Summary
Interstitial fibrosis (IF) and tubular atrophy (TA) are integral parts of chronic allograft dysfunction and represent in the new classification a separate entity with or without the identification of a specific etiology. Loss of kidney graft function with IF/TA is one of the causes of most kidney allograft losses. Despite progress in immunosuppression, chronic allograft dysfunction remains the main clinical challenge for improving long-term graft survival. The sustained damage to the allograft does not represent a single entity, but the summated effects of tissue injury from several pathogenic insults as well as the kidney’s healing response, modified by alloimmunity and immunosuppression.
A major challenge in the future of kidney transplantation includes the study of chronic allograft dysfunction pathogenesis to identify early markers of disease progression as well as potential therapeutics pathways.
Keywords: kidney transplantation, chronic allograft injury, long-term outcomes, biomarkers
Introduction
Kidney transplantation (KTx) is the treatment of choice for end-stage renal disease. The incidence of acute rejection and graft loss in the first year after renal transplantation has decreased markedly in the past 20 years, mainly as consequence of the introduction of more potent immunosuppressive protocols [1–4]. However, chronic allograft dysfunction remains an important cause of loss of graft function post-KTx [1–5].
With close to 5,000 kidney transplants failing per year in the United States alone, kidney transplant failure is now a leading cause of end-stage renal disease [6]. Furthermore, the causes of late allograft loss are not well defined. One common phenotype is loss of function with tubular atrophy (TA) and interstitial fibrosis (IF), a set of findings termed chronic allograft nephropathy or ‘CAN.’ The histopathological signs of CAN (interstitial fibrosis, tubular atrophy, glomerulopathy and vasculopathy) are nonspecific; consequently, the 2007 Banff classification [7] replaced the term CAN for ‘interstitial fibrosis and tubular atrophy without evidence of any specific etiology’. In this review, we use the term ‘loss of kidney graft function with TA and IF’ to refer to the previously mentioned condition.
Dilemma: Magnitude and Consequences
At the end of 2006, 103,312 patients had a functioning kidney transplant compared with 64,779 in 1998, an increase of 59% [6]. Unadjusted patient survival rates at five years were 91% for recipients of living donor kidneys, 83% for non-extended criteria donor (ECD) deceased donor kidneys, and 70% for ECD kidney transplants. Kidney allograft survival followed the same pattern as that seen for recipient survival. Graft survival was best for recipients of living donor kidneys, intermediate for non-ECD transplants, and lowest for ECD transplants. At five years, the unadjusted graft survival rate was 81% for living donor, 71% for non-ECD, and 55% for ECD transplants [6]. The number of candidates on the kidney transplant waiting list at year-end increased from 40,825 to 76,070 (86%) between 1998 and 2007 [6]. Organ shortage and continuously growing waiting lists are key problems in organ transplantation. To address donor shortage an increasing number of marginal donor organs have been used in recent years [8]. However, despite numerous studies both organ quality and long-term outcomes remain difficult to predict [2, 4, 8].
Graft loss after the first year post-transplantation can be due to combined factors that may include immunological and non-immunological factors [9–11]. The original nephron mass present in the graft at transplantation time is mostly dependent on the donor quality. Among deceased donor kidneys, donor factors are estimated to account for 35–45% of the variability in early allograft function [12–14] (Figure 1). After kidney transplantation and as a consequence of surgery itself, preservation methods, ischemia, acute rejection and other stressors, the number of initial nephrons in the donor kidney will be reduced. Ordinarily, 6 months after post-transplantation, the number of nephrons is established resulting in an allograft with normal function. However, for an important number of grafts, the loss of nephrons continues, reflecting continued graft injury. This condition leads to fibrosis and atrophy, with the final event of loss of graft function [4]. It is unclear why certain kidneys maintain a stable function and others develop injury and inevitable progression to fibrosis and loss of kidney function with IF/TA.
Figure 1:
The cellular and molecular mechanisms underlying these histopathological hallmarks remain obscure. Immunological and non-immunological factors have been associated with the progression to chronic allograft dysfunction. Many cell-cell, cell-matrix, and intracellular pathways have been implicated in the complex pathogenesis of CAN. CAN is characterized by slowly progressive graft dysfunction ultimately leading to chronic renal failure.
The early appearance of loss of kidney function with IF/TA is an independent risk factor for long term allograft failure [15, 16]. Previous researchers have reported the frequency of early subclinical rejection at 45.7% at three months [17]. Others have reported the frequency of histological findings associated with loss of kidney function with IF/TA in protocol biopsies to be 66.7% among all patients receiving a deceased donor organ at 2 years post-KTx [18].
A biopsy of a chronically failing kidney transplant usually shows nonspecific or end-stage changes, so the relative contributions of preexisting disease in the allograft and immunologic and non-immunologic factors become difficult to distinguish. Furthermore, a number of clinical risk factors have been correlated with renal allograft failure [19–21]. Long-term function and failure of the transplanted kidney reflects the overall cumulative injury, resulting from immune and non-immune mechanisms such as brain death, donor age, ischemia and reperfusion injury, nephron loss, allo-antigens, or infections causing ongoing renal tissue stress and inflammation leading to loss of kidney function with IF/TA over time [16–22].
The mechanisms of nephron loss resulting in graft dysfunction are several, comprising both immunologic factors such as acute and chronic antibody- or T-cell-mediated rejection and non-immunologic components. T-cell recognition of alloantigen is the key primary event that initiates allograft rejection, although several other factors may contribute to acute and chronic allograft dysfunction. T-cells recognize alloantigen via two distinct yet non-mutually exclusive pathways: the direct and indirect pathways of allorecognition [23, 24]. In the direct pathway, alloreactive T cells recognize intact allo-major histocompatibility complex (MHC) molecules on the surface of donor cells, while in the indirect pathway, T-cells recognize alloantigens in the form of peptides after processing and presentation by self-antigen presenting cells (APC’s). It has been suggested that the direct pathway of allorecognition predominantly mediates acute allograft rejection, while the indirect pathway mediates chronic rejection of vascularized grafts [23, 24].
Broadly, two overall phases of allograft injury are observed in sequential biopsy studies: early tubulointerstitial damage [25–29] followed by later microvascular and glomerular changes with progressive fibrosis and atrophy [27, 28, 29, 30]. Most tubular atrophy and fibrosis begins soon after transplantation following injury from ischemia-reperfusion, acute and subclinical rejection, calcineurin nephrotoxicity or BK nephropathy. Further tubular injury is determined by residual subclinical inflammation and calcineurin inhibitor nephrotoxicity, with subsequent appendage of glomerular, microvascular and capillary abnormalities.
Immunosuppression: Role in long-term outcomes
Regardless of ongoing advances in immunosuppression and supportive therapy, there has been modest improvement in the long-term deceased donor kidney graft survival [6, 31]. As it was previously described, there are many factors, including immunological and non-immunological, that might be associated with long-term outcomes. Among the non-immunological factors, toxicity by immunosuppression drugs, especially calcineurin inhibitor (CNI) toxicity is perhaps one of the leading causes of graft dysfunction. It indicates that the choice of immunosuppressive therapy has also a considerable impact on the long-term results. Combining CNIs and corticosteroids offers potent immunosuppression, but may also cause side effects leading to progressive graft dysfunction or an increased risk of death [32, 33]
New immunosuppressive strategies might include inhibitors of mTOR, a downstream effector of phosphatidylinositol-3 kinase that provides the signal for cell proliferation by phosphorylating a cascade of kinases. Mammalian target of rapamycin (mTOR) inhibitors include sirolimus (SRL) and everolimus. SRL is a macrocyclic lactone antibiotic produced from Streptomyces hygroscopicus; everolimus is a derivative of SRL [34]. Recent trials have shown that it is possible to minimize the dose or withdraw CNIs a few weeks after KTx when they are combined with mTOR inhibitors. Moreover, it has been reported that their combination may diminish or avoid the use of corticosteroids. Few studies have explored the possibility of minimizing CNIs while using mTOR inhibitors. A randomized controlled trial (RCT) evaluated the withdrawal of cyclosporine A (CsA) a few weeks after KTx. Patients (N=525) received CsA, sirolimus and steroids for 3 months and were then randomized to continue triple therapy or to stop CsA while increasing the sirolimus dose (95 patients were not randomized because of delayed graft function or rejection). Protocol-biopsies were performed at engraftment and after 12 and 36 months and 484 biopsies were blindly assessed by two pathologists using the Chronic Allograft Damage Index (CADI). After 36 months, the mean CADI score of patients with serial biopsies was significantly lower in those treated with sirolimus and steroids, as was the mean TA score [35]. After 4 years, the mean glomerular filtration rate (GFR) for any quartile of the patients receiving sirolimus and steroids was significantly higher than that of the patients on triple therapy. The benefit was more pronounced if the baseline GFR was ≤45 ml/min. However, the rates of mortality and graft loss were not significantly different between the two groups [36].
Moroever, mTOR inhibitors also decrease the replication rate of cytomegalovirus inside host cells, preclude transplant vasculopathy, and exert anti-oncogenic activity. All of these characteristics offer future opportunities to reduce the risk of long-term allograft failure. However, both types of mTOR inhibitors (SRL and everolimus) are known to cause bone marrow suppression, dyslipidemia, delayed wound healing and at times its use is associated with acute tubular injury.
Even when experimental and clinical studies have shown that mTOR inhibitors may facilitate to solve some imperative problems related to post-KTx immunosuppression more clinical results are needed before changing therapeutic conducts. However, the introduction of mTOR inhibitors may allow the prevention of rejection while minimizing the doses of corticosteroids and CNIs, the agents mainly responsible for causing substantial side effects in renal transplant recipients. Early detection of IF and TA offers the opportunity for replacement of the CNI with mTOR inhibitors. Early detection of CNI-associated graft damage is critical to prevent progressive nephron loss [33].
Cellular and molecular mechanisms underlying chronic allograft nephropathy and graft dysfunction
Loss of kidney function with IF/TA is pleomorphic with a mixed histology, pathophysiology and differential rates of progression. This entity should be considered as a non-specific final event of tubulointerstitial, microvascular and glomerular damage resulting from a variety of insults to the transplanted kidney [1, 2, 10, 11]. It seems likely that epithelial-to-mesenchymal transition (EMT) contributes to IF and TA [11, 37].
Chronic tubulointerstitial lesions progress rapidly during the first months after transplantation. The prevalence of IF/TA in patients receiving a calcineurin inhibitor–based regimen is approximately 40%, 60%, and 90% at 3 months, 2 years and 5 years, respectively. These lesions progress silently, and IF/TA precedes functional deterioration [17].
EMT is an important event in native [37–42] and transplant kidney injury, including chronic allograft IF/TA [11, 43–45]. During EMT, tubular epithelial cells are transformed into myofibroblasts through a stepwise process including loss of cell-cell adhesion and E-cadherin expression, de novo α-smooth muscle actin (α-SMA) expression, actin reorganization, tubular basement membrane disruption, cell migration and fibroblast invasion with production of profibrotic molecules such as collagen type I and III, and fibronectin [38].
Early tubulointerstitial damage appears to result from ischemia-reperfusion injury, acute tubular necrosis, acute and subclinical rejection and calcineurin inhibitor (CNI) nephrotoxicity, superimposed upon donor abnormalities [11, 15, 20]. Later, microvascular and glomerular injury increases frequently as a result of CNI nephrotoxicity, but also from other factors including hypertension, immune-mediated vascular hyperplasia, transplant glomerulopathy and rarely from recurrent or de novo glomerulonephritis. Additional mechanisms of loss of kidney function with IF/TA include internal structural disruption of the kidney, cortical ischemia, and failure to resolve chronic inflammation.
Vascular injury, proliferation, and progression to fibrosis seem critically important in the development of loss of kidney function with IF/TA. The histopathological features of loss of kidney function with IF/TA can be portrayed by proliferative changes of the vascular wall leading to a narrowing of renal arteries, which may contribute to a relative hypoxia of the tissue. Afterward, fibroblasts will contribute to fibrotic scarring in the vascular wall. Loss of kidney function with IF/TA is also characterized by the occurrence of fibrosis in the interstitial tissue along with tubular atrophy [43–47]. The glomeruli are also often affected by various degrees of glomerulosclerosis. Additionally, an inflammatory infiltrate is frequently found consisting of invading lymphocytes and monocytes-macrophages into virtually all affected compartments of the kidney [46–50]. These histopathological changes operate together, contributing to a gradual progression of renal allograft dysfunction.
Differentially expressed genes in chronic allograft nephropathy: Moving from single gene studies to microarray assay reactions
The cellular and molecular mechanisms underlying the histopathological characteristics of loss of kidney function with IF/TA remain obscure. Many cell-cell, cell-matrix, and intracellular pathways have been implicated in the complex pathogenesis of loss of kidney function with IF/TA. Conventional kidney function measurements like serum creatinine and glomerular filtration rates used to predict loss of kidney function with IF/TA have poor predictive values [51] and a reliable diagnosis requires a transplant biopsy [52, 53]. While it is critical to examine structural changes prior to graft loss, predicting graft outcomes strictly based on the kidney biopsy is difficult. Moreover, this invasive procedure has significant costs and risks for patients. Thus, there is an imperative clinical necessity to identify minimally invasive biomarkers that are able to identify early stages of loss of kidney function with IF/TA at a time when changes in therapy may modify outcomes. Different genes have been recently associated to the biology of loss of kidney function with IF/TA [54–56] as an essential initial step to advance the understanding of both immune and non-immune injury mechanisms (Figure 1).
Regardless of the insult causing tissue injury following transplantation, the response to injury consists of both a proliferative response and an inflammatory response, culminating in extracellular matrix deposition. Transforming growth factor beta (TGF-β) has long been recognized as a mediator of loss of kidney function with IF/TA in both humans [54, 56–58] and rodents [59]. TGF-β is a multifunctional peptide that stimulates the synthesis of individual extracellular matrix components, and blocks matrix degradation by stimulating protease inhibitors such as plasminogen activator inhibitor-1 (PAI-1) [60]. Moreover, there is strong evidence to suggest that the intrarenal renin-angiotensin (RAS) system can be activated in loss of kidney function with IF/TA [61–63].
A recently established downstream effector of TGF-β is connective tissue growth factor (CTGF). CTGF can induce cell proliferation, collagen synthesis and chemotaxis in a variety of cells [64]. Recent reports suggest that different domains in CTGF mediate collagen synthesis and myofibroblast differentiation [65]. In vivo, CTGF appears to have pro-sclerotic properties in rodent models of fibrotic kidney disease [66, 67] and in human kidney disease [68]. More recently, CTGF has been linked to chronic graft injury in rat kidney allografts during viral infection [69] and in rejecting mouse kidney allografts [70]. This response is not kidney specific, as it was shown by a study of heart allografts with chronic rejection where over expression of TGF-β was accompanied by elevated CTGF expression within the graft [71]. Moreover, Cheng et al. [72] identified up regulated gene expression of CTGF in human biopsies with loss of kidney function with IF/TA. Interestingly, the authors also found serum CTGF levels to be significantly higher in recipients compared to normal, healthy, non-transplanted individuals.
Fibronectin (FN) is a major component of the extra cellular matrix (ECM). FN has been reported to undergo alternative splicing and produce several isoforms including the extra domain-B (ED-B) containing the embryonic isoform. Siddiqui et al. [73] investigated ED-B FN expression in loss of kidney function with IF/TA and its relationship with endothelins (ET). Allografts were performed between Fisher 344 to Lewis rats. Grafts were harvested at 30, 90 and 140 days after transplant for histological and gene expression analyses with respect to ED-B FN, ET-1 and TGF-β mRNA. ED-B FN protein levels were also assessed by immunohistochemical analysis. Moreover, the authors analyzed human renal allograft biopsies. Results obtained from both human and rat renal allograft tissues suggest that expression of ED-B FN is up regulated in loss of kidney function with IF/TA and such up regulation is mediated via ET-1 and its interaction with TGF-β [73].
Clearly, more experimental and clinical studies are needed to elucidate the vascular protection/ restitution necessary for long-term allograft survival. Renal graft vasculopathy as well as glomerular sclerosis and vascular intimal proliferations are the major characteristics of loss of kidney function with IF/TA. Allograft vascular endothelial cells are the first biological interface between the transplanted donor organ and circulating immunocompetent recipient cells [74]. Chronic allograft nephropathy has been associated with the expression of adhesion molecules, chemokines, and growth factors such as VEGF [75] that have the capacity to mediate endothelial progenitor cells (EPC) recruitment [74]. In a recent publication [76], results suggested that the abnormal expression and reciprocal regulation of angiogenic factors as Angiopoietin 1, Angiopoietin 2, and Tie2 may play important roles in the development of loss of kidney function with IF/TA in rat renal allografts.
While different genes have been associated to the biology of loss of kidney function with IF/TA [54–60], it is essential to advance the understanding of the responses of the allograft to both immune and non-immune injury mechanisms.
Microarray is a powerful technology that measures the expression levels of’ thousands of genes simultaneously and can be an important tool in elucidating patterns for mechanism, diagnosis, prognosis, and treatment of complex, multifactorial diseases, such as loss of kidney function with IF/TA [77, 78]. Microarray technology is based on the principle of complementary, single-stranded, nucleic acid sequences forming double-stranded hybrids; thus, in essence, it is a high-throughput Southern blot, where thousands of single-stranded sequences that are complementary to target sequences are synthesized, or spotted on to a small glass or membrane support [79]. Initially designed to measure the transcriptional levels of RNA transcripts derived from thousands of genes within a genome in a single experiment, this technology has made it possible to relate physiological cell states to gene-expression patterns for studying tumors, disease progression, cellular response to stimuli, drug target identification and transplant injury mechanisms [77–79].
Microarrays are expected to play a critical role in organ transplant in issues related to the identification of molecular mechanisms for acute rejection, chronic injury, effect of drug toxicity and tolerance. Most importantly, microarrays are expected to help identify biomarkers, preferably minimal or noninvasive, in clinical diagnosis for acute rejection, tolerance induction, and early progression to chronic allograft nephropathy [77].
To find appropriate therapeutic strategies, more detailed insights into the molecular pathogenesis of loss of graft function with IF/TA are essential. Transcriptional changes may be detectable prior to histologically apparent fibrosis, and discrimination of inflammatory infiltrates according to the group of expressed genes, promises to both improve diagnoses and optimize treatment strategies.
Microarrays are being utilized to also understand long-term outcomes of organ transplantation in the form of immune-mediated organ injury (Table-1). Recently, Flechner et al. [80] showed different profiles for transplant patients with normal graft function compared with transplant patients with acute rejection using microarray technology. Sarwal et al. [81] evaluated the gene-expression profiles of 67 allograft-biopsy samples from kidney transplant patients observing specific patterns associated with acute rejection. However, all samples from patients with loss of kidney function with IF/TA were grouped in the same cluster showing a similar profiling. In addition, a study using microarrays of kidney biopsies identified 10 genes correlated with the risk of developing chronic rejection [82]. However, these differentially expressed genes were not confirmed in our preliminary study, probably due to differences in the timing of biopsies and/or the immunosuppressive treatment. Another possible reason for the differences between the studies may be the limited number of biopsy samples assayed.
Table-1.
Recent publications including kidney transplant patients with chronic allograft nephropathy for microarray analysis.
| Study | Biological specimens | Patients | Findings |
|---|---|---|---|
| Sarwal et al. [81] | Allograft kidney biopsies | 67 biopsy samples, including 18 kidney transplant patients with chronic allograft nephropathy | The authors found consistent differences among the gene-expression patterns associated with acute rejection, nephrotoxic effects of drugs, chronic allograft nephropathy, and normal kidneys. High homogeneity in biopsies with IF/TA. This study was more concentrated in acute rejection analysis. |
| Flechner et al. [80] | Allograft kidney biopsies | 41 kidney transplant patients tested by DNA microarrays | Gene expression profiles from kidneys with higher Banff chronic allograft nephropathy (CAN) scores confirmed significant up-regulation of genes responsible for immune/inflammation and fibrosis/tissue remodeling. At 2 years the sirolimus-treated recipients have better renal function, a diminished prevalence of CAN and down-regulated expression of genes responsible for progression of CAN. All may provide for an alternative natural history with improved graft survival. |
| Hotchkiss et al. [84] | Allograft kidney biopsies | 16 kidney transplant biopsy samples with CAN by high-density oligonucleotide microarrays, comparing to six normal transplant biopsies. | 112 genes were down regulated in CAN samples. There was differential expression of profibrotic and growth factors that while transforming growth factor-beta induced factor, thrombospondin 1, and platelet derived growth factor-C were up-regulated, vascular endothelial growth factor, epidermal growth factor, and fibroblast growth factors 1 and 9 were down regulated. |
| Mas et al. [83] | Allograft kidney biopsies, PBMC | Renal tissue from kidney transplant patients (KTP) with CAN (n=11) and normal kidneys (NK; n=7) were studied using microarrays. | 728 probe sets were differentially expressed. Genes related to fibrosis and extracellular matrix deposition (i.e., TGF-β, laminin, gamma 2, metalloproteinases-9, and collagen type IX alpha 3) were up-regulated. Genes related to immunoglobulins, B cells, T-cell receptor, nuclear factor of activated T cells, and cytokine and chemokines receptors were also upregulated. EGFR and growth factor receptor activity (FGFR)2 were downregulated in CAN samples. AGT, EGFR, and TGF-β levels were statistical different in urine but not in blood samples of CAN patients when compared to KTx with SRF (P<0.001, P=0.04, and P<0.001, respectively). |
| Scherer et al. [92] | Allograft kidney biopsies | To evaluate events occurring before the overt onset of IF/TA, gene expression profiling of 3-month protocol biopsies from patients with IF/TA was performed in a patient group (n = 8) who developed mild IF/TA [chronic allograft nephropathy (CAN) grade I, by the Banff scoring system] in the subsequent 6-month protocol biopsy (‘progressors’), and in 12 patients without IF/TA at 6 months (‘non-progressors’). | Compared to the non-progressors, the 3-month biopsies of the progressor group showed overexpression of several genes that are important in the T- and B-cell activation and immune response. Genes involved in pro-fibrotic processes were identified in the biopsies of the progressors that preceded the observed IF/TA at 6 months. Furthermore, several genes with transporter and metabolic functions were underrepresented in the progressors in the 3-month biopsies. |
| Kurian et al. [107] | Peripheral blood | DNA microarrays, tandem mass spectroscopy proteomics and bioinformatics were used to identify genomic and proteomic markers of mild and moderate/severe CAN in peripheral blood of two distinct cohorts (n = 77 total) of kidney transplant patients with biopsy-documented histology. | Gene expression profiles reveal over 2400 genes for mild CAN, and over 700 for moderate/severe CAN. A consensus analysis reveals 393 (mild) and 63 (moderate/severe) final candidates as CAN markers with predictive accuracy of 80% (mild) and 92% (moderate/severe). Proteomic profiles show over 500 candidates each, for both stages of CAN including 302 proteins unique to mild and 509 unique to moderate/severe CAN. |
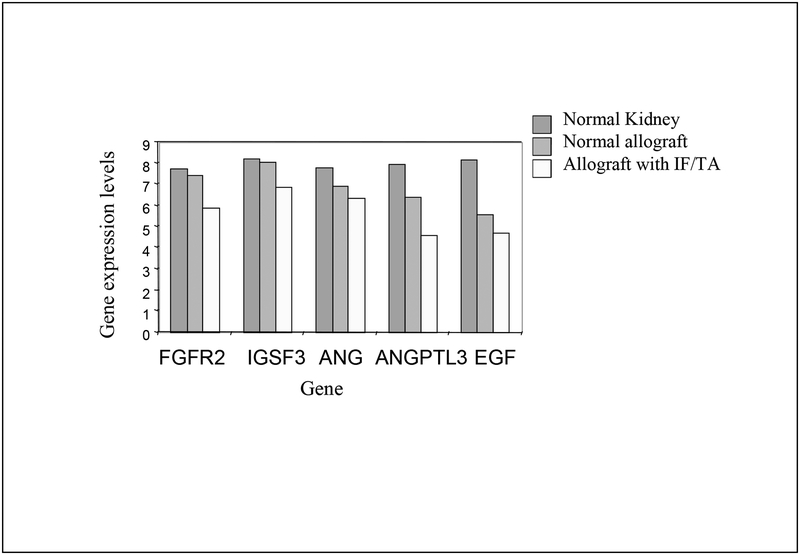
| Maluf et al. [85] | Kidney biopsies from normal kidneys (n = 24), normal allografts (n = 6), and allografts with IF/TA (n = 17) were analyzed using high-density oligonucleotide microarray | Gene ontology classified the differentially expressed genes as related to immune response, inflammation, and matrix deposition. Chemokines (CX), CX receptor (for example, CCL5 and CXCR4), interleukin, and interleukin receptor (for example, IL-8 and IL10RA) genes were overexpressed in IF/TA samples compared with normal allografts and normal kidneys. Genes involved in apoptosis (for example, CASP4 and CASP5) were importantly overexpressed in IF/TA. Genes related to angiogenesis (for example, ANGPTL3, ANGPT2, and VEGF) were downregulated in IF/TA. Genes related to matrix production-deposition were upregulated in IF/TA. A distinctive gene expression pattern was observed in IF/TA samples compared with normal allografts and normal kidneys. We were able to establish a trend in gene expression for genes involved in different pathways among the studied groups. The top-scored networks were related to immune response, inflammation, and cell-to-cell interaction, showing the importance of chronic inflammation in progressive graft deterioration. |
PBMC, peripheral mononuclear cells, IF/TA, interstitial fibrosis and tubular athrophy; KTx, kidney transplant patients; SRF, stable renal function.
Underlying triggers for loss of kidney function with IF/TA may in fact be impossible to decipher when a graft with established injury is sampled. As we an others have published in previous studies [83, 84], an extensive homogeneity of genomic responses is seen at this time. The signature of increased fibrogenesis may in fact be simply reflecting the current injury mechanism rather than the trigger. Many of the pathways involved in chronic injury and fibrogenesis might be regulated very early in the course of the injury when the downstream effects of these alterations are still not evident by pathology. This suggests a valuable role of early graft sampling by microarray technology, prior to the onset of chronic pathology, to identify triggers and early molecular markers for loss of kidney function with IF/TA disease progression. Thus, sequential genomic sampling of the graft and periphery (i.e., blood, urine) during early post-transplantation may be the approach most likely to result in the greatest yield for mechanistic understanding.
Some limitations in studies using microarrays include among others (specifically in the transplant patient group) the heterogeneity of graft specimens, timing of sample collection (i.e., protocol biopsy performed at dissimilar times in different studies), influence of donor characteristics, immunosuppressant drugs, and sample collection and preparation. All these factors are critical and complicate inferences regarding the underlying mechanisms of graft injury, rejection, and immune regulation [79].
Portraying progression to loss of graft function with IF/TA
Histopathological evaluation of biopsies is an indispensable method for the detection of allograft injury, although it tends to suffer from the subjectivity of the reviewer [46–50]. However, evaluation of molecular changes, by using DNA microarrays, may add important aspects to the complexity and an improvement of classification and diagnosis [77–79] and may help to identify critical molecules and pathways related to graft injury.
A major impediment in the study of loss of kidney function with IF/TA has been the lack of prospective, longitudinal histological data from studies of human tissues with chronic graft dysfunction. Thus, we recently evaluated how the gene expression changed from normal donor kidneys, normal allografts, and IF/TA samples [85]. Using Jonckheere-Terpstra test for trend analysis, we were able to establish a trend in gene expression for genes involved in the different pathways leading to progressive graft deterioration with loss of kidney function with IF/TA. Normal native kidneys, normal allografts, and IF/TA samples were used for the analysis.
Activation of the humoral branch of the immunological response was observed. However, as we reported recently [83, 85], genes related to both B- and T- cells were involved in IF/TA sample profiles, and a predominantly immunologic pattern and/or response was not clearly identified. Moreover, when samples for different groups were compared for identifying specific cell types and/or immunological branch predominance, a positive trend leading to IF/TA was also observed, reaffirming our initial results.
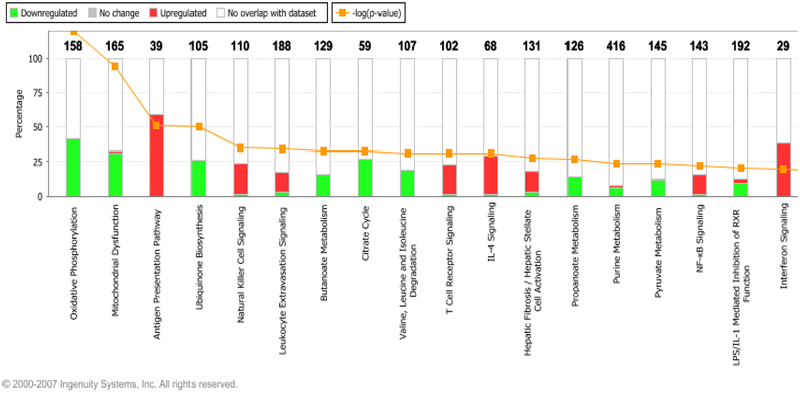
Interesting, a negative trend in expression of genes related to angiogenesis in IF/TA samples was observed (Figure 2). In chronic inflammation, where tissue destruction and mononuclear cell infiltration are dominant, the persistent delivery and local expression of angiogenic factors can serve to sustain the angiogenesis response [86, 87]. In physiological conditions, angiogenesis is thought to facilitate the repair of injured tissues and to restore oxygenation. In diseases such as glomerulonephritis, ischemic nephropathy, and tubulointerstitial fibrosis, and in the aging process, accelerated attrition of the microvasculature (as a result of inefficient delivery of angiogenic factors) results in ongoing and persistent hypoxia, which can result in further tissue destruction [88, 89]. Angiogenesis might play an important role in IF/TA progression and deserves further evaluation and study as it has been previously demonstrated in rat models [76]. Finally, as was previously shown, expression of matrix production and deposition related gene expression was increased in kidneys with IF/TA [57–60, 68–73]. From the analysis of the canonical pathways, we observed that antigen presentation pathway, leukocyte extravasation signaling, and natural killer cell signaling were among the more important identified pathways (Figure 3). We are conscious about the limitation related to inter-individual variation, but this study design provided an initial step for the analysis of the more important pathways leading to progressive graft deterioration associated to loss of kidney function with IF/TA.
Figure 2:
Negative trend in gene expression for genes involved in angiogenesis in kidney allograft samples with IF/TA.
Figure 3:
Top canonical pathways identified for the analysis of IF/TA samples. Over expressed genes: red boxes, under expressed genes: green boxes. The numbers over the boxes indicated the number of genes that are part of the specific pathway identified as differentially expressed in the dataset (www.ingenuity.com) [200].
Evidence suggests that the first few months after transplantation are critical in the development of IF/TA and that protocol biopsies may be a valuable means of detecting early signs of chronic allograft damage that have yet to become clinically apparent. In particular, early protocol biopsies have shown that the presence of tubulointerstitial damage and vascular chronic damage are powerful predictors of allograft survival [90]. Furthermore, serial protocol biopsies have shown that both interstitial and vascular chronic damage rapidly increase during the first 6 months after transplantation, then slowly thereafter [91]. Scherer et al. [92] in a recent publication found a panel of significant changes in the transcriptome that preceded the histological phenotype of IF/TA by 3 months. Specifically, transcriptional levels of genes involved in complement activation, leukocyte homing, T- and B-cell infiltration and activation, beginning pro-fibrotic events, functional changes in specific metabolic and transport properties of the nephron, or in EMT, were significantly different in early protocol biopsies of patients with consequent progression to IF/TA. These findings are in concordance with our previous reports [83, 85] and further corroborate the assumption that inflammation that is quantitatively below the diagnostic threshold of acute rejection is involved in early stages of progressive renal allograft damage [93].
Role of Donor Kidney Biopsies in Renal Transplantation: How much the transcriptomics of donor biopsies at time-zero can tell us about short and long term function?
It is recommended that all donor kidneys should be examined by a pre-transplantation or post-perfusion biopsy [94]. Histological evaluation of the donor biopsy provides a baseline with which future biopsies may be compared. Pre-existing lesions such as capillary thrombosis, arteriolosclerosis, glomerulosclerosis and interstitial fibrosis can be documented so that the occurrence of the same lesions in post-transplantation biopsies is not misinterpreted as evidence of tacrolimus/cyclosporine toxicity or chronic allograft nephropathy. A donor kidney biopsy is mandatory in clinical settings such as an older donor (age>55), hypertension, diabetes mellitus, acute tubular necrosis, disseminated intravascular coagulation, or unexplained rise in serum creatinine prior to death [94].
Interpretation of a donor kidney biopsy includes a semi-quantitative assessment of the degree of glomerulosclerosis, arteriosclerosis, and interstitial fibrosis present. However, the extent of acceptable chronic changes within the donor kidney has not yet been clearly established. It is a common practice not to use kidneys with more than 20% sclerotic glomeruli [95]. In most transplant centers, surgeons are cautious to use any kidney with more than mild interstitial fibrosis (greater than 25% of cortical area affected) or mild arteriosclerosis (greater than 25% luminal occlusion).
Analysis of individual organ quality and outcomes requires integration of the complexity of the donor/recipient variables and events. In an initial study, Hauser et al. [96] determined the genome-wide gene-expression pattern using cDNA microarrays in three groups of 36 donor kidney biopsies including living donor kidneys with primary function, deceased donor kidneys with primary function and deceased donor kidneys with biopsy proven acute renal failure. The authors retrieved 132 genes that significantly separated living from deceased kidneys, and 48 genes that classified the donor kidneys according to their post-transplant course. The main functional roles of these genes were cell communication, apoptosis and inflammation. In particular, members of the complement cascade were activated in cadaveric, but not in living donor kidneys. The current study revealed a large cumulative effect of donor factors reflected by a distinct gene-expression profile and some individual donor and recipient factors on early graft function. Two distinct gene-expression patterns, which could clearly separate living from deceased donors and primary graft function from post-ischemic acute renal failure, were observed for the data analysis [96].
Because there is evidence showing that donor characteristics as well post-reperfusion changes might be related to the long-term graft outcomes, we studied gene expression profiles in 42 paired kidney biopsies samples from 21 unique deceased donors donor biopsies at pre-implantation and at 60 minutes post-reperfusion [97]. Gene expression profiling was performed using high-density oligonucleotide microarrays. A total of 2,405 probe sets (corresponding to 1,947 unique genes) were significantly differentially expressed comparing the paired pre-implantation to post-reperfusion samples. Core analysis was performed to interpret the data set in the context of biological processes, pathways and molecular networks. From this analysis, 59 networks with a score higher than 15 were identified. The top functions for the higher scored networks included DNA replication, recombination, and repair; connective tissue development; and cell-to-cell interaction. We identified 34 probe sets (28 genes) significantly associated with warm ischemia time in the post-reperfusion biopsies. The top canonical pathways affected were calcium signaling, death receptor signaling, and IL-4 signaling. The top genes over expressed in longer warm ischemia times were GREM1, LMOD1, MEF2A, ING1 (involved in cytokine activity, p38 MAPK signaling, negative cell proliferation) while the genes down regulated with longer warm ischemia times included HLADQB1, SUB1, TIPRL, XAF1, TH1L, OSGEP, CTSC (involved in proteolysis, IL-4 signaling, and apoptosis). Further evaluation of the identified genes, might provide insights about the effect of ischemia/reperfusion in the kidney grafts [97]. Moreover, because transplant function in the recipient is largely dependent on donor factors, the gene expression patterns in biopsy at time zero may provide critical information about short and long term graft function when associated with clinical parameters. In an unsupervised analysis of these biopsies we observed that samples clustered in two different groups. We analyzed donor and recipient characteristics between the two groups as well as cold ischemia time, warm ischemia time, and kidney function at 1 week, 1 month, and 6 months post-KTx. We observed a trend in significance in ischemia time between groups (P=0.06).
Tissue injury of the allograft produced by ischemia is a critical early preoperative insult that enhances the risk of acute tubular necrosis and delayed graft function (DGF). Also, reperfusion injury, endothelial dysfunction, and renovascular resistance alterations contribute to early tubular injury and ultimately tubular necrosis [98]. Finally, leukocyte infiltration of the injured tissue increases the risk for acute rejection, further predisposing these kidneys to progressive atrophic and fibrotic changes in the tubulointerstitial compartment [99], which is a major cause for late graft loss [100]. DGF is thus an important clinical and pathophysiological entity that has been associated with long-term graft outcomes, and as consequence. It is of critical importance to better understand its molecular mechanisms and diminish its rate. To evaluate differences in gene expression patterns between kidneys that developed DGF vs. kidney that did not, gene expression in donor biopsy grafts at time zero were evaluated [101]. Gene expression profiling was performed on donor kidney tissues from 33 deceased donor (DD) kidneys with the use of microarrays. DD were classified as grafts with immediate function (non-DGF; n=21) and grafts with DGF (n=12). DGF was defined as a dialysis requirement in the first week after transplantation. Demographic donor and recipient information was collected. The robust-multiarray average method was used to estimate probe set expression summaries. Logistic regression was used to identify genes significantly associated with DGF development. Patients were followed for 3 months after KTx. Thirty-eight probe sets (n=36 genes) were univariably differentially expressed in DD with DGF when compared with DD with non-DGF (alpha=0.001). Sixty-nine probe sets (n=65 genes) were differentially expressed in DD kidneys with DGF when compared with DD with non-DGF after adjusting for cold ischemia time (alpha=0.001). Gene ontology terms classified the over expressed genes in DD with DGF as principally related to cell cycle/growth, signal transduction, immune response, and metabolism. TNFRSF1B was over expressed in DD with DGF. Cold ischemia time was a predictor of DGF independently of the preservation method. We also identified a set of 36 candidate genes for DGF in DD, with genes involved in the inflammatory response being the more important.
Mueller et al [102], showed microarray results of 87 consecutive implantation biopsies taken post-reperfusion in 42 DD kidneys and 45 living (LD) donor kidneys. Unsupervised analysis separated the 87 kidneys into three groups: LD, DD1 and DD2. Kidneys in DD2 had a greater incidence of DGF (38.1 vs. 9.5%, p<0.05) than those in DD1. However, clinical and histopathological risk scores did not discriminate the two identified groups (DD1 and DD2). A total of 1051 transcripts were differentially expressed between DD1 and DD2, but no transcripts separated DGF from immediate graft function (adjusted p< 0.01). Principal components analysis revealed a continuum from LD to DD1 to DD2, i.e. from best to poorest functioning kidneys. Within DD kidneys, the odds ratio for DGF was significantly increased with a transcriptome-based score and recipient age (p<0.03) but not with clinical or histopathologic scores. The authors concluded that the transcriptome reflects kidney quality and susceptibility to DGF better than available clinical and histopathological scoring systems.
Recently (103), we studied kidney biopsies from 49 DD kidneys using microarrays. Patients were followed-up prospectively for 1 year post-KTx. The patients were classified by allografts with normal function vs. impaired function (Glomerular Filtration Rate (GFR)<45 vs. GFR≥45 ml/min/1.73 m2) at 1 year post-KTx. To simultaneously model delayed graft function (DGF)(yes/no) and graft function at 1 year post-KTx using donor gene expression as the independent predictor variable, the two outcomes were combined to form a bivariate response matrix and for each probe set, a vector generalized linear model was fit. Differential expression profiles were analyzed on the level of molecular and biological function using Interaction Networks and Functional Analysis [200]. Genes related with renal damage (AGTR1, C1QA, PLAT) and damage of renal tube (TLR4) were present in the list. 79 probe sets predictive of the multivariate response (DGF and graft function at 1 year post-KTx) were significant (P<0.01). The top molecular and cellular functions included cellular growth and proliferation, cellular development, and cell cycle. The top identified predictive of outcome genes included ATM, BAT3, CAP2B, CD44, CEACAM8, CITED2, and DUSP6. This preliminary study showed that the combination of donor, recipient characteristics, and set of molecular markers in the pre-implantation biopsy might provide useful information about post-transplant long-term graft outcomes.
These described studies provide an initial and essential step in the elucidation of possible pathways involved in early renal allograft injury and functionality. Well-controlled human studies in organ transplantation, supported by robust data analyses and independent validation sets, should identify novel target molecules and molecular pathways of injury and progression to chronic allograft dysfunction and help to discovery novel preventative therapeutic opportunities in transplantation [104].
Disease progression biomarker discovery: Integration of protocol biopsy, gene expression profiling, and clinical parameters.
Identifying surrogate markers of graft outcome in kidney transplantation is a critical task for the transplant community. Rapidly evolving technologies for genomics have created new opportunities to develop minimally invasive biomarkers. Recent studies have identified genes that are differentially expressed at the mRNA level in kidney biopsies in the presence of loss of kidney function with IF/TA [65, 66, 72–76]. The limitation of these studies is that they require an invasive transplant biopsy. There is accumulating evidence that noninvasive immune monitoring may be useful in the early period after renal transplant [105, 106], particularly with regard to predicting the presence of acute rejection. It is less clear whether loss of kidney function with IF/TA is also associated with consistent changes in peripheral blood or urine cells [103].
For establishing a relationship between biopsy and urine-blood findings, we evaluated three of the differentially expressed markers in kidney biopsies with IF/TA in urine and blood samples from the same patients [107]. From this analysis we observed that the gene expression levels of the studied markers in urine samples were more representative of the gene expression in the kidney biopsies than peripheral blood mononuclear cells (PBMC) samples. These results showed that the evaluation of molecular markers in urine samples could represent an invaluable resource for monitoring kidney transplant recipients. Also, we performed whole gene expression analysis in paired biopsy-blood samples of patients with biopsies with IF/TA (Figure 4).
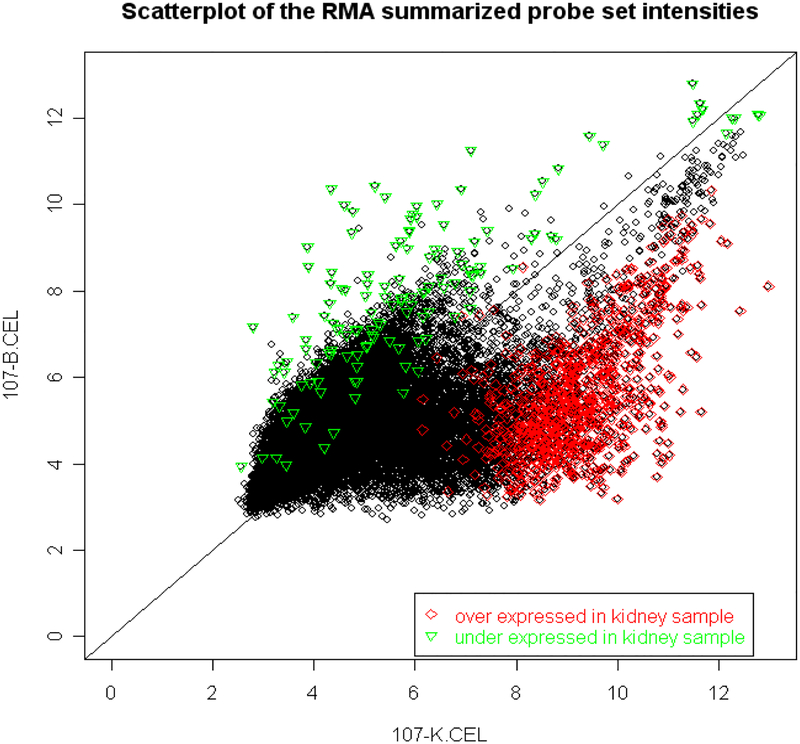
Figure 4:
Two Affymetrix microarrays (HGU133Av2) were run for two samples (peripheral blood and kidney biopsy) from the same patient (ID 107). Samples were collected at the biopsy time and the biopsy showed IF/TA. In comparing gene expression of the kidney sample to the blood sample, 902 probe sets were found to be over expressed, 136 probe sets were under expressed (False Discovery Rate (FDR) <5%). The scatter plot of the intensities for these probe sets is shown in which green points represent over expressed probe sets and red points represent under expressed probe sets in the kidney sample.
In a recent publication, Kurian et al. [108] identified several unique signatures of transcript and protein biomarkers with high predictive accuracies for mild and moderate/severe loss of kidney function with IF/TA using peripheral blood. The results showed several hundred mRNA and proteomic biomarkers in peripheral blood defining unique proteogenomic signatures and demonstrated correlations with histologically mild (80% class prediction accuracy for top 50 gene candidates) and moderate/severe loss of kidney function with IF/TA (92% class prediction accuracy for top 50 gene candidates). The authors recognized the complications in the genetic and clinical diversity of transplant patients, multiple clinical centers, the cellular complexity of peripheral blood, and the impact of factors such as immunosuppression, environment and time post-transplant. The authors also suggested that because the study was based on profiling subjects with biopsy-proven IF/TA, the evidence presented supports the conclusion that the identified candidate genes are diagnostic.
Moreover, a recent publication showed the potential of molecular profiling for evaluating new possible therapeutic conducts in kidney transplant patients with IF/TA [109]. However, recent microarray studies for diagnosis of rejection, and comparable studies in cancer, suggest that a set of less than 20 genes provides adequate power to identify disease classes with a diagnostic accuracy of >80% [95, 110–112]. Reduction to the most predictive genes will support biological credibility and allow for transition of high-throughput technology to alternative methods such as real-time PCR (QPCR). These alternatives to high-throughput methods will allow large sample size evaluation for biomarker-validation (cost effective technologies) and will provide a more dynamic monitoring of patients and grafts.
Before a biomarker can be considered a good surrogate that would be clinically useful for early diagnosis and disease prognosis, careful design studies need to be performed for development and validation of these biomarkers. Prospective studies with appropriate number of patients are needed for validating these initial results.
Proteomic-based approaches and chronic allograft dysfunction
Advances have been performed in discerning molecular pathways associated with allograft injury, especially in acute rejection. However, these findings might be of limited diagnostic values. First, changes in gene expression levels do not always result in changes in the corresponding protein levels. Moreover, even when changes in gene expression levels result in changes in protein levels, the changes observed in the allograft kidney tissue may not correlate well with those in the peripheral blood and/or urine.
The main difference between genomics and proteomics is that the genome is a static entity that goes relatively unchanged from day to day, whereas the proteome is a dynamic collection of proteins that demonstrates significant variation between individuals, between cell types, and between entities of the same type but under different pathologic or physiologic conditions [113]. Characterization of the allograft, urine and plasma proteome is the next logical step toward investigation of kidney recipients with progression to IF/TA. Protein signatures will be useful tools in the development of disease biomarkers. Understanding the proteome, the structure and function of each protein and the complexities of protein-protein interactions will be critical for developing the most effective diagnostic techniques and disease treatments in the future [113].
Advances in analytical instrumentation, as well as technology for protein purification and identification, have driven proteomics research to a different approach where comprehensive protein databases for individual conditions can be used to characterize individual patients and disease states [114].
Thus far, the few research studies published using an unbiased proteomic approach to identify protein biomarkers for renal allograft monitoring [115–119] have been focused in subclinical rejection and acute rejection. Interestingly, each group found a different pattern of protein biomarkers in urine samples that were associated with allograft rejection. This discrepancy is not surprising, as each study had differences in disease definition, sample collection/handling, protocol for protein separation/visualization and data analysis.
In a recent publication [119], the authors used mass spectrometry to analyze differences in the urinary polypeptide patterns of 32 patients with chronic allograft dysfunction (14 with pure interstitial fibrosis and tubular atrophy and 18 with chronic active antibody-mediated rejection) and 18 control subjects (eight stable recipients and 10 healthy control subjects). Unsupervised hierarchical clustering showed a good association of samples in concordance with the biomedical conditions. The proteome signature of the pure interstitial fibrosis and tubular atrophy group differed from that of the chronic active antibody-mediated rejection group. These results were confirmed in an independent validation set. The 14 protein ions that best discriminated between these two groups correctly identified 100% of the patients with pure interstitial fibrosis and tubular atrophy and 100% of the patients with chronic active antibody-mediated rejection. These results raise the expectations of the proteomic-based approaches to identify novel biomarkers.
The accomplishment of any biomarker development study will be related to the patient cohorts used for the initial analysis and subsequent validation, as well as the commitment to design prospective trials implementing promising biomarkers into patient management to determine their clinical utility [120].
Expert commentary
Chronic allograft nephropathy, now defined as interstitial fibrosis and tubular atrophy not otherwise specified, is a near universal finding in transplant kidney biopsies by the end of the first decade post-transplantation (9). After excluding death with functioning graft, caused by cardiovascular disease or malignancy, chronic allograft nephropathy is the leading cause of graft failure (9, 28). Subclinical cellular or humoral rejection, and chronic calcineurin inhibitor toxicity have been described as some of the more important factors associated with progression to allograft function deterioration.
While significant progress has been achieved in short-term outcomes, long-term survival has only marginally improved. Adaptation of immunosuppressive drugs to the individual needs of every patient at every time point after transplant will be essential to improve long-term outcomes. Thus, assays are required that detect allograft injury very early, which implies frequent noninvasive measurements (e.g. in urine or serum).
Moreover, recent advances in genomics and proteomics have provided the basis for a better understanding of the molecular pathways involved in the disease progression that can lead to new biomarker and therapy discovery.
Five-year view
Loss of kidney function with IF/TA is the final event of cumulative damage, where a series of time-dependent immune and non-immune mechanisms converge and injure the allograft, leading to chronic interstitial fibrosis and tubular atrophy. Allograft damage is common, progressive, time-dependent, and clinically important. Therapeutic strategies are needed to provide primary prevention of this process. A better understanding of the molecular events involved in disease evolution could lead to the discovery of markers of disease initiation and progression as well as new targets for drug development.
The quality of diagnostic tools is defined by their sensitivity and specificity. In general, biomarker patterns will confer significantly more information than a single measurement and enable better specificity and sensitivity. Novel screening technologies in the fields of genetics, genomics, protein profiling (proteomics), and biochemical profiling (metabolomics) allow for an unbiased or “systems biology” approach to the study loss of kidney function with IF/TA. Proteomics and metabolomics offer a nonbiased suite of tools to address pathophysiologic mechanisms from various levels by integrating signal transduction, cellular metabolism, and phenotype analysis. The integration of the information resulting from these systems is an important ongoing area that still under development but that will result in important advances in biomarker discovery in the transplantation area [121, 122].
However, the implementation of potential biomarkers as clinical markers is not an easy undertaking and will require the concerted effort of the immunologists, molecular biologists, transplantation specialists, geneticists, and experts in bioinformatics. Rigorous prospective validation studies will be needed using large sets of independent patient samples.
Key issues.
Kidney transplantation has evolved as the treatment of choice in patients with end-stage renal failure.
The development of loss of kidney function with IF/TA, defined as the progressive development of interstitial fibrosis, tubular atrophy, occlusive vascular changes and glomerulosclerosis is a major barrier for long-term kidney allograft survival.
Many of the pathways involved in chronic injury and fibrogenesis might be regulated very early in the course of the injury when the downstream effects of these alterations are still not evident by pathology, suggesting an additional role of early graft sampling by microarray technology, prior to the onset of chronic pathology, to identify triggers and early molecular markers for loss of kidney function with IF/TA disease progression.
A prospective approach to monitor molecular changes in transplant patients could be used to predict progression to chronic graft dysfunction. Moreover, if predicting progression to chronic allograft dysfunction can be carried out in peripheral blood and/or urine samples alone, then the potential for a minimally invasive strategy would be realized. The development of assays that will allow transplant physicians to detect allograft dysfunction progression and predict long-term outcomes is vital for the success of transplantation.
Before a biomarker can be considered as a good surrogate that would be clinically useful for early diagnosis and disease prognosis, careful design studies need to be performed for development and validation of these biomarkers.
Financial disclosure
The research results included in this report were partially supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant, R01DK080074. The authors declare that they have no competing interests.
Footnotes
Website: 200-(http://www.ingenuity.com)
Contributor Information
Valeria R Mas, Molecular Transplant Research Laboratory. Transplant Division, Department of Surgery, Molecular Medicine Research Building, Virginia Commonwealth University, 1220 E. Broad Street, Richmond, VA, 23298.
Kellie J Archer, Department of Biostatistics & Massey Cancer Center, Virginia Commonwealth University, 730 East Broad St., 3-022, Richmond, VA 23298-0032.
Mariano Scian, Molecular Transplant Research Laboratory. Transplant Division, Department of Surgery, Molecular Medicine Research Building, Virginia Commonwealth University, 1220 E. Broad Street, Richmond, VA, 23298.
Daniel G Maluf, Transplant Division, Virginia Commonwealth University Medical Center, Medical College of Virginia Hospitals, 1200 East Broad Street, West Hospital, 9th Fl, South Wing, PO Box 980254, Richmond, VA, 23298.
References
Papers of special note have been high-lighted as:
* = of interest
** = of considerable interest
- 1-.Hariharan S, Johnson CP, Bresnahan BA, Taranto SE, McIntosh MJ, Stablein D. Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med 342(9), 605–612 (2000). [DOI] [PubMed] [Google Scholar]
- 2-.Meier-Kriesche HU, Schold JD, Srinivas TR, Kaplan B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am J Transplant 4(3), 378–383 (2004). [DOI] [PubMed] [Google Scholar]
- 3-.European Mycophenolate Mofetil Study Group. Placebo-controlled study of mycophenolate mofetil combined with cyclosporin and corticosteroids for prevention of acute rejection. Lancet 345(8961), 1321–1325 (1995). [PubMed] [Google Scholar]
- 4-.Halloran PF, Langone AJ, Helderman JH, Kaplan B. Assessing long-term nephron loss: is it time to kick the CAN grading system? Am J Transplant. 4(11), 1729–1730 (2004). [DOI] [PubMed] [Google Scholar]
- 5-.Solez K, Colvin RB, Racusen LC, et al. Banff ‘05 Meeting Report: differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy (‘CAN’). Am J Transplant. 7(3), 518–726 (2007). [DOI] [PubMed] [Google Scholar]
- 6-.2008 Annual Report of the U.S Organ Procurement and Transplantation Network and the Scientific Registry of Transplant Recipients: Transplant Data 1998–2007. U.S. Department of Health and Human Services, Health Resources and Services Administration, Healthcare Systems Bureau, Division of Transplantation, Rockville, MD. [Google Scholar]
- 7-.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 8(4), 753–760 (2008). [DOI] [PubMed] [Google Scholar]
- 8-.Womer KL, Kaplan B. Recent development in kidney transplantation- a critical assessment. Am J Transplant 9 (6), 1265–1271 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9-.Nankivell BJ, Chapman JR. Chronic allograft nephropathy: current concepts and future directions. Transplantation. 81(5), 643–54 (2006). [DOI] [PubMed] [Google Scholar]; * This interesting review provides a very detailed description of the pathology, risk factors, utility of actual diagnosis tool, possible new therapeutic strategies and future challenges.
- 10-.Robertson H, Ali S, McDonnell BJ, Burt AD, Kirby JA. Chronic renal allograft dysfunction: The role of T cell-mediated tubular epithelial to mesenchymal cell transition. J Am Soc Nephrol. 15(2), 390–397 (2004). 15(2), 390–397 (2004). [DOI] [PubMed] [Google Scholar]
- 11-.Racusen LC, Solez K, Colvin R. Fibrosis and atrophy in the renal allograft: Interim report and new directions. Am J Transplant 2(3), 203–206 (2002). [DOI] [PubMed] [Google Scholar]
- 12-.Collini A, Kalmar P, Dhamo A, Ruggieri G, Carmellini M. Renal transplant from very old donors: how far can we go? Transplantation. 87(12), 1830–1836 (2009). [DOI] [PubMed] [Google Scholar]
- 13-.Vadivel N, Tullius SG, Chandraker A. Chronic allograft nephropathy. Semin Nephrol. 27(4), 414–429 (2007). [DOI] [PubMed] [Google Scholar]
- 14-.Roodnat JI, van Riemsdijk IC, Mulder PG, et al. The superior results of living donor renal transplantation are not completely caused by selection or short cold ischemia time: a single center, multivariate analysis. Transplantation 75 (12), 2014–2018 (2003). [DOI] [PubMed] [Google Scholar]
- 15-.Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med 349(24), 2326–2333 (2003). [DOI] [PubMed] [Google Scholar]
- 16-.Serón D Interstitial fibrosis and tubular atrophy in renal allograft protocol biopsies as a surrogate of graft survival. Transplant Proc. 41(2), 769–770 (2009). [DOI] [PubMed] [Google Scholar]
- 17-.Solez K, Vincenti F, Filo RS. Histopathologic findings from 2-year protocol biopsies from a U.S. multicenter kidney transplant trial comparing tarolimus versus cyclosporine: a report of the FK506 Kidney Transplant Study Group. Transplantation 66(12):1736–1740 (1998). [DOI] [PubMed] [Google Scholar]
- 18-.Vincenti F, Larsen C, Durrbach A, et al. Belatacept Study Group. Costimulation blockade with belatacept in renal transplantation. N Engl J Med. 353(8), 770–781 (2005). [DOI] [PubMed] [Google Scholar]
- 19-.Morrissey PE, Yango AF. Renal transplantation: older recipients and donors. Clin Geriatr Med. 22(3), 687–707 (2006). [DOI] [PubMed] [Google Scholar]
- 20-.Keith DS, Cantarovich M, Paraskevas S, Tchervenkov J. Recipient age and risk of chronic allograft nephropathy in primary deceased donor kidney transplant. Transpl Int. 19(8), 649–656 (2006). [DOI] [PubMed] [Google Scholar]
- 21-.Moreso F, Ibernon M, Goma M, et al. Subclinical rejection associated with chronic allograft nephropathy in protocol biopsies as a risk factor for late graft loss. Am J Transplant. 6(4), 747–52 (2006). [DOI] [PubMed] [Google Scholar]
- 22-.Freese P, Svalander CT, Molne J, Norden G, Nyberg G. Chronic allograft nephropathy--biopsy findings and outcome. Nephrol Dial Transplant. 16(12), 2401–2416 (2001). [DOI] [PubMed] [Google Scholar]
- 23-.Sayegh MH & Turka LA. The role of T-cell costimulatory activation pathways in transplant rejection. N Engl J Med 338, 1813–1821 (1998). [DOI] [PubMed] [Google Scholar]
- 24-.Womer KL, Stone JR, Murphy B, et al. Indirect allorecognition of donor class I and II major histocompatibility complex peptides promotes the development of transplant vasculopathy. J Am Soc Nephrol 12: 2500–2506 (2001). [DOI] [PubMed] [Google Scholar]
- 25-.Isoniemi H, Taskinen E, Hayry P. Histological chronic allograft damage index accurately predicts chronic renal allograft rejection. Transplantation 58 (11), 1195–1198 (1994). [PubMed] [Google Scholar]
- 26-.Nankivell BJ, Borrows RJ, Fung CL, et al. Delta analysis of posttransplantation tubulointerstitial damage. Transplantation 78 (3), 434–441 (2004). [DOI] [PubMed] [Google Scholar]; ** Very interesting description of the mechanisms involved in tubulointerstitial damage after kidney transplantation.
- 27-.Kuypers DR, Chapman JR, O’Connell PJ, et al. Predictors of renal transplant histology at three months. Transplantation 67 (9), 1222–1230 (1999). [DOI] [PubMed] [Google Scholar]
- 28-.Cosio FG, Pelletier RP, Sedmak DD, et al. Pathologic classification of chronic allograft nephropathy: pathogenic and prognostic implications. Transplantation 67 (5), 690–696 (1999). [DOI] [PubMed] [Google Scholar]
- 29-.Weir MR, Wali RK. Minimizing the risk of chronic allograft nephropathy. Transplantation. 87(8 Suppl):S14–18 (2009). [DOI] [PubMed] [Google Scholar]
- 30-.Sijpkens YW, Doxiadis II, van Kemenade FJ, et al. Chronic rejection with or without transplant vasculopathy. Clin Transplant 17 (3), 163–170 (2003). [DOI] [PubMed] [Google Scholar]
- 31-.Meier-Kriesche HU, Schold JD, Kaplan B. Long-term renal allograft survival: have we made significant progress or is it time to rethink our analytic and therapeutic strategies? Am J Transplant 4(8), 1289–1295 (2004). [DOI] [PubMed] [Google Scholar]
- 32-.Ponticelli C Can mTOR inhibitors reduce the risk of late kidney allograft failure? Transpl Int. 21(1), 2–10 (2008). [DOI] [PubMed] [Google Scholar]
- 33-.Wali RK, Weir MR. Chronic allograft dysfunction: can we use mammalian target of rapamycin inhibitors to replace calcineurin inhibitors to preserve graft function? Curr Opin Organ Transplant. 13(6), 614–621 (2008). [DOI] [PubMed] [Google Scholar]
- 34-.Halloran PF. Immunosuppressive drugs for kidney transplantation. N Engl J Med 351 (26), 2715–2729 (2004). [DOI] [PubMed] [Google Scholar]
- 35-.Mota A, Arias M, Taskinen EI, et al. Sirolimus-based therapy following early cyclosporine withdrawal provides significantly improved renal histology and function at 3 years. Am J Transplant 4(6), 953–961 (2004). [DOI] [PubMed] [Google Scholar]
- 36-.Russ G, Segoloni G, Oberbauer R, et al. Superior outcomes in renal transplantation after early cyclosporine withdrawal and sirolimus maintenance therapy, regardless of baseline renal function. Transplantation 80(9), 1204–1211 (2005). [DOI] [PubMed] [Google Scholar]
- 37-.Bedi S, Vidyasagar A, Djamali A. Epithelial-to-mesenchymal transition and chronic allograft tubulointerstitial fibrosis. Transplant Rev (Orlando). 22(1), 1–5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38-.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 110 (3), 341–350 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39-.Liu Y Epithelial to mesenchymal transition in renal fibrogenesis: pathologic significance, molecular mechanism, and therapeutic intervention. J Am Soc Nephrol. 15 (1), 1–12 (2004). [DOI] [PubMed] [Google Scholar]
- 40-.Zeisberg EM, Potenta SE, Sugimoto H, Zeisberg M, Kalluri R. Fibroblasts in kidney fibrosis emerge via endothelial-to-mesenchymal transition. J Am Soc Nephrol. 19(12), 2282–2287 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41-.Yang J, Liu Y. Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am J Pathol. 159(4),1465–75 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42-.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 112(12), 1776–1784 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43-.Djamali A, Reese S, Yracheta J, Oberley T, Hullett D, Becker B. Epithelial-to-Mesenchymal Transition and Oxidative Stress in Chronic Allograft Nephropathy. Am J Transplant. 5(3), 500–509 (2005). [DOI] [PubMed] [Google Scholar]
- 44-.Vongwiwatana A, Tasanarong A, Rayner DC, Melk A, Halloran PF. Epithelial to mesenchymal transition during late deterioration of human kidney transplants: the role of tubular cells in fibrogenesis. Am J Transplant. 5(6),1367–1374 (2005). [DOI] [PubMed] [Google Scholar]
- 45-.Hertig A, Verine J, Mougenot B, et al. Risk Factors for Early Epithelial to Mesenchymal Transition in Renal Grafts. Am J Transplant. 6(12), 2937–2946 (2006). [DOI] [PubMed] [Google Scholar]
- 46-.Serón D, Moreso F, Fulladosa X, Hueso M, Carrera M, Grinyó JM. Reliability of chronic allograft nephropathy diagnosis in sequential protocol biopsies, Kidney Int 61(2), 727–733 (2002). [DOI] [PubMed] [Google Scholar]
- 47-.Chapman JR. Longitudinal analysis of chronic allograft nephropathy: clinicopathologic correlations. Kidney Int Suppl. (99), S108–S112 (2005). [DOI] [PubMed] [Google Scholar]
- 48-.Ortiz F, Paavonen T, Tornroth T, et al. Predictors of renal allograft histologic damage progression. J Am Soc Nephrol. 16(3), 817–824 (2005). [DOI] [PubMed] [Google Scholar]
- 49-.Schwarz A, Mengel M, Gwinner W, et al. Risk factors for chronic allograft nephropathy after renal transplantation: a protocol biopsy study. Kidney Int. 67(1), 341–348 (2005). [DOI] [PubMed] [Google Scholar]
- 50-.Vallejos A, Alperovich G, Moreso F, et al. Resistive index and chronic allograft nephropathy evaluated in protocol biopsies as predictors of graft outcome. Nephrol Dial Transplant. 20 (11), 2511–2516 (2005). [DOI] [PubMed] [Google Scholar]
- 51-.Botev R, Mallié JP, Couchoud C, et al. Estimating glomerular filtration rate: Cockcroft-Gault and Modification of Diet in Renal Disease formulas compared to renal inulin clearance. Clin J Am Soc Nephrol. 4(5), 899–906 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52-.Pöge U, Gerhardt T, Stoffel-Wagner B, et al. Can modifications of the MDRD formula improve the estimation of glomerular filtration rate in renal allograft recipients? Nephrol Dial Transplant. 22(12), 3610–3615 (2007). [DOI] [PubMed] [Google Scholar]
- 53-.Gera M, Slezak JM, Rule AD, Larson TS, Stegall MD, Cosio FG. Assessment of changes in kidney allograft function using creatinine-based estimates of glomerular filtration rate. Am J Transplant. 7(4), 880–7 (2007). [DOI] [PubMed] [Google Scholar]
- 54-.Mas V, Diller A, Albano S, et al. Intragraft expression of transforming growth factor-beta 1 by a novel quantitative reverse transcription polymerase chain reaction ELISA in long lasting kidney recipients. Transplantation. 70(4), 612–616 (2000). [DOI] [PubMed] [Google Scholar]
- 55-.Robertson H, Wong WK, Talbot D, Burt AD, Kirby JA. Tubulitis after renal transplantation: Demonstration of an association between CD103+ T cells, transforming growth factor beta1 expression and rejection grade. Transplantation 71(2), 306–313 (2001). [DOI] [PubMed] [Google Scholar]
- 56-.Campistol JM, Iñigo P, Larios S, Bescos M, Oppenheimer F. Role of transforming growth factor-beta1 in the progression of chronic allograft nephropathy. Nephrol Dial Transplant. 16 Suppl 1:114–6 (2001). [DOI] [PubMed] [Google Scholar]
- 57-.Harris S, Coupes BM, Roberts SA, Roberts IS, Short CD, Brenchley PE. TGF-beta1 in chronic allograft nephropathy following renal transplantation. J Nephrol. 20(2), 177–185 (2007). [PubMed] [Google Scholar]
- 58-.Lee S, Kim DJ, Park MG, et al. Expression of transforming growth factor-beta1 and hypoxia-inducible factor-1alpha in renal transplantation. Transplant Proc. 40(7), 2147–2148 (2008). [DOI] [PubMed] [Google Scholar]
- 59-.Pilmore HL, Yan Y, Eris JM, Hennessy A, McCaughan GW, Bishop GA. Time course of upregulation of fibrogenic growth factors and cellular infiltration in a rodent model of chronic renal allograft rejection. Transpl Immunol. 10(4), 245–54 (2002). [DOI] [PubMed] [Google Scholar]
- 60-.Otsuka G, Stempien-Otero A, Frutkin AD, Dichek DA. Mechanisms of TGF-beta1-induced intimal growth: plasminogen-independent activities of plasminogen activator inhibitor-1 and heterogeneous origin of intimal cells. Circ Res. 100(9), 1300–1307 (2007). [DOI] [PubMed] [Google Scholar]
- 61-.Huang Y, Noble NA, Zhang J, Xu C, Border WA. Renin-stimulated TGF-beta1 expression is regulated by a mitogen-activated protein kinase in mesangial cells. Kidney Int. 72(1):45–52 (2007). [DOI] [PubMed] [Google Scholar]
- 62-.Mas V, Alvarellos T, Giraudo C, Massari P, De Boccardo G. Intragraft messenger RNA expression of angiotensinogen: relationship with transforming growth factor beta-1 and chronic allograft nephropathy in kidney transplant patients. Transplantation. 74(5), 718–721 (2002). [DOI] [PubMed] [Google Scholar]
- 63-.Becker BN, Jacobson LM, Hullett DA, et al. Type 2 angiotensin II receptor expression in human renal allografts: an association with chronic allograft nephropathy. Clin Nephrol. 57(1):19–26 (2002). [DOI] [PubMed] [Google Scholar]
- 64-.Garrett Q, Khaw PT, Blalock TD, Schultz GS, Grotendorst GR, Daniels JT. Involvement of CTGF in TGF-beta1-stimulation of myofibroblast differentiation and collagen matrix contraction in the presence of mechanical stress. Invest Ophthalmol Vis Sci. 45(4), 1109–1116 (2004). [DOI] [PubMed] [Google Scholar]
- 65-.Grotendorst GR, Duncan MR. Individual domains of connective tissue growth factor regulate fibroblast proliferation and myofibroblast differentiation. FASEB J 2005; 19, 729–738. [DOI] [PubMed] [Google Scholar]
- 66-.Ito Y, Goldschmeding R, Bende R et al. Kinetics of connective tissue growth factor expression during experimental proliferative glomerulonephritis. J Am Soc Nephrol 12(3), 472–484 (2001). [DOI] [PubMed] [Google Scholar]
- 67-.Yokoi H, Mukoyama M, Sugawara A et al. Role of connective tissue growth factor in fibronectin expression and tubulointerstitial fibrosis. Am J Physiol Renal Physiol 282(5), F933–F942 (2002). [DOI] [PubMed] [Google Scholar]
- 68-.Okada H, Kikuta T, Kobayashi T, et al. Connective tissue growth factor expressed in tubular epithelium plays a pivotal role in renal fibrogenesis. J Am Soc Nephrol. 16(1), 133–43 (2005). [DOI] [PubMed] [Google Scholar]
- 69-.Inkinen K, Soots A, Krogerus L, Loginov R, Bruggeman C, Lautenschlager I. Cytomegalovirus enhance expression of growth factors during the development of chronic allograft nephropathy in rats. Transpl Int 18(6), 743–9 (2005). [DOI] [PubMed] [Google Scholar]
- 70-.Franceschini N, Cheng O, Zhang X, Ruiz P, Mannon RB. Inhibition of prolyl-4-hydroxylase ameliorates chronic rejection of mouse kidney allografts. Am J Transplant 3(4), 396–402 (2003). [DOI] [PubMed] [Google Scholar]
- 71-.Csencsits K, Wood SC, Lu G, et al. Transforming growth factor beta-induced connective tissue growth factor and chronic allograft rejection. Am J Transplant 6(5), 959–966 (2006). [DOI] [PubMed] [Google Scholar]
- 72-.Cheng O, Thuillier R, Sampson E, et al. Connective tissue growth factor is a biomarker and mediator of kidney allograft fibrosis. Am J Transplant. 6(10, 2292–306 (2006). [DOI] [PubMed] [Google Scholar]
- 73-.Siddiqui I, Khan ZA, Lian D, et al. Endothelin-mediated oncofetal fibronectin expression in chronic allograft nephropathy. Transplantation. 82(3), 406–414 (2006). [DOI] [PubMed] [Google Scholar]
- 74-.Reinders ME, Rabelink TJ, Briscoe DM. Angiogenesis and endothelial cell repair in renal disease and allograft rejection. J Am Soc Nephrol. 17(4), 932–942 (2006). [DOI] [PubMed] [Google Scholar]; ** Extensive review of the role of angiogenesis in renal disease with especial emphasis in chronic allograft nephropathy.
- 75-.Malmström NK, Kallio EA, Rintala JM, et al. Vascular endothelial growth factor in chronic rat allograft nephropathy. Transpl Immunol. 9(2), 136–144 (2008). [DOI] [PubMed] [Google Scholar]
- 76-.Ma X, Lu YP, Yang L, et al. Expressions of Angiopoietin-1, Angiopoietin-2, and Tie2 and their roles in rat renal allografts with chronic allograft nephropathy. Transplant Proc. 40(8), 2795–2799 (2008). [DOI] [PubMed] [Google Scholar]
- 77-.Mansfield ES, Sarwal MM. Arraying the orchestration of allograft pathology. Am J Transplant 4(6), 853–862 (2004). [DOI] [PubMed] [Google Scholar]
- 78-.Halloran PF, Einecke G. Microarrays and transcriptome analysis in renal transplantation. Nat Clin Pract Nephrol. 2(1), 2–3 (2006). [DOI] [PubMed] [Google Scholar]
- 79-.Ying L, Sarwal M. In praise of arrays. Pediatr Nephrol. 24(9):1643–1659 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80-.Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant 4(11), 1475–1485 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81-.Sarwal M, Fernandez-Fresnedo G, Rodrigo E, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med 349(2):125–38 (2003). [DOI] [PubMed] [Google Scholar]; ** Pioneer article in gene expression analysis in kidney allograft biopsies and identification of distinctive patterns in acute rejection, nephrotoxic effects of drugs, chronic allograft nephropathy, and normal kidneys. Extensive analysis of different pathways associated with acute rejection in kidney biopsies post-kidney transplantation.
- 82-.Scherer A, Krause A, Walker JR, Korn A, Niese D, Raulf F. Early prognosis of the development of renal chronic allograft rejection by gene expression profiling of human protocol biopsies. Transplantation 75(8):1323–1330 (2003). [DOI] [PubMed] [Google Scholar]
- 83-.Mas V, Maluf D, Archer K, et al. Establishing the molecular pathways involved in chronic allograft nephropathy for testing new noninvasive diagnostic markers. Transplantation. 83(4), 448–457 (2007). [DOI] [PubMed] [Google Scholar]
- 84-.Hotchkiss H, Chu TT, Hancock WW, et al. Differential expression of profibrotic and growth factors in chronic allograft nephropathy. Transplantation 81(3), 342–349 (2006). [DOI] [PubMed] [Google Scholar]
- 85-.Maluf DG, Mas VR, Archer KJ, et al. Molecular pathways involved in loss of kidney graft function with tubular atrophy and interstitial fibrosis. Mol Med. 14(5–6), 276–285 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86-.Libby P, Zhao DX. Allograft arteriosclerosis and immune-driven angiogenesis. Circulation. 107(9), 1237–1239 (2003). [DOI] [PubMed] [Google Scholar]
- 87-.Reinders ME, Briscoe DM. Angiogenesis and allograft rejection. Graft. 5, 96–98 (2002). [Google Scholar]
- 88-.Kang DH, Kanellis J, Hugo C, et al. Role of the microvascular endothelium in progressive renal disease. J Am Soc Nephrol. 13(3), 806–16 (2002). [DOI] [PubMed] [Google Scholar]
- 89-.Kang DH, Hughes J, Mazzali M, Schreiner GF, Johnson RJ. Impaired angiogenesis in the aging kidney: vascular endothelial growth factor and thrombospondin-1 in renal disease. Am J Kidney Dis. 12(7):1448–1457. [DOI] [PubMed] [Google Scholar]
- 90-.Seron D, Moreso F, Ramon JM, Hueso M, Condom E, Fulladosa X. Protocol renal allograft biopsies and the design of clinical trials aimed to prevent or treat chronic allograft nephropathy. Transplantation 69(9) 1849–1855 (2000). [DOI] [PubMed] [Google Scholar]
- 91-.Nicholson ML, McCulloch TA, Harper SJ, et al. Early measurement of interstitial fibrosis predicts long-term renal function and graft survival in renal transplantation. Br J Surg 83 (8), 1082–1085 (1996). [DOI] [PubMed] [Google Scholar]
- 92-.Scherer A, Gwinner W, Mengel M, et al. Transcriptome changes in renal allograft protocol biopsies at 3 months precede the onset of interstitial fibrosis/tubular atrophy (IF/TA) at 6 months. Nephrol Dial Transplant. 24(8):2567–2575 (2009). [DOI] [PubMed] [Google Scholar]
- 93-.Mengel M, Gwinner W, Schwarz A, et al. Infiltrates in protocol biopsies from renal allografts. Am J Transplant 7(2), 356–365 (2007). [DOI] [PubMed] [Google Scholar]
- 94-.Randhawa P Role of donor kidney biopsies in renal transplantation. Transplantation. 71(10), 1361–1365 (2001). [DOI] [PubMed] [Google Scholar]
- 95-.Gaber LW, Moore LW, Alloway RR, Amiri MH, Vera SR, Gaber AO. Glomerulosclerosis as a determinant of posttransplant function of older donor renal allografts. Transplantation. 60(4), 334–339 (1995). [DOI] [PubMed] [Google Scholar]
- 96-.Hauser P, Schwarz C, Mitterbauer C, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab Invest. 84(3), 353–361 (2004). [DOI] [PubMed] [Google Scholar]
- 97-.Yanek K, Maluf D, Archer K, et al. Molecular Profiles in Donor Kidney Biopsies Associated with Warm Ischemia Time. Abstract, Presented at: American Transplant Congress, Boston, MA May 31-June 3 2009. [Google Scholar]
- 98-.Perico N, Cattaneo D, Sayegh MH, Remuzzi G. Delayed graft function in kidney transplantation. Lancet 364(9447), 1814–1927 (2004). [DOI] [PubMed] [Google Scholar]
- 99-.Azuma H, Nadeau K, Takada M, Mackenzie HS, Tilney NL. Cellular and molecular predictors of chronic renal dysfunction after initial ischemia/reperfusion injury of a single kidney. Transplantation 64(2), 190–7 (1997). [DOI] [PubMed] [Google Scholar]
- 100-.Chapman JR, O’Connell PJ, Nankivell BJ. Chronic renal allograft dysfunction. J Am Soc Nephrol 16(10):3015–26 (2005). [DOI] [PubMed] [Google Scholar]
- 101-.Mas VR, Archer KJ, Yanek K, et al. Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation. 85(4), 626–635 (2008). [DOI] [PubMed] [Google Scholar]
- 102-.Mueller TF, Reeve J, Jhangri GS, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am J Transplant. 8(1), 78–85 (2008). [DOI] [PubMed] [Google Scholar]
- 103-.Maluf D, Mas V, Archer K, et al. Time-zero biopsies in kidney transplantation: Gene expression data predict the short and long term outcomes post-transplantation. Abstract, Presented at: American Transplant Congress, Boston, MA May 31-June 3 2009. [Google Scholar]
- 104-.Naesens M, Sarwal M. Looking into the crystal chip: can microarrays predict graft function? Transplantation. 85(4), 499–500 (2008). [DOI] [PubMed] [Google Scholar]
- 105-.Tatapudi RR, Muthukumar T, Dadhania D, et al. Noninvasive detection of renal allograft inflammation by measurements of mRNA for IP-10 and CXCR3 in urine. Kidney Int 65(6):2390–2397 (2004). [DOI] [PubMed] [Google Scholar]
- 106-.Magee CC, Denton MD, Womer KL, et al. Assessment by flow cytometry of intracellular cytokine production in the peripheral blood cells of renal transplant recipients. Clin Transplant 18(4), 395–401 (2004). [DOI] [PubMed] [Google Scholar]
- 107-.Mas VR, Mas LA, Archer KJ, et al. Evaluation of gene panel mRNAs in urine samples of kidney transplant recipients as a non-invasive tool of graft function. Mol Med. 13(5–6):315–24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108-.Kurian SM, Heilman R, Mondala TS, et al. Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS One. 4(7):e6212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Very interesting article about the potential of non-invasive markers identified in peripheral blood as earlier non-invasive markers for monitoring progression to chronic allograft nephropathy.
- 109-.Flechner SM, Kurian SM, Solez K, et al. De novo kidney transplantation without use of calcineurin inhibitors preserves renal structure and function at two years. Am J Transplant. 4(11), 1776–1785 (2004). [DOI] [PubMed] [Google Scholar]
- 110-.Malumbres R, Chen J, Tibshirani R, et al. Paraffin-based 6-gene model predicts outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Blood 111(12), 5509–5514 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111-.Bunnag S, Einecke G, Reeve J, et al. Diagnosing rejection in renal transplants: a comparison of molecular- and histopathology-based approaches. Am Soc Nephrol. 20(5):1149–60 (2009). [DOI] [PubMed] [Google Scholar]
- 112-.Bullinger L, Döhner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med 350(16), 1605–1616 (2004). [DOI] [PubMed] [Google Scholar]
- 113-.Huber LA. Is proteomics heading in the wrong direction? Nat Rev Mol Cell Biol. 4(1), 74–80 (2003). [DOI] [PubMed] [Google Scholar]
- 114-.Aebersold R A mass spectrometric journey into protein and proteome research. J Am Soc Mass Spectrom. 14, 685–695 (2003). [DOI] [PubMed] [Google Scholar]
- 115-.O’Riordan E, Orlova TN, Podust VN, et al. Characterization of urinary peptide biomarkers of acute rejection in renal allografts. Am J Transplant. 7(4), 930–940 (2007). [DOI] [PubMed] [Google Scholar]
- 116-.Clarke W, Silverman BC, Zhang Z, et al. Characterization of renal allograft rejection by urinary proteomic analysis. Ann. Surg 237(5), 660–665 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117-.Wittke S, Haubitz M, Walden M, et al. Detection of acute tubulointerstitial rejection by proteomic analysis of urinary samples in renal transplant recipients. Am. J. Transplant 5(10), 2479–2488 (2005). [DOI] [PubMed] [Google Scholar]
- 118-.Schaub S, Rush D, Wilkins J, et al. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J. Am. So. Nephrol 15(1), 219–227 (2004) [DOI] [PubMed] [Google Scholar]
- 119-.Quintana LF, Solé-Gonzalez A, Kalko SG, et al. Proteomics to detect biomarkers for chronic allograft dysfunction. J Am Soc Nephrol.20(2), 428–435 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120-.Schaub S, Wilkins JA, Rush D, Nickerson P. Developing a tool for noninvasive monitoring of renal allografts. Expert Rev Proteomics. 3(5), 497–509 (2006). [DOI] [PubMed] [Google Scholar]
- 121-.Christians U, Schmitz V, Schöning W, et al. Toxicodynamic therapeutic drug monitoring of immunosuppressants: promises, reality, and challenges. Ther Drug Monit. 30(2), 151–158 (2008). [DOI] [PubMed] [Google Scholar]
- 122-.Mayr M, Madhu B, Xu Q. Proteomics and metabolomics combined in cardiovascular research. Trends Cardiovasc Med. 17(2), 43–48 (2007). [DOI] [PubMed] [Google Scholar]