Abstract
Background.
Due to the shortage of organs for transplantation, procurement of kidneys from extended criteria donors (ECD) is inevitable. Frequently, donors infected with hepatitis C virus (HCV) are used. To elucidate an initial compromise of molecular pathways in HCV graft, gene expression (GE) profiles were evaluated.
Methods.
Twenty-four donor allograft biopsies (N=12 HCV (+) and N=12 HCV (–)) were collected at pre-implantation time and profiled using microarrays. Donors were age-race-gender, cold ischemia and warm ischemia time matched between groups. Probe level data were read into the R programming environment using the affy Bioconductor package and the robust multiarray average method was used to obtain probe set expression summaries. To identify probe sets exhibiting differential expression, a two sample t-test was performed. Molecular and biological functions were analyzed using Interaction Networks and Functional Analysis.
Results.
Fifty-eight probe sets were differentially expressed between HCV (+) vs. HCV (–) donors (p<0.001). The molecular functions associated with the two top scored networks from the analysis of the differentially expressed genes were connective tissue development and function and tissue morphology (score 34), cell death, cell signaling, cellular assembly and organization (score 32). Among the differentially affected top canonical pathways we found role of RIG1-like receptors in antiviral innate immunity (p-value<0.001), natural killer cell signaling (p-value=0.007), IL8 signaling (p-value=0.048), interferon signaling (p-value=0.0 11) (INFA21, INFGR1, MED14), ILK signaling (p-value=0.001) and apoptosis signaling.
Conclusions.
A unique GE pattern was identified in HCV (+) kidney grafts. Innate immune system and inflammatory pathways were the most affected.
Keywords: kidney transplant, hepatitis C virus, marginal donors, gene expression
Introduction
Chronic infection with hepatitis C virus (HCV) is an important global health problem that affects a large part of the world population (1). The prevalence of HCV is significantly higher in end stage renal disease (ESRD) patients as compared to the general population (2, 3), likely as consequence of the fact that ESRD patients are at a higher risk for exposure to HCV infection because of frequent surgeries (i.e., vascular access), blood transfusions, and the potential for nosocomial transmission (4, 9).
In recent years, the increasing number of patients on hemodialysis waiting for transplantation has forced the transplant community to identify alternatives ways for increasing the donor pool. With the growing number of HCV positive patients and a higher mortality in the waiting list in the US (10), better allocation and more aggressive utilization of kidneys is necessary to optimize the use of this scarce resource and minimize patients dying waiting for an organ.
Several strategies are in use to increase the donor pool, the use of extended criteria donors (ECD), the use of donors after cardiac death (DCD) and the use of organs from HCV positive donors have steadily increased in the last decades (10). However, the use of kidney grafts from HCV (+) donors (HCV+ graft) remains controversial, mainly because has been suggested that may provide worse graft survival when compared with graft from HCV negative donors (11–17). Moreover, the patho-physiological mechanisms involved in this accelerated path to graft dysfunction are unclear.
Microarray is a powerful technology that detects thousands of genes simultaneously and can be an important tool in elucidating patterns for mechanism, diagnosis, prognosis, and treatment of complex, multifactorial diseases, such as acute cellular rejection, HCV disease, graft fibrosis development, among others (18, 19). This technology has made it possible to relate physiological cell states to gene-expression patterns for studying tumors, disease progression, cellular response to stimuli, drug target identification and transplant injury mechanisms (18–20).
In the present study, we aimed to elucidate an initial compromise in the molecular profiles of HCV positive graft biopsies at pre-implantation time that might be associated with outcomes post-transplantation.
Results
Donor and kidney biopsy characteristics.
Twenty-four deceased donor kidney biopsies (N=12 from HCV positive (HCV (+)) and N=12 from HCV negative (HCV (–)) donors), were studied. Donor graft biopsies were obtained at pre-implantation time (post cold ischemia time). A sample was sent for pathological evaluation and a second sample was storage in RNAlater for gene expression profiling. Donor’s demographics and surgical characteristics were no statically different between groups and are shown in Table 1.
Table 1.
Characteristics of donors between groups
| HCV (−) | HCV (+) | P-value | |
|---|---|---|---|
| Donor Age (years) | 0.98 | ||
| Mean | 48.1 | 48.0 | |
| SD | 9.5 | 9.3 | |
| Range | (32, 61) | (31, 62) | |
| Donor Race | 75 | 66.7 | 0.5 |
| (% Caucasian) | (9/12) | (8/12) | |
| Donor Gender | 58.3 | 66.7 | 0.5 |
| (% Male) | (7/12) | (8/12) | |
| Cold ischemia time | 0.06 | ||
| (minutes) | |||
| Mean | 1,129.5 | 1,632.3 | |
| SD | 423.6 | 749.1 | |
| Range | (576, 1,980) | (856, 3,611) | |
| Delayed graft | 16.7 | 58.3 | 0.045 |
| function (%Yes) | (2/12) | (7/12) | |
In addition, as a consequence of the frequent longer ischemia time associated with HCV (+) donor grafts (mainly because these organs are procured out of state or in a different OPTN), 10 out of 12 donor kidneys underwent pump perfusion preservation before transplantation (83.3%). However, we also matched the use of pump in the HCV (–) group (9 out of 12, 75%, P=0.342). Moreover, there was not statistical significantly difference in the time on pump between groups (455.8 ± 581.0 vs. 605.9 ± 450.3 minutes, P=0.42).
Histological evaluation was performed by the same patho-nephrologist. At pre-implantation time, biopsies of HCV (+) kidney grafts demonstrated a mean glomerulosclerosis (GSC) of 4% (range 0–12). Mild interstitial fibrosis (IF) was present in 20% of the grafts and mild tubular atrophy (TA) in 24%. HCV (–) grafts demonstrated a mean of GSC of 6% (range=0–10). Also, IF was present in 25% of the grafts and TA in 24%.
Analysis of gene expression profiles between HCV (+) and HCV (–) donor biopsies.
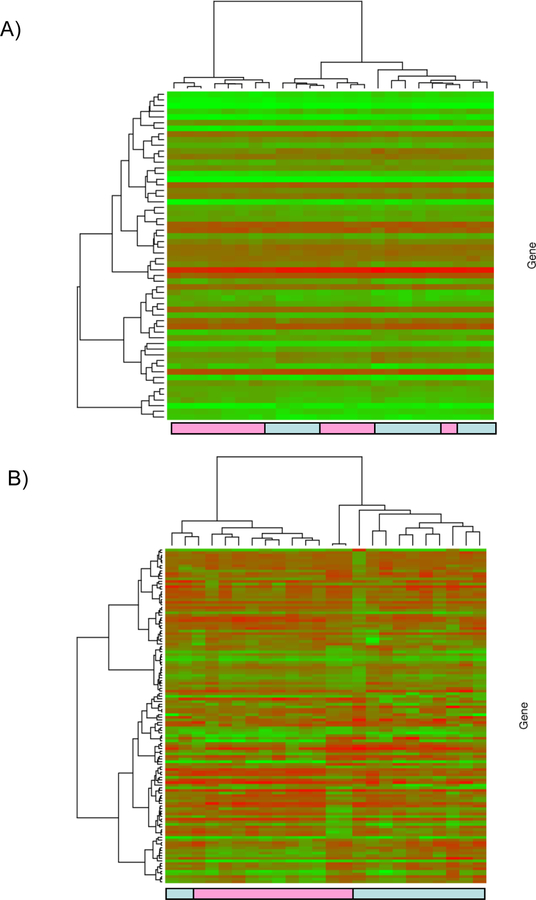
Fifty-eight probe sets were significantly differentially expressed between HCV (+) versus HCV (–) donor biopsies at the P<0.001 level. Thereafter, the gene expression dataset was restricted to the 58 significant probe sets. Hierarchical clustering Ward’s method and 1-|ρ| as the distance measure where ρ is Pearson’s correlation was applied. In addition to the supervised clustering, an unsupervised clustering was performed where the gene expression data were first filtered by retaining only probe sets called present across all arrays and having a standard deviation in the 90th percentile or greater. The heatmap and associated dendograms were plotted as a graphical representation of the significant genes for each analysis (Figure 1). However, when a FDR<15% was applied as a cut-off for significance, 614 probesets were identified as significant differentially expressed (Supplemental Table-1).
Figure 1: Supervised cluster analysis.
The gene expression dataset was restricted to the 58 significant probe sets. Hierarchical clustering Ward’s method and 1-|ρ| as the distance measure where ρ is Pearson’s correlation was applied. The heatmap and associated dendograms were plotted as a graphical representation of the significant genes (Figure 1A). Unsupervised cluster analysis. the gene expression data were first filtered by retaining only probe sets called present across all arrays and having a standard deviation in the 90th percentile or greater. Hierarchical clustering Ward’s method and 1-|ρ| as the distance measure where ρ is Pearson’s correlation was applied. The heatmap and associated dendograms for the unsupervised clustering appear in Figure 1 B. Light blue boxes: HCV (+) donor kidney samples, Pink boxes: HCV (–) donor samples.
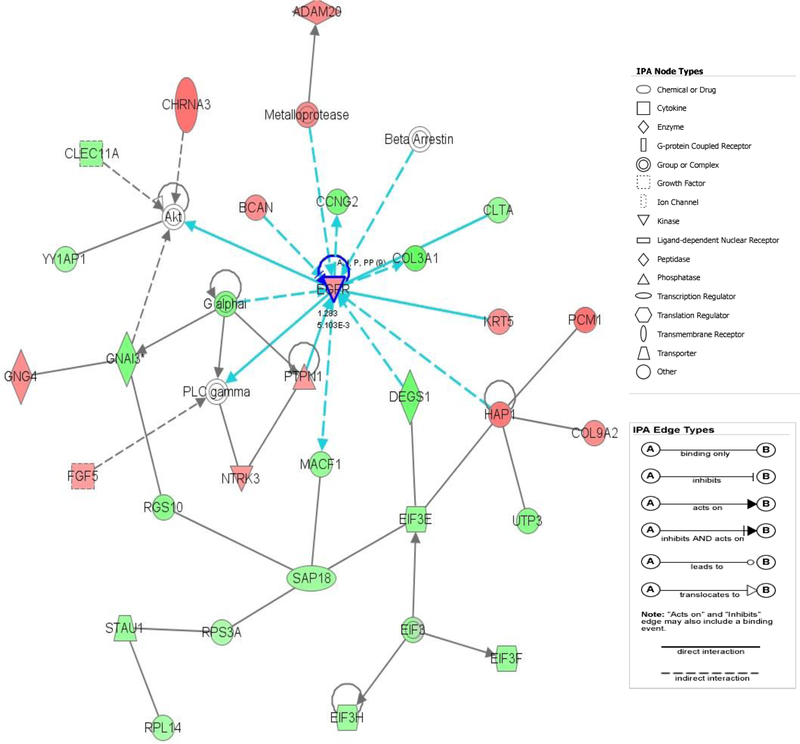
Core analysis was performed to interpret the data set in the context of biological processes, pathways and molecular networks. A total of 25 networks with a score≥10 were identified from the analysis of the differentially expressed genes between groups. Among the top five scored networks, the associated network functions included protein synthesis, cell cycle, cellular growth and proliferation (network 1, score 40), cellular assembly and organization, carbohydrate metabolism, cellular movement (network 4, score 34), post-translation modification, inflammatory disease, renal nephritis (network 5, score 33). The Figure 2 shows the top scored network.
Figure 2:
The top-scoring network of interactions among the 58 probe sets identified as significantly differentially expressed when comparing HCV (+) vs. HCV (–) donors. The probe sets were subsequently analyzed using the Ingenuity pathway analysis software (https://analysis.ingenuity.com). This software is designed to identify dynamically generated biological networks, global canonical pathways and global functions. Interconnection of significant functional networks, where protein nodes in different shades of red and green or white depending on being up-regulated and down-regulated or no-change, respectively, in HCV (+) kidney donor samples. [Blue] For canonical pathways, molecules that are members of the network being examined are outlined in blue.
The top molecular and cellular functions identified as associated with the differentially expressed genes (Table 2) included gene expression, cell death, cellular development, lipid metabolism, and small molecule biochemistry.
Table 2.
Differentially expressed genes between HCV (+) and HCV (–) donor kidney grafts
| Affy IF | Gene Symbol | P-value | Mean Expression HCV (+) | Mean Expression HCV (–) |
|---|---|---|---|---|
| 200023_s_at | EIF3F | 0.00056 | 10.04 | 10.46 |
| 200066_at | IK | 0.00019 | 9.07 | 9.57 |
| 200820_at | PSMD8 | 0.00039 | 9.39 | 9.77 |
| 200910_at | CCT3 | 0.00017 | 9.51 | 9.92 |
| 200943_at | HMGN1 | 0.00012 | 10.27 | 10.74 |
| 201301_s_at | ANXA4 | 0.00059 | 10.61 | 11.13 |
| 201888_s_at | IL13RA1 | 0.00071 | 6.19 | 7.00 |
| 202273_at | PDGFRB | 0.00027 | 7.13 | 7.70 |
| 202535_at | FADD | 0.00094 | 7.46 | 7.95 |
| 202596_at | ENSA | 0.00015 | 9.32 | 9.77 |
| 202659_at | PSMB10 | 0.00061 | 6.79 | 7.51 |
| 203154_s_at | PAK4 | 0.00039 | 8.91 | 8.64 |
| 203414_at | MMD | 0.00038 | 6.54 | 7.47 |
| 203561_at | FCGR2A | 0.00057 | 5.60 | 6.32 |
| 204050_s_at | CLTA | 0.00021 | 10.15 | 10.62 |
| 204559_s_at | LSM7 | 0.00084 | 8.62 | 9.13 |
| 204780_s_at | FAS | 0.00078 | 7.25 | 8.12 |
| 204875_s_at | GMDS | 0.00073 | 6.21 | 6.71 |
| 206833_s_at | ACYP2 | 0.00098 | 7.04 | 7.53 |
| 206868_at | STARD8 | 0.00095 | 7.87 | 7.43 |
| 206954_at | WIT1 | 0.00090 | 5.72 | 5.38 |
| 207423_s_at | ADAM20 | 0.00019 | 5.62 | 5.23 |
| 208635_x_at | NACA | 0.00050 | 12.48 | 12.67 |
| 208708_x_at | EIF5 | 0.00070 | 8.34 | 9.10 |
| 208921_s_at | SRI | 0.00080 | 9.42 | 9.95 |
| 209067_s_at | HNRPDL | 0.00065 | 9.19 | 10.02 |
| 209089_at | RAB5A | 0.00036 | 9.75 | 10.23 |
| 209102_s_at | HBP1 | 0.00058 | 9.16 | 9.70 |
| 209310_s_at | CASP4 | 0.00036 | 6.70 | 7.45 |
| 209486_at | UTP3 | 0.00094 | 7.86 | 8.48 |
| 209719_x_at | SERPINB3 | 0.00037 | 4.99 | 4.76 |
| 209997_x_at | PCM1 | 0.00024 | 6.41 | 5.92 |
| 210102_at | VWA5A | 0.00065 | 7.34 | 8.02 |
| 210221_at | CHRNA3 | 0.00019 | 6.14 | 5.63 |
| 211460_at | NA | 0.00097 | 5.55 | 5.12 |
| 211653_x_at | AKR1C2 | 0.00096 | 10.03 | 10.78 |
| 212055_at | C18orf10 | 0.00050 | 7.89 | 8.40 |
| 212117_at | NA | 0.00040 | 7.88 | 8.47 |
| 212785_s_at | LARP7 | 0.00074 | 8.02 | 8.54 |
| 213152_s_at | SFRS2B | 0.00025 | 7.62 | 8.10 |
| 213535_s_at | UBE2I | 0.00058 | 7.43 | 7.85 |
| 214512_s_at | SUB1 | 0.00049 | 9.46 | 10.02 |
| 215095_at | ESD | 0.00078 | 5.84 | 5.34 |
| 215411_s_at | TRAF3IP2 | 0.00071 | 6.90 | 7.31 |
| 216072_at | NA | 0.00055 | 5.03 | 4.84 |
| 216194_s_at | TBCB | 0.00011 | 8.67 | 9.20 |
| 217850_at | GNL3 | 0.00061 | 7.26 | 7.69 |
| 218194_at | REXO2 | 0.00060 | 10.32 | 10.71 |
| 218238_at | GTPBP4 | 0.00022 | 5.57 | 6.06 |
| 218291_at | ROBLD3 | 0.00047 | 8.37 | 8.72 |
| 219194_at | SEMA4G | 0.00094 | 7.25 | 6.65 |
| 219862_s_at | NARF | 0.00042 | 7.37 | 7.76 |
| 220073_s_at | PLEKHG6 | 0.00097 | 6.20 | 5.93 |
| 220221_at | VPS13D | 0.00088 | 5.58 | 5.15 |
| 221240_s_at | B3GNT4 | 0.00071 | 7.32 | 6.87 |
| 36004_at | IKBKG | 0.00068 | 7.99 | 7.63 |
| 36829_at | PER1 | 0.00005 | 8.63 | 8.23 |
| 48612_at | N4BP1 | 0.00098 | 9.23 | 8.86 |
Role of IG1-like receptors in antiviral innate immunity (P-value<0.001), represented the top canonical pathway. The expression of CASP10, IFNA21, and IKBK6 genes was up-regulated in the HCV (+) donor kidneys. Moreover, two additional canonical pathways also associated with innate immune response (crosstalk between dendritic cells and natural killer cells (P=0.003) and natural killer cell signaling (P=0.007)), were identified as up-regulated in HCV (+) donor graft tissues. Specifically, KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL5A, KIR2DS1genes (natural killer cell signaling) were over-expressed in HCV (+) donor tissues when compared with HCV (–). Moreover, from the specific analysis of the canonical pathways associated with cellular immune response, IL8 signaling (EGFR, FIGF, GNG4, MAPK8, IKBKG, PIK3C3, TEK) and interferon signaling genes (INFA21, INFGR1, MED14) were also up-regulated in HCV (+) donors.
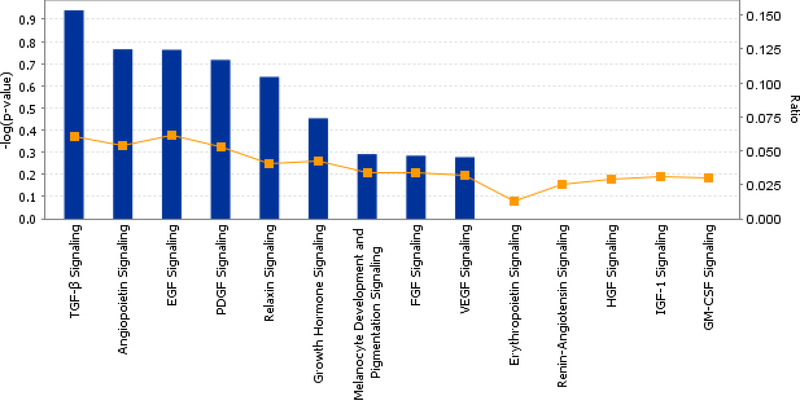
Furthermore, from the analysis of growth factor signaling, we observed that TGF-beta signaling (ACVR1B, BMP7, HNF4A, MAPK8), angiopoietin signaling (IKBKG, PAK3, PIK3C3, TEK), EGF signaling (ACTN2, FIGF, PIK3C3), and FGF signaling (FGF5, MAPK8, PIK3C3) were up-regulated in kidney biopsy samples from HCV (+) donors (Figure 3).
Figure 3: Top canonical pathways.
Growth factor signaling identified for the analysis of HCV (+) kidney donor samples. From an expression data analysis, global canonical pathways are displayed on the canonical pathways tab.
There was a borderline significant difference between the HCV (+) vs. HCV (–) donors with respect to CIT. Therefore, the analysis was restricted to patients having a CIT <1500 minutes where there was no significant difference between the two groups (P=0.27). To identify probe sets exhibiting differential expression between HCV and non-HCV donor biopsies, a two sample t-test was performed (P-value<0.001). Thirty-three probe sets were significantly differentially expressed between HCV (+) vs. HCV(–) donors at the p<0.001 level (Supplemental Table 2). The heatmap and dendrogram from the hierarchical clustering procedure applied to the 33 significant probe sets appears on the Supplemental Figure 1.
Clinically, we observed an increased incidence of DGF in HCV (+) grafts (58.3% versus 16.7%) (Table-1).
Gene expression quantitation using QPCR.
We selected the genes used for the microarray assay validation using QPCR by two strategies: 1) genes were in the top of the differentially expressed genes; and/or 2) the genes were among the top scoring networks and pathways when using Interaction Networks and Functional Analysis. Expression levels of the FADD, CASP10, IFNA21, KIR3LD2, and EGFR mRNAs were further confirmed using QPCR. These genes were identified as significantly differentially expressed between different tissues types. The results from the microarray were reproduced by QPCR (r=0.82, P<0.001; r=0.71, P=0.002; r=0.65, P<0.001; r=0.75, P=0.001; and r=0.7, P<0.001 respectively).
Discussion
The use of grafts from donors with positive HCV serology is becoming more routinely practice in transplant centers. However, possibly due to incomplete or unclear data related with these grafts outcomes, large amount of kidney grafts from HCV (+) donors are discarded every year (21). Despite that some reports describe a reduced graft survival in recipients of HCV (+) grafts, the cause or patho-physiological mechanisms behind these findings continue to be unclear. Furthermore, a better understanding of the mechanisms and processes leading to accelerated graft dysfunction in HCV (+) grafts will open opportunities for therapeutic approaches.
Epidemiologic (22), clinical (23), and experimental (24) data had suggested that HCV infection may induce glomerular damage in native kidneys. A high seroprevalence of anti-HCV has been observed in patients with glomerulonephritis in several countries. Most evidence suggests that glomerular injury results from the deposition of circulating immune complexes containing HCV antibodies, HCV antigens and complement mainly C3 within the sub-endothelium and mesangium (25).
D’Amico et al. (26) showed that, in Europe, almost all membranoproliferative glomerulonephritis associated with HCV infection has a concomitant type II cryoglobulinaemia, which suggests that the monoclonal immunoglobin (IG) Mk compound of cryoglobulins has a fundamental role in the pathogenesis of HCV-associated glomerulonephritis. However, HCV has also been implicated in the development of membranoproliferative (24) and membranous (27) glomerulonephritis in patients without cryoglobulinemia. Roth et al. (28) have reported five cases of de novo membranoproliferative glomerulonephritis in HCV-infected renal allograft recipients. Although these authors suggested that it was an immune complex nephritis, they failed to show direct evidence for cryoglobulinemia and circulating immune complexes.
Additionally, it has been published (29) that chronic HCV infection in renal allograft recipients is associated with reduced activated helper/induced T lymphocytes and natural killer cells, but increased suppressor/cytotoxic T cells in peripheral blood.
Forty-three kidney transplants performed at our institution between January 2000 and December 2004 were tracked prospectively (30). A group of 52 contemporaneous HCV(+)/ESRD patients listed, but never transplanted, was also analyzed. HCV-negative transplanted patients were used as the control group. Patient survival post-transplantation was 81.4% and 68.5% at 1 and 3 years in the HCV (+) group, and 97.1% and 92.9% at 1 and 3 years in the HCV (–) group, respectively (P=0.001). Graft survival was 81.2% and 64.1% at 1 and 3 years in the HCV+ group, and 93.2% and 84.1% at 1 and 3 years post-transplantation in the HCV (–) group (P=0.01). Patient survival was superior in HCV (+) patients undergoing transplantation vs. remaining on dialysis. It was not the goal of this study to evaluate clinical graft or recipient outcomes, but patient survival was 100% at one year in both groups, and graft survival was 100% in both groups also. These numbers may be affected for the small sample size and short follow-up, but understanding the biology behind HCV donor is critical.
However, regardless of multiple attempts to provide pieces in this complicated puzzle, many questions about pathogenesis and pathological mechanisms about the effect of the virus in the kidney of HCV infected patients continue to be unanswered.
We propose that the evaluation of gene expression patterns in HCV (+) donor kidneys might provide critical insight of the initial molecular compromise associated with the virus infection and might represent the first step of further evaluations of HCV grafts post-kidney transplantation.. To our knowledge, this is the first report of differential gene expression profiling in HCV (+) kidney graft compared with a matched group of HCV (–) deceased donor kidneys. It needs to be noted that our study represents whole tissue gene expression analysis and the analysis can not be dissected in the partial contribution of each component of the kidney allograft. We are aware of the limitation that sample cellular heterogeneity introduces to the analysis. Kidney tissue samples used for microarray analysis contain a mixture of different cells and cell infiltrates. However, in this study we aimed to evaluate the global differences that characterize HCV (+) donor at the transplantation time.
In the present analysis, a distinctive gene expression pattern was identified in HCV (+) when compared with non-HCV kidney donor biopsies. Among the top canonical pathways affected in HCV (+) grafts we found RIG1-like receptors associated genes and natural killer cell signaling. RIG 1- like receptors plays an important role in antiviral innate immunity (31). Similarly, natural killer cells are an important component of the innate immune response against many viruses due to their ability to lyses virus-infected cells and to secrete cytokines that inhibit viral replication and activate and recruit cells of the adaptive immune response (32, 33). The high incidence of recurrent chronic HCV after a course of antiviral therapy reveals the urgency of determining the extrahepatic reservoirs of HCV (39). The most important extrahepatic site for the virus is peripheral blood mononuclear cells (PBMC) (34, 35). Assuming that HCV replication occurs in PMCs, these cells can be considered as a potential reservoir of HCV infection. There is evidence that HCV is detectable just in the tissues of the involved organs (36). Thus, most systemic manifestations may be directly associated with extrahepatic HCV replication. IFN and IFN inducible genes were also up-regulated in HCV (+) kidney donor tissues. Overall, our findings showed a gene expression in HCV (+) donor kidneys associated with an ongoing host response to the virus that it is already present in the kidney before transplantation.
The contribution of apoptosis and underlying mechanisms in liver injury in HCV-infected transplant recipients has been the subject of investigation in a small number of studies. These studies provided information limited to findings in liver allografts (37, 38), whereas in the setting of renal transplant, data are lacking. Genes associated with apoptosis were differentially expressed in the HCV (+) kidneys, with over-expression of CASP10 in these kidneys. Caspases are a family of cysteine proteases that act in concert in a cascade triggered by apoptosis signaling. The effect of the apoptosis may affect the immediate outcome of the grafts as shown in other studies, especially associated with liver transplantation (39–41). There is little information about the effect of apoptosis in kidney allograft in HCV recipients (42). The over-expression of genes associated with apoptosis in HCV (+) donors may explain the increased incidence of DGF observed in HCV(+) kidneys. Induction to apoptosis may suggest that graft from HCV(+) donors are less tolerant to cold ischemia time and may suffer from more cell death, acute tubular necrosis and associated DGF (43, 44), especially when considering that the studied groups were cold ischemia time matched.
Other findings included affection of IL 12 and TGFB signaling. IL 12 signaling is critical as a linker between the innate immunity and adaptive immunity, capable of TH1 (T Helper Type-1) differentiation and IFN-Gamma release by T-Cells and NK cells. Moreover, IL-18 acts as a synergist with IL-12 in some of their effects, especially in the induction of IFN-Gamma production and inhibition of angiogenesis.
Over expression of growth factor signaling was observed in HCV (+) donor biopsies. Transforming growth factor beta (TGF-beta 1) pathway and over-expression of most of its genes (over-expression of ACVR1B, BMP7, HNF4A, MAPK8) was found in HCV (+) grafts. TGF-beta 1 is a well known key modulator of glomerulosclerosis, tubulointerstitial fibrosis in the kidney (45–48), that has been associated with long term graft function. TGF-beta 1 is a key regulator of cell proliferation, differentiation, apoptosis, immune response, and extracellular matrix remodeling depending on the physiologic context (45–47).
The present study provides preliminary information about characteristic gene expression patterns present in the kidney grafts of HCV (+) donors at the pre-implantation time. A predominant pattern of increased innate immune response and response to virus indicates an ongoing host response to the virus in the graft. Moreover, activation of growth factor signaling might associate with fibrosis progression post-kidney transplantation. Further studies to evaluate the role of these findings in the graft recipient and their effects in long-term outcomes are needed.
Patients and Methods
Patients and samples.
The study was conducted at Virginia Commonwealth University after IRB approval was obtained (VCU-IRB Protocol #HM11454). Twenty four patients received a kidney transplant at the Hume-Lee Transplant center and consented to participate in this study. Patients received a deceased donor kidney transplant between January 2007 and July 2009. All HCV patients (HCV (+) group N=12) had recent HCV positive testing (serology and PCR viral load) and received a primary kidney transplant from a deceased donor with confirmed HCV positive serology. A matching group of non-HCV primary kidney recipients (N=12), that received a HCV negative (HCV (–) graft during the same period of time was used for comparison purposes. These patients are part of an ongoing prospective study including protocol biopsies and 48 months post-transplant follow-up (R01DK080074).
Patients with re-transplantation, multiple transplants or graft from living donors were excluded from this study. Renal allograft tissue was obtained on the backbench just before implantation (after cold ischemia time) using an 18-gauge biopsy needle. All the biopsies had histological evaluation by the same renal pathologist. Kidney recipients were followed during the first 12 months after kidney transplantation and complete donor, surgical, recipient information was collected. Delayed graft function (DGF) was defined as the need for dialysis in the first week after kidney transplantation.
RNA Isolation, cDNA Synthesis, and In Vitro Transcription (IVT) for Labeled cRNA Probe.
With minor modifications, the sample preparation protocol follows the Affymetrix GeneChip® Expression Analysis Manual (Santa Clara, CA) as it was previously published (48, 49). The labeled cRNA was chemically fragmented and then hybridized to U133A 2.0 GeneChip probe arrays (Affymetrix, Santa Clara, CA). The array image was generated by the high-resolution GeneChip® Scanner 3000 by Affymetrix® (Affymetrix, Santa Clara, CA). All CEL files were processed using Affymetrix GeneChip Operating Software version 1.4, and appropriate Bioconductor packages (50) available in the R programming environment (51).
Data analysis.
Gene expression microarray data for twelve HCV (+) donor biopsies were available. Twelve microarrays corresponding HCV-matched donors to the HCV (+) donors were included for comparison. Probe level data were read into the R programming environment using the affy Bioconductor package and the robust multiarray average method was used to obtain probe set expression summaries. Prior to statistical analysis all control probe sets were removed. Furthermore, probe sets declared absent by the detection call algorithm for all 24 arrays were also removed leaving 15,988 probe sets.
Interaction Networks and Functional Analysis.
Gene ontology and gene interaction analyses were executed using Ingenuity Pathways Analysis tools 7.0 (http://www.ingenuity.com).
Statistical analysis.
Descriptive statistics were reported for demographic and clinical variables, including proportions for categorical variables and mean ± standard deviation for continuous variables.
(See “Supplemental Methods” for QPCR reactions and quality control)
Supplementary Material
Acknowledgments
The research results included in this report were supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant, R01DK080074.
Abbreviations
- MDS
classical multidimensional scaling
- DCD
donation after cardiac death
- ESRD
end stage renal disease
- ECD
extended criteria donor
- HCV
Hepatitis C Virus
- IVT
In Vitro Transcription
- QPCR
quantitative reverse transcriptase-“real time” PCR
- QC
quality control
Footnotes
Financial disclosure
The authors declare that they have no competing interests.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 2005; 5: 567. [DOI] [PubMed] [Google Scholar]
- 2.Pereira BJ, Levey AS. Hepatitis C virus infection in dialysis and renal transplantation. Kidney Int 1997; 51: 981. [DOI] [PubMed] [Google Scholar]
- 3.Meyers CM, Seeff LB, Stehman-Breen CO, Hoofnagle JH. Hepatitis C and renal disease: an update. Am J Kidney Dis 2003; 42: 631. [DOI] [PubMed] [Google Scholar]
- 4.Fabrizi F, Poordad FF, Martin P. Hepatitis C infection and the patient with end-stage renal disease. Hepatology 2002; 36: 3. [DOI] [PubMed] [Google Scholar]
- 5.Finelli L, Miller JT, Tokars JI, Alter MJ, Arduino MJ. National surveillance of dialysis-associated diseases in the United States, 2002. Semin Dial 2005; 18: 52. [DOI] [PubMed] [Google Scholar]
- 6.Tokars JI, Miller ER, Alter MJ, Arduino MJ. National surveillance of dialysis associated diseases in the United States, 1995. ASAIO J 1998; 44: 98. [DOI] [PubMed] [Google Scholar]
- 7.Fissell RB, Bragg-Gresham JL, Woods JD, et al. Patterns of hepatitis C prevalence and seroconversion in hemodialysis units from three continents: the DOPPS. Kidney Int 2004; 65: 2335. [DOI] [PubMed] [Google Scholar]
- 8.Jadoul M, Poignet JL, Geddes C, et al. The changing epidemiology of hepatitis C virus (HCV) infection in haemodialysis: European multicentre study. Nephrol Dial Transplant 2004; 19: 904. [DOI] [PubMed] [Google Scholar]
- 9.Fehr T, Ambuhl PM. Chronic hepatitis virus infections in patients on renal replacement therapy. Nephrol Dial Transplant 2004; 19: 1049. [DOI] [PubMed] [Google Scholar]
- 10.USRDS, Annual Data Report. 2009 [Google Scholar]
- 11.Kasprzyk T, Kwiatkowski A, Wszola M, et al. Long term results of kidney transplantation from HCV-positive donors. Transplant Procc 2007; 39: 2701. [DOI] [PubMed] [Google Scholar]
- 12.Fabrizi F, Bunnapradist S, Lunghi G, Martin P. Transplantation in kidneys from HCV-positive donors: a safe strategy? J Nephrol. 2003; 16: 617. [PubMed] [Google Scholar]
- 13.Gentil MA, Rocha JL, Rodríguez Algarra G, et al. Impaired kidney transplant survival in patients with antibodies to hepatitis C virus. Nephrol Dial Transplant. 1999; 14: 2455. [DOI] [PubMed] [Google Scholar]
- 14.Mathurin P, Mouquet C, Poynard T, et al. Impact of hepatitis B and C virus on kidney transplantation outcome. Hepatology. 1999; 29: 257. [DOI] [PubMed] [Google Scholar]
- 15.Batty DS Jr, Swanson SJ, Kirk AD, et al. Hepatitis C virus seropositivity at the time of renal transplantation in the United States: associated factors and patient survival. Am J Transplant. 2001; 1: 179. [PubMed] [Google Scholar]
- 16.Abbott KC, Lentine KL, Bucci JR, Agodoa LY, Peters TG, Schnitzler MA. The impact of transplantation with deceased donor hepatitis C-positive kidneys on survival in wait-listed long-term dialysis patients. Am J Transplant. 2004; 4:203. [DOI] [PubMed] [Google Scholar]
- 17.Fabrizi Fabrizio, Messa Piergiorgio, Martin Paul. Current status of renal transplantation fromHCV-positive donors. Int J Artif Organs. 2009; 32: 251. [DOI] [PubMed] [Google Scholar]
- 18.Mansfield ES, Sarwal MM. Arraying the orchestration of allograft pathology. Am J Transplant 2004; 4: 853. [DOI] [PubMed] [Google Scholar]
- 19.Halloran PF, Einecke G. Microarrays and transcriptome analysis in renal transplantation. Nat Clin Pract Nephrol. 2006; 2: 2. [DOI] [PubMed] [Google Scholar]
- 20.Ying L, Sarwal M. In praise of arrays. Pediatr Nephrol. 2009; 24:1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kucirka LM, Singer AL, Ros RL, et al. Underutilization of hepatitis C-positive kidneys for hepatitis C-positive recipients. Am J Transplant. 2010; 10:1238. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Valdecasas J, Bernal C, Garcia F, et al. Epidemiology of hepatitis C virus infection in patients with renal disease. J Am Soc Nephrol 1994; 5:186. [DOI] [PubMed] [Google Scholar]
- 23.Johnson RJ, Gretch DR, Yamabe H, et al. Membranoproliferative glomerulonephritis associated with hepatitis C virus infection. N Engl J Med 1993; 328 : 465. [DOI] [PubMed] [Google Scholar]
- 24.Fornasieri A, Li M, Armelloni S, et al. Glomerulonephritis induced by human IgMk-IgG cryoglobulins in mice. Lab Invest 1993; 69: 531. [PubMed] [Google Scholar]
- 25.Ayodele OE, Salako BL. Hepatitis C virus (HCV) and chronic renal disease. Afr J Med Med Sci. 2003; 32: 287. [PubMed] [Google Scholar]
- 26.D’Amico G, Fornasieri A. Cryoglobulinemic glomerrulonephritis: A Membranoproliferative glomerulonephritis induced by hepatitis C virus. Am J Kidney Dis 1995; 25:361. [DOI] [PubMed] [Google Scholar]
- 27.Stehman-Breen C, Alpers CE, Couser WG, et al. Hepatitis C virus associated membranous glomerulonephritis. Clin Nephrol 1995; 44:141. [PubMed] [Google Scholar]
- 28.Roth D, Cirocco R, Zucker K, et al. De novo membranoproliferative glomerulonephritis in hepatitis C virus infected renal allograft recipients. Transplantation 1995; 59: 1676. [DOI] [PubMed] [Google Scholar]
- 29.Chan TM, Ho SK, Lai CL, Cheng IK, Lai KN. Lymphocyte subsets in renal allograft recipients with chronic hepatitis C virus infection. Nephrol Dial Transplant. 1999; 14: 717. [DOI] [PubMed] [Google Scholar]
- 30.Maluf DG, Fisher RA, King AL, et al. Hepatitis C virus infection and kidney transplantation: predictors of patient and graft survival. Transplantation. 2007;83 :853. [DOI] [PubMed] [Google Scholar]
- 31.Kanto T, Hayashi N. Innate immunity in hepatitis C virus infection: Interplay among dendritic cells, natural killer cells and natural killer T cells. Hepatol Res. 2007; 37: S319. [DOI] [PubMed] [Google Scholar]
- 32.O’Neill LA, Bowie AG. Sensing and signaling in antiviral innate immunity. Curr Biol. 2010; 20: R328. [DOI] [PubMed] [Google Scholar]
- 33.Radkowski M, Gallegos-Orozco J, Jablonska J, et al. Persistence of hepatitis C virus in patients successfully treated for chronic hepatitis C. Hepatology 2005; 41: 106. [DOI] [PubMed] [Google Scholar]
- 34.Barría MI, Vera-Otarola J, León U, et al. Influence of extrahepatic viral infection on the natural history of hepatitis C. Ann Hepatol. 2008; 7:136. [PubMed] [Google Scholar]
- 35.Blackard JT, Kemmer N, Sherman KE. Extrahepatic replication of HCV: insights into clinical manifestations and biological consequences. Hepatology. 2006; 44:15. [DOI] [PubMed] [Google Scholar]
- 36.Radkowski M, Wang LF, Vargas HE, Rakela J, Laskus T. Detection of hepatitis C virus replication in peripheral blood mononuclear cells after orthotopic liver transplantation. Transplantation 1998; 66: 664. [DOI] [PubMed] [Google Scholar]
- 37.Di Martino V, Brenot C, Samuel D, et al. Influence of liver hepatitis C virus RNA and hepatitis C virus genotype on FAS-mediated apoptosis after liver transplantation for hepatitis C. Transplantation. 2000; 70: 1390. [DOI] [PubMed] [Google Scholar]
- 38.Hsu EC, Hsi B, Hirota-Tsuchihara M, Ruland J, et al. Modified apoptotic molecule (BID) reduces hepatitis C virus infection in mice with chimeric human livers. Nat Biotechnol. 2003; 21: 519. [DOI] [PubMed] [Google Scholar]
- 39.Ballardini G, De Raffele E, Groff P, et al. Timing of reinfection and mechanisms of hepatocellular damage in transplanted hepatitis C virus-reinfected liver. Liver Transpl. 2002; 8:10. [DOI] [PubMed] [Google Scholar]
- 40.Delladetsima I, Psichogiou M, Alexandrou P, et al. Apoptosis and hepatitis C virus infection in renal transplant recipients. Am J Clin Pathol. 2008; 129: 744. [DOI] [PubMed] [Google Scholar]
- 41.Drognitz Oliver, Obermaier Robert, Liu Xuemei, et al. Effects of Organ Preservation, Ischemia Time and Caspase Inhibition on Apoptosis and Microcirculation in Rat Pancreas Transplantation. Am J Transplant. 2004; 4:1042. [DOI] [PubMed] [Google Scholar]
- 42.Song SW, Guo KJ, Shi R, Cheng Y, Liu YF. Pretreatment with calcitonin gene-related peptide attenuates hepatic ischemia/reperfusion injury in rats. Transplant Proc. 2009; 41: 1493. [DOI] [PubMed] [Google Scholar]
- 43.Bottinger EP. TGF-beta in renal injury and disease. Semin Nephrol 2007; 27: 309. [DOI] [PubMed] [Google Scholar]
- 44.Tyler JR, Robertson H, Booth TA, et al. Chronic allograft nephropathy: Intraepithelial signals generated by transforming growth factor-beta and bone morphogenetic protein-7. Am J Transplant 2006; 6: 1367. [DOI] [PubMed] [Google Scholar]
- 45.Bottinger EP, Bitzer M. TGF-beta signaling in renal disease. J Am Soc Nephrol 2002; 13: 2600. [DOI] [PubMed] [Google Scholar]
- 46.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- 48.Mas VR, Archer KJ, Yanek K, et al. Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation. 2008; 85:626. [DOI] [PubMed] [Google Scholar]
- 49.Maluf DG, Mas VR, Archer KJ, et al. Molecular pathways involved in loss of kidney graft function with tubular atrophy and interstitial fibrosis. Mol Med. 2008; 14:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irizarry RA, Hobbs B, Collin F, et al. Exploration, 21normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249. [DOI] [PubMed] [Google Scholar]
- 51.Gautier L, Cope L, Bolstad BM, AIrizarry R. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20: 307. [DOI] [PubMed] [Google Scholar]
- 52.Archer KJ, Dumur CI, Joel SE, Ramakrishnan V. Assessing quality of hybridized RNA in Affymetrix GeneChip experiments using mixed effects models. Biostatistics 2006; 7: 198. [DOI] [PubMed] [Google Scholar]
- 53.Archer K. An application for assessing quality of RNA hybridized to Affymetrix GeneChips. Bioinformatics 2006; 22: 2699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.