered to be very high to extremely high, whereas an odds ratio below 0.78 is considered to be very low to extremely low. The dotted lines in Fig. 1 represent these conventional thresholds for crude determination of over- or under-representation. Yet the exact significance of each dinucleotide composition was evaluated by the P-values using the Monte Carlo estimation (see Supplementary Table S1).
Figure 1.
Dinucleotide Monte Carlo odds ratios in influenza A viruses in boxplot. The odds ratios were calculated for all the sixteen dinucleotide motifs by the order as shown in the legend. The odds ratios over 1.23 or below 0.78 were considered significantly over-represented or under-represented (dotted lines), respectively. The lower and upper hinges in the boxplot correspond to the first and third quartiles of the data, the upper/lower whisker extends from the hinge to the largest/smallest value no further than 1.5×IQR from the hinge. Data beyond the end of the whiskers are plotted individually.
The usages of different dinucleotide motifs vary greatly. Large variations can be observed in all the studied sequences, irrespective of the origin of viral segment or virus subtype (P < 0.001 for all forty-two sequence datasets, Kruskal−Wallis rank sum test). Although the odds ratios/usages of different dinucleotides are distinct from each other, the usages of certain individual dinucleotides are relatively similar across different segments or different subtypes. For example, the CpG dinucleotide always has an extremely low usage regardless of the subtype or segment that the sequence belongs to. We also observed that the dinucleotide usage biases are highly determined by the nature of viral segment, rather than the host origin or subtype of these viruses (Fig. 1). This observation is further supported by the correspondence analysis (Wong et al. 2010) where the boundaries of the sequence space become clearer when separated by segments (see Supplementary Fig. S1), confirming that the ancestor sequences of these genes have pronounced founder effects on viral sequence features.
Amongst all forty-two different dinucleotide sequence sets, four dinucleotides were found to have highly biased usage in all analyses (CpG, UpA, UpG, and CpA; P < 0.05 for all forty-two sequence sets). The CpG and UpA dinucleotides were significantly under-represented, whereas the UpG and CpA dinucleotides were over-represented in all cases. The mean P-value of GpA was also found to be <0.05 in eleven sequence sets, but no other dinucleotide has extensive significant usage pattern (see Supplementary Fig. S2). One should note that the Monte Carlo method excluded the potential bias from the codon usage bias or the conserved amino acid coding capacity.
3.2 CpG decreases in Segments 1, 3, 4, and 5 after 1977 in human seasonal H1 virus, and CpG usage of avian and human virus in P (polymerase) genes and M gene varies
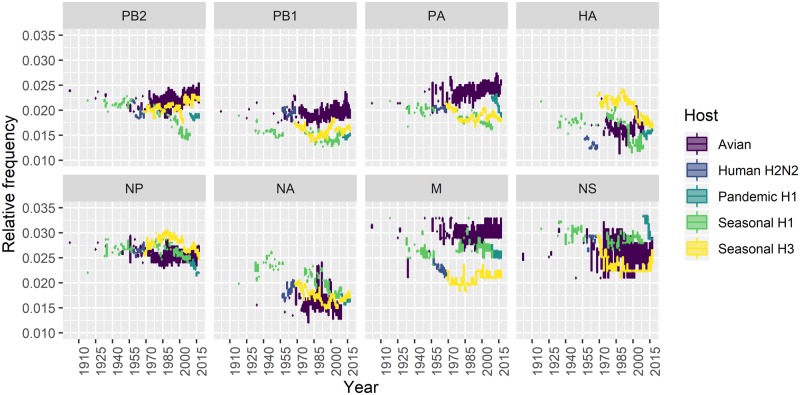
We next investigated the temporal dynamics of the four dinucleotide motifs that have extreme high or low frequencies. CpG is the most well-known under-represented dinucleotide that has been studied for a long time, while the evolution pattern of UpA, CpA, or UpG has been less well studied. We plotted the distribution of the CpG relative frequency in time series between 1908 and 2017, by different segments and different groups of sequences (Fig. 2). We observed different patterns of CpG dynamics between segments and found remarkable divergence between virus groups.
Figure 2.
Evolution of the relative frequency of CpG dinucleotide in influenza A virus by segment in boxplot.
For human seasonal H1 which circulated between 1918−57 and 1977−2009, we observed that the CpG frequencies in these two periods were different. The CpG frequency of seasonal H1 virus remained rather stable for all eight segments during the first period, however, its usage in Segments 1, 3, 4, 5, 6, and 7 (i.e. PB2, PA, HA, NP, NA, and M) dropped significantly during the second period (P < 0.01, Pearson’s correlation test, outliers exceed 1.5 ×IQR dropped). The negative associations were strong (correlation coefficient from -0.9 to -0.5) in Segments 1, 3, 4, and 5. Further linear regression analysis suggested a strong linear relationship between CpG usage and time in Segment 1 (adjusted R2: 0.7198; Supplementary Fig. S3). Although the data for Segments 3, 4, and 5 also showed observable negative linear association, they failed to meet the assumptions of the linear model mainly due to unequal variance. Such negative associations were also detected in Segments 4 and 5 of human seasonal H3 virus (correlation coefficient =-0.79 and -0.68), however, no significant declining trend was observed on the other six segments. On the contrary, Segments 1 and 5 even showed moderate to strong positive correlations between CpG usage and time (correlation coefficient = 0.56 and 0.40). No major trend could be identified in human seasonal H2 and human pandemic H1N1 viruses, due to the relatively short periods of circulation in humans.
Although there was no consistent increase or decrease of UpA, CpA, or UpG in the entire seasonal H1 viral genome (data not shown), noticeable changes of these dinucleotide motifs were detected only in some specific segments (e.g. increase of CpA in H1N1 NA segment, correlation coefficient = 0.88, P < 0.01 and decrease of CpA in H1N1 PB1 segment, correlation coefficient = -0.77, P < 0.01; Supplementary Fig. S4). These observations suggest that there are segment-specific selection pressures against or for certain dinucleotide motifs in some human influenza virus subtypes.
We also observed that human and avian influenza viruses might have remarkable differences in viral segments in terms of dinucleotide relative frequencies (see Supplementary Fig. S4). For example, human influenza viruses have lower CpG relative frequency than avian influenza virus in P genes (PB2, PB1, and PA) and M gene, but they have higher UpA relative frequency in P, NP, and M genes than those of avian viruses. No obvious difference between human and avian was found for CpA and UpG.
3.3 CpG reduction of H1 virus took place at the 3-1 and 2-3 positions
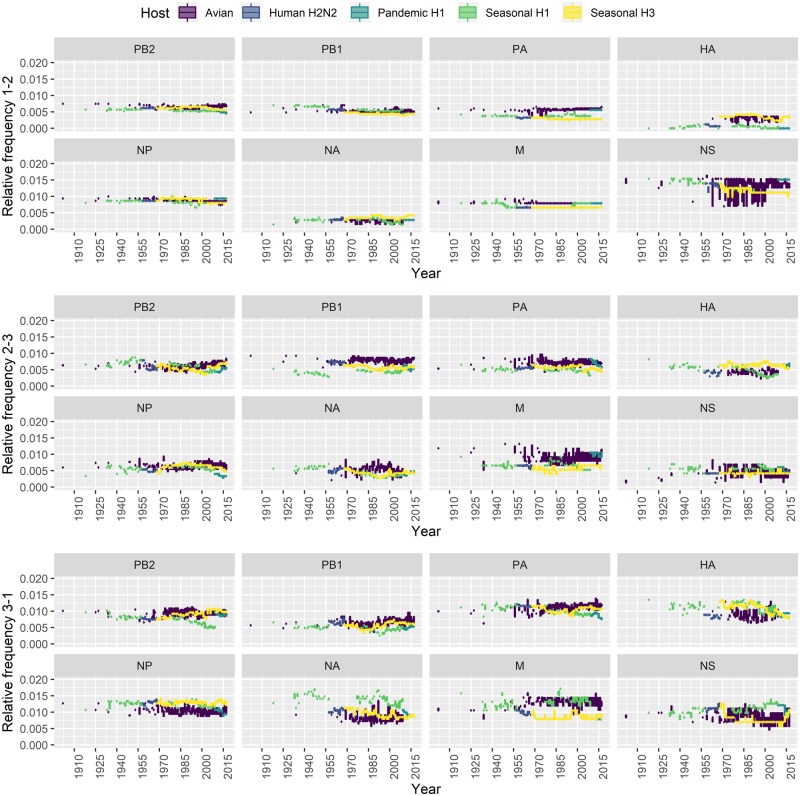
Dinucleotides were characterized into three categories according to their positions in the codons. Dinucleotide12 is the motif formed by the first two bases of a codon, dinucleotide23 consists of the last two bases of a codon, and dinucleotide31 is the motif at the codon−codon junctions.
We investigated the evolution dynamics of CpG in seasonal H1 virus at different codon positions and found that the decrease of CpG usage occurred primarily at the 3-1 position and secondarily at the 2-3 position, while the CpG frequency of dinucleotide12 was stable over time in all segments (Fig. 3). For example, the CpG frequency of dinucleotide31 in Segment 1 of H1 virus dropped from 0.75 to 0.5 per cent, and the CpG frequency of dinucleotide23 decreased from ∼0.65 to 0.5 per cent. Comparable patterns were also found in Segments 4 and 6. Interestingly, the reduced CpG frequency in Segments 1, 4, and 6 at the 3-1 position coincided with the increase of UpG at similar scale (see Supplementary Fig. S5c), which prompted us to further explore the interactions between the four dinucleotide extremes.
Figure 3.
Evolution of CpG frequency at different codon positions (top: 1-2; middle: 2-3; bottom: 3-1).
3.4 The depletion of CpG dinucleotide through silent mutations in H1 evolution directly led to over-representation of UpG and CpA
Given that the frequency of CpG dinucleotide in human H1 virus decreased over years, we continued to examine the outcome of this continuous change in this human subtype. Annual consensus on gene sequences of human H1 virus after 1977 were generated to study the point mutations responsible for elimination of CpG dinucleotide in evolution. Phylogeny of these consensus sequences from different groups were also analyzed for comparison between the consecutive yearly consensuses (see Supplementary Fig. S6).
Comparisons between the consensus sequences of two consecutive years suggested CpG is more likely to mutate to UpG and CpA. In human H1 virus, 67.2 per cent of the point mutations existed at the third bases of the codons, and 75.4 per cent of the CpG mutations (e.g. CpG to other dinucleotides) involved the third bases (data not shown). We found that a large proportion of CpG dinucleotides mutated to UpG (171, 39.3%) and CpA (164, 37.7%), totally accounting for nearly 80 per cent of the CpG substitutes (Table 1). Totally, 155 (90.6%) of the CpG to UpG mutations were mutations at the third bases of the codons, i.e. CpG to UpG at the 3-1 position. In addition, 65.9 per cent of the CpG to CpA mutations were at the 2-3 position. These results were in line with our above result that CpG reduction took place at 3-1 positions.
Table 1.
The substitutes of CpG in human H1N1 virus evolution.
| Mutant | UpG (39.3%) | CpA (37.7%) | ApG (17.0%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP position | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 |
| Count | 9 | 7 | 155 | 41 | 15 | 108 | 28 | 3 | 43 |
We also investigated the mutations of UpA in human H1N1, which is another universally under-represented dinucleotide motif. Similarly, UpA were more likely mutated to UpG (264, 41.6%) or CpA (191, 30.1%) (Table 2), mainly U2pA3 to U2pG3 and U3pA1 to C3pA1. The mutations of CpG and UpA in evolution explain UpG and CpA over-representation.
Table 2.
The substitutes of UpA in human H1N1 virus evolution.
| Mutant | UpG (41.6%) | CpA (30.1%) | UpC (9.0%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP position | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 |
| Count | 82 | 3 | 179 | 13 | 20 | 158 | 15 | 3 | 39 |
3.5 The dinucleotide preference directly resulted in significant synonymous codon usage bias
We have discovered that the point mutations at the third bases of the codons played a role in forming dinucleotide usage bias. These mutations could probably be the consequence of a strong selection of dinucleotide preference. Besides dinucleotide preference, codon usage bias was also shown to exert pressure on the virus evolution. However, the relationship between dinucleotide preference and codon usage bias in influenza virus evolution is yet to be fully determined. If the dinucleotide selection is independent of codon usage bias, one would expect that the dinucleotide usage bias existed not only at the position 2-3 of a codon but also at the position 3-1 between codons, which has been partially proven by the above results. In order to examine how the dinucleotide preference affects the synonymous codon usage in IAV, we performed the following analysis. We selected twelve codons coding for six amino acids (Phenylalanine, Tyrosine, Histidine, Asparagine, Aspartic acid, and Cysteine) in this analysis. These codons are all synonymous pairs with different nucleotides at the third base of the codon (C/U). The C/U ratio at the third base indicated the synonymous codon usage bias. Meanwhile, this ratio can also be affected by the dinucleotide preference at the 3-1 positions. Therefore, by investigating whether these C/U frequencies were significantly affected by the dinucleotide 3-1, we can estimate the influence from dinucleotide usage preference to codon usage bias.
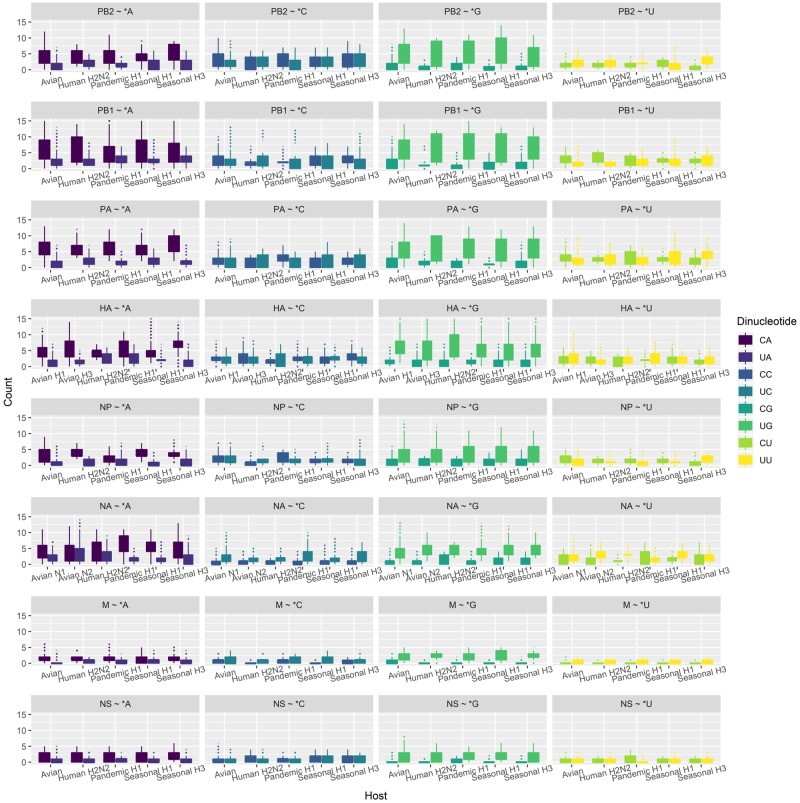
For every virus group in every segment, we plotted the counts of the eight possible dinucleotides (CpA, CpC, CpG, CpU, UpA, UpC, UpG, and UpU) in every sequence (Fig. 4), and Mann−Whitney test was performed to check the significance of the difference between four dinucleotide pairs (CpA/UpA, CpC/UpC, CpG/UpG, and CpU/UpU). The difference is considered significant if: (1) the P-value <0.01 in the Mann−Whitney test and (2) the difference between the mean values is greater than one (to control minor but statistically significant difference).
Figure 4.
The 3-1 dinucleotide composition of the U/C-ended synonymous codons in boxplot. UpA and CpG are much less represented than their corresponding counterparts, CpA and UpG, respectively.
Our results showed that the CpG/UpG ratio was significantly biased in all forty-two groups of sequences, and CpA/UpA was significantly different in thirty-five of forty-two groups. In contrast, the difference of CpC/UpC and CpU/UpU pairs were only found to be significant in nine and six groups, receptively (Table 3). We found that when the second codon in two serial codons starts with a G nucleotide at the first base, the third base of the preceding codon is selected towards U rather than C, resulting in a CpG under-representation and UpG over-representation (Fig. 4). This significant effect on codon usage bias from dinucleotide selection was also observed for UpA under-representation and CpA over-representation. Conversely, when CpG or UpA is not involved, these U/C-ended synonymous codon usage biases disappear. We also conducted a similar study on amino acids that have and only have four degenerated codons ended with A, C, G, or U (Ala, Gly, Pro, Thr, and Val) and obtained the same conclusion (Supplementary Fig. S7). These results suggest that the dinucleotide preference we identified has direct effect on the codon usage bias and dinucleotide preference poses a significant selection pressure on IAV in evolution.
Table 3.
The mean 3-1 dinucleotide occurrence of the U/C-ended synonymous codons.a
| Segment | Host | *pA |
*pC |
*pG |
*pU |
||||
|---|---|---|---|---|---|---|---|---|---|
| CpA | UpA | CpC | UpC | CpG | UpG | CpU | UpU | ||
| PB2 | Avian | 4.17 | 1.56 | 2.84 | 2.22 | 1.39 | 4.42 | 1.63 | 1.8 |
| PB2 | Human H2N2 | 3.98 | 1.85 | 2.26 | 2.89 | 0.78 | 4.9 | 1.26 | 2.07 |
| PB2 | Pandemic H1 | 4.69 | 1.22 | 2.98 | 2.18 | 1.11 | 4.66 | 1.51 | 1.83 |
| PB2 | Seasonal H1 | 3.97 | 1.77 | 2.39 | 2.57 | 0.49 | 5.51 | 1.83 | 1.7 |
| PB2 | Seasonal H3 | 4.64 | 1.51 | 2.5 | 2.55 | 1.17 | 4.32 | 0.74 | 2.59 |
| PB1 | Avian | 6.61 | 2.36 | 2.51 | 2.81 | 1.64 | 6.16 | 2.66 | 1.71 |
| PB1 | Human H2N2 | 6.57 | 2.5 | 2.2 | 2.99 | 1.76 | 6.06 | 2.98 | 1.36 |
| PB1 | Pandemic H1 | 6.54 | 2.8 | 2.26 | 2.92 | 1.03 | 6.63 | 2.4 | 2.31 |
| PB1 | Seasonal H1 | 6.46 | 2.98 | 2.58 | 2.3 | 0.85 | 7.02 | 2.47 | 2.68 |
| PB1 | Seasonal H3 | 6.41 | 2.84 | 2.63 | 2.4 | 1.56 | 5.95 | 2.52 | 2.21 |
| PA | Avian | 6.19 | 1.54 | 2.35 | 1.98 | 1.53 | 6.1 | 3.27 | 2.44 |
| PA | Human H2N2 | 6.19 | 2.16 | 1.57 | 2.35 | 1.49 | 6.54 | 2.83 | 3.16 |
| PA | Pandemic H1 | 6.01 | 1.59 | 3.09 | 1.58 | 1.12 | 6.87 | 3 | 2.5 |
| PA | Seasonal H1 | 6.14 | 1.97 | 1.48 | 2.49 | 1.02 | 6.57 | 2.73 | 3.57 |
| PA | Seasonal H3 | 6.78 | 1.91 | 1.99 | 1.69 | 1.69 | 5.91 | 2.01 | 4.08 |
| HA | Avian H1 | 4.65 | 1.49 | 2.58 | 2.37 | 1.58 | 6.54 | 2.38 | 2.46 |
| HA | Avian H3 | 6.06 | 1.33 | 2.83 | 2.42 | 1.47 | 6.6 | 2.1 | 1.43 |
| HA | Human H2N2 | 4.45 | 2.23 | 1.88 | 2.37 | 1.58 | 6.45 | 1.53 | 1.8 |
| HA | Pandemic H1 | 5 | 2.21 | 2.53 | 2.41 | 1.43 | 6.02 | 2.02 | 2.32 |
| HA | Seasonal H1 | 4.68 | 1.98 | 2.28 | 2.57 | 1.24 | 6.79 | 2.34 | 1.8 |
| HA | Seasonal H3 | 6.4 | 1.49 | 3.33 | 1.91 | 1.56 | 6.37 | 1.3 | 1.84 |
| NP | Avian | 2.96 | 0.77 | 2.01 | 1.65 | 1.37 | 3.88 | 1.66 | 1 |
| NP | Human H2N2 | 3.84 | 0.78 | 1.03 | 2.12 | 1.4 | 3.96 | 1.31 | 1.19 |
| NP | Pandemic H1 | 2.49 | 1.39 | 2.08 | 1.25 | 0.83 | 4.61 | 1.67 | 1.06 |
| NP | Seasonal H1 | 3.74 | 0.54 | 1.33 | 1.83 | 1.16 | 4.35 | 1.37 | 1.19 |
| NP | Seasonal H3 | 3.37 | 0.98 | 1.61 | 1.66 | 1.39 | 3.93 | 0.92 | 1.79 |
| NA | Avian N1 | 4.22 | 2.07 | 0.6 | 2.15 | 0.88 | 4.6 | 1.65 | 2.16 |
| NA | Avian N2 | 4.16 | 3.04 | 0.74 | 1.53 | 0.86 | 4.95 | 1.41 | 2.79 |
| NA | Human H2N2 | 4.35 | 2.87 | 0.61 | 1.9 | 1.37 | 4.59 | 1.23 | 3.02 |
| NA | Pandemic H1 | 5.15 | 1.68 | 0.54 | 2.55 | 0.81 | 4.84 | 2.46 | 1.51 |
| NA | Seasonal H1 | 5.33 | 1.86 | 0.74 | 2 | 1.06 | 4.28 | 1.46 | 2.71 |
| NA | Seasonal H3 | 4.4 | 2.27 | 0.72 | 2.34 | 1.1 | 4.84 | 1.43 | 2.03 |
| M | Avian | 1.75 | 0.26 | 0.61 | 0.76 | 0.33 | 1.98 | 0.16 | 0.36 |
| M | Human H2N2 | 1.43 | 0.74 | 0.17 | 0.97 | 0.18 | 2.14 | 0.18 | 0.32 |
| M | Pandemic H1 | 1.82 | 0.31 | 0.67 | 0.8 | 0.17 | 2.07 | 0 | 0.5 |
| M | Seasonal H1 | 1.12 | 0.92 | 0.18 | 1.14 | 0.24 | 2.09 | 0.16 | 0.34 |
| M | Seasonal H3 | 1.51 | 0.66 | 0.28 | 0.89 | 0.05 | 2.28 | 0.1 | 0.4 |
| NS | Avian | 1.41 | 0.71 | 0.77 | 0.69 | 0.28 | 1.77 | 0.57 | 0.34 |
| NS | Human H2N2 | 1.42 | 0.72 | 1 | 0.6 | 0.19 | 1.54 | 0.34 | 0.56 |
| NS | Pandemic H1 | 1.7 | 0.73 | 0.71 | 0.56 | 0.33 | 1.67 | 0.74 | 0.17 |
| NS | Seasonal H1 | 1.47 | 0.62 | 0.95 | 0.9 | 0.17 | 1.81 | 0.37 | 0.48 |
| NS | Seasonal H3 | 1.69 | 0.65 | 0.74 | 0.82 | 0.17 | 1.59 | 0.34 | 0.5 |
The dinucleotide pairs with significant difference were shown in bold (P < 0.05 in Mann−Whitney U test and, difference of mean values >1).
4. Discussion
The dinucleotide composition of IAV genome has been known to be bias for many years, still this bias and the consequence of such phenomenon are yet to be fully understood. The goal of this study was to examine the dinucleotide bias by different segments and virus groups with an up-to-date sequence database. In addition, this study traced the mutations of specific dinucleotide motifs in order to identify any specific mutation patterns and determine the consequences of such mutational biases in the influenza virus evolution.
For each studied real sequence, we generated 1,000 simulated sequences for analysis. Unlike tradition methods, our Monte Carlo simulated sequences were synonymous alternatives of studied sequences. This was achieved by reshuffling the codons of each real sequence. This approach can identify dinucleotides with an extreme high- or low-frequency, while maintaining the codon bias and amino acid constraints of the studied sequences. Thus, the simulated sequences would be in a sequence space that is more relevant to the actual studied sequences. In contrast, previous traditional dinucleotide studies of IAV did not consider the above sequence constrains at amino acid level. Most of these previous studies simply used the deduced nucleotide composition of influenza viral genome to generate simulated sequences. We therefore reason that our simulated sequences would provide a better reference dataset for studying the evolution of influenza virus.
The dinucleotide usage varied greatly between segments, while the overall pattern of the same segment was largely conserved across viruses from different host origins. This result extends the previous observation in animal RNA viruses that the family which a virus belongs to has stronger impacts on the dinucleotide composition than those from its corresponding host (Di Giallonardo et al. 2017).
Our data confirmed four dinucleotide extremes among all eight segments in different subtypes of IAV. These are CpG, UpA, UpG, and CpA, the first two of which were found under-represented and the latter two were found over-represented. Similar observation could also be observed in avian HA H2 sequences (Supplementary Fig. S8). This result is in line with the previous reports using limited data or conventional measurements (Greenbaum et al. 2008; Atkinson et al. 2014; Gaunt et al. 2016). Among these four dinucleotides, CpG was found to be the most biased one (with most extreme Monte Carlo odds ratios), there were also significant evolutionary directions and noticeable differences between human and avian samples found in CpG frequency, but not for UpG or CpA. Therefore, CpG was applied as a proxy for further studying the details in dinucleotide dynamics in our study.
The dinucleotide frequency of CpG has continued to evolve in human H1 virus for decades. In addition to the previous findings that human influenza A H1N1 virus evolved in a direction selected to reduce the CpG frequency (Greenbaum et al. 2008), we discovered the CpG depletion of human seasonal H1 virus mainly occurred in Segments 1, 3, 4, and 5 after 1977. Although the CpG frequency was already universally low in all segments, a significant temporal decrease of CpG frequency was found in Segments 1, 3, 4, 5, 6, and 7. In contrast, Segments 2 and 8 showed no significant increasing or decreasing trend over time. The discrepancy between the changes in CpG usage of different segments cannot be explained by the differences in the function or location of the proteins, nor the differences in mRNA expression level (Hatada et al. 1989; Russell, Trapnell, and Bloom 2018). HA and NA are two antigenic proteins that evolve much more rapidly than other influenza proteins (Lyons and Lauring 2018), but the NA segment of human H1N1 surprisingly did not have a high reduction of CpG frequency as we observed in the PB2, PA or NP segment (see Supplementary Fig. S3). This suggested that there are other mechanisms contributed to the unequal evolution rate in different genes.
The 3-1 and 2-3 positions played a pivotal role in temporal CpG depletion in seasonal H1 virus (Fig. 3). The CpG frequencies at positions 1-2 and 2-3 are lower than the frequency at position 3-1. This may be because position 3-1 allows more variations as the dinucleotide motif at this position is less restricted by the amino acid constraint. The higher relative frequency of CpG at position 3-1 may in turn contribute to its more obvious decrease during the studied period. Comparison between consecutive yearly consensus sequences reveal the outcome of CpG depletion. We found a large proportion of CpG mutated to UpG and CpA via silent mutations at the third base of the codons. Similarly, UpA is also significantly under-represented in IAV, and its mutations mainly led to UpG or CpA substitutions. It is worth noting that the most common mutational substitutions of CpG and UpA, i.e. UpG and CpA, were exactly the two over-represented dinucleotides in IAV. It is likely that the over-representation of UpG and CpA in IAV may be a by-product during evolution that selected against CpG and UpA, or at least, the level of over-representation was amplified by CpG and UpA under-representation. We hypothesize that there are close relationships among these four dinucleotide extremes, mainly driven by the CpG and UpA avoidance in evolution. This hypothesis is further supported by recent findings that CpG/UpA dinucleotide frequencies are the key determinants of host immune response to virus infection (Jimenez-Baranda et al. 2011; Atkinson et al. 2014; Gaunt et al. 2016), which can be a strong selective pressure during virus evolution. In this setting, the depletion of CpG and UpA in evolution serves as a trigger in forming biased dinucleotide composition, taking synonymous mutations as a proxy, directly resulting in UpG and CpA over-representation.
The interplay between dinucleotide preference and codon usage bias was also estimated in our study. Codon usage bias was found evident in influenza virus (Wong et al. 2010), but how it interacts with dinucleotide bias remained unclear. The dinucleotide bias at codon positions 3-1 may have weaker impacts on codon usage bias in RNA virus than codon positions 2-3 (Belalov and Lukashev 2013), however, we observed preference on 3-1 dinucleotides that can directly result in significant synonymous codon usage bias. We found that the preference of U/C-ended synonymous codons was greatly affected by the 3-1 dinucleotides, in other words, it is the newly formed 3-1 dinucleotides that determine the fitness of the U/C-ended synonymous codons. The third base in a codon with an adenine and/or guanine at this position significantly affected the U/C preference of the first base of preceding codon, ensuring the over- and under-representation of CpA/UpG and UpA/CpG. Accordingly, this bias was merely for codons that start with cytosine and uracil, as CpC/UpC/CpU/UpU were not under strong selection pressure (Table 3).
Understanding the origin and basis of biased dinucleotide composition in IAV is essential to recognize its genetic variation during the long period of evolution. Frequencies of specific dinucleotides can potentially be targets for pattern recognition receptors that trigger the host immune response. Our results provide insights regarding forming of dinucleotide extremes and their impacts on codon usage bias. These findings add information to the existing knowledge of the IAV genomic signature and help understand the evolutionary history of IAV. Such observations, when coupled with future experimental analysis, can elucidate the selection pressures that shape the virus genome.
Data availability
The accession numbers of the dataset are provided at https://github.com/Koohoko/Dinucleotide-Evolutionary-Dynamics-in-Influenza-A-Virus.
Funding
This project is supported by RGC General Research Fund (Project No. 17112117) and Research Grant Council of Hong Kong (Project No. T11-705/14N). L.L.M.P. is supported by Croucher Foundation.
Conflict of interest: None declared.
Supplementary Material
References
- Atkinson N. J. et al. (2014) ‘The Influence of CpG and UpA Dinucleotide Frequencies on RNA Virus Replication and Characterization of the Innate Cellular Pathways Underlying Virus Attenuation and Enhanced Replication’, Nucleic Acids Research, 42: 4527–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belalov I. S., Lukashev A. N. (2013) ‘Causes and Implications of Codon Usage Bias in RNA Viruses’, PLoS One, 8: e56642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Campbell A. M., Karlin S. (1992) ‘Over- and Under-representation of Short Oligonucleotides in DNA Sequences’, Proceedings of the National Academy of Sciences of the United States of America, 89: 1358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D., Lobry J. R. (2007) ‘SeqinR 1.0-2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis’, in Bastolla Uet al. (eds) Structural Approaches to Sequence Evolution: Molecules, Networks, Populations, pp. 207–32. New York: Springer. [Google Scholar]
- Cheng X. et al. (2013) ‘CpG Usage in RNA Viruses: Data and Hypotheses’, PLoS One, 8: e74109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fros J. J. et al. (2017) ‘CpG and UpA Dinucleotides in Both Coding and Non-coding Regions of Echovirus 7 Inhibit Replication Initiation Post-Entry’, Elife, 6: e12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E. et al. (2016) ‘Elevation of CpG Frequencies in Influenza a Genome Attenuates Pathogenicity but Enhances Host Response to Infection’, Elife, 5: e12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giallonardo F. et al. (2017) ‘Dinucleotide Composition in Animal RNA Viruses Is Shaped More by Virus Family than by Host Species’, Journal of Virology, 91: e02381–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum B. D. et al. (2008) ‘Patterns of Evolution and Host Gene Mimicry in Influenza and Other RNA Viruses’, PLoS Pathogens, 4: e1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada E. et al. (1989) ‘Control of Influenza Virus Gene Expression: Quantitative Analysis of Each Viral RNA Species in Infected Cells’, The Journal of Biochemistry, 105: 537–46. [DOI] [PubMed] [Google Scholar]
- Jimenez-Baranda S. et al. (2011) ‘Oligonucleotide Motifs That Disappear during the Evolution of Influenza Virus in Humans Increase Alpha Interferon Secretion by Plasmacytoid Dendritic Cells’, Journal of Virology, 85: 3893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Burge C. (1995) ‘Dinucleotide Relative Abundance Extremes: A Genomic Signature’, Trends in Genetics, 11: 283–90. [DOI] [PubMed] [Google Scholar]
- Karlin S., Cardon L. R. (1994) ‘Computational DNA Sequence Analysis’, Annual Review of Microbiology, 48: 619–54. [DOI] [PubMed] [Google Scholar]
- Karlin S., Doerfler W., Cardon L. R. (1994) ‘Why Is CpG Suppressed in the Genomes of Virtually All Small Eukaryotic Viruses but Not in Those of Large Eukaryotic Viruses?’ Journal of Virology, 68: 2889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Mrázek J. (1997) ‘Compositional Differences Within and Between Eukaryotic Genomes’, Proceedings of the National Academy of Sciences of the United States of America, 94: 10227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kuyl A. C., Berkhout B. (2012) ‘The Biased Nucleotide Composition of the HIV Genome: A Constant Factor in a Highly Variable Virus’, Retrovirology, 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons D., Lauring A. (2018) ‘Mutation and Epistasis in Influenza Virus Evolution’, Viruses, 10: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña E. A., and Slate E. H. (2006) ‘Global Validation of Linear Model Assumptions’, Journal of the American Statistical Association, 101: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B., Trapnell C., Bloom J. D. (2018) ‘Extreme Heterogeneity of Influenza Virus Infection in Single Cells’, Elife, 7: e32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpaer E. G., Mullins J. I. (1990) ‘Selection Against CpG Dinucleotides in Lentiviral Genes: A Possible Role of Methylation in Regulation of Viral Expression’, Nucleic Acids Research, 18: 5793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M. A. et al. (2017) ‘CG Dinucleotide Suppression Enables Antiviral Defence Targeting Non-Self RNA’, Nature, 550: 124–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch F. et al. (2014) ‘RNA Virus Attenuation by Codon Pair Deoptimisation Is an Artefact of Increases in CpG/UpA Dinucleotide Frequencies’, Elife, 3: e04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2005) ‘The Spectrum of Genomic Signatures: From Dinucleotides to Chaos Game Representation’, Gene, 346: 173–85. [DOI] [PubMed] [Google Scholar]
- Willner D., Thurber R. V., Rohwer F. (2009) ‘Metagenomic Signatures of 86 Microbial and Viral Metagenomes’, Environmental Microbiology, 11: 1752–66. [DOI] [PubMed] [Google Scholar]
- Wong E. H. et al. (2010) ‘Codon Usage Bias and the Evolution of Influenza a Viruses. Codon Usage Biases of Influenza Virus’, BMC Evolutionary Biology, 10: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer S. M., Burke D. S. (2009) ‘Historical Perspective—Emergence of Influenza A (H1N1) Viruses’, The New England Journal of Medicine, 361: 279–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers of the dataset are provided at https://github.com/Koohoko/Dinucleotide-Evolutionary-Dynamics-in-Influenza-A-Virus.